- Cellular and Molecular Neurobiology & Drug Targeting Laboratory, Department of Zoology, Indira Gandhi National Tribal University, Amarkantak, Madhya Pradesh, India
Memory formation is associated with constant modifications of neuronal networks and synaptic plasticity gene expression in response to different environmental stimuli and experiences. Dysregulation of synaptic plasticity gene expression affects memory during aging and neurodegenerative diseases. Covalent modifications such as methylation on DNA and acetylation on histones regulate the transcription of synaptic plasticity genes. Changes in these epigenetic marks correlated with alteration of synaptic plasticity gene expression and memory formation during aging. These epigenetic modifications, in turn, are regulated by physiology and metabolism. Steroid hormone estrogen and metabolites such as S-adenosyl methionine and acetyl CoA directly impact DNA and histones’ methylation and acetylation levels. Thus, the decline of estrogen levels or imbalance of these metabolites affects gene expression and underlying brain functions. In the present review, we discussed the importance of DNA methylation and histone acetylation on chromatin modifications, regulation of synaptic plasticity gene expression and memory consolidation, and modulation of these epigenetic marks by epigenetic modifiers such as phytochemicals and vitamins. Further, understanding the molecular mechanisms that modulate these epigenetic modifications will help develop recovery approaches.
Introduction
The brain is the most complex and dynamic organ of an organism. It controls numerous vital functions, such as decision-making, cognition, learning, and memory. Memory is a higher-order brain function that stores and recalls previously acquired experiences, facts, and information. This acquired information is stabilized in the form of long-term memory by strengthening the synaptic connections, and it requires the expression of memory-linked synaptic plasticity genes (Kandel, 2001). The expression of these synaptic plasticity genes is altered during aging and neuropathological conditions, which affects learning and memory (Hermann et al., 2014). Previous research revealed that epigenetic mechanisms play an essential role in the regulation of these synaptic plasticity genes and dysregulation of which can lead to memory impairment during aging and age-associated neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) (Singh et al., 2017).
Epigenetic modifications
Chromatin is a complex interaction of DNA and histones that packs the large size of DNA inside the nucleus. The nucleosome is the smaller unit of the chromatin and is made of 147 bp DNA wrapped around a histone octamer core. The linker DNA is associated with a linker histone H1 (Handy et al., 2011). Chromatin is present in two states. During the condensed state, it inhibits the interaction of transcription factors on gene promoters and does not allow gene transcription. On the other hand, the relaxed state allows the interaction of transcription factors on gene promoters and allows gene transcription. The most important factors that regulate the condensation and relaxation of chromatin are the covalent modifications of DNA and histone proteins and thus influence gene expression (Lossi et al., 2024). The covalent modifications found on DNA are methylation and hydroxymethylation. Similarly, the most common post-translational modifications found on the n-terminal tail of histones are acetylation, phosphorylation, methylation, etc. These covalent modifications on DNA and histone alter their interactions in the nucleosome, thus regulating chromatin’s condensation and relaxation and thereby regulating gene expression (Kelly et al., 2010).
DNA methylation
The methylation of DNA takes place on the cytosine (C) residue followed by a guanine (G) residue called CpG island and regulates gene expression at the transcriptional level. During this modification, a methyl (CH3) group is transferred from S-adenosyl methionine to the fifth carbon of cytosine (5 mC) in a CpG island (Gao et al., 2018). The transfer of methyl group on DNA is catalyzed by DNA methyltransferases (DNMTs). DNMTs are classified as de novo methyltransferase (DNMT3a and DNMT3b) which transfers a methyl group on a previously unmethylated CpG island on DNA and maintenance methyltransferase (DNMT1) which transfers a methyl group on a hemimethylated CpG site that arises as a result of DNA replication and maintains the DNA methylation pattern similar to pre-replication (Moore et al., 2013). The epigenetic mark, DNA methylation is found on the CpG sites on the promoter of a gene affecting the binding of transcription factors, and regulating gene expression. Also, the cytosine methylation on the CpG island at the promoter recruits a group of proteins called methyl-binding proteins. This interaction of cytosine methylation and methyl-binding proteins forms the repressor complex and suppresses gene transcription (Lu et al., 2013). DNA methylation also activates the expression of genes by recruiting activator complexes at their promoter region (Chahrour et al., 2008).
DNA methylation and memory
Covalent modification of DNA regulates synaptic plasticity genes’ expression and underlying neuronal functions such as learning and memory. Levenson et al. (2004) first examined the role of DNA methylation and DNMTs in regulating synaptic plasticity and memory formation. To check the role of DNA methylation on synaptic plasticity gene expression, they treated hippocampal slices with DNMT inhibitor zebularine, and observed decreased CpG methylation levels at the promoter of reelin and BDNF. Further, the treatment of hippocampal slices with phorbol-12,13-diacetate, an activator of protein kinase C upregulated DNMT3a expression in the CA1 of the hippocampus. They also reported that zebularine or 5-aza-2-deoxycytidine, an inhibitor of DNA methylation, treatment impaired LTP in brain slice culture, which suggests that basal activity DNMTs are required for learning and memory.
To investigate further, reports from the same group checked the involvement of DNMTs during contextual fear memory. They observed that the expression of de novo DNMTs DNMT3a and 3b increased in the hippocampus 30 min post contextual fear paradigm in adult rats. However, the level of maintenance methyl transferase DNMT1 remained unchanged. The fear memory formation was impaired when DNA methylation was inhibited in these animals after contextual fear conditioning. Further, they checked the methylation level at the promoter of protein phosphatase 1 (PP1), which negatively regulates memory and reelin, which is important for learning and memory. Interestingly, they observed hypermethylation at the promoter of PP1 decreased its expression, and hypomethylation at the promoter of reelin increased its expression after fear memory formation. This suggests that DNA methylation marks are dynamic in nature and important for hippocampal-dependent memory formation during normal physiological condition (Miller and Sweatt, 2007).
Knockout studies in animal models showed that DNMTs play an important role in synaptic plasticity and memory formation. Conditional knockout (CKO) of DNMT1 in the forebrain of mice showed impaired barrel cortex development, the brain region important for processing touch inputs. Also, a patch clamp study showed impaired induction of LTP in DNMT1 KO brain slices, suggesting DNMT1 is important not only for development, however, also for synaptic plasticity (Golshani et al., 2005). CKO of DNMT1 and DNMT3a mice showed around 10% reduction in hippocampal volume and the volume of the neurons, mainly in the dentate gyrus region, with impaired LTP induction and enhanced LTD. DKO mice show impaired spatial memory and fear memory consolidation (Feng et al., 2010). Similarly, Liu et al. (2011) investigated the effect of DNMT1 haploinsufficiency in heterozygous DNMT1 ± mice during aging. They observed that the heterozygous DNMT1 ± mice showed lowered DNA methylation in the cerebral cortex and hippocampus during aging compared to age-match control. They observed a positive correlation between hypomethylation and impaired spatial memory during aging in DNMT1 ± mice as compared to the control.
DNA methylation during aging and neurodegeneration
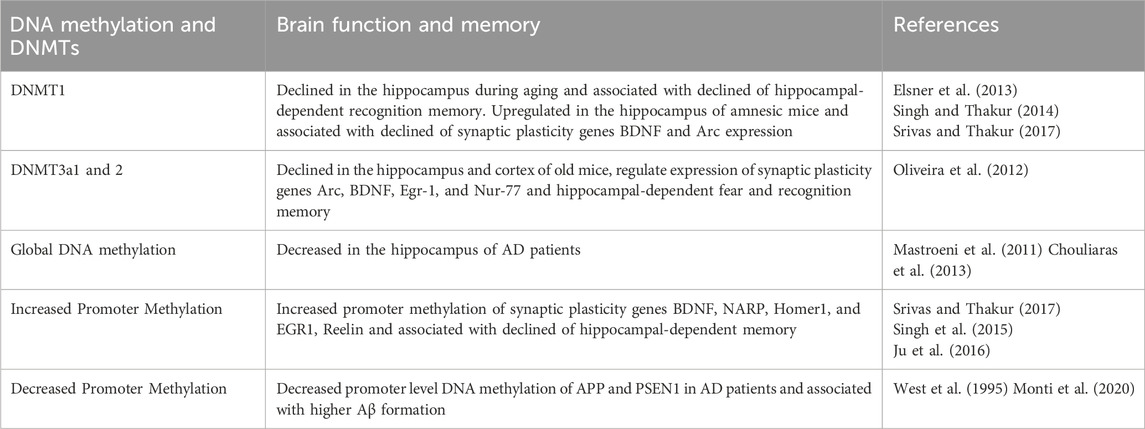
Several reports show that an imbalance of epigenetic changes, such as DNA methylation, is observed during aging and different neuropathological conditions (Table 1). Elsner et al. (2013) reported that the expression of DNMT1 but not DNMT3b declined in the hippocampus of old rats compared to young rats. Similarly, Singh and Thakur (2014) also reported that the level of DNMT1 but not DNMT3a and DNMT3b declined in the cortex and hippocampus of old compared to young and adult mice. Further, the decline of DNMT1 level is associated with impaired recognition memory in old mice. Mastroeni et al. (2011) reported that the expression of DNMT1 declined in Alzheimer’s disease (AD) patients as compared to age-matched healthy control. Further, the decline of DNMT1 level is associated with a reduction of total cytosine methylation in AD patients. In humans, DNMT1 mutation causes hereditary sensory and autonomic neuropathy (HSAN1). Mutation of DNMT1 leads to abnormal DNA methylation levels, resulting in neurodegeneration, hearing loss, and dementia in patients. At the molecular level, DNMT1 mutation causes aberrant heterochromatin formation during the G2 phase of the cell cycle, global-level hypomethylation, and local-level hypermethylation (Klein et al., 2011). Oliveira et al. (2012) observed that the Dnmt3a1 and Dnmt3a2 expression decreased in the hippocampus and cortex of old mice. CKO of DNMT3a2 reduced the expression of Arc, BDNF, Egr-1, and Nur-77 and hippocampal-dependent fear memory and recognition memory. Further, rescuing of DNMT3a2 expression improved cognitive functions in old mice. Further, age-dependent impairment in spatial memory consolidation is associated with increased promoter-specific methylation of ARC and EGR1 genes and decreased expression of synaptic plasticity genes in the CA1 and DG regions (Penner et al., 2011; Penner et al., 2016). Genome wide methylation studies showed that he global level of Cytosine methylation decreased in the hippocampus of AD patients as compared to age matched control (Chouliaras et al., 2013; Zocher, 2024). Similarly, decreased promoter level DNA methylation of APP and PSEN1 gene leads to their higher expression and Aβ formation in AD patients (West et al., 1995; Monti et al., 2020). PD is a neurodegenerative disease associated with the accumulation of α-synuclein (α-syn) protein (Negi et al., 2024). Analysis of postmortem brain sample of PD patients showed hypomethylation of SNCA gene promoter associated with upregulation of α-syn expression (Jowaed et al., 2010). On the other hand, promoter hypermethylation of PGC-1α associated with impaired mitochondrial biogenesis in the mice model of PD (Su et al., 2015).
Scopolamine is a M1 muscarinic antagonist used to induce amnesia animal models. Singh et al. (2015) reported that the expression of DNMT1 upregulated in the hippocampus of scopolamine-induced amnesic mice and associated with the decline expression of synaptic plasticity genes BDNF and Arc. Similarly, Srivas and Thakur (2017) also reported that the methylation level at the promoter of synaptic plasticity genes NARP, Homer1, and EGR1 increased, and their transcription decreased in the hippocampus of scopolamine-induced amnesic mice. Further, treatment of DNMTs inhibitor 5-aza-2′-deoxycytidine, reduced promoter level methylation of BDNF and Arc, upregulated their expression, and improved memory in amnesic mice. Sevoflurane is a general anesthetic that induces neurobehavioral abnormalities in rodents (Kameyama et al., 2022). Ju et al. (2016) reported that repeated exposure to sevoflurane decreased dendritic spines, impaired hippocampal-dependent spatial and fear memory in rats, and was associated with upregulation in the expression of DNMTs. Further, they observed that the downregulation of synaptic plasticity genes BDNF and Reelin was due to hypermethylation at their promoter regions. Further, pre-treatment of DNMTs inhibitor 5-AZA rescued the sevoflurane-induced memory impairment. These reports suggest that DNMTs are crucial during development and essential in regulating memory consolidation during aging and different physiological and pathological conditions.
Histone acetylation
Histone proteins undergo several covalent modifications after the translation. These modifications include acetylation, phosphorylation, ubiquitination, methylation, etc. Among these, histone acetylation is correlated with active gene expression, while deacetylation leads to gene repression (Ramazi et al., 2020). Two groups of enzymes that regulate the acetylation level on the N terminal of the tail of histones are histone acetyltransferases (HATs) and histone deacetylases (HDACs). HATs transfer an acetyl group (−COCH₃) from the acetyl CoA to the histones and HDACs remove the acetyl group from the histones. Acetylation on histone reduces its interaction with DNA and results in relaxed or open chromatin, which allows the binding of transcription factors on the gene’s promoter region and, thus, active gene transcription (Clayton et al., 2006). On the contrary, deacetylation increases the interaction between histones and DNA, resulting in a closed or condensed chromatin, which inhibits the binding of transcription factors leading to gene repression. HATs are mostly transcriptional activators as they positively regulate gene expression. They are divided into the GNAT family, p300/CBP family, MYST family, Transcription-related HATs, and nuclear receptor-associated HATs (Selvi et al., 2010). The p300/CBP family is one of the vital histone lysine transferases and a transcription activator. This HAT family regulates many functions, including neurogenesis, neuronal development and differentiation, and memory formation (Partanen et al., 1999; Bedford and Brindle, 2012; Chatterjee et al., 2013; Alarcón et al., 2004; Barrett et al., 2011; Crump et al., 2011; Lopez-Atalaya et al., 2011). Deacetylases are classified into class I, II, III, and IV HDACs based on their localization in the cells. The class I includes HDAC1, 2, 3, and 8, and primarily present in the nucleus. They mostly regulate gene expression and act as transcriptional repressors. HDAC2 is one of the most important classes of HDACs studied extensively about the regulation of synaptic plasticity genes and memory (Kilgore et al., 2010).
Histone acetylation and memory
CREB binding protein (CBP) is an important HAT that performs its role as a transcriptional activator of many synaptic plasticity genes and helps in memory consolidation. Mice with CBP CKO showed normal neuronal morphology but lowered histone acetylation levels and reduced long-term associative and recognition memory formation. Further, they reported downregulating synaptic plasticity genes such as NMDARs, AMPARs, PSD95, etc (Chen et al., 2010). CKO of CBP in the forebrain of mice reduced histone acetylation at H2A, H2B, H3, and H4 and impaired recognition memory (Valor et al., 2011). Similarly, mice with CKO of CBP in the medial prefrontal cortex showed impaired long-term spatial, object location, and fear memory (Vieira and Korzus, 2015; Korzus et al., 2004). Guan et al. (2009) reported that HDAC2 overexpression decreased spine density, histone H3K14/K12 acetylation, negatively regulated synaptic plasticity gene expression such as BDNF, GluR, EGR1, etc., and impaired memory consolidation. However, no changes in synaptic plasticity gene expression, and memory were observed when HDAC 1 was overexpressed. Hsiao et al. (2017) reported that HDAC2 knockout APP/PS1 mice showed improved freezing behavior. Further, the binding of HDAC2 decreased at the promoter of synaptic plasticity gene BDNF which results increased BDNF gene expression. This showed that HDAC2 is an important regulator of gene expression and learning, and memory.
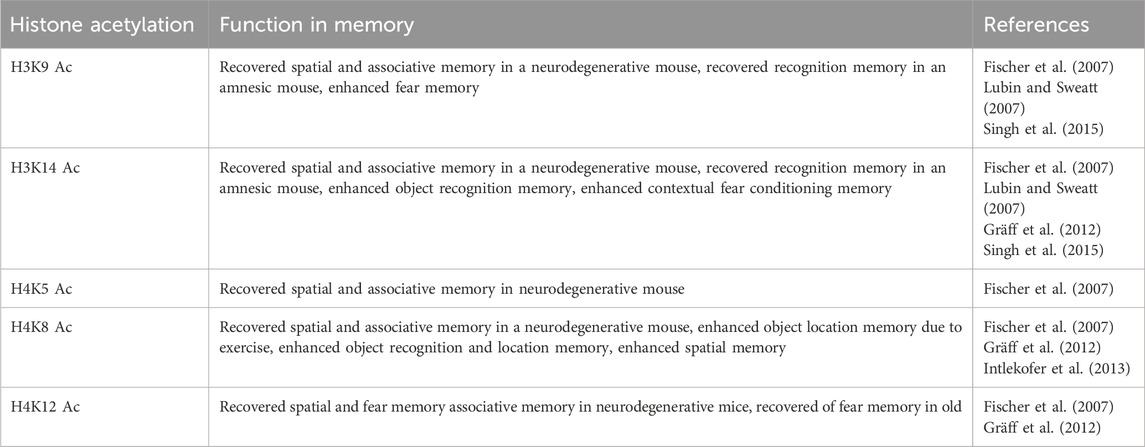
Histone acetylation is dependent on neuronal activity and is essential for regulating synaptic plasticity gene expression and memory consolidation (Table 2). Levenson et al. (2008) reported that acetylation on H3 is essential for long-term hippocampal-dependent memory formation. Fischer et al. (2007) reported that acetylation at H3K9/14 and H4K5/8/12 in the hippocampus increased upon environmental enrichment and is associated with improved memory consolidation in CK-p25 mice. Acetylation on the cortex and hippocampus is very dynamic. Gräff et al. (2012) reported that acetylation at H3K14 and H4K5 increased rapidly and transiently in the hippocampus however increased slowly and persistently in the cortex after a novel object recognition test. This difference in acetylation pattern may be due to their difference in functions during memory consolidation.
Histone acetylation during aging and neurodegeneration
Total histone acetylation level and gene promoter-specific histone level decreased during aging and neurodegenerative diseases. This change in acetylation level is a result of alteration in HATs and HDACs expression. Chung et al. (2002) reported that the CBP immunoreactivity decreased significantly in the cerebral cortex and the CA1 and DG region of the hippocampus in old rats as compared to the adult. Further, the decline of CBP is mainly observed in the pyramidal layer as well as in the granule cell layer and the polymorphic layer of CA1/CA3 and DG region of the hippocampus respectively. Giralt et al. (2012) reported that the expression of CBP decreased and was associated with lowered acetylation level at H3 and memory impairment in HD mice model. Further, treatment of C646, a selective inhibitor of p300/CBP, decreased histone acetylation level and impaired recognition and fear memory consolidation (Mitchnick et al., 2016; Maddox et al., 2013). On the other hand, CBP over-expression rescued spatial memory and increased the expression of BDNF in the AD mouse model (Caccamo et al., 2010). CBP positively regulates several synaptic plasticity gene expressions such as ARC, BDNF, c-FOS, EGR-1, etc., and thereby helps in memory consolidation. On the other hand, the downregulation of CBP is associated with the downregulation of these synaptic plasticity genes and memory impairment (Wood et al., 2005; Wood et al., 2006).
Gräff et al. (2012) reported that the expression of HDAC2 increased in the brains of AD patients and the hippocampus of CK-p25 mice, a mouse model of neurodegeneration. Further, the knockdown of HDAC2 rescued histone acetylation, ARC, BDNF, and EGR1 expression and memory consolidation in CK-p25 mice. Similarly, Singh and Thakur (2014) reported that HDAC2 level upregulated in the hippocampus and negatively correlated with recognition memory consolidation in old mice. Reports from the same group also showed that upregulation of HDAC2 is also associated with reduced acetylation levels at the promoter of BDNF and Arc. Inhibition with sodium butyrate (NaB) or knockdown of HDAC2 in the hippocampus, rescued histone acetylation level, gene expression, and memory (Singh and Thakur, 2018). Singh et al. (2015) reported that the level of HDAC2 was upregulated in the hippocampus of scopolamine-induced amnesic mice and negatively regulated synaptic plasticity gene expression and hippocampal-dependent memory consolidation. Aging, neurodegeneration, and amnesic conditions are associated with decreased total histone acetylation levels at H3 and H4 as well as at the promoter of synaptic plasticity genes and thus memory impairment (Gräff et al., 2012; Singh et al., 2015; Singh and Thakur, 2018).
Several reports showed that acetylation at H3K9 and H3K27 increased in the brain of AD patients and upregulated the expression of AD related gene and underlying neurodegeneration (Marzi et al., 2018; Klein et al., 2019; Nativio et al., 2020). Similarly, Nativio et al. (2018) reported that acetylation at H3K16 position decreased in the brain of AD patients as compared to cognitively normal aged individuals. Dysregulation of histone acetylation is also observed in the PD patients. Genome wide acetylation analysis in PD postmortem brain sample showed increased H3K27 acetylation and higher expression of gene associated with PD pathology such as SNCA, MAPT and PRKN (Toker et al., 2021). Similarly, higher histone acetylation level was observed in the brain sample of PD patients as well as cellular model of PD and MPTP treated mice brain. This increased in histone acetylation in PD brain samples associated with decreased HDAC1,2 and 6 (Park et al., 2016). In a comparison study between APP/PS1 and wild type mice during aging, McClarty et al. (2024) reported that impaired recognition and spatial memory in old (18 months) wild type mice and mid aged (12 months) and old (18 months) APP/PS1 mice associated with increased HDAC2 in the hippocampus and HDAC3 in the pre frontal cortex. This increased in HDACs decreased H3K9 acetylation level and synaptic plasticity gene expression. On the other hand, different behavioral paradigms such as environmental enrichment, contextual fear conditioning, and object location memory is associated with increased total histone acetylation at H3 and H4 as well as at the promoter of synaptic plasticity genes during different physiological conditions such as aging, neurodegeneration, etc. (Intlekofer et al., 2013; Lubin and Sweatt, 2007; Fischer et al., 2007; Wang et al., 2020).
Regulation of DNA methylation and histone acetylation
These epigenetic marks on DNA and histones are modulated by physiology and metabolism. Folate/vitamin B12 pathway, acetyl CoA metabolism as well as hormone-like estrogen 17β-estradiol (E2) plays important roles in regulating DNA methylation and histone acetylation (Fortress et al., 2014; Frick et al., 2015). Alteration in metabolic pathways and hormonal levels during aging and neurodegenerative diseases affects DNA methylation and histone acetylation levels and, thus, gene expression and underlying brain functions (Singh and Paramanik, 2022).
The methyl group transferred to the CpG is donated by S-adenyl-methionine (SAM). The synthesis of SAM is catalyzed by methionine adenosyltransferase enzymes using the amino acid methionine and ATP (Cantoni, 1953). After removal of a methyl group, SAM is converted to S-adenosyl-homocysteine (SAH) and finally to homocysteine. This homocysteine is then recycled methionine by a vitamin B12-dependent enzyme methionine synthase with the help of 5-methyltetrahydrofolate, a folate derivative. Thus, methionine, vitamin B12, and folate are essential to maintain the methylation level. SAH is a potent inhibitor of DNMTs, therefore, accumulation of SAH affects the DNA methylation process. In a long-term population-based study, Mihara et al. (2022) reported that a high SAM/SAH ratio is associated with a lower risk of dementia and death. Using a folate and vitamin B12-deprived media to mimic AD-like conditions, Fuso et al. (2005) reported that the methylation at the promoter of APP and Presenilin1 decreased, and their expression increased in neuroblastoma cell lines. The upregulation of APP and Presenilin1 leads to the production of higher amyloid β levels. Further, administration of SAM in the deprived media reversed promoter methylation, APP, and Presenilin1 expression, and the reduced amyloid β level. Similarly, they also showed that deprivation of vitamin B12, B6, and folate dietary deficiency leads to hyperhomocysteinemia due to alteration of SAM and SAH level in TgCRND8 and 129Sv mice. The imbalance of SAM and SAH is associated with the upregulation of PS1 and BACE with higher amyloid β deposition and cognitive impairment (Fuso et al., 2008). This showed that an imbalance in SAM and SAH levels decreased methylation at the promoter of APP, PS1, and BACE that leads to higher amyloid β deposition, a characteristic feature of AD. To check the effect of early-life SAM supplementation on AD symptoms in an AD mice mouse model, Raia et al. (2023) reported that perinatal supplementation of SAM repressed PS1 expression and amyloid β deposition in adult AD mice. Further, the effect of perinatal SAM supplementation is similar to long-term post-weaning supplementation of SAM regarding AD symptom manifestation in adult AD mice. This showed early life SAM supplementation is beneficial and can lead to healthy aging.
The acetyl group on histone acetylation is donated by Acetyl CoA, a product of glucose metabolism. Metabolic enzymes like Pyruvate dehydrogenase complex (PDC) and ATP citrate lyase (ACL) regulate the synthesis of Acetyl CoA and are thus involved in chromatin remodelling and gene expression (Brinton, 2008; Biju et al., 2024). Wellen et al. (2009) reported that ATP citrate lyase (ACL), a key metabolic enzyme that converts citrate into acetyl-CoA in the cytoplasm and is an essential link between cellular metabolism and histone acetylation level in the nucleus. Further, the metabolic enzyme PDC consists of three enzymes Pyruvate Dehydrogenase (E1), Dihydrolipoyl transacetylase (E2), and Dihydrolipoyl dehydrogenase (E3) is first to convert pyruvate into Acetyl CoA in the mitochondria which then used in the citric acid cycle to generate ATP and Citrate which ACL then uses to synthesize Acetyl CoA, Acetyl group donor for histone (Choudhary et al., 2014). Alterations in the expression and activities of these metabolic enzymes are found in aging and neurodegenerative diseases. Perry et al. (1980) reported that the ACL and PDC activity decreased in the post-mortem brain tissue of AD patients. Similarly, Sorbi et al. (1983) observed that PDC activity decreased the brain of AD and Huntington disease patients. Further, Wellen et al. (2009) established that ATP-citrate lyase and citrate are essential for metabolism and histone acetylation. Jiang et al. (2008) elucidated the metabolic profile in SAMP8 mice, a rodent model, to study age-associated memory impairment and neurodegeneration. They reported that the level of many metabolites that are responsible for cellular metabolism, including citrate and pyruvate, is reduced in the serum. Further, the alteration of these metabolites is higher in females than males. This may be one reason that females are more vulnerable to neurodegenerative diseases. Peleg et al. (2010) also reported that the citrate level is crucial for nuclear histone acetylation and gene expression. The reduction of citrate level impaired histone acetylation level, gene expression, and memory in old age. Short-chain fatty acid such as butyrate is synthesized as a result of the fermentation of dietary fiber by the microbes in the intestine. Further, dietary supplementation of high fiber diet or butyrate producing microbe improve cognitive functions, reduced anxiety and stress as compared to low fiber diet in human subjects and animal models (Khan et al., 2015; Bourassa et al., 2016). In a “MitoPark” PD mice model, mitochondrial stress associated with increased H4K12 acetylation and degeneration of DAergic neurons. This change in H4K12 acetylation level is due to imbalance of HATs and HDACs (Huang et al., 2024).
Sex steroid hormone, such as estrogen 17β-estradiol (E2), plays a crucial role in learning and memory by regulating hippocampal neuronal morphology, plasticity, and memory in different models, and its decline after menopause severely increases cognitive declined and chance of neurodegenerative during aging (Frick, 2009; Pike et al., 2009; Viña and Lloret, 2010; Zárate et al., 2017; Singh and Paramanik, 2022). Initial research in the field of E2-mediated epigenetic regulation showed E2-induced DNA demethylation and histone acetylation in the brains of young and adult rats as compared to old rats (Thakur et al., 1978; Thakur and Kanungo, 1981). These changes in DNA methylation and histone acetylation were associated with higher gene transcription in young and adults when compared to old rats (Kanungo and Thakur, 1979). Estrogen-mediated signaling pathways help mitochondria to enhance aerobic respiration through the coupling of glycolysis to the Krebs cycle and ATP synthesis in hippocampal and cortical neurons. Apart from mitochondrial function and bioenergetics, E2 regulates the expression of enzymes like DNMTs, HDAC2, CBP, PDC, and ACL and thus indirectly regulates chromatin remodelling and gene expression (Pearce and Balnave, 1976; Brinton, 2008). Zhao et al. (2010) observed that hippocampal administration of E2 increased the expression of DNMT3a and DNMT3b and improved recognition memory in mice. Further, this effect of E2 was diminished when mice co-treated with 5-aza-2′-deoxycytidine. Further, they also reported that intrahippocampal administration of E2 decreased HDAC2 expression, increased H3/H4 acetylation level, and improved recognition memory. Thus, E2 plays an important role in regulating histone acetylation modifications and synaptic plasticity gene expression and improves cognitive performance in aged as well as AD mouse models (Frick et al., 2002; Heikkinen et al., 2004; Frye et al., 2005).
Therapeutic potential of epigenetic modifiers
Epigenetic modifications such as DNA methylaton and histone acetylation are reversible in nature. Therefore, these modifications are suitable for drug targeting. Several drugs, small molecules as well as plant derived molecules and herbal formulations are known to be target chromatin modifying enzymes and underlying epigenetic modifications.
Phytoestrogens are a group of plant derived compounds having similar chemical structure to steroid hormone estrogen. Similar to estrogen, phytoestrogens also shown to modulate the DNA methylation and histone PTMs. Genistein is a plant polyphenols, most abundantly found in soy and soy-based product. Due to its structural similarities with estrogen, genistein is also known as phytoestrogen. Studies in animal model suggests that genistein is neuroprotective in nature and help in learning and memory. Bagheri et al. (2012) reported that genistein treatment reduced the Aβ1-40 plaque formation in the hippocampus and improved learning and memory in AD rat model. Khodamoradi et al. (2017) reported that intraperitoneal administration of soy extract containing genistein improved long term potentiation, learning and memory in ovariectomized rats as compared to control rats. Studies in different cancer cell line suggests geneistein also found to be regulate epigenetic modification such as DNA methylation and histone acetylation. Genestein treatment alters the expression of chromatin modifying enzymes. The expression of DNMTs (DNMT1, DNMT3a and DNMT3b) and HDACs (HDAC1, HDAC5 and HDAC6) downregulated while HATs (CIITA and ESCO2) upregulated in geneistein treated HeLa cells. Also geneistein treatment decreased the activity of DNMTs and HDACs as well as global DNA CpG methylation level (Sundaram et al., 2019). Geneistein treatment decreased DNMT activity, DNMT1 expression and global DNA methylation. In slilico analysis showed genestein inhibit the activity of DNMT1 by interacting directly with its catalytic domain Xie et al. (2014).
Epigallocatechin-3-gallate is a polyphenols found in tea. Fang et al. (2007) reported that Epigallocatechin-3-gallate inhibits DNMTs that leading to promoter specific hypomethylation and increased gene expression in esophageal cancer cell line. Apart from acts as DNMT inhibitor, Epigallocatechin-3-gallate also inhibits HDACs and decreased the expression of APP in neuronal cell (Hu et al., 2015). Epigallocatechin-3-gallate treatment inhibited HDAC1 activity, downregulated APP expression and decreased Aβ level in AD mice model (Chang et al., 2015). Curcumin is polyphenolic compound found in the turmaric. Due to its anti-inflammatory and antioxidant properties turmaric is commonly used in tradiational Indian ayurvedic medicine (Hatami et al., 2019; Sharifi-Rad et al., 2020). Apart from these, curcumin also modulates chromatin modifying enzymes and underlying gene expression (Abdul-Rahman et al., 2024). Several studies showed that curcumin inhibits DNMTs activity and expression; and thus decreased methylation level in cancer cell (Liu et al., 2009; Yu et al., 2013; Hassan et al., 2015). Lu et al. (2014) reported that curcumin inhibits HAT/P300, decreased promoter level histone H3 acetylation and the expression of PS1 and BACE in cellular model of AD. Dysregulation of SAM level altered expression and accumulation of APP and phosphorylated Tau, which in turn increased Aβ secretion in AD cellular model (Sontag et al., 2007). Similarly, Fuso et al. (2007) reported that folic acid, vitamin B12 and B6 deprivation reduced SAM level and increased AD related genes and Aβ secretion in neuroblastoma cell line. Further, supplementation SAM reverse the AD related gene expression and Aβ secretion. Lee et al. (2024) reported that supplementation of folic acid and vitamin B12 for 6 months improved Mini-Mental State Examination scored in AD patients as compared to controls. These result suggest phytochemicals and small molecules regulate expression and activities of chromatin modifying enzymes and plays important role in recovery of brain functions and learing and memory in during aging and associated pathologies.
Conclusion
Due to their dynamic nature, DNA methylation and histone acetylation levels alter during different physiological conditions including aging, which inhibits gene expression and impaired synaptic plasticity. Reports also showed that these epigenetic marks modulated by physiology and metabolic processes and dietary supplementation epigenetic modifiers are beneficial in improving chromatin modifications and brain functions (Figure 1). Growing evidence showed the presence of PDC in the nucleus, synthesizes Acetyl CoA, and donates the acetyl group for histone acetylation. This nuclear PDC is translocated directly from the mitochondrial matrix to the nucleus (de Boer and Houten, 2014; Ng and Tang, 2014; Sutendra et al., 2014). Therefore, it will be essential to study the translocation of PDC from mitochondria as mitochondria are mostly affected during aging and neurodegenerative diseases. Further, these epigenetic marks, gene expression, and memory can be reversed by applying enzyme inhibitors. Phytochemicals such as phytoestrogens, polyphenols and as well as B vitamins plays important role in regulating the expression and activities chromatin modifying enzymes, global and promoter level DNA methylation, histone acetylation and gene expression. Further, many phytochemicals and small moleules also showed chromatin modifying activities but they have not been explored in brain or neuronal cells. As chromatin-modifying enzymes regulate diverse functions apart from regulating synaptic plasticity, the application of these inhibitors showed many side effects (Subramanian et al., 2010; Zhang et al., 2022). Thus, the designing of specific inhibitors is also needed apart from supplementation to rescue epigenetic dysregulation and memory during neurodegenerative diseases.
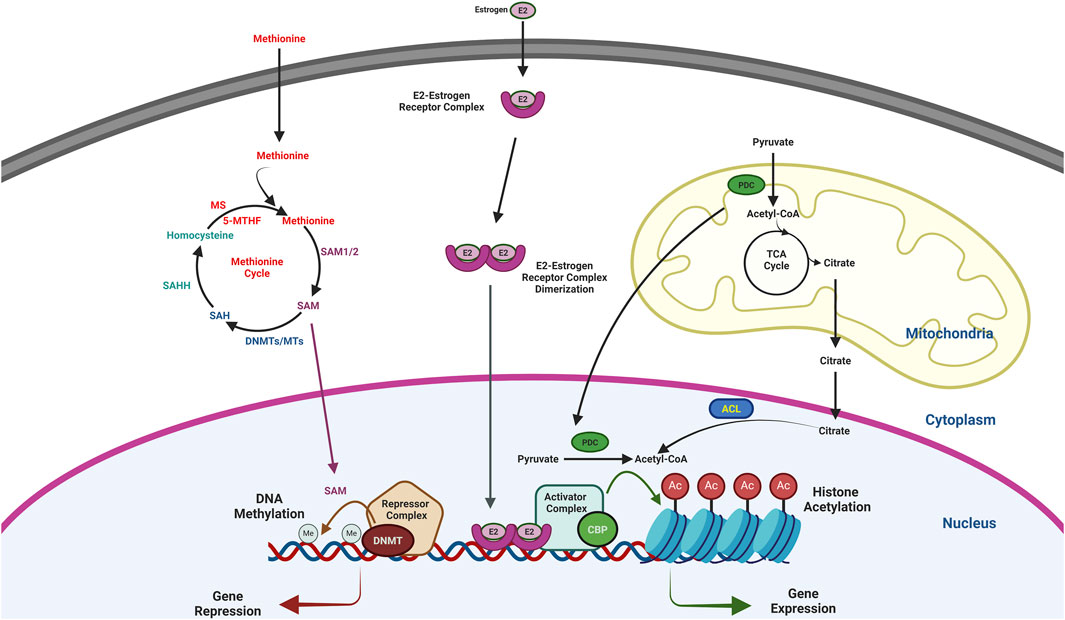
Figure 1. Schematic diagram showing the regulation of gene expression by DNA methylation and histone acetylation. DNMT, a part of the repressor complex methylates DNA that inhibit gene expression. On the other hand, CBP, a part of activator complex acetylates histone that activate gene expression. SAM, which is the substrate of DNA methylation is a product of Methionine cycle. After donating the methyl group, SAM converted to SAH and then homocysteine to continue the metheonine cycle. Further, Acetyl-CoA, the substrate for the histone acetylation is mainly synthesized by PDC in the nucleus and ACL by using the citrate, a product of TCA cycle in the mitochondria. Therefore, SAM and Acetyl-CoA play an important role in regulating the epigenetic modifications and gene expression. E2 also plays a crucial role in regulating histone acetylation and gene expression. E2-ER dimer after translocate to the nucleus recruits the activator complex and helps in histone acetylation and gene expression.
Author contributions
PS: Conceptualization, Writing–original draft, Writing–review and editing. VP: Conceptualization, Funding acquisition, Supervision, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. PS acknowledges Department of Biotechnology (DBT), Government of India for the DBT-Research Associate Fellowship (Award Letter No. DBT-RA/2021/January/N/807). VP acknowledges Indian Council of Medical Research (Grant No-2021-11648) New Delhi for financial support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdul-Rahman, T., Awuah, W. A., Mikhailova, T., Kalmanovich, J., Mehta, A., Ng, J. C., et al. (2024). Antioxidant, anti-inflammatory and epigenetic potential of curcumin in Alzheimer's disease. Biofactors 50, 693–708. doi:10.1002/biof.2039
Alarcón, J. M., Malleret, G., Touzani, K., Vronskaya, S., Ishii, S., Kandel, E. R., et al. (2004). Chromatin acetylation, memory, and LTP are impaired in CBP+/- mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 42, 947–959. doi:10.1016/j.neuron.2004.05.021
Bagheri, M., Roghani, M., Joghataei, M. T., and Mohseni, S. (2012). Genistein inhibits aggregation of exogenous amyloid-beta₁₋₄₀ and alleviates astrogliosis in the hippocampus of rats. Brain Res. 1429, 145–154. doi:10.1016/j.brainres.2011.10.020
Barrett, R. M., Malvaez, M., Kramar, E., Matheos, D. P., Arrizon, A., Cabrera, S. M., et al. (2011). Hippocampal focal knockout of CBP affects specific histone modifications, long-term potentiation, and long-term memory. Neuropsychopharmacology 36, 1545–1556. doi:10.1038/npp.2011.61
Bedford, D. C., and Brindle, P. K. (2012). Is histone acetylation the most important physiological function for CBP and p300? Aging 4, 247–255. doi:10.18632/aging.100453
Biju, A. K., Chennam Lakshmikumar, R. R., and Rengasamy, K. R. R. (2024). ATP-Citrate Lyase (ACLY): an extensive investigation from molecular insight to therapeutic implications. Nat. Resour. Hum. Health 4, 208–229. doi:10.53365/nrfhh/189500
Bourassa, M. W., Alim, I., Bultman, S. J., and Ratan, R. R. (2016). Butyrate, neuroepigenetics and the gut microbiome: can a high fiber diet improve brain health? Neurosci. Lett. 625, 56–63. doi:10.1016/j.neulet.2016.02.009
Brinton, R. D. (2008). The healthy cell bias of estrogen action: mitochondrial bioenergetics and neurological implications. Trends Neurosci. 31, 529–537. doi:10.1016/j.tins.2008.07.003
Caccamo, A., Maldonado, M. A., Bokov, A. F., Majumder, S., and Oddo, S. (2010). CBP gene transfer increases BDNF levels and ameliorates learning and memory deficits in a mouse model of Alzheimer's disease. Proc. Nat. Acad. Sci. 107, 22687–22692. doi:10.1073/pnas.1012851108
Cantoni, G. L. (1953). S-Adenosylmethionine; a new intermediate formed enzymatically from L-methionine and adenosinetriphosphate. J. Biol. Chem. 204, 403–416. doi:10.1016/s0021-9258(18)66148-4
Chahrour, M., Jung, S. Y., Shaw, C., Zhou, X., Wong, S. T., Qin, J., et al. (2008). MeCP2, a key contributor to neurological disease, activates and represses transcription. Sci. (New York, N.Y.) 320, 1224–1229. doi:10.1126/science.1153252
Chang, X., Rong, C., Chen, Y., Yang, C., Hu, Q., Mo, Y., et al. (2015). (-)-Epigallocatechin-3-gallate attenuates cognitive deterioration in Alzheimer's disease model mice by upregulating neprilysin expression. Exp. Cell Res. 334, 136–145. doi:10.1016/j.yexcr.2015.04.004
Chatterjee, S., Mizar, P., Cassel, R., Neidl, R., Selvi, B. R., Mohankrishna, D. V., et al. (2013). A novel activator of CBP/p300 acetyltransferases promotes neurogenesis and extends memory duration in adult mice. J. Neurosci. 33, 10698–10712. doi:10.1523/JNEUROSCI.5772-12.2013
Chen, G., Zou, X., Watanabe, H., van Deursen, J. M., and Shen, J. (2010). CREB binding protein is required for both short-term and long-term memory formation. J. Neurosci. 30, 13066–13077. doi:10.1523/JNEUROSCI.2378-10.2010
Choudhary, C., Weinert, B. T., Nishida, Y., Verdin, E., and Mann, M. (2014). The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550. doi:10.1038/nrm3841
Chouliaras, L., Mastroeni, D., Delvaux, E., Grover, A., Kenis, G., Hof, P. R., et al. (2013). Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol. Aging 34, 2091–2099. doi:10.1016/j.neurobiolaging.2013.02.021
Chung, Y. H., Kim, E. J., Shin, C. M., Joo, K. M., Kim, M. J., Woo, H. W., et al. (2002). Age-related changes in CREB binding protein immunoreactivity in the cerebral cortex and hippocampus of rats. Brain Res. 956, 312–318. doi:10.1016/s0006-8993(02)03562-x
Clayton, A. L., Hazzalin, C. A., and Mahadevan, L. C. (2006). Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell. 23, 289–296. doi:10.1016/j.molcel.2006.06.017
Crump, N. T., Hazzalin, C. A., Bowers, E. M., Alani, R. M., Cole, P. A., and Mahadevan, L. C. (2011). Dynamic acetylation of all lysine-4 trimethylated histone H3 is evolutionarily conserved and mediated by p300/CBP. Proc. Nat. Acad. Sci. 108, 7814–7819. doi:10.1073/pnas.1100099108
de Boer, V. C., and Houten, S. M. (2014). A mitochondrial expatriate: nuclear pyruvate dehydrogenase. Cell 158, 9–10. doi:10.1016/j.cell.2014.06.018
Elsner, V. R., Lovatel, G. A., Moysés, F., Bertoldi, K., Spindler, C., Cechinel, L. R., et al. (2013). Exercise induces age-dependent changes on epigenetic parameters in rat hippocampus: a preliminary study. Exp. Gerontol. 48, 136–139. doi:10.1016/j.exger.2012.11.011
Fang, M., Chen, D., and Yang, C. S. (2007). Dietary polyphenols may affect DNA methylation. J. Nutr. 223S-228S. doi:10.1093/jn/137.1.223S
Feng, J., Zhou, Y., Campbell, S. L., Le, T., Li, E., Sweatt, J. D., et al. (2010). Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 13, 423–430. doi:10.1038/nn.2514
Fischer, A., Sananbenesi, F., Wang, X., Dobbin, M., and Tsai, L. H. (2007). Recovery of learning and memory is associated with chromatin remodelling. Nature 447, 178–182. doi:10.1038/nature05772
Fortress, A. M., Kim, J., Poole, R. L., Gould, T. J., and Frick, K. M. (2014). 17β-Estradiol regulates histone alterations associated with memory consolidation and increases Bdnf promoter acetylation in middle-aged female mice. Learn. Mem. (Cold Spring Harb. N.Y.) 21, 457–467. doi:10.1101/lm.034033.113
Frick, K. M. (2009). Estrogens and age-related memory decline in rodents: what have we learned and where do we go from here? Horm. Behav. 55, 2–23. doi:10.1016/j.yhbeh.2008.08.015
Frick, K. M., Fernandez, S. M., and Bulinski, S. C. (2002). Estrogen replacement improves spatial reference memory and increases hippocampal synaptophysin in aged female mice. Neuroscience 115, 547–558. doi:10.1016/s0306-4522(02)00377-9
Frick, K. M., Kim, J., Tuscher, J. J., and Fortress, A. M. (2015). Sex steroid hormones matter for learning and memory: estrogenic regulation of hippocampal function in male and female rodents. Learn. Mem. 22, 472–493. doi:10.1101/lm.037267.114
Frye, C. A., Rhodes, M. E., and Dudek, B. (2005). Estradiol to aged female or male mice improves learning in inhibitory avoidance and water maze tasks. Brain Res. 1036, 101–108. doi:10.1016/j.brainres.2004.12.014
Fuso, A., Cavallaro, R. A., Zampelli, A., D'Anselmi, F., Piscopo, P., Confaloni, A., et al. (2007). gamma-Secretase is differentially modulated by alterations of homocysteine cycle in neuroblastoma and glioblastoma cells. J. Alzheimers Dis. 11, 275–290. doi:10.3233/jad-2007-11303
Fuso, A., Nicolia, V., Cavallaro, R. A., Ricceri, L., D'Anselmi, F., Coluccia, P., et al. (2008). B-vitamin deprivation induces hyperhomocysteinemia and brain S-adenosylhomocysteine, depletes brain S-adenosylmethionine, and enhances PS1 and BACE expression and amyloid-beta deposition in mice. Mol. Cell Neurosci. 37, 731–746. doi:10.1016/j.mcn.2007.12.018
Fuso, A., Seminara, L., Cavallaro, R. A., D'Anselmi, F., and Scarpa, S. (2005). S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol. Cell Neurosci. 28, 195–204. doi:10.1016/j.mcn.2004.09.007
Gao, J., Cahill, C. M., Huang, X., Roffman, J. L., Lamon-Fava, S., Fava, M., et al. (2018). S-adenosyl methionine and transmethylation pathways in neuropsychiatric diseases throughout life. Neurotherapeutics 15, 156–175. doi:10.1007/s13311-017-0593-0
Giralt, A., Puigdellívol, M., Carretón, O., Paoletti, P., Valero, J., Parra-Damas, A., et al. (2012). Long-term memory deficits in Huntington's disease are associated with reduced CBP histone acetylase activity. Hum. Mol. Gen. 21, 1203–1216. doi:10.1093/hmg/ddr552
Golshani, P., Hutnick, L., Schweizer, F., and Fan, G. (2005). Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Rel Sys 3, 227–233. doi:10.1017/S1472928807000222
Gräff, J., Rei, D., Guan, J. S., Wang, W. Y., Seo, J., Hennig, K. M., et al. (2012). An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature 483, 222–226. doi:10.1038/nature10849
Guan, J. S., Haggarty, S. J., Giacometti, E., Dannenberg, J. H., Joseph, N., Gao, J., et al. (2009). HDAC2 negatively regulates memory formation and synaptic plasticity. Nature 459, 55–60. doi:10.1038/nature07925
Handy, D. E., Castro, R., and Loscalzo, J. (2011). Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation 123, 2145–2156. doi:10.1161/CIRCULATIONAHA.110.956839
Hassan, H. E., Carlson, S., Abdallah, I., Buttolph, T., Glass, K. C., and Fandy, T. E. (2015). Curcumin and dimethoxycurcumin induced epigenetic changes in leukemia cells. Pharm. Res. 32, 863–875. doi:10.1007/s11095-014-1502-4
Hatami, M., Abdolahi, M., Soveyd, N., Djalali, M., Togha, M., and Honarvar, N. M. (2019). Molecular mechanisms of curcumin in neuroinflammatory disorders: a Mini review of current evidences. Immune Disord. Drug Targets 19, 247–258. doi:10.2174/1871530319666181129103056
Heikkinen, T., Puoliväli, J., and Tanila, H. (2004). Effects of long-term ovariectomy and estrogen treatment on maze learning in aged mice. Exp. Gerontol. 39, 1277–1283. doi:10.1016/j.exger.2004.05.005
Hermann, P. M., Watson, S. N., and Wildering, W. C. (2014). Phospholipase A2 - nexus of aging, oxidative stress, neuronal excitability, and functional decline of the aging nervous system? Insights from a snail model system of neuronal aging and age-associated memory impairment. Front. Gen. 5, 419. doi:10.3389/fgene.2014.00419
Hsiao, Y. H., Hung, H. C., Yu, Y. J., Su, C. L., Chen, S. H., and Gean, P. W. (2017). Co-housing reverses memory decline by epigenetic regulation of brain-derived neurotrophic factor expression in an animal model of Alzheimer's disease. Learn. Mem. 141, 1–8. doi:10.1016/j.nlm.2017.02.020
Hu, Q., Chang, X., Yan, R., Rong, C., Yang, C., Cheng, S., et al. (2015). (-)-Epigallocatechin-3-gallate induces cancer cell apoptosis via acetylation of amyloid precursor protein. Med. Oncol. 32, 390. doi:10.1007/s12032-014-0390-0
Huang, M., Jin, H., Anantharam, V., Kanthasamy, A., and Kanthasamy, A. G. (2024). Mitochondrial stress-induced H4K12 hyperacetylation dysregulates transcription in Parkinson's disease. Front. Cell Neurosci. 18, 1422362. doi:10.3389/fncel.2024.1422362
Intlekofer, K. A., Berchtold, N. C., Malvaez, M., Carlos, A. J., McQuown, S. C., Cunningham, M. J., et al. (2013). Exercise and sodium butyrate transform a subthreshold learning event into long-term memory via a brain-derived neurotrophic factor dependent mechanism. Neuropsychopharmacology 38, 2027–2034. doi:10.1038/npp.2013.104
Jiang, N., Yan, X., Zhou, W., Zhang, Q., Chen, H., Zhang, Y., et al. (2008). NMR-based metabonomic investigations into the metabolic profile of the senescence-accelerated mouse. J. Proteome Res. 7, 3678–3686. doi:10.1021/pr800439b
Jowaed, A., Schmitt, I., Kaut, O., and Wüllner, U. (2010). Methylation regulates alpha-synuclein expression and is decreased in Parkinson's disease patients' brains. J. Neurosci. 30, 6355–6359. doi:10.1523/JNEUROSCI.6119-09.2010
Ju, L. S., Jia, M., Sun, J., Sun, X. R., Zhang, H., Ji, M. H., et al. (2016). Hypermethylation of hippocampal synaptic plasticity-related genes is involved in neonatal sevoflurane exposure-induced cognitive impairments in rats. Neurotox. Res. 29, 243–255. doi:10.1007/s12640-015-9585-1
Kameyama, A., Asai, H., Nomoto, M., Ohno, S., Ghandour, K., Ohkawa, N., et al. (2022). Sevoflurane-induced amnesia is associated with inhibition of hippocampal cell ensemble activity after learning. Biol. Open 11, bio059666. doi:10.1242/bio.059666
Kandel, E. R. (2001). The molecular biology of memory storage: a dialogue between genes and synapses. Science 294, 1030–1038. doi:10.1126/science.1067020
Kanungo, M. S., and Thakur, M. K. (1979). Modulation of acetylation of histones and transcription of chromatin by butyric acid and 17beta-estradiol in the brain of rats of various ages. Biochem. Biophys. Res. Commun. 87, 266–271. doi:10.1016/0006-291x(79)91675-9
Kelly, T. K., De Carvalho, D. D., and Jones, P. A. (2010). Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 28, 1069–1078. doi:10.1038/nbt.1678
Khan, N. A., Raine, L. B., Drollette, E. S., Scudder, M. R., Kramer, A. F., and Hillman, C. H. (2015). Dietary fiber is positively associated with cognitive control among prepubertal children. J. Nutr. 145, 143–149. doi:10.3945/jn.114.198457
Khodamoradi, M., Asadi-Shekaari, M., Esmaeili-Mahani, S., Sharififar, F., and Sheibani, V. (2017). Effects of hydroalcoholic extract of soy on learning, memory and synaptic plasticity deficits induced by seizure in ovariectomized rats. Basic Clin. Neurosci. 8, 395–403. doi:10.18869/nirp.bcn.8.5.395
Kilgore, M., Miller, C. A., Fass, D. M., Hennig, K. M., Haggarty, S. J., Sweatt, J. D., et al. (2010). Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology 35, 870–880. doi:10.1038/npp.2009.197
Klein, C. J., Botuyan, M. V., Wu, Y., Ward, C. J., Nicholson, G. A., Hammans, S., et al. (2011). Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat. Gen. 43, 595–600. doi:10.1038/ng.830
Klein, H. U., McCabe, C., Gjoneska, E., Sullivan, S. E., Kaskow, B. J., Tang, A., et al. (2019). Epigenome-wide study uncovers large-scale changes in histone acetylation driven by tau pathology in aging and Alzheimer's human brains. Nat. Neurosci. 22, 37–46. doi:10.1038/s41593-018-0291-1
Korzus, E., Rosenfeld, M. G., and Mayford, M. (2004). CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron 42, 961–972. doi:10.1016/j.neuron.2004.06.002
Lee, C. Y., Chan, L., Hu, C. J., Hong, C. T., and Chen, J. H. (2024). Role of vitamin B12 and folic acid in treatment of Alzheimer's disease: a meta-analysis of randomized control trials. Aging (Albany NY) 16, 7856–7869. doi:10.18632/aging.205788
Levenson, J. M., O'Riordan, K. J., Brown, K. D., Trinh, M. A., Molfese, D. L., and Sweatt, J. D. (2004). Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 279, 40545–40559. doi:10.1074/jbc.M402229200
Levenson, J. M., Qiu, S., and Weeber, E. J. (2008). The role of reelin in adult synaptic function and the genetic and epigenetic regulation of the reelin gene. Biochimi Biophys. 1779, 422–431. doi:10.1016/j.bbagrm.2008.01.001
Liu, L., van Groen, T., Kadish, I., Li, Y., Wang, D., James, S. R., et al. (2011). Insufficient DNA methylation affects healthy aging and promotes age-related health problems. Clin. Epigen. 2, 349–360. doi:10.1007/s13148-011-0042-6
Liu, Z., Xie, Z., Jones, W., Pavlovicz, R. E., Liu, S., Yu, J., et al. (2009). Curcumin is a potent DNA hypomethylation agent. Bioorg. Med. Chem. Lett. 19, 706–709. doi:10.1016/j.bmcl.2008.12.041
Lopez-Atalaya, J. P., Ciccarelli, A., Viosca, J., Valor, L. M., Jimenez-Minchan, M., Canals, S., et al. (2011). CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO. J. 30, 4287–4298. doi:10.1038/emboj.2011.299
Lossi, L., Castagna, C., and Merighi, A. (2024). An overview of the epigenetic modifications in the brain under normal and pathological conditions. Int. J. Mol. Sci. 25, 3881. doi:10.3390/ijms25073881
Lu, H., Liu, X., Deng, Y., and Qing, H. (2013). DNA methylation, a hand behind neurodegenerative diseases. Front. Aging Neurosci. 5, 85. doi:10.3389/fnagi.2013.00085
Lu, X., Deng, Y., Yu, D., Cao, H., Wang, L., Liu, L., et al. (2014). Histone acetyltransferase p300 mediates histone acetylation of PS1 and BACE1 in a cellular model of Alzheimer's disease. PLoS One 9, e103067. doi:10.1371/journal.pone.0103067
Lubin, F. D., and Sweatt, J. D. (2007). The IkappaB kinase regulates chromatin structure during reconsolidation of conditioned fear memories. Neuron 55, 942–957. doi:10.1016/j.neuron.2007.07.039
Maddox, S. A., Watts, C. S., and Schafe, G. E. (2013). p300/CBP histone acetyltransferase activity is required for newly acquired and reactivated fear memories in the lateral amygdala. Learn. Mem. 20, 109–119. doi:10.1101/lm.029157.112
Marzi, S. J., Leung, S. K., Ribarska, T., Hannon, E., Smith, A. R., Pishva, E., et al. (2018). A histone acetylome-wide association study of Alzheimer's disease identifies disease-associated H3K27ac differences in the entorhinal cortex. Nat. Neurosci. 21, 1618–1627. doi:10.1038/s41593-018-0253-7
Mastroeni, D., Grover, A., Delvaux, E., Whiteside, C., Coleman, P. D., and Rogers, J. (2011). Epigenetic mechanisms in Alzheimer's disease. Neurobiol. Aging. 32, 1161–1180. doi:10.1016/j.neurobiolaging.2010.08.017
McClarty, B. M., Rodriguez, G., and Dong, H. (2024). Class 1 histone deacetylases differentially modulate memory and synaptic genes in a spatial and temporal manner in aged and APP/PS1 mice. Brain Res. 1837, 148951. doi:10.1016/j.brainres.2024.148951
Mihara, A., Ohara, T., Hata, J., Chen, S., Honda, T., Tamrakar, S., et al. (2022). Association of serum s-adenosylmethionine, s-adenosylhomocysteine, and their ratio with the risk of dementia and death in a community. Sci. Rep. 12, 12427. doi:10.1038/s41598-022-16242-y
Miller, C. A., and Sweatt, J. D. (2007). Covalent modification of DNA regulates memory formation. Neuron 53, 857–869. doi:10.1016/j.neuron.2007.02.022
Mitchnick, K. A., Creighton, S. D., Cloke, J. M., Wolter, M., Zaika, O., Christen, B., et al. (2016). Dissociable roles for histone acetyltransferases p300 and PCAF in hippocampus and perirhinal cortex-mediated object memory. Genes, Brain, Behav. 15, 542–557. doi:10.1111/gbb.12303
Monti, N., Cavallaro, R. A., Stoccoro, A., Nicolia, V., Scarpa, S., Kovacs, G. G., et al. (2020). CpG and non-CpG Presenilin1 methylation pattern in course of neurodevelopment and neurodegeneration is associated with gene expression in human and murine brain. Epigenetics 15, 781–799. doi:10.1080/15592294.2020.1722917
Moore, L. D., Le, T., and Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38. doi:10.1038/npp.2012.112
Nativio, R., Donahue, G., Berson, A., Lan, Y., Amlie-Wolf, A., Tuzer, F., et al. (2018). Dysregulation of the epigenetic landscape of normal aging in Alzheimer's disease. Nat. Neurosci. 21, 497–505. doi:10.1038/s41593-018-0101-9
Nativio, R., Lan, Y., Donahue, G., Sidoli, S., Berson, A., Srinivasan, A. R., et al. (2020). An integrated multi-omics approach identifies epigenetic alterations associated with Alzheimer's disease. Nat. Genet. 52, 1024–1035. doi:10.1038/s41588-020-0696-0
Negi, S., Khuran, N., and Duggal, N. (2024). The misfolding mystery: α-synuclein and the pathogenesis of Parkinson’s disease. Neurochem Int. 177, 105760. doi:10.1016/j.neuint.2024.105760
Ng, F., and Tang, B. L. (2014). Pyruvate dehydrogenase complex (PDC) export from the mitochondrial matrix. Mol. Memb. Biol. 31, 207–210. doi:10.3109/09687688.2014.987183
Oliveira, A. M., Hemstedt, T. J., and Bading, H. (2012). Rescue of aging-associated decline in Dnmt3a2 expression restores cognitive abilities. Nat. Neurosci. 15, 1111–1113. doi:10.1038/nn.3151
Park, G., Tan, J., Garcia, G., Kang, Y., Salvesen, G., and Zhang, Z. (2016). Regulation of histone acetylation by autophagy in Parkinson disease. J. Biol. Chem. 291, 3531–3540. doi:10.1074/jbc.M115.675488
Partanen, A., Motoyama, J., and Hui, C. C. (1999). Developmentally regulated expression of the transcriptional cofactors/histone acetyltransferases CBP and p300 during mouse embryogenesis. Int. J. Dev. Biol. 43, 487–494.
Pearce, J., and Balnave, D. (1976). The effects of estradiol administration of the hepatic activities of some enzymes of carbohydrate, amino acid and lipid metabolism in the immature pullet. Horm. Met. Res. 8, 181–183. doi:10.1055/s-0028-1093656
Peleg, S., Sananbenesi, F., Zovoilis, A., Burkhardt, S., Bahari-Javan, S., Agis-Balboa, R. C., et al. (2010). Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 328, 753–756. doi:10.1126/science.1186088
Penner, M. R., Parrish, R. R., Hoang, L. T., Roth, T. L., Lubin, F. D., and Barnes, C. A. (2016). Age-related changes in Egr1 transcription and DNA methylation within the hippocampus. Hippocampus 26, 1008–1020. doi:10.1002/hipo.22583
Penner, M. R., Roth, T. L., Chawla, M. K., Hoang, L. T., Roth, E. D., Lubin, F. D., et al. (2011). Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging. 32, 2198–2210. doi:10.1016/j.neurobiolaging.2010.01.009
Perry, E. K., Perry, R. H., Tomlinson, B. E., Blessed, G., and Gibson, P. H. (1980). Coenzyme A-acetylating enzymes in Alzheimer's disease: possible cholinergic 'compartment' of pyruvate dehydrogenase. Neurosci. Lett. 18, 105–110. doi:10.1016/0304-3940(80)90220-7
Pike, C. J., Carroll, J. C., Rosario, E. R., and Barron, A. M. (2009). Protective actions of sex steroid hormones in Alzheimer's disease. Front. Neuroendocr. 30, 239–258. doi:10.1016/j.yfrne.2009.04.015
Raia, T., Armeli, F., Cavallaro, R. A., Ferraguti, G., Businaro, R., Lucarelli, M., et al. (2023). Perinatal S-adenosylmethionine supplementation represses PSEN1 expression by the cellular epigenetic memory of CpG and non-CpG methylation in adult TgCRD8 mice. Int. J. Mol. Sci. 24, 11675. doi:10.3390/ijms241411675
Ramazi, S., Allahverdi, A., and Zahiri, J. (2020). Evaluation of post-translational modifications in histone proteins: a review on histone modification defects in developmental and neurological disorders. J. Biosci. 45, 135. doi:10.1007/s12038-020-00099-2
Selvi, B. R., Cassel, J. C., Kundu, T. K., and Boutillier, A. L. (2010). Tuning acetylation levels with HAT activators: therapeutic strategy in neurodegenerative diseases. Biochim. Biophys. Acta. 1799, 840–853. doi:10.1016/j.bbagrm.2010.08.012
Sharifi-Rad, J., Rayess, Y. E., Rizk, A. A., Sadaka, C., Zgheib, R., Zam, W., et al. (2020). Turmeric and its major compound curcumin on health: bioactive effects and safety profiles for food, pharmaceutical, biotechnological and medicinal applications. Front. Pharmacol. 11, 01021. doi:10.3389/fphar.2020.01021
Singh, P., Konar, A., Kumar, A., Srivas, S., and Thakur, M. K. (2015). Hippocampal chromatin-modifying enzymes are pivotal for scopolamine-induced synaptic plasticity gene expression changes and memory impairment. J. Neurochem. 134, 642–651. doi:10.1111/jnc.13171
Singh, P., and Paramanik, V. (2022). Neuromodulating roles of estrogen and phytoestrogens in cognitive therapeutics through epigenetic modifications during aging. Front. Aging Neurosci. 14, 945076. doi:10.3389/fnagi.2022.945076
Singh, P., Srivas, S., and Thakur, M. K. (2017). Epigenetic regulation of memory-therapeutic potential for disorders. Curr. Neuropharmacol. 15, 1208–1221. doi:10.2174/1570159X15666170404144522
Singh, P., and Thakur, M. K. (2014). Reduced recognition memory is correlated with decrease in DNA methyltransferase1 and increase in histone deacetylase2 protein expression in old male mice. Biogerontology 15, 339–346. doi:10.1007/s10522-014-9504-5
Singh, P., and Thakur, M. K. (2018). Histone deacetylase 2 inhibition attenuates downregulation of hippocampal plasticity gene expression during aging. Mol. Neurobiol. 55, 2432–2442. doi:10.1007/s12035-017-0490-x
Sontag, E., Nunbhakdi-Craig, V., Sontag, J. M., Diaz-Arrastia, R., Ogris, E., and Dayal, S. (2007). Protein phosphatase 2A methyltransferase links homocysteine metabolism with tau and amyloid precursor protein regulation. J Neurosci. 27 (11), 2751–9. doi:10.1523/JNEUROSCI.3316-06.2007
Sorbi, S., Bird, E. D., and Blass, J. P. (1983). Decreased pyruvate dehydrogenase complex activity in Huntington and Alzheimer brain. Ann. Neurol. 13, 72–78. doi:10.1002/ana.410130116
Srivas, S., and Thakur, M. K. (2017). Epigenetic regulation of neuronal immediate early genes is associated with decline in their expression and memory consolidation in scopolamine-induced amnesic mice. Mol. Neurobiol. 54, 5107–5119. doi:10.1007/s12035-016-0047-4
Su, X., Chu, Y., Kordower, J. H., Li, B., Cao, H., Huang, L., et al. (2015). PGC-1α promoter methylation in Parkinson's disease. PLoS One 10, e0134087. doi:10.1371/journal.pone.0134087
Subramanian, S., Bates, S. E., Wright, J. J., Espinoza-Delgado, I., and Piekarz, R. L. (2010). Clinical toxicities of histone deacetylase inhibitors. Pharmaceuticals 3, 2751–2767. doi:10.3390/ph3092751
Sundaram, M. K., Unni, S., Somvanshi, P., Bhardwaj, T., Mandal, R. K., Hussain, A., et al. (2019). Genistein modulates signaling pathways and targets several epigenetic markers in HeLa cells. Genes (Basel) 10, 955. doi:10.3390/genes10120955
Sutendra, G., Kinnaird, A., Dromparis, P., Paulin, R., Stenson, T. H., Haromy, A., et al. (2014). A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84–97. doi:10.1016/j.cell.2014.04.046
Thakur, M. K., Das, R., and Kanungo, M. S. (1978). Modulation of acetylation of chromosomal proteins of the brain of rats of various ages by epinephrine and estradiol. Biochem. Biophys. Res. Commun. 81, 828–831. doi:10.1016/0006-291x(78)91426-2
Thakur, M. K., and Kanungo, M. S. (1981). Methylation of chromosomal proteins and DNA of rat brain and its modulation by estradiol and calcium during aging. Exp. Gerontol. 16, 331–336. doi:10.1016/0531-5565(81)90052-8
Toker, L., Tran, G. T., Sundaresan, J., Tysnes, O. B., Alves, G., Haugarvoll, K., et al. (2021). Genome-wide histone acetylation analysis reveals altered transcriptional regulation in the Parkinson's disease brain. Mol. Neurodegener. 16, 31. doi:10.1186/s13024-021-00450-7
Valor, L. M., Pulopulos, M. M., Jimenez-Minchan, M., Olivares, R., Lutz, B., and Barco, A. (2011). Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation but does not affect cell viability. J. Neurosci. 31, 1652–1663. doi:10.1523/JNEUROSCI.4737-10.2011
Vieira, P. A., and Korzus, E. (2015). CBP-Dependent memory consolidation in the prefrontal cortex supports object-location learning. Hippocampus 25, 1532–1540. doi:10.1002/hipo.22473
Viña, J., and Lloret, A. (2010). Why women have more Alzheimer's disease than men: gender and mitochondrial toxicity of amyloid-beta peptide. J. Alz. Dis 2, S527–S533. doi:10.3233/jad-2010-100501
Wang, X., Meng, Z. X., Chen, Y. Z., Li, Y. P., Zhou, H. Y., Yang, M., et al. (2020). Enriched environment enhances histone acetylation of NMDA receptor in the hippocampus and improves cognitive dysfunction in aged mice. Neur. Regen. Res. 15, 2327–2334. doi:10.4103/1673-5374.285005
Wellen, K. E., Hatzivassiliou, G., Sachdeva, U. M., Bui, T. V., Cross, J. R., and Thompson, C. B. (2009). ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080. doi:10.1126/science.1164097
West, R. L., Lee, J. M., and Maroun, L. E. (1995). Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer's disease patient. J. Mol. Neurosci. 6, 141–146. doi:10.1007/BF02736773
Wood, M. A., Attner, M. A., Oliveira, A. M., Brindle, P. K., and Abel, T. (2006). A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn. Mem. 13, 609–617. doi:10.1101/lm.213906
Wood, M. A., Kaplan, M. P., Park, A., Blanchard, E. J., Oliveira, A. M., Lombardi, T. L., et al. (2005). Transgenic mice expressing a truncated form of CREB-binding protein (CBP) exhibit deficits in hippocampal synaptic plasticity and memory storage. Learn. Mem. 12, 111–119. doi:10.1101/lm.86605
Xie, Q., Bai, Q., Zou, L. Y., Zhang, Q. Y., Zhou, Y., Chang, H., et al. (2014). Genistein inhibits DNA methylation and increases expression of tumor suppressor genes in human breast cancer cells. Genes Chromosom. Cancer 53, 422–431. doi:10.1002/gcc.22154
Yu, J., Peng, Y., Wu, L. C., Xie, Z., Deng, Y., Hughes, T., et al. (2013). Curcumin down-regulates DNA methyltransferase 1 and plays an anti-leukemic role in acute myeloid leukemia. PLoS One 8, e55934. doi:10.1371/journal.pone.0055934
Zárate, S., Stevnsner, T., and Gredilla, R. (2017). Role of estrogen and other sex hormones in brain aging. Neuroprotection and DNA repair. Front. Aging Neurosci. 9, 430. doi:10.3389/fnagi.2017.00430
Zhang, Z., Wang, G., Li, Y., Lei, D., Xiang, J., Ouyang, L., et al. (2022). Recent progress in DNA methyltransferase inhibitors as anticancer agents. Front. Pharmacol. 13, 1072651. doi:10.3389/fphar.2022.1072651
Zhao, Z., Fan, L., and Frick, K. M. (2010). Epigenetic alterations regulate estradiol-induced enhancement of memory consolidation. Proc. Nat. Acad. Sci. 107, 5605–5610. doi:10.1073/pnas.0910578107
Keywords: DNA methylation, histone acetylation, learning and memory, aging, phytochemials
Citation: Singh P and Paramanik V (2025) DNA methylation, histone acetylation in the regulation of memory and its modulation during aging. Front. Aging 5:1480932. doi: 10.3389/fragi.2024.1480932
Received: 14 August 2024; Accepted: 27 November 2024;
Published: 06 January 2025.
Edited by:
John Tower, University of Southern California, United StatesReviewed by:
Tiyash Parira, Florida International University, United StatesMaria Florencia Tevy, University of Chile, Chile
Copyright © 2025 Singh and Paramanik. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Vijay Paramanik, dmlqYXlwYXJhbWFuaWtAZ21haWwuY29t
 Padmanabh Singh
Padmanabh Singh Vijay Paramanik
Vijay Paramanik