- Section for Evolutionary Hologenomics, Globe Institute, University of Copenhagen, Copenhagen, Denmark
The aim of this systematic review is to determine whether Deep Learning (DL) algorithms can provide a clinically feasible alternative to classic algorithms for synthetic Computer Tomography (sCT). The following categories are presented in this study: MR-based treatment planning and synthetic CT generation techniques. Generation of synthetic CT images based on Cone Beam CT images. Low-dose CT to High-dose CT generation. Attenuation correction for PET images. To perform appropriate database searches, we reviewed journal articles published between January 2018 and June 2023. Current methodology, study strategies, and results with relevant clinical applications were analyzed as we outlined the state-of-the-art of deep learning based approaches to inter-modality and intra-modality image synthesis. This was accomplished by contrasting the provided methodologies with traditional research approaches. The key contributions of each category were highlighted, specific challenges were identified, and accomplishments were summarized. As a final step, the statistics of all the cited works from various aspects were analyzed, which revealed that DL-based sCTs have achieved considerable popularity, while also showing the potential of this technology. In order to assess the clinical readiness of the presented methods, we examined the current status of DL-based sCT generation.
1 Introduction
Image synthesis is an active area of research with broad applications in radiation oncology and radiotherapy (RT). This technology allows clinicians to bypass or replace imaging procedures if time, labor, or expense constraints prevent acquisition; there are certain circumstances when it is not advisable to use ionizing radiation; or there are instances when image registration can introduce unacceptable uncertainty between images of different imaging modalities. There have been many exciting clinical applications that have been developed as a result of these benefits, including the planning of RT with Magnetic Resonance Imaging (MRI) and the use of Positron Emission Tomography (PET)/MRI in tandem with RT treatment.
In recent decades, image synthesis has been investigated in relation to its potential applications. Traditionally, image conversion from one modality to another is carried out using models with explicit human-defined rules, which require adaptive parameter tuning on a case-by-case basis in order to achieve optimal results. Additionally, these models have varied characteristics based on the unique attributes of the imaging modalities involved, resulting in a variety of complex methodologies that are application-specific. In the case of anatomical imaging and functional imaging, it is particularly challenging to construct such models. It is for this reason that most of these studies employ Computed Tomography (CT synthesis from MRI) as the primary imaging tool.
It is now possible to combine image synthesis with other imaging modalities such as PET and Cone-Beam CT (CBCT) as a result of rapid advances in machine learning (ML) and computer vision over the last two decades (1). ML and Artificial Intelligence (AI) have been dominated for several years by deep learning (DL) as a broad sub-field within ML. To extract useful features from images, DL algorithms employ neural networks containing many layers and a large number of neurons.
Many networks have been proposed to achieve better performance in various applications. Data-driven approaches to image intensity mapping are commonly used by DL-based image synthesis methods. Generally, a network learns how to map the input to its target through a training stage, followed by a prediction stage where the target is synthesized from the input. In contrast to conventional model-based methods, a DL-based method can be generalized to multiple pairs of image modalities without requiring significant adjustments. By utilizing this approach, rapid translation to various imaging modalities is possible, allowing clinically relevant synthesis to be produced. Despite the effort required in collecting and curating data during network training, the prediction process usually takes only a few seconds. In medical imaging and RT, DL-based methods have demonstrated great promise because of these advantages.
In the domain of RT, MRI is preferred over CT for patient positioning and Organ at Risk (OAR) delineation (2–6) due to its better capacity to differentiate soft tissues (7). In RT conventionally, the primary imaging modality is CT. MRI is fused by deformable enrollment with CT scans because they deliver precise and high-resolution anatomy which is needed for dose calculations (2) for RT. However, residual mis-registration and variations in patient setup may introduce systematic errors that might influence the accuracy of the entire treatment. The point of MRI only RT is to eliminate the CT scans from the workflow and in its place use MR image(s) alone.
MRI-based treatments are getting very common because of the advancement of MR-guided treatment methods, e.g., MRI-linac (8). Here, online versatile RT utilizing MRI can be performed, exploiting the functional data and anatomy supplied by the modality (9) and reducing the registration error (2, 10, 11). Additionally, MR only RT can also help us protect the patient from the ionizing radiations and may decrease treatment cost (12) and workload (13).
Furthermore, similar techniques have been proposed to improve the quality of CBCT by converting a different imaging modality into sCT. Photon and proton therapy are effectively utilized using CBCT in image-guided adaptive radiotherapy (IGART). Despite this, the reconstruction of the image suffers from several artifacts such as shading, streaking, and cupping due to the severe scatter noise and truncated projections. As a result of these reasons, online adaptation of treatment plans does not commonly utilize daily CBCT. By converting CBCT to CT, it should be possible to compute accurate doses and provide patients with a better quality of treatment.
Furthermore, sCT estimation plays a significant role in PET attenuation correction (AC). The photon AC map from CT is often necessary for accurate PET quantification. A solution to the MRAC issue has been proposed to address this issue with the new hybrid PET/MRI scanners. The derivation of sCT from uncorrected PET can provide additional benefits to stand-alone PET scanners.
We present an in-depth review of emerging DL-based methods and applications for synthesising medical images and their applications in RT in this review. This review categorized recent literature according to their DL methods and highlighted their contributions. A survey of clinical applications is presented along with an assessment of relevant limitations and challenges. We conclude with a summary of recent trends and future directions.
2 Materials & methods
We looked through the Scopus, PubMed and ScienceDirect electronic databases from January 2018 to Jun 2023 utilizing the associated keywords:((“radiotherapy” OR “radiation therapy” OR “MR-only radipotherapy” OR “proton therapy” OR “oncology” OR “imaging” OR “radiology” OR “healthcare” OR “CBCT” OR “cone-beam CT” OR “Low dose CT” OR “PET” OR “MRI” OR “attenuation correction” OR “attenuation map”) AND (“synthetic CT” OR “sCT” OR “pseudo CT” OR “pseudoCT” OR “CT substitute”) AND (“deep learning” OR “convolutional neural network” OR “CNN” OR “GAN” OR “Generative Adversarial Network” OR artificial intelligence)). We just selected original research papers written in English excluding the review papers. This review was conducted based on the PRISMA guidelines. The screening criteria is given in the Figure 1.
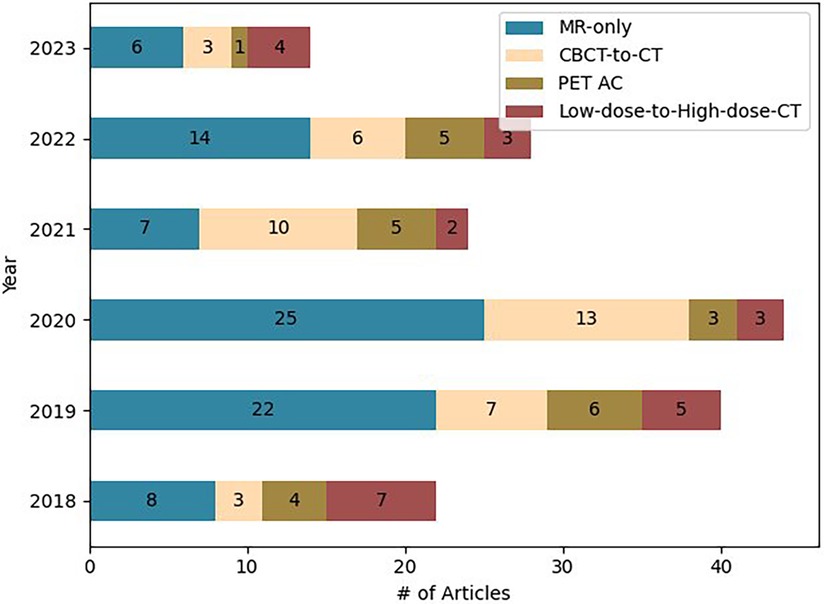
For each paper, we screened: Magnetic Resonance (MR) devices, MR images and sequences, number of patients, dataset split details (training, validation, and testing set), pre and post-processing of dataset, Deep learning (DL) technique utilized, loss functions, metrics used for the image comparison and dose evaluation. Figure 2 provides the information regarding the articles selected for this study.
2.1 Deep learning in medical images
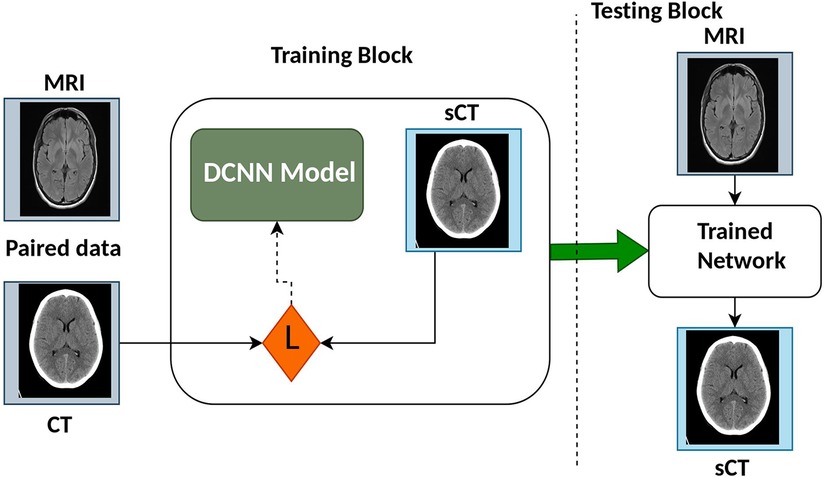
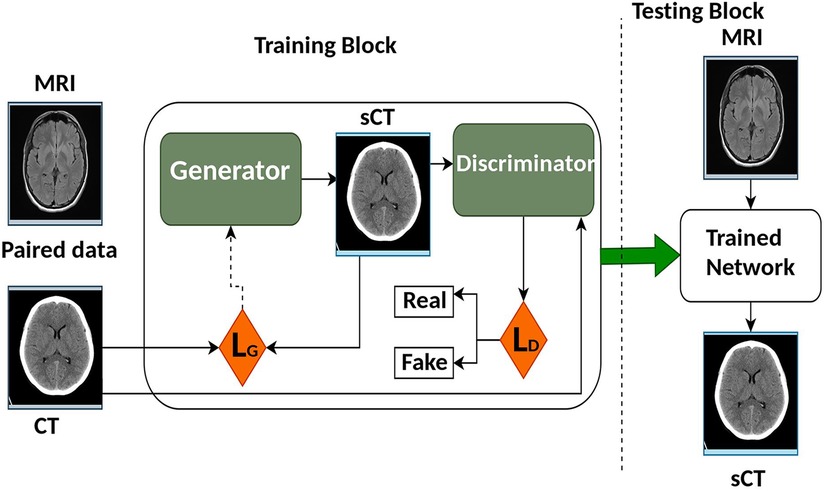
Deep Learning (DL) is a specialized subset of machine learning (ML) that focuses on deep neural networks and automated feature extraction. It has achieved remarkable success in tasks with large datasets, but it comes with higher computational requirements and challenges in interpretability compared to traditional ML methods. The choice between DL and ML depends on the specific problem, dataset size, and computational resources available. Recent reviews provide further insight into DL network architectures for medical imaging and RT (14–20). The synthetic Computed Tomography (sCT) generation using DL methods generally utilizes Convolution Neural Network (CNN)/Deep Convolution Neural Network (DCNN) or Generative Adversarial Network (GAN) and variants. Figure 3 shows the architecture of some CNN/DCNN and GAN networks.
2.1.1 Convolution neural network (CNN)
Convolution Neural Network (CNN) is a famous class of deep neural networks utilizing a bunch of convolution filters for distinguishing image features. A CNN comprises an input layer, several hidden layers and an output layer.
CNN take an input image/feature vector (one information node for every passage) and change it through a progression of a series of hidden layers, regularly utilizing nonlinear activation functions. Each hidden layer is likewise comprised of a bunch of neurons, where every neuron is completely associated with all neurons in the previous layer. The last layer of a neural network (i.e., the “output layer”) is likewise completely associated and addresses the last result classification of the network. Several types of layers are utilized to build a CNN but the most common ones include:
• Convolutional (Conv) - These layers apply a convolution to the information, passing the outcome to the following layer. A convolution changes over every one of the pixels in its open field into a single value, resulting in a vector.
• Activation (ACT or RELU, where we use the same or the actual activation function) – The decision of activation function in the hidden layer will control how well the network model learns the training dataset. The decision of enactment work in the result layer will characterize the kind of predictions the model can make. Nonlinear activation functions (Rectified Linear Units (ReLU) (21), Leaky-RELU (22), Parametric-ReLU (PreLU) or exponential linear unit (ELU) (23)) play a crucial role in discriminative capabilities of the deep neural networks. The ReLU layer protects the information and is a commonly utilized activation layer because of its computational minimalism, authentic sparsity, and linearity.
• Pooling (POOL) - These layers are used to reduce the dimension (subsampling) of the feature maps. It decreases the number of parameters to learn and computation in the network. The pooling layer sums up the features present in a region of the feature map produced by a convolution layer. It stabilizes the learning process and also reduces the training epochs required.
• Fully connected (FC) – These layers are used to connect all the inputs from a layer with the activation function of the next layer.
• Batch normalization (BN) (24) - This layer permits each layer of the network to learn more freely. It is utilized to standardize the result of the previous layers.
• Dropout – This layer is used to prevent overfitting in the model. During each step of training time, it set the input units to 0 randomly.
• Softmax – This is the last layer in a neural network that performs multi-class characterization.
During training stage, the model attempts to limit a true capacity called loss function, which is a intensity based similarity estimation between real image and the generated image. Figure 4 presents the architecture of CNN models commonly utilized for synthetic image generation. In the literature, the variations of CNN model incorporate convolution encoder-decoder (CED) (25), DCNN (26), Fully convolutional network (FCN) (27), U-Net (28–43), ResNet (44), SE-ResNet (45), and DenseNet (46).
The CNN network comprises combined encoder and decoder networks. CNN has been broadly utilized in DL literature due to its groundbreaking results (47–49). In the encoding part, it uses the method of downsampling to translate the low-level features map to a high-level features map. In the decoding part, the transposed convolution layer’s function is to translate the high-level feature maps to low-level feature maps to generate the synthetic image. The encoder part of the network utilizes a bunch of consolidated 2D convolution for distinguishing image features, followed by normalization, activation function and max pooling.
The decoder part utilizes transposed convolutional layers to join the feature and spatial information from the encoding part, followed by concatenation, up-sampling, and convolutional layers with a ReLU activation function.
The most notable and well-known CNN model is the U-shaped CNN (U-Net) architecture proposed by Ronneberger et al. (50). The U-Net architecture has direct skip connections between the encoder and decoder that helps in extracting and reconstructing the image features.
2.1.2 Generative adversarial network (GAN)
Generative adversarial network (GAN) was first introduced in 2014 by Goodfellow et al. (51). It improved the quality of image generation as compared to the previous Convolution Neural Network (CNN) models. The architecture of GAN as shown in Figure 5 trains two separate neural networks, the generator (G) and the discriminator (D). G attempts to create synthetic images while D on the other side decides if that image looks like the real image or not (52, 53). GAN presents an information-driven regularizer, it tries to improve itself and guarantees that the learned features bring the outcome close to the ground truth.
In basic GAN architectures, D and G are executed as Multi-Layered Perceptrons (MLPs). U-Net is the most used architecture as the G for GAN. Another frequently used G in GANs is the ResNet, as highlighted in the work by Emami et al. (54). ResNet stands out for its ease of optimization and its ability to reliably produce the desired results.
For the D part of the GAN, PatchGAN is used and it comprises six convolutional layers with different filters but the same kernel size and stride, trailed by five fully connected layers. For activation purposes, ReLU is utilized and for the convolution layer, batch normalization is utilized. The dropout layer is added to the fully connected layers, and in the last fully connected layer, a sigmoid function is utilized. The traditional GAN model uses adversarial loss () as the cost function and it helps the network to produce better-looking sCT images with less blurry features (55, 56) compared to the images generated by other CNN models.
The discriminator attempts to boost it while the generator attempts to limit it as mentioned in the equation. Where, D(x) is the discriminator’s estimate if that real data instance x is real, and is the expected value over all real data and random instances respectively. G(z) is genertor’s output over noise (z) while D(G(z)) estimate if a fake instance is real. The formula derives from the cross-entropy between the real and generated distributions. The generator can’t directly affect the log(D(x)) term in the function, so, for the generator, minimizing the loss is equivalent to minimizing log(1 - D(G(z))) given a discriminator.
The most common variants of GAN used for synthetic image generation are Conditional GAN (cGAN) and cycle-GAN. The first cGAN architecture to generate synthetic Computed Tomography (sCT) to Magnetic Resonance Images (MRI) was proposed by Emami et al. (54). Unlike standard GAN, both the G and D of cGAN perceive the input image dataset. This approach tends to be more accurate as compared to previous approaches. Unlike standard GAN, several studies have been proposed to include SE-ResNet (41, 43), U-Net (44, 45, 57), DenseNet (46) and Embedded Net (26) as Generator for the cGANs. Evaluation of all four G, Embedded Net, DenseNet, SE ResNet and U-Net in cGAN is proposed by Fetty et al. (58) to generate synthetic images from MR T2 weighted images.
Several studies used a cGAN architecture to generate sCT from MRI (35, 41–43, 57, 59–69).
Cycle-GAN are commonly used to train Deep Convolutional Neural Networks (DCNN) to translate image-to-image. Cycle-GAN consists of two G and two D. In synthetic image generation using cycle-GAN, one G is used to generate sCT from MRI and the other to generate sMRI from sCT. A cyclic loss function is used to learn concurrently the features between the two modalities. The unpaired dataset is used to learn the mapping between two modalities and in some cases, it outperforms GANs using paired datasets (55).
2.1.3 Loss functions used in deep learning models
Loss functions play an important role in guiding model training. Different loss functions are used based on the requirement and network configuration. L1 norm (70) and L2 (62, 69) are used frequently used to avoid overfitting and control complexity of model. L1 as compared to L2 is used more often due to its robustness to outliers in training data and it tends to perform better for image generation tasks. An image fidelity loss is commonly calculated by subtracting the average squared difference between the predicted and actual image, which is commonly referred to as Mean Square Error (MSE). It is imperative to maintain fine details when structural similarity index (SSIM) loss is applied while cross entropy loss is widely used for classification problem. Adversarial Loss is utilized by Generative Adversarial Networks to create realistic images. Some combination of different loss functions are also utilized in GANs and other models to improve the model accuracy (40, 41, 58, 64).
2.2 Dataset, dataset size & training dataset
The challenging part of the DL-based approaches applied on image synthesis is the paucity of datasets available for training and testing the different methods. Several studies are conducted with a minimum of 10 patients. Studies have also suggested that a higher number of images in the dataset can improve the performance of Deep Learning (DL) models. To improve model performance, diversity of training datasets is required. The images used for most of the studies were taken from adult patients. For training the model, most of the studies were conducted using paired datasets (where the images from both the modalities are given as input to learn the features) and very few studies used unpaired datasets. Some studies also compared the results on paired datasets over unpaired datasets (29, 60, 71, 72). Most commonly used networks were 2D networks, where 3D images were sliced into 2D images for training the network. Multiple configurations were also investigated in some studies (40, 73) described in this review. The most popular architecture for the image synthesis was Generative Adversarial Network (GAN), followed by U-Net and other Convolution Neural Networks (CNN). For the generator (G) of the GAN, mostly U-Net was used. Data augmentation is also used to train the network with different features and properties using small samples within the training dataset. Some conventional data augmentation techniques (19, 30, 63) such as rotation, translation, noise addition and deformations can be used with the training dataset. In this review, several training strategies were utilized: single-fold validation, k-fold cross-validation and leave-one-out validation. For single-fold cross-validation, the dataset is divided into two sets: one for training and the other for testing. For k-fold cross-validation, the dataset is separated into k number of subsets. For each training, one k subset is utilized for the testing phase and the remaining k subsets for the training phase. Leave-one-out validation is equivalent to k-fold validation with k being the number of samples in the training dataset.
2.3 Evaluation metrics
In the literature, several metrics are reported based on the image similarity or intensity, accuracy based on the geometry and evaluation of the dose for the radio therapy (RT). The metrics used in the literature are provided in Table 1. To evaluate the quality of the synthetic image based on voxels, the most used similarity metrics are Mean Absolute Error (MAE), Structural Similarity (SSIM), and Peak Signal to Noise Ratio (PSNR).
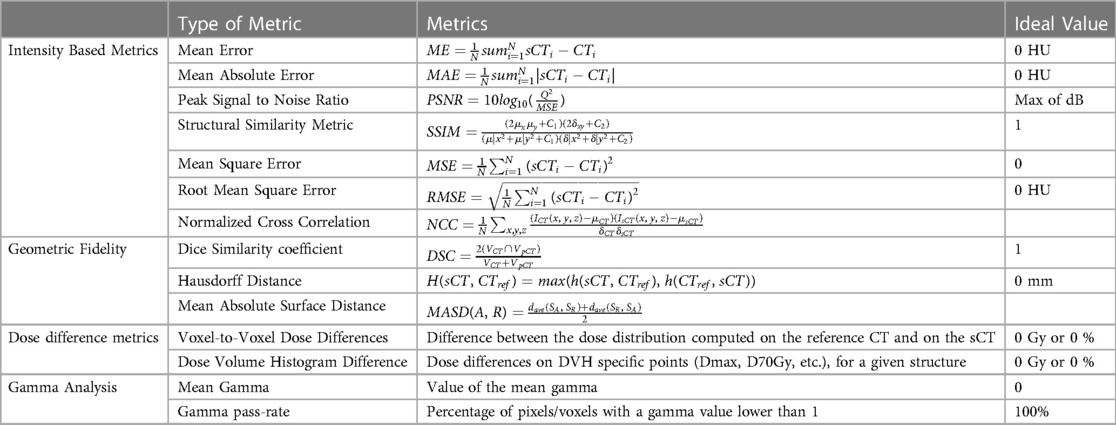
Table 1. Metrics reported in literature for synthetic image analysis using ground truth as reference.
Besides voxel-based metrics, geometric accuracy can also be assessed by comparing delineated structures with corresponding voxel-based metrics. In terms of evaluating the accuracy of depicting specific tissue classes and structures, the Dice Similarity Coefficient (DSC) is a commonly used metric. DSC is calculated after applying morphological operations to binary masks and applying a threshold to Computed Tomography (CT) and synthetic CT (sCT). In addition to the Hausdorff distance, the mean absolute surface distance can be used to assess the segmentation accuracy, as it measures the distance between two contour sets (74).
A comparison of dose calculation between sCT and CT is generally performed using specific regions of interest (ROI) for both photon (f) and proton (p) RT. The most commonly used voxel-based metric dose difference (DD) is calculated by taking the average dose () of the ROI and redistributing it across the whole body, target, or other structures of interest. DD is expressed as a percentage of the prescribed dose (%) or the maximum dose (Gy), either relative to it or an absolute value. DD is directly correlated to the dose pass rate, which is the percentage of voxels with DD below a specified threshold.
Gamma analysis can be conducted in both 2D and 3D, offering a combined evaluation of dose and spatial factors. However, this process involves the configuration of multiple parameters, such as dose criteria, distance-to-agreement criteria, and dose thresholds. It’s important to note that there is no standardized approach for interpreting and comparing gamma index outcomes across various studies. The results can significantly differ due to variations in parameters, grid sizes, and voxel resolutions (75, 76). As a result, the gamma pass rate (GPR) is typically expressed as the percentage of voxels within a region of interest (ROI) that meet a specific threshold based on the reference dose distribution.
The dose-volume histogram (DVH) is a tool used routinely in clinical practice. As a general rule, clinically significant DVH points are reported in an evaluation of sCT. Also, range shift (RS) is considered in proton RT. In this case, the ideal range is determined as the distance from the distal dose fall-off () point at which the dose is at 80% of the maximum (77). As well as absolute RS error (RSe) expressed as the shift in the prescribed range relative to the actual beam direction (), relative RS error (%RS) can also be specified.
3 Results
3.1 MR to synthetic CT (sCT) generation for radiotherapy
A significant amount of research has been published in this field on the problem of Magnetic Resonance Image (MRI) to synthetic Computer Tomography (sCT) image synthesis as one of the first applications utilizing Deep learning (DL) for medical image analysis. The results for this section is provided in Table 2 CT acquisition is being replaced by MR-based CT synthesis primarily for clinical reasons (78). Despite recent improvements in sCT imaging, they are still inconclusive as diagnostic tools. The tool is also valuable for non-diagnostic settings, such as treatment planning and PET Attenuation Correction (AC).
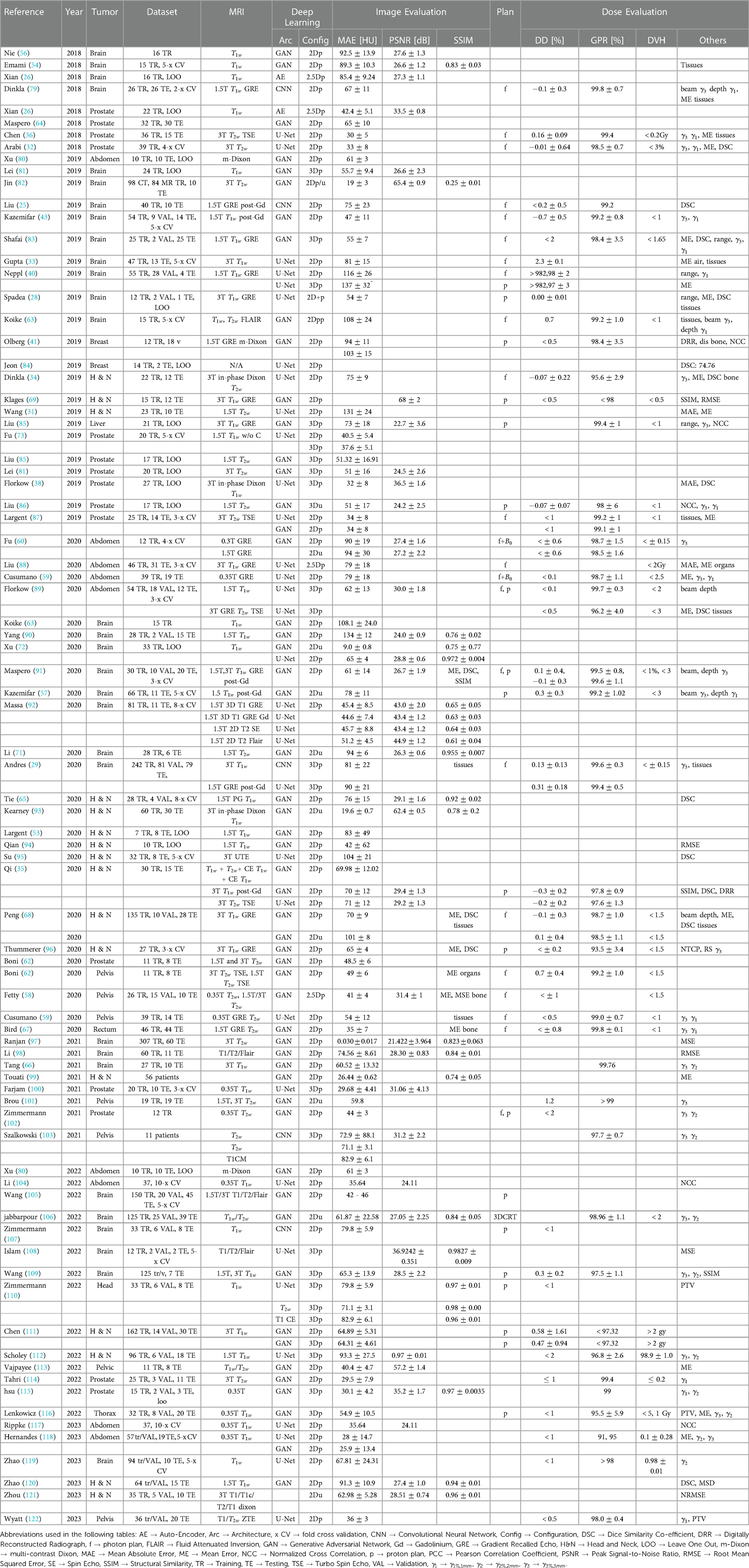
Table 2. Overview of sCT generation methods for MR-only RT with image-based and dose difference evaluation.
Radio therapy (RT) workflows commonly utilize MR and CT imaging for treatment planning on many patients. CT images provide electron density maps for dose calculation and reference images to position the patient prior to treatment. MR images offer excellent tissue contrast to diagnose gross tumours and organs at risk (OARs). Using image registration, treatment planning is performed by propagating MR contours to CT images. In addition to time and expense costs for the patient, combining both modalities contributes to systematic image fusion errors. Furthermore, CT may also expose patients to non-negligible doses of ionizing radiation (123), especially those requiring re-simulation. MRI-based treatment planning workflows would therefore be highly desirable instead of CT scans. Additionally, there is a growing demand for MRI exclusively for RT as MR linear accelerator (MR-linac) technology emerges.
Due to the lack of a one-to-one relationship between MR voxel intensity and CT’s Hounsfield Unit (HU is a quantitative measure to represent the radio density of tissues, helping in the differentiation of structures based on their properties), intensity-based calibration methods fail to deliver accurate and consistent results. CT imaging differs from MRI because in CT, air is dark and bone is bright. While translating MR images to CT, MR images are typically segmented into several classes of materials (e.g., air, soft tissue, bone) and then assigned CT HU values (11, 124–128) or registered to an atlas with known CT HU values (129–131). Segmentation and registration are the main components of both of these methods, which introduce significant errors due to ambiguous boundaries between bone and air, for instance, and significant inter-patient variations.
In literature, nearly all studies reported the image quality of their sCT using mean absolute error (MAE), peak signal-to-noise ratio (PSNR) and structural similarity (SSIM) metrics for CT synthesis applications in RT. Many studies also calculated the dose from the original treatment plan. Approximately 1% of the dose was different, which is small compared to the uncertainty associated with the total dose over the entire treatment course (5%).
In RT, DL-based methods generate relatively minor improvements in dosimetric accuracy compared to image accuracy and may not be clinically relevant. VMAT (Volumetric Modulated Arc Therapy) plans offer greater flexibility in dose calculations, particularly when dealing with image inaccuracies, especially in uniform areas like the brain. In VMAT, random image inaccuracies tend to balance out within an arc, but it’s worth noting that there’s a non-linear relationship between random image inaccuracies and dosimetric errors.
According to Liu et al. (85), most of the dose difference caused by sCTs occurs at the distal end of the proton beam due to errors along the beam path on the planning CT. As a result, the tumour could be substantially underdosed or Organs at risk overdosed. According to Liu et al. (85, 86), the largest absolute difference observed among patients with liver cancer is 0.56 cm, while for those with prostate cancer, the mean absolute difference is 0.75 cm. Besides assessing dosimetric accuracy for treatment planning, geometric fidelity is another essential consideration. Despite this, there are very few studies assessing sCT positioning accuracy. It has also been investigated whether sCT can work in proton therapy for prostate (86), liver (85), and brain cancers (83).
3.2 CBCT to synthetic CT (sCT) generation for radiotherapy
Synthetic Computed Tomography (sCT) using Cone beam CT (CBCT) is a physics problem that is governed by the same principles of x-ray attenuation and back projection. However, their application in clinical practice differs. So, we consider them as two distinct imaging modalities in this review. By comparing anatomic landmark displacements from the treatment planning CT images and CBCT images, image-guided radio therapy (IGRT) is used to check for patient setup errors and interfraction motion (132). More demanding applications of CBCT have been proposed with increased adoption of adaptive RT techniques, such as daily dose estimation and auto-contouring based on deformable image registration obtained through simulation with CT images (133, 134). The results for this section is provided in Table 3.
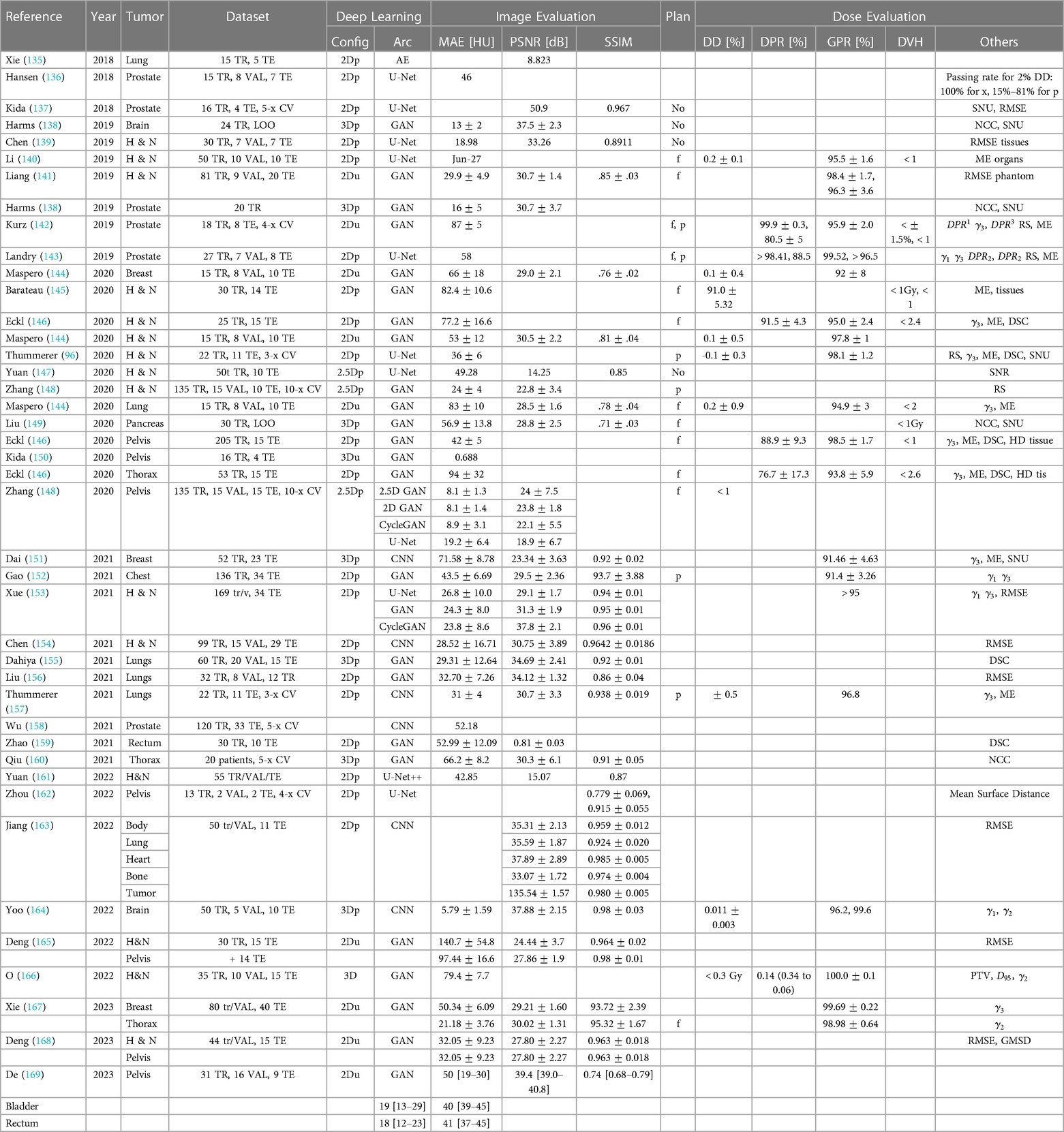
Table 3. Overview of sCT generation methods for RT with CBCT image-based and dose difference evaluation.
CBCT scanners generate a cone-shaped x-ray beam that is incident on a flat panel detector, unlike CT scanners with fan-shaped x-ray beams and multi-slice detectors. The flat panel detector offers a wide coverage along the -axis and high spatial resolution but also suffers from decreased signal due to scattered x-rays coming from the whole body. This results in significant quantitative CT errors as a result of severe streaking and cupping artifacts. When utilizing images for dose calculations, these errors introduce challenges in the calibration of Hounsfield Units (HU) to electron densities. HU represents the radiodensity of tissues in computed tomography (CT) scans.
CBCT can also suffer from degraded image contrast and bone suppression (170). As CBCT images are significantly degraded, they cannot be used for quantitative RT. CBCT Hounsfield Unit (HU) can be corrected and restored relative to CT using Deep learning (DL) based approaches, as shown in Table. The CBCT image is created through a combination of hundreds of projections in different directions. Before volume reconstruction, few studies applied neural networks to 2D projections, namely, the projection images. The CBCT volume was reconstructed from the improved quality projection images. Another approach relies on reconstructed CBCT images as inputs and produces sCT images with enhanced image quality. Utilizing projection domain methods for training with an extensive dataset of over 300 2D projection images offers the advantage of achieving a desired level of proficiency with a reduced number of training iterations compared to conventional image domain methods, which typically require approximately 100 iterations to achieve similar competence. CBCT images also suffer from artifacts such as cupping and streaking caused by scattering, whereas projection images are easier to learn for neural networks. Further, images have higher artifactual variation between patients, so much so that image domain methods rarely train models on non-anthropomorphic phantoms since the data collected is useless. However, in the projection domain, there is little variation in image features.
Therefore, Nomura et al. (171) showed that non-anthropomorphic phantom projections can also be used to learn to scatter distribution features that characterize anthropomorphic phantom projections. As a result, the neural network learned how to relate scatter distribution to objective thickness in the projection domain. Image scatter artifacts have a much more complicated relationship to objective appearance and cannot be easily learned. As the ground truth in the reviewed studies is often the corresponding CT images/projections from the same patient, CBCT images/projections are typically used while training. CT and CBCT are often out of geometric agreement, and registration reduces artifacts caused by the mismatch. As part of a pancreas study, Liu et al. (149) compared CBCT/CT training data rigidly and deformably registered. The researchers found that sCT created from rigidly registered training data produced lower noise and better organ boundaries compared with deformably registered CT (56.89 * 13.84 HU, ). As Kurz et al. (142) have demonstrated, generating sCT with satisfactory quality can be achieved without using pixel-wise loss functions in a cycle-GAN.
According to Hansen et al. (136) and Landry et al. (143), the registration step can be bypassed by correcting CBCTs first by conventional methods and then using the corrected CBCTs as ground truth. The corrected CBCTs do not require registration because the geometry of the corrected CBCTs remains the same as the original CBCT. However, CBCT generating methods in this setting limit the quality of sCT. Study findings suggest that DL-based methods have better image quality than conventional CBCT correction methods on the same datasets (96, 135, 137). They found that Adrian’s U-Net based method was more accurate and better suited to registering bone geometry than an image-based method or a deformable method. A comparison of Harms et al.’s (138) sCT to real CT study also demonstrated reduced noise and an improved subjective similarity. Corrective methods that are conventional are designed to improve only one specific aspect of image quality. DL-based methods, on the other hand, can modify every aspect of image quality to simulate CT, including noise level, which typically is not considered in conventional methods. Cycle-GAN outperformed both GAN and U-Net in several studies comparing the same patient datasets.
An analysis of 135 pelvic patients with 2.5D conditional GAN was conducted by Zhang et al. (148). Additional 15 pelvic and 10 H & N patients were analyzed afterwards. In both testing groups, the network predicted sCT at similar MAEs, showing that pre-trained models can be transferred to varying anatomical regions. In addition to different GAN architectures, the researchers compared U-Net configurations and found that it was statistically worse than any GAN configuration. The cycle-GAN has been tested with unpaired training in three works (141, 142, 144). A study performed the unsupervised training comparison of cycle-GAN, DCGAN (172), and PGGAN (173), where the first performed better in terms of image similarity and dose agreement.
The dosimetric accuracy of sCTs is significantly improved over that of original CBCTs, and an approach is used to calculate photon dose based on sCT. Select dose-volume histogram (DVH) metrics and dose or gamma differences have been investigated as a basis for evaluating sCT feasibility in VMAT planning at various body sites. According to Liu et al. (156) local dosimetric errors are large in areas with severe artifacts. These artifacts and dosimetric errors were successfully mitigated using sCT. With proton planning, it is more difficult to achieve acceptable dosimetric accuracy due to the range shift, which can be up to 5 mm (174).
3.3 PET attenuation correction
For PET Attenuation Correction (AC), the influence of synthetic Computed Tomography (sCT) error on PET quantification has been analyzed. It is extremely difficult to specify an error tolerance beyond which clinical decision making is affected; however, it has generally been accepted that quantitative errors of 10% or less rarely affect diagnostic imaging decisions (175). Most of the methods proposed in the studies met this criterion, based on their average relative biases. It should be noted however, that because of variations among study objects, there may be a bias exceeding 10% in some volumes of interest (VOI) for some patients (176, 177), suggesting that when interpreting results, it is important to take into account the standard deviation of the bias as well as the mean, since the proposed methods may not have good local performance for some patients. The results for this section is provided in Table 4. As an alternative to providing the mean and standard deviation in demonstrating the performance of the proposed methods, listing or plotting all the data points, or at least their range, would ultimately prove more useful (178). Being made up of high density and atomic number, bone has the most capacity for attenuation, and its accuracy on sCT has a huge impact on the final results of attenuation-corrected PET. It is more common for PET AC to evaluate the geometric accuracy of bone on sCT than radio therapy (RT). It has been shown that more accurate CT images generated by learning based methods result in more accurate PET AC (179–181).
Deep learning (DL) based methods, designed to produce more precise sCT images, lead to enhanced accuracy in PET AC. Several studies have demonstrated the substantial improvements achieved by these methods. In contrast, PET AC using classical CT synthesis approaches exhibited an average bias of approximately 5% when compared to selected VOIs, while DL-based methods exhibited a reduced bias of around 2% in the same comparison (183, 184).
A 3D patch cycle-GAN was trained with unregistered MR/CT pairs, compared to atlas-based MRAC and CNNs with registered pairs by Gong et al. (190). A comparison of DL methods to atlas MRAC revealed that both performed better in DSC and MAE. CNN and cycle-GAN did not differ significantly in their performance in DSC and MAE. According to their research, cycle-GAN is able to avoid the challenge of training on perfectly aligned datasets, but more data is needed to improve its performance.
It was examined whether different network configurations (VGG-16 (48), VGG-19 and ResNet (44)) can be used as a benchmark with a 2D conditional GAN that receives either two Dixon inputs (water and fat) or four Dixon inputs (water, fat, in-phase, and opposed). When four inputs are used in the GAN, results are more accurate than the VGG-19 and the ResNet.
Several authors have proposed that the sCT could be obtained directly from diagnostic imaging, - or -weighted, by using standalone MRI scanners(32, 184) or hybrid machines (185).
Bradshaw et al. (185) a three CNN trained on Gradient Echo (GRE) and Turbo Spin Echo (TSE) MRI sequences, specifically, the and sequences. The CNN was trained to predict tissue segmentation across distinct classes, including air, water, fat, and bone. Subsequently, the model’s performance was compared with the default Magnetic Resonance Attenuation Correction (MRAC) method commonly employed by scanners. PET reconstruction had substantially lower RMSE when calculated with DL method and / input. Recent studies have investigated a CNN with input either or Dixon and multiple echo UTE (mUTE) on a brain patient cohort, and found that it outperformed the others. A CNN was trained on 1.5 T diagnostic GRE data of 30 patients in Liu et al. (184). A total of ten patients from the same cohort were used and their results are reported in the following table. Using a 3 T MRI/PET scanner, they then predicted the pathology for five patients ( GRE), and calculated the error (), resulting in a 1% error rate. The authors concluded that DL-based approaches are flexible and suitable for handling heterogeneous datasets acquired using many scanner types.
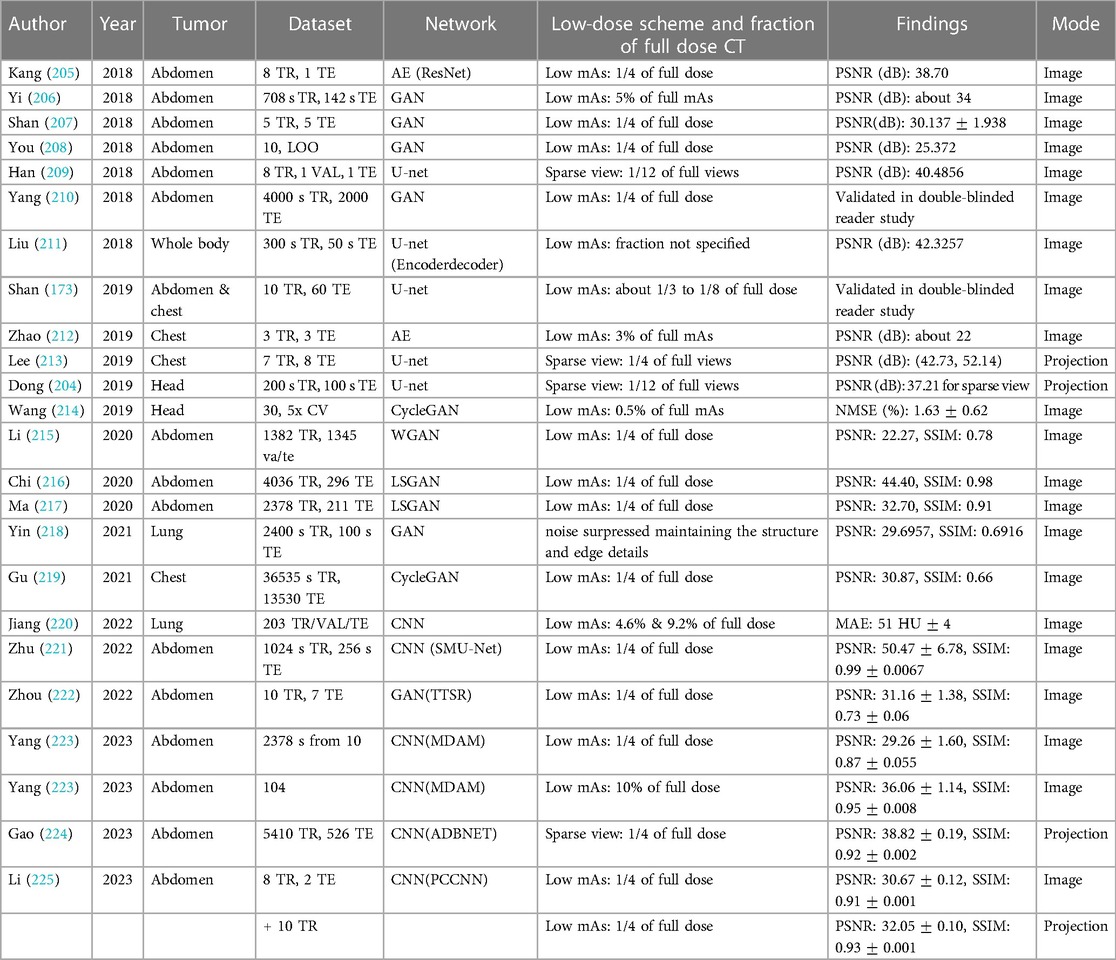
3.4 Low dose CT to full dose synthetic CT (sCT)
The data-driven approach to automatically learning image features and model parameters makes deep learning (DL) an attractive option for low-dose Computed Tomography (LDCT) restoration. The existing literature primarily discusses two approaches for LDCT image enhancement. The results for this section is provided in Table 5. Some methods focus on direct image translation from LDCT to full-dose CT (FDCT), while others involve a two-step process. In the latter approach, DL is utilized to restore the sinogram, followed by image reconstruction using Filtered Back Projection (FBP). The proposed method by Dong et al. (204) reduces lower-resolution edges of objects with better down-sampling artifacts than an image-based one.
There is a possibility that projection-based methods do not directly detect prediction errors, while image-based methods do. During the reconstruction process, the predicted error on the sinogram will be compensated for, and the outcome will be the average of all the sinograms. These models are more error-resistant because of their projection-based nature. The network may be encoded with a mapping from polar to Cartesian coordinates for direct mapping from the projection domain to the image domain.
In their progressive method, Shan et al. (173) generated a sequence of denoised images at different levels of noise by iteratively denoising the input LDCT. Rather than directly mapping LDCT or FDCT images, Kang et al. (226) mapped their wavelet coefficients. Better recovery of structures was achieved with wavelet transformations compared to direct mapping. DL-based methods are less time consuming than iterative reconstruction methods and do not require prior knowledge of energy spectrum. The LDCT model reported by Wang et al. (214) was trained on an average personal computer in 1 minute and generated an entire 3D volume from denoised images. Due to the resource-intensive nature of traditional iterative reconstruction methods, their implementation is limited on personal computers, especially when slice thickness and field of view (FOV) are small.
Numerous studies have conducted comparisons between traditional iterative methods and state-of-the-art DL-based techniques. Among these advanced methods, Total Variation (TV) regularization has received attention. TV-based techniques, while known to sometimes over smooth images and create uneven textures, excel in preserving fine structures and maintaining image texture similarity to FDCT scans. Utilizing analytical optimization objectives in deep learning enhances image quality while preserving texture, resulting in predictions that closely align with the ground truth, as represented by FDCT images. This improvement is quantitatively measured through metrics such as peak signal-to-noise ratio (PSNR), structural similarity (SSIM), and mean absolute error (MAE). A double-blinded reader study conducted by Shan et al. (173) proved their DL-based method performed similarly to three commercially available iterative algorithms for noise suppression and structural fidelity. Almost all the studies reviewed used their restored FDCT images for diagnostic purposes. This method is particularly suitable for adaptive RT where re-scanning and planning throughout a treatment course is common, as Wang et al. (214) evaluated it in the context of RT treatment planning. Planning CT requires accurate Hounsfield Unit (HU) and dose calculation accuracy vs diagnostic CT, which emphasizes high resolution and low contrast. When a dose of 21 Gy is prescribed, the average difference in dose volume histogram (DVH) metrics between original FDCT and synthetic FDCT is less than 0.1 Gy (). Although the training and testing strategies may differ among these studies, the results are similar. Most of the reviewed studies used the dataset from the AAPM 2016 LDCT Grand Challenge (227). Because LDCT does not contain any clinical data, it is also used as an example of Poisson noise or a downsampled sinogram in many other studies. There are a few exceptions, such as Yi et al. (206) who used piglets, and Shan et al. (173) who used LDCTs from real patients. It is therefore important to evaluate these methods against actual LDCT datasets since simulated noise may not accurately reflect the properties of true noise and potential artifacts.
4 Discussion
In many domains of biomedical research and clinical treatment, imaging has become a necessary component. Radiologists identify and quantify tumors from Magnetic Resonance Image (MRI) and Computed Tomography (CT) scans, and neuro-scientists detect regional metabolic brain activity from Positron Emission Tomography (PET) and functional MRI scans. Biologists study cells and generate 3D confocal microscopy data sets, virologists generate 3D reconstructions of viruses from micrographs, radiologists identify and quantify tumors from MRI and CT scans, and neuro-scientists detect regional metabolic brain activity from PET and functional MRI scans. In contrast to traditional digital image processing and computer vision approaches that need many MRI modalities to properly show all areas. There are few novel Deep Learning (DL) approaches available (discussed in literature) for generating brain sCT images that only requires one MRI pulse sequence to accurately display all regions (43, 228, 229).
DL-based image synthesis is a young and rapidly developing field, with all of the studies evaluated published within the past five years. There is much literature on DL-based image synthesis. Future studies need to address specific unanswered questions. Since GPU memory constraints prevented training on three-dimensional (3D) slices, some DL algorithms were trained on two-dimensional (2D) slices. Unlike 3D loss functions, 2D loss functions do not consider continuity in the third dimension, thus making slices appear discontinuous. In addition to using 3D patches to train models that exploit 3D spatial information more effectively, they can also extract features from larger-scale images (34, 81). A 2D and 3D model was examined using the exact U-Net implementation by Fu et al. (73). The study’s findings suggest that a 3D sCT offered more accurate results with smaller MAE. In the absence of additional data, the model might use many adjacent slices to gather additional 3D context or generate independent networks for each of the three orthogonal 2D planes (230).
A DL-based approach can produce images that are more realistic improve quantitative metrics. Depending on the technology, it can take from an hour to days to train a model using DL-based approaches. A synthetic image for a new patient can be generated within seconds or minutes after training a model. Our study reviews the feasibility of using various imaging methods to build CTs using DL-based methods. It has become possible to train large datasets and translate images in seconds due to higher computing capabilities. DL’s clinical applications are made simpler by fast image-to-image translation, proving the method’s usefulness.
1. MR based RT: There are many types of sCT generation approaches, but MR only RT with DL is the most prevalent. The eighty two studies in this review demonstrate that DL algorithms effectively produce sCT from MRI data. Many methods of training and combinations have been proposed. The pelvis and the head and neck can be treated using photon radiotherapy (RT) and proton therapy, which achieve high image similarity and dosimetry accuracy. As part of the feasibility phase of testing, application of DL algorithms to abdominal and thoracic positions with significant motion are showing promise (37, 41, 59, 60, 86, 89, 104, 116–118, 185, 231). The MR-only simulation of pediatric patients could be extremely beneficial when their simulations are repeated since they are more radiation-sensitive than adults.It is necessary to confirm the geometrical accuracy of sCT before it can be used for clinical planning, mainly if MRI or sCT is used to replace CT for position verification. So far, research on DL-based sCT has been limited to a few studies. There have only been two studies that used CBCT and digitally reconstructed radiography to assess their alignments: Gupta et al. (33) for brain cancer and Olberg et al. (41) for breast cancer. The accuracy of sCT produced with standard 3T techniques has been extensively investigated, notably for geometric accuracy. Research is critical to enhancing the clinical application of sCT (232–234).DL-based sCT generation may reduce the duration of treatment in MR-guided RT, (235–239) because solitary MRI allows daily image guidance and plan modification. It is essential to assess the accuracy of dose calculation in a magnetic field before using it clinically. The current state of research on this topic is limited to studies on abdominal and pelvic tumors (59) and they have only considered low-strength magnetic fields. Recently, Groot Koerkamp et al. (240) reported the first dosimetric study demonstrating DDs for breast cancer patients treated with DL-based sCT. It is encouraging that the results were positive, but we recommend further study of other anatomical sites and magnetic field strengths.
2. CBCT to CT: CBCT imaging is an integral part of the daily patient setup for photon and proton RT. Due to scattering and reconstruction abnormalities, it is not routinely used to adjust daily plans and recalculate doses. This problem can be addressed in several ways (241), including image registration (242), scatter correction (243), a look-up table to rescale HU intensities (244), and histogram matching. In contrast to image registration and analytical adjustments, converting CBCT to sCT enhanced image quality. CBCT-to-CT conversion presents a challenge for clinical use because of the two imaging technologies’ different fields of view (FOV). This is usually overcome by cropping, registering, and resampling the volume to a smaller CBCT size than planned.However, the small field of view presents some challenges. For missing information (145), some have suggested assigning water equivalent density to the CT body contours. The sCT patch can also be sewed directly to the intended CT, guaranteeing that the whole dose volume will be covered. This stage is essential for online adaptive RT, especially in areas with a high degree of motion, as Liu et al. hypothesized in their work on pancreatic cancer (149). There is currently no consensus on whether improving CBCT quality with synthesis and reconstruction is the optimal approach. In preliminary experiments, training convolutional networks for reconstruction resulted in greater generalizability to diverse anatomy.
3. PET attenuation Correction (AC): sCTs generated in this category are derived either from MRIs or PETs that have not been corrected. Attenuation maps in MRI/PET hybrid acquisitions are currently inaccurate due to limitations in attenuation map construction. DL-based sCT has always been more consistent than commercially available MRAC. This review suggests that using deep learning for synthetic CT (sCT) can overcome most of the challenges associated with current AC methods. Although there has been a consistent number of studies in this field over the past few years, the specific factors and trends in these studies vary. These studies focus primarily on translating images into CT. Alternatively, Shiri et al. (245) studied the most significant number of patients to date (1150 patients split into 900 pieces of training, 100 validations, and 150 testings). This field could benefit from DL’s direct-map prediction capabilities in the future.
5 Trends in deep learning
5.1 Application
Deep Learning (DL) approaches, including supervised, semi-supervised, unsupervised, and reinforcement learning may tackle a wide range of issues. Computer vision and digital image processing applications have been divided into three groups by some researchers: structural scenarios, non-structural scenarios, and miscellaneous application situations. The term “structural scenario” refers to a circumstance in which data is processed in relational structures that are clear, such as physical systems and chemical structures. The term “non-structural scenario” refers to a circumstance in which data is not structured, e.g., images and texts with ambiguous patterns.
For clinicians who manage the search for representative images, it does not matter that how many times the data is reproduced. CT scanning, ultrasound, and MRI are all used in x-ray imaging. Physicians may examine the body’s obscure or concealed third dimension in this manner.
5.1.1 Image registration
Synthetic images can be used for diverse tasks downstream, revealing many possibilities. Intricate processes like image registration can be simplified with synthetic images generated using cutting-edge techniques. Chen et al. (192) have demonstrated in their work that synthetic images can facilitate streamlined workflows when it comes to registration by acting like reliable substitutes for real-world images in streamlined workflows.
Image alignment is essential for cross-domain image registration, where synthetic image generation is used to create images. In feature-based supervised registration of 3D multimodal images, deep learning has been used in several ways. To predict registration parameters, researchers have primarily used deep regression models (246–248). As well as being used in pre-processing, deep learning has also been used in the process of determining control points, which are then used to determine the registration parameters based on the information that is acquired from the deep learning technique.
An AIRNet (affine image registration network) model was developed by Chee and Wu (249) to predict the parameters of affine transformations between 2D and 3D images. An intra-patient T1 and T2 MRI image of the head was transformed using a deep learning regression model by Sloan et al. (250). According to Liu et al. (251), multi-modal medical image registration can be performed using synthetic image generation and deep learning. For rigid-body medical image registration, Zou et al. (252) implemented feature extraction and interest/control points-based deep learning models.
CT synthesis using MR-based technology also proves to be promising for radiation treatment planning and PET attenuation correction. In deformable registration when significant geometric distortion is allowed, direct registration between CT and MR images is even less reliable because of disparate image contrast. By replacing MRI with synthetic CT images, McKenzie et al. (253) reduced an inter-modality registration problem to intra-modality registration in the head and neck by using a CycleGAN-based method. CBCT technology is being increasingly adopted to improve the quality of radiation therapy, including higher diagnostic accuracy and better auto-contouring based on improvements in image registration, deformable image registration (DIR) and simulator analysis of CT images (133, 134). These capabilities are being offered in an increasing number of applications.
5.1.2 Image augmentation
A synthetic image can also enhance training sets in supervised learning applications. As Frid et al. (254) demonstrated, synthesized data augmentation can be a productive tool for improving model performance and robustness, which is one of the critical challenges of training deep learning models on limited datasets. In addition to extending synthetic images in downstream tasks across a broad range of domains, these investigations also shed light on the transformative role that synthetic images can play in optimizing complex processes in diverse domains.
A method for increasing the size of existing databases is known as data augmentation. A synthetic set of data is typically generated from the original database data. A synthetic image is created from the original dataset by using a particular method and generating a certain number of synthetic images from it. The former question has given rise to numerous methods, many of which are aimed at addressing it, including generative adversarial networks (255), random cropping (256), geometric transformations (257, 258), mixing images (259), and neural style transfers (260).
To improve the network’s generalizability and reduce overfitting, data augmentation is heavily used in deep neural network training nowadays. There are currently no data augmentation operations that can cover all variations of the data, as they are all manually designed operations, such as rotation and color jittering. The search space of Cubuk et al. (261) was still restricted to basic handcrafted image processing operations when they proposed to learn an augmentation policy with reinforcement learning. As a result, GANs are much more flexible for augmenting the training data, as they can sample the whole distribution of data (262). In styleGAN, realistic face images can be generated with unprecedented detail. Using this technique, images of pathology classes with sufficient numbers of cases could be generated from chest x-ray datasets. Medical data distribution is well known to be highly skewed with common diseases accounting for the majority of data. Rheumatoid arthritis, sickle cell disease, and other rare diseases cannot be adequately trained. The long tail of these diseases can be detected by radiologists. It is also anticipated that GANs will be used for the purpose of synthesizing cases and circumstances with uncommon pathologies. This will be done by conditionally generating information with medical experts supplying the conditioned information either on the basis of text descriptions or drawings.
5.1.3 Datasets, open-source libraries and tools
Computer vision techniques are evaluated using a variety of datasets and standards in different branches, including medical imaging (healthcare), agriculture, surveillance, sports and automotive etc. The implementation of DL in computer vision (medical imaging) is limited by a relatively small training dataset and a huge imaging volume. Example datasets include CT medical images (CT images from cancer imaging archive with contrast and patient age), Deep Lesion (contains 32,120 axial CT slices from 10,594 CT scans of 4,427 unique patients), OASIS Brain (Open Access Series of Imaging Studies dataset for normal aging and Alzheimer’s Disease), MRNet (dataset consists of 1,370 knee MRI) and IVDM3Seg (3D multi-modal MRI datasets of in-vitro diagnostics of the lower spine). Some open-source libraries have been established by certain research organizations and researchers, which comprise both common and classic computer vision techniques e.g., OpenCV, SimpleCV and TensorFlow etc (176, 263, 264).
MIPAV (Medical Image Processing, Analysis, and Visualization) is a java-based tool that allows for quantitative analysis and visualization of medical images from a variety of modalities, including PET, MRI, and CT. FSL (FMRIB Software Library) encompasses an extensive array of analysis tools designed for processing FMRI, MRI, and DTI brain imaging data (265). AFNI (Analysis of Functional Neuro Images) is a Python-based application that analyzes and displays data from different MRI modalities, including anatomical, functional MRI (FMRI), and diffusion weighted (DW) data (176, 263, 264).
5.1.4 Predictive analytics and therapy using computer vision
The use of computer vision in surgery and the treatment of certain illnesses has demonstrated to be quite beneficial especially in the field of surgery. Three-dimensional (3D) modeling and rapid prototyping technologies have lately helped medical imaging modalities such as CT and MRI. Human activity recognition (HAR) is also one of the most well-studied computer vision challenges. S. Zhang et al. (266) provide an overview of several HAR techniques as well as their evolution with traditional Chinese literature.
In vision-based activity recognition, the authors emphasize developments in image representation methodologies and classification algorithms. Common representation approaches include global representations, local representations, and depth-based representations. They divide and describe human activities into three levels, in that order: action primitives, actions/activities, and interactions. They also offer a description of the HAR application’s classification techniques (266, 267).
5.2 Diffusion models
Developing realistic and high-fidelity images is a challenge that has seen a paradigm shift with the emergence of diffusion models. Intuitive patterns and dependencies within image data can be captured using these probability distribution models based on probability distributions. According to recent studies, diffusion models can produce diverse and realistic samples more effectively than traditional generative models, as demonstrated in work by Dhariwal et al. (268). Diffusion models are robust and versatile tools for image synthesis since they can consider the underlying uncertainty in pixel values. This article intends to shed light on the potential of diffusion models to redefine the landscape of image synthesis in various domains, drawing inspiration from recent developments and applications in multiple fields.
Integrating diffusion models can profoundly advance diagnostic and therapeutic applications of diffusion models in medical imaging. According to Hung et al. (269), diffusion models can capture nuanced variations in medical images, enhancing the realism of synthesized medical data. This article aims to demonstrate how diffusion models can be used to address challenges like data scarcity and to create realistic synthetic datasets based on image synthesis. Utilizing diffusion models is a critical trend in medical imaging as synthesis data is increasingly used for training machine learning models, resulting in improved diagnostic accuracy and treatment planning. This article examines diffusion models in the context of current advancements and future possibilities in medical imaging.
5.3 Open issues
There are studies in medical imaging research that demonstrate accuracy of above 95%. Though, we are concerned with more than simply the accuracy of a classifier. Because false negatives and false positives in medical imaging may have catastrophic effects. This is one of the reasons why, despite their high performance, stand-alone decision systems are not widely used. In this section, we will describe many potential research topics and open concerns for computer vision in medical imaging.
Imaging Modality: Medical imaging modalities are classified according to how images are generated. In radiology, a modality is a phrase that refers to a certain kind of imaging, such as CT scanning, ultrasound, radiation (x-rays), and MRI. X-ray machines, which are made up of a single x-ray source and produce two-dimensional images, are examples of radiation-generated images. In literature, medical imaging modalities algorithms have received a great attention, but it is critical that the medical imaging modalities algorithms, be designed to retain high performance.
Generative Medical Image Synthesis: Inspired by the GAN, because of its capacity to generate data without explicitly modeling the probability density function, GANs have gotten a lot of interest in the computer vision field. If diagnostic images are to be utilized in a publication or put into the public domain, patient’s agreement could be necessary, depending on institutional rules. GANs are commonly used in the medical imaging for image synthesis. This helps to address the privacy concerns around diagnostic medical images as well as the lack of positive instances for each disease. Another barrier to the implementation of supervised training techniques is the lack of professionals who can annotate medical images (52).
Interpretability/Explainability in Medical Image Analysis: An explanation of the machine learning (ML) algorithm can be described as interpretability. Various computer vision algorithms achieve outstanding results at the cost of greater complexity. As a result, they become less interpretable, perhaps leading to distrust. DL-based approaches, have shown to be quite successful for several medical diagnostic tasks, outperforming human specialists in certain cases. However, the algorithms’ black-box nature has limited their clinical use. Recent explainability studies have attempted to demonstrate the characteristics that have the most impact on a model’s choice. Furthermore, interpretability findings are often based on a comparison of explanations with domain knowledge. As a result, objective, quantitative, and systematic assessment procedures are required (270). Finally, AI safety in healthcare is intimately linked to interpretability and explainability.
6 Conclusion
This study includes a broad overview of computer vision techniques as well as a complete assessment of medical imaging with respect to CT, CBCT, PET and MRI techniques. We looked at current digital image processing techniques with respect to medical imaging. We did our best to emphasize both the potential and the obstacles that this medical imaging application industry faces in the healthcare field. Our goal is to uncover the important need for computer vision algorithms in the clinical and theoretical context of medical imaging. This special research article discusses a few recent advancements in computer vision related to medical images and clinical applications.
In conclusion, this study presents a glimpse of computer vision in healthcare applications using medical images. Hopefully, future computer vision, analysis techniques, and ML of medical images will benefit from this paper. However, even though these works outperform conventional and state-of-the-art approaches, there are still limitations and challenges for computer vision and different algorithms and processing techniques of medical images. In addition, we discuss some potential future research areas in the sCT generation. We really hope that this survey proves to be useful. We believe that this survey will aid scholars and practitioners in their computer vision, medical imaging and related research and development.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
MS: Writing – original draft, writing – review & editing. SG: Funding acquisition, Methodology, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article.
This project received funding from the European Union's Horizon 2020 research and innovation programme (952914) (FindingPheno). SG also received support from the Danish National Research Foundation award DNRF143 ‘A Center for Evolutionary Hologenomics’.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Barragán-Montero A, Javaid U, Valdés G, Nguyen D, Desbordes P, Macq B, et al. Artificial intelligence, machine learning for medical imaging: a technology review. Phys Med. (2021) 83:242–56. doi: 10.1016/j.ejmp.2021.04.016
2. Nyholm T, Jonsson J. Counterpoint: opportunities, challenges of a magnetic resonance imaging–only radiotherapy work flow. Semin Radiat Oncol. (2014) 24(3):175–80. doi: 10.1016/j.semradonc.2014.02.005
3. Beaton L, Bandula S, Gaze MN, Sharma RA. How rapid advances in imaging are defining the future of precision radiation oncology. Br J Cancer. (2019) 120(8):779–90. doi: 10.1038/s41416-019-0412-y
4. Verellen D, De Ridder M, Linthout N, Tournel K, Soete G, Storme G. Innovations, advances in radiation technology. Nat Rev Cancer. (2007) 7:949–60. doi: 10.1038/nrc2288
5. Jaffray DA. Image-guided radiotherapy: from current concept to future perspectives. Nat Rev Clin Oncol. (2012) 9(12):688–99. doi: 10.1038/nrclinonc.2012.194
6. Seco J, Spadea MF. Imaging in particle therapy: state of the art and future perspective. Acta Oncol. (2015) 54(9):1254–8. doi: 10.3109/0284186X.2015.1075665
7. Dirix P, Haustermans K, Vandecaveye V. The value of magnetic resonance imaging for radiotherapy planning. Semin Radiat Oncol. (2014) 24(3):151–9. doi: 10.1016/j.semradonc.2014.02.003
8. Lagendijk JJ, Raaymakers BW, Van Vulpen M. The magnetic resonance imaging–linac system. Semin Radiat Oncol. (2014) 24(3):207–9. doi: 10.1016/j.semradonc.2014.02.009
9. Kupelian P, Sonke J-J. Magnetic resonance–guided adaptive radiotherapy: a solution to the future. Semin Radiat Oncol. (2014) 24(3):227–32. doi: 10.1016/j.semradonc.2014.02.013
10. Fraass B, McShan D, Diaz R, Ten Haken R, Aisen A, Gebarski S, et al. Integration of magnetic resonance imaging into radiation therapy treatment planning: I. technical considerations. Int J Radiat Oncol* Biol* Phys. (1987) 13(12):1897–908. doi: 10.1016/0360-3016(87)90358-0
11. Lee YK, Bollet M, Charles-Edwards G, Flower MA, Leach MO, McNair H, et al. Radiotherapy treatment planning of prostate cancer using magnetic resonance imaging alone. Radiother Oncol. (2003) 66(2):203–16. doi: 10.1016/S0167-8140(02)00440-1
12. Owrangi AM, Greer PB, Glide-Hurst CK. MRI-only treatment planning: benefits and challenges. Phys Med Biol. (2018) 63(5):05TR01. doi: 10.1088/1361-6560/aaaca4
13. Karlsson M, Karlsson MG, Nyholm T, Amies C, Zackrisson B. Dedicated magnetic resonance imaging in the radiotherapy clinic. Int J Radiat Oncol* Biol* Phys. (2009) 74(2):644–51. doi: 10.1016/j.ijrobp.2009.01.065
14. Meyer P, Noblet V, Mazzara C, Lallement A. Survey on deep learning for radiotherapy. Comput Biol Med. (2018) 98:126–46. doi: 10.1016/j.compbiomed.2018.05.018
15. Wang T, Lei Y, Fu Y, Wynne JF, Curran WJ, Liu T, et al. A review on medical imaging synthesis using deep learning and its clinical applications. J Appl Clin Med Phys. (2021) 22(1):11–36. doi: 10.1002/acm2.13121
16. Spadea MF, Maspero M, Zaffino P, Seco J. Deep learning based synthetic-ct generation in radiotherapy and pet: a review. Med Phys. (2021) 48(11):6537–66. doi: 10.1002/mp.15150
17. Kazeminia S, Baur C, Kuijper A, van Ginneken B, Navab N, Albarqouni S, et al. GANs for medical image analysis. Artif Intell Med. (2020) 109:101938. doi: 10.1016/j.artmed.2020.101938
18. L. G. K. T. B. BE. Setio aaa ciompi f ghafoorian m van der laak ja van ginneken b sánchez ci a survey on deep learning in medical image analysis. Med Image Anal. (2017) 42(1995):60. doi: 10.1016/j.media.2017.07.005
19. Zhou SK, Greenspan H, Davatzikos C, Duncan JS, Van Ginneken B, Madabhushi A, et al. A review of deep learning in medical imaging: imaging traits, technology trends, case studies with progress highlights, and future promises. Proc IEEE. (2021) 109(5). doi: 10.1109/2FJPROC.2021.3054390
20. Shen C, Nguyen D, Zhou Z, Jiang SB, Dong B, Jia X. An introduction to deep learning in medical physics: advantages, potential, challenges. Phys Med Biol. (2020) 65(5):05TR01. doi: 10.1088/1361-6560/ab6f51
21. Nair V, Hinton GE. Rectified linear units improve restricted Boltzmann machines. In: Icml (2010).
22. Maas AL, Hannun AY, Ng AY, et al. Rectifier nonlinearities improve neural network acoustic models. Proc. icml. (2013) 30(1):3.
23. Clevert D-A, Unterthiner T, Hochreiter S. Fast, accurate deep network learning by exponential linear units (elus). arXiv [Preprint]. arXiv:1511.07289 (2015).
24. Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International Conference on Machine Learning (2015). p. 448–56.
25. Liu F, Yadav P, Baschnagel AM, McMillan AB. MR-based treatment planning in radiation therapy using a deep learning approach. J Appl Clin Med Phys. (2019) 20(3):105–14. doi: 10.1002/acm2.12554
26. Xiang L, Wang Q, Nie D, Zhang L, Jin X, Qiao Y, et al. Deep embedding convolutional neural network for synthesizing CT image from T1-weighted MR image. Med Image Anal. (2018) 47:31–44. doi: 10.1016/j.media.2018.03.011
27. Nie D, Cao X, Gao Y, Wang L, Shen D. Estimating CT image from MRI data using 3D fully convolutional networks. Deep Learn Data Label Med Appl. (2016) 10008:170–8. doi: 10.1007/978-3-319-46976-8_18
28. Spadea MF, Pileggi G, Zaffino P, Salome P, Catana C, Izquierdo-Garcia D, et al. Deep convolution neural network (DCNN) multiplane approach to synthetic CT generation from MR images—application in brain proton therapy. Int J Radiat Oncol* Biol* Phys. (2019) 105(3):495–503. doi: 10.1016/j.ijrobp.2019.06.2535
29. Andres EA, Fidon L, Vakalopoulou M, Lerousseau M, Carré A, Sun R, et al. Dosimetry-driven quality measure of brain pseudo computed tomography generated from deep learning for MRI-only radiation therapy treatment planning. Int J Radiat Oncol* Biol* Phys. (2020) 108(3):813–23. doi: 10.1016/j.ijrobp.2020.05.006
30. Han X. MR-based synthetic CT generation using a deep convolutional neural network method. Med Phys. (2017) 44(4):1408–19. doi: 10.1002/mp.12155
31. Wang Y, Liu C, Zhang X, Deng W. Synthetic CT generation based on t2 weighted MRI of nasopharyngeal carcinoma (NPC) using a deep convolutional neural network (DCNN). front oncol 2019; 9: 1333 (2019).
32. Arabi H, Dowling JA, Burgos N, Han X, Greer PB, Koutsouvelis N, et al. Comparative study of algorithms for synthetic CT generation from MRI: consequences for MRI-guided radiation planning in the pelvic region. Med Phys. (2018) 45(11):5218–33. doi: 10.1002/mp.13187
33. Gupta D, Kim M, Vineberg KA, Balter JM. Generation of synthetic CT images from MRI for treatment planning, patient positioning using a 3-channel U-net trained on sagittal images. Front Oncol. (2019) 9:964. doi: 10.3389/fonc.2019.00964
34. Dinkla AM, Florkow MC, Maspero M, Savenije MH, Zijlstra F, Doornaert PA, et al. Dosimetric evaluation of synthetic CT for head, neck radiotherapy generated by a patch-based three-dimensional convolutional neural network. Med Phys. (2019) 46(9):4095–104. doi: 10.1002/mp.13663
35. Qi M, Li Y, Wu A, Jia Q, Li B, Sun W, et al. Multi-sequence MR image-based synthetic CT generation using a generative adversarial network for head and neck MRI-only radiotherapy. Med Phys. (2020) 47(4):1880–94. doi: 10.1002/mp.14075
36. Chen S, Qin A, Zhou D. Yan di technical note: U-net-generated synthetic CT images for magnetic resonance imaging-only prostate intensity-modulated radiation therapy treatment planning. Med Phys. (2018) 45(12):5659–65. doi: 10.1002/mp.13247
37. Florkow MC, Zijlstra F, Kerkmeijer LG, Maspero M, van den Berg CA, van Stralen M, et al. The impact of MRI-CT registration errors on deep learning-based synthetic CT generation. Med Imaging 2019: Image Process. (2019) 10949:831–7. doi: 10.1117/12.2512747
38. Florkow MC, Zijlstra F, Willemsen K, Maspero M, van den Berg CA, Kerkmeijer LG, et al. Deep learning–based MR-to-CT synthesis: the influence of varying gradient echo–based MR images as input channels. Magn Reson Med. (2020) 83(4):1429–41. doi: 10.1002/mrm.28008
39. Stadelmann JV, Schulz H, van der Heide UA, Renisch S. Pseudo-ct image generation from mdixon MRI images using fully convolutional neural networks. Med Imaging 2019: Biomed Appl Mol Struct Funct Imaging. (2019) 10953:109530Z. doi: 10.1117/12.2512741
40. Neppl S, Landry G, Kurz C, Hansen DC, Hoyle B, Stöcklein S, et al. Evaluation of proton and photon dose distributions recalculated on 2D and 3D Unet-generated pseudoCTs from T1-weighted MR head scans. Acta Oncol. (2019) 58(10):1429–34. doi: 10.1080/0284186X.2019.1630754
41. Olberg S, Zhang H, Kennedy WR, Chun J, Rodriguez V, Zoberi I, et al. Synthetic CT reconstruction using a deep spatial pyramid convolutional framework for MR-only breast radiotherapy. Med Phys. (2019) 46(9):4135–47. doi: 10.1002/mp.13716
42. Li W, Li Y, Qin W, Liang X, Xu J, Xiong J, et al. Magnetic resonance image (MRI) synthesis from brain computed tomography (CT) images based on deep learning methods for magnetic resonance (MR)-guided radiotherapy. Quant Imaging Med Surg. (2020) 10(6):1223. doi: 10.21037/qims-19-885
43. Kazemifar S, McGuire S, Timmerman R, Wardak Z, Nguyen D, Park Y, et al. MRI-only brain radiotherapy: assessing the dosimetric accuracy of synthetic CT images generated using a deep learning approach. Radiother Oncol. (2019) 136:56–63. doi: 10.1016/j.radonc.2019.03.026
44. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2016). p. 770–8.
45. Gong E, Pauly JM, Wintermark M, Zaharchuk G. Deep learning enables reduced gadolinium dose for contrast-enhanced brain MRI. J Magn Reson Imaging. (2018) 48(2):330–40. doi: 10.1002/jmri.25970
46. Huang G, Liu Z, Weinberger KQ. Densely connected convolutional networks. corr abs/1608.06993 (2016) (2015).
47. Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Commun ACM. (2017) 60(6):84–90. doi: 10.1145/3065386
48. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv [Preprint]. arXiv:1409.1556 (2014).
49. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, et al. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2015). p. 1–9.
50. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (2015>). p. 234–241.
51. Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, et al. Generative adversarial networks. arxiv: 14062661 (2014). avilable at: https://arxiv.org/abs/1406.2661.
52. Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: a review. Med Image Anal. (2019) 58:101552. doi: 10.1016/j.media.2019.101552
53. Largent A, Marage L, Gicquiau I, Nunes J-C, Reynaert N, Castelli J, et al. Head-and-neck MRI-only radiotherapy treatment planning: From acquisition in treatment position to pseudo-CT generation. Cancer/Radiothér. (2020) 24(4):288–97. doi: 10.1016/j.canrad.2020.01.008
54. Emami H, Dong M, Nejad-Davarani SP, Glide-Hurst CK. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med Phys. (2018) 45(8):3627–36. doi: 10.1002/mp.13047
55. Wolterink JM, Dinkla AM, Savenije MH, Seevinck PR, van den Berg CA, Išgum I. Deep MR to CT synthesis using unpaired data. In: International Workshop on Simulation and Synthesis in Medical Imaging (2017). p. 14–23 (
56. Nie D, Trullo R, Lian J, Petitjean C, Ruan S, Wang Q, et al. Medical image synthesis with context-aware generative adversarial networks. In: International Conference on Medical Image Computing and Computer-Assisted Intervention (2017). p. 417–25.
57. Kazemifar S, Barragán Montero AM, Souris K, Rivas ST, Timmerman R, Park YK, et al. Dosimetric evaluation of synthetic CT generated with GANs for MRI-only proton therapy treatment planning of brain tumors. J Appl Clin Med Phys. (2020) 21(5):76–86. doi: 10.1002/acm2.12856
58. Fetty L, Löfstedt T, Heilemann G, Furtado H, Nesvacil N, Nyholm T, et al. Investigating conditional GAN performance with different generator architectures, an ensemble model, and different MR scanners for MR-SCT conversion. Phys Med Biol. (2020) 65(10):105004. doi: 10.1088/1361-6560/ab857b
59. Cusumano D, Lenkowicz J, Votta C, Boldrini L, Placidi L, Catucci F, et al. A deep learning approach to generate synthetic CT in low field MR-guided adaptive radiotherapy for abdominal and pelvic cases. Radiother Oncol. (2020) 153:205–12. doi: 10.1016/j.radonc.2020.10.018
60. Fu J, Singhrao K, Cao M, Yu V, Santhanam AP, Yang Y, et al. Generation of abdominal synthetic CTs from 0.35 T MR images using generative adversarial networks for MR-only liver radiotherapy. Biomed Phys Eng Express. (2020) 6(1):015033. doi: 10.1088/2057-1976/ab6e1f
61. Mao X, Li Q, Xie H, Lau RY, Wang Z, Paul Smolley S. Least squares generative adversarial networks. In: Proceedings of the IEEE International Conference on Computer Vision (2017). p. 2794–802.
62. Boni KNB, Klein J, Vanquin L, Wagner A, Lacornerie T, Pasquier D, et al. MR to CT synthesis with multicenter data in the pelvic area using a conditional generative adversarial network. Phys Med Biol. (2020) 65(7):075002. doi: 10.1088/1361-6560/ab7633
63. Koike Y, Akino Y, Sumida I, Shiomi H, Mizuno H, Yagi M, et al. Feasibility of synthetic computed tomography generated with an adversarial network for multi-sequence magnetic resonance-based brain radiotherapy. J Radiat Res. (2020) 61(1):92–103. doi: 10.1093/jrr/rrz063
64. Maspero M, Savenije MH, Dinkla AM, Seevinck PR, Intven MP, Jurgenliemk-Schulz IM, et al. Dose evaluation of fast synthetic-ct generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys Med Biol. (2018) 63(18):185001. doi: 10.1088/1361-6560/aada6d
65. Tie X, Lam S-K, Zhang Y, Lee K-H, Au K-H, Cai J. Pseudo-CT generation from multi-parametric MRI using a novel multi-channel multi-path conditional generative adversarial network for nasopharyngeal carcinoma patients. Med Phys. (2020) 47(4):1750–62. doi: 10.1002/mp.14062
66. Tang B, Wu F, Fu Y, Wang X, Wang P, Orlandini LC, et al. Dosimetric evaluation of synthetic CT image generated using a neural network for MR-only brain radiotherapy. J Appl Clin Med Phys. (2021) 22(3):55–62. doi: 10.1002/acm2.13176
67. Bird D, Nix MG, McCallum H, Teo M, Gilbert A, Casanova N, et al. Multicentre, deep learning, synthetic-CT generation for ano-rectal MR-only radiotherapy treatment planning. Radiother Oncol. (2021) 156:23–8. doi: 10.1016/j.radonc.2020.11.027
68. Peng Y, Chen S, Qin A, Chen M, Gao X, Liu Y, et al. Magnetic resonance-based synthetic computed tomography images generated using generative adversarial networks for nasopharyngeal carcinoma radiotherapy treatment planning. Radiother Oncol. (2020) 150:217–24. doi: 10.1016/j.radonc.2020.06.049
69. Klages P, Benslimane I, Riyahi S, Jiang J, Hunt M, Deasy J, et al. Comparison of patch-based conditional generative adversarial neural net models with emphasis on model robustness for use in head and neck cases for MR-only planning. arXiv [Preprint]. arXiv:1902.00536 (2019).
70. Dodge Y. An introduction to L1-norm based statistical data analysis. Comput Stat Data Anal. (1987) 5(4):239–53. doi: 10.1016/0167-9473(87)90048-X
71. Li Y, Li W, Xiong J, Xia J, Xie Y. Comparison of supervised and unsupervised deep learning methods for medical image synthesis between computed tomography and magnetic resonance images. Biomed Res Int. (2020) 2020:5193707. doi: 10.1155/2020/5193707
72. Xu L, Zeng X, Zhang H, Li W, Lei J, Huang Z. BPGAN: Bidirectional CT-to-MRI prediction using multi-generative multi-adversarial nets with spectral normalization and localization. Neural Netw. (2020) 128:82–96. doi: 10.1016/j.neunet.2020.05.001
73. Fu J, Yang Y, Singhrao K, Ruan D, Chu F-I, Low DA, et al. Deep learning approaches using 2D and 3D convolutional neural networks for generating male pelvic synthetic computed tomography from magnetic resonance imaging. Med Phys. (2019) 46(9):3788–98. doi: 10.1002/mp.13672
74. Vaassen F, Hazelaar C, Vaniqui A, Gooding M, van der Heyden B, Canters R, et al. Evaluation of measures for assessing time-saving of automatic organ-at-risk segmentation in radiotherapy. Phys Imaging Radiat Oncol. (2020) 13:1–6. doi: 10.1016/j.phro.2019.12.001
75. Clasie BM, Sharp GC, Seco J, Flanz JB, Kooy HM. Numerical solutions of the -index in two and three dimensions. Phys Med Biol. (2012) 57(21):6981. doi: 10.1088/0031-9155/57/21/6981
76. Hussein M, Clark C, Nisbet A. Challenges in calculation of the gamma index in radiotherapy–towards good practice. Phys Med. (2017) 36:1–11. doi: 10.1016/j.ejmp.2017.03.001
77. Paganetti H. Range uncertainties in proton therapy and the role of monte carlo simulations. Phys Med Biol. (2012) 57(11):R99. doi: 10.1088/0031-9155/57/11/R99
78. Yang X, Lei Y, Shu H-K, Rossi P, Mao H, Shim H, et al. Pseudo CT estimation from MRI using patch-based random forest. Med Imaging 2017: Image Process. (2017) 10133:101332Q. doi: 10.1117/2F12.2253936
79. Dinkla AM, Wolterink JM, Maspero M, Savenije MH, Verhoeff JJ, Seravalli E, et al. MR-only brain radiation therapy: dosimetric evaluation of synthetic CTs generated by a dilated convolutional neural network. Int J Radiat Oncol* Biol* Phys. (2018) 102(4):801–12. doi: 10.1016/j.ijrobp.2018.05.058
80. Xu K, Cao J, Xia K, Yang H, Zhu J, Wu C, et al. Multichannel residual conditional GAN-leveraged abdominal pseudo-CT generation via Dixon MR images. IEEE Access. (2019) 7:163823–30. doi: 10.1109/ACCESS.2019.2951924
81. Lei Y, Harms J, Wang T, Liu Y, Shu H-K, Jani AB, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys. (2019) 46(8):3565–81. doi: 10.1002/mp.13617
82. Jin C-B, Kim H, Liu M, Jung W, Joo S, Park E, et al. Deep CT to MR synthesis using paired and unpaired data. Sensors. (2019) 19(10):2361. doi: 10.3390/s19102361
83. Shafai-Erfani G, Lei Y, Liu Y, Wang Y, Wang T, Zhong J, et al. MRI-based proton treatment planning for base of skull tumors. Int J Part Ther. (2019) 6(2):12–25. doi: 10.14338/IJPT-19-00062.1
84. Jeon W, An HJ, Kim J-i., Park JM, Kim H, Shin KH, et al. Preliminary application of synthetic computed tomography image generation from magnetic resonance image using deep-learning in breast cancer patients. J Radiat Prot Res. (2019) 44(4):149–55. doi: 10.14407/jrpr.2019.44.4.149
85. Liu Y, Lei Y, Wang Y, Wang T, Ren L, Lin L, et al. MRI-based treatment planning for proton radiotherapy: dosimetric validation of a deep learning-based liver synthetic CT generation method. Phys Med Biol. (2019) 64(14):145015. doi: 10.1088/1361-6560/ab25bc
86. Liu Y, Lei Y, Wang Y, Shafai-Erfani G, Wang T, Tian S, et al. Evaluation of a deep learning-based pelvic synthetic CT generation technique for MRI-based prostate proton treatment planning. Phys Med Biol. (2019) 64(20):205022. doi: 10.1088/1361-6560/ab41af
87. Largent A, Barateau A, Nunes J-C, Mylona E, Castelli J, Lafond C, et al. Comparison of deep learning-based and patch-based methods for pseudo-CT generation in MRI-based prostate dose planning. Int J Radiat Oncol* Biol* Phys. (2019) 105(5):1137–50. doi: 10.1016/j.ijrobp.2019.08.049
88. Liu L, Johansson A, Cao Y, Dow J, Lawrence TS, Balter JM. Abdominal synthetic CT generation from MR Dixon images using a U-net trained with ‘semi-synthetic’ CT data. Phys Med Biol. (2020) 65(12):125001. doi: 10.1088/1361-6560/ab8cd2
89. Florkow MC, Guerreiro F, Zijlstra F, Seravalli E, Janssens GO, Maduro JH, et al. Deep learning-enabled MRI-only photon and proton therapy treatment planning for paediatric abdominal tumours. Radiother Oncol. (2020) 153:220–7. doi: 10.1016/j.radonc.2020.09.056
90. Yang H, Sun J, Carass A, Zhao C, Lee J, Prince JL, et al. Unsupervised MR-to-CT synthesis using structure-constrained CycleGAN. IEEE Trans Med Imaging. (2020) 39(12):4249–61. doi: 10.1109/TMI.2020.3015379
91. Maspero M, Bentvelzen LG, Savenije MH, Guerreiro F, Seravalli E, Janssens GO, et al. Deep learning-based synthetic CT generation for paediatric brain MR-only photon and proton radiotherapy. Radiother Oncol. (2020) 153:197–204. doi: 10.1016/j.radonc.2020.09.029
92. Massa HA, Johnson JM, McMillan AB. Comparison of deep learning synthesis of synthetic CTs using clinical MRI inputs. Phys Med Biol. (2020) 65(23):23NT03. doi: 10.1088/1361-6560/abc5cb
93. Kearney V, Ziemer BP, Perry A, Wang T, Chan JW, Ma L, et al. Attention-aware discrimination for MR-to-CT image translation using cycle-consistent generative adversarial networks. Radiol Artif Intell. (2020) 2(2):e190027. doi: 10.1148/ryai.2020190027
94. Qian P, Xu K, Wang T, Zheng Q, Yang H, Baydoun A, et al. Estimating CT from MR abdominal images using novel generative adversarial networks. J Grid Comput. (2020) 18(2):211–26. doi: 10.1007/s10723-020-09513-3
95. Su P, Guo S, Roys S, Maier F, Bhat H, Melhem E, et al. Transcranial MR imaging–guided focused ultrasound interventions using deep learning synthesized CT. Am J Neuroradiol. (2020) 41(10):1841–8. doi: 10.3174/ajnr.A6758
96. Thummerer A, Zaffino P, Meijers A, Marmitt GG, Seco J, Steenbakkers RJ, et al. Comparison of CBCT based synthetic CT methods suitable for proton dose calculations in adaptive proton therapy. Phys Med Biol. (2020) 65(9):095002. doi: 10.1088/1361-6560/ab7d54
97. Ranjan A, Lalwani D, Misra R. GAN for synthesizing CT from T2-weighted MRI data towards MR-guided radiation treatment. Magn Reson Mater Phys Biol Med. (2022) 35(3):449–57. doi: 10.1007/s10334-021-00974-5
98. Li W, Kazemifar S, Bai T, Nguyen D, Weng Y, Li Y, et al. Synthesizing CT images from MR images with deep learning: model generalization for different datasets through transfer learning. Biomed Phys Eng Express. (2021) 7(2):025020. doi: 10.1088/2057-1976/abe3a7
99. Touati R, Le WT, Kadoury S. A feature invariant generative adversarial network for head and neck mri/ct image synthesis. Phys Med Biol. (2021) 66(9):095001. doi: 10.1088/1361-6560/abf1bb
100. Farjam R, Nagar H, Kathy Zhou X, Ouellette D, Chiara Formenti S, DeWyngaert JK. Deep learning-based synthetic CT generation for MR-only radiotherapy of prostate cancer patients with 0.35 T MRI linear accelerator. J Appl Clin Med Phys. (2021) 22(8):93–104. doi: 10.1002/acm2.13327
101. Brou Boni KN, Klein J, Gulyban A, Reynaert N, Pasquier D. Improving generalization in MR-to-CT synthesis in radiotherapy by using an augmented cycle generative adversarial network with unpaired data. Med Phys. (2021) 48(6):3003–10. doi: 10.1002/mp.14866
102. Zimmermann L, Buschmann M, Herrmann H, Heilemann G, Kuess P, Goldner G, et al. An MR-only acquisition and artificial intelligence based image-processing protocol for photon and proton therapy using a low field mr. Z Med Phys. (2021) 31(1):78–88. doi: 10.1016/j.zemedi.2020.10.004
103. Szalkowski G, Nie D, Zhu T, Yap P-T, Lian J. Synthetic digital reconstructed radiographs for MR-only robotic stereotactic radiation therapy: a proof of concept. Comput Biol Med. (2021) 138:104917. doi: 10.1016/j.compbiomed.2021.104917
104. Li X, Yadav P, McMillan AB. Synthetic computed tomography generation from 0.35 T magnetic resonance images for magnetic resonance–only radiation therapy planning using perceptual loss models. Pract Radiat Oncol. (2022) 12(1):e40–8. doi: 10.1016/j.prro.2021.08.007
105. Wang C, Uh J, Patni T, Merchant T, Li Y, Hua C-h., et al. Toward MR-only proton therapy planning for pediatric brain tumors: synthesis of relative proton stopping power images with multiple sequence MRI and development of an online quality assurance tool. Med Phys. (2022) 49(3):1559–70. doi: 10.1002/mp.15479
106. Jabbarpour A, Mahdavi SR, Sadr AV, Esmaili G, Shiri I, Zaidi H. Unsupervised pseudo CT generation using heterogenous multicentric ct/mr images and CycleGAN: dosimetric assessment for 3D conformal radiotherapy. Comput Biol Med. (2022) 143:105277. doi: 10.1016/j.compbiomed.2022.105277
107. Zimmermann L, Knäusl B, Stock M, Lütgendorf-Caucig C, Georg D, Kuess P. An MRI sequence independent convolutional neural network for synthetic head CT generation in proton therapy. Z Med Phys. (2022) 32(2):218–27. doi: 10.1016/j.zemedi.2021.10.003
108. Islam KT, Wijewickrema S, O’Leary S. A deep learning framework for segmenting brain tumors using MRI and synthetically generated CT images. Sensors. (2022) 22(2):523. doi: 10.3390/s22020523
109. Wang C, Uh J, Merchant TE, Hua C-h., Acharya S. Facilitating MR-guided adaptive proton therapy in children using deep learning-based synthetic ct. Int J Part Ther. (2022) 8(3):11–20. doi: 10.14338/IJPT-20-00099.1
110. Zimmermann L, Knäusl B, Stock M, Lütgendorf-Caucig C, Georg D, Kuess P. An MRI sequence independent convolutional neural network for synthetic head CT generation in proton therapy. Z Med Phys. (2022) 32(2):218–27. doi: 10.1016/j.zemedi.2021.10.003
111. Chen S, Peng Y, Qin A, Liu Y, Zhao C, Deng X, et al. MR-based synthetic CT image for intensity-modulated proton treatment planning of nasopharyngeal carcinoma patients. Acta Oncol. (2022) 61(11):1417–24. doi: 10.1080/0284186X.2022.2140017
112. Scholey JE, Rajagopal A, Vasquez EG, Sudhyadhom A, Larson PEZ. Generation of synthetic megavoltage CT for MRI-only radiotherapy treatment planning using a 3D deep convolutional neural network. Med Phys. (2022) 49(10):6622–34. doi: 10.1002/mp.15876
113. Vajpayee R, Agrawal V, Krishnamurthi G. Structurally-constrained optical-flow-guided adversarial generation of synthetic CT for MR-only radiotherapy treatment planning. Sci Rep. (2022) 12(1):14855. doi: 10.1038/s41598-022-18256-y
114. Tahri S, Barateau A, Cadin C, Chourak H, Ribault S, Nozahic F, et al. A high-performance method of deep learning for prostate MR-only radiotherapy planning using an optimized Pix2Pix architecture. Phys Med. (2022) 103:108–18. doi: 10.1016/j.ejmp.2022.10.003
115. Hsu S-H, Han Z, Leeman JE, Hu Y-H, Mak RH, Sudhyadhom A. Synthetic CT generation for MRI-guided adaptive radiotherapy in prostate cancer. Front Oncol. (2022) 12:969463. doi: 10.3389/fonc.2022.969463
116. Lenkowicz J, Votta C, Nardini M, Quaranta F, Catucci F, Boldrini L, et al. A deep learning approach to generate synthetic CT in low field MR-guided radiotherapy for lung cases. Radiother Oncol. (2022) 176:31–8. doi: 10.1016/j.radonc.2022.08.028
117. Rippke C, Renkamp CK, Stahl-Arnsberger C, Miltner A, Buchele C, Hörner-Rieber J, et al. A body mass index-based method for “MR-only” abdominal MR-guided adaptive radiotherapy. Z Med Phys. (2023). doi: 10.1016/j.zemedi.2022.12.001
118. Hernandez AG, Fau P, Wojak J, Mailleux H, Benkreira M, Rapacchi S, et al. Synthetic computed tomography generation for abdominal adaptive radiotherapy using low-field magnetic resonance imaging. Phys Imaging Radiat Oncol. (2023) 25:100425. doi: 10.1016/j.phro.2023.100425
119. Zhao S, Geng C, Guo C, Tian F, Tang X. SARU: A self-attention resunet to generate synthetic CT images for MR-only BNCT treatment planning. Med Phys. (2023) 50(1):117–27. doi: 10.1002/mp.15986
120. Zhao Y, Wang H, Yu C, Court LE, Wang X, Wang Q, et al. Compensation cycle consistent generative adversarial networks (Comp-GAN) for synthetic CT generation from MR scans with truncated anatomy. Med Phys. (2023) 50(7):4399–414. doi: 10.1002/mp.16246
121. Zhou X, Cai W, Cai J, Xiao F, Qi M, Liu J, et al. Multimodality MRI synchronous construction based deep learning framework for MRI-guided radiotherapy synthetic CT generation. Comput Biol Med. (2023) 162:107054. doi: 10.1016/j.compbiomed.2023.107054
122. Wyatt JJ, Kaushik S, Cozzini C, Pearson RA, Petit S, Capala M, et al. Comprehensive dose evaluation of a deep learning based synthetic computed tomography algorithm for pelvic magnetic resonance-only radiotherapy. Radiother Oncol. (2023) 184:109692. doi: 10.1016/j.radonc.2023.109692
123. Devic S. MRI simulation for radiotherapy treatment planning. Med Phys. (2012) 39(11):6701–11. doi: 10.1118/1.4758068
124. Jonsson JH, Karlsson MG, Karlsson M, Nyholm T. Treatment planning using MRI data: an analysis of the dose calculation accuracy for different treatment regions. Radiat Oncol. (2010) 5(1):1–8. doi: 10.1186/1748-717X-5-62
125. Lambert J, Greer PB, Menk F, Patterson J, Parker J, Dahl K, et al. MRI-guided prostate radiation therapy planning: investigation of dosimetric accuracy of MRI-based dose planning. Radiother Oncol. (2011) 98(3):330–4. doi: 10.1016/j.radonc.2011.01.012
126. Kristensen BH, Laursen FJ, Løgager V, Geertsen PF, Krarup-Hansen A. Dosimetric and geometric evaluation of an open low-field magnetic resonance simulator for radiotherapy treatment planning of brain tumours. Radiother Oncol. (2008) 87(1):100–9. doi: 10.1016/j.radonc.2008.01.014
127. Johansson A, Karlsson M, Nyholm T. CT substitute derived from MRI sequences with ultrashort echo time. Med Phys. (2011) 38(5):2708–14. doi: 10.1118/1.3578928
128. Hsu S-H, Cao Y, Huang K, Feng M, Balter JM. Investigation of a method for generating synthetic CT models from MRI scans of the head and neck for radiation therapy. Phys Med Biol. (2013) 58(23):8419. doi: 10.1088/0031-9155/58/23/8419
129. Dowling JA, Lambert J, Parker J, Salvado O, Fripp J, Capp A, et al. An atlas-based electron density mapping method for magnetic resonance imaging (MRI)-alone treatment planning and adaptive MRI-based prostate radiation therapy. Int J Radiat Oncol* Biol* Phys. (2012) 83(1):5–11. doi: 10.1016/j.ijrobp.2011.11.056
130. Uh J, Merchant TE, Li Y, Li X, Hua C. MRI-based treatment planning with pseudo CT generated through atlas registration. Med Phys. (2014) 41(5):051711. doi: 10.1118/1.4873315
131. Sjölund J, Forsberg D, Andersson M, Knutsson H. Generating patient specific pseudo-CT of the head from MR using atlas-based regression. Phys Med Biol. (2015) 60(2):825. doi: 10.1088/0031-9155/60/2/825
132. Barney BM, Lee RJ, Handrahan D, Welsh KT, Cook JT, Sause WT. Image-guided radiotherapy (IGRT) for prostate cancer comparing kV imaging of fiducial markers with cone beam computed tomography (CBCT). Int J Radiat Oncol* Biol* Phys. (2011) 80(1):301–5. doi: 10.1016/j.ijrobp.2010.06.007
133. Zhu L, Xie Y, Wang J, Xing L. Scatter correction for cone-beam CT in radiation therapy. Med Phys. (2009) 36(6Part1):2258–68. doi: 10.1118/1.3130047
134. de la Zerda A, Armbruster B, Xing L. Formulating adaptive radiation therapy (ART) treatment planning into a closed-loop control framework. Phys Med Biol. (2007) 52(14):4137. doi: 10.1088/0031-9155/52/14/008
135. Xie S, Yang C, Zhang Z, Li H. Scatter artifacts removal using learning-based method for CBCT in IGRT system. IEEE Access. (2018) 6:78031–7. doi: 10.1109/ACCESS.2018.2884704
136. Hansen DC, Landry G, Kamp F, Li M, Belka C, Parodi K, et al. Scatternet: a convolutional neural network for cone-beam CT intensity correction. Med Phys. (2018) 45(11):4916–26. doi: 10.1002/mp.13175
137. Kida S, Nakamoto T, Nakano M, Nawa K, Haga A, Kotoku J, et al. Cone beam computed tomography image quality improvement using a deep convolutional neural network. Cureus. (2018) 10(4):e2548. doi: 10.7759/2Fcureus.2548
138. Harms J, Lei Y, Wang T, Zhang R, Zhou J, Tang X, et al. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys. (2019) 46(9):3998–4009. doi: 10.1002/mp.13656
139. Chen L, Liang X, Shen C, Jiang S, Wang J. Synthetic CT generation from CBCT images via deep learning. Med Phys. (2020) 47(3):1115–25. doi: 10.1002/mp.13978
140. Li Y, Zhu J, Liu Z, Teng J, Xie Q, Zhang L, et al. A preliminary study of using a deep convolution neural network to generate synthesized CT images based on CBCT for adaptive radiotherapy of nasopharyngeal carcinoma. Phys Med Biol. (2019) 64(14):145010. doi: 10.1088/1361-6560/ab2770
141. Liang X, Chen L, Nguyen D, Zhou Z, Gu X, Yang M, et al. Generating synthesized computed tomography (CT) from cone-beam computed tomography (CBCT) using CycleGAN for adaptive radiation therapy. Phys Med Biol. (2019) 64(12):125002. doi: 10.1088/1361-6560/ab22f9
142. Kurz C, Maspero M, Savenije MH, Landry G, Kamp F, Pinto M, et al. CBCT correction using a cycle-consistent generative adversarial network and unpaired training to enable photon and proton dose calculation. Phys Med Biol. (2019) 64(22):225004. doi: 10.1088/1361-6560/ab4d8c
143. Landry G, Hansen D, Kamp F, Li M, Hoyle B, Weller J, et al. Comparing Unet training with three different datasets to correct CBCT images for prostate radiotherapy dose calculations. Phys Med Biol. (2019) 64(3):035011. doi: 10.1088/1361-6560/aaf496
144. Maspero M, Houweling AC, Savenije MH, van Heijst TC, Verhoeff JJ, Kotte AN, et al. A single neural network for cone-beam computed tomography-based radiotherapy of head-and-neck, lung and breast cancer. Phys Imaging Radiat Oncol. (2020) 14:24–31. doi: 10.1016/j.phro.2020.04.002
145. Barateau A, De Crevoisier R, Largent A, Mylona E, Perichon N, Castelli J, et al. Comparison of CBCT-based dose calculation methods in head and neck cancer radiotherapy: from hounsfield unit to density calibration curve to deep learning. Med Phys. (2020) 47(10):4683–93. doi: 10.1002/mp.14387
146. Eckl M, Hoppen L, Sarria GR, Boda-Heggemann J, Simeonova-Chergou A, Steil V, et al. Evaluation of a cycle-generative adversarial network-based cone-beam CT to synthetic CT conversion algorithm for adaptive radiation therapy. Phys Med. (2020) 80:308–16. doi: 10.1016/j.ejmp.2020.11.007
147. Yuan N, Dyer B, Rao S, Chen Q, Benedict S, Shang L, et al. Convolutional neural network enhancement of fast-scan low-dose cone-beam CT images for head and neck radiotherapy. Phys Med Biol. (2020) 65(3):035003. doi: 10.1088/1361-6560/ab6240
148. Zhang Y, Yue N, Su M-Y, Liu B, Ding Y, Zhou Y, et al. Improving CBCT quality to CT level using deep learning with generative adversarial network. Med Phys. (2021) 48(6):2816–26. doi: 10.1002/mp.14624
149. Liu Y, Lei Y, Wang T, Fu Y, Tang X, Curran WJ, et al. CBCT-based synthetic CT generation using deep-attention CycleGAN for pancreatic adaptive radiotherapy. Med Phys. (2020) 47(6):2472–83. doi: 10.1002/mp.14121
150. Kida S, Kaji S, Nawa K, Imae T, Nakamoto T, Ozaki S, et al. Visual enhancement of cone-beam CT by use of CycleGAN. Med Phys. (2020) 47(3):998–1010. doi: 10.1002/mp.13963
151. Dai Z, Zhang Y, Zhu L, Tan J, Yang G, Zhang B, et al. Geometric and dosimetric evaluation of deep learning-based automatic delineation on CBCT-synthesized CT and planning CT for breast cancer adaptive radiotherapy: A multi-institutional study. Front Oncol. (2021) 11:725507. doi: 10.3389/fonc.2021.725507
152. Gao L, Xie K, Wu X, Lu Z, Li C, Sun J, et al. Generating synthetic CT from low-dose cone-beam CT by using generative adversarial networks for adaptive radiotherapy. Radiat Oncol. (2021) 16(1):1–16. doi: 10.1186/s13014-021-01928-w
153. Xue X, Ding Y, Shi J, Hao X, Li X, Li D, et al. Cone beam CT (CBCT) based synthetic CT generation using deep learning methods for dose calculation of nasopharyngeal carcinoma radiotherapy. Technol Cancer Res Treat. (2021) 20:15330338211062415. doi: 10.1177/15330338211062415
154. Chen L, Liang X, Shen C, Nguyen D, Jiang S, Wang J. Synthetic CT generation from CBCT images via unsupervised deep learning. Phys Med Biol. (2021) 66(11):115019. doi: 10.1088/1361-6560/ac01b6
155. Dahiya N, Alam SR, Zhang P, Zhang S-Y, Li T, Yezzi A, et al. Multitask 3D CBCT-to-CT translation and organs-at-risk segmentation using physics-based data augmentation. Med Phys. (2021) 48(9):5130–41. doi: 10.1002/mp.15083
156. Liu J, Yan H, Cheng H, Liu J, Sun P, Wang B, et al. CBCT-based synthetic CT generation using generative adversarial networks with disentangled representation. Quant Imaging Med Surg. (2021) 11(12):4820. doi: 10.21037/qims-20-1056
157. Thummerer A, Seller Oria C, Zaffino P, Meijers A, Guterres Marmitt G, Wijsman R, et al. Clinical suitability of deep learning based synthetic CTs for adaptive proton therapy of lung cancer. Med Phys. (2021) 48(12):7673–84. doi: 10.1002/mp.15333
158. Wu W, Qu J, Cai J, Yang R. Multi-resolution residual deep neural network for improving pelvic CBCT image quality. Med Phys. (2022) 49(3):1522–34. doi: 10.1002/mp.15460
159. Zhao J, Chen Z, Wang J, Xia F, Peng J, Hu Y, et al. MV CBCT-based synthetic CT generation using a deep learning method for rectal cancer adaptive radiotherapy. Front Oncol. (2021) 11:1733. doi: 10.3389/fonc.2021.655325
160. Qiu RL, Lei Y, Shelton J, Higgins K, Bradley JD, Curran WJ, et al. Deep learning-based thoracic CBCT correction with histogram matching. Biomed Phys Eng Express. (2021) 7(6):065040. doi: 10.1088/2057-1976/ac3055
161. Yuan N, Rao S, Chen Q, Sensoy L, Qi J, Rong Y. Head and neck synthetic CT generated from ultra-low-dose cone-beam CT following image gently protocol using deep neural network. Med Phys. (2022) 49(5):3263–77. doi: 10.1002/mp.15585
162. Zhou H, Cao M, Min Y, Yoon S, Kishan A, Ruan D. Ensemble learning and tensor regularization for cone-beam computed tomography-based pelvic organ segmentation. Med Phys. (2022) 49(3):1660–72. doi: 10.1002/mp.15475
163. Jiang Z, Zhang Z, Chang Y, Ge Y, Yin F-F, Ren L. Enhancement of 4-D cone-beam computed tomography (4D-CBCT) using a dual-encoder convolutional neural network (DeCNN). IEEE Trans Radiat Plasma Med Sci. (2021) 6(2):222–30. doi: 10.1109/TRPMS.2021.3133510
164. Yoo SK, Kim H, Choi BS, Park I, Kim JS. Generation and evaluation of synthetic computed tomography (CT) from cone-beam CT (CBCT) by incorporating feature-driven loss into intensity-based loss functions in deep convolutional neural network. Cancers. (2022) 14(18):4534. doi: 10.3390/cancers14184534
165. Deng L, Hu J, Wang J, Huang S, Yang X. Synthetic CT generation based on CBCT using respath-CycleGAN. Med Phys. (2022) 49(8):5317–29. doi: 10.1002/mp.15684
166. O’Hara CJ, Bird D, Al-Qaisieh B, Speight R. Assessment of CBCT–based synthetic CT generation accuracy for adaptive radiotherapy planning. J Appl Clin Med Phys. (2022) 23(11):e13737.
167. Xie K, Gao L, Xi Q, Zhang H, Zhang S, Zhang F, et al. New technique and application of truncated CBCT processing in adaptive radiotherapy for breast cancer. Comput Methods Programs Biomed. (2023) 231:107393. doi: 10.1016/j.cmpb.2023.107393
168. Deng L, Ji Y, Huang S, Yang X, Wang J. Synthetic CT generation from CBCT using double-chain-CycleGAN. Comput Biol Med. (2023) 161:106889. doi: 10.1016/j.compbiomed.2023.106889
169. de Hond YJ, Kerckhaert CE, van Eijnatten MA, van Haaren PM, Hurkmans CW, Tijssen RH. Anatomical evaluation of deep-learning synthetic computed tomography images generated from male pelvis cone-beam computed tomography. Phys Imaging Radiat Oncol. (2023) 25:100416. doi: 10.1016/j.phro.2023.100416
170. Hou J, Guerrero M, Chen W, D’Souza WD. Deformable planning CT to cone-beam CT image registration in head-and-neck cancer. Med Phys. (2011) 38(4):2088–94. doi: 10.1118/1.3554647
171. Nomura Y, Xu Q, Shirato H, Shimizu S, Xing L. Projection-domain scatter correction for cone beam computed tomography using a residual convolutional neural network. Med Phys. (2019) 46(7):3142–55. doi: 10.1002/mp.13583
172. Radford A, Metz L, Chintala S. Unsupervised representation learning with deep convolutional generative adversarial networks. arXiv [Preprint]. arXiv:1511.06434 (2015).
173. Shan H, Padole A, Homayounieh F, Kruger U, Khera RD, Nitiwarangkul C, et al. Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction. Nat Mach Intell. (2019) 1(6):269–76. doi: 10.1038/s42256-019-0057-9
174. Liu R, Sun B, Zhang T, Williamson JF, O’Sullivan JA, Zhao T. Dosimetric impact of range uncertainty in passive scattering proton therapy. J Appl Clin Med Phys. (2021) 22(5):6–14. doi: 10.1002/acm2.13179
175. Hofmann M, Steinke F, Scheel V, Charpiat G, Farquhar J, Aschoff P, et al. MRI-based attenuation correction for PET/MRI: a novel approach combining pattern recognition and atlas registration. J Nucl Med. (2008) 49(11):1875–83. doi: 10.2967/jnumed.107.049353
176. Leynes AP, Yang J, Wiesinger F, Kaushik SS, Shanbhag DD, Seo Y, et al. Direct pseudoCT generation for pelvis PET/MRI attenuation correction using deep convolutional neural networks with multi-parametric MRI: zero echo-time and Dixon deep pseudoCT (ZeDD-CT). J Nucl Med. (2017) 59(5):852–8. doi: 10.2967/jnumed.117.198051
177. Blanc-Durand P, Khalifé M, Sgard B, Kaushik S, Soret M, Tiss A, et al. Attenuation correction using 3D deep convolutional neural network for brain 18F-FDG PET/MR: Comparison with atlas, ZTE and CT based attenuation correction. PLoS one. (2019) 14(10):e0223141. doi: 10.1371/journal.pone.0223141
178. Yang X, Fei B. Multiscale segmentation of the skull in MR images for MRI-based attenuation correction of combined MR/PET. J Am Med Inform Assoc. (2013) 20(6):1037–45. doi: 10.1136/amiajnl-2012-001544
179. Torrado-Carvajal A, Vera-Olmos J, Izquierdo-Garcia D, Catalano OA, Morales MA, Margolin J, et al. Dixon-VIBE deep learning (DIVIDE) pseudo-CT synthesis for pelvis PET/MR attenuation correction. J Nucl Med. (2019) 60(3):429–35. doi: 10.2967/jnumed.118.209288
180. Gong K, Yang J, Kim K, El Fakhri G, Seo Y, Li Q. Attenuation correction for brain PET imaging using deep neural network based on Dixon and ZTE MR images. Phys Med Biol. (2018) 63(12):125011. doi: 10.1088/1361-6560/aac763
181. Arabi H, Zeng G, Zheng G, Zaidi H. Novel adversarial semantic structure deep learning for MRI-guided attenuation correction in brain PET/MRI. Eur J Nucl Med Mol Imaging. (2019) 46(13):2746–59. doi: 10.1007/s00259-019-04380-x
182. Liu F, Jang H, Kijowski R, Zhao G, Bradshaw T, McMillan AB. A deep learning approach for 18 F-FDG PET attenuation correction. EJNMMI Phys. (2018) 5(1):1–15. doi: 10.1186/s40658-018-0225-8
183. Jang H, Liu F, Zhao G, Bradshaw T, McMillan AB. Deep learning based MRAC using rapid ultrashort echo time imaging. Med Phys. (2018) 45(8):3697–704. doi: 10.1002/mp.12964
184. Liu F, Jang H, Kijowski R, Bradshaw T, McMillan AB. Deep learning MR imaging–based attenuation correction for PET/MR imaging. Radiology. (2018) 286(2):676–84. doi: 10.1148/radiol.2017170700
185. Bradshaw TJ, Zhao G, Jang H, Liu F, McMillan AB. Feasibility of deep learning–based PET/MR attenuation correction in the pelvis using only diagnostic MR images. Tomography. (2018) 4(3):138–47. doi: 10.18383/j.tom.2018.00016
186. Dong X, Wang T, Lei Y, Higgins K, Liu T, Curran WJ, et al. Synthetic CT generation from non-attenuation corrected PET images for whole-body PET imaging. Phys Med Biol. (2019) 64(21):215016. doi: 10.1088/1361-6560/ab4eb7
187. Ladefoged CN, Marner L, Hindsholm A, Law I, Højgaard L, Andersen FL. Deep learning based attenuation correction of PET/MRI in pediatric brain tumor patients: evaluation in a clinical setting. Front Neurosci. (2019) 12:1005. doi: 10.3389/fnins.2018.01005
188. Spuhler KD, Gardus J, Gao Y, DeLorenzo C, Parsey R, Huang C. Synthesis of patient-specific transmission data for PET attenuation correction for PET/MRI neuroimaging using a convolutional neural network. J Nucl Med. (2019) 60(4):555–60. doi: 10.2967/jnumed.118.214320
189. Armanious K, Hepp T, Küstner T, Dittmann H, Nikolaou K, La Fougère C, et al. Independent attenuation correction of whole body [18 F] FDG-PET using a deep learning approach with generative adversarial networks. EJNMMI Res. (2020) 10(1):1–9. doi: 10.1186/s13550-020-00644-y
190. Gong K, Yang J, Larson PE, Behr SC, Hope TA, Seo Y, et al. MR-based attenuation correction for brain PET using 3-D cycle-consistent adversarial network. IEEE Trans Radiat Plasma Med Sci. (2020) 5(2):185–92. doi: 10.1109/TRPMS.2020.3006844
191. Baydoun A, Xu K, Yang H, Zhou F, Heo JU, Jones RS, et al. Dixon-based thorax synthetic CT generation using generative adversarial network. Intell Based Med. (2020) 3:100010. doi: 10.1016/j.ibmed.2020.100010
192. Chen Y, Ying C, Binkley MM, Juttukonda MR, Flores S, Laforest R, et al. Deep learning-based T1-enhanced selection of linear attenuation coefficients (DL-TESLA) for PET/MR attenuation correction in dementia neuroimaging. Magn Reson Med. (2021) 86(1):499–513. doi: 10.1002/mrm.28689
193. Kläser K, Varsavsky T, Markiewicz P, Vercauteren T, Hammers A, Atkinson D, et al. Imitation learning for improved 3D PET/MR attenuation correction. Med Image Anal. (2021) 71:102079. doi: 10.1016/j.media.2021.102079
194. Gong K, Han PK, Johnson KA, El Fakhri G, Ma C, Li Q. Attenuation correction using deep learning and integrated UTE/multi-echo Dixon sequence: evaluation in amyloid and tau PET imaging. Eur J Nucl Med Mol Imaging. (2021) 48(5):1351–61. doi: 10.1007/s00259-020-05061-w
195. Jiang C, Zhang X, Zhang N, Zhang Q, Zhou C, Yuan J, et al. Synthesizing PET/MR (T1-weighted) images from non-attenuation-corrected PET images. Phys Med Biol. (2021) 66(13):135006. doi: 10.1088/1361-6560/ac08b2
196. Pozaruk A, Pawar K, Li S, Carey A, Cheng J, Sudarshan VP, et al. Augmented deep learning model for improved quantitative accuracy of MR-based PET attenuation correction in PSMA PET-MRI prostate imaging. Eur J Nucl Med Mol Imaging. (2021) 48(1):9–20. doi: 10.1007/s00259-020-04816-9
197. Ahangari S, Beck Olin A, Kinggård Federspiel M, Jakoby B, Andersen TL, Hansen AE, et al. A deep learning-based whole-body solution for PET/MRI attenuation correction. EJNMMI Phys. (2022) 9(1):1–15. doi: 10.1186/s40658-022-00486-8
198. Li Q, Zhu X, Zou S, Zhang N, Liu X, Yang Y, et al. Eliminating CT radiation for clinical PET examination using deep learning. Eur J Radiol. (2022) 154:110422. doi: 10.1016/j.ejrad.2022.110422
199. Olin AB, Hansen AE, Rasmussen JH, Jakoby B, Berthelsen AK, Ladefoged CN, et al. Deep learning for Dixon MRI-based attenuation correction in PET/MRI of head and neck cancer patients. EJNMMI Phys. (2022) 9(1):20. doi: 10.1186/s40658-022-00449-z
200. Shi L, Zhang J, Toyonaga T, Shao D, Onofrey JA, Lu Y. Deep learning-based attenuation map generation with simultaneously reconstructed PET activity and attenuation and low-dose application. Phys Med Biol. (2022) 68:035014. doi: 10.1088/1361-6560/acaf49
201. Arabi H, Zaidi H. MRI-guided attenuation correction in torso PET/MRI: Assessment of segmentation-, atlas-, and deep learning-based approaches in the presence of outliers. Magn Reson Med. (2022) 87(2):686–701. doi: 10.1002/mrm.29003
202. Ladefoged CN, Andersen FL, Andersen TL, Anderberg L, Engkebølle C, Madsen K, et al. Deepdixon synthetic CT for [18F] FET PET/MRI attenuation correction of post-surgery glioma patients with metal implants. Front Neurosci. (2023) 17:1142383. doi: 10.3389/fnins.2023.1142383
203. Rajagopal A, Natsuaki Y, Wangerin K, Hamdi M, An H, Sunderland JJ, et al. Synthetic PET via domain translation of 3-D MRI. IEEE Trans Radiat Plasma Med Sci. (2022) 7(4):333–43. doi: 10.1109/TRPMS.2022.3223275
204. Dong J, Fu J, He Z. A deep learning reconstruction framework for x-ray computed tomography with incomplete data. PLoS one. (2019) 14(11):e0224426. doi: 10.1371/journal.pone.0224426
205. Kang E, Chang W, Yoo J, Ye JC. Deep convolutional framelet denosing for low-dose CT via wavelet residual network. IEEE Trans Med Imaging. (2018) 37(6):1358–69. doi: 10.1109/TMI.2018.2823756
206. Yi X, Babyn P. Sharpness-aware low-dose CT denoising using conditional generative adversarial network. J Digit Imaging. (2018) 31(5):655–69. doi: 10.1007/s10278-018-0056-0
207. Shan H, Zhang Y, Yang Q, Kruger U, Kalra MK, Sun L, et al. 3-D convolutional encoder-decoder network for low-dose CT via transfer learning from a 2-D trained network. IEEE Trans Med Imaging. (2018) 37(6):1522–34. doi: 10.1109/TMI.2018.2832217
208. You C, Yang Q, Shan H, Gjesteby L, Li G, Ju S, et al. Structurally-sensitive multi-scale deep neural network for low-dose CT denoising. IEEE Access. (2018) 6:41839–55. doi: 10.1109/ACCESS.2018.2858196
209. Han Y, Ye JC. Framing U-net via deep convolutional framelets: application to sparse-view CT. IEEE Trans Med Imaging. (2018) 37(6):1418–29. doi: 10.1109/TMI.2018.2823768
210. Yang Q, Yan P, Zhang Y, Yu H, Shi Y, Mou X, et al. Low-dose CT image denoising using a generative adversarial network with wasserstein distance and perceptual loss. IEEE Trans Med Imaging. (2018) 37(6):1348–57. doi: 10.1109/TMI.2018.2827462
211. Liu Y, Zhang Y. Low-dose CT restoration via stacked sparse denoising autoencoders. Neurocomputing. (2018) 284:80–9. doi: 10.1016/j.neucom.2018.01.015
212. Zhao T, McNitt-Gray M, Ruan D. A convolutional neural network for ultra-low-dose CT denoising and emphysema screening. Med Phys. (2019) 46(9):3941–50. doi: 10.1002/mp.13666
213. Lee H, Lee J, Kim H, Cho B, Cho S. Deep-neural-network-based sinogram synthesis for sparse-view CT image reconstruction. IEEE Trans Radiat Plasma Med Sci. (2018) 3(2):109–19. doi: 10.1109/TRPMS.2018.2867611
214. Wang T, Lei Y, Tian Z, Dong X, Liu Y, Jiang X, et al. Deep learning-based image quality improvement for low-dose computed tomography simulation in radiation therapy. J Med Imaging. (2019) 6(4):043504. doi: 10.1117/1.jmi.6.4.043504
215. Li M, Hsu W, Xie X, Cong J, Gao W. SACNN: self-attention convolutional neural network for low-dose CT denoising with self-supervised perceptual loss network. IEEE Trans Med Imaging. (2020) 39(7):2289–301. doi: 10.1109/TMI.2020.2968472
216. Chi J, Wu C, Yu X, Ji P, Chu H. Single low-dose CT image denoising using a generative adversarial network with modified U-net generator and multi-level discriminator. IEEE Access. (2020) 8:133470–87. doi: 10.1109/ACCESS.2020.3006512
217. Ma Y, Wei B, Feng P, He P, Guo X, Wang G. Low-dose CT image denoising using a generative adversarial network with a hybrid loss function for noise learning. IEEE Access. (2020) 8:67519–29. doi: 10.1109/ACCESS.2020.2986388
218. Yin Z, Xia K, He Z, Zhang J, Wang S, Zu B. Unpaired image denoising via wasserstein gan in low-dose CT image with multi-perceptual loss and fidelity loss. Symmetry. (2021) 13(1):126. doi: 10.3390/sym13010126
219. Gu J, Ye JC. AdaIN-based tunable CycleGAN for efficient unsupervised low-dose CT denoising. IEEE Trans Comput Imaging. (2021) 7:73–85. doi: 10.1109/TCI.2021.3050266
220. Jiang B, Li N, Shi X, Zhang S, Li J, de Bock GH, et al. Deep learning reconstruction shows better lung nodule detection for ultra–low-dose chest ct. Radiology. (2022) 303(1):202–12. doi: 10.1148/radiol.210551
221. Zhu M, Mao Z, Li D, Wang Y, Zeng D, Bian Z, et al. Structure-preserved meta-learning uniting network for improving low-dose CT quality. Phys Med Biol. (2022) 67(24):245015. doi: 10.1088/1361-6560/aca194
222. Zhou S, Yu L, Jin M. Texture transformer super-resolution for low-dose computed tomography. Biomed Phys Eng Express. (2022) 8(6):065024. doi: 10.1088/2057-1976/ac9da7
223. Yang M, Wang J, Zhang Z, Li J, Liu L. Transfer learning framework for low-dose CT reconstruction based on marginal distribution adaptation in multiscale. Med Phys. (2023) 50(3):1450–65. doi: 10.1002/mp.16027
224. Gao X, Su T, Zhang Y, Zhu J, Tan Y, Cui H, et al. Attention-based dual-branch deep network for sparse-view computed tomography image reconstruction. Quant Imaging Med Surg. (2023) 13(3):1360. doi: 10.21037/qims-22-609
225. Li Q, Li R, Li S, Wang T, Cheng Y, Zhang S, et al. Unpaired low-dose computed tomography image denoising using a progressive cyclical convolutional neural network. Med Phys. (2024) 51(2):1289–312. doi: 10.1002/mp.16331
226. Kang E, Min J, Ye JC. A deep convolutional neural network using directional wavelets for low-dose x-ray CT reconstruction. Med Phys. (2017) 44(10):e360–75. doi: 10.1002/mp.12344
227. McCollough CH, Bartley AC, Carter RE, Chen B, Drees TA, Edwards P, et al. Low-dose CT for the detection and classification of metastatic liver lesions: results of the 2016 low dose CT grand challenge. Med Phys. (2017) 44(10):e339–52. doi: 10.1002/mp.12345
228. Johnstone E, Wyatt JJ, Henry AM, Short SC, Sebag-Montefiore D, Murray L, et al. Systematic review of synthetic computed tomography generation methodologies for use in magnetic resonance imaging–only radiation therapy. Int J Radiat Oncol* Biol* Phys. (2018) 100(1):199–217. doi: 10.1016/j.ijrobp.2017.08.043
229. Gholamiankhah F, Mostafapour S, Arabi H. Deep learning-based synthetic CT generation from MR images: comparison of generative adversarial and residual neural networks. arXiv [Preprint]. arXiv:2103.01609 (2021).
230. Schilling KG, Blaber J, Huo Y, Newton A, Hansen C, Nath V, et al. Synthesized b0 for diffusion distortion correction (Synb0-Disco). Magn Reson Imaging. (2019) 64:62–70. doi: 10.1016/j.mri.2019.05.008
231. Reinhold JC, Dewey BE, Carass A, Prince JL. Evaluating the impact of intensity normalization on MR image synthesis. Med Imaging 2019: Image Process. (2019) 10949:890–8. doi: 10.1117/2F12.2513089
232. Walker A, Metcalfe P, Liney G, Batumalai V, Dundas K, Glide-Hurst C, et al. MRI geometric distortion: Impact on tangential whole-breast IMRT. J Appl Clin Med Phys. (2016) 17(5):7–19. doi: 10.1120/jacmp.v17i5.6242
233. Gustafsson C, Nordström F, Persson E, Brynolfsson J, Olsson L. Assessment of dosimetric impact of system specific geometric distortion in an MRI only based radiotherapy workflow for prostate. Phys Med Biol. (2017) 62(8):2976. doi: 10.1088/1361-6560/aa5fa2
234. Maspero M, Tyyger MD, Tijssen RH, Seevinck PR, Intven MP, van den Berg CA. Feasibility of magnetic resonance imaging-only rectum radiotherapy with a commercial synthetic computed tomography generation solution. Phys Imaging Radiat Oncol. (2018) 7:58–64. doi: 10.1016/j.phro.2018.09.002
235. Lagendijk JJ, Raaymakers BW, Raaijmakers AJ, Overweg J, Brown KJ, Kerkhof EM, et al. MRI/linac integration. Radiother Oncol. (2008) 86(1):25–9. doi: 10.1016/j.radonc.2007.10.034
236. Fallone BG. The rotating biplanar linac–magnetic resonance imaging system. Semin Radiat Oncol. (2014) 24(3):200–2. doi: 10.1016/j.semradonc.2014.02.011
237. Mutic S, Dempsey JF. The viewray system: magnetic resonance–guided and controlled radiotherapy. Semin Radiat Oncol. (2014) 24(3):196–9. doi: 10.1016/j.semradonc.2014.02.008
238. Keall PJ, Barton M, Crozier S, et al. The australian magnetic resonance imaging–linac program. Semin Radiat Oncol. (2014) 24(3):203–6. doi: 10.1016/j.semradonc.2014.02.015
239. Jaffray DA, Carlone MC, Milosevic MF, Breen SL, Stanescu T, Rink A, et al. A facility for magnetic resonance–guided radiation therapy. Semin Radiat Oncol. (2014) 24(3):193–5. doi: 10.1016/j.semradonc.2014.02.012
240. Koerkamp MG, de Hond Y, Maspero M, Kontaxis C, Mandija S, Vasmel J, et al. Synthetic CT for single-fraction neoadjuvant partial breast irradiation on an MRI-linac. Phys Med Biol. (2021) 66(8):085010. doi: 10.1088/1361-6560/abf1ba
241. Arai K, Kadoya N, Kato T, Endo H, Komori S, Abe Y, et al. Feasibility of CBCT-based proton dose calculation using a histogram-matching algorithm in proton beam therapy. Phys Med. (2017) 33:68–76. doi: 10.1016/j.ejmp.2016.12.006
242. Peroni M, Ciardo D, Spadea MF, Riboldi M, Comi S, Alterio D, et al. Automatic segmentation and online virtualCT in head-and-neck adaptive radiation therapy. Int J Radiat Oncol* Biol* Phys. (2012) 84(3):e427–33. doi: 10.1016/j.ijrobp.2012.04.003
243. Park Y-K, Sharp GC, Phillips J, Winey BA. Proton dose calculation on scatter-corrected CBCT image: feasibility study for adaptive proton therapy. Med Phys. (2015) 42(8):4449–59. doi: 10.1118/1.4923179
244. Kurz C, Nijhuis R, Reiner M, Ganswindt U, Thieke C, Belka C, et al. Feasibility of automated proton therapy plan adaptation for head and neck tumors using cone beam CT images. Radiat Oncol. (2016) 11(1):1–9. doi: 10.1186/s13014-016-0641-7
245. Shiri I, Arabi H, Geramifar P, Hajianfar G, Ghafarian P, Rahmim A, et al. Deep-JASC: joint attenuation and scatter correction in whole-body 18F-FDG PET using a deep residual network. Eur J Nucl Med Mol Imaging. (2020) 47(11):2533–48. doi: 10.1007/s00259-020-04852-5
246. De Vos BD, Berendsen FF, Viergever MA, Sokooti H, Staring M, Išgum I. A deep learning framework for unsupervised affine and deformable image registration. Med Image Anal. (2019) 52:128–43. doi: 10.1016/j.media.2018.11.010
247. Yang X, Kwitt R, Niethammer M. Fast predictive image registration. In: Deep Learning and Data Labeling for Medical Applications: First International Workshop, LABELS 2016, and Second International Workshop, DLMIA 2016, Held in Conjunction with MICCAI 2016, Athens, Greece, October 21, 2016, Proceedings 1. Springer (2016). p. 48–57.
248. Miao S, Wang ZJ, Liao R. A CNN regression approach for real-time 2D/3D registration. IEEE Trans Med Imaging. (2016) 35(5):1352–63. doi: 10.1109/TMI.2016.2521800
249. Chee E, Wu Z. Airnet: Self-supervised affine registration for 3D medical images using neural networks. arXiv [Preprint]. arXiv:1810.02583 (2018).
250. Sloan JM, Goatman KA, Siebert JP. Learning rigid image registration-utilizing convolutional neural networks for medical image registration (2018)
251. Liu X, Jiang D, Wang M, Song Z. Image synthesis-based multi-modal image registration framework by using deep fully convolutional networks. Med Biol Eng Comput. (2019) 57:1037–48. doi: 10.1007/s11517-018-1924-y
252. Zou M, Hu J, Zhang H, Wu X, He J, Xu Z, et al. Rigid medical image registration using learning-based interest points and features. Comput Mater Continua. (2019) 60(2):511–25. doi: 10.32604/cmc.2019.05912
253. McKenzie EM, Santhanam A, Ruan D, O’Connor D, Cao M, Sheng K. Multimodality image registration in the head-and-neck using a deep learning-derived synthetic CT as a bridge. Med Phys. (2020) 47(3):1094–104. doi: 10.1002/mp.13976
254. Frid-Adar M, Klang E, Amitai M, Goldberger J, Greenspan H. Synthetic data augmentation using GAN for improved liver lesion classification. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE (2018). p. 289–93.
255. Mahapatra D, Ge Z. Training data independent image registration using generative adversarial networks and domain adaptation. Pattern Recognit. (2020) 100:107109. doi: 10.1016/j.patcog.2019.107109
256. Park S, Lee S-b., Park J. Data augmentation method for improving the accuracy of human pose estimation with cropped images. Pattern Recognit Lett. (2020) 136:244–50. doi: 10.1016/j.patrec.2020.06.015
257. Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. (2019) 6(1):1–48. doi: 10.1186/s40537-019-0197-0
258. Kim EK, Lee H, Kim JY, Kim S. Data augmentation method by applying color perturbation of inverse PSNR and geometric transformations for object recognition based on deep learning. Appl Sci. (2020) 10(11):3755. doi: 10.3390/app10113755
259. Lin W-H, Zhong J-X, Liu S, Li T, Li G. Roimix: proposal-fusion among multiple images for underwater object detection. In: ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE (2020). p. 2588–92.
260. Anicet Zanini R, Luna Colombini E. Parkinson’s disease EMG data augmentation and simulation with DCGANs and style transfer. Sensors. (2020) 20(9):2605. doi: 10.3390/s20092605
261. Cubuk ED, Zoph B, Mane D, Vasudevan V, Le QV. Autoaugment: learning augmentation policies from data. arXiv [Preprint]. arXiv:1805.09501 (2018).
262. Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A, et al. GAN augmentation: augmenting training data using generative adversarial networks. arXiv [Preprint]. arXiv:1810.10863 (2018).
263. Zhang D, Shi N, Peng C, Aziz A, Zhao W, Xia F. MAM: a metaphor-based approach for mental illness detection. In: International Conference on Computational Science (2021). p. 570–83.
264. Kraus KM, Jäkel O, Niebuhr NI, Pfaffenberger A. Generation of synthetic CT data using patient specific daily MR image data and image registration. Phys Med Biol. (2017) 62(4):1358. doi: 10.1088/1361-6560/aa5200
265. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. (2012) 62(2):782–90. doi: 10.1016/j.neuroimage.2011.09.015. 20 YEARS OF fMRI.21979382
266. Zhang S, Wei Z, Nie J, Huang L, Wang S, Li Z. A review on human activity recognition using vision-based method. J Healthc Eng. (2017) 2017:3090343. doi: 10.1155/2017/3090343
267. Gargiulo P, Árnadóttir Í, Gíslason M, Edmunds K, Ólafsson I. New directions in 3D medical modeling: 3D-printing anatomy and functions in neurosurgical planning. J Healthc Eng. (2017) 2017:1439643. doi: 10.1155/2017/1439643
269. Hung ALY, Zhao K, Zheng H, Yan R, Raman SS, Terzopoulos D, et al. Med-cDiff: conditional medical image generation with diffusion models. Bioengineering. (2023) 10(11):1258. doi: 10.3390/bioengineering10111258
Keywords: deep learning, convolutional neural network, radiotherapy, synthetic CT, photon therapy, proton therapy, generative adversarial network
Citation: Sherwani MK and Gopalakrishnan S (2024) A systematic literature review: deep learning techniques for synthetic medical image generation and their applications in radiotherapy. Front. Radiol. 4:1385742. doi: 10.3389/fradi.2024.1385742
Received: 13 February 2024; Accepted: 11 March 2024;
Published: 27 March 2024.
Edited by:
Jax Luo, Harvard Medical School, United StatesReviewed by:
Zhennong Chen, Harvard Medical School, United StatesJian Wang, Harvard Medical School, United States
© 2024 Sherwani and Gopalakrishnan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moiz Khan Sherwani bW9pei5zaGVyd2FuaUBzdW5kLmt1LmRr
 Moiz Khan Sherwani
Moiz Khan Sherwani Shyam Gopalakrishnan
Shyam Gopalakrishnan