- 1The Russell H. Morgan Department of Radiology & Radiological Science, The Johns Hopkins Medical Institutions, Baltimore, MD, United States
- 2Department of Orthopaedic Surgery, Johns Hopkins University School of Medicine, Baltimore, MD, United States
- 3Department of Oncology, Johns Hopkins University School of Medicine, Baltimore, MD, United States
With the recent developments in deep learning and the rapid growth of convolutional neural networks, artificial intelligence has shown promise as a tool that can transform several aspects of the musculoskeletal imaging cycle. Its applications can involve both interpretive and non-interpretive tasks such as the ordering of imaging, scheduling, protocoling, image acquisition, report generation and communication of findings. However, artificial intelligence tools still face a number of challenges that can hinder effective implementation into clinical practice. The purpose of this review is to explore both the successes and limitations of artificial intelligence applications throughout the muscuskeletal imaging cycle and to highlight how these applications can help enhance the service radiologists deliver to their patients, resulting in increased efficiency as well as improved patient and provider satisfaction.
Introduction
Radiological imaging has come to play a central role in the diagnosis and management of different muscuskeletal (MSK) disorders, and both technological improvements and increased access to medical imaging have led to a rise in the utilization of common MSK imaging modalities (1, 2). As such, there is a growing need for technical innovations that can help optimize workflow and increase productivity, especially in radiology practices that are witnessing higher volumes of increasingly complex cases (3).
Artificial intelligence (AI), or the development of computer systems that can mimic human intelligence when performing human tasks, is rapidly expanding in the field of diagnostic imaging and could potentially help improve workflow efficiency (4). AI is a broad term that encompasses numerous techniques, and recent advances in the field have transformed this technology into a powerful tool with several promising applications. Nested within AI is machine learning (ML), a subfield that gives computers the ability to learn and adapt by drawing inferences from patterns in data without following explicit instructions (5). ML uses observations from data to create algorithms and subsequently makes use of these algorithms to determine future output, with the goal of designing a system that can automatically learn without any human intervention. Deep learning (DL) is an even more specialized subfield within ML that uses multiple processing layers to progressively extract higher-level features from raw input presented in the form of large datasets, and the recent development of DL with convolutional neural networks (CNN) is an important technological advancement apt at solving image-based problems with reportedly outstanding performance in several key aspects of medical imaging (Figure 1) (6–8). CNNs are widely used in computer vision; they represent feedforward neural networks with multiple layers of non-linear transformations between inputs and outputs and can be programmed to classify an image or objects according to their features (output) by means of a training dataset with numerous images or objects (input) (4).
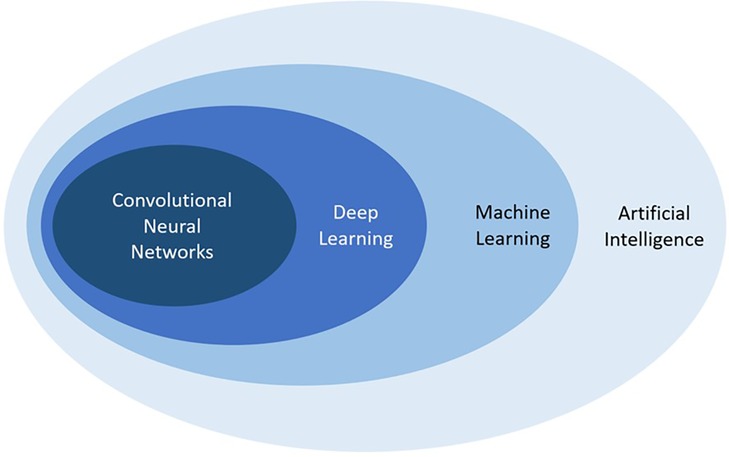
Figure 1. Schematic representation demonstrating the relationship between artificial intelligence, machine learning, deep learning, and convolutional neural networks, all subfields of each other.
For AI models to be developed, large data sets with high-quality images and annotations are needed for both training of a model and validation of its performance, and, given that developers are usually not located within medical practices or hospital systems and therefore do not have access to such data, image sharing between the two becomes necessary. This multi-step process requires collaboration between clinicians and developers and, following approval from the responsible ethical committees, begins with image de-identification, storage, and resampling of resolutions (9). Images must then be appropriately labeled with ground truth definitions, and, depending on the outcome of interest, this can involve several different steps such as manual labeling of images, data extraction from medical charts and pathology reports, and detection of imaging findings from radiology reports or by radiologists' re-review of the imaging findings (9). Typically, data sets used for training are larger than data sets used for validation and testing, and, although logistically challenging in many cases, images should ideally be obtained from multiple diverse sources to increase representation of different populations and ensure generalizability of the model's performance (9).
To understand how images are used for training AI and DL models, it is important to understand the architecture of the neural networks often employed in such models. The basic building block of a deep neural network is a node, which can be considered analogous to a neuron, and one neural network is comprised of several weighted nodes arranged into layers and connected through weighted connections (10). Training data is fed to a network at an initial input layer and then propagated throughout all layers of the model: each layer performs both linear (e.g., weighted additions) and non-linear (e.g., thresholding) mathematical computations from input received from the previous layer and feeds the output to the next layer, which then performs the same computations until one final output layer is reached (10). The model then provides a prediction, which is compared to the ground truth label previously assigned. Discrepancies between the two are fed back into the network through backward propagation and gradient descent: nodal weights and connections are adjusted accordingly, and the model is refined with every data point from the training set (Figure 2). Once the model is sufficiently refined, a validation set is typically used to evaluate the model's generalizability and further refine predictions, after which the model is then tested using a final test set with unseen data to simulate and assess real-life performance (10). The size of the data sets needed for training, validation, and testing can vary depending on the outcome and/or the targeted population (with larger sets needed for populations with more diversity and heterogeneity) but generally follows a ratio of 80:10:10 or 70:15:15, respectively (9).
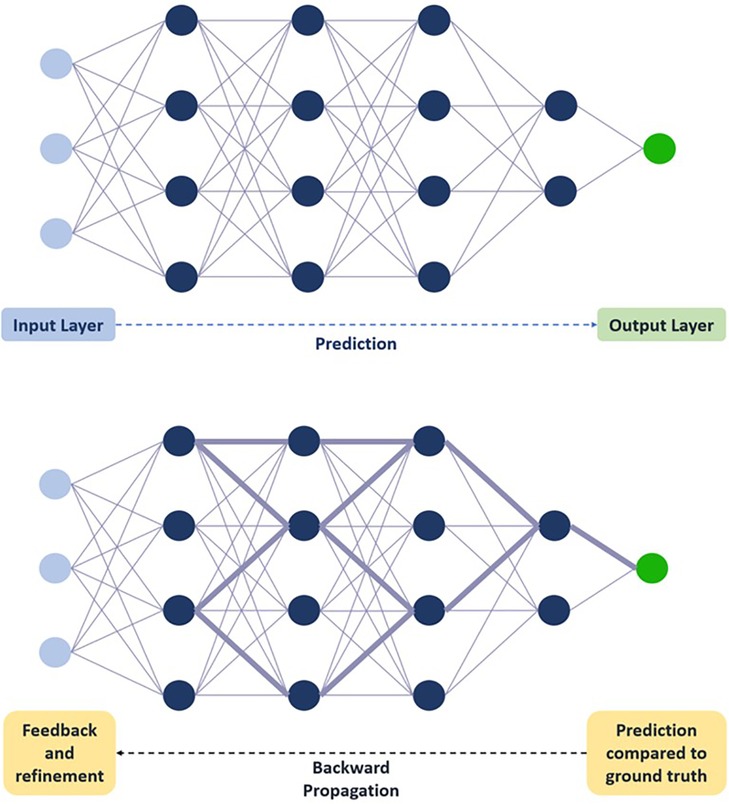
Figure 2. Schematic representation of deep neural network training. Training data is fed to the network at the initial input layer and propagated through subsequent layers for a prediction to be made at the final output layer. Prediction is compared to ground truth, and feedback through backward propagation leads to progressive refinement of weights. Circles represent nodes. Lines connecting circles represent weighted connections, with thickness correlating with weight magnitude. Dashed arrow represents flow of information through network.
With its rapid and exponential growth, AI has the potential to significantly strengthen several steps of the MSK imaging value chain and offer applications that extend beyond imaging interpretation to assist with non-interpretive tasks such as patient scheduling, optimal protocoling, image acquisition, and data sharing (11). AI can theoretically improve an MSK radiologist's ability to respond to the increasing workload of high-volume practices and continue delivering high-quality care by allotting more time for demanding tasks and minimizing time spent on more routine and less complex functions. However, AI is not without its pitfalls, and overutilization of this resource can pose multiple problems relating to medical errors, bias and inequality, data availability, and privacy concerns (12). The purpose of this review is to highlight the different applications AI is presently offering or can potentially offer throughout the MSK radiology imaging cycle and to discuss risks, limitations, and future directions of this important technology.
Prominent AI applications
Image appropriateness and protocoling
The first step in the MSK imaging process is to order the appropriate imaging test, the responsibility of which falls on the referring clinician or provider confronted with a wide range of available modalities. AI, and ML in particular, could help facilitate the process: ML algorithms could be used to generate holistic clinical decision support systems that can consider various aspects from a patient's medical chart such as symptomatology, laboratory test results, physical examination findings, and previous imaging to recommend the modality best suited to address the clinical query in question (13, 14).
Protocoling comes next, and once an imaging modality is chosen, the MSK radiologist or trainee is usually responsible for ensuring that scans are performed correctly. Choosing the right protocol is crucial to reaching a proper diagnosis and optimizing patient care but can prove arduous and time-consuming for the radiologist tasked with several other responsibilities; as such, several recent studies have looked into how DL can be of assistance. Lee assessed the feasibility of using short-text classification to develop a CNN classifier capable of determining whether MRI scans should be completed following a routine or tumor protocol and, after comparing CNN-derived protocols to those determined by MSK radiologists, reported an area under the curve (AUC) of 0.977 and an accuracy of 94.2% (15). Similarly, Trivedi et al. developed and validated a DL-based natural language classifier capable of automatically determining the need for intravenous contrast for MSK-specific MRI protocols based on the free-text clinical indication of the study and reported up to 90% agreement with human-based decisions (16). Although these studies show promising results, MSK imaging protocols are complex and diverse, given that MSK as a field encompasses localized and systemic diseases from neck to toe. More investigations could potentially explore the use of other composite classifiers such as medical history, prior imaging protocols, scanner-specific data, contrast information, and radiation exposure to help with protocoling decisions (13).
Scheduling
Given the rise in the use of medical imaging, adherence to set schedules has become more important for radiology practices, especially in the MSK setting where advanced and sometimes lengthy examinations such as MRI and CT are frequently used. No-shows or appointment cancellations can be a significant burden on practices and also represent missed opportunities for other patients to be scanned (17). There has been a growing interest in how AI can help optimize scheduling in various medical practices, and ML algorithms with predictive frameworks have been successfully used to predict missed appointments in diabetes clinics as well as urban, academic, and underserved settings (18, 19). Various ML predictive models have also been used to predict imaging no-shows effectively (20, 21), and Chong et al. demonstrated how using a pre-trained CNN with a predictive framework to predict MRI no-shows and accordingly send out proactive reminders to patients resulted in a reduced appointment no-show rate from 19.3% to 15.9% (22). ML could also help maximize patient throughput; Muelly et al. developed a feed-forward neural network that can make use of patient demographics and dynamic block lengths to estimate average MRI scan durations, resulting in decreased wait times, improved patient satisfaction, and optimized schedule fill rates (23).
Image acquisition
Magnetic resonance imaging acquisition
Given the critical need but lengthy nature of MRI scans in MSK imaging, there has always been an interest in reducing MRI acquisition times in order to decrease patient discomfort and improve scanner efficiency. Previous attempts at MRI acceleration focused on parallel imaging and compressed sensing, both of which operate by subsampling k-space and reducing the number of phase-encoding lines acquired during a scan, ultimately resulting in less data being collected (24, 25). While efficient, these two techniques suffer from reduced image quality and increased artifacts in the reconstructed images, leading to less diagnostic imaging. ML has been proposed as a possible solution that can help mitigate the limitations of accelerated imaging by using subsampled k-space data to generate up-sampled high-resolution output images comparable to images generated from otherwise fully sampled k-space data (26). Using high quality MR images, Wang et al. trained a CNN to restore fine structural details on brain images obtained from zero-filled k-space data and were able to generate images of diagnostic quality comparable to images from a fully sampled k-space but with a fivefold increase in acquisition speed (27). Hammernik et al. were able to achieve a fourfold increase in knee MRI acquisition speed by using a DL technique that created high-quality reconstructions of under-sampled data (28), while Chaudhari et al. successfully made use of a CNN to output thin-slice knee images from thicker slices, thereby improving spatial resolution and image quality (29). Similarly, Wu et al. developed an eightfold-accelerated DL model capable of up-sampling sparsely sampled MRI data to output images with minimal artifacts and a permissible signal-to-noise ratio (30). In one study by Roh et al., DL-accelerated turbo spin echo sequences were assessed for their ability to depict acute fractures of the radius in patients wearing a splint and were shown to be effective for both increasing acquisition speed by a factor of 2 as well as improving image quality when compared to standard sequences (31). Studies are still ongoing, with AI-driven 10-fold accelerated MRI increasingly becoming within reach (32) and other exciting ML applications being explored such as the production of MR images from CT images (33) and the post-processing of a single MRI acquisition to obtain other planes and tissue weightings (34). One such advance in MSK imaging is the synthetic construction of fat-suppressed imaging from non-fat-suppressed imaging (35).
Computed tomography
Unlike MRI, CT exposes patients to ionizing radiation, and ML has shown promise as a tool that can help reduce the radiation dose of a CT scan while maintaining a high quality of images (36). The premise is similar to ML applications for MRI acquisitions, whereby the goal is to reconstruct images of diagnostic quality using lower-quality source data or reduced quantities of source data. Cross et al. demonstrated how CT images acquired at a low radiation dose and reconstructed in part using an artificial neural network were found to be similar to or improved compared to images obtained using standard radiation doses by more than 90% of the readers in the study (37). Other AI developments can also help enhance image quality by decreasing artifacts related to different factors, as demonstrated in the study by Zhang and Yu where a CNN trained to merge original- and corrected-image data was capable of suppressing metal artifacts and preserving anatomical structural integrity near metallic implants (38).
Image presentation
In radiology practices that use a Picture Archiving and Communication System (PACS), radiologists often spend a considerable amount of time manipulating image displays and toggling between sequences and viewing panes to display different imaging features in several anatomic planes. This is known as the hanging protocol and constitutes another venue that can be enhanced through the use of AI to afford radiologists more productivity and efficiency. A study by Kitamura showed how ML techniques using DenseNet-based neural network models can successfully optimize hanging protocols of lumbar spine x-rays by considering several parameters such as dynamic position and rotation correction (39). Moreover, one PACS vendor is currently using ML-based algorithms to learn a radiologist's preferences when viewing examinations, record orientations of the sequences most commonly used, suggest displays for future similar studies, and incorporate adaptations following every correction, all in an effort to improve the workflow in the reading room (40).
Image interpretation
Although AI can assist MSK radiologists with several steps of the imaging cycle, it is AI's ability to help with image interpretation, arguably a radiologist's most important responsibility, that has garnered the most attention in recent years. The next section discusses different ways AI and ML can help radiologists with MSK imaging interpretations to diagnose different conditions with greater efficiency. Table 1 provides a summary of AI tools with such applications.
Fractures
Automated fracture detection using AI can be helpful not only to radiologists but also to other clinicians (such as overnight emergency department personnel) who might not always have access to radiology services and would sometimes have to rely on their own preliminary fracture diagnosis. DL techniques have been gaining increasing attention over the past few years in their ability to detect fractures on images, as this can increase diagnostic reliability and reduce the rate of medical errors.
Some studies have shown that CNNs can outperform orthopedic surgeons when it comes to the detection of upper limb and ankle fractures on radiographs (41, 42). Additionally, multiple studies have shown promise when assessing the competence of AI in detecting both axial and appendicular skeletal fractures on radiographic and CT images (26, 43, 44, 97, 98), with one CNN model by Cheng et al. achieving an AUC of 0.98 and an accuracy of 91% for radiographic hip fracture detection (45). Rajpurkar et al. trained a 169-layer DenseNet baseline model to detect and localize fractures using a large dataset of MSK radiographs containing 40,561 manually-labeled images; when tested on a set of 207 studies, the model successfully detected finger and wrist abnormalities with an AUC of 0.929 but was less competent at detecting abnormalities of the shoulder, humerus, elbow, forearm, and hand (46). Since then, their large dataset was made publicly available under the name MURA to encourage public submissions and improve fracture detection rates of the original study (99). With all the collective efforts being made to improve AI-assisted fracture detection, models are no longer just objects of research studies but have been implemented into clinical practice. Presently, Gleamer BoneView (Gleamer, Paris, France) is an FDA-approved commercially available software that can help detect fractures on radiographs and is the only AI fracture detection software to have FDA clearance for use in both adults and pediatric patients over two years of age (100).
However, despite all these promising applications, AI-assisted fracture detection still has a key limitation: each CNN model must be specifically trained on the body part being assessed using large numbers of properly labeled images, whereas humans can transfer their knowledge of one body part to another. Moreover, models can be less reliable when trying to detect less obvious fractures such as a non-displaced femoral neck fracture (98), and most models report the output in a binary fashion (fracture present or not present) without providing an in depth description of the lesion or other related findings.
Osteoarthritis
Several studies have looked into how AI can assist radiologists in evaluating images for the presence and grading of osteoarthritis. Xue et al. fine-tuned a CNN model using a set of 420 hip radiographs to detect hip osteoarthritis using a binary system and reported a performance akin to that of a radiologist with ten years of experience (47). Tiulpin et al. took advantage of the large publicly available Osteoarthritis Initiative (OAI) and Multicenter Osteoarthritis Study (MOST) datasets to train and test a CNN model to automatically score knee osteoarthritis severity using to the Kellgren-Lawrence grading scale and reported promising results with an AUC of 0.80 (48). Interestingly, the probability distribution of KL grades was also reported to show when predicted probabilities may be comparable across two contiguous grades, rendering the model's performance more illustrative of real-life practice where arthritis severity may represent the transition between two adjacent grades instead of being neatly tiered at one single level. This was also done in a study by Antony et al. where, in an attempt to circumvent the limitation of a finite and discreet scale, knee osteoarthritis grading was redefined as a regression with continuous variables (49). Although osteoarthritis assessment has been traditionally done using radiographs, AI can also augment the quantitative and qualitative assessment of cartilage on MRI to render the evaluation of osteoarthritis more accurate, and several studies have worked on developing models capable of successfully detecting cartilage lesions and staging cartilage degenerative changes (50, 51).
Bone age
Radiographic assessment of bone age is important for pediatricians to assess the skeletal maturity and growth of a child, and efforts have been put into using AI to automate bone age assessment and avoid the use of the inflexible and error-prone traditional methods such as the Greulich-Pyle atlas and the Tanner-Whitehouse method (101, 102). The Radiological Society of North America Pediatric Bone Age Machine Learning Challenge freely provided ML developers with a dataset containing over 14,000 hand radiographs and used competitions to promote collaborative effort into designing tools competent at automating bone age assessment (52). With over 100 submissions, the winning algorithm was designed by the University of Toronto's Cicero and Bilbily who used Google's Inception V3 network for pixel information, concatenated the architecture with sex information, and added layers after concatenation for data augmentation (52). The ultimate goal would be to provide radiologists with a tool that can help them assess bone age rather than perform the task independently. Tajmir et al. revealed how radiologists assisted by AI software when assessing bone age perform better than an unaided AI model, a single radiologist working independently, and a group of expert radiologists working together (53). Moreover, Kim et al. showed how the use of AI software can reduce reading times by approximately 30%, from 1.8 to 1.38 min per study (54). Presently, BoneXpert is a commercially-available widely-used software developed by Visiana that provides automated bone assessment by delineating the distal epiphyses of several hand bones, with at least eight needed for computation (55). Using the Greulich and Pyle or Tanner-Whitehouse standards, skeletal maturity is assessed with a precision of 0.17 years, reportedly nearly three times better than human performance (14, 56, 57).
Bone fragility
Imaging is often used for the evaluation of osteoporosis, a bone disorder characterized by a decreased bone mineral density (BMD), as bone strength assessment is fundamental for clinical decision making and therapy monitoring. Several studies have coupled ML support vector machines with methods of evaluating trabecular bone microarchitecture to automate and improve quantitative bone imaging and assessment (58, 59). In one study, Yang et al. used DL algorithms to combine BMD data from dual-energy x-ray absorptiometry (DXA) with bone microarchitecture data from multi-detector CT in an attempt to predict proximal femur failure loads; analysis revealed that trabecular bone characterization and ML methods, when coupled with conventional DXA BMD data, can appreciably enhance biomechanical strength prediction (58). Huber et al. applied similar ML methods to predict proximal tibial trabecular bone strength using MRI data instead and concluded that combining ML techniques with data on bone structure can enhance MRI assessment of bone quality (59).
In the same vein, ML algorithms have been employed in an attempt to predict osteoporotic fractures from MRI data (103), with one study making use of a CNN to automate segmentation of the proximal femur and facilitate the measurement of bone quality on MRI (60). Research has also focused on developing tools that can offer opportunistic screening and assessment of bone fragility, with one study looking at a system that can evaluate bone quality on dental panoramic radiographs (61) and another describing a DL system that can measure BMD on low-dose chest CT performed for lung cancer screening (62). With all those recent developments, AI is showing promise as a tool that can help with osteoporosis diagnosis; however, further refinement of such models is still needed to better automate the objective assessment of osteoporosis, its progression, and its response to therapy (104).
Spine imaging
Given that MSK radiologists spend a considerable amount of time looking at spine imaging, efforts have been made to develop ML algorithms that can automate tasks related to spine imaging interpretation and decrease the amount of time needed to interpret individual scans (105). Multiple studies have presented AI tools that can successfully detect and label spinal vertebrae as well as intervertebral discs on MRI and CT images (63–65), obviating the need for human manual labeling and streamlining the review of images. Information from these models can be used to automate other processes, as demonstrated by Jamaludin et al. who, after presenting a model that could label vertebral bodies and intervertebral discs on MRI with a 95.6% accuracy, used a CNN to successfully provide radiologist-level assessment of several other findings such as disc narrowing, central canal stenosis, spondylolisthesis, and end plate defects (66).
In addition to that, researchers have focused on designing models that can automate segmentation of the vertebrae, with one study making use of a U-Net architecture to segment the six lumbar intervertebral disc levels (67) and another adopting an iterative instance approach whereby information on one segmented vertebra is used to iteratively detect the following one (64). In the former study, Lu et al. also trained their model to automate spinal and foraminal stenosis grading using a large dataset obtained from 4,075 patients and reported an accuracy of 80% for grading spinal stenosis and 78% for grading neural foraminal stenosis (67). To increase concordance between automated segmentation outputs and ground truth labels, generative adversarial networks have also been used, with one resultant model concurrently segmenting the neural foramen, the vertebral bodies, and intervertebral discs (68).
Advancements in this line of research are ongoing, supported by large publicly available datasets such as SpineWeb and the MICCAI 2018 Challenge on Automatic Intervertebral Disc Localization and Segmentation dataset. The Pulse platform (NuVasive, San Diego, California, USA) is a recent FDA-approved spinal surgical automation platform that combines multiple technologies to provide intraoperative assessment during spine surgeries and can help with tasks such as neuromonitoring of nerves, improvement of screw placement, and minimizing intraoperative radiation exposure (106). Other spine imaging applications could include automating radiographic measurements of spinal alignment (69, 70) and using CNNs to distinguish tuberculous from pyogenic spondylitis (71). However, despite the promising results of all these recent developments, further research is still needed, and studies are often hindered by several limitations such as the lack of a consistent gold standard for entities where radiologists may exhibit high variability in interpretation (107).
Muscuskeletal oncology
AI can potentially have several applications in MSK oncology and may be able to help radiologists detect metastatic bone lesions, determine their origin, and assess progression and treatment response. Using CT texture analysis, Acar et al. developed an ML model with an AUC reaching up to 0.76 when differentiating metastatic bone lesions from sclerotic bone lesions with complete response in patients with prostate cancer (72). To determine tumor origin on contrast-enhanced MRI, Lang et al. used DL methods and radiomics to devise a model that successfully differentiated between spinal metastatic lesions from the lung and other origin sites with a high accuracy reaching 0.81 (73). In addition, AI can potentially help with the assessment of primary musculoskeletal tumors. For example, two studies making use of ML techniques and radiomics demonstrated how lipoma and liposarcoma could be differentiated on MRI with expert-level performance (74) and how the histopathological grades of soft tissue sarcomas can be pre-operatively and non-invasively predicted on fat-suppressed T2-weighted imaging with an accuracy reaching 0.88 (75). AI might also serve other proposed roles, such as assisting clinicians in predicting tumor recurrence as well monitoring post-treatment tumor changes on imaging (76, 77).
Cruciate ligaments and menisci
Several studies have evaluated the performance of AI models when detecting meniscal injuries and ligamentous tears of the knee. Bien et al. trained a CNN model using a set of 1,130 training and 120 validation MRI exams to recognize meniscal and anterior cruciate ligament (ACL) tears, reporting an AUC of 0.847 for meniscal tears and an AUC of 0.937 for ACL tears on the internal validation set and 0.824 on the external validation set (78). Using arthroscopy as the reference, Liu et al. trained a CNN to isolate ACL lacerations with an AUC of 0.98 and a sensitivity of 96% (51, 79), and Ma et al. trained a CNN to diagnose meniscal injury, reporting a an accuracy of 85.6% for anterior horn injury detection and 92% for posterior horn injury detection, a performance comparable to a chief physician (80).
Isolation of individual joint structures might help enhance model performance, as demonstrated by Chang et al. who, after isolating the ACL on coronal proton density 2D MRI using CNN U-Net, subsequently used a CNN classifier to evaluate the isolated ACLs for the presence of pathology and reported an AUC of 0.97 and a sensitivity of 100% (81). When testing a CNN model for meniscal segmentation on fat-suppressed MRI sequences, Pedoia et al. reported a sensitivity reaching 90%, a specificity reaching 82%, and an AUC reaching 0.89 (50). Likewise, Couteaux et al., Roblot et al., and Lassau et al. all reported similar performances, with AUC values for meniscal tear detection reaching 0.9 in all three studies (82, 83, 108).
Quantitative analysis: segmentation and radiomics
Segmentation, or the process of delineating anatomic structures, can be time-consuming but is nevertheless important for evaluating the potential degeneration of or damage to segmented structures and the resultant decline in their functionality. Semi-automated segmentation software are currently being applied in clinical cardiac and prostate MRI, but such software make use of algorithms with manually designed hand-engineered features and thus require manual adjustments to the computer-generated contours (26). As such, interest has shifted to fully automating segmentation processes using CNN, which can have a profound impact on a radiologist's functionality and efficiency in the reading room. Performance of segmentation algorithms is often assessed with a dice coefficient to assess the similarity of a segmentation to its ground truth by reporting the percentage overlap between the two regions, and a dice score of 0.95 is usually indicative of a successful algorithm (109). Recent research has heavily focused on knee segmentation, with Liu et al. designing a model that successfully segmented the different structures of the knee using a CNN combined with a 3D deformable modeling approach (84). Using both T1-rho weighted and 3D double-echo steady-state images, Norman et al. also evaluated a DL model for automated segmentation of knee cartilage and menisci but with simultaneous evaluation of cartilage relaxometry and morphology; they found the model to be adept at generating accurate segmentations and morphologic characterizations when compared to manual segmentations (85). DL techniques can have applications outside the knee as well, as demonstrated by Deniz et al. who used similar methods but shifted attention to the segmentation of the proximal femur, reporting a CNN algorithm with a dice similarity score reaching 0.95 (60). Other venues are also being explored, with AI tools showing promise in neurography segmentation (86) as well as muscle segmentation in osteoarthritis patients to help with muscular trophism evaluation (87).
Besides segmentation, AI may also have applications in radiomics, which is an emerging field in medicine that treats medical images not only as pictures intended solely for visual interpretation but also as a source of diverse quantitative characteristics extracted as mineable data that can be used for pattern identification to eventually assist with decision support, characterization, and prediction of disease processes (110). Spatial distribution of signal intensities and information on pixel interrelationships are mathematically extracted to provide and quantify textural information, which in turn can be used for quantitative imaging biomarker discovery and validation for a number of different conditions such as acute and chronic injuries, spinal abnormalities, and neoplasms (111). By uncovering imperceptible patterns in medical imaging, radiomics-bases predictive models can play different roles such as providing a detailed description of disease burden, identifying relationships between phenotypes and outcomes, and predicting diagnosis and prognosis for certain diseases, ultimately playing a key role in improving precision medicine and personalized patient management (112). ML models can identify and gather imaging characteristics such as the distribution of signal intensities and the spatial relationship of pixels that are not easily discernible with visual interpretation and that can help improve clinical care (113, 114). When testing different ML-augmented radiomics models for preoperative differentiation of sacral chordomas from sacral giant cell tumors on 3D CT; Yin et al. found contrast- enhanced CT features more optimal than non-enhanced features for helping identify the histology of the sacral tumor in question (88). In one retrospective study, Gitto et al. assessed the diagnostic performance of ML-enhanced radiomics-based MRI for the classification and differentiation of atypical lipomatous tumors of the extremities from other benign lipomas, reporting a sensitivity of 92%, a specificity of 33%, and no statistically significant difference when compared to qualitative image assessment performed by a radiologist with 7 years of experience (89). Research into the field is ongoing, and although radiomics has shown promise as a powerful and innovative tool that can help with the evaluation of different types of cancers, more research is needed to fully explore the full scope of its applications (115).
Other miscellaneous applications
Several research studies have looked into other potential applications of AI such as joint space evaluation in rheumatoid arthritis (90, 91), epidural mass detection on CT scans (92), rotator cuff pathology detection (116), femoroacetabular impingement and hip dysplasia detection (93), sex determination using CT imaging of the sacrum and coccyx (94) or hand radiographs (95), and assessment of Achilles tendon healing (117). In addition to that, AI applications can have multiple applications in MSK ultrasound (US), including but not limited to segmentation of US images (96), quantitative analysis of skeletal muscles (118), and detection of pediatric conditions such as wrist fractures and developmental dysplasia of the hip (4, 119). Research is still ongoing, and additional repetitive and time-consuming tasks might be tackled in coming years in an attempt to automate more processes and thus accelerate the process of imaging interpretation.
Results reporting
AI can have several applications that can revolutionize the production of radiology reports and the communication of findings between physicians. Speech recognition, which has already transformed the writing of reports, could be further optimized with DL methods (120). Language processing systems can also be applied, as shown by Do et al. who presented a system capable of recognizing anatomy data from reports generated with speech recognition software to concurrently extract information on possible fractures (121) and Tan et al. who presented a system capable of scanning x-ray and MRI radiology reports to identify lumbar spine imaging findings that could be related to low back pain (122). Natural language processing (NLP) refers to the use of a computer to analyze and interpret human language. Although NLP systems are not entirely novel, recent advances in ML and neural networks have revolutionized this technology, subsequently turning it into a tool that can help with data extraction from radiology reports (123). At their core, NLP systems operate using a multistep approach, beginning with a preprocessing step whereby reports are broken down into different subsets and processing steps during which text from specific sections or differently-weighted sections is split into sentences and words (a process known as tokenization) (124). Word normalization and syntactic analysis follow, whereby spelling mistakes are fixed, medical abbreviations are fully expanded, and word roots are identified with the goal of determining grammatical structures and linking words to semantic concepts (such as symptom or disease), thus assigning meaning to the data (124). The textual features extracted are then processed by an automatic classifier using ML applications to solve the ultimate task assigned to the system (such as information extraction from reports), and ML applications have to be trained on a set of manually-annotated reports, which can be split into a training set and a validation set, both of which are needed to develop the system and assess its performance (124).
Such tools could play a number of roles, such as suggesting management recommendations to radiologists during the dictation of a report or assisting with research purposes by establishing links between different radiological findings and resultant symptomatology or prognosis. Additionally, ML applications may extend to extracting follow-up recommendations from reports, thus ensuring the adequate management of reported key findings (125).
Limitations
Although AI shows several promising applications across the entire MSK imaging cycle, this technology is still facing a number of challenges and limitations when it comes to both development of AI tools and implementation into clinical practice. Large datasets are needed to develop successful DL tools: tasks or diagnoses for which such datasets are not available might be challenging to automate, and data can be fragmented across many different systems, thus increasing the risk of errors, decreasing the comprehensiveness of datasets, and increasing the expenses of gathering complete data. Moreover, challenges in establishing reference standards, such as irregularities in contouring lesions, diagnostic uncertainties, as well as inconsistencies in human performance and labeling, can all reduce performance and hinder development. DL models being developed are usually trained to perform one single task, whereas patients seen clinically might have a number of etiologies and conditions that require complex simultaneous interpretations.
Given that large amounts of data need to be collected for the development of successful algorithms, issues pertaining to privacy and ownership of such data arise: patients may be concerned that collection of such data is a violation of privacy, especially if an AI model can predict private information about a patient without having received that information and subsequently make it available to third parties (such as life insurance companies). Large datasets can be problematic in a different way: they may be more representative of a specific subset of the population rather than the whole population and could also reflect underlying biases and inequalities in the health system. As such, algorithms trained using such datasets may propagate systemic biases and inequalities that are already present and may not be suitable for treating all patients but rather the subset with the most representation in the training dataset.
Evidently, AI models can and will make mistakes, resulting in errors and injuries to patients being treated using the model. Although medical errors are sometimes inevitable in the medical field and can occur irrespective of the use of AI, the danger of AI-related mistakes is that an underlying problem in one system might result in injuries to thousands of patients if that system becomes widespread (whereas errors from a single human provider will affect the limited number of patients being treated by that provider). Additionally, with errors arises the issue of accountability: models often do not disclose the statistical rationale behind the elaboration of their tasks, making it hard to identify the cause of the error or understand the rationale behind the final output of an algorithm and limiting implementation into medical settings. To catch errors and refine algorithms, post-implementation evaluation, maintenance, and performance monitoring of implemented AI tools is just as vital as pre-implementation development processes to the success of a model. However, such monitoring can prove to be labor-intensive, especially for smaller practices that will inevitably experience workflow disruptions due to a lack of dedicated informatics resources and an increase in the radiologists' burden (126).
Conclusion
AI, ML and DL have the potential to significantly augment several aspects of the MSK imaging chain, with applications in the ordering of imaging, scheduling, protocoling, acquisition and presentation, image interpretation, as well as report generation and communication of findings. Although research into this technology is showing very promising results, development of tools still faces a number of challenges that impede successful implementation into clinical practice. The ultimate goal is not to design a completely independent system that replaces the need for human expertise but rather to equip radiologists and medical professionals with tools that can automate certain functions and thus alleviate some of the increasing responsibilities radiologists face, affording them more time to focus on more demanding and complex tasks. Radiologists and AI algorithms working hand in hand have the potential to increase the value provided to patients by improving imaging quality and efficiency, patient centricity, and diagnostic accuracy, all of which can greatly enhance both patient and provider satisfaction.
Author contributions
PD has made significant contributions to the conception and design of the work and drafting and revision. He provides approval for the publication of content and agrees to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. LF has made significant contributions to the conception and design of the work and drafting and critical revision for important intellectual content. She provides approval for the publication of content and agrees to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Farrell TP, Adams NC, Walsh JP, Hynes J, Eustace SK, Kavanagh E. Musculoskeletal imaging: current practice and future directions. Semin Musculoskelet Radiol. (2018) 22(5):564–81. doi: 10.1055/s-0038-1672193
2. Harkey P, Duszak R, Gyftopoulos S, Rosenkrantz AB. Who refers musculoskeletal extremity imaging examinations to radiologists? Am J Roentgenol. (2018) 210(4):834–41. doi: 10.2214/AJR.17.18591
3. Doshi AM, Moore WH, Kim DC, Rosenkrantz AB, Fefferman NR, Ostrow DL, et al. Informatics solutions for driving an effective and efficient radiology practice. Radiographics. (2018) 38(6):1810–22. doi: 10.1148/rg.2018180037
4. D’Angelo T, Caudo D, Blandino A, Albrecht MH, Vogl TJ, Gruenewald LD, et al. Artificial intelligence, machine learning and deep learning in musculoskeletal imaging: current applications. J Clin Ultrasound. (2022) 50(9):1414–31. doi: 10.1002/jcu.23321
5. Do S, Song KD, Chung JW. Basics of deep learning: a radiologist’s guide to understanding published radiology articles on deep learning. Korean J Radiol. (2020) 21(1):33–41. doi: 10.3348/kjr.2019.0312
6. Choy G, Khalilzadeh O, Michalski M, Do S, Samir AE, Pianykh OS, et al. Current applications and future impact of machine learning in radiology. Radiology. (2018) 288(2):318–28. doi: 10.1148/radiol.2018171820
7. Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E. Convolutional neural networks for radiologic images: a radiologist’s guide. Radiology. (2019) 290(3):590–606. doi: 10.1148/radiol.2018180547
8. Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. RadioGraphics. (2017) 37(7):2113–31. doi: 10.1148/rg.2017170077
9. Willemink MJ, Koszek WA, Hardell C, Wu J, Fleischmann D, Harvey H, et al. Preparing medical imaging data for machine learning. Radiology. (2020) 295(1):4–15. doi: 10.1148/radiol.2020192224
10. Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z für Med Phys. (2019) 29(2):102–27. doi: 10.1016/j.zemedi.2018.11.002
11. Richardson ML, Garwood ER, Lee Y, Li MD, Lo HS, Nagaraju A, et al. Noninterpretive uses of artificial intelligence in radiology. Acad Radiol. (2021) 28(9):1225–35. doi: 10.1016/j.acra.2020.01.012
12. Saw SN, Ng KH. Current challenges of implementing artificial intelligence in medical imaging. Phys Med. (2022) 100:12–7. doi: 10.1016/j.ejmp.2022.06.003
13. Gyftopoulos S, Lin D, Knoll F, Doshi AM, Rodrigues TC, Recht MP. Artificial intelligence in musculoskeletal imaging: current status and future directions. AJR Am J Roentgenol. (2019) 213(3):506–13. doi: 10.2214/AJR.19.21117
14. Gorelik N, Gyftopoulos S. Applications of artificial intelligence in musculoskeletal imaging: from the request to the report. Can Assoc Radiol J. (2021) 72(1):45–59. doi: 10.1177/0846537120947148
15. Lee YH. Efficiency improvement in a busy radiology practice: determination of musculoskeletal magnetic resonance imaging protocol using deep-learning convolutional neural networks. J Digit Imaging. (2018) 31(5):604–10. doi: 10.1007/s10278-018-0066-y
16. Trivedi H, Mesterhazy J, Laguna B, Vu T, Sohn JH. Automatic determination of the need for intravenous contrast in musculoskeletal MRI examinations using IBM watson’s natural language processing algorithm. J Digit Imaging. (2018) 31(2):245–51. doi: 10.1007/s10278-017-0021-3
17. Mieloszyk RJ, Rosenbaum JI, Hall CS, Raghavan UN, Bhargava P. The financial burden of missed appointments: uncaptured revenue due to outpatient no-shows in radiology. Curr Probl Diagn Radiol. (2018) 47(5):285–6. doi: 10.1067/j.cpradiol.2018.06.001
18. Kurasawa H, Hayashi K, Fujino A, Takasugi K, Haga T, Waki K, et al. Machine-learning-based prediction of a missed scheduled clinical appointment by patients with diabetes. J Diabetes Sci Technol. (2016) 10(3):730–6. doi: 10.1177/1932296815614866
19. Torres O, Rothberg MB, Garb J, Ogunneye O, Onyema J, Higgins T. Risk factor model to predict a missed clinic appointment in an urban, academic, and underserved setting. Popul Health Manag. (2015) 18(2):131–6. doi: 10.1089/pop.2014.0047
20. Nelson A, Herron D, Rees G, Nachev P. Predicting scheduled hospital attendance with artificial intelligence. NPJ Digit Med. (2019) 2(1):1–7. doi: 10.1038/s41746-019-0103-3
21. Harvey HB, Liu C, Ai J, Jaworsky C, Guerrier CE, Flores E, et al. Predicting no-shows in radiology using regression modeling of data available in the electronic medical record. J Am Coll Radiol. (2017) 14(10):1303–9. doi: 10.1016/j.jacr.2017.05.007
22. Chong LR, Tsai KT, Lee LL, Foo SG, Chang PC. Artificial intelligence predictive analytics in the management of outpatient MRI appointment no-shows. AJR Am J Roentgenol. (2020) 215(5):1155–62. doi: 10.2214/AJR.19.22594
23. (ISMRM 2017) Using machine learning with dynamic exam block lengths to decrease patient wait time and optimize MRI schedule fill rate. Available at: https://archive.ismrm.org/2017/4343.html (Cited June 14, 2023).
24. Jaspan ON, Fleysher R, Lipton ML. Compressed sensing MRI: a review of the clinical literature. Br J Radiol. (2015) 88(1056):20150487. doi: 10.1259/bjr.20150487
25. Glockner JF, Hu HH, Stanley DW, Angelos L, King K. Parallel MR imaging: a user’s guide. Radiographics. (2005) 25(5):1279–97. doi: 10.1148/rg.255045202
26. Chea P, Mandell JC. Current applications and future directions of deep learning in musculoskeletal radiology. Skeletal Radiol. (2020) 49(2):183–97. doi: 10.1007/s00256-019-03284-z
27. Wang S, Su Z, Ying L, Peng X, Zhu S, Liang F, et al. Accelerating magnetic resonance imaging via deep learning. Proc IEEE Int Symp Biomed Imaging. (2016) 2016:514–7. doi: 10.1109/ISBI.2016.7493320
28. Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. (2018) 79(6):3055–71. doi: 10.1002/mrm.26977
29. Chaudhari AS, Fang Z, Kogan F, Wood J, Stevens KJ, Gibbons EK, et al. Super-resolution musculoskeletal MRI using deep learning. Magn Reson Med. (2018) 80(5):2139–54. doi: 10.1002/mrm.27178
30. Wu Y, Ma Y, Capaldi DP, Liu J, Zhao W, Du J, et al. Incorporating prior knowledge via volumetric deep residual network to optimize the reconstruction of sparsely sampled MRI. Magn Reson Imaging. (2020) 66:93–103. doi: 10.1016/j.mri.2019.03.012
31. Roh S, Park JI, Kim GY, Yoo HJ, Nickel D, Koerzdoerfer G, et al. Feasibility and clinical usefulness of deep learning-accelerated MRI for acute painful fracture patients wearing a splint: a prospective comparative study. PLoS One. (2023) 18(6):e0287903. doi: 10.1371/journal.pone.0287903
32. Lin DJ, Walter SS, Fritz J. Artificial intelligence–driven ultra-fast superresolution MRI : 10-fold accelerated musculoskeletal turbo spin Echo MRI within reach. Invest Radiol. (2023) 58(1):28–42. doi: 10.1097/RLI.0000000000000928
33. Lee JH, Han IH, Kim DH, Yu S, Lee IS, Song YS, et al. Spine computed tomography to magnetic resonance image synthesis using generative adversarial networks : a preliminary study. J Korean Neurosurg Soc. (2020) 63(3):386–96. doi: 10.3340/jkns.2019.0084
34. Galbusera F, Bassani T, Casaroli G, Gitto S, Zanchetta E, Costa F, et al. Generative models: an upcoming innovation in musculoskeletal radiology? A preliminary test in spine imaging. Eur Radiol Exp. (2018) 2:29. doi: 10.1186/s41747-018-0060-7
35. Fayad LM, Parekh VS, de Castro Luna R, Ko CC, Tank D, Fritz J, et al. A deep learning system for synthetic knee magnetic resonance imaging: is artificial intelligence-based fat-suppressed imaging feasible? Invest Radiol. (2021) 56(6):357–68. doi: 10.1097/RLI.0000000000000751
36. Kambadakone A. Artificial intelligence and CT image reconstruction: potential of a new era in radiation dose reduction. J Am Coll Radiol. (2020) 17(5):649–51. doi: 10.1016/j.jacr.2019.12.025
38. Zhang Y, Yu H. Convolutional neural network based metal artifact reduction in x-ray computed tomography. IEEE Trans Med Imaging. (2018) 37(6):1370–81. doi: 10.1109/TMI.2018.2823083
39. Kitamura G. Hanging protocol optimization of lumbar spine radiographs with machine learning. Skeletal Radiol. (2021) 50(9):1809–19. doi: 10.1007/s00256-021-03733-8
40. Intelligent Tools for a Productive Radiologist Workflow, Universal Viewer Smart Reading Protocols. Available at: https://healthimaging.com/sponsored/1155/ge-healthcare/intelligent-tools-productive-radiologist-workflow-universal-viewer (Cited June 14, 2023).
41. Chung SW, Han SS, Lee JW, Oh KS, Kim NR, Yoon JP, et al. Automated detection and classification of the proximal humerus fracture by using deep learning algorithm. Acta Orthop. (2018) 89(4):468–73. doi: 10.1080/17453674.2018.1453714
42. Olczak J, Fahlberg N, Maki A, Razavian AS, Jilert A, Stark A, et al. Artificial intelligence for analyzing orthopedic trauma radiographs. Acta Orthop. (2017) 88(6):581–6. doi: 10.1080/17453674.2017.1344459
43. Yu JS, Yu SM, Erdal BS, Demirer M, Gupta V, Bigelow M, et al. Detection and localisation of hip fractures on anteroposterior radiographs with artificial intelligence: proof of concept. Clin Radiol. (2020) 75(3):237.e1–e9. doi: 10.1016/j.crad.2019.10.022
44. Tomita N, Cheung YY, Hassanpour S. Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Comput Biol Med. (2018) 98:8–15. doi: 10.1016/j.compbiomed.2018.05.011
45. Cheng CT, Ho TY, Lee TY, Chang CC, Chou CC, Chen CC, et al. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur Radiol. (2019) 29(10):5469–77. doi: 10.1007/s00330-019-06167-y
46. Rajpurkar P, Irvin J, Bagul A, Ding D, Duan T, Mehta H, et al. MURA: Large Dataset for Abnormality Detection in Musculoskeletal Radiographs. arXiv (2018). Available at: http://arxiv.org/abs/1712.06957 (Cited June 15, 2023).
47. Xue Y, Zhang R, Deng Y, Chen K, Jiang T. A preliminary examination of the diagnostic value of deep learning in hip osteoarthritis. PLoS One. (2017) 12(6):e0178992. doi: 10.1371/journal.pone.0178992
48. Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S. Automatic knee osteoarthritis diagnosis from plain radiographs: a deep learning-based approach. Sci Rep. (2018) 8(1):1727. doi: 10.1038/s41598-018-20132-7
49. Antony J, McGuinness K, Connor NEO, Moran K. Quantifying Radiographic Knee Osteoarthritis Severity using Deep Convolutional Neural Networks. arXiv (2016) Available at: http://arxiv.org/abs/1609.02469 (Cited June 15, 2023).
50. Pedoia V, Norman B, Mehany SN, Bucknor MD, Link TM, Majumdar S. 3D convolutional neural networks for detection and severity staging of meniscus and PFJ cartilage morphological degenerative changes in osteoarthritis and anterior cruciate ligament subjects. J Magn Reson Imaging. (2019) 49(2):400–10. doi: 10.1002/jmri.26246
51. Liu F, Zhou Z, Samsonov A, Blankenbaker D, Larison W, Kanarek A, et al. Deep learning approach for evaluating knee MR images: achieving high diagnostic performance for cartilage lesion detection. Radiology. (2018) 289(1):160–9. doi: 10.1148/radiol.2018172986
52. Halabi SS, Prevedello LM, Kalpathy-Cramer J, Mamonov AB, Bilbily A, Cicero M, et al. The RSNA pediatric bone age machine learning challenge. Radiology. (2019) 290(2):498–503. doi: 10.1148/radiol.2018180736
53. Tajmir SH, Lee H, Shailam R, Gale HI, Nguyen JC, Westra SJ, et al. Artificial intelligence-assisted interpretation of bone age radiographs improves accuracy and decreases variability. Skeletal Radiol. (2019) 48(2):275–83. doi: 10.1007/s00256-018-3033-2
54. Kim JR, Shim WH, Yoon HM, Hong SH, Lee JS, Cho YA, et al. Computerized bone age estimation using deep learning based program: evaluation of the accuracy and efficiency. AJR Am J Roentgenol. (2017) 209(6):1374–80. doi: 10.2214/AJR.17.18224
55. Thodberg HH, Kreiborg S, Juul A, Pedersen KD. The BoneXpert method for automated determination of skeletal maturity. IEEE Trans Med Imaging. (2009) 28(1):52–66. doi: 10.1109/TMI.2008.926067
56. Martin DD, Calder AD, Ranke MB, Binder G, Thodberg HH. Accuracy and self-validation of automated bone age determination. Sci Rep. (2022) 12(1):6388. doi: 10.1038/s41598-022-10292-y
57. Maratova K, Zemkova D, Sedlak P, Pavlikova M, Amaratunga SA, Krasnicanova H, et al. A comprehensive validation study of the latest version of BoneXpert on a large cohort of Caucasian children and adolescents. Front Endocrinol (Lausanne). (2023) 14. Available at: https://www.frontiersin.org/articles/10.3389/fendo.2023.1130580 doi: 10.3389/fendo.2023.1130580
58. Yang CC, Nagarajan MB, Huber MB, Carballido-Gamio J, Bauer JS, Baum T, et al. Improving bone strength prediction in human proximal femur specimens through geometrical characterization of trabecular bone microarchitecture and support vector regression. J Electron Imaging. (2014) 23(1):013013. doi: 10.1117/1.JEI.23.1.013013
59. Huber MB, Lancianese SL, Nagarajan MB, Ikpot IZ, Lerner AL, Wismuller A. Prediction of biomechanical properties of trabecular bone in MR images with geometric features and support vector regression. IEEE Trans Biomed Eng. (2011) 58(6):1820–6. doi: 10.1109/TBME.2011.2119484
60. Deniz CM, Xiang S, Hallyburton RS, Welbeck A, Babb JS, Honig S, et al. Segmentation of the proximal femur from MR images using deep convolutional neural networks. Sci Rep. (2018) 8(1):16485. doi: 10.1038/s41598-018-34817-6
61. Lee JS, Adhikari S, Liu L, Jeong HG, Kim H, Yoon SJ. Osteoporosis detection in panoramic radiographs using a deep convolutional neural network-based computer-assisted diagnosis system: a preliminary study. Dentomaxillofac Radiol. (2019) 48(1):20170344. doi: 10.1259/dmfr.20170344
62. Pan Y, Shi D, Wang H, Chen T, Cui D, Cheng X, et al. Automatic opportunistic osteoporosis screening using low-dose chest computed tomography scans obtained for lung cancer screening. Eur Radiol. (2020) 30(7):4107–16. doi: 10.1007/s00330-020-06679-y
63. Jimenez-Pastor A, Alberich-Bayarri A, Fos-Guarinos B, Garcia-Castro F, Garcia-Juan D, Glocker B, et al. Automated vertebrae localization and identification by decision forests and image-based refinement on real-world CT data. Radiol Med. (2020) 125(1):48–56. doi: 10.1007/s11547-019-01079-9
64. Lessmann N, van Ginneken B, de Jong PA, Išgum I. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med Image Anal. (2019) 53:142–55. doi: 10.1016/j.media.2019.02.005
65. Wimmer M, Major D, Novikov AA, Bühler K. Fully automatic cross-modality localization and labeling of vertebral bodies and intervertebral discs in 3D spinal images. Int J Comput Assist Radiol Surg. (2018) 13(10):1591–603. doi: 10.1007/s11548-018-1818-3
66. Jamaludin A, Lootus M, Kadir T, Zisserman A, Urban J, Battié MC, et al. ISSLS PRIZE IN BIOENGINEERING SCIENCE 2017: automation of reading of radiological features from magnetic resonance images (MRIs) of the lumbar spine without human intervention is comparable with an expert radiologist. Eur Spine J. (2017) 26(5):1374–83. doi: 10.1007/s00586-017-4956-3
67. Lu JT, Pedemonte S, Bizzo B, Doyle S, Andriole KP, Michalski MH, et al. DeepSPINE: Automated Lumbar Vertebral Segmentation, Disc-level Designation, and Spinal Stenosis Grading Using Deep Learning. arXiv (2018). Available at: http://arxiv.org/abs/1807.10215 (Cited June 17, 2023).
68. Han Z, Wei B, Mercado A, Leung S, Li S. Spine-GAN: semantic segmentation of multiple spinal structures. Med Image Anal. (2018) 50:23–35. doi: 10.1016/j.media.2018.08.005
69. Pan Y, Chen Q, Chen T, Wang H, Zhu X, Fang Z, et al. Evaluation of a computer-aided method for measuring the cobb angle on chest x-rays. Eur Spine J. (2019) 28(12):3035–43. doi: 10.1007/s00586-019-06115-w
70. Weng CH, Wang CL, Huang YJ, Yeh YC, Fu CJ, Yeh CY, et al. Artificial intelligence for automatic measurement of sagittal vertical axis using ResUNet framework. J Clin Med. (2019) 8(11):1826. doi: 10.3390/jcm8111826
71. Kim K, Kim S, Lee YH, Lee SH, Lee HS, Kim S. Performance of the deep convolutional neural network based magnetic resonance image scoring algorithm for differentiating between tuberculous and pyogenic spondylitis. Sci Rep. (2018) 8(1):13124. doi: 10.1038/s41598-018-31486-3
72. Acar E, Leblebici A, Ellidokuz BE, Başbınar Y, Kaya GÇ. Machine learning for differentiating metastatic and completely responded sclerotic bone lesion in prostate cancer: a retrospective radiomics study. Br J Radiol. (2019) 92(1101):20190286. doi: 10.1259/bjr.20190286
73. Lang N, Zhang Y, Zhang E, Zhang J, Chow D, Chang P, et al. Differentiation of spinal metastases originated from lung and other cancers using radiomics and deep learning based on DCE-MRI. Magn Reson Imaging. (2019) 64:4–12. doi: 10.1016/j.mri.2019.02.013
74. Malinauskaite I, Hofmeister J, Burgermeister S, Neroladaki A, Hamard M, Montet X, et al. Radiomics and machine learning differentiate soft-tissue lipoma and liposarcoma better than musculoskeletal radiologists. Sarcoma. (2020) 2020:e7163453. doi: 10.1155/2020/7163453
75. Zhang Y, Zhu Y, Shi X, Tao J, Cui J, Dai Y, et al. Soft tissue sarcomas: preoperative predictive histopathological grading based on radiomics of MRI. Acad Radiol. (2019) 26(9):1262–8. doi: 10.1016/j.acra.2018.09.025
76. He Y, Guo J, Ding X, van Ooijen PMA, Zhang Y, Chen A, et al. Convolutional neural network to predict the local recurrence of giant cell tumor of bone after curettage based on pre-surgery magnetic resonance images. Eur Radiol. (2019) 29(10):5441–51. doi: 10.1007/s00330-019-06082-2
77. Blackledge MD, Winfield JM, Miah A, Strauss D, Thway K, Morgan VA, et al. Supervised machine-learning enables segmentation and evaluation of heterogeneous post-treatment changes in multi-parametric MRI of soft-tissue sarcoma. Front Oncol. (2019) 9:941. doi: 10.3389/fonc.2019.00941
78. Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: development and retrospective validation of MRNet. PLoS Med. (2018) 15(11):e1002699. doi: 10.1371/journal.pmed.1002699
79. Liu F, Guan B, Zhou Z, Samsonov A, Rosas H, Lian K, et al. Fully automated diagnosis of anterior cruciate ligament tears on knee MR images by using deep learning. Radiology: Artificial Intelligence. (2019) 1(3):180091. doi: 10.1148/ryai.2019180091
80. Ma Y, Qin Y, Liang C, Li X, Li M, Wang R, et al. Visual cascaded-progressive convolutional neural network (C-PCNN) for diagnosis of Meniscus injury. Diagnostics. (2023) 13(12):2049. doi: 10.3390/diagnostics13122049
81. Chang PD, Wong TT, Rasiej MJ. Deep learning for detection of complete anterior cruciate ligament tear. J Digit Imaging. (2019) 32(6):980–6. doi: 10.1007/s10278-019-00193-4
82. Couteaux V, Si-Mohamed S, Nempont O, Lefevre T, Popoff A, Pizaine G, et al. Automatic knee meniscus tear detection and orientation classification with mask-RCNN. Diagn Interv Imaging. (2019) 100(4):235–42. doi: 10.1016/j.diii.2019.03.002
83. Roblot V, Giret Y, Bou Antoun M, Morillot C, Chassin X, Cotten A, et al. Artificial intelligence to diagnose meniscus tears on MRI. Diagn Interv Imaging. (2019) 100(4):243–9. doi: 10.1016/j.diii.2019.02.007
84. Liu F, Zhou Z, Jang H, Samsonov A, Zhao G, Kijowski R. Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magn Reson Med. (2018) 79(4):2379–91. doi: 10.1002/mrm.26841
85. Norman B, Pedoia V, Majumdar S. Use of 2D U-net convolutional neural networks for automated cartilage and Meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. (2018) 288(1):177–85. doi: 10.1148/radiol.2018172322
86. Balsiger F, Steindel C, Arn M, Wagner B, Grunder L, El-Koussy M, et al. Segmentation of peripheral nerves from magnetic resonance neurography: a fully-automatic, deep learning-based approach. Front Neurol. (2018) 9:777. doi: 10.3389/fneur.2018.00777
87. Kemnitz J, Baumgartner CF, Eckstein F, Chaudhari A, Ruhdorfer A, Wirth W, et al. Clinical evaluation of fully automated thigh muscle and adipose tissue segmentation using a U-net deep learning architecture in context of osteoarthritic knee pain. MAGMA. (2020) 33(4):483–93. doi: 10.1007/s10334-019-00816-5
88. Yin P, Mao N, Zhao C, Wu J, Sun C, Chen L, et al. Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur Radiol. (2019) 29(4):1841–7. doi: 10.1007/s00330-018-5730-6
89. Gitto S, Interlenghi M, Cuocolo R, Salvatore C, Giannetta V, Badalyan J, et al. MRI radiomics-based machine learning for classification of deep-seated lipoma and atypical lipomatous tumor of the extremities. Radiol Med. (2023) 128(8):989–98. doi: 10.1007/s11547-023-01657-y
90. Pfeil A, Renz DM, Hansch A, Kainberger F, Lehmann G, Malich A, et al. The usefulness of computer-aided joint space analysis in the assessment of rheumatoid arthritis. Joint Bone Spine. (2013) 80(4):380–5. doi: 10.1016/j.jbspin.2012.10.022
91. Langs G, Peloschek P, Bischof H, Kainberger F. Model-based erosion spotting and visualization in rheumatoid arthritis. Acad Radiol. (2007) 14(10):1179–88. doi: 10.1016/j.acra.2007.06.013
92. Liu J, Pattanaik S, Yao J, Turkbey E, Zhang W, Zhang X, et al. Computer aided detection of epidural masses on computed tomography scans. Comput Med Imaging Graph. (2014) 38(7):606–12. doi: 10.1016/j.compmedimag.2014.04.007
93. Stotter C, Klestil T, Röder C, Reuter P, Chen K, Emprechtinger R, et al. Deep learning for fully automated radiographic measurements of the pelvis and hip. Diagnostics. (2023) 13(3):497. doi: 10.3390/diagnostics13030497
94. Etli Y, Asirdizer M, Hekimoglu Y, Keskin S, Yavuz A. Sex estimation from sacrum and coccyx with discriminant analyses and neural networks in an equally distributed population by age and sex. Forensic Sci Int. (2019) 303:109955. doi: 10.1016/j.forsciint.2019.109955
95. Yune S, Lee H, Kim M, Tajmir SH, Gee MS, Do S. Beyond human perception: sexual dimorphism in hand and wrist radiographs is discernible by a deep learning model. J Digit Imaging. (2019) 32(4):665–71. doi: 10.1007/s10278-018-0148-x
96. Bowness J, Varsou O, Turbitt L, Burkett-St Laurent D. Identifying anatomical structures on ultrasound: assistive artificial intelligence in ultrasound-guided regional anesthesia. Clinical Anatomy. (2021) 34(5):802–9. doi: 10.1002/ca.23742
97. Gundry M, Knapp K, Meertens R, Meakin JR. Computer-aided detection in musculoskeletal projection radiography: a systematic review. Radiography (Lond). (2018) 24(2):165–74. doi: 10.1016/j.radi.2017.11.002
98. Langerhuizen DWG, Janssen SJ, Mallee WH, van den Bekerom MPJ, Ring D, Kerkhoffs GMMJ, et al. What are the applications and limitations of artificial intelligence for fracture detection and classification in orthopaedic trauma imaging? A systematic review. Clin Orthop Relat Res. (2019) 477(11):2482–91. doi: 10.1097/CORR.0000000000000848
99. MURA Dataset: Towards Radiologist-Level Abnormality Detection in Musculoskeletal Radiographs. Available at: https://stanfordmlgroup.github.io/competitions/mura/ (Cited June 15, 2023).
100. Diagnostic Imaging. Gleamer’s BoneView Gains FDA Clearance for AI-Powered Pediatric Fracture Detection (2023). Available at: https://www.diagnosticimaging.com/view/gleamer-boneview-fda-clearance-for-ai-pediatric-fracture-detection (Cited Jun 15, 2023).
101. Greulich WW, Pyle SI. Radiographic atlas of skeletal development of the hand and wrist. Stanford, CA: Stanford University Press (1959). p. 288.
102. Tanner JM, Healy MJR, Cameron N, Goldstein H. Assessment of skeletal maturity and prediction of adult height (TW3 method). London, UK: W.B. Saunders (2001). p. 110.
103. Ferizi U, Besser H, Hysi P, Jacobs J, Rajapakse CS, Chen C, et al. Artificial intelligence applied to osteoporosis: a performance comparison of machine learning algorithms in predicting fragility fractures from MRI data. J Magn Reson Imaging. (2019) 49(4):1029–38. doi: 10.1002/jmri.26280
104. Ferizi U, Honig S, Chang G. Artificial intelligence, osteoporosis and fragility fractures. Curr Opin Rheumatol. (2019) 31(4):368–75. doi: 10.1097/BOR.0000000000000607
105. Galbusera F, Casaroli G, Bassani T. Artificial intelligence and machine learning in spine research. JOR Spine. (2019) 2(1):e1044. doi: 10.1002/jsp2.1044
106. NuVasive. The Pulse platform. Available at: https://www.nuvasive.com/surgical-solutions/pulse-3/ (Cited July 18, 2023).
107. Miskin N, Gaviola GC, Huang RY, Kim CJ, Lee TC, Small KM, et al. Intra- and intersubspecialty variability in lumbar spine MRI interpretation: a multireader study comparing musculoskeletal radiologists and neuroradiologists. Curr Probl Diagn Radiol. (2020) 49(3):182–7. doi: 10.1067/j.cpradiol.2019.05.003
108. Lassau N, Estienne T, de Vomecourt P, Azoulay M, Cagnol J, Garcia G, et al. Five simultaneous artificial intelligence data challenges on ultrasound, CT, and MRI. Diagn Interv Imaging. (2019) 100(4):199–209. doi: 10.1016/j.diii.2019.02.001
109. Keles E, Irmakci I, Bagci U. Musculoskeletal MR image segmentation with artificial intelligence. Adv Clin Radiol. (2022) 4(1):179–88. doi: 10.1016/j.yacr.2022.04.010
110. Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. (2016) 278(2):563–77. doi: 10.1148/radiol.2015151169
111. van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging—“how-to” guide and critical reflection. Insights Imaging. (2020) 11:91. doi: 10.1186/s13244-020-00887-2
112. Khalvati F, Zhang Y, Wong A, Haider MA. Radiomics. In: Narayan R, editors. Encyclopedia of biomedical engineering. Oxford: Elsevier (2019). p. 597–603. Available at: https://www.sciencedirect.com/science/article/pii/B9780128012383999641 (Cited July 18, 2023).
113. McBee MP, Awan OA, Colucci AT, Ghobadi CW, Kadom N, Kansagra AP, et al. Deep learning in radiology. Acad Radiol. (2018) 25(11):1472–80. doi: 10.1016/j.acra.2018.02.018
114. Fritz B, Yi PH, Kijowski R, Fritz J. Radiomics and deep learning for disease detection in musculoskeletal radiology : an overview of novel MRI- and CT-based approaches. Invest Radiol. (2023) 58(1):3–13. doi: 10.1097/RLI.0000000000000907
115. Klontzas ME, Triantafyllou M, Leventis D, Koltsakis E, Kalarakis G, Tzortzakakis A, et al. Radiomics analysis for multiple myeloma: a systematic review with radiomics quality scoring. Diagnostics. (2023) 13(12):2021. doi: 10.3390/diagnostics13122021
116. Zhan H, Teng F, Liu Z, Yi Z, He J, Chen Y, et al. Artificial intelligence aids detection of rotator cuff pathology: a systematic review. Arthroscopy. (2023):S0749-8063(23)00471-1. doi: 10.1016/j.arthro.2023.06.018
117. Kapiński N, Zieliński J, Borucki BA, Trzciński T, Ciszkowska-Łysoń B, Zdanowicz U, et al. Monitoring of the achilles tendon healing process: can artificial intelligence be helpful? Acta Bioeng Biomech. (2019) 21(1):103–11.
118. McKendrick M, Yang S, McLeod GA. The use of artificial intelligence and robotics in regional anaesthesia. Anaesthesia. (2021) 76(Suppl 1):171–81. doi: 10.1111/anae.15274
119. Ghasseminia S, Lim AKS, Concepcion NDP, Kirschner D, Teo YM, Dulai S, et al. Interobserver variability of hip dysplasia indices on sweep ultrasound for novices, experts, and artificial intelligence. J Pediatr Orthop. (2022) 42(4):e315–23. doi: 10.1097/BPO.0000000000002065
120. Hannun A, Case C, Casper J, Catanzaro B, Diamos G, Elsen E, et al. Deep Speech: Scaling up end-to-end speech recognition. arXiv (2014). Available at: http://arxiv.org/abs/1412.5567 (Cited June 18, 2023).
121. Do BH, Wu AS, Maley J, Biswal S. Automatic retrieval of bone fracture knowledge using natural language processing. J Digit Imaging. (2013) 26(4):709–13. doi: 10.1007/s10278-012-9531-1
122. Tan WK, Hassanpour S, Heagerty PJ, Rundell SD, Suri P, Huhdanpaa HT, et al. Comparison of natural language processing rules-based and machine-learning systems to identify lumbar spine imaging findings related to low back pain. Acad Radiol. (2018) 25(11):1422–32. doi: 10.1016/j.acra.2018.03.008
123. Mozayan A, Fabbri AR, Maneevese M, Tocino I, Chheang S. Practical guide to natural language processing for radiology. RadioGraphics. (2021) 41(5):1446–53. doi: 10.1148/rg.2021200113
124. Pons E, Braun LMM, Hunink MGM, Kors JA. Natural language processing in radiology: a systematic review. Radiology. (2016) 279(2):329–43. doi: 10.1148/radiol.16142770
125. Carrodeguas E, Lacson R, Swanson W, Khorasani R. Use of machine learning to identify follow-up recommendations in radiology reports. J Am Coll Radiol. (2019) 16(3):336–43. doi: 10.1016/j.jacr.2018.10.020
Keywords: artificial intelligence, machine learning, neural networks, musculoskeletal imaging, image interpretation, automation
Citation: Debs P and Fayad LM (2023) The promise and limitations of artificial intelligence in musculoskeletal imaging. Front. Radiol. 3:1242902. doi: 10.3389/fradi.2023.1242902
Received: 19 June 2023; Accepted: 26 July 2023;
Published: 7 August 2023.
Edited by:
Brandon K. K. Fields, University of California, San Francisco, United StatesReviewed by:
Ajit Mahale, KMC MANGALORE MAHE Manipal India, IndiaMichał Strzelecki, Lodz University of Technology, Poland
© 2023 Debs and Fayad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura M. Fayad bGZheWFkMUBqaG1pLmVkdQ==
Abbreviations ACL, anterior cruciate ligament; AI, artificial intelligence; AUC, area under the curve; BMD, bone mineral density; CNN, convolutional neural network; CT, computed tomography; DL, deep learning; DXA, dual-energy x-ray absorptiometry; ML, machine learning; MRI, magnetic resonance imaging; MSK, muscuskeletal; PACS, picture archiving and communication systems.
 Patrick Debs
Patrick Debs Laura M. Fayad1,2,3*
Laura M. Fayad1,2,3*