- 1Department of Data and Information, The Children’s Hospital Zhejiang University School of Medicine, Hangzhou, China
- 2Medicine Engineering and Information Research Institute for Children's Health, Sino-Finland Joint AI Laboratory for Child Health of Zhejiang Province, Hangzhou, China
- 3Medicine Engineering and Information Research Institute for Children's Health, National Clinical Research Center for Child Health, Hangzhou, China
- 4Department of Scientific Research, Huiying Medical Technology (Beijing) Co., Ltd, Beijing, China
- 5Department of Neurology, The Warren Alpert Medical School of Brown University, Providence, RI, United States
- 6Department of Scientific Research, Avaintec Oy Company, Helsinki, Finland
- 7Department of Radiology, Children’s Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 8Department of Information Standardization Research,National Institute of Hospital Administration, NHC, Beijing, China
- 9Polytechnic Institute, Zhejiang University, Hangzhou, China
Background: Chest x-ray (CXR) is widely applied for the detection and diagnosis of children's lung diseases. Lung field segmentation in digital CXR images is a key section of many computer-aided diagnosis systems.
Objective: In this study, we propose a method based on deep learning to improve the lung segmentation quality and accuracy of children's multi-center CXR images.
Methods: The novelty of the proposed method is the combination of merits of TransUNet and ResUNet. The former can provide a self-attention module improving the feature learning ability of the model, while the latter can avoid the problem of network degradation.
Results: Applied on the test set containing multi-center data, our model achieved a Dice score of 0.9822.
Conclusions: This novel lung segmentation method proposed in this work based on TransResUNet is better than other existing medical image segmentation networks.
1. Introduction
With the development of modern medicine, the importance of medical imaging has become increasingly prominent. Among routine imaging examinations, x-ray examination, especially chest film examination, occupies a large proportion in children's photography due to its advantages of low dose radiation, convenient examination and low cost, accounting for more than 40% of all imaging diagnoses, of which infant chest photography accounts for more than 60% of the whole pediatric x-ray examination (1, 2). Therefore, chest x-ray has become one of the most commonly used medical diagnostic methods to detect children's lung diseases. Lung segmentation is the process of accurately identifying lung field regions and boundaries from surrounding thoracic tissue and is therefore an important first step in lung image analysis for many clinical decision support systems, such as detection and diagnosis of Covid-19 (3–5), classification of pneumonia (6), detectionof tuberculosis (7), etc. Correct identification of lung fields enables further computational analysis of these anatomical regions, such as extraction of clinically relevant features to train a machine learning algorithm for the detection of disease and abnormalities. These computational methodologies can assist physicians with making a timely, accurate medical diagnosis to improve the quality of care and outcome for patients. Lung diseases are the leading cause of death among children in many countries, mainly including bronchiolitis, bronchitis, bronchopneumonia, interstitial pneumonia, lobar pneumonia and pneumothorax. But the diagnosis of these diseases relies on the precise segmentation of the lungs in children's chest x-rays (9). Due to the immature development of the anatomical and physiological structures of preschool children, and the common thymus shadow and transverse heart in infants and young children, children's chest x-ray images generally have image problems such as the unclear boundary of lung field and severe inter-organization interference, which brings great difficulties to lung tissue segmentation (8, 9). Therefore, improving the segmentation quality and accuracy of children's lung x-ray images is of great significance to improve the diagnosis of chest x-ray film.
Despite the presence of many variations in medical image segmentation methods, these methods are broadly divided into the following five categories: medical image segmentation based on region growing arithmetic, medical image segmentation based on the deformation models, medical image segmentation based on thresholds, medical image segmentation based on graph theory, and medical image segmentation based on artificial intelligence. The traditional image segmentation method relies on artificial means to extract and select information such as edges, colors, and textures in the image. In addition to consuming considerable energy resources and workers' time, it also requires a high degree of expertise to extract useful feature information, which no longer meets the practical application requirements of medical image segmentation and recognition (10–13). With the increasing requirements for the performance of segmentation in medical imaging in recent years, Deep learning has already been extensively applied to segmentation in medical imaging. The U-Net (14) proposed in 2015 demonstrates the advantages of accurate segmentation of small targets and its scalable architecture based on encoder-decoder architecture. It achieved significant success in medical image segmentation. Many researchers have made improvements on its basis. Ummadi V (15) summarized the application of UNet and its variants in the field of medical image segmentation, including UNet++ (16), R2UNet (17), AttUNet (18), etc. In addition, Literature (19–22) also proposed or use UNet+++, ResUNet, ResUNet+, DenseResUNet, etc. for medical image segmentation. Literature et al. (23, 24) proposed 3D UNet and VNet for segmenting 3D medical images and achieved good performance. Recently, Vision transformers have achieved decent performance on computer vision tasks. Dosovitskiy et al. (25) use a pure transformer to achieve state-of-the-art performance on image segmentation data. Literature (26–29) proposed transformer-based models for the task of image segmentation. Chen et al. (27) proposed TransUNet by combining UNet and transformer for medical image segmentation. Literature (28, 29) proposed TransBTS and UNETR, which are based transformers, for 3D medical image segmentation.
In this study, we developed a novel method based on TransResUNet for the segmentation of lung areas from chest x-ray images. The TransResUNet utilized here can extract more valid information at the encoder stage and makes use of self-attention from the input feature maps. In addition, we also employ gray-scale truncation to normalize the data, and ecological operations to optimize the segmentation of the model. Experimental results on the children's x-ray dataset demonstrate the superiority of our method against other existing methods for lung segmentation.
2. Materials and methods
The development of the proposed model mainy includes three steps. Firstly, the image preprocessing step is to normalize the collected x-ray images. The second step is the training of the lung segmentation model based on the TransResUNet with TransUNet (27) and ResUNet (30) as the core to obtain the lung area. Then, in the image post-processing step, the segmentation performance of the model can be further improved.
2.1. Image preprocessing
CXR images used to train the segmentaion model in this work were collected from different machines with different parameter settings, which resulted in the inconsistent pixel value ranges of these CXR images. If the normalization operation is used directly to these unprocessed CXR images, there will be great differences in the grayscale of the processed images, as shown in Figure 1A.
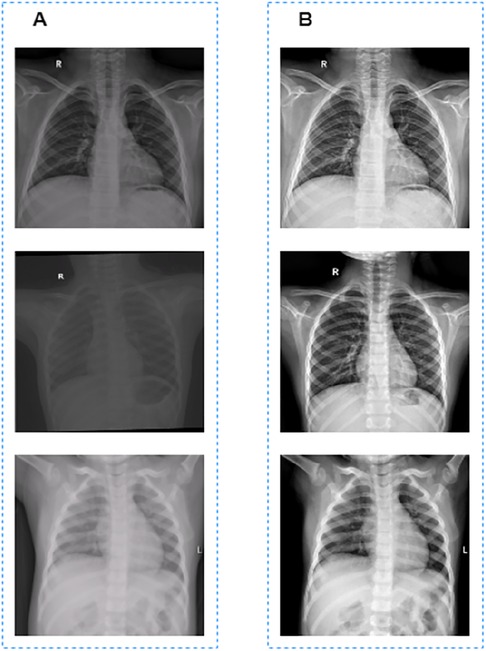
Figure 1. Image preprocessing results. (A) Images before grayscale truncation. (B) Images after grayscale truncation.
To solve this problem, we used the grayscale truncation method to preprocess the images. First, for an image with the size of x*y, we selected the 1/4 central area of the image, that is . Then the maximum and minimum values of the central area were obtained. The whole image were then grayscale truncated use [] with the formula as follows:
The grayscale truncated image is shown in Figure 1B. Finally, the truncated images were normalized to [0, 1] for model training and validation.
2.2. Model traning
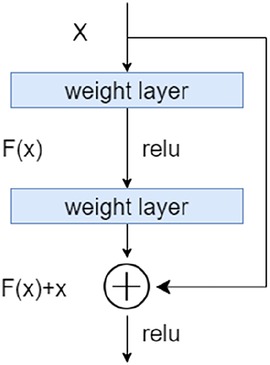
Our TransResUNet model combined TransUNet and ResUNet, which were separately reported in previous works (27, 30), and further refined the combination. The original TransUNet was an encoder-decoder architecture that integrate the advantages of both Transformers and UNet. This model used CNN-Transformer as an encoder to extract global contexts and then used a UNet decoder to achieve precise localization. The TransUNet model showed satisfying performances in many medical image segmentation tasks (31, 32). The original ResUNet was a semantic segmentation neural network that combines the strengths of residual learning and UNet. This model used residual units to ease the training of deep networks. Here, we proposed a new encoder-decoder architecture called TransResUNet which equipped the classical TransUNet with residual learning unit. The model is shown in Figure 2.
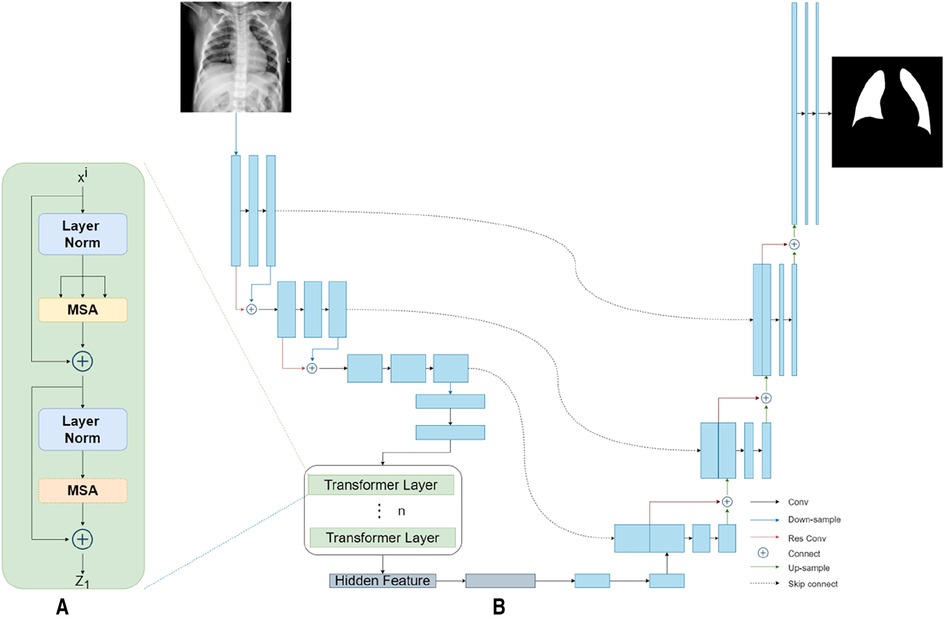
Figure 2. The architecture of TransResUNet. (A) Depicts the deconstruction schematic of Transformer Layer. (B) Depicts the proposed model.
2.2.1. The residual learning
To address the degradation problem in deep networks, we used a Res learning block before each downsampling and upsampling. As shown in Figure 3, the Res learning block connected neurons in non-adjacent layers by passing the neurons of the adjacent layers, thereby weakened the strong connections between the adjacent layers.
2.2.2. CNN-Transformer hybrid as encoder
Different from the common transformer model, the CNN-Transformer hybrid model was used here as the encoder. Firstly, we used the ResUNet downsampling the input image three times continuously to obtain the corresponding feature matrix . The feature matrix were then flattened into a sequence of 2D patches according to a reported method (25). Afterwards, in order to obtain the spatial information between patches, the location information of those 2D patches were embeded. Finally, the patches with position information were inputted into the transformer encoder, which contains 12 transformer layers with each layer composing of Multihead Self-Attention (MSA) and Multi-Layer Perceptron (MLP) blocks. The output of the transformer encoder was the feature matrix .
2.2.3. ResUNet as decoder
In the model reported here, ResUNet was chosen as Decoder. The feature matrix and were concatenated and then unsampled twice. The feature matrix obtained from each upsampling process was concatenated with the downsampling matrix of the same size. Finally, the prediction result of each point in the input image were obtained by the segmentation model through the fully connection layer.
2.3. Image post-processing
There are two major problems in the segmentation results outputted directly from the model (Figure 4A): one is that the non-lung areas with gray levels close to that of the lung areas were identified as lungs; the other is that a small section in the lung area could not be recognized as lungs by the model. In order to further optimize the segmentation results, we removed the misidentified part by connected domain, and then used the ecological opening operation to complete the missing section of the whole lung filed. The final segamentation result after post-processing is shown in Figure 4B.
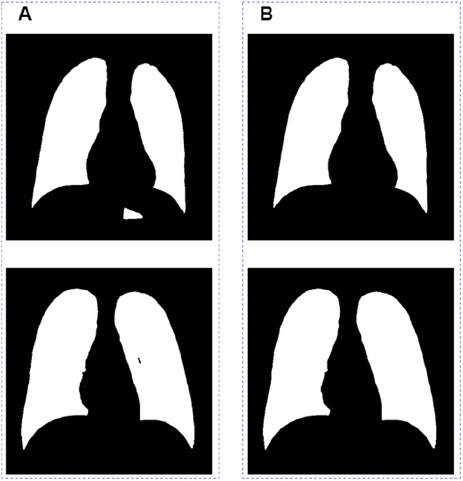
Figure 4. Image post-processing results. (A) Images before grayscale truncate. (B) Images after grayscale truncate.
3. Results and discussion
3.1. Datasets
3.1.1. X-ray scanning method
GE Definium 6000 machine was used to scan the lungs. The infants are generally photographed in a lying position. The patient is placed in a supine front-back position, with the head advanced, and the arms abducted at right angles to the body. One attendant stands on the side of the child's head and gently presses the two upper arms, the other attendant stands on the side of the child's feet and presses the abdomen and knees. The target film distance shall be 90 cm, and no filter shall be used. The older children are photographed in a standing position. The child shall be in a standing front-back position, the upper jaw is slightly raised, and the arms are abducted and placed on both sides of the body. The attendants stand on both sides of the patient, fix the child's arms and then take an x-ray. The upper end of the film shall exceed the shoulder by 2 fingers, and the target film distance shall be 180 cm. No filter shall be used. During the filming, both the patients and the attendants should take necessary protection. Before scanning, pay attention to removing metal ornaments and foreign objects to avoid artifacts.
3.1.2. X-ray scanning parameters
GE Definium 6000 machine was used with the horizontal photography parameters set at 55 kV, 100 mA, 6.3 mAs, 31.5 mSec, and the vertical photography parameters set at 60 kV, 300 mA, 6.3 mAs, 31.5 mSec.
3.1.3. Data set
In order to verify the performance of the proposed model, we conducted a method test on the chest x-ray image dataset of children. All data comes from the Children's Hospital of Zhejiang University School of Medicine. The children's data set includes a total of 1,158 cases. Manual segmentation was performed by two physicians with 5 years of clinical experience, and the segmentation results were used as ground truch
3.2. Evaluation metrics
We use the accuracy AC, sensitivity SE, specificity SP of the lung region and Dice coefficient (DI) to evaluate the accuracy of segmentation results. The first three are calculated by four variables: true positive TP, true negative TN, false positive FP, and false negative FN. The calculation formula of the Dice coefficient is as follows:
where e represents the ground truth and f represents the segmentation result.
3.3. Implementation details
We implement model in PyTorch. All training, testing and verification experiments are completed on Ubuntu 16.04 server. The basic configuration is: CPU Intel E5-1650 3.50 GHz, 64G DDR4 memory, the graphics card is RTX 2080Ti. All annotation work is done in RadCloud (Huiying Medical Technology (Beijing) Co., Ltd). All models were trained with the batch size of 2, using the RMSprop optimizer (33) with an initial learning rate of 0.001 for 20 epochs. All images were reshaped to 512*512. We use R50-ViT-B_16 (25) architecture with L = 12 layers and an embedding size of K = 768. We split the data into train, validation and test with a ratio of 70:10:20, and their numbers are 810, 116, and 232, respectively.
3.4. Quantitative evaluations
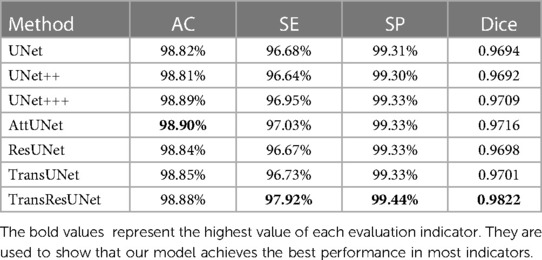
We selected UNet (14), UNet++ (16), UNet+++ (19), AttUNet (18), ResUNet (32), TransUNet (27), and TransResUNet for training and testing on the dataset. As shown in Table 1, TransResUNet outperforms the state-of-the-art methods in our test datasets. The overall average Dice score of TransResUNet is 98.02% which outperforms the second by 1.06%. The overall average AC, SE and SP of all models are very close because the lung area has a certain grayscale difference from the surrounding area. Nonetheless, TransResUNet achieves the highest scores on both SE and SP, indicating that it performs well for the lung segmentation task.
3.5. Qualitative results
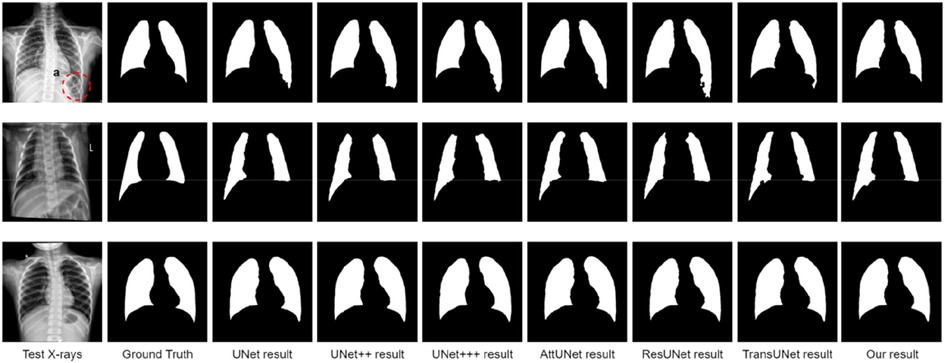
Qualitative lung segmentation comparisons are presented in Figure 5. TransResUNet shows improved lung segmentation performance. In Figure 5, the first row and second row demonstrate a clear segmentation against surrounding tissues. For example, the area (Figure 5A) below the lungs in the first row indicates that TransResUNet has sufficient learning for global information. For areas with clear lung edges, such as the third rows in Figure 5, the segmentation performance of each model is relatively close. However, like other models, our model is not smooth at the edges because the edges of the lungs are occluded by the bright ribs, but this does not reflect the segmentation performance of the main lung regions.
4. Conclusion
In this work, a novel lung segmentation method based on TransResUNet in chest x-ray images has been reported. First of all, we truncated the raw x-ray images with the central region gray value range of the images to reduce the inconsistence caused by different machine acquisitions settings. When developing the segmentation model, we used the encoder part of ResUNet to replace the encoder part of classical TransUNet, thereby improving the feature extraction ability of the model in the downsampling stage. Finally, ecological operations were used to further optimize the results of segmentation. The experimental results demonstrated that the methods of this paper outperform the original TransUNet model and other reviously reported models. In the future, we will consider eliminating the rib region before segmentation, so as to improve the segmentation performance of the model, so that the model can be better applied to clinical diagnosis and research on other lung diseases.
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding authors.
Ethics statement
This study was approved by the Medical Ethics Committee of Children's Hospital (Approval Letter of IRB/EC, 2020-IRB-058) and waived the need for written informed consent from patients, as long as the data of the patient remained anonymous. All of the methods were carried out in accordance with the Declaration of Helsinki. Written informed consent to participate in this study was provided by the participants’ legal guardian/next of kin. Written informed consent was not obtained from the individual(s), nor the minor(s)' legal guardian/next of kin, for the publication of any potentially identifiable images or data included in this article.
Author contributions
GY and LC made great contributions on proposing and designing the integration of AI and medicine. JH and ZY primarily focused on the AI research and applications concerning lung segmentation. LS investigated and summarized the reviewed studies using active learning framework. ZL and CS was primarily responsible for collecting related research and preparing scrub data. PK, JL, TS, CS, and XM co-wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was partially supported by the National Key R&D Program of China (grant number 2019YFE0126200) and the National Natural Science Foundation of China (grant number 62076218).
Conflict of interest
ZY was employed by Huiying Medical Technology (Beijing) Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. An FP, Liu ZW. Medical image segmentation algorithm based on feedback mechanism CNN. Contrast Media Mol Imaging. (2019) 2019:13. doi: 10.1155/2019/6134942
2. Antani S. Automated detection of lung diseases in chest X-rays[C]. A Report to the Board of Scientific Counselors, Bethesda, The United States of America (2015). p. 1-21. Available at: https://lhncbc.nlm.nih.gov/system/files/pub9126.pdf.
3. Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Kashem SBA, et al. Exploring the effect of image enhancement techniques on COVID-19 detection using chest x-ray images. Comput Biol Med. (2021) 132:104319. doi: 10.1016/j.compbiomed.2021.104319
4. Vidal PL, de Moura J, Novo J, Ortega M. Multi-stage transfer learning for lung segmentation using portable x-ray devices for patients with COVID-19. Expert Syst Appl. (2021) 173:114677. doi: 10.1016/j.eswa.2021.114677
5. Teixeira LO, Pereira RM, Bertolini D, Oliveira LS, Nanni L, Cavalcanti GDC, et al. Impact of lung segmentation on the diagnosis and explanation of COVID-19 in chest x-ray images. Sensors. (2021) 21(21):7116. doi: 10.3390/s21217116
6. Ikechukwu AV, Murali S, Deepu R, Shivamurthy RC. ResNet-50 vs VGG-19 vs training from scratch: a comparative analysis of the segmentation and classification of pneumonia from chest x-ray images. Global Transitions Proceedings. (2021) 2(2):375–81. doi: 10.1016/j.gltp.2021.08.027
7. Ayaz M, Shaukat F, Raja G. Ensemble learning based automatic detection of tuberculosis in chest x-ray images using hybrid feature descriptors. Phys Eng Sci Med. (2021) 44(1):183–94. doi: 10.1007/s13246-020-00966-0
8. Jaeger S, Karargyris A, Candemir S, Folio L, Sielgelman J, Callaghan F, et al. Automatic tuberculosis screening using chest radiographs. IEEE Trans Med Imaging. (2014) 33(2):233–45. doi: 10.1109/TMI.2013.2284099
9. Chen KC, Yu HR, Chen WS, Lin WC, Lu HS. Diagnosis of common pulmonary diseases in children by x-ray images and deep learning. Sci Rep. (2020) 10:1–9. doi: 10.1038/s41598-019-56847-4
10. Xue Y, Xu T, Zhang H, Long R, Huang XL. Segan: adversarial network with multi-scale l 1 loss for medical image segmentation. Neuroinformatics. (2018) 16(6):1–10. doi: 10.1007/s12021-018-9377-x
11. Zhang WL, Li RJ, Deng HT, Wang L, Lin WL, ji SW, et al. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage. (2015) 108:214–24. doi: 10.1016/j.neuroimage.2014.12.061
12. Shen D, Wu G, Suk HI. Deep learning in medical image analysis. Annu Rev Biomed Eng. (2017) 19(1):221–48. doi: 10.1146/annurev-bioeng-071516-044442
13. Reamaroon N, Sjoding MW, Derksen H, Sabeti E, Gryak J, Barbaro RP, et al. Robust segmentation of lung in chest x-ray: applications in analysis of acute respiratory distress syndrome. BMC Med Imaging. (2020) 20:116. doi: 10.1186/s12880-020-00514-y
14. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. Medical Image Computing and Computer- AssistedIntervention-MICCAl2015-18th International Conference Munich Germany. Munich: Springer International Publishing (2015).
15. Ummadi V. U-Net and its variants for medical image segmentation : a short review[J]. Computer Vision and Pattern Recognition (cs.CV) (2022). arXiv preprint arXiv: 2204.08470. doi: 10.48550/arXiv.2204.08470
16. Zhou Z, Siddiquee M, Tajbakhsh N, Liang J. Unet++: a nested UNet architecture for medical image segmentation[J]. DLMIA, Granada, Spain (2018) 11045:3-11. doi: doi: 10.1007/978-3-030-00889-5_1
17. Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation [J]. J. Med. Imaging. (2018) 6(1):014006. doi: 10.1117/1.JMI.6.1.014006
18. Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: learning where to look for the pancreas[J]. Medical Imaging with Deep Learning, Amsterdam, Netherlands (2018). doi: 10.48550/arXiv.1804.03999
19. Huang H, Lin L, Tong R, Hu H, Zhang QW, Iwamoto Y, et al. UNet 3+: a full-scale connected UNet for medical image segmentation. arXiv (2020).
20. Liu Z, Yuan H. An res-unet method for pulmonary artery segmentation of CT images. J Phys Conf Ser. (2021) 1924(1):012018. doi: 10.1088/1742-6596/1924/1/012018
21. Zhang TT, Jin PJ. Spatial-temporal map vehicle trajectory detection using dynamic mode decomposition and Res-UNet+ neural networks[J]. Computer Vision and Pattern Recognition (cs.CV), (2021). arXiv preprint arXiv:2201.04755. doi: doi: 10.48550/arXiv.2201.04755
22. Kiran I, Raza B, Ijaz A, Khan MA. DenseRes-Unet: segmentation of overlapped/clustered nuclei from multi organ histopathology images. Comput Biol Med. (2022) 143:105267. doi: 10.1016/compbiomed2022.105267
23. Çiçek Ö, zgün Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. Cham: Springer (2016).
24. Milletari F, Navab N, Ahmadi SA. V-Net: fully convolutional neural networks for volumetric medical image segmentation[C]. 2016 Fourth International Conference on 3D Vision (3DV), Stanford, The United States of America (2016) p. 565-571. doi: 10.1109/3DV.2016.79
25. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai XH, Unterthiner T. An image is worth 16x16 words: transformers for image recognition at scale[C]. International Conference on Learning Representations (ICLR), ElECTR NETWORK (2021). p. 1-22. doi: 10.48550/arXiv.2010.11929
26. Zheng S, Lu J, Zhao Hs, Zhu XT, Luo ZK, Wang YB, et al. Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers[C]. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), ElECTR NETWORK (2021). p. 6877-6886. doi: 10.1109/CVPR46437.2021.00681
27. Chen JN, Lu YY, Yu QH, Luo XD, Adeli E, Wang Y, et al. TransUNet: transformers make strong encoders for medical image segmentation[J]. Computer Vision and Pattern Recognition (cs.CV) (2021). arXiv preprint arXiv:2201.04755. pp: 1-13. doi: doi: 10.48550/arXiv.2102.04306
28. Valanarasu J, Oza P, Hacihaliloglu I, Patel VM. Medical transformer: gated axial-attention for medical image segmentation[C]. International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), ElECTR NETWORK (2021) (12901): 36-46. doi: 10.1007/978-3-030-87193-2_4
29. Zhang Y, Liu H, Hu Q. TransFuse: fusing transformers and CNNs for medical image segmentation[C]. International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), ElECTR NETWORK (2021) (12901): 14-24. doi: 10.1007/978-3-030-87193-2_2
30. Wang W, Chen C, Ding M, Yu H, Zha S, Li JY. TransBTS: multimodal brain tumor segmentation using transformer[C]. Medical Image Computing and Computer Assisted Intervention-MICCAI 2021, Strasbourg, France (2021) :109-119. doi: 10.1007/978-3-030-87193-2_11.
31. Hatamizadeh A, Tang YC, Nath V, Yang D, Myronenko A, Landman B, et al. UNETR: transformers for 3D medical image segmentation[C]. 22nd IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI (2022) : 1748-1758. doi: 10.1109/WACV51458.2022.00181
32. Zhang Z, Liu Q, Wang Y. Road extraction by deep residual UNet[J]. IEEE Geoscience and Remote Sensing Letters (2018) 15(5): 749-753. doi: 10.1109/LGRS.2018.2802944
Keywords: children, lung segmentation, TransResUNet, chest x-ray, multi-center
Citation: Chen L, Yu Z, Huang J, Shu L, Kuosmanen P, Shen C, Ma X, Li J, Sun C, Li Z, Shu T and Yu G (2023) Development of lung segmentation method in x-ray images of children based on TransResUNet. Front. Radiol. 3:1190745. doi: 10.3389/fradi.2023.1190745
Received: 21 March 2023; Accepted: 6 April 2023;
Published: 27 April 2023.
Edited by:
Zhicheng Zhang, ByteDance, ChinaReviewed by:
Xiaokun Liang, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences (CAS), ChinaXinping Shao, Hangzhou Dianzi University, China
© 2023 Chen, Yu, Huang, Shu, Kuosmanen, Shen, Ma, Li, Sun, Li, Shu and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Shu bmN0aW5ndGluZ0AxMjYuY29t Gang Yu eXVnYm1lQHpqdS5lZHUuY24=
†These authors have contributed equally to this work
 Lingdong Chen
Lingdong Chen Zhuo Yu
Zhuo Yu Jian Huang
Jian Huang Liqi Shu
Liqi Shu Pekka Kuosmanen2,6
Pekka Kuosmanen2,6 Chen Shen
Chen Shen Xiaohui Ma
Xiaohui Ma