- Department of Biochemistry & Microbiology, North South University, Dhaka, Bangladesh
Citrobacter spp. are Gram-negative, non-spore forming, rod-shaped, facultative anaerobic bacteria from the Enterobacteriaceae family often found in soil, sewage, sludge, water, food, and the intestinal tracts of animals and humans. Several members of Citrobacter spp. especially C. freundii, C. koseri, C. braakii are frequently detected in newborn illnesses, urinary tract infections, and patients with severe underlying conditions, including hypertension, diabetes, cancer, and respiratory infections, or those who are immunocompromised. Strains of Citrobacter spp. can spread vertically or horizontally from carriers or other hospital sources and thus cause nosocomial infections in hospital settings. A total of 19 Citrobacter genomospecies have been recognized based on genomics. It has been noted that the Citrobacter genus acquired antimicrobial resistance and virulence, including invasion, colonization, biofilm formation, and toxin production. The recent emergence and spread of antimicrobial resistance to β-lactams, carbapenems, fluoroquinolones, aminoglycosides, and colistin in Citrobacter spp. through chromosomal and plasmid-mediated resistance limits the empiric treatment options. Therefore, combination therapy involving costly and potentially hazardous antibiotics poses significant challenges in treating Citrobacter infections. Here we summarized the nomenclature of Citrobacter spp., clinical manifestations, epidemiology, pathogenesis, antibiotic resistance mechanisms, and treatments from various clinical samples. This review will expand our knowledge of the genomics and epidemiology of Citrobacter spp., enabling improved control of infections and the spread of these organisms.
Introduction
Citrobacter spp. accounts for 3-6% of all isolates from the Enterobacteriaceae family, which causes nosocomial infections (Doran, 1999; Borenshtein and Schauer, 2006; Maraki et al., 2017; Rostamzad et al., 2019; Nair et al., 2020; Aguirre-Sánchez et al., 2023; Heljanko et al., 2023) Citrobacter spp. are found in soil, sewage sludge water, food, and the intestinal tracts of animals and humans (Cong’En et al., 2014; Forsythe et al., 2015). Citrobacter is considered an opportunistic nosocomial pathogen that is commonly associated with urinary tract infections (UTIs), bloodstream infections, intra-abdominal sepsis, brain abscesses, pneumonia, and other neonatal infections such as meningitis, neonatal sepsis, joint infections, or common bacteremia (Ashish et al., 2012; Maraki et al., 2017; Hawaldar and Sadhna, 2019; Räisänen et al., 2021). It is evident that the two prominent opportunistic pathogens, C. koseri and C. freundii, account for most of the Citrobacter infections where more than 80% of patients were found to have underlying medical conditions including diabetes, cardiovascular disease, renal disease, leukemia, neurologic disease, or abnormalities of the urinary tracts (Mohanty et al., 2007; Liu et al., 2018c; Chen and Ji, 2019; Lalaoui et al., 2019; Lee et al., 2019; Khan et al., 2020; Dominguez Céspedes and Céspedes Fonseca, 2022; Hua et al., 2022; Ramachandran et al., 2022).
Citrobacter spp., mainly C. freundii, has started to cause various diseases, and they are also becoming increasingly resistant to several types of antibiotics (Liu et al., 2017). C. freundii is often considered more resistant than C. koseri to β-lactam antibiotics, including amoxicillin, amoxicillin-clavulanate, ampicillin, first- and second-generation cephalosporins. The incidence of antibiotic-resistant Citrobacter isolates has been reported increasingly worldwide(Osei Sekyere and Reta, 2021). Despite advances in diagnostic methods and antibiotic therapy, Citrobacter infections are considered fatal, with case-fatality rates of 30% and death rates of 33-48% in neonates (Pepperell et al., 2002; Hewitt et al., 2021). Surviving infants can experience significant central nervous system (CNS) damage, including severe intellectual disability, hemiplegia, and seizures. Phylogenomic research revealed that, regardless of boundaries, a single strain gradually evolved during diffusion from host to host (Osei Sekyere and Reta, 2021).
This review provides insight into the nomenclature, clinical manifestations, pathogenesis, antibiotic resistance mechanism, and possible treatment options for infections caused by clinical strains of Citrobacter spp.
Classification and nomenclature
In 1928, Braak identified two bacterial strains that could convert glycerol to trimethylene glycol without oxygen. The strains were named “Bacterium freundii” in remembrance of Freund’s 1881 discovery that found trimethylene glycol was a by-product of glycerol fermentation (Braak, 1928). Later, Werkman and Gillen proposed the genus Citrobacter in 1932 (Werkman and Gillen, 1932). As the name implies, members of the genus Citrobacter utilize citrate as the primary carbon source and produce acid and gas due to the fermentation of glucose and numerous other carbohydrates (Brenner et al., 1999). However, in the following years, many synonyms have been proposed to describe such organisms, including Escherichia freundii and Salmonella ballerup (Fritsche, 1964; Schoch et al., 2020). But finally, in 1958, the International Subcommittee on the Taxonomy of the Enterobacteriaceae agreed to recognize the name “Citrobacter freundii” for this heterogenous group of bacteria, followed by the recognition of another two groups of bacteria similar to C. freundii The first group has been named as “C. koseri,” “C. diversus” or “Levinea malonatica” and the second group has been designated as “L. amalonaticus”. In 1993, C. diversus was formally named as C. koseri, granted by the Judicial Commission of the International Committee on Systematic Bacteriology (Borenshtein and Schauer, 2006; Schoch et al., 2020).
Bacterial classification is essential for microbial diversity, diagnostic purposes, serotyping, and scientific studies. Citrobacter isolates have been identified based on biochemical and carbon source utilization tests, and detailed biotypes have been reported by Brenner et al. (Brenner et al., 1999). Taxonomically it is evident that the genus Citrobacter is most closely related to Salmonella and Escherichia coli (Katzenellenbogen et al., 2017; Schoch et al., 2020). A total of 19 Citrobacter genomospecies have been recognized based on DNA relatedness to date (Table 1; Figure 1) (Borenshtein and Schauer, 2006; Oberhettinger et al., 2020; Ribeiro et al., 2021; Wang et al., 2021). Additionally, based on the lipopolysaccharide (LPS) O antigen, a total of 43 Citrobacter O-serogroups have been elucidated and also a total of 20 chemo groups have been classified according to the sugar composition of their lipopolysaccharides (LPS) (Keleti et al., 1971; Gross and Rowe, 1974; Knirel et al., 2002). Such OPS is crucial for serological classification of bacterial strains cross-reactivity between Citrobacter and other Enterobacteriaceae spp. This, in turn, will provide further insights in understanding the mechanism of antibiotic susceptibility patterns of Citrobacter spp, to cause infections (Katzenellenbogen et al., 2017).
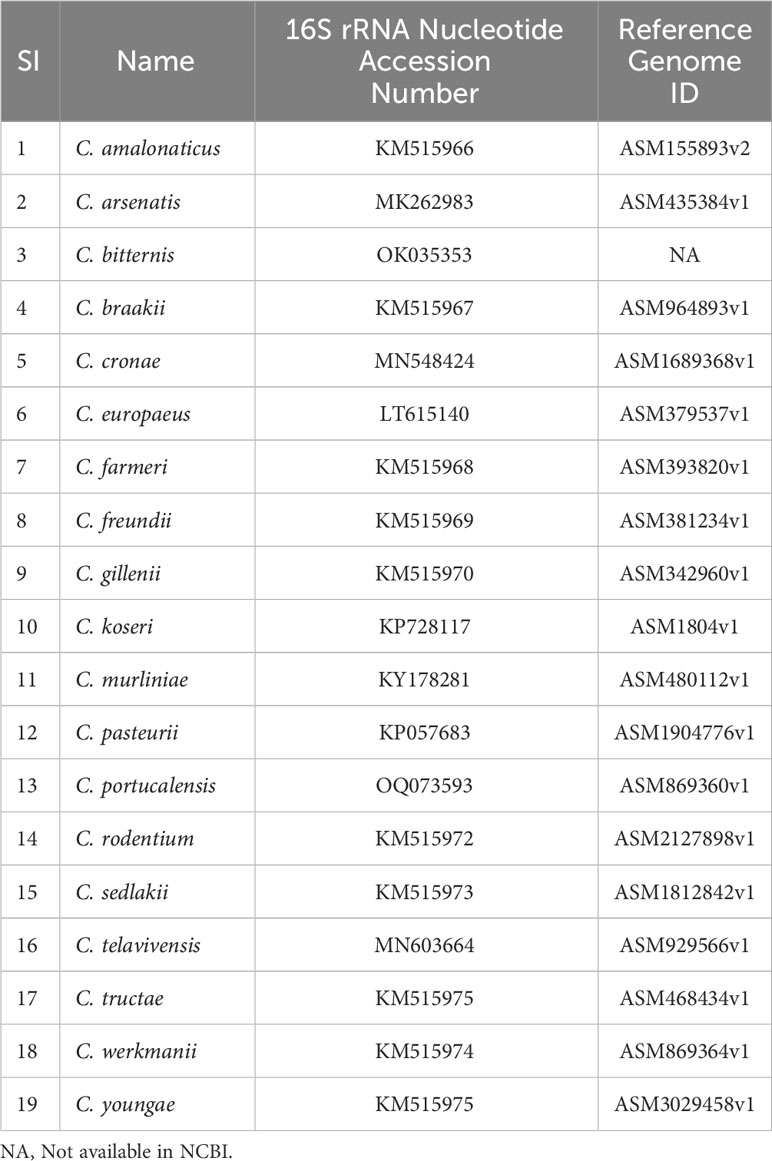
Table 1 19 Citrobacter spp. and their associated NCBI accession number of 16S rRNA gene sequences and Reference Genome ID.
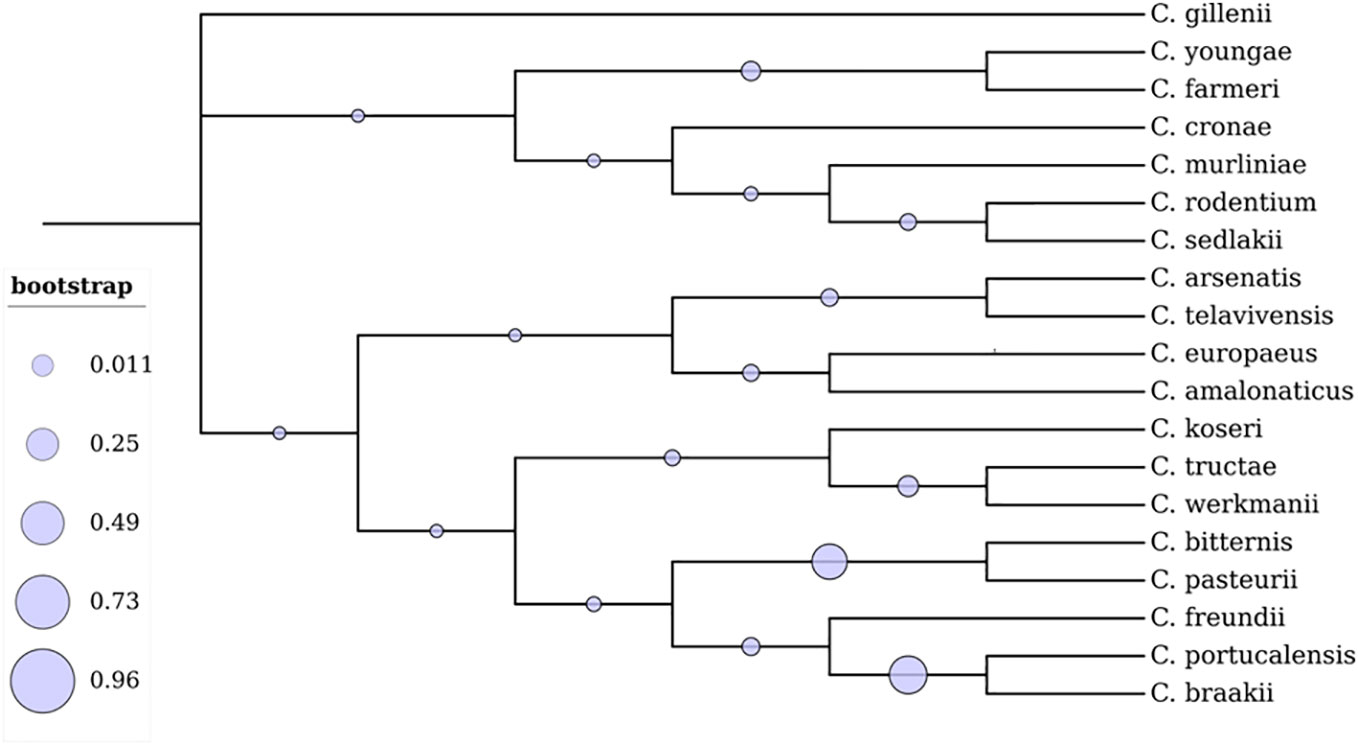
Figure 1 Phylogenetic analysis of 19 Citrobacter spp. The branches display the percentage of trees where related groups of species clustered together. The evolutionary tree was constructed using the Maximum Likelihood technique in the MEGA X software (Kumar et al., 2018). The tree was visualized using Interactive Tree of Life (iTOL).
Clinical manifestations
A number of Citrobacter spp. are opportunistic pathogens known to be the causative agent of severe infections, sepsis, respiratory infections, UTIs, keratitis, and meningitis, particularly in high-risk populations like infants and immunocompromised adults (Doran, 1999; dos Santos et al., 2015; Garcia et al., 2016; Jiménez et al., 2017; Emery et al., 2020; Urbinati et al., 2023). Furthermore, skin infections like folliculitis, cellulitis, hives, ulcers, and necrotizing fasciitis were also associated with Citrobacter spp. (Mohanty et al., 2007; Raia et al., 2015; Licata et al., 2021; Moussa et al., 2023). Among the Citrobacter spp. C. freundii and C. koseri are frequently isolated mostly from UTIs in addition to surgical wounds, respiratory infections, and gastrointestinal infections (Mohanty et al., 2007; Metri et al., 2013; Gajdács and Urbán, 2019; Hossain et al., 2021). The clinical manifestation associated with Citrobacter spp. is addressed below in detail.
Bacteremia
Bacteremia caused by Citrobacter spp. can be nosocomial or community-acquired (Doran, 1999; Lai et al., 2010; Dziri et al., 2022). Also, infections caused by Citrobacter spp. are commonly linked to polymicrobial bacteremia (Shih et al., 1996; Hashimoto et al., 2021). Diabetics, hypertension, cancer, and liver cirrhosis are the frequent underlying medical conditions associated with Citrobacter spp. infections (Lai et al., 2010; Hashimoto et al., 2021; Casas-Martínez et al., 2023). Therefore, the primary sites of infection and prognosis of Citrobacter bacteremia are still under investigation.
The frequent initial symptom of bacteremia caused by Citrobacter spp. in patients is fever with shivering. However, few individuals experience hypothermia or average body temperature (Hashimoto et al., 2021; Hua et al., 2022). A small group of patients also experienced hypotension, oliguria, and impaired mental status. Jaundice, ileus, abdominal pain, and/or gastrointestinal bleeding are other indicators of bacteremia caused by the Citrobacter spp. (Lai et al., 2010; Hewitt et al., 2021). Numerous significant health problems from Citrobacter bacteremia, including liver dysfunction, respiratory failure, renal dysfunction, and thrombocytopenia, are also reported (Shih et al., 1996). Additionally, C. koseri has been recently isolated from transfusion-related bacteremia and keratitis (Emery et al., 2020; Urbinati et al., 2023).
Meningitis
The role of Citrobacter spp. in infants and adult meningitis following central nervous system (CNS) abscesses is well documented (Doran, 1999; Cuadros et al., 2013; Chen and Ji, 2019; Hewitt et al., 2021; Bonasoni et al., 2022). C. koseri has a significant preference for the CNS in the first two months of life, whereas it also frequently causes meningitis and brain abscesses, brain adhesions, encephalitis, and pneumocephalus (Doran, 1999; Vaz Marecos et al., 2012; Cuadros et al., 2013; Hewitt et al., 2021; Bonasoni et al., 2022). Additionally, strains of C. freundii can cause meningitis in adults and infants (Plakkal et al., 2013; Lien et al., 2018).
Infections caused by Citrobacter spp. spread horizontally through hospital settings or vertically through mother-to-child contact; however, the source of the infection is primarily unclear (Doran, 1999; Plakkal et al., 2013; Hewitt et al., 2021). In infants, if the symptoms appear in the first days of life, vertical transmission is most likely the cause of infection. However, isolation of the pathogens from the mother is rare (Bonasoni et al., 2022). C. koseri infection in newborns can be divided into early onset (5-12 days of age) and late-onset (4-5 weeks of age) (Doran, 1999). Compared to 1% of cases from other sources, brain abscesses occur in about 80% of cases of C. koseri meningitis. The mortality rate for C. koseri meningitis is around 30%, and more than 80% of such cases also have neurological sequelae (Bonasoni et al., 2022). Fever, altered consciousness, and headache were the typical clinical manifestations of meningitis; therefore, computerized tomography (CT) scans and cerebrospinal fluid cultures were necessary to detect meningitis and its causative agent (Liu et al., 2015a; Lien et al., 2018). Furthermore, it has also been reported the involvement of C. braakii, C. amalonaticus, and C. sedlakii in meningitis (Lai et al., 2010; Hirai et al., 2016; Tripathi et al., 2020).
Epidemiology
Recently, a genome-wide epidemiological investigation of 686 Citrobacter strains from 67 countries was reported (Osei Sekyere and Reta, 2021). In this analysis, only C. freundii had a multi-locus sequence typing (MLST) scheme among the studied strains. In that study, C. freundii had 84 distinct clones or sequence types (STs). The study reported the most prevalent clones were ST100, ST22, ST62, ST11, ST299, ST8, ST114, and ST98 (Osei Sekyere and Reta, 2021). They also observed that three clades of Citrobacter spp. are predominant in different parts of the world. In Clade A, C. freundii was primarily distributed in Europe, North America, and South East Asia, C. koseri is present only in the USA, and C. amalonaticus was observed in North America and South Korea. In clade B, C. freundii was identified worldwide; C. koseri was identified in the USA, Europe, China, Malaysia, and Malawi, whereas C. amalonaticus was identified in France, Malaysia, and the USA. Clade C only contained C. amalonaticus, found in Malaysia, Switzerland, and the USA (Osei Sekyere and Reta, 2021).
Another six-year epidemiological investigation on short-term bloodstream infections associated with peripheral venous catheters was conducted in 14 Middle Eastern nations, and the results showed that Citrobacter spp. was responsible for 1% of the infections(Rosenthal et al., 2020). Additionally, a meta-analysis that mostly comprised publications published in Iran between 2012 and 2018 found that the antibiotic sensitivity patterns of C. freundii varied by region (Rostamzad et al., 2019). This review also includes major investigations of Citrobacter spp. from various clinical samples from different countries (Table 2).
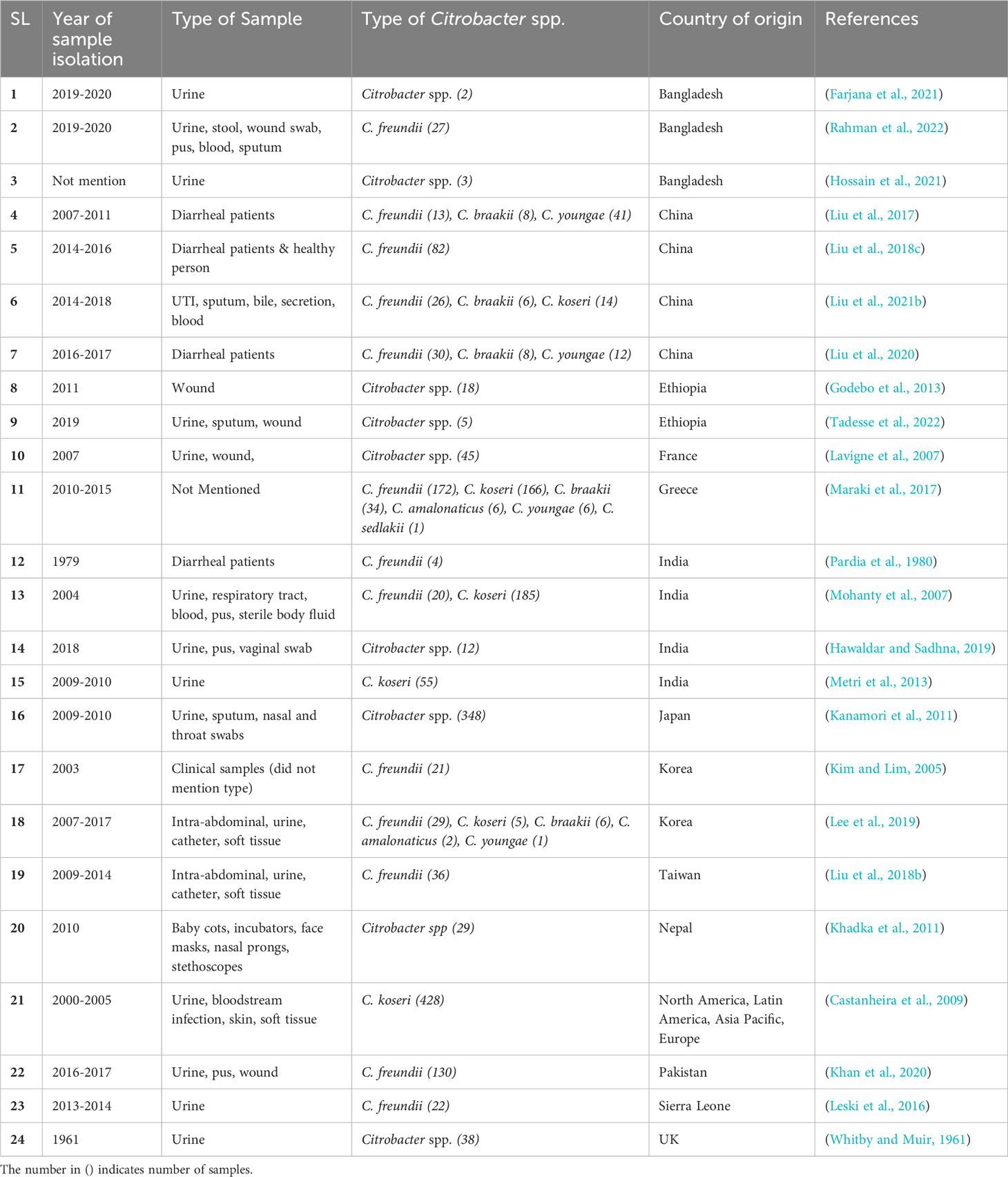
Table 2 A list of major studies of Citrobacter spp. isolated from various clinical samples worldwide.
Recently, carbapenem-resistant Citrobacter spp., especially C. freundii strains, were frequently isolated from Europe (Arana et al., 2017; Räisänen et al., 2021; Yao et al., 2021). In Spain, from 2013-2015, an increasing number (53%) of carbapenemase-producing Citrobacter spp. were isolated (Arana et al., 2017). During the years 2017, 2018, and 2019, Citrobacter spp. constituted 10%, 17%, and 14% of the total carbapenemase-producing strains identified in Germany, respectively (Yao et al., 2021). Furthermore, genomic and epidemiologic studies performed between 2000-2018 in the USA found carbapenem-resistant Citrobacter spp. increased from 4% to 10% (Babiker et al., 2020).
Pathogenesis
The majority of the infections caused by Citrobacter spp. are primarily associated with C. koseri and C. freundii isolated from human clinical specimens (Samonis et al., 2009). However, only experimental and in silico serotyping systems for Citrobacter detection have been developed to manage infections caused by it (Qian et al., 2018). It has been proposed that the affinity of C. koseri for nerve tissues and its propensity to induce meningitis and abscesses were related to a unique 32 kilodalton (kDa) outer-membrane protein (Southern and Bagby, 1977; Kline et al., 1988; Li et al., 1990).
In C. koseri, major virulence factors were associated with flagellar apparatus biosynthesis and iron uptake. C. koseri was found to have a High Pathogenicity Island (HPI) gene cluster, similar to a highly pathogenic Yersinia strain, enabling iron uptake in iron-deficient environments. The presence of such HPI could explain the remarkable pathogenic effects of C. koseri on the CNS. In contrast, C. freundii and C. braakii contained genes encoding the VI capsule polysaccharide. This VI capsule polysaccharide contributes to evading host defenses by Salmonella typhi, possibly leading to the higher pathogenic potential of C. freundii and C. braakii (Yuan et al., 2019). Furthermore, flagellar apparatus, tad pilus, and type IV pilus were unique to Citrobacter spp., whereas types II, III, IV, V, & VI secretion systems were found in some, but not all strains. Also, C. koseri lacked several of these secretion systems and the tad pilus, which are thought to be critical for colonizing human environments. The study also revealed three classes of Type VI secretion system (T6SS) genes, acquired through horizontal gene transfer, with distinct functions in biofilm formation (T6SS-1), colonization, survival, or invasion (T6SS-2), and antibacterial activity (T6SS-3) of Citrobacter spp. In C. koseri strains, only T6SS-2 genes were identified (Yuan et al., 2019).
The mechanism of infection and pathogenesis by C. freundii is known to occur through the T6SS and its effectors Hemolysin-coregulated protein (Hcp) family proteins, comprises Hcp-1 and Hcp-2, which are localized in the bacterial outer membrane and prevent phagocytosis by macrophage along with Vgr family orthologs (Zheng and Leung, 2007; Liu et al., 2015b; Aubert et al., 2016; Liu et al., 2021a). The T6SS effector Hcp-2 triggers IL-1β secretion via Nucleotide Oligomerization Domain - Like Receptor Family, Pyrin Domain Containing-3 (NLRP3)-dependent activation of caspase 1. Caspase 1 cleaves the gasdermin–N domain (GSDMD) to mediate the pyroptosis of macrophages (Yuan et al., 2019).
Several C. freundii strains were found to carry virulence factors including Shiga-like toxins and heat-stable toxins or virulence islands, thus associated with diarrhoea and food poisoning in humans. Shiga-like Toxin -II (SLT-II) has been reported in seven C. freundii strains, which had the same degree of cytotoxicity as the E. coli SLT-IIvhc control strain (Liu et al., 2020).
Another study revealed the presence of 3 virulence genes (hcp, msgA, and, rtx) contributed by 152 intact prophages associated with C. freundii strains (Jabeen et al., 2023). The most predominantly distributed MsgA plays a role in the biofilm formation and antibiotic resistance in Bacillus subtilis and Staphylococcus aureus (Branda et al., 2004; Lindsay, 2014). Thus, it might be assumed to contribute to biofilm formation and antibiotic resistance in C. freundii. The same study also identified the RTX as the second highly distributed virulence gene among those intact prophages, which has not been identified in Citrobacter spp. yet. The presence of such protein with hemolytic activity in some prophages may confer new virulence factors and/or antibiotic resistance genes required for bacterial pathogenesis and beneficial traits like increased fitness to the host (Boyd, 2012; Bobay et al., 2014).
The distribution of virulence proteins across the reference genomes of 18 different Citrobacter spp. have been investigated against the virulence factor data base (VFDB) (Liu et al., 2022). The genome IDs of the total of 18 Citrobacter spp. has been derived from National Center for Biotechnology Information (NCBI) as shown in Table 1.The heat map, generated using SRplot (https://www.bioinformatics.com.cn/en) (Figure 2), analysis of a total of 118 virulence proteins reveals that all the Citrobacter spp. examined are associated with virulence traits conferring immunomodulation, regulation and antimicrobial activity with the exception in C. koseri, and C. rodentium strains. Additionally, 78% of the studied Citrobacter spp. are found to have virulence factors associated to regulation and invasion.
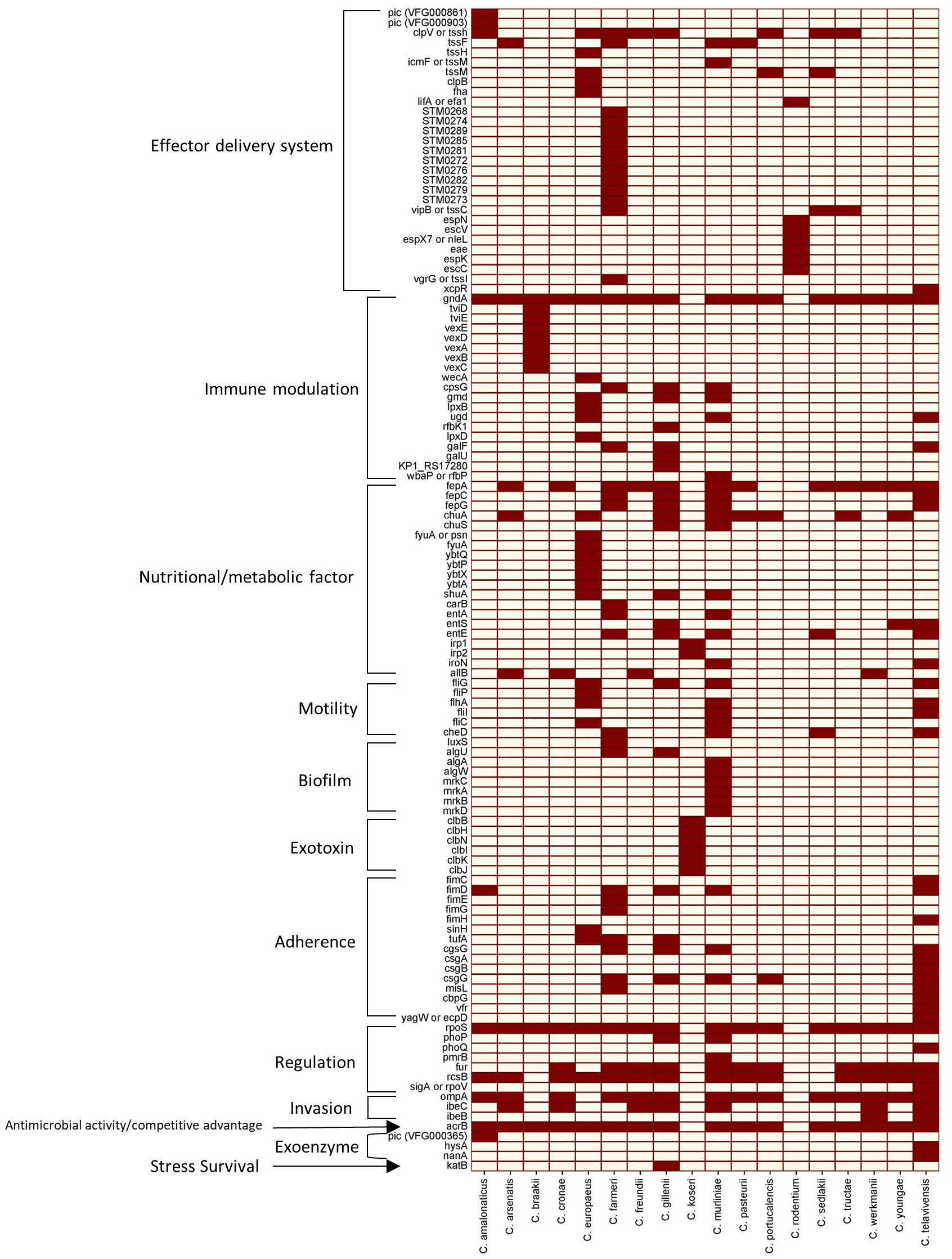
Figure 2 Heat map of the 118 virulence proteins across 18 different Citrobacter spp. studied. Dark brown squares denote the presence of the genes, and grey squares denote the absence of the genes listed.
Antibiotic resistance
Antibiotic resistance in Citrobacter spp. become a growing public health concern. Citrobacter spp. possess several antibiotic-resistant determinants encoded either in the plasmid or chromosome (Pepperell et al., 2002; Poire et al., 2011; Liu et al., 2017; Liu et al., 2018b; Liu et al., 2020; Shinu, 2022; Huang et al., 2023). They also have an increased propensity to take up genetic material conferring antibiotic resistance from other related or unrelated species (Frenk et al., 2021). The presence of a total of 86 naturally occurring antibiotic-resistant determinant genes identified in the reference genomes of 18 Citrobacter spp. is shown in a heatmap according to the Comprehensive Antibiotic Resistance Database (CARD) Resistance Gene Identifier (Figure 3). They mostly rely on antibiotic efflux and target alteration to confer resistance to the antibiotics they are exposed to, except for C. sedlakii and C. youngae, which carry numerous other antibiotic-resistant determinants.
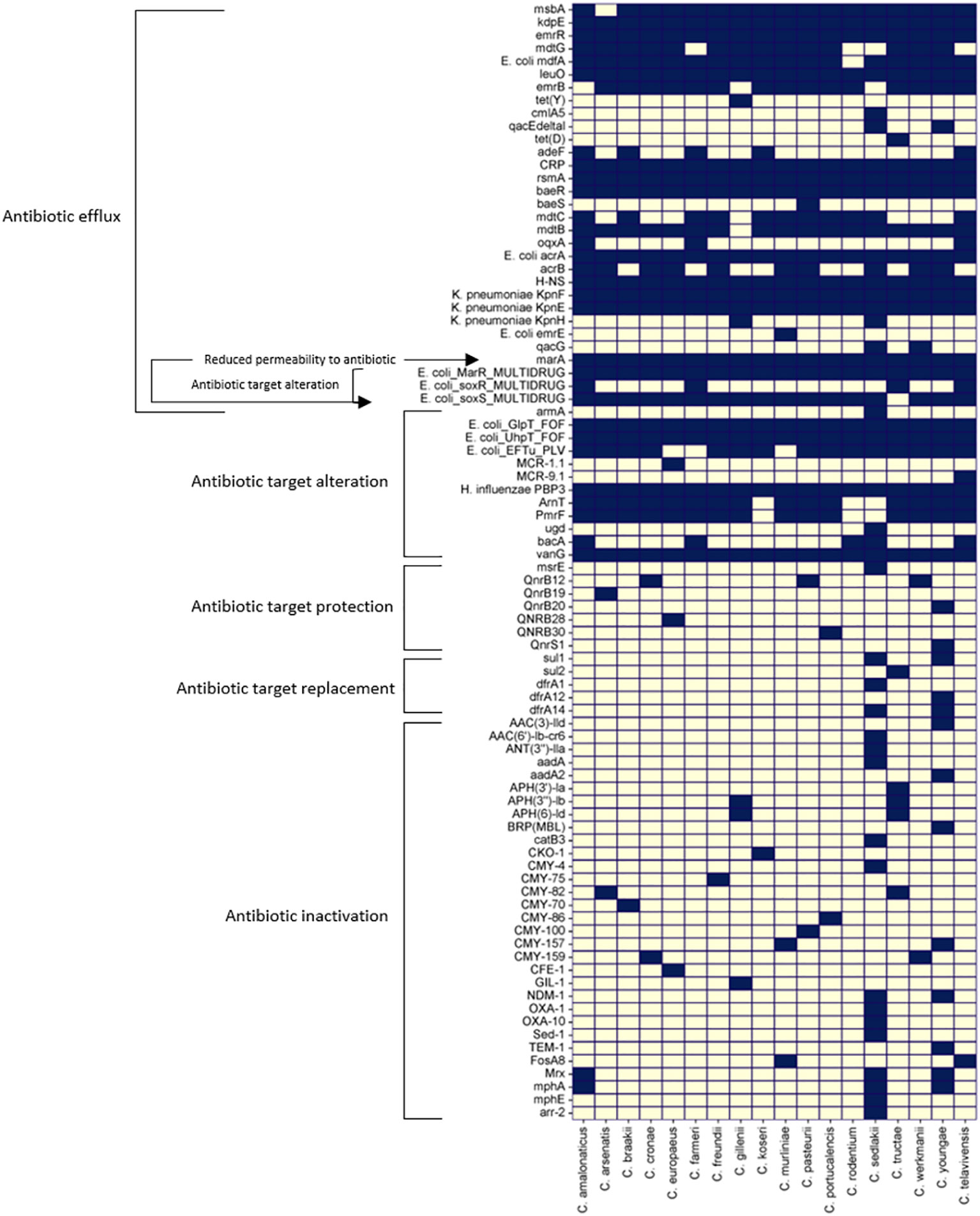
Figure 3 Heat map of 86 antibiotic resistant genes across the reference genomes of 18 different Citrobacter spp. studied were predicted using CARD database (Alcock et al., 2020) Dark blue squares denote the presence of the genes and grey squares denote the absence of the genes listed.
In this section of the review, we have summarized recent antibiotic resistance trends and mechanisms of Citrobacter spp.
There are several ways through which Citrobacter spp. can be resistant to fluoroquinolone antibiotics. Several studies have reported mutations in quinolone resistance-determining regions (QRDR) in Citrobacter spp., especially C. freundii, isolated from different sources and geographic regions (Weigel et al., 1998; Minarini, 2012; Kotb et al., 2019). For example, a common mutation in codon 59 (T59I) of gyrA was found in quinolone-resistant C. freundii isolates from China (Liu et al., 2021b). Besides, mutations such as S83L and D87N in gyrA and S80I in parC have also been detected in C. freundii isolates from various countries (Weigel et al., 1998; Minarini, 2012; Kotb et al., 2019).
However, some Citrobacter spp. have acquired resistance to quinolones through plasmids. One of the mechanisms of plasmid-mediated quinolone resistance (PMQR) in Citrobacter spp. is the expression of qnr genes. The qnr encodes the pentapeptide repeat family protein that binds to and protects DNA gyrase and topoisomerase IV from quinolone inhibition. The qnrB is the most common and diverse among Citrobacter spp. out of six qnr families (qnrA, qnrB, qnrC, qnrD, qnrS, and qnrVC) (Park et al., 2007; Jacoby et al., 2011). In Citrobacter spp., almost two-thirds of the alleles have been reported as qnrB, and several were shown to be located on the chromosome (Jacoby and Hooper, 2013). Second to qnrB, it has also been reported that Citrobacter spp. harbour qnrS in isolates producing blaCTX-M-2 or blaCTX-M-15 genes (Kanamori et al., 2011; Liu et al., 2018c). The qnrS are usually associated with transposable elements on plasmids; are often incorporated into sul1-type integrons, which are genetic platforms for antibiotic resistance gene capture and expression (Jacoby et al., 2014).
Furthermore, PMQR in Citrobacter spp. is the modification of quinolones by a variant of the aminoglycoside acetyltransferase AAC(6’)-Ib. This enzyme can acetylate quinolones with an amino nitrogen target, such as ciprofloxacin and norfloxacin, and reduce their antibacterial activity. This variant AAC(6’)-Ib-cr has two amino acid substitutions (W102R and D179Y) that enhance its ability to acetylate quinolones (Robicsek et al., 2006). The aac(6’)-Ib-cr gene is often found on plasmids with other resistance genes, such as blaCTX-M (Perilli et al., 2009; Liu et al., 2011; Goudarzi and Fazeli, 2017).
Lastly, in Citrobacter spp., PMQR increases quinolone efflux via plasmid-encoded pumps OqxAB and QepAB, which belong to the major facilitator superfamily (MFS) and the resistance-nodulation-cell division (RND) family, respectively. They can extrude many substrates, including quinolones, from the bacterial cell. The qepAB and oqxAB genes are also frequently linked with other resistance genes on plasmids or integrons (Jacoby et al., 2014).
PMQR in Citrobacter spp. poses a serious threat to public health, as it can compromise the efficacy of quinolones, which are important drugs for treating infections caused by MDR bacteria. Moreover, PMQR can facilitate the selection of higher-level resistance by chromosomal mutations in DNA gyrase and topoisomerase IV (Strahilevitz et al., 2009). The prevalence and diversity of quinolone resistance in Citrobacter spp. is an emerging problem in this genus and warrants further surveillance and molecular characterization.
The recent rise of carbapenem-resistant Citrobacter spp. cause severe public health concerns worldwide. Studies on carbapenem-resistant Citrobacter spp. found that the Inc family of plasmids was the primary group that carries genes for carbapenem resistance (Cao et al., 2021; Dziri et al., 2022; Ju et al., 2022); and was one of the main reasons for the rapid dissemination of carbapenem-resistant Citrobacter spp. around the world.
A report of an extremely drug-resistant strain of C. freundii was identified in a patient from India (Poire et al., 2011). This strain produced the NDM-1 enzyme, which is known to confer resistance to carbapenem antibiotics. In addition to the NDM-1 enzyme, the strain also produced several other ESBL-producing genes, including blaOXA-1, blaOXA-9, blaOXA-10, blaOXA181, blaTEM-1, blaVIM-4, and blaCMY genes indicating the potential for rapid dissemination of multidrug resistance among bacterial populations (Poire et al., 2011). The IncX3 plasmid pZY-NDM1 was also reported by another study harboring the blaNDM-1 gene in a C. portucalensis clinical strain (Cao et al., 2021). The co-occurrence of multiple antibiotic resistance determinant genes greatly limits the treatment options for infections. In a UTI isolate, C. amalonaticus was found resistant to carbapenems and colistin conferred by blaNDM-1 and mcr-1.5, respectively. blaNDM-1 and mcr-1.5 co-occurred in separate plasmids of type 1 IncC2 and incompatibility group Incl2, respectively. The isolate showed reduced susceptibility to carbapenems, 3rd and 4th-generation cephalosporins, aminoglycoside, trimethoprim-sulfamethoxazole, and colistin (Faccone et al., 2019). Moreover, the blaNDM-1, found to coexist with armA in the C. sedlakii strain, isolated from the same patient, is another example of conferring resistance to a broad range of antibiotics, including carbapenems and aminoglycosides via horizontal transfer (Moser et al., 2021). Co-existence of blaNDM-1 with blaSHV-12 on the same transferrable IncX3 plasmid pZY-NDM1 in C. freundii and co-production of NDM-1 and OXA-10 in C. braakii isolate on different plasmids was also reported (Zhang et al., 2021; Han et al., 2022).
Two decades ago, the most prevalent isolate found was C. freundii, followed by C. werkmanii, C. koseri, and C. farmeri, where most of the C. freundii isolates harboring blaKPC-3 genes, followed by a few blaKPC-2 and blaNDM-1 genes (Babiker et al., 2020). This study also reported the presence of the blaKPC-3 gene in C. farmeri and C. werkmanii. A similar study detected plasmid-borne blaNDM-1, blaCMY-48, blaCTX-M-15, blaOXA-10, blaOXA-1, blaTEM-1B in a South African extensively drug-resistant (XDR) strain of C. freundii (Ramsamy et al., 2020). Unlike efflux genes, most resistant determining genes are in plasmids in this strain. Citrobacter spp. isolates have developed resistance to carbapenems due to the spread of carbapenemases such as NDM, VIM-1, OXA-48, and VIM-2. Carbapenem-resistant C. freundii carrying the blaNDM-1 gene has been increasingly reported in countries such as China, India, Denmark, and South Africa (Yang et al., 2018). In contrast, C. freundii strains that are positive for VIM-1 and VIM-2 have been documented in Europe (Gaibani et al., 2013; Porres-Osante et al., 2014; Santos et al., 2017).
Carbapenem-resistant Citrobacter isolates are a diverse group of bacteria that can acquire carbapenem resistance through horizontal gene transfer. They do not usually form a single clonal complex, but sometimes isolates from different hospitals can be genetically similar, suggesting the potential for clonal spread (Yao et al., 2021). Carbapenem resistance can also be acquired through chromosomal mutations. In a Citrobacter freundii strain, carbapenem resistance was conferred by marA, soxS, and mutations in penicillin-binding proteins (PBP3) (Yap et al., 2020).
The prevalence of various aminoglycoside-modifying enzymes (AME) in Citrobacter spp. depending on the geographic region and the type of infection. Among the AMEs, the most prevalent was aminoglycoside-N-acetyltransferases (AACs). Since its discovery, the most common AAC found globally in several studies is aac(6’)-ib-cr, followed by aac(6’)-II and aac(3’)-II enzymes in Citrobacter spp. (Jiang et al., 2019; Zhou et al., 2019; Babiker et al., 2020; Ramsamy et al., 2020; Cao et al., 2021; Yao et al., 2021; Zhang et al., 2021; Han et al., 2022). AAC(6’)-I enzymes are highly active in inactivating amikacin and gentamicin (C1a & C2), whereas AAC(6’)-II enzymes do not modify amikacin but modify all three types of gentamicin (C1, C1a, and C2) (Woloj et al., 1986; Rather et al., 1992; Shaw et al., 1993). Furthermore, AAC(3’)-II enzymes are active against gentamicin, netilmicin, tobramycin, sisomicin, 2′-N-ethylnetilmicin, 6′-N-ethylnetilmicin, and dibekacin (Shaw et al., 1993).
After AACs, the 2nd most prevalent AME is the aminoglycoside-O-phosphotransferases (APH), mainly aph(3′′)-Ib, aph(6)-Id followed by aph(3’)-Ia (Babiker et al., 2020; Ramsamy et al., 2020; Cao et al., 2021; Zhang et al., 2021). APH(3′′) confers resistance against streptomycin, whereas the APH(3′) group of enzymes is responsible for the resistance profile against kanamycin, neomycin, paromomycin, ribostamycin, lividomycin and is usually found in large plasmids or as part of transposon (Vakulenko and Mobashery, 2003).
The least prevalent AME in Citrobacter spp. belongs to aminoglycoside-O-necleotidyl transferases (ANT), and only a few ANT genes, ant(2”)-Ia, also known as aadB, was found in an IncA/C2 plasmid (Babiker et al., 2020). These genes conferred resistance to gentamicin, tobramycin, dibekacin, sisomicin, and kanamycin (Vakulenko and Mobashery, 2003). A number of studies reported that few ANT(3”) enzymes, encoded by aadA (aadA1, aadA2, aadA16), conferred resistance to spectinomycin and streptomycin in Citrobacter spp. (Babiker et al., 2020; Ramsamy et al., 2020; Moser et al., 2021; Han et al., 2022; Luo et al., 2022). The presence of streptomycin inactivating genes (strA, strB) was also reported in Citrobacter spp. (Yao et al., 2021; Luo et al., 2022).
In two studies, aminoglycoside resistance caused by target methylation was observed in C. sedlakii and C. portucalensis. This resistance was due to the presence of the armA gene in C. sedlakii and both armA and rmtC genes in C. portucalensis (Moser et al., 2021; Luo et al., 2022). In C. portucalensis, armA were coharbored with aacA4cr, aphA1 blaSHV-12 gene on a separate IncC group plasmid, whereas rmtC gene was coharboured with IncFII : FIB plasmid-borne blaNDM-1 gene (Luo et al., 2022). In addition, the involvement of efflux genes, baeR and kdpE, was reported in a clinical MDR C. freundii strain showing resistance against gentamicin and tobramycin (Yap et al., 2020).
The emergence and dissemination of plasmid-mediated colistin resistance in Citrobacter spp., particularly mediated by the mobilized colistin resistance (mcr) genes, is a growing concern. Several variants of mcr have been recently reported in many spp. of Citrobacter, including mcr-1 in C. freundii (Li et al., 2017; Yan Hu et al., 2017) and mcr-1 in C. braakii (Sennati et al., 2017; Liu et al., 2018a), and mcr-1.5 in C. amalonaticus (Faccone et al., 2019), mcr-3.5 in C. sedlakii and C. amalonaticus (Phuadraksa et al., 2023), mcr-9 (Bitar et al., 2020) in C. freundii and, chromosomal mcr-9 in C. telavivensis (Ribeiro et al., 2021). In Citrobacter spp., mcr can co-occur with genes conferring resistance to β-lactams (Faccone et al., 2019) and aminoglycosides (Ju et al., 2022). This may result in developing MDR strains that are challenging to manage using currently available antibiotics.
Citrobacter spp. can acquire antibiotic-resistant determinants from related and non-related species. The existing carbapenemase gene repertoire in Enterobacteriaceae can mutate, evolve, and be transferred horizontally. The IncC plasmid-borne carbapenem-resistant determinants blaOXA-900 are located in C. freundii. This gene is believed to have originated from a distinct Gram-negative bacterium Shewanella, a marine environmental extremophile (Frenk et al., 2021). In C. koseri, a single blaKPC-82 (a blaKPC-2 variant) conferring resistance to β -lactam/β -lactam inhibitor combination was carried in a transposon integrated chromosomally. The transposon was initially harbored by a plasmid acquired by C. koseri from S. marcescens within the same host (Lebreton et al., 2021).
Citrobacter spp. are equipped with a repertoire of antibiotic efflux genes conferring intrinsic resistance and are highly efficient in horizontal gene transfer for acquired resistance within and outside its genus Citrobacter. Therefore, to identify the cause of antibiotic resistance in clinical settings, it is essential to look for common chromosomal mutations and include relevant plasmid-mediated antibiotic-resistant determinants for each class of antibiotics.
Treatment
Various types of antibiotics, including monobactams, aminoglycosides, carbapenems, cephalosporins, sulfonamides, nitrofuran, chloramphenicol, quinolones, and colistin are used for the treatment of Citrobacter infections (Doran, 1999; McPherson et al., 2008; Samonis et al., 2009; Deveci and Coban, 2014; Hrbacek et al., 2020; Jiménez-Guerra et al., 2020; Chavan et al., 2021; Gogry et al., 2021). However, multidrug-resistant Citrobacter strains limit the use of empirical antibiotics. Various studies have found that lately, Citrobacter spp. are more sensitive to meropenem, imipenem, colistin, Tigecycline, piperacillin/tazobactam, and cefoperazone/sulbactam (Hawaldar and Sadhna, 2019; Lee et al., 2019; Yao et al., 2021). Therefore, those drugs might be a good option for treating Citrobacter infections. In addition, In-vitro studies have found that silver nanoparticles can be used to treat Citrobacter infections (Abady et al., 2021). Likewise, a recent study has found that phage-antibiotic combined treatment against C. amalonaticus indicated that a sublethal concentration of phages used as an adjuvant with antibiotics could be an effective therapeutic strategy (Manohar et al., 2022). Therefore, the combination of phages and antibiotics can be tested in the future on other types of Citrobacter spp., especially C. freundii and C. koseri.
Conclusion
Infections caused by various MDR Citrobacter spp. are widespread, seriously threatening public health worldwide. The presence of virulence genes and prophages in the Citrobacter spp. accounts for its increased virulence in urinary, respiratory, and intra-gastrointestinal tract infections and other complications. Moreover, the emergence of MDR Citrobacter spp. resulted in difficult-to-treat infections in humans revealed, as reflected by the epidemiological studies. The information provided in this review paper will offer great benefits in addressing the health burden of Citrobacter spp.
Author contributions
IJ: Writing – original draft, Writing – review & editing. SI: Writing – original draft, Writing – review & editing. AH: Writing – original draft. ZT: Writing – original draft. SS: Conceptualization, Writing – original draft, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abady N. R., Abdul S., Salman K. (2021). Antimicrobial effect of silver nanosilver particles on multi-drug resistant isolates of kluyvera cryocrescens and Citrobacter freundii. Ann. Romanian Soc. Cell Biol. 25, 6687–6701.
Aguirre-Sánchez J. R., Quiñones B., Ortiz-Muñoz J. A., Prieto-Alvarado R., Vega-López I. F., Martínez-Urtaza J., et al. (2023). Comparative genomic analyses of virulence and antimicrobial resistance in Citrobacter werkmanii, an emerging opportunistic pathogen. Microorganisms 11, 1–17. doi: 10.3390/microorganisms11082114
Alcock B. P., Raphenya A. R., Lau T. T. Y., Tsang K. K., Bouchard M., Edalatmand A., et al. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Arana D. M., Ortega A., González-Barberá E., Lara N., Bautista V., Gómez-Ruíz D., et al. (2017). Carbapenem-resistant Citrobacter spp. isolated in Spain from 2013 to 2015 produced a variety of carbapenemases including VIM-1, OXA-48, KPC-2, NDM-1 and VIM-2. J. Antimicrob. Chemother. 72, 3283–3287. doi: 10.1093/jac/dkx325
Ashish K., Singh N., Aggarwal A., Khanna M. (2012). The antibiotic resistance pattern in citro-bacter species: an emerging nosocomial pathogen in a tertiary care hospital. J. Clin. Diagn. Res. 6, 642–644.
Aubert D. F., Xu H., Yang J., Shi X., Gao W., Li L., et al. (2016). A burkholderia type VI effector deamidates rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 19, 664–674. doi: 10.1016/j.chom.2016.04.004
Babiker A., Evans D. R., Griffith M. P., McElheny C. L., Hassan M., Clarke L. G., et al. (2020). Clinical and genomic epidemiology of carbapenem- nonsusceptible Citrobacter spp. At a tertiary health care center over 2 decades. J. Clin. Microbiol. 58, e00275-20. doi: 10.1128/JCM.00275-20
Bitar I., Papagiannitsis C. C., Kraftova L., Chudejova K., Mattioni Marchetti V., Hrabak J. (2020). Detection of five mcr-9-carrying enterobacterales isolates in four czech hospitals. mSphere 5, e01008-20. doi: 10.1128/MSPHERE.01008-20
Bobay L.-M. M., Touchon M., Rocha E. P. C. (2014). Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. 111, 12127–12132. doi: 10.1073/pnas.1405336111
Bonasoni M. P., Comitini G., Pati M., Russello G., Vizzini L., Bardaro M., et al. (2022). Second trimester fetal loss due to Citrobacter koseri infection: a rare cause of preterm premature rupture of membranes (PPROM). Diagnostics 12, 10–15. doi: 10.3390/diagnostics12010159
Borenshtein D., Schauer D. B. (2006). “The genus Citrobacter,” in The prokaryotes (New York, NY: Springer New York), 90–98. doi: 10.1007/0-387-30746-X_5
Boyd E. F. (2012). “Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions,” in Advances in virus research (USA: Academic Press Inc), 91–118. doi: 10.1016/B978-0-12-394621-8.00014-5
Braak H. R. (1928). Onderzoekingen over Vergisting van Glycerine [thesis]. (Delft, Netherlands: W.D. Meinema-Utitgeurer). 1928, 166.
Branda S. S., González-Pastor J. E., Dervyn E., Ehrlich S. D., Losick R., Kolter R. (2004). Genes involved in formation of structured multicellular communities by Bacillus subtilis. J. Bacteriol. 186, 3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004
Brenner D. J., O’Hara C. M., Grimont P. A. D., Janda J. M., Falsen E., Aldova E., et al. (1999). Biochemical identification of Citrobacter species defined by DNA hybridization and description of Citrobacter gillenii sp. nov. (formerly Citrobacter genomospecies 10) and Citrobacter murliniae sp. nov. (formerly Citrobacter genomospecies 11). J. Clin. Microbiol. 37, 2619–2624. doi: 10.1128/jcm.37.8.2619-2624.1999
Cao X., Xie H., Huang D., Zhou W., Liu Y., Shen H., et al. (2021). Detection of a clinical carbapenem-resistant Citrobacter portucalensis strain and the dissemination of C. portucalensis in clinical settings. J. Glob. Antimicrob. Resist. 27, 79–81. doi: 10.1016/j.jgar.2021.04.027
Casas-Martínez M. R., Rodríguez-Rubio H. A., Bonilla-Suastegui A., López-Rodríguez R., Serrano-Rubio A., Montes-Aguilar O. J., et al. (2023). Citrobacter koseri: A rare cause of an epidural spinal abscess. Surg. Neurol. Int. 14. doi: 10.25259/SNI_147_2023
Castanheira M., Debbia E., Marchese A., Jones R. N. (2009). Emergence of a plasmid mediated blaVIM-1 in Citrobacter koseri: Report from the SENTRY antimicrobial surveillance program (Italy). J. Chemother. 21, 98–100. doi: 10.1179/joc.2009.21.1.98
Chavan R., Naphade B., Waykar B., Bhagwat S. (2021). In vitro activity of fosfomycin and nitrofurantoin against contemporary enterobacterales pathogens isolated from Indian tertiary care hospitals. Microb. Drug Resist. 27, 678–684. doi: 10.1089/mdr.2020.0200
Chen D., Ji Y. (2019). New insights into Citrobacter freundii sepsis in neonates. Pediatr. Int. 61, 375–380. doi: 10.1111/ped.13715
Cong’En J. H., Miah M., Sünkel-Laing B., Emmanuel J. (2014). Endogenous endophthalmitis caused by Citrobacter koseri originating from a renal abscess. BMJ Case Rep. 1–5. doi: 10.1136/bcr-2014-204095
Cuadros E. N., Castilla C. Y., Algarra C. M., Peérez D. M., López B. R., Martín F. J. G., et al. (2013). Medical and neurosurgical management of Citrobacter koseri, a rare cause of neonatal meningitis. J. Med. Microbiol. 63, 144–147. doi: 10.1099/jmm.0.063586-0
Deveci A., Coban A. Y. (2014). Optimum management of Citrobacter koseri infection. Expert Rev. Anti Infect. Ther. 12, 1137–1142. doi: 10.1586/14787210.2014.944505
Dominguez Céspedes L., Céspedes Fonseca Y. M. (2022). Antimicrobial susceptibility of Citrobacter Koseri isolated on clinical samples of hospitalized patients. J. Microbiol. Exp. 10, 54–57. doi: 10.15406/jmen.2022.10.00353
Doran T. I. (1999). The role of Citrobacter in clinical disease of children: Review. Clin. Infect. Dis. 28, 384–394. doi: 10.1086/515106
dos Santos G. S., Solidônio E. G., Costa M. C. V. V., Melo R. O. A., de Souza I. F. A. C., Silva G. R., et al. (2015). Study of the enterobacteriaceae group CESP (Citrobacter, enterobacter, serratia, providencia, morganella and hafnia): a review. Battle Against Microb. Pathog. Basic Sci. Technol. Adv. Educ. Programs, 794–805.
Dziri R., Kuşkucu M. A., Arfaoui A., Fethi M., Ifaoui S., Bellaaj R., et al. (2022). Whole genome sequencing of a Citrobacter freundii strain isolated from the hospital environment: an extremely multiresistant NDM-1 and VIM-48 coproducing isolate. Microb. Drug Resist. 28, 18–22. doi: 10.1089/mdr.2020.0417
Emery A., Marpaux N., Naegelen C., Valot B., Morel P., Hocquet D. (2020). Genotypic study of Citrobacter koseri, an emergent platelet contaminant since 2012 in France. Transfusion 60, 245–249. doi: 10.1111/trf.15617
Faccone D., Albornoz E., Tijet N., Biondi E., Gomez S., Pasterán F., et al. (2019). Characterization of a multidrug resistant Citrobacter amalonaticus clinical isolate harboring blaNDM-1 and mcr-1.5 genes. Infect. Genet. Evol. 67, 51–54. doi: 10.1016/j.meegid.2018.10.020
Farjana N. E., Islam M. A., Zerin T., Begum M. A. (2021). Bacterial association in urinary tract infection and their drug resistance among patients in Rajshahi, Bangladesh. Int. J. Community Med. Public Heal. 8, 2144–2149 doi: 10.18203/2394-6040.ijcmph20211730
Forsythe S. J., Abbott S. L., Pitout J. (2015). “Klebsiella, enterobacter, Citrobacter, cronobacter, serratia, plesiomonas, and other enterobacteriaceae,” in Manual of clinical microbiology (John Wiley & Sons, Ltd), 714–737. doi: 10.1128/9781555817381.ch38
Frenk S., Rakovitsky N., Kon H., Rov R., Abramov S., Lurie-Weinberger M. N., et al. (2021). Oxa-900, a novel oxa sub-family carbapenemase identified in Citrobacter freundii, evades detection by commercial molecular diagnostics tests. Microorganisms 9, 1898. doi: 10.3390/microorganisms9091898
Fritsche D. (1964). Contribution to the problems of Salmonella diagnosis: biochemical separation of Citrobacter (E. freundii inclusive of Ballerup-Bethesda) from the Salmonella group with the aid of the demonstration of lysin decarboxylase. Zentralbl. Bakteriol. Orig. 194, 188–192.
Gaibani P., Ambretti S., Farruggia P., Bua G., Berlingeri A., Tamburini M. V., et al. (2013). Outbreak of Citrobacter freundii carrying Vim-1 in an italian hospital, identified during the carbapenemases screening actions, June 2012. Int. J. Infect. Dis. 17, e714–e717. doi: 10.1016/j.ijid.2013.02.007
Gajdács M., Urbán E. (2019). Resistance trends and epidemiology of citrobacter-enterobacter-serratia in urinary tract infections of inpatients and outpatients (RECESUTI): A 10-year survey. Med. 55, 1–13. doi: 10.3390/medicina55060285
Garcia V., Abat C., Moal V., Rolain J. M. (2016). Citrobacter amalonaticus human urinary tract infections, Marseille, France. New Microbes New Infect. 11, 1. doi: 10.1016/J.NMNI.2016.01.003
Godebo G., Kibru G., Tassew H. (2013). Multidrug-resistant bacterial isolates in infected wounds at Jimma University Specialized Hospital, Ethiopia. Ann. Clin. Microbiol. Antimicrob. 12, 17. doi: 10.1186/1476-0711-12-17
Gogry F. A., Siddiqui M. T., Sultan I., Haq Q. M. R. (2021). Current update on intrinsic and acquired colistin resistance mechanisms in bacteria. Front. Med. 8. doi: 10.3389/fmed.2021.677720
Goudarzi M., Fazeli M. (2017). Quinolone resistance determinants qnr, qep, and aac(6’)-Ib-cr in extended-spectrum B-lactamase producing escherichia coli isolated from urinary tract infections in Tehran, Iran. Shiraz E Med. J. 18, 1–7. doi: 10.5812/semj.44498
Gross R. J., Rowe B. (1974). The serology of Citrobacter koseri, levinea malonatica, and levinea amalonatica. J. Med. Microbiol. 7, 155–161. doi: 10.1099/00222615-7-2-155
Han H., Zhao Z., Lin Y., Lin B., Xu H., Zheng B. (2022). Co-production of NDM-1 and OXA-10 β-lactamase in Citrobacter braakii strain causing urinary tract infection. Infect. Drug Resist. 15, 127–133. doi: 10.2147/IDR.S347943
Hashimoto M., Mogi K., Sakurai M., Sakata T., Tani K., Takahara Y. (2021). Rupture of a dissecting thoracoabdominal aortic aneurysm due to Citrobacter freundii infection. Clin. Case Rep. 9, 1–5. doi: 10.1002/ccr3.4719
Hawaldar R., Sadhna S. (2019). Prevalence and drug resistance pattern of Citrobacter sps – A retrospective study. Indian J. Microbiol. Res. 6, 142–145. doi: 10.18231/j.ijmr.2019.030
Heljanko V., Johansson V., Räisänen K., Anttila V. J., Lyytikäinen O., Jalava J., et al. (2023). Genomic epidemiology of nosocomial carbapenemase-producing Citrobacter freundii in sewerage systems in the Helsinki metropolitan area, Finland. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1165751
Hewitt M. K., Klowak J. A., Pernica J. M., Leung J. (2021). Citrobacter koseri meningitis and septicemia in a neonate with borderline fever at home. Cmaj 193, E1592–E1594. doi: 10.1503/cmaj.210285
Hirai J., Uechi K., Hagihara M., Sakanashi D., Kinjo T., Haranaga S., et al. (2016). Bacteremia due to Citrobacter braakii: A case report and literature review. J. Infect. Chemother. 22, 819–821. doi: 10.1016/j.jiac.2016.07.003
Hossain I., Bhowmik S., Uddin M. S., Devnath P., Akter A., Eti L. N., et al. (2021). Prevalence of urinary tract infections, associated risk factors, and antibiotic resistance pattern of uropathogens in young women at Noakhali, Bangladesh. Asian J. Med. Biol. Res. 7, 202–213. doi: 10.3329/ajmbr.v7i2.55000
Hrbacek J., Cermak P., Zachoval R. (2020). Current antibiotic resistance trends of uropathogens in central europe: Survey from a tertiary hospital urology department 2011–2019. Antibiotics 9, 1–11. doi: 10.3390/antibiotics9090630
Hua D. T., Lo J., Do H. Q., Pham C. D. (2022). A case of Citrobacter koseri renal abscess and review of the literature. SAGE Open Med. Case Rep. 10, 0–4. doi: 10.1177/2050313X221135347
Huang J., Zhao J., Yi M., Yuan Y., Xia P., Yang B., et al. (2023). Emergence of Tigecycline and Carbapenem-Resistant Citrobacter freundii Co-Carrying tmexCD1-toprJ1, blaKPC-2, and blaNDM-1 from a Sepsis Patient. Infect. Drug Resist. 16, 5855–5868. doi: 10.2147/IDR.S426148
Jabeen I., Mahamud S., Islam S., Lamisa A., Anjum A., Oishy S., et al. (2023). Genomic identification and characterization of prophages associated with Citrobacter freundii strains. J. Adv. Biotechnol. Exp. Ther. 6, 648. doi: 10.5455/jabet.2023.d156
Jacoby G. A., Griffin C. M., Hooper D. C. (2011). Citrobacter spp. as a source of qnrB alleles. Antimicrob. Agents Chemother. 55, 4979–4984. doi: 10.1128/AAC.05187-11
Jacoby G. A., Hooper D. C. (2013). Phylogenetic analysis of chromosomally determined qnr and related proteins. Antimicrob. Agents Chemother. 57, 1930–1934. doi: 10.1128/AAC.02080-12
Jacoby G. A., Strahilevitz J., Hooper D. C. (2014). Plasmid-mediated quinolone resistance. Microbiol. Spectr. 2. doi: 10.1128/microbiolspec.PLAS-0006-2013
Jiang X., Cui X., Liu W., Xu H., Zheng B. (2019). Genetic characterization of a novel sequence type of multidrug-resistant Citrobacter freundii strain recovered from wastewater treatment plant. Infect. Drug Resist. 12, 2775–2779. doi: 10.2147/IDR.S213525
Jiménez A., Castro J. G., Munoz-Price L. S., De Pascale D., Shimose L., Mustapha M. M., et al. (2017). Outbreak of Klebsiella pneumoniae Carbapenemase-Producing Citrobacter freundii at a Tertiary Acute Care Facility in Miami, Florida. Infect. Control Hosp. Epidemiol. 38, 320–326. doi: 10.1017/ice.2016.273
Jiménez-Guerra G., Borrego-Jiménez J., Gutiérrez-Soto B., Expósito-Ruiz M., Navarro-Marí J. M., Gutiérrez-Fernández J. (2020). Susceptibility evolution to antibiotics of Enterobacter cloacae, Morganella morganii, Klebsiella aerogenes and Citrobacter freundii involved in urinary tract infections: An 11-year epidemiological surveillance study. Enfermedades Infecc. y Microbiol. Clin. (English ed.) 38, 166–169. doi: 10.1016/j.eimce.2019.07.003
Ju X., Wang S., Yang X., Han W., Cai C., Wu Y., et al. (2022). Epidemiology and Molecular Characteristics of mcr-9 in Citrobacter spp. from Healthy Individuals and Patients in China. Microbiol. Spectr. 10, e0134622. doi: 10.1128/spectrum.01346-22
Kanamori H., Yano H., Hirakata Y., Endo S., Arai K., Ogawa M., et al. (2011). High prevalence of extended-spectrum $β$-lactamases and qnr determinants in Citrobacter species from Japan: Dissemination of CTX-M-2. J. Antimicrob. Chemother. 66, 2255–2262. doi: 10.1093/jac/dkr283
Katzenellenbogen E., Staniszewska M., Kocharova N. A., Mieszała M., Korzeniowska-Kowal A., Górska S., et al. (2017). Re-classification within the serogroups O3 and O8 of Citrobacter strains. BMC Microbiol. 17, 1–8. doi: 10.1186/s12866-017-1078-3
Keleti J., Lüderitz O., Mlynaršék D., Sedlák J. (1971). Immunochemical studies on Citrobacter O antigens (Lipopolysaccharides). Eur. J. Biochem. 20, 237–244. doi: 10.1111/j.1432-1033.1971.tb01386.x
Khadka S. B., Thapa B., Mahat K. (2011). Nosocomial Citrobacter infection in neonatal intensive care unit in a hospital of Nepal. J. Nepal Paediatr. Soc 31, 105–109. doi: 10.3126/jnps.v31i2.4094
Khan S., Taj R., Rehman N., Ullah A., Khan I., Rahman S. (2020). Incidence and antibiogram of β Lactamases-producing citrobacter freundii recovered from clinical isolates in Peshawar, Pakistan. Pak. J. Zool. 52, 1877–1882. doi: 10.17582/journal.pjz/20181118151126
Kim J., Lim Y.-M. (2005). Prevalence of Derepressed AmpC Mutants and Extended-Spectrum β-Lactamase Producers among Clinical Isolates of Citrobacter freundii , Enterobacter spp., and Serratia marcescens in Korea: Dissemination of CTX-M-3, TEM-52, and SHV-12. J. Clin. Microbiol. 43, 2452–2455. doi: 10.1128/JCM.43.5.2452-2455.2005
Kline M. W., Kaplan S. L., Hawkins E. P., Mason E. O. (1988). Pathogenesis of brain abscess formation in an infant rat model of Citrobacter diversus bacteremia and meningitis. J. Infect. Dis. 157, 106–112. doi: 10.1093/infdis/157.1.106
Knirel Y. A., Kocharova N. A., Bystrova O. V., Katzenellenbogen E., Gamian A. (2002). Structures and serology of the O-specific polysaccharides of bacteria of the genus Citrobacter. Arch. Immunol. Ther. Exp. (Warsz). 50, 379–391.
Kotb D. N., Mahdy W. K., Mahmoud M. S., Khairy R. M. M. (2019). Impact of co-existence of PMQR genes and QRDR mutations on fluoroquinolones resistance in Enterobacteriaceae strains isolated from community and hospital acquired UTIs. BMC Infect. Dis. 19, 1–8. doi: 10.1186/s12879-019-4606-y
Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lai C. C., Tan C. K., Lin S. H., Liu W. L., Liao C. H., Huang Y. T., et al. (2010). Bacteraemia caused by non-freundii, non-koseri Citrobacter species in Taiwan. J. Hosp. Infect. 76, 332–335. doi: 10.1016/j.jhin.2010.06.006
Lalaoui R., Djukovic A., Bakour S., Hadjadj L., Sanz J., Salavert M., et al. (2019). Genomic characterization of Citrobacter freundii strains coproducing OXA-48 and VIM-1 carbapenemase enzymes isolated in leukemic patient in Spain. Antimicrob. Resist. Infect. Control 8, 4–9. doi: 10.1186/s13756-019-0630-3
Lavigne J. P., Defez C., Bouziges N., Mahamat A., Sotto A. (2007). Clinical and molecular epidemiology of multidrug-resistant Citrobacter spp. infections in a French university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 26, 439–441. doi: 10.1007/s10096-007-0315-3
Lebreton F., Corey B. W., McElheny C. L., Iovleva A., Preston L., Margulieux K. R., et al. (2021). Characterization of KPC-82, a KPC-2 variant conferring resistance to ceftazidime-avibactam in a carbapenem-nonsusceptible clinical isolate of Citrobacter koseri. Antimicrob. Agents Chemother. 65, e00150-21. doi: 10.1128/AAC.00150-21
Lee R., Choi S. M., Jo S. J., Lee J., Cho S. Y., Kim S. H., et al. (2019). Clinical characteristics and antimicrobial susceptibility trends in Citrobacter bacteremia: An 11-year single-center experience. Infect. Chemother. 51, 1–9. doi: 10.3947/ic.2019.51.1.1
Leski T. A., Taitt C. R., Bangura U., Stockelman M. G., Ansumana R., Cooper W. H., et al. (2016). High prevalence of multidrug resistant Enterobacteriaceae isolated from outpatient urine samples but not the hospital environment in Bo, Sierra Leone. BMC Infect. Dis. 16, 1–9. doi: 10.1186/s12879-016-1495-1
Li X. P., Fang L. X., Jiang P., Pan D., Xia J., Liao X. P., et al. (2017). Emergence of the colistin resistance gene mcr-1 in Citrobacter freundii. Int. J. Antimicrob. Agents 49, 786–787. doi: 10.1016/J.IJANTIMICAG.2017.04.004
Li J., Musser J. M., Beltran P., Kline M. W., Selander R. K. (1990). Genotypic heterogeneity of strains of Citrobacter diversus expressing a 32-kilodalton outer membrane protein associated with neonatal meningitis. J. Clin. Microbiol. 28, 1760–1765. doi: 10.1128/jcm.28.8.1760-1765.1990
Licata G., De Rosa A., Gambardella A., Calabrese G., Argenziano G., Della Rocca M. T., et al. (2021). Bullous Erysipelas caused by Citrobacter koseri. J. Clin. Aesthet. Dermatol. 14, 12.
Lien C. Y., Lee J. J., Chien C. C., Huang C. R., Lu C. H., Chang W. N. (2018). Clinical characteristics of Citrobacter meningitis in adults: High incidence in patients with a postneurosurgical state and strains not susceptible to third-generation cephalosporins. J. Clin. Neurosci. 54, 83–87. doi: 10.1016/j.jocn.2018.06.019
Lindsay J. A. (2014). Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int. J. Med. Microbiol. 304, 103–109. doi: 10.1016/j.ijmm.2013.11.010
Liu W., Bai L., Zhou X., Liu L. L. L., Chen D., Liu L. L. L., et al. (2018c). Genetic diversity, multidrug resistance, and virulence of Citrobacter freundii from diarrheal patients and healthy individuals. Front. Cell. Infect. Microbiol. 8. doi: 10.3389/fcimb.2018.00233
Liu H. W., Chang C. J., Hsieh C. T. (2015a). Brain abscess caused by Citrobacter koseri infection in an adult. Neurosciences 20, 170–172. doi: 10.17712/nsj.2015.2.20140749
Liu L., Hao S., Lan R., Wang G., Xiao D., Sun H., et al. (2015b). The type VI secretion system modulates flagellar gene expression and secretion in Citrobacter freundii and contributes to adhesion and cytotoxicity to host cells. Infect. Immun. 83, 2596–2604. doi: 10.1128/IAI.03071-14
Liu L., Lan R., Liu L., Wang Y., Zhang Y., Wang Y., et al. (2017). Antimicrobial resistance and cytotoxicity of Citrobacter spp. in Maanshan Anhui Province, China. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.01357
Liu L., Qin L., Hao S., Lan R., Xu B., Guo Y., et al. (2020). Lineage, antimicrobial resistance and virulence of Citrobacter spp. Pathogens 9, 195. doi: 10.3390/pathogens9030195
Liu L., Song L., Deng R., Lan R., Jin W., Tran Van Nhieu G., et al. (2021a). Citrobacter freundii Activation of NLRP3 Inflammasome via the Type VI Secretion System. J. Infect. Dis. 223, 2174–2185. doi: 10.1093/infdis/jiaa692
Liu B. T., Wang X. M., Liao X. P., Sun J., Zhu H. Q., Chen X. Y., et al. (2011). Plasmid-mediated quinolone resistance determinants oqxAB and aac(6’)-ib-cr and extended-spectrum $β$-lactamase gene blaCTX-M-24 co-located on the same plasmid in one Escherichia coli strain from China. J. Antimicrob. Chemother. 66, 1638–1639. doi: 10.1093/jac/dkr172
Liu L. H., Wang N. Y., Wu A. Y. J., Lin C. C., Lee C. M., Liu C. P. (2018b). Citrobacter freundii bacteremia: Risk factors of mortality and prevalence of resistance genes. J. Microbiol. Immunol. Infect. 51, 565–572. doi: 10.1016/j.jmii.2016.08.016
Liu J., Yang Y., Li Y., Liu D., Tuo H., Wang H., et al. (2018a). Isolation of an IncP-1 plasmid harbouring mcr-1 from a chicken isolate of Citrobacter braakii in China. Int. J. Antimicrob. Agents 51, 936–940. doi: 10.1016/J.IJANTIMICAG.2017.12.030
Liu L., Zhang L., Zhou H., Yuan M., Hu D., Wang Y., et al. (2021b). Antimicrobial resistance and molecular characterization of citrobacter spp. Causing extraintestinal infections. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.737636
Liu B., Zheng D., Zhou S., Chen L., Yang J. (2022). VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Luo X., Yu L., Feng J., Zhang J., Zheng C., Hu D., et al. (2022). Emergence of Extensively Drug-Resistant ST170 Citrobacter portucalensis with Plasmids pK218-KPC, pK218-NDM, and pK218-SHV from a Tertiary Hospital, China. Microbiol. Spectr. 10, 1–13. doi: 10.1128/spectrum.02510-22
Manohar P., Madurantakam Royam M., Loh B., Bozdogan B., Nachimuthu R., Leptihn S. (2022). Synergistic Effects of Phage-Antibiotic Combinations against Citrobacter amalonaticus. ACS Infect. Dis. 8, 59–65. doi: 10.1021/acsinfecdis.1c00117
Maraki S., Vardakas K. Z., Mavromanolaki V. E., Kyriakidou M., Spais G., Kofteridis D. P., et al. (2017). In vitro susceptibility and resistance phenotypes in contemporary Citrobacter isolates in a University Hospital in Crete, Greece. Infect. Dis. (Auckl). 49, 532–539. doi: 10.1080/23744235.2017.1297896
McPherson C., Gal P., Ransom J. L. (2008). Treatment of Citrobacter koseri infection with ciprofloxacin and cefotaxime in a preterm infant. Ann. Pharmacother. 42, 1134–1138. doi: 10.1345/APH.1L008
Metri B. C., Jyothi P., Peerapur B. V. (2013). Antibiotic resistance in Citrobacter spp. isolated from urinary tract infection. Urol. Ann. 5, 312–313. doi: 10.4103/0974-7796.120295
Minarini L. (2012). Mutations in the quinolone resistance-determining regions of. Braz. J. Microbiol. 43, 1309–1314.
Mohanty S., Singhal R., Sood S., Dhawan B., Kapil A., Das B. K. (2007). Citrobacter infections in a tertiary care hospital in Northern India. J. Infect. 54, 58–64. doi: 10.1016/j.jinf.2006.01.015
Moser A. I., Keller P. M., Campos-Madueno E. I., Poirel L., Nordmann P., Endimiani A. (2021). A Patient With Multiple Carbapenemase Producers Including an Unusual Citrobacter sedlakii Hosting an IncC blaNDM-1- and armA-carrying Plasmid. Pathog. Immun. 6, 119–134. doi: 10.20411/PAI.V6I2.482
Moussa S., Mamadou G., Binta G., Mamoudou D., Yannick M. N., Alimata K., et al. (2023). Citrobacter freundii skin infection in an immunocompetent subject simulating varicella: a treatment-resistant case article information. Dermatology 8, 1–3. doi: 10.17140/DRMTOJ-8-152
Nair G., Lakshminarayana M., Nagaraj L., Kumar M. N., Thilak S. A. (2020). Urinary infections and Citrobacter: An unpleasant scenario. Int. J. Adv. Community Med. 3, 249–253. doi: 10.33545/comed.2020.v3.i1d.217
Oberhettinger P., Schüle L., Marschal M., Bezdan D., Ossowski S., Dörfel D., et al. (2020). Description of Citrobacter cronae sp. Nov., isolated from human rectal swabs and stool samples. Int. J. Syst. Evol. Microbiol. 70, 2998–3003. doi: 10.1099/ijsem.0.004100
Osei Sekyere J., Reta M. A. (2021). Global evolutionary epidemiology and resistome dynamics of Citrobacter species, Enterobacter hormaechei, Klebsiella variicola, and Proteeae clones. Environ. Microbiol. 23, 7412–7431. doi: 10.1111/1462-2920.15387
Pardia S. N., Verma I. C., Deb M., Bhujwala R. A. (1980). An outbreak of diarrhea due to Citrobacter freundii in a neonatal special care nursery. Indian J. Pediatr. 47, 81–84. doi: 10.1007/BF02900180
Park Y. J., Yu J. K., Lee S., Oh E. J., Woo G. J. (2007). Prevalence and diversity of qnr alleles in AmpC-producing Enterobacter cloacae, Enterobacter aerogenes, Citrobacter freundii and Serratia marcescens: A multicentre study from Korea. J. Antimicrob. Chemother. 60, 868–871. doi: 10.1093/jac/dkm266
Pepperell C., Kus J. V., Gardam M. A., Humar A., Burrows L. L. (2002). Low-virulence Citrobacter species encode resistance to multiple antimicrobials. Antimicrob. Agents Chemother. 46, 3555–3560. doi: 10.1128/AAC.46.11.3555-3560.2002
Perilli M., Forcella C., Celenza G., Frascaria P., Segatore B., Pellegrini C., et al. (2009). Evidence for qnrB1 and aac(6′)-Ib-cr in CTX-M-15–producing uropathogenic Enterobacteriaceae in an Italian teaching hospital. Diagn. Microbiol. Infect. Dis. 64, 90–93. doi: 10.1016/j.diagmicrobio.2009.01.009
Phuadraksa T., Wichit S., Songtawee N., Tantimavanich S., Isarankura-Na-Ayudhya C., Yainoy S. (2023). Emergence of plasmid-mediated colistin resistance mcr-3.5 gene in Citrobacter amalonaticus and Citrobacter sedlakii isolated from healthy individual in Thailand. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.1067572
Plakkal N., Soraisham A. S., Amin H. (2013). Citrobacter freundii brain abscess in a preterm infant: A case report and literature review. Pediatr. Neonatol. 54, 137–140. doi: 10.1016/j.pedneo.2012.10.004
Poire L., Ros A., Carricajo A., Berthelot P., Pozzetto B., Bernabeu S., et al. (2011). Extremely drug-resistant citrobacter freundii isolate producing NDM-1 and other carbapenemases identified in a patient returning from India. Antimicrob. Agents Chemother. 55, 447–448. doi: 10.1128/AAC.01305-10
Porres-Osante N., Estepa V., Seral C., Rojo-Bezares B., Salvo S., Algarate S., et al. (2014). First description of a blaVIM-2-carrying Citrobacter freundii isolate in Spain. Antimicrob. Agents Chemother. 58, 6331–6332. doi: 10.1128/AAC.03168-14
Qian C., Du Y., Li H., Wu P., Wang L., Wei Y., et al. (2018). Development of rapid and simple experimental and in silico serotyping systems for Citrobacter. Future Microbiol. 13, 1511–1522. doi: 10.2217/fmb-2018-0187
Rahman A., Styczynski A., Khaleque A., Hossain S. A., Sadique A., Hossain A., et al. (2022). Genomic landscape of prominent XDR Acinetobacter clonal complexes from Dhaka, Bangladesh. BMC Genomics 23. doi: 10.1186/s12864-022-08991-x
Raia D. D., Barbareschi M., Veraldi S. (2015). Citrobacter koseri folliculitis of the face. Infection 43, 595–597. doi: 10.1007/s15010-015-0734-5
Räisänen K., Sarvikivi E., Arifulla D., Pietikäinen R., Forsblom-Helander B., Tarkka E., et al. (2021). Three clusters of carbapenemase-producing Citrobacter freundii in Finland 2016-20. J. Antimicrob. Chemother. 76, 2697–2701. doi: 10.1093/jac/dkab209
Ramachandran K., Patel Y., Shetty A. P., Shanmuganathan R. (2022). Citrobacter koseri as a rare cause of hematogenous pyogenic spondylodiscitis in young adult – A case report. J. Orthop. Rep. 1, 51–54. doi: 10.1016/j.jorep.2022.03.005
Ramsamy Y., Mlisana K. P., Amoako D. G., Allam M., Ismail A., Singh R., et al. (2020). Pathogenomic analysis of a novel extensively drug-resistant Citrobacter freundii isolate carrying a blandm-1 carbapenemase in South Africa. Pathogens 9, 89. doi: 10.3390/pathogens9020089
Rather P. N., Munayyer H., Mann P. A., Hare R. S., Miller G. H., Shaw K. J. (1992). Genetic analysis of bacterial acetyltransferases: identification of amino acids determining the specificities of the aminoglycoside 6’-N-acetyltransferase Ib and IIa proteins. J. Bacteriol. 174, 3196–3203. doi: 10.1128/JB.174.10.3196-3203.1992
Ribeiro T. G., Izdebski R., Urbanowicz Pawełand Carmeli Y., Gniadkowski M., Peixe L., Urbanowicz P., et al. (2021). Citrobacter telavivum sp. nov. with chromosomal mcr-9 from hospitalized patients. Eur. J. Clin. Microbiol. Infect. Dis. 40, 123–131. doi: 10.1007/s10096-020-04003-6
Robicsek A., Strahilevitz J., Jacoby G. A., Macielag M., Abbanat D., Chi H. P., et al. (2006). Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12, 83–88. doi: 10.1038/nm1347
Rosenthal V. D., Belkebir S., Zand F., Afeef M., Tanzi V. L., Al-Abdely H. M., et al. (2020). Six-year multicenter study on short-term peripheral venous catheters-related bloodstream infection rates in 246 intensive units of 83 hospitals in 52 cities of 14 countries of Middle East: Bahrain, Egypt, Iran, Jordan, Kingdom of Saudi Arabia, Kuwait, Leb. J. Infect. Public Health 13, 1134–1141. doi: 10.1016/j.jiph.2020.03.012
Rostamzad A., Abozar A. C. K., Omidi M., Hatamnia A. A. (2019). Antibiotic resistance of Citrobacter freundii in clinical isolates: A systematic review and meta-analysis. J. Basic Res. Med. Sci. 6, 58–63.
Samonis G., Karageorgopoulos D. E., Kofteridis D. P., Matthaiou D. K., Sidiropoulou V., Maraki S., et al. (2009). Citrobacter infections in a general hospital: characteristics and outcomes. Eur. J. Clin. Microbiol. Infect. Dis. 28, 61–68. doi: 10.1007/s10096-008-0598-z
Santos C., Ramalheira E., Da Silva G., Mendo S. (2017). Genetically unrelated multidrug- and carbapenem-resistant Citrobacter freundii detected in outpatients admitted to a Portuguese hospital. J. Glob. Antimicrob. Resist. 8, 18–22. doi: 10.1016/J.JGAR.2016.09.010
Schoch C. L., Ciufo S., Domrachev M., Hotton C. L., Kannan S., Khovanskaya R., et al. (2020). NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020. doi: 10.1093/database/baaa062
Sennati S., Di Pilato V. D., Riccobono E., Maggio T., Villagran A. L., Pallecchi L., et al. (2017). Citrobacter braakii carrying plasmid-borne mcr-1 colistin resistance gene from ready-to-eat food from a market in the Chaco region of Bolivia. J. Antimicrob. Chemother. 72, 2127–2129. doi: 10.1093/JAC/DKX078
Shaw K. J., Rather P. N., Hare R. S., Miller G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138. doi: 10.1128/MR.57.1.138-163.1993
Shih C. C., Chen Y. C., Chang S. C., Luh K. T., Hsieh W. C. (1996). Bacteremia due to Citrobacter species: Significance of primary intraabdominal infection. Clin. Infect. Dis. 23, 543–549. doi: 10.1093/clinids/23.3.543
Shinu P. (2022). Antimicrobial resistance, phenotypic characteristics, and biofilm production in Citrobacter freundii isolates obtained from urinary tract infections. J. Pharmacol. Pharmacother. 13, 375–381. doi: 10.1177/0976500X221147747
Southern P. M., Bagby M. K. (1977). Antimicrobial susceptibility patterns (Antibiograms) as an aid in differentiating citrobacter species. Am. J. Clin. Pathol. 67, 187–189. doi: 10.1093/ajcp/67.2.187
Strahilevitz J., Jacoby G. A., Hooper D. C., Robicsek A. (2009). Plasmid-mediated quinolone resistance: A multifaceted threat. Clin. Microbiol. Rev. 22, 664–689. doi: 10.1128/CMR.00016-09
Tadesse S., Mulu W., Genet C., Kibret M., Belete M. A. (2022). Emergence of high prevalence of extended-spectrum beta-lactamase and carbapenemase-producing enterobacteriaceae species among patients in Northwestern Ethiopia Region. BioMed. Res. Int. 2022, 1–9. doi: 10.1155/2022/5727638
Tripathi S., Chauhan A., Kumar M., Shukla S., Srivastava C. (2020). Citrobacter sedlakii – A rare cause of meningitis and brain abscess in a neonate. Indian J. Child Health 7, 468–470. doi: 10.32677/IJCH.2020.v07.i11.0010
Urbinati F., Rocha-de-Lossada C., García-Montesinos J., García-Lorente M., Borroni D., Pardo-Ruiz R., et al. (2023). Citrobacter koseri as emergent microorganism in infectious keratitis. J. Fr. Ophtalmol. 46, e30–e33. doi: 10.1016/J.JFO.2022.06.011
Vakulenko S. B., Mobashery S. (2003). Versatility of aminoglycosides and prospects for their future. Clin. Microbiol. Rev. 16, 430. doi: 10.1128/CMR.16.3.430-450.2003
Vaz Marecos C., Ferreira M., Ferreira M. M., Barroso M. R. (2012). Sepsis, meningitis and cerebral abscesses caused by Citrobacter koseri. BMJ Case Rep., 1–5. doi: 10.1136/bcr.10.2011.4941
Wang H., Hou H., Huang J. (2021). Citrobacter arsenatis sp. nov., an arsenate-reducing bacterium isolated from freshwater sediment. Antonie van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 114, 1285–1292. doi: 10.1007/s10482-021-01601-y
Weigel L. M., Steward C. D., Tenover F. C. (1998). gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrob. Agents Chemother. 42, 2661–2667. doi: 10.1128/aac.42.10.2661
Werkman C. H., Gillen G. F. (1932). Bacteria producing trimethylene glycol. J. Bacteriol. 23, 167–182. doi: 10.1128/jb.23.2.167-182.1932
Whitby J. L., Muir G. G. (1961). Bacteriological studies of urinary tract infection. Br. J. Urol. 33, 130–134. doi: 10.1111/j.1464-410X.1961.tb11595.x
Woloj M., Tolmasky M. E., Roberts M. C., Crosa J. H. (1986). Plasmid-encoded amikacin resistance in multiresistant strains of Klebsiella pneumoniae isolated from neonates with meningitis. Antimicrob. Agents Chemother. 29, 315. doi: 10.1128/AAC.29.2.315
Yang L., Li P. P., Liang B., Hu X., Li J., Xie J., et al. (2018). Multidrug-resistant Citrobacter freundii ST139 co-producing NDM-1 and CMY-152 from China. Sci. Rep. 8, 1–7. doi: 10.1038/s41598-018-28879-9
Yan Hu Y., ling Wang Y., ling Sun Q., Huang Z. X., Wang H. Y., Zhang R., et al. (2017). Colistin resistance gene mcr-1 in gut flora of children. Int. J. Antimicrob. Agents 50, 593–597. doi: 10.1016/J.IJANTIMICAG.2017.06.011
Yao Y., Falgenhauer L., Falgenhauer J., Hauri A. M., Heinmüller P., Domann E., et al. (2021). Carbapenem-resistant Citrobacter spp. as an emerging concern in the hospital-setting: results from a genome-based regional surveillance study. Front. Cell. Infect. Microbiol. 11. doi: 10.3389/fcimb.2021.744431
Yap P. S. X., Ahmad Kamar A., Chong C. W., Yap I. K. S., Teh C. S. J. (2020). Whole genome analysis of multidrug resistant Citrobacter freundii B9-C2 isolated from preterm neonate’s stool in the first week. J. Glob. Antimicrob. Resist. 21, 246–251. doi: 10.1016/j.jgar.2020.03.024
Yuan C., Yin Z., Wang J., Qian C., Wei Y., Zhang S., et al. (2019). Comparative genomic analysis of Citrobacter and key genes essential for the pathogenicity of citrobacter koseri. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02774
Zhang T., Lin Y., Li P. P., Li Z., Liu X., Li J., et al. (2021). Characterization of plasmid co-harboring ndm-1 and shv-12 from a multidrug-resistant Citrobacter freundii strain zt01-0079 in China. Infect. Drug Resist. 14, 947–952. doi: 10.2147/IDR.S301736
Zheng J., Leung K. Y. (2007). Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66, 1192–1206. doi: 10.1111/j.1365-2958.2007.05993.x
Keywords: Citrobacter spp., epidemiology, pathogenesis, multidrug resistance, treatment
Citation: Jabeen I, Islam S, Hassan AKMI, Tasnim Z and Shuvo SR (2023) A brief insight into Citrobacter species - a growing threat to public health. Front. Antibiot. 2:1276982. doi: 10.3389/frabi.2023.1276982
Received: 13 August 2023; Accepted: 30 October 2023;
Published: 05 December 2023.
Edited by:
Latiful Bari, University of Dhaka, BangladeshReviewed by:
Mohammad Sholeh, Pasteur Institute of Iran (PII), IranItziar Chapartegui-González, Karolinska Institutet (KI), Sweden
Copyright © 2023 Jabeen, Islam, Hassan, Tasnim and Shuvo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabbir R. Shuvo, c2FiYmlyLnNodXZvQG5vcnRoc291dGguZWR1
 Ishrat Jabeen
Ishrat Jabeen Sohidul Islam
Sohidul Islam Zerin Tasnim
Zerin Tasnim Sabbir R. Shuvo
Sabbir R. Shuvo