- 1Department of Biomedical Science, College of Veterinary Medicine and Animal Science, University of Gondar, Gondar, Ethiopia
- 2Department of Veterinary Epidemiology and Public Health, College of Veterinary Medicine and Animal Science, University of Gondar, Gondar, Ethiopia
- 3Department of Veterinary Pharmacy, College of Veterinary Medicine and Animal Science, University of Gondar, Gondar, Ethiopia
- 4Department of Veterinary Clinical Medicine, College of Veterinary Medicine and Animal Science, University of Gondar, Gondar, Ethiopia
Background: Staphylococcus aureus (S. aureus) infecting animals and humans via close contact, handling, or consuming contaminated products is a growing public health concern. In Ethiopia, it is important to examine the overall prevalence of S. aureus, patterns of multidrug resistance, and potential risks in human-animal interface settings. Thus, this review was conducted to estimate the pooled prevalence of S. aureus, its multidrug resistance, and potential risk factors for worker-animal-working equipment interactions.
Methods: This systematic review and meta-analysis were carried out by the PRISMA guidelines. The research articles were searched from PubMed, HINARI, Web of Sciences, and Google Scholar databases.
Results: This meta-analysis included 13 independent articles and 52 dependent studies. In total, 5,329 humans, 5,475 animals, and 5,119 samples of working equipment were analyzed. The pooled prevalence of S. aureus at the interfaces between humans, animals, and working equipment was 22%, there was a high level of heterogeneity (I2 = 94%: p < 0.01). The overall pooled prevalence of S. aureus in dairy farm sources was 23% (95% CI, 17–30%) compared to 18% in abattoirs. The pooled prevalence of S. aureus was estimated to be 25% for human sources, 23% for animal sources, and 19% for working equipment. The total multidrug resistance (MDR) rate was 27%. The present study illustrates that a predominant antimicrobials comprising ampicillin, penicillin, chloramphenicol, tetracycline, and ciprofloxacin, accounts for the development of resistance in S. aureus strains, with a prevalence of 72%. According to the qualitative assessment of potential risk factors, animal age, worker education, lactation stage, and hand washing by milkers influenced the circulation of S. aureus at animal-worker and working equipment interfaces.
Conclusion: The pooled prevalence of S. aureus at the interface of human,-and animal-working equipment was quantified at 22%. S. aureus was found in humans, animals, and equipment at nearly the same rate. The results of this study demonstrate that S. aureus is hazardous and circulates among animals, workers, and equipment: farmers, animal owners, employees, and the public need to be educated about S. aureus. Moreover, animals and work equipment should be included in the control and prevention of S. aureus infection.
Introduction
In pastoral environments characterized by close human-animal interactions, Staphylococcus aureus has emerged as a zoonotic concern of public health importance due to the prevalence of multidrug-resistant microorganisms. It has long been recognized as a significant concern in terms of public health (1). This bacterium, which exhibits a positive Gram stain, displays a vast geographic dispersion, as evidenced by investigations conducted in diverse regions across the globe (1, 2). The frequency of food products contaminated with S. aureus varies, with cereals having the highest frequency and meat and bean products having the lowest. The hospital and community settings have been the sites of methicillin-resistant S. aureus (MRSA) strains, with several dominant clones reported in various countries (3, 4). Antibiotic-resistant genotypes and host epidemiology have combined to cause the spread of community-associated MRSA lineages, changing the transmission dynamics and resulting in sustained transmission in regional population centers (1, 5).
In addition to causing zoonotic diseases, it can lead to mastitis, bacteremia, and food-borne poisoning in humans (1, 6). Staphylococcal enterotoxins are a prominent source of food-borne outbreaks that are frequently linked to S. aureus (5, 7). Since S. aureus frequently co-occurs with humans, care must be taken to lower the possibility of contamination when preparing food. The presence of biofilm-related medical equipment infections is also a significant issue. Currently, methicillin-resistant S. aureus infections have become endemic worldwide (1, 2, 7).
Research has indicated a significant frequency of S. aureus colonization among employees in various work environments, such as bakeries, pig farms, fire departments, and healthcare facilities (4). Investigations into the presence of S. aureus in cows, their handlers, and their immediate environment have also revealed a diversity of clonal lineages and suggested a potential route of transmission between handlers and cows (4, 8). As it has been pointed out in the different studies, there is a potential occupational contact with swine, such as that experienced by hog slaughterhouse workers, influences nasal S. aureus colonization (9, 10). Among the antibiotic resistance genes, variations in lineage distribution and virulence factors of S. aureus have been studied in pig workers compared to non-workers. Due to their close contact with livestock treated with antibiotics, pig workers are at higher risk of carrying antibiotic-resistant S. aureus (11). Methicillin-susceptible S. aureus (MSSA) strains with variable virulence factors and antibiotic-resistance genes have been detected in swine populations (11, 12). Furthermore, research on the epidemiology and genetic relatedness of the mecA gene in S. aureus strains isolated from pets, contact persons with pets, and veterinary clinical environments has revealed the increasing prevalence of methicillin-resistant S. aureus (MRSA) strains at animal-human-environment interfaces (5).
Staphylococcal Cassette Chromosome mec (SCCmec) is indeed a critical genetic element responsible for the multidrug resistance in S. aureus strains, particularly methicillin-resistant S. aureus (MRSA). SCCmec is a mobile genetic element that carries the mecA gene, which encodes penicillin-binding protein 2a (PBP2a). PBP2a has a low affinity for β-lactam antibiotics, including methicillin and other β-lactam antibiotics, rendering the bacteria resistant to these drugs (13). Methicillin-resistant S. aureus is connected to hospitals, and the population is most commonly associated with type IV SCCmec elements, which are categorized into 15 categories (14). The whole-genome sequence of the SCCmec IVd (2B) subtype, one of several subtypes of SCCmec type IV strains, is still missing (13). Additionally, S. aureus has multiple transmissible genes, including plasmids and transposons. These genes spread antibiotic resistance genes between S. aureus strains that confer resistance to various antibiotics through mechanisms like enzymatic inactivation, altered target sites, and reduced intracellular drug accumulation (15). Little is known about the processes that facilitate the acquisition of foreign genes and the dynamics of their transfer between hosts (16).
Workers may be susceptible to colonization by S. aureus in a variety of work contexts. There is a high incidence of S. aureus colonization among healthcare workers (HCWs) in several nations, including Indonesia (17), Portugal (18), Ecuador (19), and Nigeria (20). Healthcare professionals who have previously worked in a hospital with a positive MRSA colonization history are at risk for S. aureus colonization (21), and other risk factors include not practicing hand hygiene, being male, being older, not washing your hands well, having a wound or skin infection, and recently using antibiotics (22–24).
Because S. aureus has a high incidence of antimicrobial resistance, it presents substantial clinical danger in Africa. Methicillin-resistant S. aureus (MRSA) is a contributing factor to hospital-associated and community-associated infections in Africa. Africa has yielded several clonal complexes (CCs), demonstrating the diversity of the clonal spread of MRSA across the continent. The three most frequently used MRSA strains are the ST5-IV [2B], ST8-IV [2B], and ST88-IV [2B] clones (3). Many CCs have MRSA that is Panton-Valentine leukocidin (PVL)-positive meticillin-resistant S. aureus and the frequency of PVL-positive MRSA varies from 0 to 77% in Africa (25). Inducible clindamycin resistance in S. aureus isolates varies from 2.9 to 44% in Africa; MRSA strains have a greater incidence of this resistance (26). Regarding the significance of antibiotic resistance in S. aureus, routine screening, prudent clindamycin usage, and molecular identification of resistance genes are advised (24, 27, 28). Kenyan community-associated MRSA (CA-MRSA) strains have been characterized by whole-genome sequencing, which has led to the discovery of rare sequence types (STs), such as ST7460 and ST7635 (28). Different spa types and clonal complexes have been identified by combining DNA microarray and spa typing to characterize S. aureus isolates from South Africa and Nigeria (3, 25, 29, 30).
Studies show that S. aureus is prevalent in a variety of contexts and is a major concern in Ethiopia. The country has a 23% overall pooled apparent prevalence of S. aureus in milk and meat samples according to a comprehensive study and meta-analysis (31). Another study that examined lines of cattle abattoirs revealed that 35.5% of the isolates were Staphylococcus, and 13.6% of those isolates were S. aureus (32). S. aureus is the most often isolated bacterium in burn wound infections, with an overall incidence of 57.8% (33). Methicillin-resistant S. aureus (MRSA) and other various strains were identified by molecular analysis of S. aureus isolates, underscoring the possibility of pandemic strains in the nation (34). Additionally, samples of cottage cheese and yoghourt included Staphylococcus species, with an overall prevalence of 14.3% and a specific prevalence of 22 and 6.5% in the former and yoghourt, respectively (35). Numerous researchers have examined the status of bacteria and the risk factors that are connected to the interface between personnel, animals, and working equipment, with a focus on Ethiopia (36–38). These results highlight the need for better sanitation habits, antibiotic stewardship, and public education to lower the prevalence of drug-resistant S. aureus infections in Ethiopia.
Thus, to support successful preventative and control initiatives, it is imperative to understand the general occurrence, multidrug resistance rate, and potential risk factors for S. aureus at the workers-animal-working utensils interface at the national level. Accordingly, this meta-analysis aimed to gather the available data and estimate the pooled prevalence of S. aureus and its antimicrobial resistance at the workers-animal-working interface in Ethiopia.
Methods
The PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-analysis) checklist (39) was used to conduct this review.
Search strategy
The literature search took place between October 2023 and November 28, 2023. A careful search technique was developed to conduct a comprehensive review of all relevant studies (B.A., and A.S). Various databases, including PubMed, Google Scholar, HINARI, Web of Science, and Snowball search engines, were used for retrieving articles, and other manual methods were also used to conduct the literature search to select the remaining included studies. This systematic review and meta-analysis used the CoCoPop (Condition, Context, and Population) and PEO (Population, Exposure, and Outcome) frameworks to search for relevant articles. The disease studied was Staphylococcus aureus (S. aureus) (Co), the context was Ethiopia (Co), and the population consisted of both animals and humans (Pop). The PubMed search strategy included Medical Subject Heading (MeSH) terms and a wide range of important keywords.
From an epidemiological perspective, S. aureus is a globally widespread, zoonotic, and transmissible bacterial infection that affects both humans and animals. The research question was formulated as follows: “What is the prevalence of S. aureus and associated risk factors at the interface between workers, animals, and equipment in Ethiopia?” The Boolean operator “AND/OR” was used for the online search, and we used this operator to identify relevant results by combining similar phrases and words. The search terms used were (Staphylococcus aureus OR S. aureus) AND (epidemiology OR prevalence OR infection rate) AND (cattle) AND people AND work equipment AND risk factors OR predisposing factors AND (Ethiopia). The language of the publications was limited to English. All identified studies were imported into End Note 20 software to avoid duplicate entries.
Operational definition
Interface
A place where two systems or subjects meet and interact.
Multidrug resistance
Resistant to three or more antimicrobial classes.
Methicillin-resistant Staphylococcus aureus
Staphylococcus aureus strains that are resistant to penicillin and β-lactams such as methicillin or oxacillin.
Community-associated MRSA infections
MRSA infections in healthy people who have not been hospitalized.
Animal
In this review stand for cattle.
Study eligibility criteria
Inclusion criteria
The inclusion criteria for this analysis specifically consisted of the following: (I) articles that presented a clear estimate of the proportion of S. aureus separately in animals and humans and implemented within the same article and study year; (II) observational studies that demonstrated the prevalence of S. aureus isolates at the human-animal interface and/or work equipment; (III) the human participants were the following populations who have direct contact with animals: dairy farm workers with frequent exposure to dairy animals, livestock owners and slaughterhouse workers, including butchers; (IV) the animal population also included domestic animals that were directly used in dairy cows and selected animals for slaughter; and (IV) the examination or sampling units had to be derived from workers (e.g., hand swabs and nasal swabs), animals (both slaughter and dairy cattle) and work equipment (including knives, milk tanks, and milking buckets).
Exclusion criteria
The following types of studies were not included in the analysis: those involving camels or other species, those with unclear or imprecise estimates of bacterial species across the affected host, review articles, Hospital workers, hospital working equipment, duplicates, summary-only studies, qualitative studies, or knowledge, attitude, and practice (KAP) questionnaire-based studies. Book chapters, case reports, editorials, short communications, opinions, or studies without original data. For studies that only used samples from dairies or slaughterhouses, the sample was excluded.
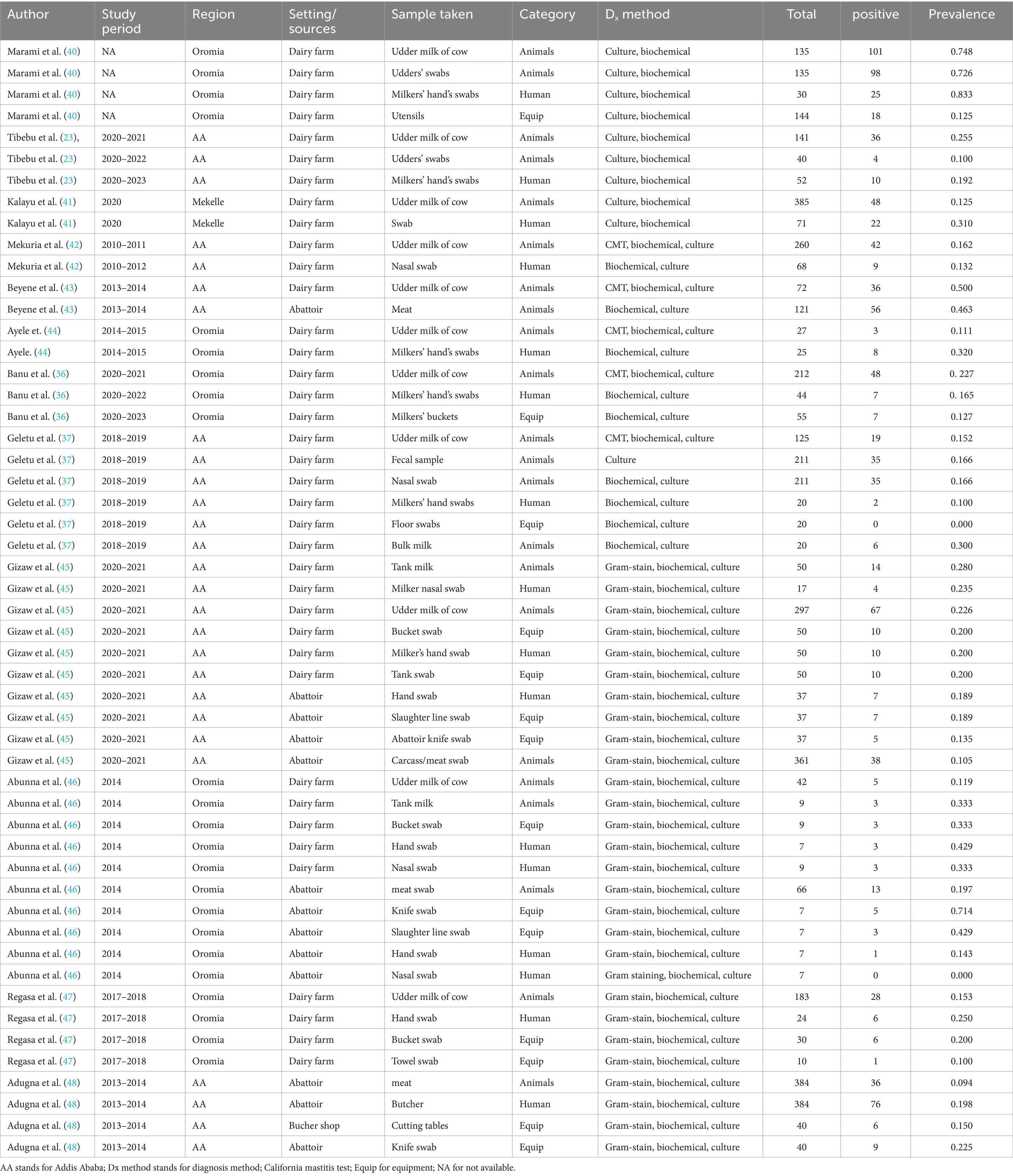
Data extraction
Data extraction was performed independently by four authors (M.D. and K.G.). The following information was collected from the included studies: the name of the first author and year of publication, study period, study design, study regions, sample source, type of sample collected, diagnostic method, overall examination, and positive events (Table 1).
Study quality assessment
An independent quality assessment was conducted by two researchers (A.S. and K.G) using the Newcastle–Ottawa Quality Assessment Scale (NOS) (Supplementary material S1).
Data synthesis and statistical analysis
To determine the pooled prevalence estimate for the S. aureus worker-animal-working equipment interfaces in the included studies, a meta-analysis with a random effects approach and a 95% confidence level were used to aggregate the studies. This meta-analysis was conducted using R software version 4.1.3 and included the overall effect size, heterogeneity, and weight of each study, as well as subgroup analysis. The Cochran Q test (reported as p value) and the inverse variance index (I2) were used to assess the degree of heterogeneity. The I2 index, estimated according to the explanation of Thompson and Higgins (49), was used to denote low, moderate, and high heterogeneity, with corresponding I2 values of 25, 50, and 75%, respectively. The presence of heterogeneity between studies was assessed using a forest plot. The forest plot diagram shows weights (both random and common effects), effect sizes, and 95% confidence intervals for individual studies (CLs). Additionally, subgroup analyses were performed to examine the prevalence of S. aureus based on the source of the pathogen (slaughterhouse or dairy farm). Small study effects and publication bias were visualized using funnel plot diagrams and Egger and Begg tests (50). Egger’s regression test was used to test funnel plot symmetry. A regression model is created with the standardized estimate of the effect size (event rate) as the dependent variable and the reciprocal of the standard error (1/SE) as the independent variable. If the intercept deviates significantly from zero, the estimate of the effect is considered biased (51).
Results
Search results
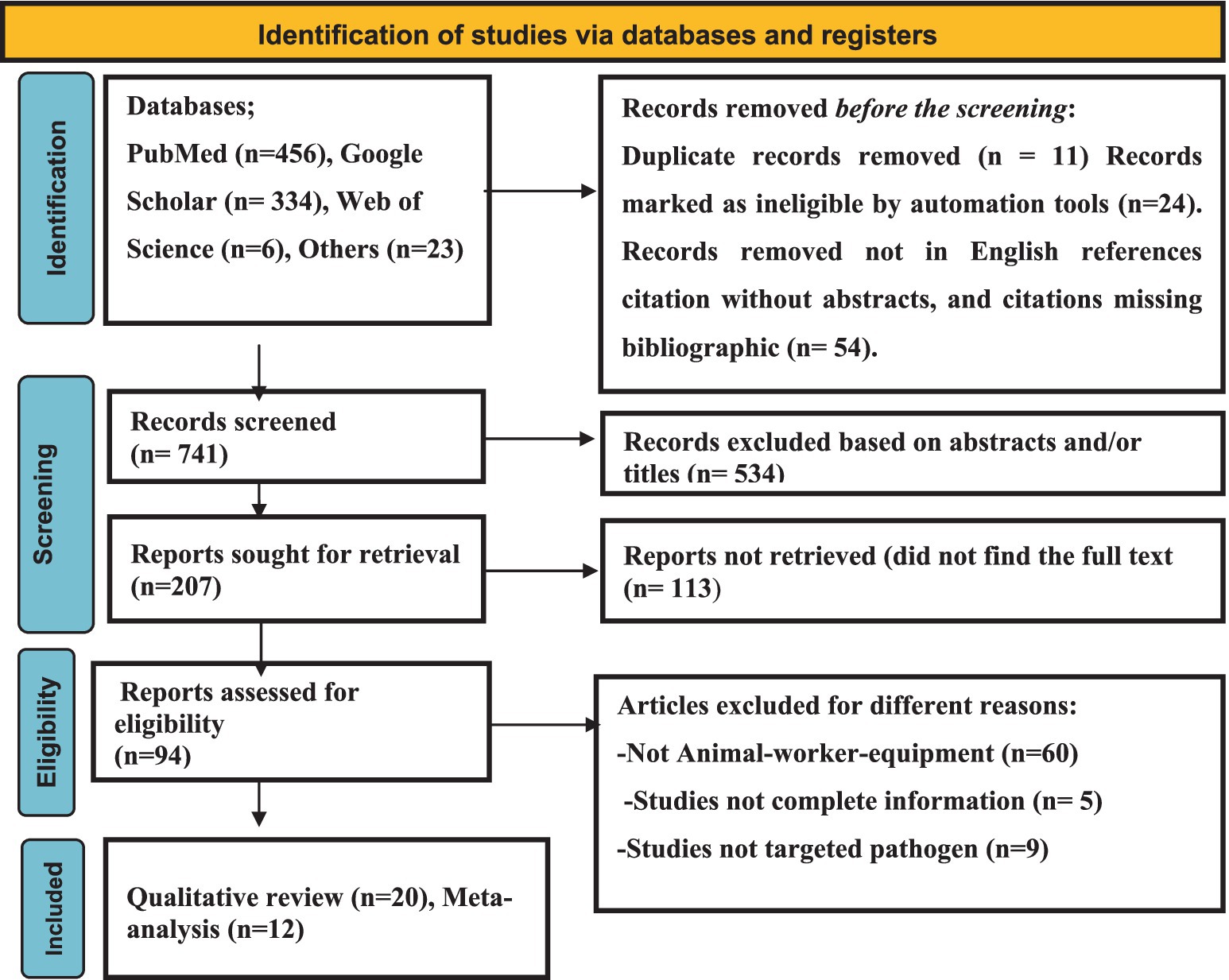
As shown in Figure 1, a total of 819 articles were browsed through different electronic databases and via other methods. A total of 11 duplicate articles were removed because they were considered ineligible for other reasons (n = 78), while 741 records were screened. A total of 534 articles were excluded through title and abstract screening. Two hundred-seven (n = 207) articles were retrieved, and 94 were evaluated for eligibility. Finally, 20 qualitative articles and 13 quantitative syntheses were included.
Characteristics of the included studies
The included studies were conducted in between 2010 and 2023 (Table 1). It should be noted that the same article was utilized multiple times due to different sampling units. The study designs employed were cross-sectional. Of the 13 articles, sex (23, 37, 42, 43, 45, 48) were conducted in Addis Ababa, the capital city of Ethiopia, and five (36, 40, 44, 46, 47) were conducted in the Oromia region. The remaining (41) two (17%) were found in the Tigray region, which is located in the northern part of the country. One of the included studies collected samples from the abattoir (meat), Bucher’s shop (equipment), and the butcher himself (human) (48), while another study collected samples from the environment (floor swabs), workers, and dairy cows (37). Most of the studies focused on collecting specimens from abattoirs, dairy settings, and workers.
Molecular characterization of isolates
Identified genes
Kalayu (41) performed the molecular characterization of isolates of S. aureus. A total of 70 isolates were examined for mecA and mecC possession, 48 of which came from the udder quarters of cows and 22 from the nares of farmers. MecA was detected in only one isolate that was taken from a farm worker’s nose. There was no mecA recorded from the cows. Furthermore, none of the isolates from humans or cows tested positive for mecC. The mecA-positive S. aureus isolates were identified as SCCmec type Iva and spa type t064 upon further examination. PCR amplification of the seven phenotypically MRSA isolates revealed that 3 (42.9%) of them were found to carry the femA gene, and 5 (71.4%) of them were found to carry mec A genes, which had molecular weights of 132 bp and 310 bp, respectively.
To further elaborate on the molecular characterization, two articles were included. Seven of the phenotypically MRSA isolates were screened for the presence of the mecA and femA genes by multiplex PCR according to the procedure described by Johnson et al. (52). The expected amplicon size for mecA was 162 bp, while that for mecC was 138 bp. in 20 (38.5%) of the isolates. However, only one (5%) of the 20 isolates was found to be positive for the mecA gene, and none of the isolates were found to carry the mecC gene. The mecA-positive isolate was obtained from a raw milk sample (53). Tigabu et al. (23) attempted to detect mecA-positive strains from dairy milk but neither found it. However, it should be noted that both studies targeted only mecA but not mecC (another gene responsible for methicillin resistance).
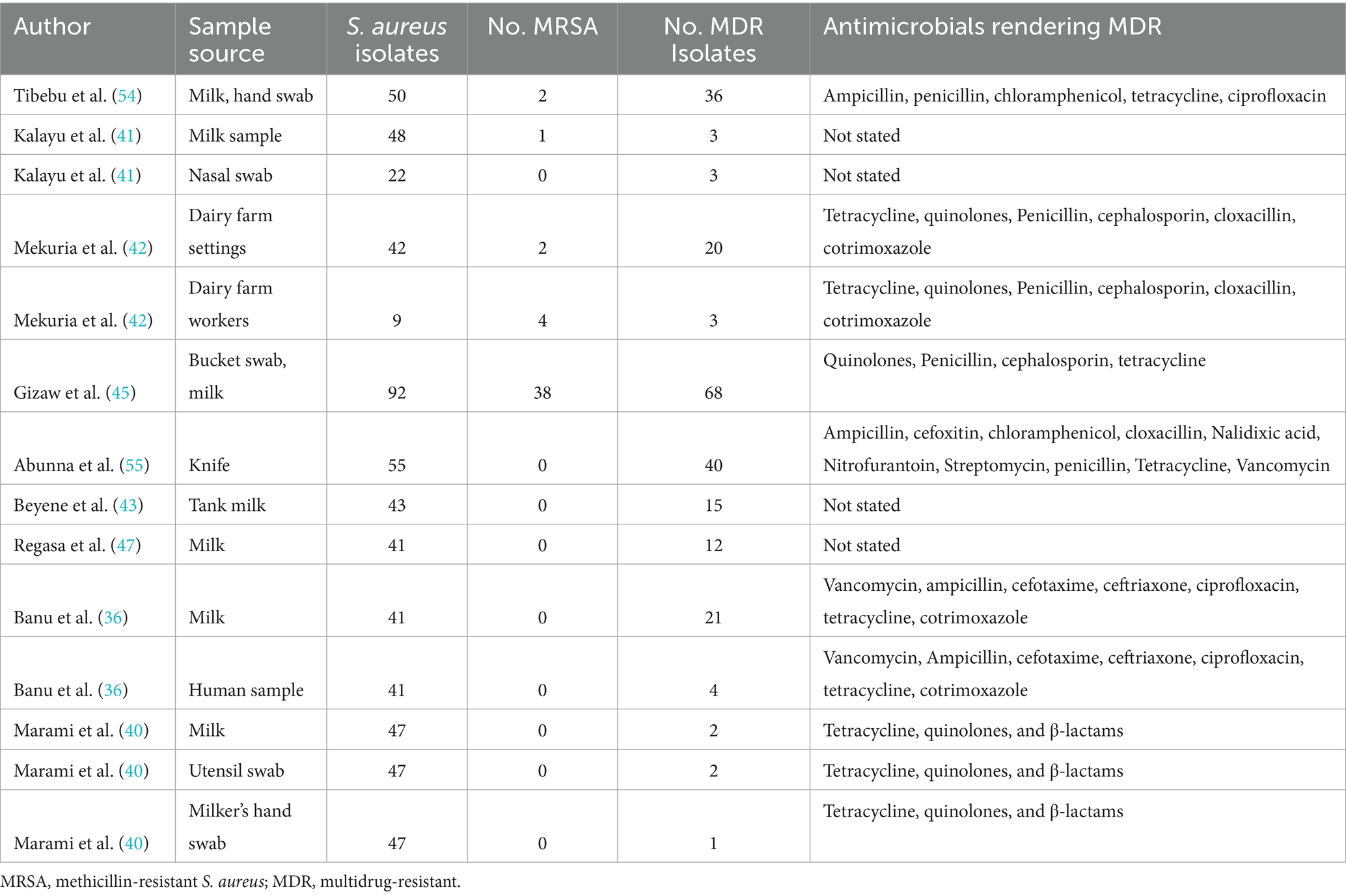
Multidrug-resistant and methicillin-resistant Staphylococcus aureus
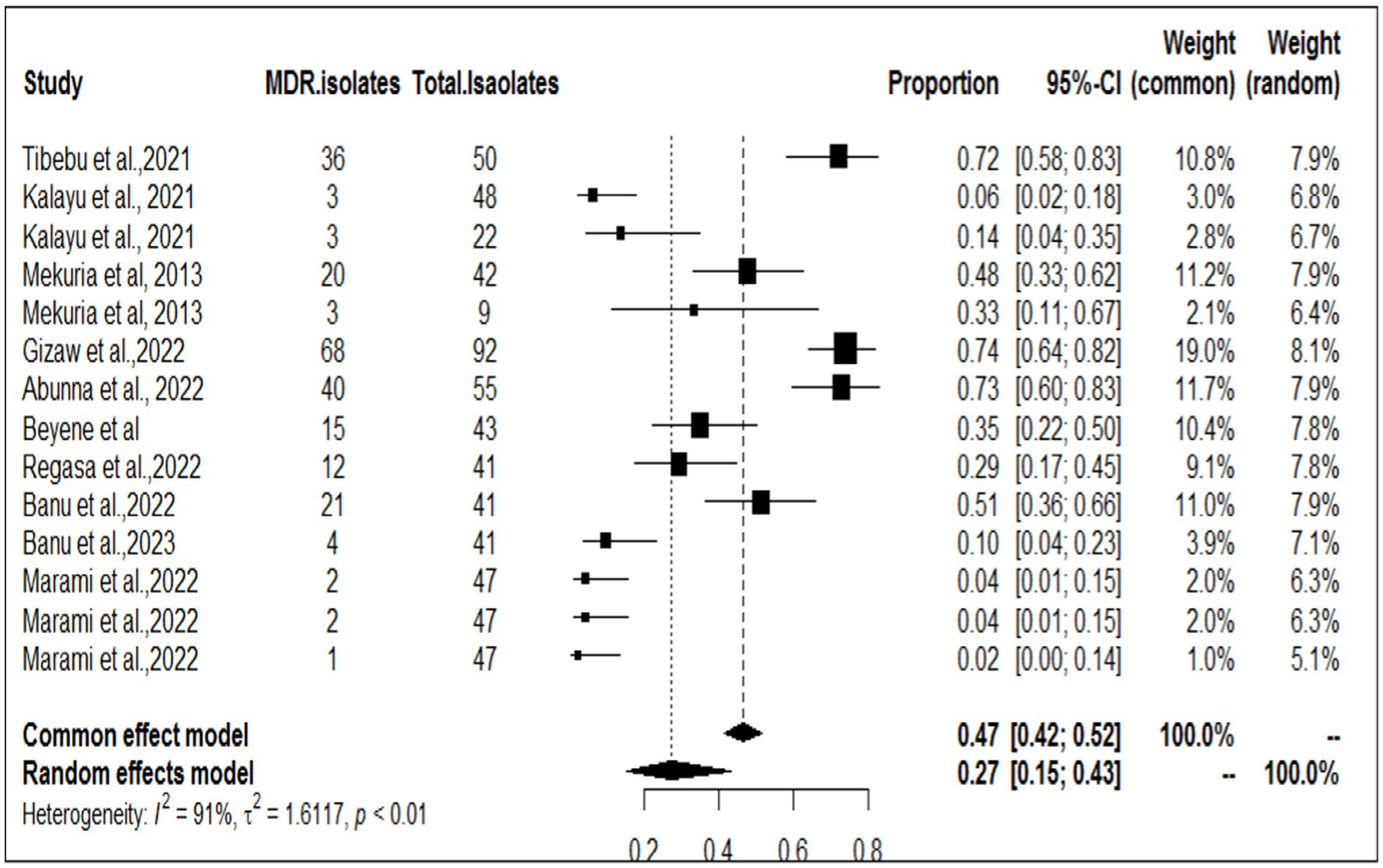
Among the studies included in this meta-analysis, nine examined the multidrug resistance profile of S. aureus, while five also investigated MERSA (Table 2). Tibebu et al. (23) reported that the highest resistance among the overall isolates was observed against penicillin, with a rate of 47 (94%), followed by ampicillin, which accounted for 46 (92%), and tetracycline, which accounted for 37 (74%). MRSA was detected in 2 (4%) isolates from cow milk samples collected from one dairy farm. Among the nine antibiotic disks used, mono-drug resistance was observed in 1 (2.08%) isolate, while the remaining isolates showed resistance to two, three, four, or five antimicrobials, accounting for 11 (22.92%), 32 (66.67%), 3 (6.25%), or 1 (2.08%) of the samples, respectively. The overall rate of MDR was 72% (ampicillin, penicillin, chloramphenicol, tetracycline, and ciprofloxacin), which was the highest proportion, followed by that reported by Gizaw et al. (56) (68/92, 67%). Among the included studies, Marami et al. (40) reported that five S. aureus isolates (two from raw milk, two from utensil swabs, and one from milker’s hand swabs) were resistant to three antimicrobial classes (tetracycline, quinolones, and β-lactams). The highest frequency of MERSA isolates was recorded by Gizaw et al. (45, 56) at (38/92, 41%). To determine the pooled resistance rate of MDR isolates of S. aureus, a meta-analysis was conducted across nine studies. The current meta-analysis revealed an overall multidrug resistance rate of 27% (95% CI, 15–43%; Figure 2) in different sampling units.
Meta-analysis and subgroup analysis
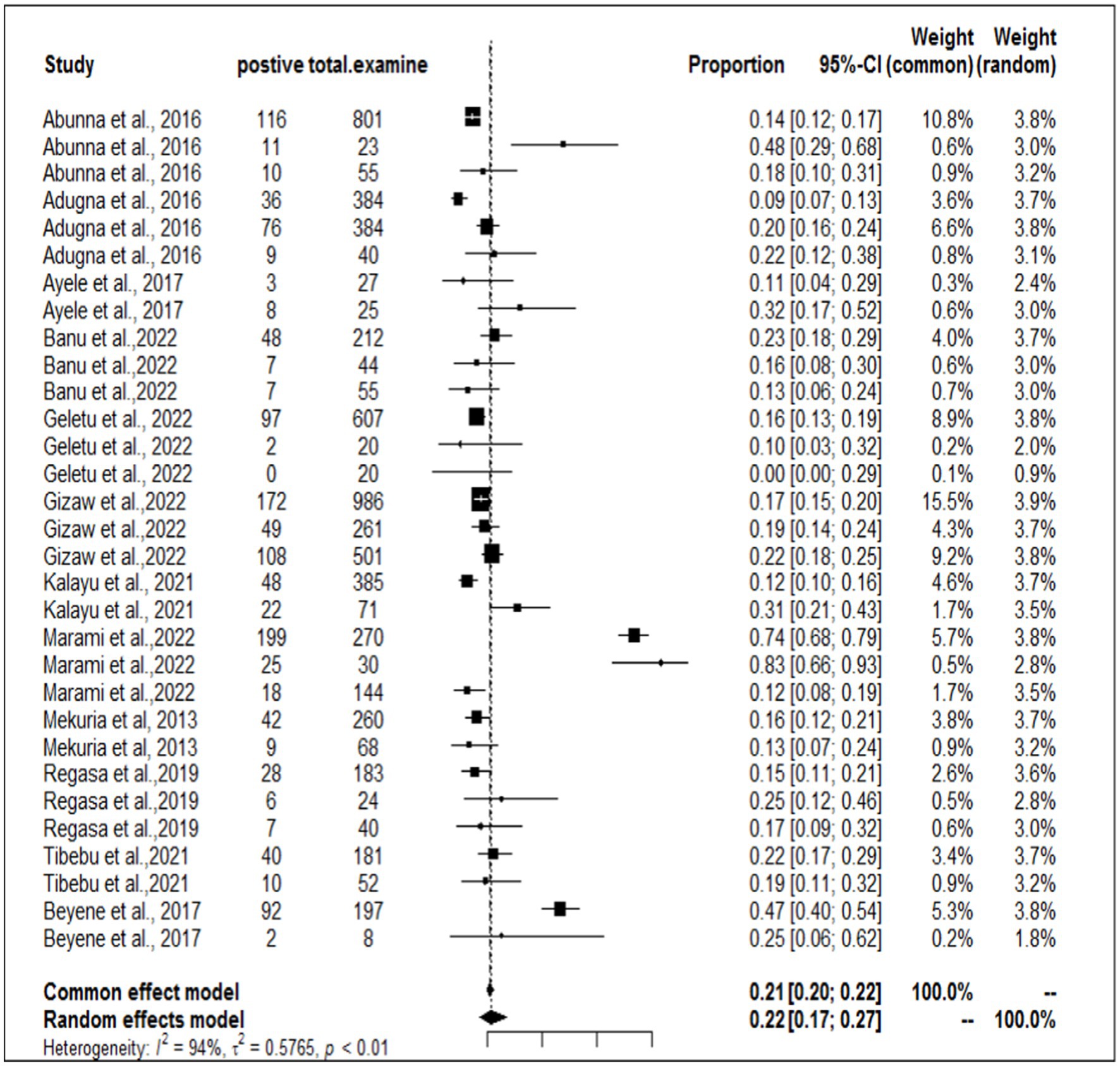
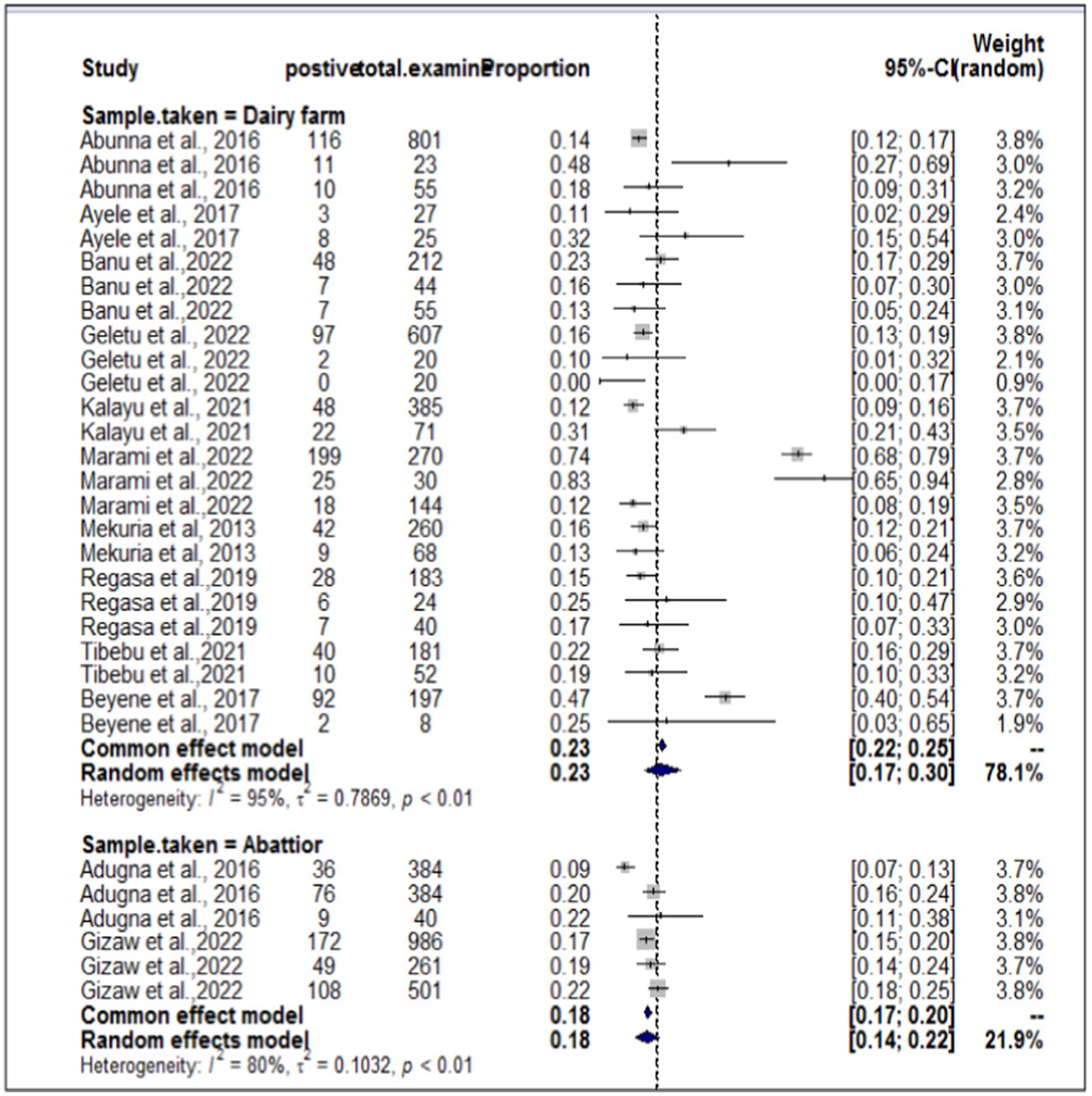
In the present meta-analysis, 13 independent articles and 31 dependent articles were included for subgroup analysis. It is important to note that certain articles were utilized multiple times due to their importance in similar years but different sampling units. The included studies demonstrated a high level of heterogeneity (I2 = 94%: τ2 = 0.5765; p < 0.01), and the estimated pooled prevalence of S. aureus at the interface between workers, animals, and working equipment was 22% (95% CI: 17–27%; Figure 3). The variability between studies was statistically significant (Q = 8139.7, DF = 30, p < 0.0001). The regression test for funnel plot asymmetry was conducted using a mixed-effects meta-regression model with standard error as the predictor (Eger’s test, b = −1.1073 (CI: −1.6238, −0.5908), z = −0.6382, p = 0.5234). Subgroup analyses were performed based on the sources of the samples (abattoir or dairy farm). The subgroup analyses of exploratory outcomes revealed greater heterogeneity across studies on dairy farms (I2 = 95%) than on abattoirs (I2 = 80%). Similarly, the subgroup analysis indicated that the overall pooled prevalence of S. aureus at the interface between animals, workers, and equipment was 23% greater for dairy farm sources (95% CI: 17–30%) than for abattoirs at 18% (95% CI: 14–22%; Figure 4).
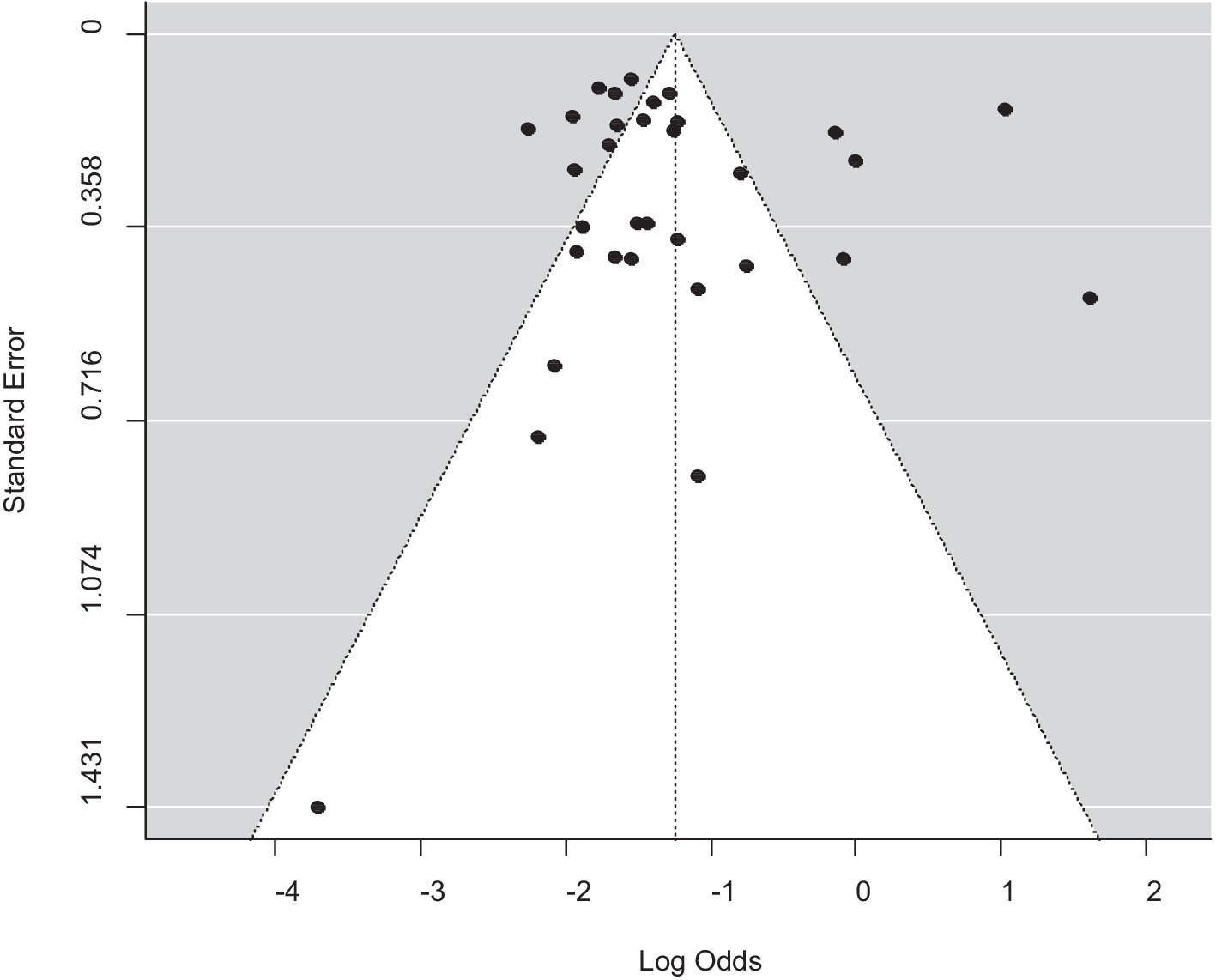
A funnel plot was used to assess publication bias, which was then supported by the Egger regression test. There was no indication of a symmetrical distribution of articles according to the funnel plot analysis (Figure 5). An analysis of funnel plot asymmetry was performed using a test for small-study effects. The results of Egger’s regression asymmetry, b = −1.1073 (CI: −1.6238, −0.5908; p = 0.5234), did not support the existence of publication bias.
Individual prevalence of Staphylococcus aureus in workers, animals, and equipment in the same article
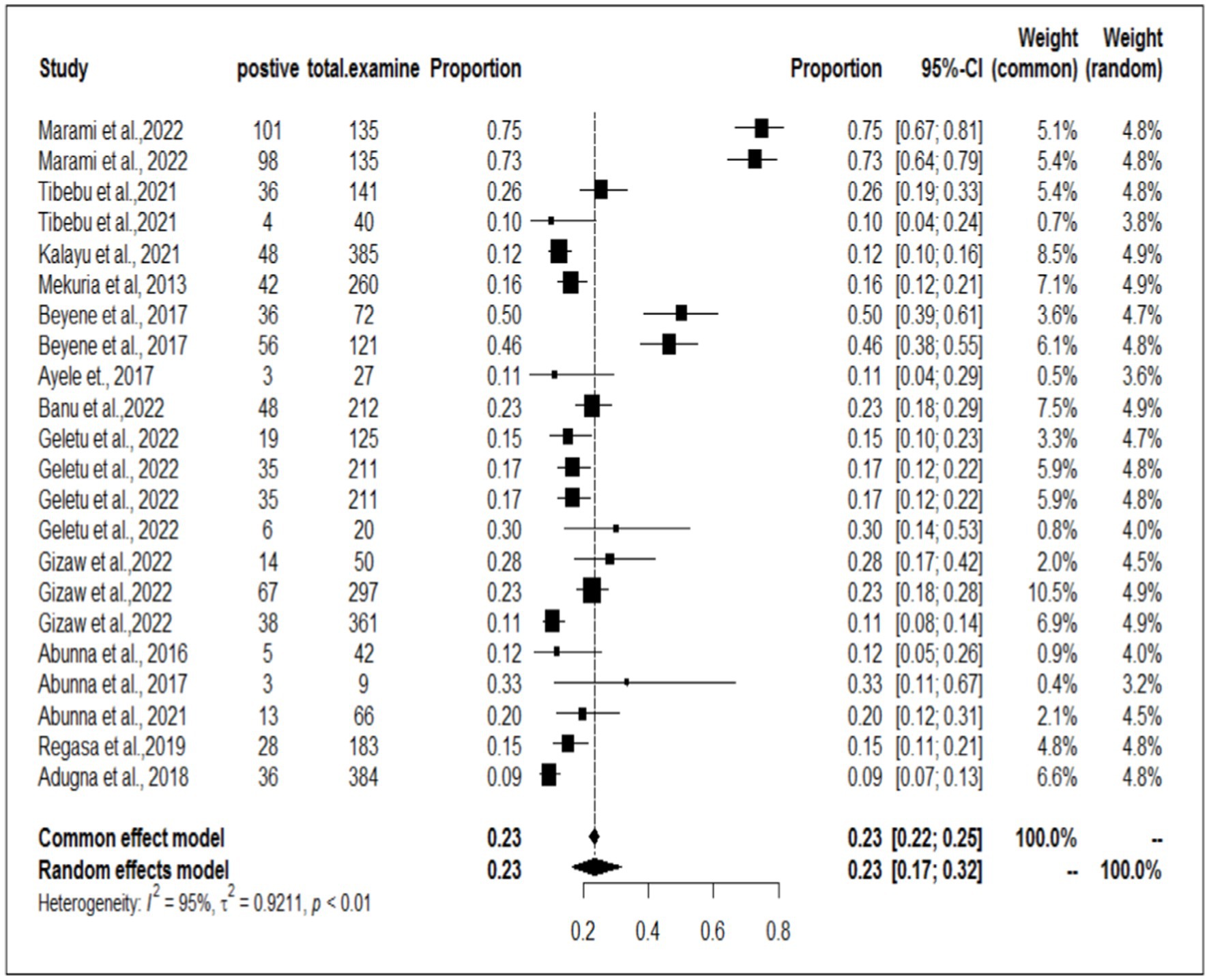
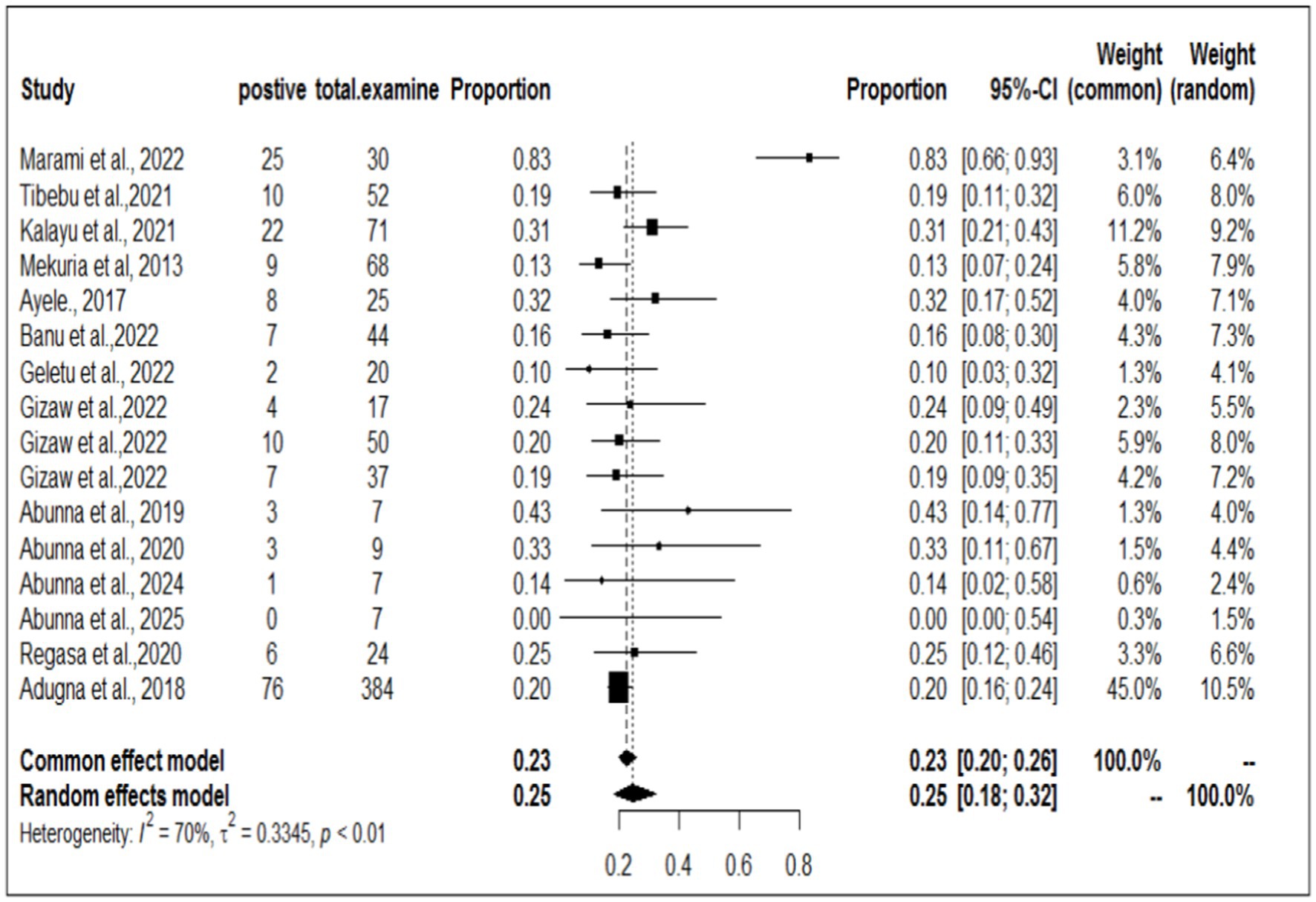
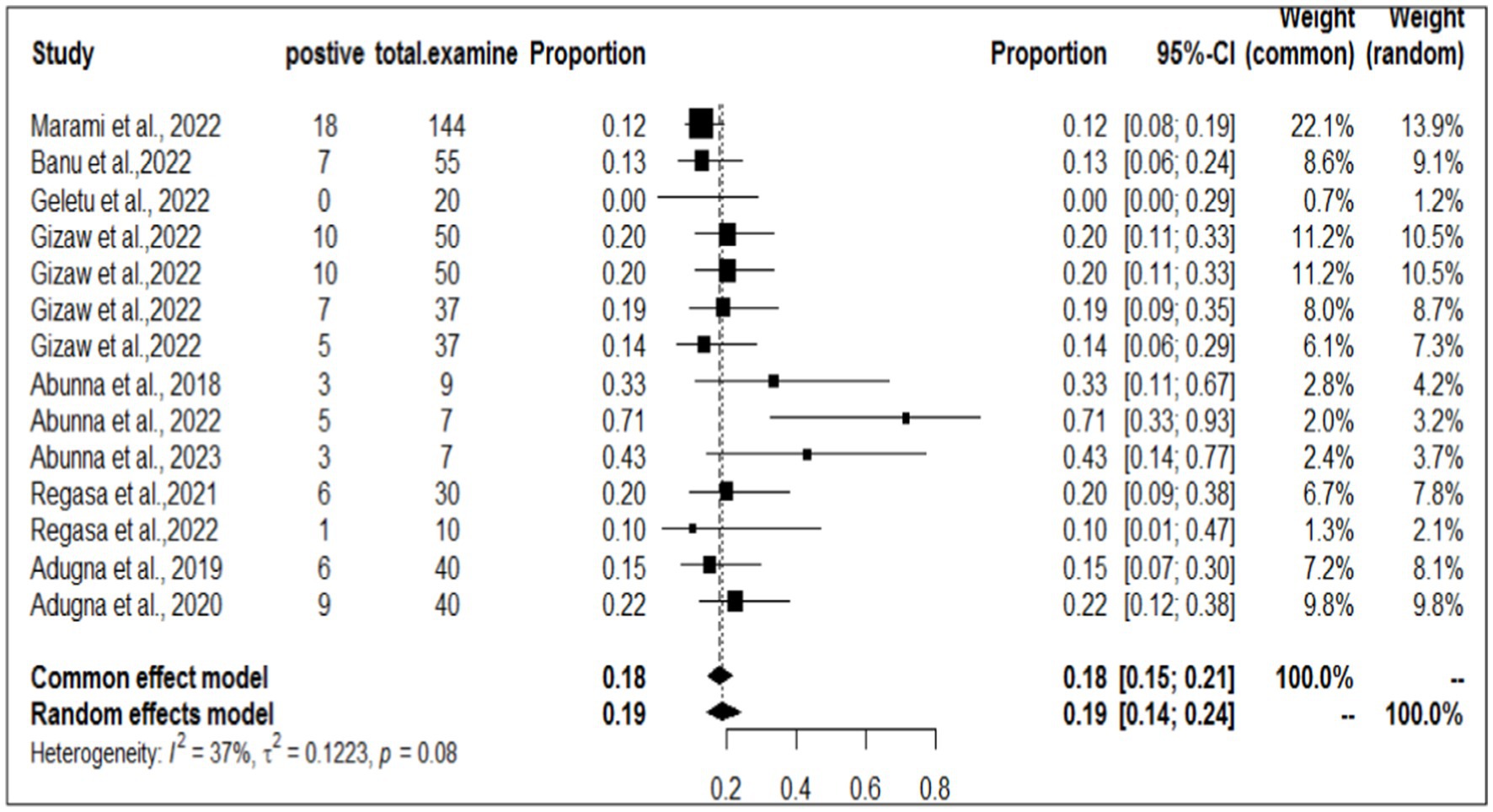
To investigate the prevalence or burden of S. aureus in each of the sampling units (animal, workers, and working equipment), a separate analysis of the articles was carried out. Some of the articles were used more than once due to different sample sources. Therefore, based on the separate meta-analysis, the pooled prevalence of S. aureus was estimated to be 25% (95% CI; 18–32%) for human/worker samples, 23% (95% CI; 17–32%) for animal sources and 19% (95% CI; 14–24%) for working equipment (Figures 6–8).
Qualitative assessment of potential factors for Staphylococcus aureus
Using logistic regression analysis, Buna’s study (36) demonstrated that milk contamination with S. aureus was considerably greater in lactating cows in the older age group, with an odds ratio of 3.91, and implied that lactating cows exhibited a 3.91 times greater risk of being affected by the organism than in the adult and younger age groups. The incidence of S. aureus in the milk of cows with numerous parities or calving also differed significantly (p = 0.001) and in the late lactation stage (p = 0.010). Additionally, there was a strong correlation (p < 0.05) between the prevalence of S. aureus in milk and hand washing before and after each milking, the frequency of cleaning dairy houses, and the farms’ drainage systems. Furthermore, there was significant variation in the prevalence of bacteria in milk according to age (p ≤ 0.001), parity (p ≤ 0.001), study area (p = 0.035), drainage quality of the milking area (p = 0.035), and management system (p = 0.035) (54). However, there was no statistically significant correlation (p > 0.05) between the prevalence of S. aureus and the study towel or udder cleaning interval.
Discussion
Staphylococcus species are prevalent foodborne bacterial pathogens that cause food poisoning in humans when ingested in contaminated foods, including dairy products. The organisms can gain access to raw milk and milk products either by direct excretion from udders with clinical and subclinical staphylococcal mastitis or by contamination from food handlers. The primary aim of interest in this meta-analysis was to estimate the pooled prevalence of S. aureus in livestock, livestock-associated workers, and working equipment interfaces in Ethiopia. Our meta-analysis revealed that there was a 22% pooled prevalence of S. aureus in the worker-animal and equipment interfaces. The significant increase in the incidence of this pathogen is alarming both for dairy farms and for public health.
The sub-analysis revealed that the pooled prevalence of S. aureus at the animal-worker-equipment interface was greater in dairy farm sources, which accounted for 23%, than in abattoir settings, which accounted for 18%. This result contradicted the findings reported by Abunna et al. (55), who reported a greater proportion of S. aureus (53.2%) in a slaughterhouse where the percentage of S. aureus was slightly greater than that in a dairy farm (44.8%). Another contradictory result was found in the study by Gizaw et al. (56), in which the occurrence of S. aureus was significantly greater in meat samples (22.7%) and considerably lower in dairy farms (11.7%) in Ethiopia. According to Tibebu et al. (23), the overall prevalence of S. aureus was estimated to be 21.46%. Of these, 25.53% were derived from cow milk, 10% from udder swabs, and 19.23% from hand swabs. On the other hand, Marami et al. (40) reported that 74% of udder milk samples, 72% of udder swabs, and 83% of milker hand swabs were positive for S. aureus.
The potential causes for this variation may stem from the inadequate management practices employed by dairy farms and their workers, as well as the insufficient attention given to dairy farms in Ethiopia. These discrepancies could arise from the use of various research methods employed in different studies conducted by multiple researchers. Another possible factor could be attributed to the characteristics of the samples, such as the presence of meat or milk, which may or may not favor the growth and proliferation of S. aureus. Moreover, the protocols for processing samples in terms of abattoir settings and dairy farms could introduce significant variations in the proportion of the pathogen in different sampling units. Like other contradictory studies in Ethiopia, the present study depicted a contrary notion in Turkey, as it was reported that meat product samples (48.7%) had a significantly greater prevalence than milk and dairy products (23.2%), implying a widespread distribution of S. aureus among diversified food items (57). A converse result might be that the Turkish dairy management system receives great attention throughout the milk and milk product processing chain and provides better treatment options for the pathogen. Moreover, this difference could be attributed to the diverse isolation and identification methods of the agent across different geographical areas as well as to global climatic changes.
The present discovery also indicated that the occurrence of S. aureus was greater in human workers (25%) than in animals (23%). The pooled prevalence of S. aureus in animals was significantly greater (23%) than that in the study conducted by Marami (40), which revealed that the prevalence of the pathogen in animals was 13.9%. Furthermore, the occurrence of S. aureus in animal workers was also found to be greater (25%) than that in Mekuria’s study (42), which reported a 15.5% prevalence of S. aureus in milk samples from dairy cows and nasal swabs of farm workers in selected dairy farms around Addis Ababa, Ethiopia. Similarly, our study demonstrated a higher result than the finding documented by Lewis and James (58), where the proportion of S. aureus accounted for 17.2% of the cattle, food chain, and human infections in Bishoftu, Ethiopia. However, this meta-analysis result was lower than that of Bendahou et al. (59, 60), who reported that 40% of S. aureus isolates were found in dairy products in North Morocco. The lower prevalence of S. aureus in the current study compared to the study in northern Morocco could be attributed to the direct collection of milk samples from cows’ udders before contact with milking utensils in this study, which may have reduced the prevalence of S. aureus (40).
Risk factors, Tibebu et al. (23) study on S. aureus in milk linked the bacteria to various possible factors, including milker age, community awareness of S. aureus, antiseptic use before and after milking, barn drainage systems, management systems, prior exposure to mastitis, and the use of drying towels for each udder separately. Only prior mastitis exposure (p < 0.05) from these linked factors to the agent had a statistically significant association with the prevalence of S. aureus. In Addition, Marami et al. (40) reported that Staphylococcus species isolates from dairy cows and smallholders were common in Central Ethiopia. The authors attempted to correlate the disease’s prevalence with several variables, including site town, breed, farm type, cow age, parity, lactation stage, blind test, eating lesion, tick infestation, floor type, and towel usage. Overall, the results of the univariate logistic regression analysis indicated that there was no significant correlation between any of the research variables in cows and the isolation of Staphylococcus. Additional research has evaluated the prevalence of this organism in relation to potential risk variables, such as sex, age, farm duty, job experience of the attendees/owners, and parity and lactation status of the cow (41). Carriage of staphylococci in the nasal passages and hands of the milkers are also potential sources of staphylococci in milk.
The prevalence and extent of antimicrobial resistance in veterinary medicine are experiencing a in the global. The spread of staphylococci that are resistant to antimicrobial agents poses a challenge to both human and animal health professionals. It is crucial to monitor and surveil foodborne pathogens and their resistance to antimicrobials in the food supply chain to effectively reduce the risks associated with food-borne hazards. In the present meta-analysis, the collective prevalence of multidrug-resistant strains was 27% in various sampling units on dairy farms and abattoirs. Our findings revealed a significantly lower prevalence than that reported in previous studies conducted in Egypt, with rates of 100, 74, and 80% reported by Seedy et al. (61), Gizaw et al. (56), and Balta et al. (62), respectively. In Korea, Moon et al. (63) reported a prevalence rate of 79%, while Beyene et al.(48)reported a rate of 34.9% in Ethiopia. However, our findings indicated a greater prevalence than that reported in other studies conducted in Ethiopia, such as the 10.42% reported by Tibebu et al. (23) and 16% by Buna et al. (36). This is a result of repeated therapeutic or indiscriminate use of these antibiotics on dairy farms and in humans for the treatment of infections.
The high prevalence of multidrug resistance of S. aureus can be attributed to the frequent use of antimicrobials, particularly microorganisms that produce β-lactamase, which inactivates penicillin and related antibiotics. The use of disinfectants containing heavy metals in washing milking equipment could potentially induce antimicrobial resistance in bacteria. It is possible to increase antimicrobial resistance via various ecological settings, by administering subtherapeutic doses, subdose antibiotics in animal feed and water, administering them at the wrong time, and administering them using the wrong route (which impacts bioavailability) (64).
The introduction and propagation of low-grade medications within a nation can also present significant obstacles (65). Additionally, the presence of antimicrobial residues in milk can contribute to the development of antimicrobial resistance in pathogens, which can then be transmitted to different individuals upon consumption (66). The abundance of multidrug-resistant Staphylococcus sp. have been found in milk from farms, meat from abattoirs, equipment, and humans poses a risk to public health.
To effectively control these pathogens at the national level in Ethiopia, veterinary professionals should provide health-related training to para-professionals, such as livestock keepers, attendants, milkers, hygiene workers, and milk collectors. To achieve optimal health outcomes for humans, animals, and the environment, national action plans should prioritize efforts to reduce antimicrobial resistance from livestock to the environment and the community.
Conclusion and future perspectives
The present meta-analysis revealed that the pooled prevalence of S. aureus at the animal-worker-working equipment interface was 22% in Ethiopia. Overall, 25, 23, and 19% of the pathogens were distributed in humans, animals, and working facilities/utensils, respectively. The current meta-analysis also revealed an overall MDR rate of 27% in different sampling units. And, the highest frequency of MRSA isolates was recorded found in the Dairy farm settings (44%). The sub-pooled results also revealed substandard practices for milk, meat, personnel, and equipment hygiene in dairy farms and abattoirs when handling, storing, and processing foods originating from livestock. The contamination rates of food of animal origin (milk and meat), personnel, and working equipment by S. aureus are approximately equal, indicating that this particular pathogen is present and circulating at the interface where humans, animals, and working utensils interact. Additionally, it can serve as a useful marker for inadequate hygiene practices and compromised animal food safety. In particular, the frequent cleaning of abattoir workers’ and milkers’ hands could be a contributing factor to the relatively low prevalence of S. aureus in abattoirs. The higher pooled prevalence of S. aureus in dairy farms, abattoir settings, and workers in this meta-analysis indicates its serious economic, animal welfare, food safety and public health problems and its association with livestock for its transmission between animals and humans in livestock farm settings. Thus, improving hygienic measures on dairy farms and working equipment as well as within abattoirs must be effective in safeguarding the public from the risk of staphylococcal food poisoning and acquiring MDR isolates. Additionally, future research should consider identifying Staphylococcus enterotoxins from milk and meat and genes responsible for AMR in Ethiopia.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
BM: Software, Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Conceptualization. KG: Data curation, Writing – review & editing, Writing – original draft, Visualization, Validation, Methodology, Investigation, Formal analysis, Conceptualization. AM: Validation, Writing – review & editing, Writing – original draft, Supervision, Data curation. MF: Visualization, Methodology, Investigation, Formal analysis, Conceptualization, Writing – review & editing, Writing – original draft, Data curation.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We thank the University of Gondar, for providing some of the materials and important pieces of training.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1403012/full#supplementary-material
Abbreviations
AMR, Antimicrobial resistance; MDR, Multidrug resistance; MERSA, Methicillin resistance Staphylococcus aureus; S. aureus, Staphylococcus aureus; CA-MRSA, Community-associated methicillin resistance Staphylococcus aureus.
References
1. Al-Mebairik, NF, El-Kersh, TA, Al-Sheikh, YA, and Marie, MAM. A review of virulence factors, pathogenesis, and antibiotic resistance in Staphylococcus aureus. Rev Med Microbiol. (2016) 27:50–6. doi: 10.1097/MRM.0000000000000067
2. Gulzar, M, and Zehra, A. Staphylococcus aureus: a brief review. Int J Vet Sci Res. (2018) 4:020–2. doi: 10.17352/ijvsr.000031
3. Breurec, S, Zriouil, S, Fall, C, Boisier, P, Brisse, S, Djibo, S, et al. Epidemiology of methicillin-resistant Staphylococcus aureus lineages in five major African towns: emergence and spread of atypical clones. Clin Microbiol Infect. (2011) 17:160–5. doi: 10.1111/j.1469-0691.2010.03219.x
4. Oliveira, K, Viegas, C, and Ribeiro, E. MRSA colonization in workers from different occupational environments—a one health approach perspective. Atmos. (2022) 13:658. doi: 10.3390/atmos13050658
5. Badawy, B, Elafify, M, Farag, AM, Moustafa, SM, Sayed-Ahmed, MZ, Moawad, AA, et al. Ecological distribution of virulent multidrug-resistant Staphylococcus aureus in livestock, environment, and dairy products. Antibiotics. (2022) 11:1651. doi: 10.3390/antibiotics11111651
6. Salazar, M. Use of antibiotics and NSAID drugs to inhibit Staphylococcus aureus biofilm formation. MDPI, section of Antibiotics. (2023).
7. Todd, EC. Bacteria: Staphylococcus aureus. American Chemical Society, ACS Infectious Diseases, Vol 9. (2023).
8. Zaher, HA, El Baz, S, Alothaim, AS, Alsalamah, SA, Alghonaim, MI, Alawam, AS, et al. Molecular Basis of Methicillin and Vancomycin Resistance in Staphylococcus aureus from Cattle, Sheep Carcasses and Slaughterhouse Workers. Antibiotics. (2023) 12:205. doi: 10.3390/antibiotics12020205
9. Nadimpalli, M, Stewart, JR, Pierce, E, Pisanic, N, Love, DC, Hall, D, et al. Livestock-associated, antibiotic-resistant Staphylococcus aureus nasal carriage and recent skin and soft tissue infection among industrial hog operation workers. PLoS One. (2016) 11:e0165713. doi: 10.1371/journal.pone.0165713
10. Ivbule, M, Miklaševičs, E, Čupāne, L, Bērziņa, L, Bālinš, A, and Valdovska, A. Presence of methicillin-resistant Staphylococcus aureus in slaughterhouse environment, pigs, carcasses, and workers. J Vet Res. (2017) 61:267–77. doi: 10.1515/jvetres-2017-0037
11. Silva, V, Correia, S, Rocha, J, Manaia, CM, Silva, A, García-Díez, J, et al. Antimicrobial resistance and clonal lineages of staphylococcus aureus from cattle: a cross-sectional study from the one health perspective. Microorganisms. (2022) 10:1–14. doi: 10.3390/microorganisms10050941
12. Dignard, C, and Leibler, JH. Recent research on occupational animal exposures and health risks: a narrative review. Curr Environ Health Rep. (2019) 6:236–46. doi: 10.1007/s40572-019-00253-5
13. John, J, Narendrakumar, L, Thomas, S, and Nelson-Sathi, S. Hybrid genome assembly and annotation of multidrug-resistant Staphylococcus aureus genotype ST672-SCCmec type IVd (2B). J Glob Antimicrob Resist. (2023) 32:74–7. doi: 10.1016/j.jgar.2022.12.011
14. Patra, B, Chakraborty, T, and Ghosh, S. Prevalence and Transmission of Multi-Drug Resistance Gene in Staphylococcus aureus. Curr Biotechnol. (2022) 11:196–211. doi: 10.2174/2211550112666221117091252
15. Stefani, S, and Goglio, A. Methicillin-resistant Staphylococcus aureus: related infections and antibiotic resistance. Int J Infect Dis. (2010) 14:S19–22. doi: 10.1016/j.ijid.2010.05.009
16. Herron-Olson, L, Fitzgerald, JR, Musser, JM, and Kapur, V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One. (2007) 2:e1120. doi: 10.1371/journal.pone.0001120
17. Santosaningsih, D, Erikawati, D, Hakim, IA, Santoso, S, Hidayat, M, Suwenda, AH, et al. Reducing transmission of methicillin-resistant Staphylococcus aureus in a surgical ward of a resource-limited hospital in Indonesia: An intervention study. Infect Prev Pract. (2019) 1:100028. doi: 10.1016/j.infpip.2019.100028
18. Conceição, T, Coelho, C, Silva, IS, de Lencastre, H, and Aires-de-Sousa, M. Staphylococcus aureus in former Portuguese colonies from Africa and the Far East: missing data to help fill the world map. Clinical Microbiology and Infection. (2015) 21:842.e1–842.e10. doi: 10.1016/j.cmi.2015.05.010
19. Baroja, I, Guerra, S, Coral-Almeida, M, Ruíz, A, Galarza, JM, de Waard, JH, et al. Methicillin-resistant Staphylococcus aureus nasal colonization among health care workers of a tertiary hospital in Ecuador and associated risk factors. Infect Drug Resist. (2021) 14:3433–40. doi: 10.2147/IDR.S326148
20. Egwuatu, C, Ogunsola, F, Egwuatu, T, and Oduyebo, O. Prevalence and risk factors for carriage of methicillin-resistant Staphylococcus aureus (MRSA) among healthcare workers in a tertiary institution in Nigeria. J Med Dent Sci. (2013) 8:9–13. doi: 10.9734/JAMMR/2021/V33I1631000
21. Sassmannshausen, R, Deurenberg, RH, Köck, R, Hendrix, R, Jurke, A, Rossen, JW, et al. MRSA prevalence and associated risk factors among health-care workers in non-outbreak situations in the Dutch-German EUREGIO. Front Microbiol. (2016) 7:1273. doi: 10.3389/fmicb.2016.01273
22. Leung, Y-h, Lai, RW-M, Chan, AC-W, Lo, JY-C, Ho, P-L, Wong, MM-H, et al. Risk factors for community-associated methicillin-resistant Staphylococcus aureus infection in Hong Kong. J Infect. (2012) 64:494–9. doi: 10.1016/j.jinf.2012.02.009
23. Tibebu, L, Belete, Y, Tigabu, E, and Tsegaye, W. Prevalence of Staphylococcus aureus, methicillin-resistant Staphylococcus aureus and potential risk factors in selected dairy farms at the interface of animal and human in Bishoftu, Ethiopia. Vet Med. (2021) 12:241–51. doi: 10.2147/VMRR.S331968
24. da Silva, LS, Andrade, YM, Oliveira, AC, Cunha, BC, Oliveira, EG, Cunha, TS, et al. Prevalence of methicillin-resistant Staphylococcus aureus colonization among healthcare workers at a tertiary care hospital in northeastern Brazil. Infect Prev Pract. (2020) 2:100084. doi: 10.1016/j.infpip.2020.100084
25. Lawal, OU, Ayobami, O, Abouelfetouh, A, Mourabit, N, Kaba, M, Egyir, B, et al. A 6-Year update on the diversity of Methicillin-Resistant Staphylococcus aureus clones in Africa: a systematic review. Front Microbiol. (2022) 13:860436. doi: 10.3389/fmicb.2022.860436
26. Ambachew, A, Gebrecherkos, T, and Ayalew, G. Prevalence and clindamycin resistance profile of Staphylococcus aureus and associated factors among patients attending the University of Gondar Comprehensive Specialized Hospital, Gondar, Northwest Ethiopia. Interdiscip Perspect Infect Dis. (2022) 2022:1–10. doi: 10.1155/2022/6503929
27. Gajdács, M. The continuing threat of methicillin-resistant Staphylococcus aureus. Antibiotics. (2019) 8:52. doi: 10.3390/antibiotics8020052
28. Nyasinga, J, Munshi, Z, Kigen, C, Nyerere, A, Musila, L, Whitelaw, A, et al. Displacement of Hospital-Acquired, Methicillin-Resistant Staphylococcus aureus Clones by Heterogeneous Community Strains in Kenya over 13 years. Microorganisms. (2024) 12:1171. doi: 10.3390/microorganisms12061171
29. Abdulgader, SM, Shittu, AO, Nicol, MP, and Kaba, M. Molecular epidemiology of Methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol. (2015) 6:348. doi: 10.3389/fmicb.2015.00348
30. Lynch, JP, and Zhanel, GG. Escalation of antimicrobial resistance among MRSA part 1: focus on global spread. Expert Rev Anti-Infect Ther. (2023) 21:99–113. doi: 10.1080/14787210.2023.2154653
31. Fayissa, N, Alemu, T, Desalegn, A, and Jirata, D. The distribution and drug resistance characteristics of methicillin resistant Staphylococcus aureous to be public and animal health burdon in Ethiopia. Research square: meta-analysis. (2022).
32. Ketena, T. Bacteriological quality, antibiotic resistance and related factor of raw cow milk from small scale producers in Hawassa Town Sidama Ethiopia. International institute for primary health care, Ethiopia: HU ; (2022).
33. Tsige, Y, Tadesse, S, Tefera, MM, Amsalu, A, Menberu, MA, and Gelaw, B. Prevalence of methicillin-resistant Staphylococcus aureus and associated risk factors among patients with wound infection at referral hospital, northeast Ethiopia. J Pathog. (2020) 2020:1–7. doi: 10.1155/2020/3168325
34. Bale, MI. Molecular characterization of methicillin-resistant Staphylococcus aureus (MRSA) isolated from blood samples of patients attending selected hospitals in Ilorin, Nigeria: Kwara State University (Nigeria); (2021), 50–57.
35. Argaw, S, Addis, M, and Degefu, H. Identification and antimicrobial resistance pattern of Staphylococci isolated from Cottage Cheese (Ayib) and Yoghurt (Ergo) in selected districts of Jimma zone, Ethiopia. Health Sci J. (2018) 12:549. doi: 10.21767/1791-809X.1000549
36. Banu, MG, and Zewdu, GE. Occurrence and antimicrobial susceptibility of Staphylococcus aureus in dairy farms and personnel in selected towns of West Shewa Zone, Oromia, Ethiopia. PLoS One. (2022) 17:e0277805. doi: 10.1371/journal.pone.0277805
37. Geletu, US, Usmael, MA, and Ibrahim, AM. Isolation, identification, and susceptibility profile of E. coli, Salmonella, and S. aureus in dairy farm and their public health implication in Central Ethiopia. Vet Med Int. (2022) 2022:1–13. doi: 10.1155/2022/1887977
38. Fesseha, H, Mathewos, M, Aliye, S, and Wolde, A. Study on prevalence of bovine mastitis and associated risk factors in dairy farms of Modjo town and suburbs, central Oromia, Ethiopia. Vet Med. (2021) 12:271–83. doi: 10.2147/VMRR.S323460
39. O'Dea, RE, Lagisz, M, Jennions, MD, Koricheva, J, Noble, DW, Parker, TH, et al. Preferred reporting items for systematic reviews and meta-analyses in ecology and evolutionary biology: a PRISMA extension. Biol Rev. (2021) 96:1695–722. doi: 10.1111/brv.12721
40. Marami, LM, Berhanu, G, Tekle, M, Agga, GE, Beyene, TJ, Tufa, TB, et al. Antimicrobial resistance of Staphylococci at animal human interface in smallholders and dairy farms in central Oromia, Ethiopia. Infect Drug Resist. (2022) 15:3767–77. doi: 10.2147/IDR.S370592
41. Kalayu, AA, Woldetsadik, DA, Woldeamanuel, Y, Wang, S-H, Gebreyes, WA, and Teferi, T. Burden and antimicrobial resistance of S. aureus in dairy farms in Mekelle, Northern Ethiopia. BMC Vet Res. (2020) 16:1–8. doi: 10.1186/s12917-020-2235-8
42. Mekuria, A, Asrat, D, Woldeamanuel, Y, and Tefera, G. Identification and antimicrobial susceptibility of Staphylococcus aureus isolated from milk samples of dairy cows and nasal swabs of farm workers in selected dairy farms around Addis Ababa, Ethiopia. Afr J Microbiol Res. (2013) 7:3501–10. doi: 10.4314/evj.v27i2.2
43. Beyene, T, Hayishe, H, Gizaw, F, Beyi, AF, Abunna, F, Mammo, B, et al. Prevalence and antimicrobial resistance profile of Staphylococcus in dairy farms, abattoir and humans in Addis Ababa, Ethiopia. BMC Res Notes. (2017) 10:1–9. doi: 10.1186/s13104-017-2487-y
44. Ayele, Y, Gutema, FD, Edao, BM, Girma, R, Tufa, TB, Beyene, TJ, et al. Assessment of Staphylococcus aureus along milk value chain and its public health importance in Sebeta, central Oromia, Ethiopia. BMC Microbiol. (2017) 17:1–7. doi: 10.1186/s12866-017-1048-9
45. Gizaw, F, Kekeba, T, Teshome, F, Kebede, M, Abreham, T, Berhe, HH, et al. Multidrug-Resistant Staphylococcus aureus Strains Thrive in Dairy and Beef Production, Processing, and Supply Lines in Five Geographical Areas in Ethiopia. Vet Sci. (2023) 10:663. doi: 10.3390/vetsci10120663
46. Abunna, F, Abriham, T, Gizaw, F, Beyene, T, Feyisa, A, Ayana, D, et al. Staphylococcus: isolation, identification and antimicrobial resistance in dairy cattle farms, municipal abattoir and personnel in and around Asella, Ethiopia. J Vet Sci Technol. (2016) 7:1–7. doi: 10.4172/2157-7579.1000383
47. Regasa, S, Mengistu, S, and Abraha, A. Milk safety assessment, isolation, and antimicrobial susceptibility profile of Staphylococcus aureus in selected dairy farms of Mukaturi and Sululta town, Oromia region, Ethiopia. Vet Med Int. (2019) 2019:1–11. doi: 10.1155/2019/3063185
48. Adugna, F, Pal, M, and Girmay, G. Prevalence and antibiogram assessment of Staphylococcus aureus in beef at municipal abattoir and butcher shops in Addis Ababa, Ethiopia. Biomed Res Int. (2018) 2018:1–7. doi: 10.1155/2018/5017685
49. Thompson, SG, and Higgins, JP. How should meta‐regression analyses be undertaken and interpreted?. Stat. Med. (2002) 21:1559–1573.
50. Jin, ZC, Zhou, XH, and He, J. Statistical methods for dealing with publication bias in meta-analysis. Stat Med. (2015) 34:343–60. doi: 10.1002/sim.6342
51. Shi, D, Song, H, Liao, X, Terry, R, and Snyder, LA. Bayesian SEM for specification search problems in testing factorial invariance. Multivar Behav Res. (2017) 52:430–44. doi: 10.1080/00273171.2017.1306432
52. Johnson, JR, Stell, AL, Scheutz, F, O'Bryan, TT, Russo, TA, Carlino, UB, et al. Analysis of the F antigen-specific papA alleles of extraintestinal pathogenic Escherichia coli using a novel multiplex PCR-based assay. Infect Immun. (2000) 68:1587–99. doi: 10.1128/IAI.68.3.1587-1599.2000
53. Lemma, F, Alemayehu, H, Stringer, A, and Eguale, T. Prevalence and antimicrobial susceptibility profile of Staphylococcus aureus in milk and traditionally processed dairy products in Addis Ababa, Ethiopia. Biomed Res Int. (2021) 2021:1–7. doi: 10.1155/2021/5576873
54. Girmay, W, Gugsa, G, Taddele, H, Tsegaye, Y, Awol, N, Ahmed, M, et al. Isolation and identification of methicillin-resistant Staphylococcus aureus (MRSA) from milk in shire dairy farms, Tigray, Ethiopia. Vet Med Int. (2020) 2020:1–7. doi: 10.1155/2020/8833973
55. Abunna, F, Adugna, B, Tufa, TB, Ayana, D, Gutema, FD, Waktole, H, et al. Detection and antimicrobial resistance of Staphylococcus species from chicken, chicken litter, and humans in Addis Ababa, Ethiopia. Vet Med Int. (2022) 2022:1–8. doi: 10.1155/2022/9084334
56. Gizaw, F, Kekeba, T, Teshome, F, Kebede, M, Abreham, T, Hayishe, H, et al. Distribution and antimicrobial resistance profile of coagulase-negative staphylococci from cattle, equipment, and personnel on dairy farm and abattoir settings. Heliyon. (2020) 6:e03606. doi: 10.1016/j.heliyon.2020.e03606
57. Guven, K, Mutlu, MB, Gulbandilar, A, and Cakir, P. Occurrence and characterization of Staphylococcus aureus isolated from meat and dairy products consumed in Turkey. J Food Saf. (2010) 30:196–212. doi: 10.1111/j.1745-4565.2009.00200.x
58. Lewis, I, and James, S. Performance standards for antimicrobial susceptibility testing. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. (2022). Wayne, PA: Clinical and Laboratory Standards Institute.
59. Bendahou, A, Lebbadi, M, Ennanei, L, Essadqui, FZ, and Abid, M. Characterization of Staphylococcus species isolated from raw milk and milk products (lben and jben) in North Morocco. J Infect Dev Ctries. (2008) 2:218–25. doi: 10.3855/jidc.266
60. Bendahou, A, Abid, M, Bouteldoun, N, Catelejine, D, and Lebbadi, M. Enterotoxigenic coagulase positive Staphylococcus in milk and milk products, lben and jben, in northern Morocco. J Infect Dev Ctries. (2009) 3:169–76. doi: 10.3855/jidc.3
61. R El-Seedy, F, A Radwan, I, H Hassan, W, and Shehata, A. Coagulase Negative Staphylococci as an emerging cause of bovine mastitis: prevalence, antimicrobial resistance and biofilm formation. Journal of Veterinary Medical. Research. (2017) 24:1–11. doi: 10.21608/jvmr.2017.43256
62. Balta, B, and Derbie, F. Nasal carriage of methicillin-resistant Staphylococcus aureus strains among inpatients of Jimma hospital, South Western Ethiopia. Ethiop J Health Sci. (2003) 13:107–115. doi: 10.10520/AJA10291857_77
63. Moon, J, Lee, A, Kang, H, Lee, E, Joo, Y, Park, YH, et al. Antibiogram and coagulase diversity in staphylococcal enterotoxin-producing Staphylococcus aureus from bovine mastitis. J Dairy Sci. (2007) 90:1716–24. doi: 10.3168/jds.2006-512
64. Silva, NCC, Guimaraes, FF, Manzi, MP, Budri, PE, Gomez-Sanz, E, Benito, D, et al. Molecular characterization and clonal diversity of methicillin-susceptible Staphylococcus aureus in milk of cows with mastitis in Brazil. J Dairy Sci. (2013) 96:6856–62. doi: 10.3168/jds.2013-6719
65. Ismael, A. Epidemiology of Bovine Mastitis and Associated Risk Factors in East Shewa Zone of Oromia Regional State, Ethiopia. J Vet Med Epidemiol. (2018) 2:1–6. doi: 10.23880/oajvsr-16000129
Keywords: animals, humans, multidrug-resistant, Staphylococcus aureus, working equipment
Citation: Mengistu BA, Getnet K, Mebratu AS and Fenta MD (2024) Occurrence, multidrug resistance and potential risk factors for Staphylococcus aureus infection at worker-animal and working equipment interfaces: a systematic review and meta-analysis of the Ethiopian literature. Front. Public Health. 12:1403012. doi: 10.3389/fpubh.2024.1403012
Edited by:
James Wabwire Oguttu, University of South Africa, South AfricaReviewed by:
Okon Okwong Kenneth, Federal Medical Center Makurdi, NigeriaMadubuike Umunna Anyanwu, University of Nigeria, Nigeria
Copyright © 2024 Mengistu, Getnet, Mebratu and Fenta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melkie Dagnaw Fenta, bWVsa2llZGFnbmF3MzUyOEBnbWFpbC5jb20=
 Bemrew Admassu Mengistu
Bemrew Admassu Mengistu Kalkidan Getnet2
Kalkidan Getnet2 Melkie Dagnaw Fenta
Melkie Dagnaw Fenta