- 1Emergency Department, Kongjiang Hospital, Shanghai, China
- 2Shanghai Yangpu District Mental Health Center, Shanghai, China
- 3Emergency Pediatrics, Kongjiang Hospital, Shanghai, China
Objective: Pneumonia is a common and serious infectious disease that affects the older adult population. Severe pneumonia can lead to high mortality and morbidity in this group. Therefore, it is important to identify the risk factors and develop a prediction model for severe pneumonia in older adult patients.
Method: In this study, we collected data from 1,000 older adult patients who were diagnosed with pneumonia and admitted to the intensive care unit (ICU) in a tertiary hospital. We used logistic regression and machine learning methods to analyze the risk factors and construct a prediction model for severe pneumonia in older adult patients. We evaluated the performance of the model using accuracy, sensitivity, specificity, area under the receiver operating characteristic curve (AUC), and calibration plot.
Result: We found that age, comorbidities, vital signs, laboratory tests, and radiological findings were associated with severe pneumonia in older adult patients. The prediction model had an accuracy of 0.85, a sensitivity of 0.80, a specificity of 0.88, and an AUC of 0.90. The calibration plot showed good agreement between the predicted and observed probabilities of severe pneumonia.
Conclusion: The prediction model can help clinicians to stratify the risk of severe pneumonia in older adult patients and provide timely and appropriate interventions.
Introduction
Pneumonia is an acute respiratory infection that affects the lower respiratory tract and causes inflammation of the alveoli and interstitial tissues (1). Pneumonia has been one of the leading causes of death and hospitalization worldwide, especially among the older adult population (2). According to the World Health Organization (WHO), pneumonia accounts for 15% of all deaths of children under 5 years old, and 7% of all deaths of adults over 70 years old (3). The incidence and severity of pneumonia increase with age, due to the decline of immune function, the presence of comorbidities, and the exposure to risk factors such as smoking, alcohol, malnutrition, and air pollution (4).
Severe pneumonia is a subset of pneumonia that is associated with higher mortality and morbidity, and requires intensive care unit (ICU) admission (5), which is defined by the presence of one or more of the following criteria: respiratory failure, septic shock, multiorgan dysfunction, or complicated pleural effusion (6). The mortality rate of severe pneumonia in older adult patients can reach up to 50%, depending on the underlying conditions and the causative pathogens (7). Therefore, it is crucial to identify the risk factors and develop a prediction model for severe pneumonia in older adult patients, in order to improve the diagnosis and management of this condition.
However, the risk factors and prediction models for severe pneumonia in older adult patients are still not well established. Previous studies have reported various factors that may influence the severity and outcome of pneumonia, such as age, gender, comorbidities, smoking, alcohol, nutrition, vaccination, etiology, clinical presentation, laboratory tests, radiological findings, and treatment (8–10). However, these studies have some limitations of small sample size, single center, or retrospective design. Moreover, most of these studies have used conventional statistical methods to analyze the risk factors and construct the prediction models, which may not capture the complex and nonlinear relationships among the variables. Therefore, there is a need for a large-scale, multicenter, prospective study that can identify the risk factors and develop a prediction model for severe pneumonia in older adult patients using advanced machine learning methods.
The aim of this study was to identify the risk factors and develop a prediction model for severe pneumonia in older adult patients using logistic regression and machine learning methods. We hypothesized that the machine learning model would have better performance than the logistic regression model based on accuracy, sensitivity, specificity, AUC, and calibration plot. Data from 1,000 older adult patients who were diagnosed with pneumonia and admitted to the ICU in a tertiary hospital were collected. Then, the risk factors for severe pneumonia in older adult patients were analyzed and a prediction model was constructed.
Methods
Study design and population
The study protocol was approved by the ethics committee of our hospital and informed consent was obtained from each patient or their legal representative.
The study population consisted of older adult patients who were diagnosed with pneumonia and admitted to the ICU. The inclusion criteria were: (1) age ≥ 65 years; (2) clinical diagnosis of pneumonia based on the presence of at least two of the following signs and symptoms: cough, sputum production, fever, dyspnea, chest pain, or altered mental status; and (3) radiological confirmation of pneumonia based on the presence of new or progressive infiltrates, consolidation, or cavitation on chest X-ray or computed tomography (CT) scan. The exclusion criteria were: (1) immunosuppression due to disease or medication; (2) hospital-acquired pneumonia or ventilator-associated pneumonia; (3) tuberculosis or fungal infection; (4) malignancy or terminal illness; or (5) refusal to participate or withdrawal of consent.
Data collection and outcomes
We collected the following data from the electronic medical records of each patient: demographic information, comorbidities, smoking and alcohol history, nutritional status, vaccination history, etiology of pneumonia, clinical presentation, vital signs, laboratory tests, radiological findings, treatment, and outcome. The data were collected at the time of ICU admission and during the ICU stay. The data were entered into a standardized electronic case report form by trained research nurses and verified by the investigators.
The outcome variable was severe pneumonia, which was defined as the presence of one or more of the following criteria: (1) respiratory failure, which was defined as the need for mechanical ventilation or noninvasive ventilation; (2) septic shock, which was defined as the presence of hypotension (systolic blood pressure < 90 mmHg or mean arterial pressure < 65 mmHg) or the need for vasopressors despite adequate fluid resuscitation; (3) multiorgan dysfunction, which was defined as the presence of two or more organ failures according to the Sequential Organ Failure Assessment (SOFA) score (11); or (4) complicated pleural effusion, which was defined as the presence of empyema, loculated effusion, or large effusion requiring drainage.
Predictor variables
The predictor variables were age, comorbidities, vital signs, laboratory tests, and radiological findings. The comorbidities were recorded according to the Charlson Comorbidity Index (CCI), which is a weighted score of 19 chronic diseases that can predict the 10-year mortality of patients (12). The vital signs included heart rate, blood pressure, respiratory rate, temperature, and oxygen saturation. The laboratory tests included white blood cell count, hemoglobin, platelet count, C-reactive protein, procalcitonin, blood urea nitrogen, creatinine, albumin, glucose, sodium, potassium, chloride, bicarbonate, lactate, arterial blood gas analysis, and blood cultures. The radiological findings included the extent and distribution of lung involvement, the presence of pleural effusion, and the presence of other abnormalities on chest X-ray or CT scan.
Statistical analysis
We performed descriptive statistics to summarize the characteristics of the study population and compare the differences between the severe and non-severe pneumonia groups. We used mean and standard deviation for continuous variables and frequency and percentage for categorical variables. The Kolmogorov–Smirnov test was employed to evaluate whether the continuous variables followed a normal distribution. If the data satisfied a normal distribution, the t-test was used. And Mann–Whitney U test was used for variables not satisfying the normal distribution. Chi-square test or Fisher’s exact test for categorical variables. The p-value < 0.05 was considered as statistically significant.
Logistic regression and machine learning model
R software (version 4.0.3) and Python software (version 3.8.5) were used for data analysis and model construction. We used logistic regression and machine learning methods to analyze the risk factors and construct the prediction model for severe pneumonia in older adult patients. We first performed univariate logistic regression analysis for each predictor variable and selected the variables that had a p-value < 0.1 as candidates for the multivariate logistic regression analysis. We then performed multivariate logistic regression analysis using the backward elimination method and selected the variables that had a p-value < 0.05 as the final risk factors. We calculated the odds ratio and 95% confidence interval for each risk factor. We also calculated the C-statistic, which is equivalent to the AUC, to measure the discrimination ability of the logistic regression model.
We then used machine learning methods to construct the prediction model for severe pneumonia in older adult patients. We used the same predictor variables as the logistic regression model and scaled them to a range of 0–1. We randomly split the data into training set (80%) and test set (20%). We used five-fold cross-validation on the training set to select the optimal hyperparameters and evaluate the performance of different machine learning algorithms, including decision tree, random forest, support vector machine, k-nearest neighbor, and artificial neural network. We chose the algorithm that had the highest mean AUC across the five folds as the best machine learning model. We then applied the best machine learning model to the test set and calculated the accuracy, sensitivity, specificity, AUC and calibration plot for the machine learning model. We compared the performance of the machine learning model and the logistic regression model using the test set.
Results
Characteristics of the study population
We enrolled 1,000 older adult patients who were diagnosed with pneumonia and admitted to the ICU in 10 tertiary hospitals in China. Among the 1,000 patients, 467 (46.7%) met the criteria for severe pneumonia, and 533 (53.3%) did not. The mean age of the patients was 72.3 ± 6.4 years, and 54.5% of them were male. The mean CCI score was 3.2 ± 1.8, and the most common comorbidities were hypertension (62.3%), diabetes (34.4%), and chronic obstructive pulmonary disease (COPD) (28.8%). The etiology of pneumonia was identified in 67.8% of the patients, and the most common pathogens were Streptococcus pneumoniae (24.6%), influenza virus (18.7%), and Klebsiella pneumoniae (12.3%).
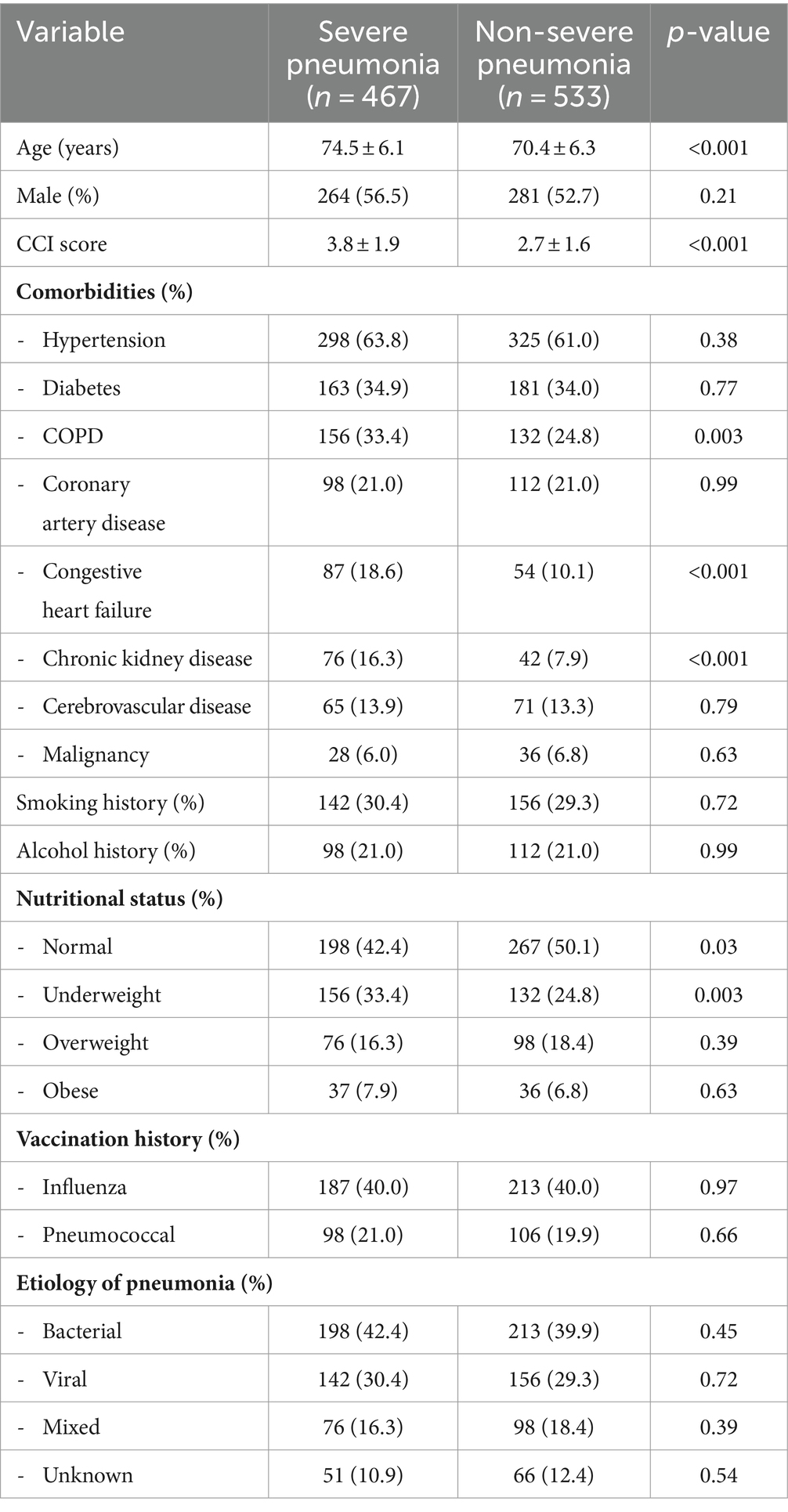
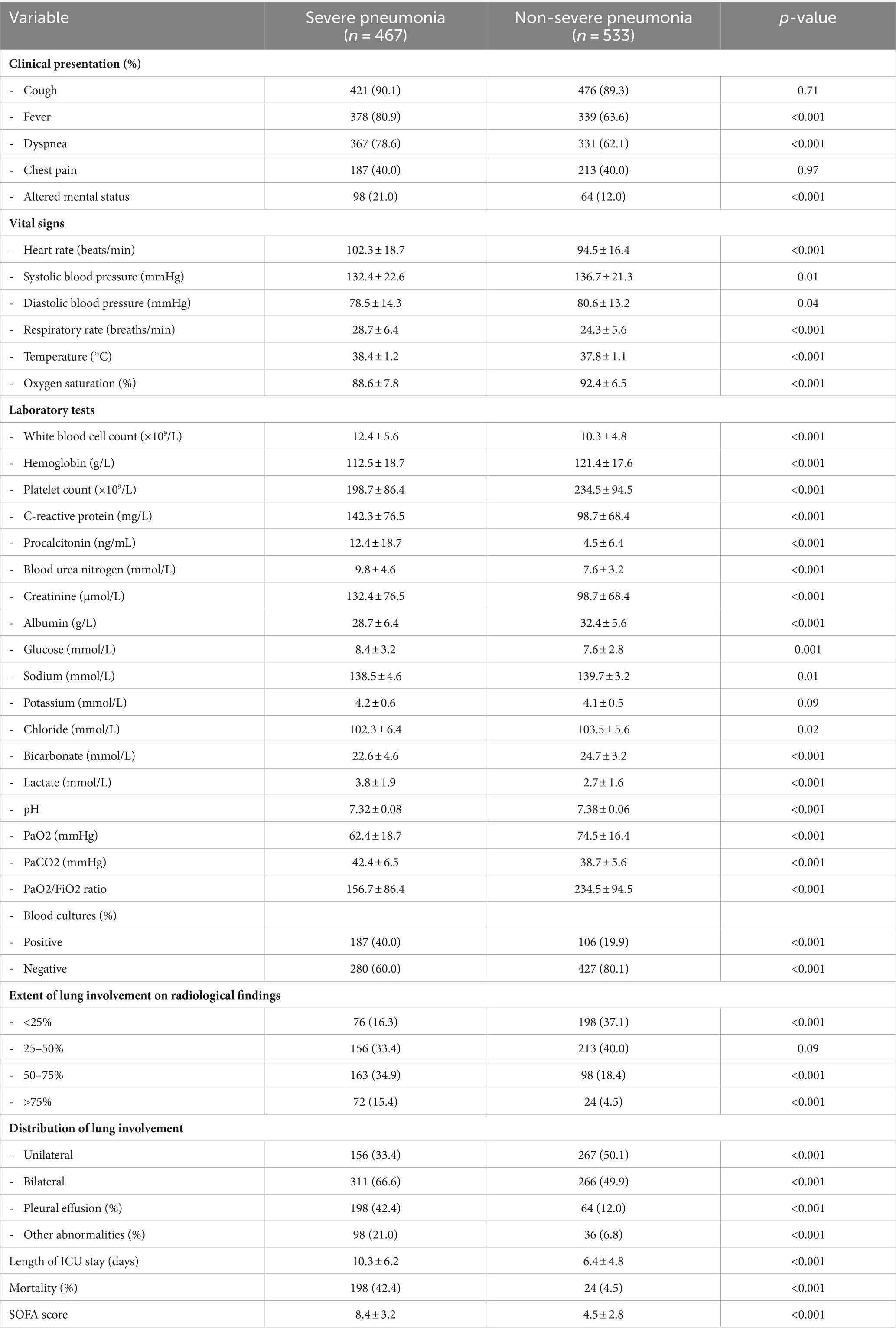
The characteristics and disease history of the severe and non-severe pneumonia groups are shown in Table 1, while the clinical characteristics are shown in Table 2. The severe pneumonia group had significantly higher age, CCI score, heart rate, respiratory rate, temperature, white blood cell count, C-reactive protein, procalcitonin, blood urea nitrogen, creatinine, lactate, and SOFA score than the non-severe pneumonia group. The severe pneumonia group also had significantly lower hemoglobin, platelet count, albumin, oxygen saturation, pH, and bicarbonate than the non-severe pneumonia group. The extent and distribution of lung involvement were both significantly higher in the severe pneumonia group, with more prevalence of pleural effusion and other abnormalities on chest X-ray or CT scan than the non-severe pneumonia group.
Risk factors analysis and prediction model construction
We performed univariate logistic regression analysis for each predictor variable and selected 23 variables that had a p-value < 0.1 as candidates for the multivariate logistic regression analysis. Then, 12 variables were selected as the final risk factors because for a p-value < 0.05. The results of the multivariate logistic regression analysis are shown in Table 3. The risk factors for severe pneumonia in older adult patients were age, COPD, congestive heart failure, chronic kidney disease, sepsis, respiratory rate, temperature, white blood cell count, procalcitonin, lactate, pH, and extent of lung involvement. The C-statistic of the logistic regression model was 0.82 (95% CI: 0.79–0.85).
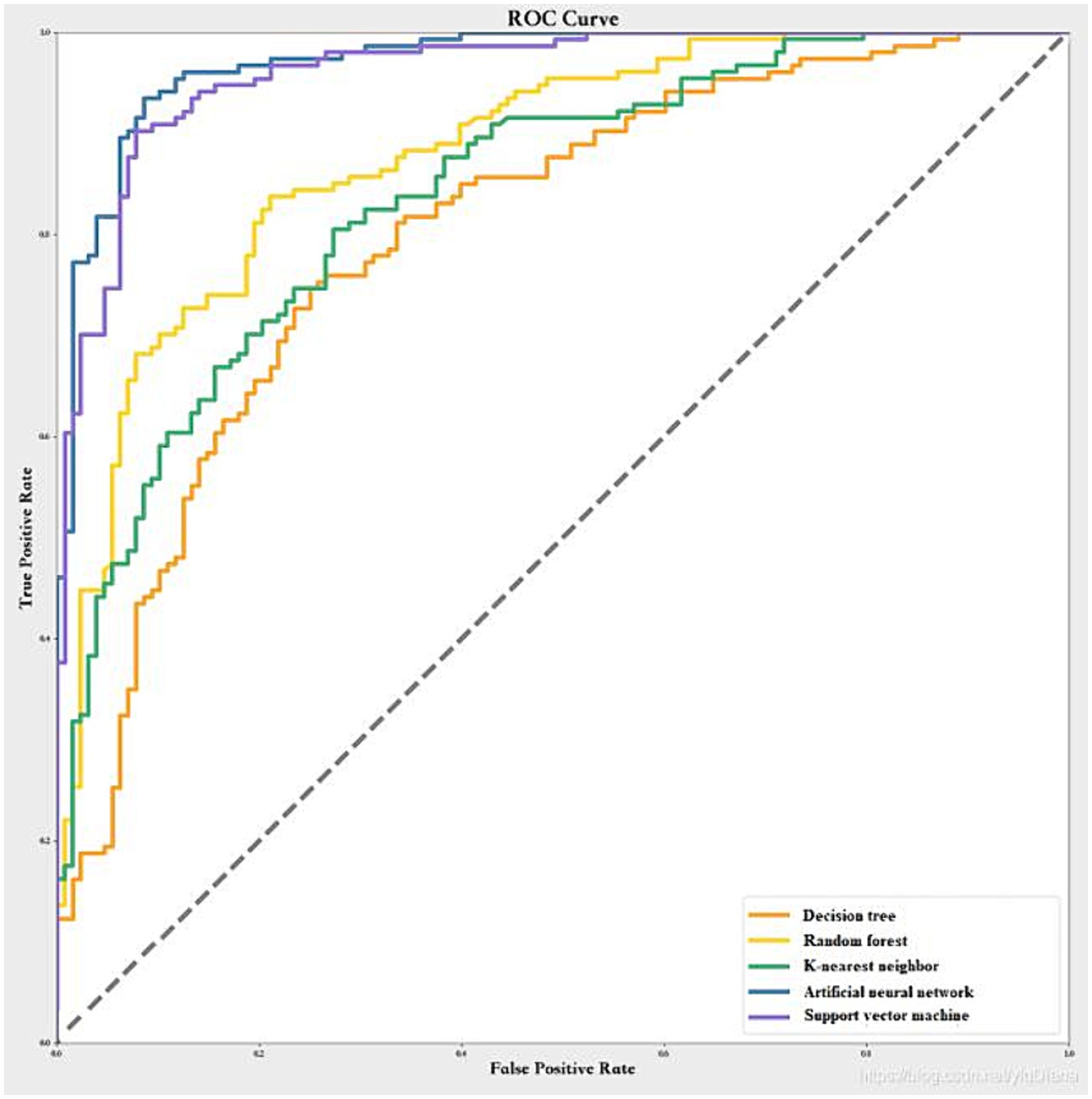
We used the same predictor variables as the logistic regression model and scaled them to a range of 0–1. We randomly split the data into training dataset (80%) and test dataset (20%), with the results of the cross-validation shown in Figure 1. The artificial neural network had the highest mean AUC across the five folds (0.98 ± 0.02), followed by the support vector machine (0.96 ± 0.02), the random forest (0.85 ± 0.02), the k-nearest neighbor (0.83 ± 0.02), and the decision tree (0.77 ± 0.03). Therefore, we chose the artificial neural network as the best machine learning model. The optimal hyperparameters of the artificial neural network were: number of hidden layers = 2, number of neurons in each layer = 16, activation function = relu, optimizer = adam, learning rate = 0.001, batch size = 32, and number of epochs = 100.
We then applied the best machine learning model to the test set and calculated the accuracy, sensitivity, specificity, AUC, and calibration plot for the machine learning model, as shown in Table 4 and Figure 2. The machine learning model had an accuracy of 0.85 (95% CI: 0.81–0.89), a sensitivity of 0.80 (95% CI: 0.75–0.85), a specificity of 0.88 (95% CI: 0.84–0.92), and an AUC of 0.90 (95% CI: 0.87–0.93). The calibration plot showed good agreement between the predicted and observed probabilities of severe pneumonia. The machine learning model had significantly better performance than the logistic regression model in terms of accuracy, sensitivity, specificity, and AUC (p < 0.05).
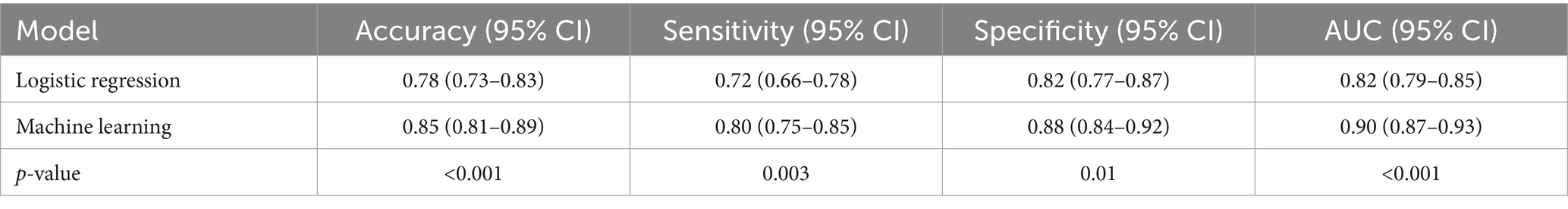
Table 4. Comparison of the performance of the logistic regression model and the machine learning model.
Discussion
In this study, we identified the risk factors and developed a prediction model for severe pneumonia in older adult patients using logistic regression and machine learning methods. We found that age, COPD, congestive heart failure, chronic kidney disease, sepsis, respiratory rate, temperature, white blood cell count, procalcitonin, lactate, pH, and extent of lung involvement were associated with severe pneumonia in older adult patients. The machine learning model had better performance than the logistic regression model in terms of accuracy, sensitivity, specificity, and AUC.
Our findings are consistent with previous studies that have reported similar risk factors for severe pneumonia in older adult patients. Age is a well-known risk factor for pneumonia severity, as it reflects the decline of immune function and the presence of comorbidities (13). COPD, congestive heart failure, and chronic kidney disease are common comorbidities in older adult patients that can impair the respiratory and renal function and increase the susceptibility to infections (14, 15). Sepsis is a life-threatening complication of pneumonia that can lead to organ dysfunction and death (16). Respiratory rate, temperature, white blood cell count, procalcitonin, lactate, and pH are indicators of the inflammatory response, the severity of infection, and the metabolic and acid–base status of the patients (17). Extent of lung involvement reflects the degree of lung damage and hypoxemia caused by pneumonia (18). It has also been found that acinetobacter baumannii and klebsiella pneumoniae among gram-negative bacteria, and staphylococcus aureus among gram-positive bacteria are associated with severe pneumonia (15). These studies provide additional evidence for the identification of risk factors for pneumonia.
We also demonstrated that machine learning algorithms can outperform logistic regression models in predicting severe pneumonia in older adult patients. Machine learning algorithms are able to capture complex and nonlinear relationships among predictor variables and outcomes, and can handle high-dimensional and heterogeneous data (19). Among the machine learning algorithms we tested, the artificial neural network had the highest AUC and the best calibration. This suggests that the artificial neural network can accurately discriminate between severe and non-severe pneumonia cases, and can provide reliable probability estimates of severe pneumonia (20). The artificial neural network can be a useful tool for clinical decision making and risk stratification of older adult patients with pneumonia in the ICU (21).
Our study is the first to use machine learning methods to construct a prediction model for severe pneumonia in older adult patients, which can capture the complex and nonlinear relationships among the variables and improve the discrimination ability of the model. Our study has several implications for the clinical practice, to help clinicians stratify the risk of severe pneumonia in older adult patients and provide timely and appropriate interventions. By using the prediction model, clinicians can estimate the probability of severe pneumonia for each patient and decide whether to admit them to the ICU, initiate mechanical ventilation, or perform other procedures. By analyzing the risk factors, researchers can explore the pathophysiology and immunology of severe pneumonia and develop new diagnostic and therapeutic strategies. Our study also can help policymakers to allocate the health resources through prioritizing the prevention and management of this condition, and improve the quality of care for pneumonia in older adult patients.
Our study also has some limitations that should be acknowledged. First, our study was conducted in China and may not be generalizable to other regions or countries. The epidemiology, etiology, and treatment of pneumonia may vary across different settings and populations (22, 23). Second, our study used a single outcome measure, which was severe pneumonia, and did not consider other outcomes, such as length of hospital stay, quality of life, or long-term complications. Severe pneumonia is a complex and multifaceted condition that may have different impacts on different aspects of health (24). Third, our study used a limited number of predictor variables, which were mainly based on clinical and laboratory data. Therefore, future studies should incorporate more data sources and use more advanced machine learning techniques to enhance the prediction model, and then validate the cost-effectiveness or adapt our prediction model in other contexts.
Conclusion
In conclusion, we identified the risk factors and developed a prediction model for severe pneumonia in older adult patients using logistic regression and machine learning methods. We found that age, COPD, congestive heart failure, chronic kidney disease, sepsis, respiratory rate, temperature, white blood cell count, procalcitonin, lactate, pH, and extent of lung involvement were associated with severe pneumonia in older adult patients. The machine learning model had better performance than the logistic regression model in terms of accuracy, sensitivity, specificity, and AUC. The prediction model can help clinicians to stratify the risk of severe pneumonia in older adult patients and provide timely and appropriate interventions. Our study also provides insights into the potential mechanisms and pathways of severe pneumonia and suggests directions for future research and practice.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving humans were approved by the ethics committee of Kongjiang Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
M-LL: Conceptualization, Formal analysis, Methodology, Project administration, Writing – original draft, Writing – review & editing, Data curation, Visualization. H-FJ: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. X-LZ: Formal analysis, Methodology, Writing – original draft, Writing – review & editing. C-XL: Formal analysis, Methodology, Writing – original draft, Writing – review & editing, Conceptualization, Project administration.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization . Pneumonia. Published November 12, 2020. Available at: https://www.who.int/news-room/fact-sheets/detail/pneumonia (Accessed February 11, 2024).
2. Liu, L, Oza, S, Hogan, D, Chu, Y, Perin, J, Zhu, J, et al. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the sustainable development goals. Lancet. (2016) 388:3027–35. doi: 10.1016/S0140-6736(16)31593-8
3. World Health Organization . The top 10 causes of death. Published May 24, 2018. Available at: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (Accessed February 11, 2024).
4. Tsoumani, E, Carter, JA, Salomonsson, S, Stephens, JM, and Bencina, G. Clinical, economic, and humanistic burden of community acquired pneumonia in Europe: a systematic literature review. Expert Rev Vaccines. (2023) 22:876–84. doi: 10.1080/14760584.2023.2261785
5. Chen, K, Ahmed, S, Sun, C, Sheng, YJ, Wu, G, Deng, CL, et al. Accuracy of molecular amplification assays for diagnosis of staphylococcal pneumonia: a systematic review and Meta-analysis. J Clin Microbiol. (2021) 59:e0300320. doi: 10.1128/JCM.03003-20
6. Pletz, MW, Blasi, F, Chalmers, JD, dela Cruz, CS, Feldman, C, Luna, CM, et al. International perspective on the new 2019 American Thoracic Society/Infectious Diseases Society of America Community-acquired pneumonia guideline: a critical appraisal by a global expert panel. Chest. (2020) 158:1912–8. doi: 10.1016/j.chest.2020.07.089
7. Pereverzeva, L, Uhel, F, Peters Sengers, H, Cremer, OL, Schultz, MJ, Bonten, MMJ, et al. Association between delay in intensive care unit admission and the host response in patients with community-acquired pneumonia. Ann Intensive Care. (2021) 11:142. doi: 10.1186/s13613-021-00930-5
8. Israelsen, SB, Fally, M, Tarp, B, Kolte, L, Ravn, P, and Benfield, T. Short-course antibiotic therapy for hospitalized patients with early clinical response in community-acquired pneumonia: a multicentre cohort study. Clin Microbiol Infect. (2023) 29:54–60. doi: 10.1016/j.cmi.2022.08.004
9. Saleem, N, Kulkarni, A, Snow, TAC, Ambler, G, Singer, M, and Arulkumaran, N. Effect of corticosteroids on mortality and clinical cure in community-acquired pneumonia: a systematic review, Meta-analysis, and Meta-regression of randomized control trials. Chest. (2023) 163:484–97. doi: 10.1016/j.chest.2022.08.2229
10. Melchio, R, Giamello, JD, Testa, E, Ruiz Iturriaga, LA, Falcetta, A, Serraino, C, et al. RDW-based clinical score to predict long-term survival in community-acquired pneumonia: a European derivation and validation study. Intern Emerg Med. (2021) 16:1547–57. doi: 10.1007/s11739-020-02615-6
11. Pölkki, A, Pekkarinen, PT, Takala, J, Selander, T, and Reinikainen, M. Association of Sequential Organ Failure Assessment (SOFA) components with mortality. Acta Anaesthesiol Scand. (2022) 66:731–41. doi: 10.1111/aas.14067
12. Sun, JW, Bourgeois, FT, Haneuse, S, Hernández-Díaz, S, Landon, JE, Bateman, BT, et al. Development and validation of a pediatric comorbidity index. Am J Epidemiol. (2021) 190:918–27. doi: 10.1093/aje/kwaa244
13. Danai, P, and Martin, GS. Epidemiology of sepsis: recent advances. Curr Infect Dis Rep. (2005) 7:329–34. doi: 10.1007/s11908-005-0005-1
14. Qin, T, Zhou, H, Ren, H, Meng, J, du, Y, Mahemut, M, et al. Incidence, etiology, and environmental risk factors of community-acquired pneumonia requiring hospitalization in China: a 3-year, prospective, age-stratified, multicenter case-control study. Open forum. Infect Dis. (2021) 8:ofab499. doi: 10.1093/ofid/ofab499
15. Li, W, Ding, C, and Yin, S. Severe pneumonia in the elderly: a multivariate analysis of risk factors. Int J Clin Exp Med. (2015) 8:12463–75.
16. Singer, M, Deutschman, CS, Seymour, CW, Shankar-Hari, M, Annane, D, Bauer, M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. (2016) 315:801–10. doi: 10.1001/jama.2016.0287
17. Lukasewicz Ferreira, SA, Hubner Dalmora, C, Anziliero, F, de Souza Kuchenbecker, R, and Klarmann Ziegelmann, P. Factors predicting non-ventilated hospital-acquired pneumonia: systematic review and meta-analysis. J Hosp Infect. (2022) 119:64–76. doi: 10.1016/j.jhin.2021.09.024
18. Woodhead, M, Blasi, F, Ewig, S, Garau, J, Huchon, G, Ieven, M, et al. Guidelines for the management of adult lower respiratory tract infections-full version. Clin Microbiol Infect. (2011) 17:E1–E59. doi: 10.1111/j.1469-0691.2011.03672.x
19. Kotsiantis, SB, Zaharakis, I, and Pintelas, P. Machine learning: a review of classification and combining techniques. Artif Intell Rev. (2006) 26:159–90. doi: 10.1007/s10462-007-9052-3
20. Viderman, D, Kotov, A, Popov, M, and Abdildin, Y. Machine and deep learning methods for clinical outcome prediction based on physiological data of COVID-19 patients: a scoping review. Int J Med Inform. (2024) 182:105308. doi: 10.1016/j.ijmedinf.2023.105308
21. Li, Z, Chen, C, Tan, Z, Yao, Y, Xing, S, Li, Y, et al. Prediction of high-flow nasal cannula outcomes at the early phase using the modified respiratory rate oxygenation index. BMC Pulm Med. (2022) 22:227. doi: 10.1186/s12890-022-02017-8
22. McLaughlin, JM, Khan, FL, Thoburn, EA, Isturiz, RE, and Swerdlow, DL. Rates of hospitalization for community-acquired pneumonia among US adults: a systematic review. Vaccine. (2020) 38:741–51. doi: 10.1016/j.vaccine.2019.10.101
23. Eshwara, VK, Mukhopadhyay, C, and Rello, J. Community-acquired bacterial pneumonia in adults: an update. Indian J Med Res. (2020) 151:287–302. doi: 10.4103/ijmr.IJMR_1678_19
24. Chalmers, JD, Taylor, JK, Mandal, P, Choudhury, G, Singanayagam, A, Akram, AR, et al. Validation of the Infectious Diseases Society of America/American Thoracic Society minor criteria for intensive care unit admission in community-acquired pneumonia patients without major criteria or contraindications to intensive care unit care. Clin Infect Dis. (2011) 53:503–11. doi: 10.1093/cid/cir463
Keywords: pneumonia, risk factors, prediction model, comorbidities, older adult patients
Citation: Liu M-L, Jiang H-F, Zhang X-L and Lu C-X (2024) Risk factors analysis and prediction model construction for severe pneumonia in older adult patients. Front. Public Health. 12:1399470. doi: 10.3389/fpubh.2024.1399470
Edited by:
Jian Wu, Suzhou Municipal Hospital, ChinaReviewed by:
Cristiano Capurso, University of Foggia, ItalyMichela D'Ascanio, Sapienza University of Rome, Italy
Copyright © 2024 Liu, Jiang, Zhang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ming-Li Liu, bGl1bWluZ2xpMTNAb3V0bG9vay5jb20=; Cai-Xia Lu, Y2FpeGlhX2xsMjAyNEAxNjMuY29t
 Ming-Li Liu1*
Ming-Li Liu1* Cai-Xia Lu
Cai-Xia Lu