
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Public Health, 10 July 2024
Sec. Life-Course Epidemiology and Social Inequalities in Health
Volume 12 - 2024 | https://doi.org/10.3389/fpubh.2024.1351732
This article is part of the Research TopicExploring Health Disparities in Black Communities: Historical Perspectives, Present Challenges, and Future DirectionsView all 3 articles
In the wake of the murder of George Floyd and the massacre in Buffalo, the editorial boards of the prominent scientific publication companies formally apologized for their journals’ historical role in advancing race science and promised to improve their standards. However, flowery commentaries cannot undo the consistent pattern of endorsing biologic differences between ethnic groups, even when discussing diseases or traits that are not considered politically charged. In this report, an exemplar is made of a recent publication claiming to identify phenotypes of atopic dermatitis that are distinct between European Americans, Asians, and African Americans. The insufficiency of the evidence and logic underlying these claims are discussed. Although devoid of malice, numerous publications continue to demonstrate how claims of biological differences between races is mainstreamed in modern scientific publications. Overall, the goal of this work is to challenge the scientific community, particularly the publication companies, to evaluate how assumptions of innate biologic disadvantage have clouded assessments of racial disparities in disease beyond the topics that are more stereotypical of race science.
In the wake of the murder of George Floyd and the massacre in Buffalo, Nature (Springer-Nature) editors formally apologized for their journals’ historical role in advancing race science (1) and promised to improve their standards (2). Other prominent journals such as Science (AAAS) and Cell (Elsevier) also appeared to grapple with their pasts (3–5). However well intentioned, such commentaries failed to address the continued pattern of endorsing biologic differences between ethnic groups, even when discussing diseases or traits that are not considered politically charged. A case study in this practice is presented by the recent work from Facheris et al. entitled “The translational revolution in atopic dermatitis: the paradigm shift from pathogenesis to treatment” (6). This manuscript appeared in Cellular & Molecular Immunology (a journal within the Springer-Nature family). Sadly, the work presented is more representative of an entrenched status quo than Thomas Kuhn’s intended definition of a shifting paradigm (7).
While the paper by Facheris et al. nicely outlines the targeted treatments in development for atopic dermatitis (AD), immune modulation for immune mediated diseases is not a paradigm shift. Yet while focusing on presenting AD from the perspective of cytokine imbalance, the authors also feed into the pattern of unintentional endorsement of race science that Nature assured its readers it would attempt to avoid (2). The authors propose generalized differences between ancestries (6, 8) exist in the molecular pathology underlying AD symptoms using primary citations which exclusively rely on racial categorization. For example, the authors claim to have “confirmed” a patient’s ancestry in a clinic visit (8) when only self-reported race or ethnicity could be assessed in such a manner. The racialized AD claims in Facheris, et al appear to have been initially presented in Czarnowski et al. 4 years prior (9). While the discussion herein focuses on the Facheris et al. and Czarnowski et al. publications in lieu of other examples (10–14), the aim is not to deride specific manuscripts but to dissect them for a teachable moment of how entrenched claims of racialized biologic determinism are unintentionally perpetuated in the scientific literature due to an uncritical assessment of the underlying evidence.
Using the de facto racial and ethnic categorizations, both Facheris and Czarnowski claim to have found molecular and biochemical classifications of AD unique to European Americans, Asians, and African Americans. Based on the citations provided by the authors, the claims of distinguishing immunologic markers between European and Asians AD were derived by contrasting two different studies, by two different groups, using different assay equipment, in two different countries, at two different points in time. Claims of Asian-specific AD was based only comparing the cytokine profiles from a Japanese population with AD (n = 42) (15) vs. a separate cohort of 51 European American (16, 17). The imperfect overlap was summarized as “European American AD cohorts feature relatively high activity of the Th2 and Th22 axes … compared to Asian and African American Cohorts” (6, 8). The only other set of studies presented as support for the existence of “the Asian AD cohort” included 21 patients “of Han Chinese descent” juxtaposed against an unrelated retrospective analysis of 107 European Americans (many of whom were both over 65 years of age and hospitalized) (18, 19).
Only one study of 15 Black and 15 White Manhattanites was used to support claims that “African American AD cohorts are characterized by an absence of Th17…” (8). Although a similar study in 18 Black Marylanders found ample Th17 signal (14), both studies failed to document a single social determinant or any of the numerous AD risk factors known to be unevenly distributed between racial groups (20). This practice is inconsistent with Nature’s updated policies on the use of racial categorization devoid of statistical adjustments for environmental exposures and social determinants of health (21).
A non-exhaustive list of exposures linked to AD would include at least the nation of birth, urbanicity of residence, distance from a major roadway, the number and type of animals and siblings living in the home, as well as exposure to: traffic related air pollution, diisocyanates, particulate matter under 2.5 microns (PM2.5), nitric oxide, hard water, phthalates, early life antibiotics, synthetic fabrics, cigarette smoke, as well as foods low in fiber or high in refined ingredients such as saturated fat, refined sugar, surfactants, oxidizers, and emulsifiers (22–38). Most of these exposures are not evenly distributed across racial groups (20). Beyond overt environmental injustices, racial differences in skin health could stem from differences in skincare product choice (39) or access to AD medications (40). Each of these factors should be assessed prior to making any racialized conclusions in data.
The perceived racial difference in AD manifestation seems limited to the phenotypic presentation of active lesions. While most image atlases of AD are skewed toward visuals of the manifestations in skin of Caucasians, online tools have been developed (41) to provide clinicians with examples of the variable presentation of AD in different skin types (accessible through the National Eczema Association).1 However, if skin pigmentation directly impacts AD risk (as opposed to being a marker of environmental injustice) one must explain the disparities within the African diaspora as much as between Africans and other ancestries (42, 43). Why would Aboriginal populations have lower rates of AD than white Australians in rural environments, but have higher rates if they move to an urbanized environment (44)? How are heavily pigmented populations in India relatively immune to melanin’s theorized AD-inducing effects (43)? The authors state “African descendent individuals, as well as Asians and Pacific Islanders, are more likely to develop AD than Caucasian individuals” (6); but how does African descent cause AD in the African descendants living in urban America but not rural Africa? Why would being Caucasian be protective in Bulgaria but deleterious in Sweden (43)? If ancestry were a key factor, why would one’s birth home be more predictive of AD risk than one’s ancestral home (45)?
Even if one believed these questions could still be answered by innate biologic differences, how many Black Americans would you need to study to make statements about all Black Americans? Fifteen Black Manhattanites are unlikely to adequately represent all five New York boroughs, let alone comment on the disease for rural Black people in Alabama. Similarly, 15 white Manhattanites should not be framed as representative of all white Americans. Presenting these racialized claims in the context of a review of targeted therapies and personalized medicine suggests that the authors envision future practice parameters segregated into separate but equal treatment algorithms.
Genetics do not explain racial disparities in AD (46, 47). The increased prevalence of AD in African American communities cannot be explained by: the allelic frequencies of FLG loss of function variants, copy number variations in FLG, the AD-polygenic score (PGS) derived from Europeans, the PGS for African ancestry, nor the PGS for pigmentation (46, 47). Taken together, the modern understanding of environmental exposures that contribute to AD require a baseline assessment of public health metrics prior to insinuating innate group differences, and especially before racializing such claims in atopy.
This manuscript was intended as a direct reply to Facheris et al. However, some may point to ongoing research into racial disparities writ large as defense of the authors’ claims. For example, the Journal of Clinical Medicine organized a special issue on “Ethnic differences in Dermatitis and Atopic Eczema and its Management” in March of 2023. Only three of the eight articles included in this special issue address differences between groups (12). One of these three citations focuses on the differences in presentation of AD in different skin colors and stresses the need to assure diverse patient cohorts in clinical trials (48). The second focuses on the environmental contributors to hand eczema that may differ by cultural practices (such as occupation or food preparation) (49). In stark contrast, the final example (12) also claims that Black American skin is devoid of Th17 cells and that European AD is distinct from Asian AD using the same flawed citations outlined above (6, 8, 18, 19). The paper (12) goes on to outline that Black patients should be given higher doses of cyclosporine by citing only an online news blog.
A recent report in JACI in Practice (50) echoed the claim that African American patients may require higher cyclosporine dosing. The 2004 review cited by JACI in Practice (51) enumerated three reports of higher cyclosporine metabolism among African Americans (52–54), two reporting no difference (55, 56), but overlooked a report of the opposite association between race and cyclosporine metabolism (57). Each of these studies enrolled patients being treated for solid organ transplants rather than AD. Each not only failed to assess a single social determinant of health, but also failed to adjust for factors known at the time to influence cyclosporine absorption such as diet, liver function, age, or concurrent medications (58). Once more, while the genetic variants referenced by modern studies as pharmacogenomic mechanisms for differing cyclosporine metabolism are not equally distributed across racial categories, race is not a functional proxy for genotyping (59). While some have attempted to argue that the correlation between race and social determinants of health indicate race is still a useful variable for statistical analysis (60), such practice represents a reliance on a flawed proxy of convenience in lieu of the effort needed to collect meaningful data. If the variable used could represent one of dozens different mechanisms (spanning sociology, psychology, hypothesized biochemistry, and more) then claims of using such information to design a targeted intervention (60) ring hollow.
Therefore, the correct phrasing would be to note that subjects with specific genotypes may require modulation of their cyclosporine dose in a race-neutral manner, as has been done successfully with other disorders (61). Doing so would accurately ascribe the need for dose modulation to the genotype, rather than racial category.
Others may defend these racialized practices by pointing to the National Institutes of Health’s (NIH) Request For Application (RFA). The assertions rest on the notion that NIH solicits research projects on racial disparities without explicitly prohibiting innate claims of biologic disadvantage in minoritized groups. To some, this is seen as tacit consent of the racialized differences in Th2 cytokine levels being valid. However, being open to further research into the mechanisms of racial disparities is not a defense of making sweeping claims about the biochemistry of the entire population of a continent using only data from one part of one nation. Claiming that a sixth generation Japanese American and someone who recently immigrated from rural China will both have similar Th22 expression based solely on their shared classification of Asian is definitionally incompatible with claims of seeing AD as a multifactorial disorder. If, however, Asian background is to be only one of many factors predicting drug response, these additional factors should be at least mentioned if not enumerated. Thankfully, a more recent review from the same group as Facheris et al. uses the more appropriate descriptor of “Japanese/Korean” instead of “Asian” (62), yet doing so continues to use the patients’ Japanese heritage as if it were a predictive variable while failing to evaluate the exposome-worth of variables that underlie the surrogate variable of ancestry.
Furthermore, openness to continued investigation does not answer the pointed questions of: what is the N value sufficient to study to justify claims of unique biology of pigmented skin?; how diverse of a cohort can be considered to be representative of Asia?; how comparable are studies that are performed years apart and using different equipment?; if a disease has identical symptoms, comorbidities, and treatment responses in populations all over the world, is it sound to predict that molecular causation would differ by skin tone?; and which environmental exposures are expected to be addressed for when evaluating AD across racial lines and national borders?
A related question would be to ask why innate biologic susceptibility to AD only manifested on a population scale after industrialization? Some have proposed that genetic variants that were beneficial in a pre-industrial era may have been rendered deleterious by exposures that were not common during human evolution (63). Yet, such framing centers disease causation on “the predisposed” in ways that mirror troubling post-WWII era of so-called “reform eugenics” (64). Even when the hypothesis of ancient DNA driving modern disease is put forward in good faith, proving such claims would require identification of the offending agent followed by mechanistic studies to verify the proposed gene–environment interaction. Furthermore, even if a toxin were shown to influence Th22 cell numbers via an allele more common among those of Han Chinese ancestry, avoidance of the toxin would remain paramount, and any imagined therapy would be targeted by genotype rather than ancestry.
One final justification of the types of claims that may be put forth by Facheris and Czarnowski is that journals which are subsidiary to the flagship publications of the publishing company should be more tolerant of claims based upon lower quality of evidence. Indeed, correlation between a journal’s prominence and its expected level of scientific veracity is a natural part of the scientific literature. However, as it applies to equity, tolerating poorly supported claims so long as they are limited to select journals suggests that the promises made by the publishers in the pages of the prestige journals were either applicable only to the lower tier journals or all together disingenuous. Furthermore, journal families are often distinguished by company logos, shared branding, and similar homepage websites. In an era of increasing concern for the potential harms of predatory journals (65, 66), branding is used by respected publishers to signal legitimacy to readers.
However, this branding is also used by bigoted online communities to endorse publications suggesting biologic determinism explains racial disparities. Although race science and eugenics are more commonly invoked for education attainment, social status, or mental health, research by several groups has demonstrated that racist online forums are the largest audience for publications purporting innate biologic differences explain racial disparities for common diseases (67–70). Such work also contributes to differential medical treatment through reinforcing the idea that biology differs between racial groups (71). Thus, all researchers should be mindful that racialized claims in their work may be dangerously misrepresented even when related to otherwise non-controversial topics like AD. Overall, a hereditarian view of AD proposes to improve care in ways that are theoretical and unlikely while provably providing aid and comfort to those wishing to advance marginalizing narratives.
Per SCOPUS, Facheris (6) and Czarnowski (9) have been cited by a total of 224 publications (only 205 of which have full text availability) for a total of at least 343 unique citations within these publications (Supplementary Table 1). A plurality of the citations (36.2%) were general comments about AD pathogenesis or symptoms or non-specific references to the existence of presented endotypes (Figure 1). 27.7% of citations similarly referenced the specific biomarkers that may define the proposed endotypes such as Th17 versus Th22 cells. However, only 6 total citations (1.7%) from 6 publications (2.9%) were focused on the pharmaceutical discussion that was the stated intent of the Facheris (6) and Czarnowski (9) reviews. Instead, 28.2% of the total citations from 37% of the publications echoed the racialized claims made by the authors (Figure 1). The means that three studies which enrolled only 88 total people from the referenced groups became the basis for 97 references to the racialized AD endotypes for African Americans and “Asians” made by the Facheris (6) and Czarnowski (9) reviews. This calculation only includes first-level citations, and thus the 97 racialized claims citing Facheris (6) and Czarnowski (9) could themselves be used as citations in other papers. These harms were compounded when 11 of the publications used the terms “race” or “racial” rather than ancestry (72–82), 5 inappropriately extrapolated from “African American” to “Black” or “African” (83–87), and 3 made the same extrapolation from “European American” to “European” (88–90) (Supplementary Table 1).
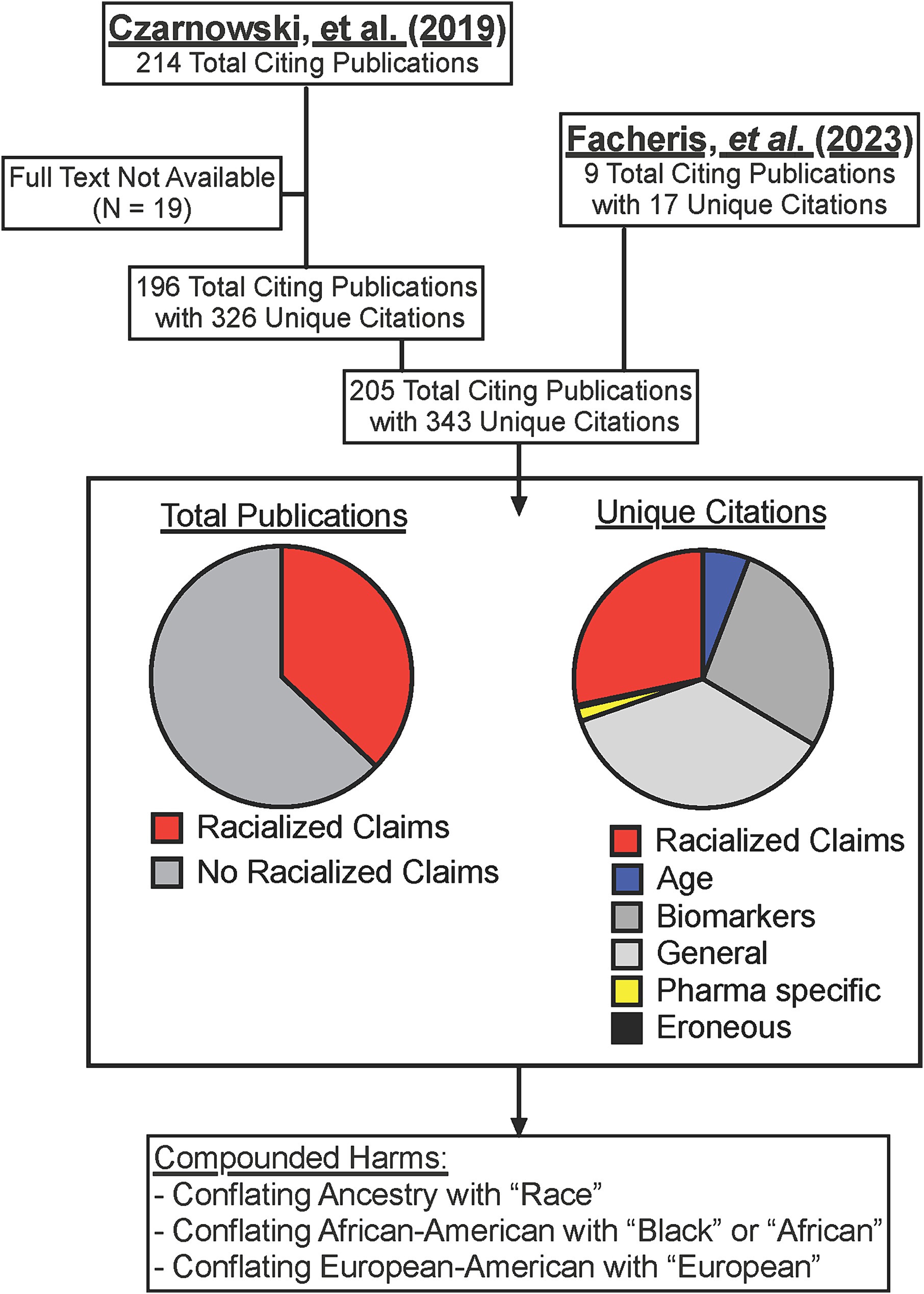
Figure 1. Dissemination of racialized claims in AD from two reviews. SCOPUS listed citations for Facheris (6) and Czarnowski (9) were collected and assessed for specific citations (some publications cited the articles more than once). Where full text was available, citations were assessed for racialized claims, or if the citation was referencing only age-related endotypes (Age), specific allergy cytokines of cells (Biomarkers), general comments on the existence of endotypes or AD symptoms (General), or comments on different prescription options in development (Pharma specific). Full citation list provided in Supplementary Table 1.
While the desire to avoid treating AD as “one size fits all” is noble, from a medical and biological perspective race is too imprecise to ever be included in “precision medicine” and too societally defined ever be appropriate for “personalized medicine.” It is likely that the authors, the reviewers, and the editors of the papers dissected herein never intended for their work to advance race science. However, extrapolating between exceedingly small cohorts and entire ancestry groups with an obliviousness to population-level environmental differences has direct ramifications for discussions of more controversial concepts. Overall, the various publishing groups will never be able to live up to the promise to avoid publishing race science until they recognize such work more often comes in the form of unintentional parroting of entrenched paradigms than overt statements of racial hierarchies. The scientific community must better adhere to reporting guidelines (21), avoid extrapolating small studies into population scales, assuring analyses are appropriately adjusted for social determinants, transparently reporting their study’s limitations, and prioritize the evidenced-based research into AD risk factors.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
IM: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Resources, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This author was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID) and the National Institutes of Health (NIH).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2024.1351732/full#supplementary-material
1. How nature contributed to science’s discriminatory legacy. Nature. (2022) 609:875–6. doi: 10.1038/d41586-022-03035-6
2. Nobles, M, Womack, C, Wonkam, A, and Wathuti, E. Science must overcome its racist legacy: Nature's guest editors speak. Nature. (2022) 606:225–7. doi: 10.1038/d41586-022-01527-z
3. Ortega, RP. Human geneticists curb use of the term ‘race’ in their papers. Science Insider Science. (2021). Available at: https://www.science.org/content/article/human-geneticists-curb-use-term-race-their-paper (Accessed May 2024).
4. Byeon, YJJ, Islamaj, R, Yeganova, L, Wilbur, WJ, Lu, Z, Brody, LC, et al. Evolving use of ancestry, ethnicity, and race in genetics research-A survey spanning seven decades. Am J Hum Genet. (2021) 108:2215–23. doi: 10.1016/j.ajhg.2021.10.008
5. S.A.O. American Society of Human Genetics Board of directors. Electronic address, on the report of the ASHG "facing our history-building an equitable future" initiative. Am J Hum Genet. (2023) 110:375–6. doi: 10.1016/j.ajhg.2023.02.006
6. Facheris, P, Jeffery, J, Del Duca, E, and Guttman-Yassky, E. The translational revolution in atopic dermatitis: the paradigm shift from pathogenesis to treatment. Cell Mol Immunol. (2023) 20:448–74. doi: 10.1038/s41423-023-00992-4
8. Sanyal, RD, Pavel, AB, Glickman, J, Chan, TC, Zheng, X, Zhang, N, et al. Atopic dermatitis in African American patients is T(H)2/T(H)22-skewed with T(H)1/T(H)17 attenuation. Ann Allergy Asthma Immunol. (2019) 122:99–110.e6. doi: 10.1016/j.anai.2018.08.024
9. Czarnowicki, T, He, H, Krueger, JG, and Guttman-Yassky, E. Atopic dermatitis endotypes and implications for targeted therapeutics. J Allergy Clin Immunol. (2019) 143:1–11. doi: 10.1016/j.jaci.2018.10.032
10. Kumar, R, Seibold, MA, and Burchard, EG. Atopic dermatitis, race, and genetics. J Allergy Clin Immunol. (2020) 145:108–10. doi: 10.1016/j.jaci.2019.11.008
11. Noda, S, Suarez-Farinas, M, Ungar, B, Kim, SJ, de Guzman Strong, C, Xu, H, et al. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. J Allergy Clin Immunol. (2015) 136:1254–64. doi: 10.1016/j.jaci.2015.08.015
12. Chiricozzi, A, Maurelli, M, Calabrese, L, Peris, K, and Girolomoni, G. Overview of atopic dermatitis in different ethnic groups. J Clin Med. (2023) 12:2701. doi: 10.3390/jcm12072701
13. Suaini, NHA, Tan, CPT, Loo, EXL, and Tham, EH. Global differences in atopic dermatitis. Pediatr Allergy Immunol. (2021) 32:23–33. doi: 10.1111/pai.13335
14. Wongvibulsin, S, Sutaria, N, Kannan, S, Alphonse, MP, Belzberg, M, Williams, KA, et al. Transcriptomic analysis of atopic dermatitis in African Americans is characterized by Th2/Th17-centered cutaneous immune activation. Sci Rep. (2021) 11:11175. doi: 10.1038/s41598-021-90105-w
15. Koga, C, Kabashima, K, Shiraishi, N, Kobayashi, M, and Tokura, Y. Possible pathogenic role of Th17 cells for atopic dermatitis. J Invest Dermatol. (2008) 128:2625–30. doi: 10.1038/jid.2008.111
16. Suarez-Farinas, M, Dhingra, N, Gittler, J, Shemer, A, Cardinale, I, de Guzman Strong, C, et al. Intrinsic atopic dermatitis shows similar TH2 and higher TH17 immune activation compared with extrinsic atopic dermatitis. J Allergy Clin Immunol. (2013) 132:361–70. doi: 10.1016/j.jaci.2013.04.046
17. Czarnowicki, T, Gonzalez, J, Shemer, A, Malajian, D, Xu, H, Zheng, X, et al. Severe atopic dermatitis is characterized by selective expansion of circulating TH2/TC2 and TH22/TC22, but not TH17/TC17, cells within the skin-homing T-cell population. J Allergy Clin Immunol. (2015) 136:104–115.e7. doi: 10.1016/j.jaci.2015.01.020
18. Chan, TC, Sanyal, RD, Pavel, AB, Glickman, J, Zheng, X, Xu, H, et al. Atopic dermatitis in Chinese patients shows T(H)2/T(H)17 skewing with psoriasiform features. J Allergy Clin Immunol. (2018) 142:1013–7. doi: 10.1016/j.jaci.2018.06.016
19. Baum, S, Porat, S, Lyakhovitsky, A, Astman, N, and Barzilai, A. Adult atopic dermatitis in hospitalized patients: comparison between those with childhood-onset and late-onset disease. Dermatology. (2019) 235:365–71. doi: 10.1159/000499708
20. Croce, EA, Levy, ML, Adamson, AS, and Matsui, EC. Reframing racial and ethnic disparities in atopic dermatitis in black and Latinx populations. J Allergy Clin Immunol. (2021) 148:1104–11. doi: 10.1016/j.jaci.2021.09.015
21. Why nature is updating its advice to authors on reporting race or ethnicity. Nature. (2023) 616:219. doi: 10.1038/d41586-023-00973-7
22. Yi, SJ, Shon, C, Min, KD, Kim, HC, Leem, JH, Kwon, HJ, et al. Association between exposure to traffic-related air pollution and prevalence of allergic diseases in children, Seoul. Korea Biomed Res Int. (2017) 2017:4216107. doi: 10.1186/s12940-020-0563-6
23. Herbarth, O, Fritz, GJ, Rehwagen, M, Richter, M, Roder, S, and Schlink, U. Association between indoor renovation activities and eczema in early childhood. Int J Hyg Environ Health. (2006) 209:241–7. doi: 10.1016/j.ijheh.2006.01.003
24. Kathuria, P, and Silverberg, JI. Association of pollution and climate with atopic eczema in US children. Pediatr Allergy Immunol. (2016) 27:478–85. doi: 10.1111/pai.12543
25. Fadadu, RP, Grimes, B, Jewell, NP, Vargo, J, Young, AT, Abuabara, K, et al. Association of Wildfire air Pollution and Health Care use for atopic dermatitis and itch. JAMA Dermatol. (2021) 157:658–66. doi: 10.1001/jamadermatol.2021.0179
26. Tham, EH, Loo, EXL, Zhu, Y, and Shek, LP. Effects of migration on allergic diseases. Int Arch Allergy Immunol. (2019) 178:128–40. doi: 10.1159/000494129
27. Penard-Morand, C, Raherison, C, Charpin, D, Kopferschmitt, C, Lavaud, F, Caillaud, D, et al. Long-term exposure to close-proximity air pollution and asthma and allergies in urban children. Eur Respir J. (2010) 36:33–40. doi: 10.1183/09031936.00116109
28. Ahn, K. The role of air pollutants in atopic dermatitis. J Allergy Clin Immunol. (2014) 134:993–9. doi: 10.1016/j.jaci.2014.09.023
29. Lee, JH, Suh, J, Kim, EH, Cho, JB, Park, HY, Kim, J, et al. Surveillance of home environment in children with atopic dermatitis: a questionnaire survey. Asia Pac Allergy. (2012) 2:59–66. doi: 10.5415/apallergy.2012.2.1.59
30. Langan, SM, Silcocks, P, and Williams, HC. What causes flares of eczema in children? Br J Dermatol. (2009) 161:640–6. doi: 10.1111/j.1365-2133.2009.09320.x
31. Miyake, Y, Tanaka, K, Fujiwara, H, Mitani, Y, Ikemi, H, Sasaki, S, et al. Residential proximity to main roads during pregnancy and the risk of allergic disorders in Japanese infants: the Osaka maternal and child health study. Pediatr Allergy Immunol. (2010) 21:22–8. doi: 10.1111/j.1399-3038.2009.00951.x
32. Mubanga, M, Lundholm, C, D'Onofrio, BM, Stratmann, M, Hedman, A, and Almqvist, C. Association of Early Life Exposure to antibiotics with risk of atopic dermatitis in Sweden. JAMA Netw Open. (2021) 4:e215245. doi: 10.1001/jamanetworkopen.2021.5245
33. Li, Y, Su, J, Luo, D, Duan, Y, Huang, Z, He, M, et al. Processed food and atopic dermatitis: A pooled analysis of three cross-sectional studies in Chinese adults. Front Nutr. (2021) 8:754663. doi: 10.3389/fnut.2021.754663
34. Richards, M, Ferber, J, Chen, H, Swor, E, Quesenberry, CP, Li, DK, et al. Caesarean delivery and the risk of atopic dermatitis in children. Clin Exp Allergy. (2020) 50:805–14. doi: 10.1111/cea.13668
35. Zeldin, J, Tran, TT, Yadav, M, Chaudhary, PP, D'Souza, BN, Ratley, G, et al. Antimony compounds associate with atopic dermatitis and influence models of itch and Dysbiosis. Environ Sci Technol Lett. (2023) 10:452–7. doi: 10.1021/acs.estlett.3c00142
36. Yadav, M, Chaudhary, PP, D'Souza, BN, Ratley, G, Spathies, J, Ganesan, S, et al. Diisocyanates influence models of atopic dermatitis through direct activation of TRPA1. PLoS One. (2023) 18:e0282569. doi: 10.1371/journal.pone.0282569
37. Zeldin, J, Chaudhary, PP, Spathies, J, Yadav, M, D'Souza, BN, Alishahedani, ME, et al. Exposure to isocyanates predicts atopic dermatitis prevalence and disrupts therapeutic pathways in commensal bacteria. Sci Adv. (2023) 9:eade8898. doi: 10.1126/sciadv.ade8898
38. Akdis, CA. Does the epithelial barrier hypothesis explain the increase in allergy, autoimmunity and other chronic conditions? Nat Rev Immunol. (2021) 21:739–51. doi: 10.1038/s41577-021-00538-7
39. Castillo, CR, Alishahedani, ME, Gough, P, Chaudhary, PP, Yadav, M, Matriz, J, et al. Assessing the effects of common topical exposures on skin bacteria associated with atopic dermatitis. Skin Health Dis. (2021) 1. doi: 10.1002/ski2.41
40. Al-Obaydi, S, Craig, TJ, and Al-Shaikhly, T. Racial and ethnic disparities in the treatment of patients with atopic dermatitis in the United States: A retrospective matched cohort study. J Allergy Clin Immunol Pract. (2023) 11:2602–2604.e1. doi: 10.1016/j.jaip.2023.05.014
41. NEA. Atopic Dermatitis Visual Guide. (2023) Available at: nationaleczema.org/visual-guide, accessed 2023.
42. Odhiambo, JA, Williams, HC, Clayton, TO, Robertson, CF, and Asher, MII.P.T.S. Group. Global variations in prevalence of eczema symptoms in children from ISAAC phase three. J Allergy Clin Immunol. (2009) 124:1251–1258.e23. doi: 10.1016/j.jaci.2009.10.009
43. Urban, K, Chu, S, Giesey, RL, Mehrmal, S, Uppal, P, Nedley, N, et al. The global, regional, and national burden of atopic dermatitis in 195 countries and territories: an ecological study from the global burden of disease study 2017. JAAD Int. (2021) 2:12–8. doi: 10.1016/j.jdin.2020.10.002
44. Ricciardo, BM, Kessaris, HL, Kumarasinghe, P, Carapetis, JR, and Bowen, AC. The burden of atopic dermatitis and bacterial skin infections among urban-living indigenous children and young people in high-income countries: A systematic review. Pediatr Dermatol. (2023) 40:35–43. doi: 10.1111/pde.15153
45. Silverberg, JI, Simpson, EL, Durkin, HG, and Joks, R. Prevalence of allergic disease in foreign-born American children. JAMA Pediatr. (2013) 167:554–60. doi: 10.1001/jamapediatrics.2013.1319
46. Abuabara, K, You, Y, Margolis, DJ, Hoffmann, TJ, Risch, N, and Jorgenson, E. Genetic ancestry does not explain increased atopic dermatitis susceptibility or worse disease control among African American subjects in 2 large US cohorts. J Allergy Clin Immunol. (2020) 145:192–198.e11. doi: 10.1016/j.jaci.2019.06.044
47. Fulton, RL, Margolis, DJ, Sockler, PG, Mitra, N, Wong, X, and Common, JE. No association of filaggrin copy number variation and atopic dermatitis risk in white and black Americans. Exp Dermatol. (2022) 31:233–6. doi: 10.1111/exd.14449
48. Bissonnette, R, Jankicevic, J, Saint-Cyr Proulx, E, and Maari, C. Ethnicity, race and skin color: challenges and opportunities for atopic dermatitis clinical trials. J Clin Med. (2023) 12:3805. doi: 10.3390/jcm12113805
49. Chai, ESX, Tey, HL, and Lim, ZV. Are there ethnic differences in hand eczema? A review. J Clin Med. (2023) 12:2232. doi: 10.3390/jcm12062232
50. Davis, CM, Flohr, C, Gupta, MR, and Koplin, JJ. Managing atopic dermatitis in patients with skin of color. J Allergy Clin Immunol Pract. (2023) 11:1376–83. doi: 10.1016/j.jaip.2023.03.041
51. Dirks, NL, Huth, B, Yates, CR, and Meibohm, B. Pharmacokinetics of immunosuppressants: a perspective on ethnic differences. Int J Clin Pharmacol Ther. (2004) 42:701–18. doi: 10.5414/CPP42701
52. Min, DI, and Ellingrod, VL. Association of the CYP3A4*1B 5′-flanking region polymorphism with cyclosporine pharmacokinetics in healthy subjects. Ther Drug Monit. (2003) 25:305–9. doi: 10.1097/00007691-200306000-00010
53. Lindholm, A, Welsh, M, Alton, C, and Kahan, BD. Demographic factors influencing cyclosporine pharmacokinetic parameters in patients with uremia: racial differences in bioavailability. Clin Pharmacol Ther. (1992) 52:359–71. doi: 10.1038/clpt.1992.156
54. Schroeder, TJ, Shah, M, Hariharan, S, and First, MR. Increased resources are required in patients with low cyclosporine bioavailability. Transplant Proc. (1996) 28:2151–5.
55. Pollak, R, Wong, RL, and Chang, CT. Cyclosporine bioavailability of Neoral and Sandimmune in white and black de novo renal transplant recipients. Neoral Study Group Ther Drug Monit. (1999) 21:661–3. doi: 10.1097/00007691-199912000-00014
56. Stein, CM, Sadeque, AJ, Murray, JJ, Wandel, C, Kim, RB, and Wood, AJ. Cyclosporine pharmacokinetics and pharmacodynamics in African American and white subjects. Clin Pharmacol Ther. (2001) 69:317–23. doi: 10.1067/mcp.2001.115073
57. Emovon, OE, Op't Holt, C, and Browne, BJ. Can a pharmacokinetic approach to immunosuppression eliminate ethnic disparities in renal allograft outcome? Clin Transpl. (2002) 16:45–8. doi: 10.1034/j.1399-0012.16.s7.6.x
58. Lindholm, A. Factors influencing the pharmacokinetics of cyclosporine in man. Ther Drug Monit. (1991) 13:465–77. doi: 10.1097/00007691-199111000-00001
59. Olafuyi, O, Parekh, N, Wright, J, and Koenig, J. Inter-ethnic differences in pharmacokinetics-is there more that unites than divides? Pharmacol Res Perspect. (2021) 9:e00890. doi: 10.1002/prp2.890
60. Borrell, LN, Elhawary, JR, Fuentes-Afflick, E, Witonsky, J, Bhakta, N, Wu, AHB, et al. Race and genetic ancestry in medicine - A time for reckoning with racism. N Engl J Med. (2021) 384:474–80. doi: 10.1056/NEJMms2029562
61. Swen, JJ, van der Wouden, CH, Manson, LE, Abdullah-Koolmees, H, Blagec, K, Blagus, T, et al. A 12-gene pharmacogenetic panel to prevent adverse drug reactions: an open-label, multicentre, controlled, cluster-randomised crossover implementation study. Lancet. (2023) 401:347–56. doi: 10.1016/S0140-6736(22)01841-4
62. Rothenberg-Lausell, C, Bar, J, Del Duca, E, and Guttman-Yassky, E. Diversity of atopic dermatitis and selection of immune targets. Ann Allergy Asthma Immunol. (2024) 132:177–86. doi: 10.1016/j.anai.2023.11.020
63. Corbett, S, Courtiol, A, Lummaa, V, Moorad, J, and Stearns, S. The transition to modernity and chronic disease: mismatch and natural selection. Nat Rev Genet. (2018) 19:419–30. doi: 10.1038/s41576-018-0012-3
64. Sear, R. Demography and the rise, apparent fall, and resurgence of eugenics. Popul Stud (Camb). (2021) 75:201–20. doi: 10.1080/00324728.2021.2009013
65. Hanson, MA, Barreiro, PG, Crosetto, P, and Brockington, D. The strain on scientific publishing. arXiv. (2023) doi: 10.48550/arXiv.2309.15884
66. Khoo, SY-S. Article processing charge hyperinflation and Price insensitivity: an open access sequel to the serials crisis. LIBER Q. (2019) 29:1–18. doi: 10.18352/lq.10280
67. Reardon, J, Lee, SS, Goering, S, Fullerton, SM, Cho, MK, Panofsky, A, et al. Trustworthiness matters: building equitable and ethical science. Cell. (2023) 186:894–8. doi: 10.1016/j.cell.2023.01.008
68. Panofsky, A, Dasgupta, K, and Iturriaga, N. How white nationalists mobilize genetics: from genetic ancestry and human biodiversity to counterscience and metapolitics. Am J Phys Anthropol. (2021) 175:387–98. doi: 10.1002/ajpa.24150
69. Carlson, J, and Harris, K. Quantifying and contextualizing the impact of bioRxiv preprints through automated social media audience segmentation. PLoS Biol. (2020) 18:e3000860. doi: 10.1371/journal.pbio.3000860
70. Lewis, ACF, Molina, SJ, Appelbaum, PS, Dauda, B, Di Rienzo, A, Fuentes, A, et al. Getting genetic ancestry right for science and society. Science. (2022) 376:250–2. doi: 10.1126/science.abm7530
71. Savage, LC, and Panofsky, A. The self-fulfilling process of clinical race correction: the case of eighth joint National Committee Recommendations. Health Equity. (2023) 7:793–802. doi: 10.1089/heq.2023.0064
72. Guttman-Yassky, E, Facheris, P, Gomez-Arias, PJ, Del Duca, E, Da Rosa, JC, Weidinger, S, et al. Effect of abrocitinib on skin biomarkers in patients with moderate-to-severe atopic dermatitis. Allergy. (2024) 79:1258–70. doi: 10.1111/all.15969
73. Simpson, EL, Gooderham, M, Wollenberg, A, Weidinger, S, Armstrong, A, Soung, J, et al. Efficacy and safety of Lebrikizumab in combination with topical corticosteroids in adolescents and adults with moderate-to-severe atopic dermatitis: A randomized clinical trial (ADhere). JAMA Dermatol. (2023) 159:182–91. doi: 10.1001/jamadermatol.2022.5534
74. Rauer, L, Reiger, M, Bhattacharyya, M, Brunner, PM, Krueger, JG, Guttman-Yassky, E, et al. Skin microbiome and its association with host cofactors in determining atopic dermatitis severity. J Eur Acad Dermatol Venereol. (2023) 37:772–82. doi: 10.1111/jdv.18776
75. Tanei, R, and Hasegawa, Y. Immunological Pathomechanisms of Spongiotic dermatitis in skin lesions of atopic dermatitis. Int J Mol Sci. (2022) 23:6682. doi: 10.3390/ijms23126682
76. Geat, D, Giovannini, M, Barlocco, EG, Pertile, R, Farina, S, Pace, M, et al. Characteristics associated with clinical response to Comano thermal spring water balneotherapy in pediatric patients with atopic dermatitis. Ital J Pediatr. (2021) 47:91. doi: 10.1186/s13052-021-00971-3
77. Holm, JG, Hurault, G, Agner, T, Clausen, ML, Kezic, S, Tanaka, RJ, et al. Immunoinflammatory biomarkers in serum are associated with disease severity in atopic dermatitis. Dermatology. (2021) 237:513–20. doi: 10.1159/000514503
78. Sadrolashrafi, K, Guo, L, Kikuchi, R, Hao, A, Yamamoto, RK, Tolson, HC, et al. An OX-Tra'Ordinary tale: the role of OX40 and OX40L in atopic dermatitis. Cells. (2024) 13:587. doi: 10.3390/cells13070587
79. Zysk, W, Sitko, K, Tukaj, S, Zaryczanska, A, and Trzeciak, M. Altered gene expression of IL-35 and IL-36alpha in the skin of patients with atopic dermatitis. Int J Mol Sci. (2023) 25:404. doi: 10.3390/ijms25010404
80. Macharadze, DS. Phenotypes of atopic dermatitis. Russian J Allergy. (2023) 20:354–65. doi: 10.36691/RJA1596
81. Adam, D, Arany, J, Toth, KF, Toth, BI, Szollosi, AG, and Olah, A. Opioidergic signaling-A neglected, yet potentially important player in atopic dermatitis. Int J Mol Sci. (2022) 23:4140. doi: 10.3390/ijms23084140
82. Szollosi, AG, Olah, A, Lisztes, E, Griger, Z, and Toth, BI. Pruritus: A sensory symptom generated in cutaneous Immuno-neuronal crosstalk. Front Pharmacol. (2022) 13:745658. doi: 10.3389/fphar.2022.745658
83. Lunjani, N, Kerbelker, T, Mdletshe, FB, Hlela, C, and O'Mahony, L. Phenotypes, endotypes and genotypes of atopic dermatitis and allergy in populations of African ancestry on the continent and diaspora. Front Allergy. (2023) 4:1203304. doi: 10.3389/falgy.2023.1203304
84. Singh, K, Valido, K, Swallow, M, Okifo, KO, Wang, A, Cohen, JM, et al. Baseline skin cytokine profiles determined by RNA in situ hybridization correlate with response to dupilumab in patients with eczematous dermatitis. J Am Acad Dermatol. (2023) 88:1094–100. doi: 10.1016/j.jaad.2022.12.052
85. Mikhaylov, D, Ungar, B, Renert-Yuval, Y, and Guttman-Yassky, E. Oral Janus kinase inhibitors for atopic dermatitis. Ann Allergy Asthma Immunol. (2023) 130:577–92. doi: 10.1016/j.anai.2023.01.020
86. Freitas, E, Gooderham, M, and Torres, T. New topical therapies in development for atopic dermatitis. Drugs. (2022) 82:843–53. doi: 10.1007/s40265-022-01722-2
87. Puar, N, Chovatiya, R, and Paller, AS. New treatments in atopic dermatitis. Ann Allergy Asthma Immunol. (2021) 126:21–31. doi: 10.1016/j.anai.2020.08.016
88. Hassoun, D, Malard, O, Barbarot, S, Magnan, A, and Colas, L. Type 2 immunity-driven diseases: towards a multidisciplinary approach. Clin Exp Allergy. (2021) 51:1538–52. doi: 10.1111/cea.14029
89. Bang, CH, Song, JY, Song, YM, Lee, JH, Park, YM, and Lee, JY. Production of IL-31 in CD45RO(+)CLA(+)H4R(+) T cells in atopic dermatitis. J Clin Med. (2021) 10:1976. doi: 10.3390/jcm10091976
Keywords: racism, race science, eczema, atopic dermatitis, allergy
Citation: Myles IA (2024) Race science without racists: how bigoted paradigms persist in allergy research. Front. Public Health. 12:1351732. doi: 10.3389/fpubh.2024.1351732
Received: 08 December 2023; Accepted: 14 June 2024;
Published: 10 July 2024.
Edited by:
Ozgur Karcioglu, University of Health Sciences, TürkiyeReviewed by:
Daniela Rodrigues, University of Coimbra, PortugalCopyright © 2024 Myles. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ian A. Myles, bXlsZXNpQG5pYWlkLm5paC5nb3Y=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.