
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health, 29 January 2024
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 11 - 2023 | https://doi.org/10.3389/fpubh.2023.1301685
This article is part of the Research TopicHigh-level Antimicrobial Resistance or Hypervirulence in Emerging and Re-emerging “Super-Bug” Foodborne Pathogens: Detection, Mechanism, and Dissemination from Omics InsightsView all 16 articles
 Tadele Shiwito Ango1*
Tadele Shiwito Ango1* Negalgn Byadgie Gelaw1
†
Negalgn Byadgie Gelaw1
† Girma Mamo Zegene1
†
Girma Mamo Zegene1
† Tizita Teshome2
†
Tizita Teshome2
† Tesfalem Getahun2
†
Tesfalem Getahun2
†Introduction: Bacterial pathogens continue to be a major cause of foodborne gastroenteritis in humans and remain a public health problem. Housemaids operating inside a kitchen could be the source of infection and may transmit disease-inflicting pathogens through contaminated hands.
Objective: This study aimed to assess the prevalence and antimicrobial susceptibility profile of bacteria isolated from the hands of housemaids in Jimma City, Ethiopia.
Methods: A laboratory-based cross-sectional study was employed among 234 housemaids. Hand swab samples from the dominant hand of the study participants were collected under sterile conditions following standard operating procedures. Then, in the laboratory, the swabs were inoculated aseptically using streak-plating methods on the growth media, such as mannitol salt agar [Staphylococcus aureus and coagulase-negative staphylococci], MacConkey agar [Klebsiella species and Proteus species], salmonella-shigella agar [Salmonella species and Shigella species], and eosin methylene blue agar [Escherichia coli (E. coli)]. In addition, a set of biochemical tests was applied to examine bacterial species. Data were double-entered into EpiData version 3.1 and then exported to the Statistical Package for Social Science (SPSS) version 26 for further analysis. Descriptive analyses were summarized using frequency and percentage.
Results: The proportion of housemaids’ hands containing one or more positive bacterial isolates was 72% (95% CI: 66.2, 77.8). The dominant bacterial isolates were Staphylococcus aureus (31.6%), Escherichia coli (21.3%), Salmonella species (1.3%), Shigella species (6.7%), Klebsiella species (23.1%) and Proteus species (14.7%). Fingernail status (AOR =15.31, 95% CI: 10.372, 22.595) and the removal of a watch, ring, and bracelet during hand washing (AOR = 20.844, 95% CI: 2.190, 9.842) were significantly associated with the prevalence of bacterial isolation. Most Staphylococcus aureus isolates were susceptible to chloramphenicol (98.6%). Escherichia coli isolates were susceptible to tetracycline (75%), ceftriaxone (79.2%), chloramphenicol (87.5%), and ceftazidime (77.1%). Eighty percent of isolated Shigella species were susceptible to chloramphenicol and gentamicin respectively. In addition, Klebsiella and Proteus species exhibited high susceptibility to chloramphenicol. However, their isolates showed resistance against a number of the tested antimicrobials. Staphylococcus aureus isolates (28.2%) were resistance to tetracycline. Moreover, One-quarter of Escherichia coli isolates were resistance to tetracycline, ceftriaxone, chloramphenicol, and ceftazidime. Whereas 46.7% and 48.5% of isolated Shigella species and Proteus species were resistance to tetracycline and ceftriaxone.
Conclusion: The hands of housemaids are important potential sources of pathogenic bacteria that would result in the potential risk of foodborne diseases. Most bacteria isolates were resistant to tetracycline, ceftriaxone, and ceftazidime. Therefore, practicing good hand hygiene helps to prevent and control the spread of antimicrobial-resistant microbes.
Enteric bacterial pathogens are a major cause of foodborne gastroenteritis in humans and remain a public health problem worldwide (1). They are common foodborne disease agents and persist as a major public health threat. A study revealed that food commodities were contaminated by food handlers or housemaids (2). Moreover, the majority of foodborne outbreak causative agents enter the body through the ingestion of contaminated food (3, 4). Banik et al. state that foodborne illnesses occurred after the entrance of those disease-causing microbes into the food supply chain (5). In 2020, the World Health Organization reported approximately 600 million cases and 420,000 deaths related to contaminated food around the world (3). According to the Centers for Disease Control and Prevention estimates, approximately 1 in 6 Americans (48 million people) get sick, 128,000 are hospitalized, and 3,000 die of foodborne diseases each year (6). However, the problem is severe in developing countries, including Ethiopia. Furthermore, it was estimated that approximately 700,000 deaths per year in Africa are caused by foodborne diseases (7). The summary report of the Federal Ministry of Health revealed that the annual incidences of foodborne illnesses ranged from 3.4 to 9.3% in Ethiopia (8). In addition, several studies have been performed to assess and estimate pathogenic bacteria and related rates of infection in Ethiopia (1, 7, 9–20).
The fecal–oral route of pathogen transmission is the most common among the other methods of infection transmission for heterogeneous pathogens (21, 22). In this sense, varieties of bacterial isolates, particularly Staphylococcus aureus, Klebsiella species, Proteus species, E. coli, Shigella species, Salmonella species, Campylobacter, Vibrio cholerae, and Streptococcus pneumonia, might be ingested that results from the hand contact with feces (5, 12–14, 17, 22–26). Furthermore, studies revealed that bacterial pathogens were the most extensively identified infectious agent for the majority of foodborne outbreak types (8, 11). Housemaids perform various daily activities and work at home; their hands quickly become contaminated with different kinds of microbes and therefore, become asymptomatic carriers of pathogens.
The public health importance of bacterial infection continues to pose a challenge to community health systems worldwide (27–29). Food poisoning (gastroenteritis), skin, ear, or sinus infections, sexually transmitted infections, bacterial pneumonia, and urinary tract infections are typical bacterial infections (6, 30). For instance, S. aureus produces toxins that cause staphylococcal food poisoning and gastroenteritis with emesis and with or without diarrhea (23, 31). Infections associated with Salmonella species and Shigella species are among the major public health problems in many countries, including Ethiopia (7, 9). For instance, Salmonella species is the most common cause of foodborne illnesses (23). Shigella causes a foodborne illness with common symptoms of diarrhea, fever, and stomach cramps (7). The estimated annual incidences of Shigella species and Salmonella species are 165 and 25 million, respectively (9). Moreover, a study disclosed that Klebsiella species cause infections at multiple sites in the bodies of people with preexisting health conditions (32).
Due to the high prevalence of antimicrobial-resistant (AMR) pathogens, advances in infection control have not completely eradicated the problem (13). In addition, the constant increase in AMR bacterial strains has become an important clinical problem (33, 34). Those AMR bacterial strains include members of Enterobacteriaceae and continue the increasing concern, which could lead to the narrowing of available therapeutic options (34, 35). Antimicrobial resistance could be determined by many contributing factors. The microbial evolution and transmission of genetic determinants of resistance between microbes enable the spread of pathogenic bacteria (34–38). In addition, the widespread and prolonged use of antibiotics leads to the emergence of resistant bacterial pathogens (13, 18, 38). Moreover, the problems of infectious diseases were worsened by the improper use of antibiotics by humans and animals, which contributed to the rise of AMR globally (13, 33–41). In sum, AMR is an emerging global challenge that results in the spread of infectious diseases that affect human populations (38, 39).
Therefore, pathogenic microbes continue to challenge the healthcare systems in developing countries, including Ethiopia (40). Evidence from studies revealed an increasing incidence of multidrug resistance in foodborne pathogens, particularly to the commonly used antimicrobial agents (40, 41). Most studies in Ethiopia have been conducted in institutions such as hospitals, mass food processing, and catering establishments (11, 13, 14, 18). There is a paucity of data showing profiles of bacteria isolated from the hands of housemaids and their antimicrobial susceptibility profile in dwellings, particularly in the study area. Therefore, the present study aimed to assess the prevalence and antimicrobial susceptibility profile of bacteria isolated from the hands of housemaids in Jimma City, Ethiopia.
The current laboratory-based cross-sectional survey was conducted in residential settings in Jimma City, Southwest Ethiopia, from April to June 2022. Jimma City is located 352 km southwest of Addis Ababa. The physical location of Jimma City lies between latitude 7°41′ N and longitude 36°50′ E. It has an average altitude of 1,780 m above sea level. It receives a mean annual rainfall of approximately 1,530 mm. The mean annual minimum and maximum temperatures of Jimma City are 14.4 and 26.7°C, respectively (42).
All housemaids who have been engaged in work in Jimma City were the source population. Those housemaids who could fulfill the eligibility criteria and were available during the data collection period were considered the study population.
Housemaids who reported having respiratory infections such as the common cold or fecal–oral diseases such as diarrhea, as well as those who had pores, skin irritation, inflammation, eczema, or scars on their palms during the data collection period, were not included in the analysis.
The sample size was determined by applying Yamane’s simplified formula for proportions. A 95% confidence interval (CI) and 0.5 or 50% proportion were assumed
where n is the sample size, N is the population size, and e is the level of precision (5%). After considering a 10% sample size (21 study subjects) with a non-response rate, the final sample size was 213 + 21 = 234.
All residential settings in Jimma City were included in the study. An inventory assessment was conducted to gather information about housemaids employed in residential settings, and data about the total number of housemaids were obtained from households and local administrations [Kebele]. In sum, 455 housemaids were engaged in residential settings in Jimma City (43). Those housemaids who were available during the data collection period were included until the sample size (234) was fulfilled. To minimize sampling bias, after convincing the heads of households, their house number was used for identification.
Data collection tools for sociodemographic data and other relevant data related to hand hygiene practices among housemaids were adapted from the World Health Organization and published articles (44–46). Whereas, hand swab sample collection procedures followed the guidelines of the Clinical and Laboratory Standards Institute (CLSI), the American Type Culture Collection, and others (47–49).
Data were collected by the data collectors after obtaining written informed consent using a pre-tested semi-structured questionnaire and observation designed to obtain sociodemographic data such as sex, age and educational status, and other relevant data related to housemaids’ hand hygiene practices such as fingernail status, frequent handwashing, handwashing method, use of soap and water for frequent handwashing, following the five steps to washing hands in the right way, removal of watch, ring, and bracelets during handwashing, and time in second to wash hands. The data were collected from the study participants after receiving their written informed consent and after receiving the ethical approval for the study from the Institutional Review Board of the Institute of Health, Jimma University.
Three data collectors with prior experience in the field and who were fluent in speaking and reading in the local language and English were hired. The data collection survey underwent 5-day training sessions on informed consent and data collection procedures from 20 to 25 March 2022. The data collectors were two individuals with Bachelor of Science degrees in medical laboratory technology and one individual with a Bachelor of Science degree in environmental health science.
For the laboratory investigation of the commensal microbes from the hands of housemaids, a swab sample was collected following standard operating procedures of CLSI, American-type culture collection, and other guidelines (47–49). In advance, hand swab samples were collected following sterile conditions for the segregation of commensal microbes.
The samples were collected and transported using sterile cotton swabs and 10 mL saline-filled sterile test containers. Following handwashing, the participant’s hands were sampled for the hand swabs by rubbing the entire surface with sterile, moistened cotton-tipped swabs. The sample was then placed or soaked in a labeled 0.85% saline solution containing sterile test tubes for microbial culturing. Nevertheless, no prior notice was given, and extra hand cleanliness was not practiced when collecting samples (18, 50). Three well-trained laboratory personnel gathered swab samples using accepted aseptic methods. Samples were transferred to Jimma University’s Department of Medical Microbiology Laboratory soon after collection. Next, in the laboratory, the samples were enhanced in a nutrient broth for a whole day to promote bacterial recovery, as handwashing has an impact on the survival of the bacteria that were collected.
The most popular techniques for identifying bacteria include the use of differential media, which makes it simpler to separate colonies of desired microorganisms from other colonies growing on the same plate, or selective media, which can prevent or reduce the growth of undesirable commensal microbes (47, 49). By providing the growth media, it is essential to grow and maintain them in carefully regulated laboratory settings. The preparation of each culture medium utilized in this investigation was performed in accordance with the manufacturer’s instructions, and aseptic culturing techniques were employed. A loop full of each hand swab sample enriched on the nutrient broth in the laboratory soon after collection was inoculated aseptically using streak-plating methods on the selective and differential such as mannitol salt agar (MSA) (S. aureus and Coagulase-negative staphylococci), MacConkey agar (MCA) (Klebsiella species and Proteus species), salmonella-shigella agar (SSA) (Salmonella species and Shigella species), and eosin methylene blue agar (EMBA) (E. coli). Then, it was incubated at 37°C for 24 h. Following an incubation period, the culture plates were inspected to see whether there was any suspected bacterial growth present (positive) or absent (negative). Biochemical and morphological testing were used to corroborate the positive laboratory results.
The single colony of bacteria grown on selective and differential media was then subcultured into nutrient agar to determine growth patterns and for further biochemical tests. Then, after obtaining pure colonies, identification of bacteria isolates was performed by using standard microbiology techniques such as the morphology of its colonies and a battery (set) of biochemical tests such as a response on catalase, coagulase, oxidase, Simon citrate agar (SCA), urease, sulfide indole motility (SIM), Kliger’s Iron Agar (KIA), and gas and hydrogen sulfide (H2S) generation (17). The isolation and identification of bacteria from the hand swabs from the hands of housemaids are shown (Figure 1).
Antimicrobial susceptibility tests were performed on Muller Hinton Agar (HIMEDIA, TITAN BIOTECH LTD, Rajasthan, India) by the disk diffusion method. The following antimicrobial drugs were used to test susceptibility: tetracycline (30 μg), ceftriaxone (30 μg), chloramphenicol (30 μg), gentamicin (10 μg), and ceftazidime (30 μg). The selection of drugs was based on availability and pieces of literature (17, 48). The selections of drugs for antimicrobial sensitivity tests were based on the standards for antimicrobial susceptibility guidelines (48), pieces of literature (13, 14, 18), and the current availability of drugs in the market that the community is accessing. The sensitivity, intermediate, and resistance of the bacterial isolates were interpreted according to CLSI guidelines (48).
All culture media used for antimicrobial susceptibility tests were prepared according to the manufacturer’s instructions, and culturing procedures were carried out aseptically. Each batch of the prepared media was checked for sterility by incubating the sample medium at 37°C for a day (14). Staphylococcus aureus ATCC25923 and E. coli ATCC25922, sensitive to all antimicrobial agents, were used as control strains (48). The zone diameter interpretive standards for the determination of antimicrobial agents are shown below (Table 1).
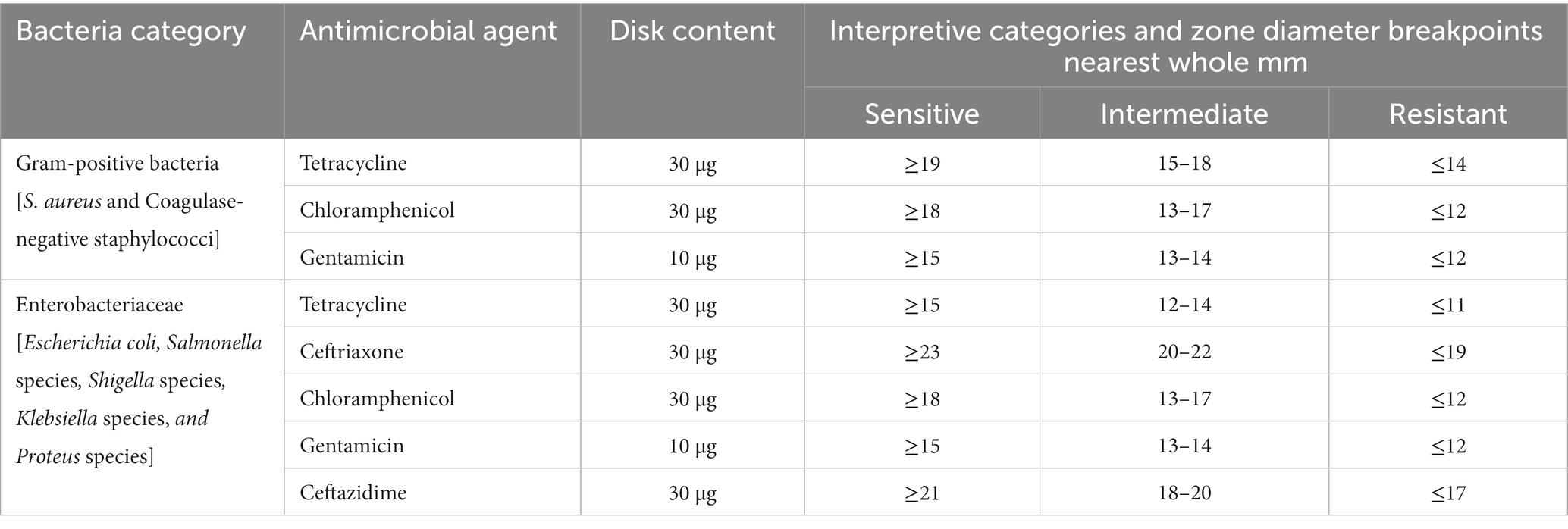
Table 1. Zone diameter interpretive standards for the determination of antimicrobial agent sensitivity and resistance tested by disk diffusion method.
The data collection questionnaire was designed, modified, and contextualized after reviewing related pieces of literature (44–46). Before data collection, a pre-test was conducted among 10% of the total sample size of the study subjects in Mizan-Aman town to determine whether any corrections were made or not.
To manage the quality of work, standard operating procedures have been strictly adhered to in laboratory tests for the investigation of commensal microbes (49). In addition, the proper functioning of the instruments utilized was checked before processing samples, and the known strains of selected organisms (S. aureus ATCC25923 and E. coli ATCC25922) were used for comparison purposes while distinguishing quality. The hand swab samples were collected aseptically, and the temperature range until the final laboratory analysis was checked. Then, the bacterial enrichment broth and growth media sterility were ensured using an autoclave and sample media storage in an incubator overnight, respectively. Interpretation of laboratory findings was confirmed by using updated microbiology guidelines such as CLSI (48).
Data were edited, cleaned, and double-entered into EpiData version 3.1 and then exported to the statistical package for social science statistics version 26 for further analysis. Descriptive analyses were summarized using frequency and percentage and presented in texts, tables, and figures. Binary logistic regression was analyzed to assess associated factors with the prevalence of bacteria isolated from the hands of housemaids. The variables with a value of p ≤ 0.25 were fitted into the multivariable analysis. A Hosmer and Lemeshow statistical test was carried out to check the goodness of fitness. Variables were selected through a backward, stepwise selection technique. The odds ratio with a respective 95% confidence interval was used to measure the strength of the association. A p value of <0.05 was considered statistically significant.
The Institute of Health Sciences at Jimma University’s IRB granted ethical clearance for the study which was carried out under reference number IHRPGS/437/22. Every respondent was asked for their informed and oral consent. Codes were used to maintain the complete confidentiality of the information collected from study participants, including their privacy. Concerning parties, such as research participants and households, would be connected to the atypical clinical result. When gathering data, personal safety measures were taken to prevent the spread of COVID-19 from the data collector to study participants and vice versa. These measures included wearing a mask, and gloves, wiping hands with sanitizer or alcohol, and washing hands with detergent.
Two hundred and twenty-five study subjects participated in this study, with a response rate of 96.2%. All the respondents were women. The age of study participants ranged from 18 to 36, with a mean age of 21.41 ± SD of (3.961). The majority, 182(81%) and 34(15%) of the study participants were between the age categories of 18–30 and 18–24 years, respectively (Figure 2). More than half (53%) of study participants attended primary school, while 7(3%) of respondents could not read or write (Figure 3).
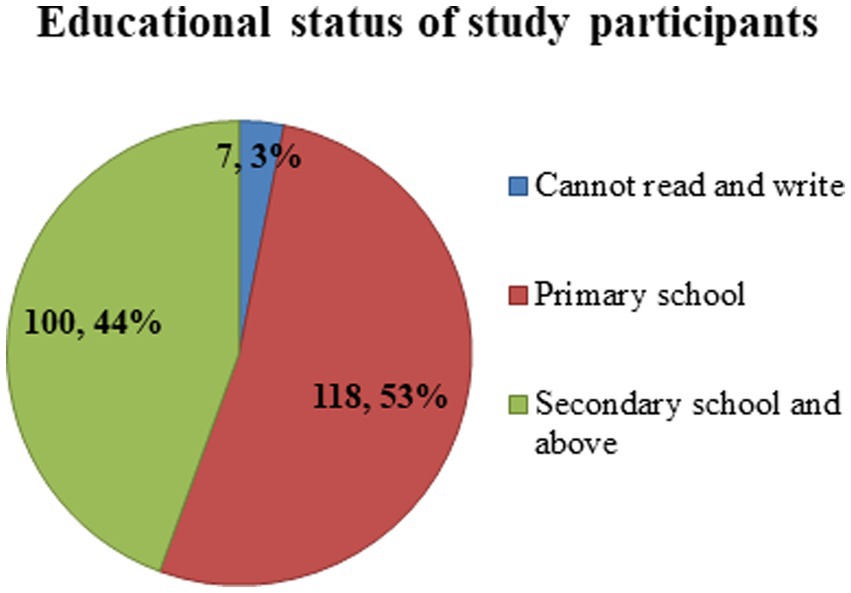
Figure 3. Educational status of housemaids (n = 225) working in Jimma City, Southwest Ethiopia, 2022.
The overall prevalence of one or more bacteria isolated from the hands of housemaids was 72% (95% CI: 66.2–77.8). The total number of bacteria isolated from hand swab samples was 224. Staphylococcus aureus 71(31.6%) was the predominant bacterial species, followed by Klebsiella species 52(23.1%) and E. coli 48(21.3%), whereas the least isolated bacteria was Coagulase-negative staphylococci (0.90%). However, bacteria were not isolated from 63(28%) of the study participants’ hands (Table 2).
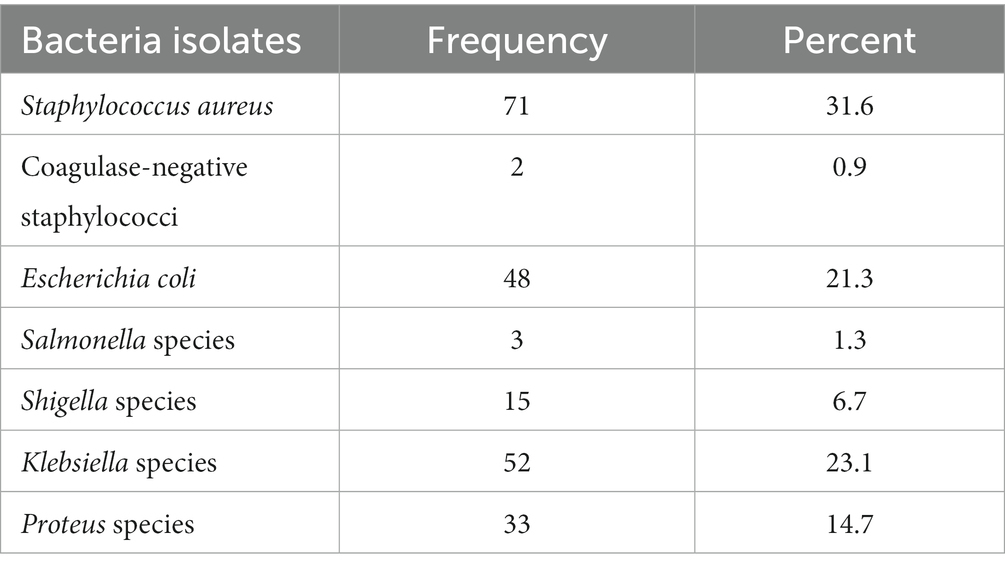
Table 2. Types of bacterial isolates from the hands of housemaids (n = 225) in Jimma City, Southwest Ethiopia, 2022.
In bivariable logistic regression, variables including fingernail status, frequent handwashing, how to wash hands, washing hands frequently with soap/other detergents, following five steps to wash hands the right way, removing a watch, ring, and bracelet during handwashing, and washing hands for 20 s were significantly associated with bacterial isolates from hands. In multivariable logistic analysis, all variables with a value of p ≤0.25 in bivariable analyses were included. Fingernail status and removing a watch, ring, and bracelet during handwashing were found to be significantly associated with bacterial isolates from the hands of housemaids, with a value of p < 0.05 (Table 3).
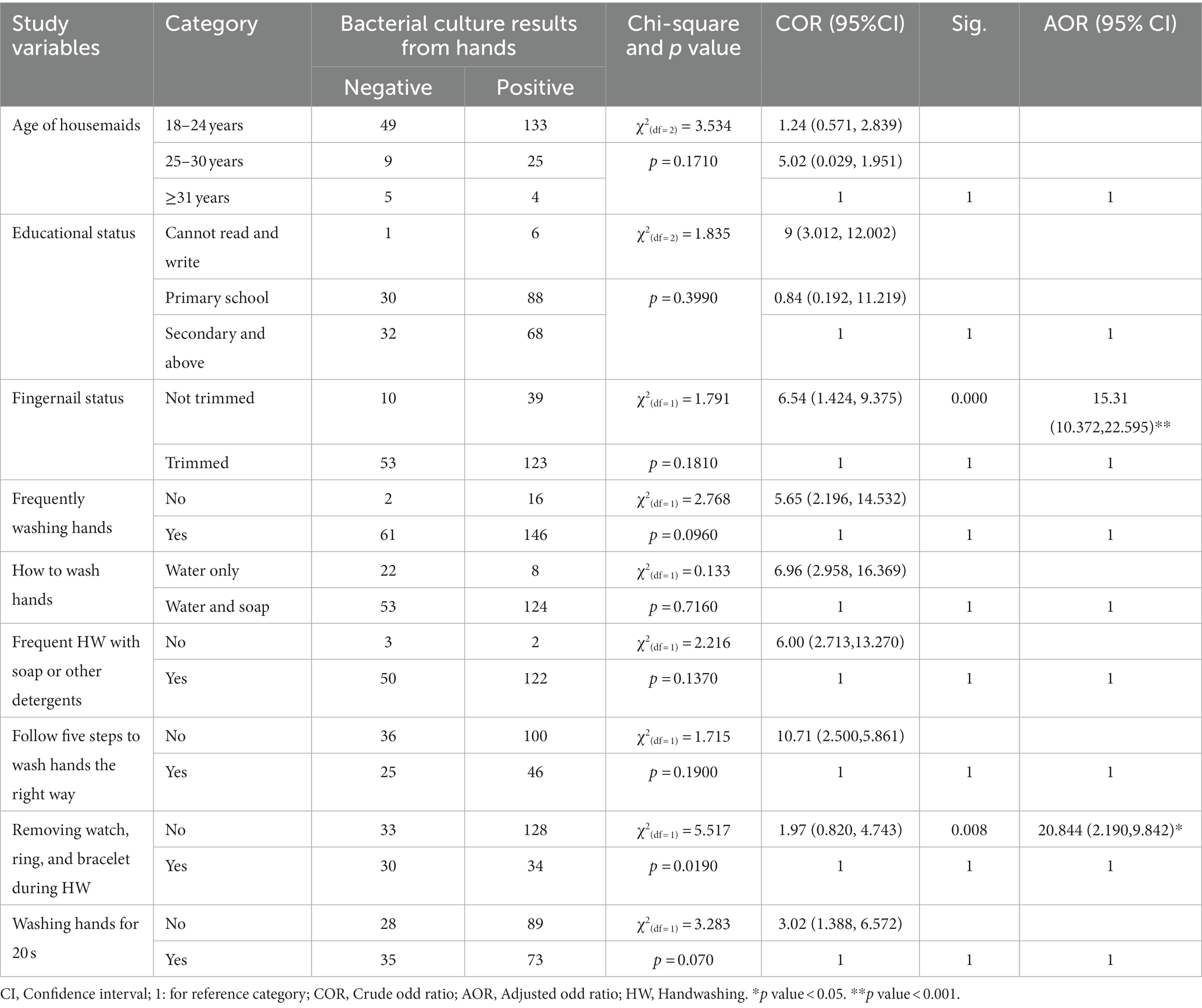
Table 3. Factors associated with the bacterial isolation from the hands of housemaids (n = 225) in Jimma City, Ethiopia, 2022.
This study suggested that housemaids who did not trim their fingernails were 15.31 times more likely to have tested positive for bacteria cultures from the hand swabs compared to housemaids who trimmed their fingernails (adjusted odd ratio = 15.31, 95% confidence interval: 10.372, 22.595). Housemaids who did not experience the removal of a watch, ring, or bracelet during handwashing had 20.844 times higher odds of positive bacteria cultures than their counterparts (adjusted odd ratio = 20.844, 95% confidence interval: 2.190, 9.842; Table 3).
The majority of Staphylococcus aureus were susceptible to chloramphenicol (n = 70; 98.6%) followed by 46 (64.8%) and 42 (59.2%) sensitive to gentamicin and tetracycline, respectively. Coagulase-negative staphylococci were sensitive to all tested antibiotics (Table 4).
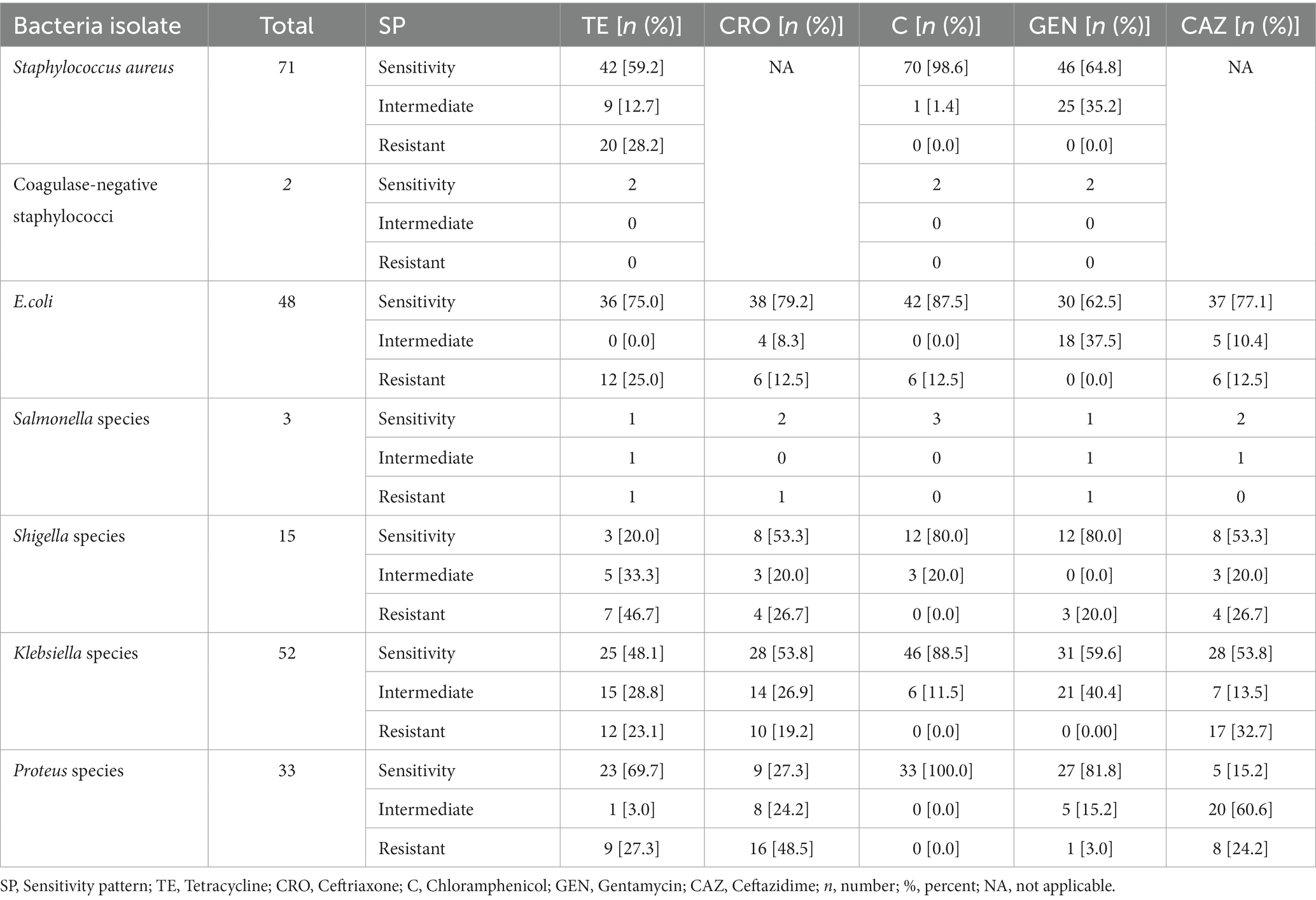
Table 4. Antimicrobial susceptibility pattern of bacteria isolates from the hands of housemaids in Jimma City, Southwest Ethiopia, 2022.
However, approximately 20 (28.2%) of isolated S. aureus were resistant to tetracycline. In addition, no resistance was reported regarding Coagulase-negative staphylococci. Regarding S. aureus isolates, no resistance was reported to chloramphenicol or gentamicin (Table 4).
For E. coli (n = 48), approximately 36 (75%), 38 (79.2%), 42 (87.5%), and 37 (77.1%) isolates were sensitive to tetracycline, ceftriaxone, chloramphenicol, and ceftazidime. For Salmonella species, all three isolates and two of them were sensitive to chloramphenicol, ceftriaxone, and ceftazidime. Approximately 80% of the Shigella isolate was sensitive to chloramphenicol and gentamicin, respectively. For Klebsiella isolates (n = 52), 46 (88.5%), 31 (59.6%), and 28 (53.8%) were sensitive to chloramphenicol, gentamicin, ceftriaxone, and ceftazidime. Moreover, 100, 81.8, and 69.7% of Proteus species were sensitive to chloramphenicol, gentamicin, and tetracycline, respectively (Table 4).
One-quarter of E. coli isolate was resistant to tetracycline, ceftriaxone, chloramphenicol, and ceftazidime, while a single isolate of Salmonella was resistant to tetracycline, ceftriaxone, and gentamicin. Most Shigella isolates were resistant to tetracycline (46.7%), ceftriaxone (26.7%), and ceftazidime (26.7%). Moreover, 16 (48.5%), 9 (27.3%), and 8 (24.2%) of isolated Proteus species were resistant to ceftriaxone, tetracycline, and ceftazidime, respectively (Table 4).
However, no resistance was reported to chloramphenicol regarding Salmonella, Shigella, Klebsiella, and Proteus species. In addition, no resistance was reported regarding gentamicin on isolates of E. coli and Proteus (Table 4).
From total bacteria isolates, no resistant chloramphenicol was recorded among six (85.7%), namely S. aureus, Coagulase-negative staphylococci, Salmonella, Shigella, Klebsiella species, and Klebsiella species Similarly, no gentamicin resistance was recorded among four (57.1%), namely S. aureus, Coagulase-negative staphylococci, E.coli, and Klebsiella species (Table 4).
Housemaids with poor hand hygiene could be potential sources of infection due to pathogenic bacteria, which can cause food contamination and, consequently, foodborne diseases that pose a potential risk to public health (18, 25). Due to the scarcity of published information, the bacterial contamination level among housemaids in Ethiopia is underexplored. Therefore, the present study was undertaken to assess the prevalence and antibiotic susceptibility profile of bacteria isolated from the hands of housemaids in Jimma City, Ethiopia.
The prevalence of bacterial isolation from the hands of housemaids was 72%. This result is comparable with a study conducted in Tripoli, Libya with the prevalence of bacterial growth (71.41%) (51). The bacteria isolation from hands could be due to poor hand hygiene practices such as untrimmed fingernails and not removing the watch, rings, and bracelets during handwashing. Thus, jewelry could lead to bacteria colonizing the hands (52). In addition, bacterial isolation from the hands of housemaids illustrates the concept of fecal contamination (53). The other reason might be the quality of the handwashing water. Pieces of evidence revealed that bacterial contamination of hands is significantly affected by handwashing water (54, 55).
However, the result is higher than the reported prevalence by a previous study performed in Iran with a bacteria isolation rate of 62.2% (25), Egypt with a positive culture for one or more microbial contaminants (60%) (56), Sudan with a carrier of pathogenic bacteria (23.2%) (24), and in different parts of Ethiopia: Jimma (49.6, 6.9, and 19.0%) (11, 15, 20), Gondar town (13. 2%) (1), Debre Markos (29.5 and 46.7%) (16, 17), and Dessie town (59.4%) (12). On the other hand, the result is lower than the study conducted in Mauritius, in which the prevalence of bacteria growth from hands was 91.0% (26), and in Ethiopia, at the University of Gondar Referral Hospital (UoGRH), the prevalence of bacterial isolation was 83.9% (13). The observed discrepancy in the bacteria isolation rate might be due to the differences in the study settings and periods, study participants, hygiene practices, and referent pressure for hygienic conditions.
In this study, S. aureus was the predominant bacterial species in the hands of housemaids, with an isolation rate of 31.6%. This result is comparable to a study conducted in the ICUs of United States medical centers (30.0%) (57) and UoGRH, Ethiopia, where the prevalence of S. aureus was 34% (13). The isolation of S. aureus could be because it is a pathogenic bacterium that is part of the normal flora of the skin and other body parts.
However, the result is lower than the study performed in Sudan (71.8%) (24), Iran (46%) (25), Nigeria (68.9%) (58), Eritrea (63.1%) (59), and Ethiopia; Addis Ababa Regional Laboratory (50.0%) (60) and Ethiopia’s Debre Markos Comprehensive Specialized Hospital (DMCSH) (46.2%) (19), but higher than the study conducted in Alexandria, Egypt (22%) (56), Eastern India (3.44%) (5), and Ethiopia; University of Gondar (16%) (14); Debre Markos (5%) (17); Gondar Town (16.5%) (61), and Jimma University main campus (23.5%) (15). The discrepancy in the isolation rate of S. aureus might be due to differences in the sociodemographic characteristics, the study periods, the study settings and participants, the working environment, and the sample size.
The majority of S. aureus was sensitive to chloramphenicol (98.6%), followed by gentamycin (64.8%) and tetracycline (59.2%). This result is higher compared to the results reported in the UoGRH with sensitivity to chloramphenicol (76.9%) (13), the Addis Ababa regional laboratory with sensitivity to chloramphenicol (53.7%) (60), and DMCSH with sensitivity to chloramphenicol (66.7%) (19). Regarding resistance, isolates of S. aureus were resistant to tetracycline (28.2%). The result is lower than the study performed by Addis Ababa Regional Laboratory (74.30%) (60), Debre Markos (54.5%) (17), and UoGRH (64.1%) (13), but the result is higher than the study conducted at the University of Gondar (21.9%) (14). Different mechanisms play a pivotal role in how S. aureus became resistant to antimicrobials. The antimicrobial resistance of S. aureus to tetracycline might be due to increased efflux, the production of β-lactamase to β-lactam-sensitive antibiotics, the presence of acetyltransferase, a decrease in accumulation of macrolide antibiotics, the expression of the mec gene, and the formation of alternative pathways for sulphonamides (62). The reason for the antimicrobial resistance could be the dissemination of the strain (31). Moreover, the antimicrobial resistance of S. aureus could be determined by a lack of access to appropriate antimicrobial susceptibility tests and bacteriological diagnosis that could lead to the misuse of antimicrobials by patients (16).
In the present study, the isolation rate of E. coli was 21.3%, which is comparable with a study conducted in India (20.68%) (5). The result is lower than the study performed in Iran (29.2%) (51), Nigeria (25.0%) (58), and Jimma University Specialized Hospital (JUSH) (25.4%) (20). However, the result is higher compared to previous studies conducted in Mauritius (0.5%) (26), in the ICUs of United States medical centers (7.1%) (57), and in the different parts of Ethiopia: Jimma University (10.9%) (15), Gondar Town (3.1 and 1.9%) (1, 61), Debre Markos (2.7%) (17), UoGRH (5.9%) (13), University of Gondar (2.67%) (14), DMCSH (3.9%) (19), and Addis Ababa Regional Laboratory (3.6%) (60). The isolation of E. coli illustrates the concept of fecal contamination in the hands of housemaids. In addition, De Alwis et al. (53) revealed that contaminated surfaces such as toilets and washrooms could be the sources of contamination of the hands when a person comes into contact.
The majority of E. coli isolates (87.5%) were sensitive to chloramphenicol. This result is lower than a study conducted in Gondar town (100%) (1); however, the result is higher compared to other studies with a sensitivity of 50% (11, 14, 17, 60), 75% (13), (66.7%) (19) and 56% to chloramphenicol (16). Similarly, approximately 79.2 and 75% of E. coli isolates were sensitive to ceftriaxone and tetracycline which is higher than the results reported in previous studies (11, 16, 17, 19, 60). However, resistance to tetracycline, ceftriaxone, chloramphenicol, and ceftazidime was reported for E. coli isolates (12.5%). It is lower than the results reported in other studies (11, 14, 16, 17, 60). A study conducted in the JUSH showed that E. coli isolates were resistant to ceftriaxone (73%) and ceftazidime (65%) (20). Another study carried out in DMCSH revealed that E. coli was resistant to tetracycline (44.4%), chloramphenicol (22.2%), and ceftazidime (33.3) (19). Escherichia coli were sensitive to chloramphenicol, which might be due to decreased levels of acetyl coenzyme A incat-expressing CM2555 cells in the presence of chloramphenicol (63). The antimicrobial resistance of E. coli to antimicrobial drugs could be due to its outer membrane and the expression of numerous efflux pumps (64). In addition, the antimicrobial resistance of E. coli might be due to the transmission of resistance genes and the unrestricted use of antimicrobials that perpetuate antimicrobial-resistant plasmids (65). Furthermore, other contributors to antimicrobial resistance are the spread of E. coli-resistant strains, overuse or inappropriate prescribing, use of antibiotics in livestock, hygiene/fecal colonization, and antibiotic resistance mechanisms (β-lactams) (64).
In the current study, the isolation rate of Shigella species was 6.70%, which is comparable to the result reported in Debre Markos University, northwest Ethiopia (5.9%) (18). The isolation of Shigella species from the hands of housemaids illustrates poor hand hygiene practices due to fecal contamination. The other reason might be due to cross-contamination of hands with surfaces such as toilets and washrooms (53). The result is lower than a study performed in Gondar town with a prevalence of 10.1% (1), but higher compared to previous studies conducted in Jimma town (0.2%) (11), the University of Gondar (2.7%) (14), and Debre Markos Referral Hospital (5.4%) (16). The variation might be due to differences in the demographic characteristics, study settings and period, study design and sampling techniques, and geographical variation of study areas.
Concerning the antimicrobial resistance profile of isolates, approximately 80% of isolated Shigella species were sensitive to chloramphenicol and gentamicin, respectively. This is comparable to a study performed in Gondar town (1) and Debre Markos University (18), but the result is higher than previous studies performed at the Debre Markos Referral Hospital (16) and the University of Gondar (14). On the other hand, approximately 46.7 and 26.7% of isolated Shigella species were resistant to tetracycline, ceftriaxone, and ceftazidime. This is consistent with reported results in Gondar town (1) and Debre Markos University (18). The result is lower than a previous study performed in Debre Markos Referral Hospital (16), Gondar town (1), and Jimma town (11), but higher than a previous study in Gondar town (1) and Jimma town (11). The resistance of Shigella species to tested antibiotics might be due to high descriptions from clinics available in the locality as well as self-medication. In addition, it might be due to genetic diversity (66).
In the present study, the isolation rate of Klebsiella species was 23.1%, which is higher than the previous study conducted in different parts of Ethiopia: Debre Markos Referral Hospital (4.3%) (16), Gondar town (1.67%) (14), Debre Markos University (2.7%) (17), UoGRH (12.5%), Gondar town (5.5%) (61), and ICUs of US medical centers (11.8%) (57). Regarding the antimicrobial susceptibility pattern of Klebsiella species, the majority of Klebsiella isolates (88.5%) were sensitive to chloramphenicol. The result is in line with a previous study conducted in Ethiopia (17). While antibiotic resistance to tetracycline, ceftriaxone, and ceftazidime among Klebsiella isolates was recorded, a study performed in different parts of Ethiopia showed antibiotic resistance of Klebsiella species regarding chloramphenicol, tetracycline, ceftriaxone, gentamicin, and ceftazidime (13, 14, 16, 17, 20). The antimicrobial resistance of Klebsiella species might be due to its strains having a 𝛽-lactam ring provided with a Zwitterionic structure (20). Another aggravating factor for antimicrobial resistance of Klebsiella species might be the self-prescribing of antibiotics by a few people due to the availability of antibiotics on the market in the study area and inappropriate use of antibiotics (60).
The isolation of Proteus species is 14.7% in the present study, which is inconsistent with Debre Markos University (1.4%) (17), JUSH (2.0%) (20), Jimma University (2.2%) (15), and UoGRH (9.6%) (13). Moreover, we observed antimicrobial resistance of Proteus species to ceftriaxone, tetracycline, and ceftazidime in the current study. It is consistent with a previous study that showed antimicrobial resistance recorded for tested drugs such as ceftriaxone, tetracycline, and ceftazidime (13, 17, 20).
In sum, sensitivity to chloramphenicol on most bacteria isolates was observed, but resistance to tetracycline, ceftriaxone, and ceftazidime among isolates E. coli, Salmonella, Shigella, Klebsiella species, and Proteus species was observed in the present study. At present, antimicrobial resistance is an emerging global challenge that results in the spread of infectious diseases that could affect human populations (38, 39). This might be due to a complex set of causes such as biological processes, human behaviors, and social factors that support the microbes to multiply, carry on, and produce harm (37). In addition, antimicrobial resistance could be due to the evolutionary processes or natural phenomena to which microbes tend to adapt (36, 37). The other reason for AMR could be the inappropriate use of drugs by the community, the use of antibiotics in animals, or the external environment (18, 38, 39). Moreover, the global connection of a large human population allows microbes to move from place to place and spread, allowing them to easily enter the environment (39).
In comparison to housemaids who clipped their fingernails, housemaids who neglected to do so were 15.31 times more likely to have positive bacteria cultures (AOR = 15.31, 95% CI: 10.372, 22.595). This result is in line with the result reported in Poland (OR: 7.1; 95% CI:1.83, 27.39) (67). The result is also supported by a study performed by Mengist et al. (17). In addition to limiting the use of proper hand hygiene, long or sharp fingernails might promote bacteria development. The likelihood of positive bacterial isolation was 20.844 times higher in housemaids who did not remove their accessories before washing their hands than their counterparts (AOR = 20.844, 95% CI: 2.190, 9.842). During handwashing, accessories must be taken off to prevent the growth and spread of harmful microorganisms. They should be well rinsed unless doing so would cause bacterial growth (53).
This study did not identify important microbial hand contaminants such as V. cholerae, Helicobacter, and Campylobacter due to constraints on resources. In addition, some antimicrobial susceptibility tests were performed in this study due to the lack of antimicrobial disks. Furthermore, a multidrug resistance test was not conducted for bacteria isolated from the hands of housemaids.
Housemaids’ hands are very important potential sources of disease-causing bacterial pathogens that would result in the potential risk of gastrointestinal tract infections. Fingernail status and the removal of accessories during handwashing were found to be significantly associated with the prevalence of bacterial isolation from the hands of housemaids. Moreover, most of the bacterial isolates were sensitive to chloramphenicol and gentamycin, while the majority of them were resistant to tetracycline, vancomycin, and ceftazidime.
Therefore, keeping fingernail status short, removing accessories during handwashing, and practicing good hand hygiene are crucial to preventing and controlling antimicrobial-resistant microbes. In addition, any community health worker, regional, national, and other stakeholders who are engaged in community health should create awareness about regular handwashing and its relevance in the prevention and reduction of pathogenic microorganisms from hands in the wider community.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The study was conducted after obtaining an ethical clearance with reference number IHRPGS/437/22 approved by the Institutional Review Board of the Institute of Health Sciences, Jimma University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
TA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. NG: Visualization, Writing – original draft, Writing – review & editing. GZ: Visualization, Writing – original draft, Writing – review & editing. TT: Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. TG: Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors would like to thank the study participants for being part of the study. In addition, the authors gratefully acknowledge the heads of households of housemaids, Jimma City municipality, and Jimma University community service directives for their collaboration. Our heartfelt thanks go to data collectors Dawit Abera, Bizuwerk Sharew, and Soressa Gershe. Finally, the authors are pleased to thank Jimma University, Institute of Health, for their approval of the study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
AMR, Antimicrobial resistance; EMB, Eosin methylene blue; KIA, Kliger iron agar; MSA, Mannitol salt agar; CLSI, Clinical Laboratory Standards Institute; SCA, Simon citrate agar; SIM, Sulfide indole motility; SSA, Salmonella–Shigella agar.
1. Getie, M, Abebe, W, and Tessema, B. Prevalence of enteric bacteria and their antimicrobial susceptibility patterns among food handlers in Gondar town, Northwest Ethiopia. Antimicrob Resist Infect Control. (2019) 8:1–6. doi: 10.1186/s13756-019-0566-7
2. Grace, D . Food safety in low and middle-income countries. Int J Environ Res Public Health. (2015) 12:10490–507. doi: 10.3390/ijerph120910490
4. Grace, D. (2015). Food safety in developing countries: An overview. A learning resource for Department for International Development (DFID) livelihoods advisers.
5. Banik, S, Chakrabarty, S, and Das, N. Hand hygiene in food handlers working in canteens of an educational institution in eastern India. Int J Commun Med Public Heal. (2020) 7:2602–6. doi: 10.18203/2394-6040.ijcmph20202983
6. Thobaben, M . Causes and prevention of foodborne illness. Home Health Care Manag Pract. (2010) 22:533–5. doi: 10.1177/1084822310376611
7. Tadesse, G, Mitiku, H, Teklemariam, Z, and Marami, D. Salmonella and Shigella among asymptomatic street food vendors in the Dire Dawa city, eastern Ethiopia: prevalence, antimicrobial susceptibility pattern, and associated factors. Environ Health Insights. (2019) 13:117863021985358–8. doi: 10.1177/1178630219853581
8. Kalekidan, T, Behailu, K, and Rediet, H. The Ethiopian perception of the food safety system. Adv Food Sci Technol. (2014) 2:260–8.
9. Mardu, F, Negash, H, Legese, H, Berhe, B, Tesfay, K, Haileslasie, H, et al. Assessment of knowledge, practice, and status of food handlers toward Salmonella, Shigella, and intestinal parasites: a cross-sectional study in Tigrai prison centers, Ethiopia. PLoS One. (2020) 15:1–13. doi: 10.1371/journal.pone.0241145
10. Mama, M, and Alemu, G. Prevalence, antimicrobial susceptibility patterns and associated risk factors of Shigella and Salmonella among food handlers in Arba Minch University, South Ethiopia. BMC Infect Dis. (2016) 16:1–7. doi: 10.1186/s12879-016-2035-8
11. Worku, T, Jejaw, A, Kannan, S, and Wondafrash, B. Isolation and antimicrobial sensitivity patterns of enteric bacterial pathogens from asymptomatic food handlers, Jimma, Ethiopia. Am J Heal Res. (2015) 3:399–406. doi: 10.11648/j.ajhr.20150306.24
12. Shiferaw, B, Gelaw, B, Assefa, A, Assefa, Y, and Addis, Z. Bacterial isolates and their antimicrobial susceptibility pattern among patients with external ocular infections at Borumeda hospital, Northeast Ethiopia. BMC Ophthalmol. (2015) 15:1–8. doi: 10.1186/s12886-015-0078-z
13. Mohammed, A, Seid, ME, Gebrecherkos, T, Tiruneh, M, and Moges, F. Bacterial isolates and their antimicrobial susceptibility patterns of wound infections among inpatients and outpatients attending the University of Gondar Referral Hospital, Northwest Ethiopia. Int J Microbiol. (2017) 2017:8953829. doi: 10.1155/2017/8953829
14. Dagnew, M, Tiruneh, M, Moges, F, and Gizachew, M. Bacterial profile and antimicrobial susceptibility pattern among food handlers at Gondar University cafeteria, Northwest Ethiopia. J Infect Dis Ther. (2013) 1:105. doi: 10.4172/2332-0877.1000105
15. Assefa, T, Tasew, H, Wondafrash, B, and Beker, J. Contamination of Bacteria and associated factors among food handlers working in the student cafeterias of Jimma University-Main campus, Jimma, south West Ethiopia. Altern Integr Med. (2015) 4:185. doi: 10.4172/2327-5162.1000185
16. Abebe, M, Tadesse, S, Meseret, G, and Derbie, A. Type of bacterial isolates and antimicrobial resistance profile from different clinical samples at a referral hospital, Northwest Ethiopia: five years data analysis. BMC Res Notes. (2019) 12:568. doi: 10.1186/s13104-019-4604-6
17. Mengist, A, Aschale, Y, and Reta, A. Bacterial and parasitic assessment from fingernails in Debre Markos, Northwest Ethiopia. Can J Infect Dis Med Microbiol. (2018) 2018:6532014. doi: 10.1155/2018/6532014
18. Mengist, A, Mengistu, G, and Reta, A. Prevalence and antimicrobial susceptibility pattern of Salmonella and Shigella among food handlers in catering establishments at Debre Markos university, Northwest Ethiopia. Int J Infect Dis. (2018) 75:74–9. doi: 10.1016/j.ijid.2018.08.008
19. Haile, Z, Mengist, HM, and Dilnessa, T. Bacterial isolates, their antimicrobial susceptibility pattern, and associated factors of external ocular infections among patients attending eye clinic at Debre Markos comprehensive specialized hospital, Northwest Ethiopia. PLoS One. (2022) 17:e0277230. doi: 10.1371/journal.pone.0277230
20. Gashe, F, Mulisa, E, Mekonnen, M, and Zeleke, G. Antimicrobial resistance profile of different clinical isolates against third-generation Cephalosporins. Aust J Pharm. (2018) 2018:1–7. doi: 10.1155/2018/5070742
21. Mekonnen, M, Aga, F, Kinati, T, and Shifera, D. Assessment of hand washing practice and associated factors among primary school children in Sebeta town Oromia regional state, Ethiopia. Health Sci J. (2018) 12:1–6. doi: 10.21767/1791-809X.1000605
22. Pickering, AJ, Boehm, AB, Mwanjali, M, and Davis, J. Efficacy of waterless hand hygiene compared with handwashing with soap: a field study in Dar Es Salaam, Tanzania. Am J Trop Med Hyg. (2010) 82:270–8. doi: 10.4269/ajtmh.2010.09-0220
23. World Health Organization (2016). Burden of Foodborne Diseases in the South-East Asia Region. New Delhi, India.
24. Ahmed, H, and Hassan, H. Bacteriological and parasitological assessment of food handlers in the Omdurman area of Sudan. J Microbiol Immunol Infect. (2010) 43:70–3. doi: 10.1016/S1684-1182(10)60010-2
25. Nasrolahei, M, Mirshafiee, S, Kholdi, S, Salehian, M, and Nasrolahei, M. Bacterial assessment of food handlers in Sari City, Mazandaran Province, north of Iran. J Infect Public Health. (2016) 10:1–6. doi: 10.1016/j.jiph.2016.03.006
26. Padaruth, SK, and Biranjia-Hurdoyal, SD. Hygiene practices and fecal contamination of the hands of children attending primary school in Mauritius. Int Health. (2015) 7:280–4. doi: 10.1093/inthealth/ihu080
27. Doron, S, and Gorbach, SL. Bacterial infections: overview. Int Encycl Public Heal. (2008) 3:273–82. doi: 10.1016/B978-012373960-5.00596-7
28. Ks, I, Lr, S, Gr, A, F, S, T, M, Ap, G, et al. Global mortality associated with 33 bacterial pathogens in 2019: a systematic analysis for the global burden of disease study 2019. Lancet. (2022) 400:2221–48. doi: 10.1016/S0140-6736(22)02185-7
29. Mahtab, T, Jyoti, A, Khusro, A, Redwan, BM, Zidan, M, Mitra, S, et al. Antibiotic resistance in microbes: history, mechanisms, therapeutic strategies, and prospects. J Infect Public Health. (2021) 14:1750–66. doi: 10.1016/j.jiph.2021.10.020
30. Cleveland Clinic (2023). Bacterial infection 1–10. Available at: https://my.clevelandclinic.org/health/diseases/24189-bacterial-infection
31. Castro, A, Santos, C, Meireles, H, Silva, J, and Teixeira, P. Food handlers as potential sources of dissemination of virulent strains of Staphylococcus aureus in the community. J Infect Public Health. (2016) 9:153–60. doi: 10.1016/j.jiph.2015.08.001
32. Chang, D, Sharma, L, Dela Cruz, CS, and Zhang, D. Clinical epidemiology, risk factors, and control strategies of Klebsiella pneumoniae infection. Front Microbiol. (2021) 12:750662. doi: 10.3389/fmicb.2021.750662
33. Adamus-Bialek, W, Zajac, E, Parniewski, P, and Kaca, W. Comparison of antibiotic resistance patterns in collections of Escherichia coli and Proteus mirabilis uropathogenic strains. Mol Biol Rep. (2013) 40:3429–35. doi: 10.1007/s11033-012-2420-3
34. Kamil, TD, and Jarjes, SF. Isolation, identification, and antibiotics susceptibility determination of Proteus species obtained from various clinical specimens in Erbil City. Polytech J. (2019) 9:86–92. doi: 10.25156/ptj.v9n2y2019.pp86-92
35. Boucher, HW, Talbot, GH, Bradley, JS, Edwards, JE, Gilbert, D, Rice, LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. (2009) 48:1–12. doi: 10.1086/595011
36. Giubilini, A, and Savulescu, J. Moral responsibility and the justification of policies to preserve antimicrobial effectiveness In: E Jamrozik and M Selgelid, editors. Ethics and Drug Resistance: Collective Responsibility for Global Public Health. Cham, Switzerland: Springer (2020). 141–54.
37. Cheah, PY, Parker, M, and Day, NPJ. Ethics and antimalarial drug resistance In: E Jamrozik and M Selgelid, editors. Ethics and Drug Resistance: Collective Responsibility for Global Public Health. Cham, Switzerland: Springer (2020). 55–73.
38. Nijsingh, N, Larsson, DGJ, de Fine, LK, and Munthe, C. Justifying antibiotic resistance interventions: uncertainty, precaution and ethics In: E Jamrozik and M Selgelid, editors. Ethics and Drug Resistance: Collective Responsibility for Global Public Health. Cham, Switzerland: Springer (2020). 357–75.
39. Michael, CA, Dominey-Howes, D, and Labbate, M. The antimicrobial resistance crisis: causes, consequences, and management. Front Public Health. (2014) 2:1–8. doi: 10.3389/fpubh.2014.00145
40. Okeke, IN, Laxminarayan, R, Bhutta, ZA, Duse, AG, Jenkins, P, Brien, TFO, et al. Antimicrobial resistance in developing countries. Part I: recent trends and current status. Lancet Infect Dis. (2005) 5:481–93. doi: 10.1016/S1473-3099(05)70189-4
41. Woodward, DL, and Rodgers, FG. Surveillance of antimicrobial resistance in salmonella, shigella and Vibrio cholerae in Latin America and the Caribbean: a collaborative project. Can J Infect Dis Med Microbiol. (2000) 11:181–6. doi: 10.1155/2000/743969
42. Tadesse, T, Deneke, Y, and Deresa, B. Seroprevalence of bovine viral diarrhea virus and its potential risk factors in dairy cattle of Jimma town, southwestern Ethiopia. J Dairy, Vet Anim Res. (2019) 8:11–7. doi: 10.15406/jdvar.2019.08.00235
43. Jimma city municipality office (2022). Population profiles of Jimma city administration, Jimma zone, Oromia.
44. Bekele, D, Tolossa, T, Tsegaye, R, and Teshome, W. The knowledge and practice towards COVID-19 pandemic prevention among residents of Ethiopia. An online cross-sectional study. PLoS One. (2021) 16:e0234585. doi: 10.1371/journal.pone.0234585
45. Engdaw, GT, Gebrehiwot, M, and Andualem, Z. Hand hygiene compliance and associated factors among health care providers in Central Gondar zone public primary hospitals, Northwest Ethiopia. Antimicrob Resist Infect Control. (2019) 8:190. doi: 10.1186/s13756-019-0634-z
47. American Type Culture Collection (2022). Introduction to microbiology. Manassas, Virginia 20110-2209.
48. CLSI . Performance Standards for Antimicrobial Susceptibility Testing. 30th edition. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2020).
49. Cheesbrough, M . District Laboratory in Tropical Practice Countries: Part 2. 2nd ed. Cambridge, UK: Cambridge University Press (2006).
50. Bargellini, A, Ferranti, G, Santangelo, M, and Venturelli, F. Hand hygiene knowledge, hand contamination and practice of Italian nursing and medical students. Epidemiol Biostat Public Health. (2014) 11:e9971. doi: 10.2427/9971
51. Alsagher, MR, Soudah, SA, Khsheba, AE, Fadel, SM, Dadiesh, MA, Houme, MA, et al. Hand washing before and after applying different hand hygiene techniques in places of public concern in Tripoli-Libya. Open Microbiol J. (2018) 12:364–75. doi: 10.2174/1874285801812010364
52. Kampf, G, and Löffler, H. Hand disinfection in hospitals-benefits and risks. JDDG. (2010) 8:978–83. doi: 10.1111/j.1610-0387.2010.07501.x
53. De Alwis, WR, Pakirisamy, P, Wai San, L, and Xiaofen, EC. A study on hand contamination and hand washing practices among medical students. ISRN Public Health. (2012) 2012:1–5. doi: 10.5402/2012/251483
54. Berhanu, L, Mereta, ST, Gume, B, Kassa, T, Berihun, G, Dadi, LS, et al. Effect of microbial quality of washing water on hand hygiene status of food handlers in Jimma town: implication for food hygiene and safety. J Multidiscip Healthc. (2021) 14:1129–34. doi: 10.2147/JMDH.S306359
55. Burton, M, Cobb, E, Donachie, P, Judah, G, Curtis, V, and Schmidt, WP. The effect of handwashing with water or soap on bacterial contamination of hands. Int J Environ Res Public Health. (2011) 8:97–104. doi: 10.3390/ijerph8010097
56. El-Kady, H . Efficacy of different alcohol-based hand disinfectants in reduction of hand contamination among food handlers in Alexandria, Eqypt. JAMB. (2018) 13:1–13. doi: 10.9734/JAMB/2018/45345
57. Sader, HS, Castanheira, M, Mendes, RE, and Flamm, RK. Frequency and antimicrobial susceptibility of gram-negative bacteria isolated from patients with pneumonia hospitalized in ICUs of US medical centres (2015-17). J Antimicrob Chemother. (2018) 73:3053–9. doi: 10.1093/jac/dky279
58. Ogba, OM, Asukwo, PE, and Otu-Bassey, IB. Assessment of bacterial carriage on the hands of primary school children in Calabar municipality, Nigeria. Biomed dermatol. (2018) 2:1–7. doi: 10.1186/s41702-017-0017-0
59. Garoy, EY, Gebreab, YB, Achila, OO, Tekeste, DG, Kesete, R, Ghirmay, R, et al. Methicillin-resistant Staphylococcus aureus (MRSA): prevalence and antimicrobial sensitivity pattern among patients-a multicenter study in Asmara, Eritrea. Can J Infect Dis Med Microbiol. (2019):8321834. doi: 10.1155/2019/8321834
60. Kitila, KT, Taddese, BD, Hailu, TK, Sori, LM, Geleto, SE, Mengistu, GZ, et al. Assessment of bacterial profile and antimicrobial resistance pattern of bacterial isolates from blood culture in Addis Ababa regional laboratory, Addis Ababa, Ethiopia. Clin Microbiol. (2018) 7:312. doi: 10.4172/2327-5073.1000312
61. Gashaw, A, Afework, K, Feleke, M, Moges, T, and Kahsay, H. Prevalence of bacteria and intestinal parasites among food-handlers in Gondar town, Northwest Ethiopia. J Health Popul Nutr. (2008) 26:451–5. doi: 10.3329/jhpn.v26i4.1887
62. Deyno, S, Fekadu, S, and Astatkie, A. Resistance of Staphylococcus aureus to antimicrobial agents in Ethiopia: a meta-analysis. Antimicrob Resist Infect Control. (2017) 6:1–15. doi: 10.1186/s13756-017-0243-7
63. Potrykus, J, and Wegrzyn, G. Chloramphenicol-sensitive Escherichia coli strain expressing the chloramphenicol acetyltransferase (cat) gene. Antimicrob Agents Chemother. (2001) 45:3610–2. doi: 10.1128/AAC.45.12.3610-3612.2001
64. Galindo-Méndez, M . Antimicrobial resistance in Escherichia coli E. coli infections—importance of early diagnosis and efficient treatment. Dermatol Int. (2020):1–20. doi: 10.5772/intechopen.93115
65. Uma, B, Prabhakar, K, Rajendran, S, Kavitha, K, and Sarayu, Y. Antibiotic sensitivity and plasmid profiles of Escherichia coli isolated from pediatric diarrhea. J Global Infect Dis. (2009) 1:107. doi: 10.4103/0974-777X.56255
66. Pakbin, B, Amani, Z, Allahyari, S, Mousavi, S, Mahmoudi, R, Brück, WM, et al. Genetic diversity and antibiotic resistance of Shigella spp. isolates from food products. Food Sci Nutr. (2021) 9:6362–71. doi: 10.1002/fsn3.2603
Keywords: bacterial isolate, antimicrobial resistance, housemaids, Jimma City, Ethiopia
Citation: Ango TS, Gelaw NB, Zegene GM, Teshome T and Getahun T (2024) Prevalence and antimicrobial susceptibility profile of bacteria isolated from the hands of housemaids in Jimma City, Ethiopia. Front. Public Health. 11:1301685. doi: 10.3389/fpubh.2023.1301685
Received: 15 November 2023; Accepted: 27 December 2023;
Published: 29 January 2024.
Edited by:
Wei Wang, China National Center for Food Safety Risk Assessment, ChinaReviewed by:
Shaoting Li, Guangdong University of Technology, ChinaCopyright © 2024 Ango, Gelaw, Zegene, Teshome and Getahun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tadele Shiwito Ango, c2hpd2l0b3QyMzUwQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.