
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Public Health , 28 December 2023
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 11 - 2023 | https://doi.org/10.3389/fpubh.2023.1290402
 Stefan Holdenrieder1*
Stefan Holdenrieder1* Carlos Eduardo Dos Santos Ferreira2
Carlos Eduardo Dos Santos Ferreira2 Jacques Izopet3
Jacques Izopet3 Elitza S. Theel4
Elitza S. Theel4 Andreas Wieser5,6,7,8
Andreas Wieser5,6,7,8Much of the global population now has some level of adaptive immunity to SARS-CoV-2 induced by exposure to the virus (natural infection), vaccination, or a combination of both (hybrid immunity). Key questions that subsequently arise relate to the duration and the level of protection an individual might expect based on their infection and vaccination history. A multi-component composite correlate of risk (CoR) could inform individuals and stakeholders about protection and aid decision making. This perspective evaluates the various elements that need to be accommodated in the development of an antibody-based composite CoR for reinfection with SARS-CoV-2 or development of severe COVID-19, including variation in exposure dose, transmission route, viral genetic variation, patient factors, and vaccination status. We provide an overview of antibody dynamics to aid exploration of the specifics of SARS-CoV-2 antibody testing. We further discuss anti-SARS-CoV-2 immunoassays, sample matrices, testing formats, frequency of sampling and the optimal time point for such sampling. While the development of a composite CoR is challenging, we provide our recommendations for each of these key areas and highlight areas that require further work to be undertaken.
The COVID-19 pandemic, caused by severe acute respiratory coronavirus 2 (SARS-CoV-2) led to unprecedented, accelerated vaccine development (1) and expansive roll-out programs (2, 3). Much of the global population now has some level of adaptive immunity to SARS-CoV-2 induced by exposure to the virus (natural infection), vaccination, or a combination of both (hybrid immunity).
Natural infection induced by, and/or vaccination against, SARS-CoV-2 leads to the development of both binding and neutralizing antibodies (nAbs) (4, 5), and the induction of T-cell responses during active immune reaction and clearance of infection (6). Key questions that subsequently arise relate to the duration and the level of protection an individual might expect based on their infection and vaccination history. Studies of those infected early in the pandemic documented that natural SARS-CoV-2 infection afforded some level of protection against reinfection in most individuals, and that subsequent reinfections were typically less severe than the primary episode (Table 1). However, SARS-CoV-2 has high rates of mutation and heavily mutated variants have emerged (21). Most significant are the “variants of concern” (VOCs) (22), and there is now ample evidence that protection against reinfection with the B.1.1.529/21 K (Omicron) variant (23, 24) is dramatically reduced compared with previous variants (Table 1).
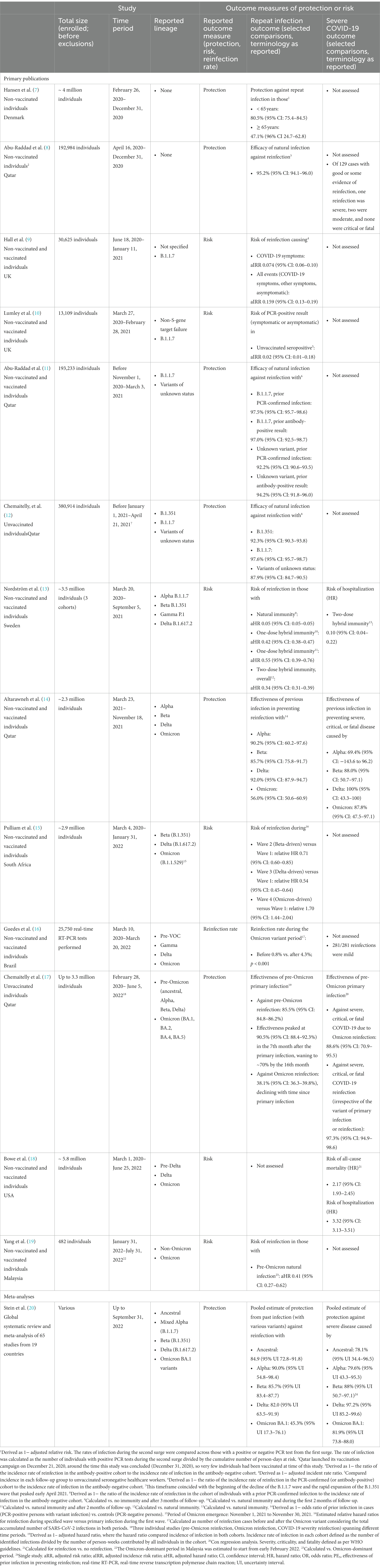
Table 1. Selection of peer-reviewed publications assessing reinfection or risk of severe COVID-19 after natural infection (ordered by study end date, earliest to most recent).
Any descriptor of immunity based on patient history will encompass a population of individuals with vastly variable exposure to vaccines and viral variants with differing orders of immune challenge intensity. Unrecognized “silent infections,” especially in Omicron-positive subjects with underlying immunity, further complicate the assessment. Therefore derivation of potential immunity based on patient history requires assistance from a surrogate composite score to inform about protection and to aid decision making.
In vaccinology, a correlate of protection (CoP) reflects a statistical non-causal relationship between an immune marker and protection after vaccination (25). Most accepted CoPs are based on antibody measurements (26) and vary depending on the clinical endpoint, for example protection from (symptomatic) infection or severe disease. In contrast, a correlate of risk (CoR) can be used as a measurement of an immunologic parameter that is correlated with a study endpoint (27) and can predict a clinical endpoint in a specified population with a defined future timeframe. Notably, antibody markers have been used as correlates of immune function in clinical trials of SARS-CoV-2 vaccine efficacy (VE) (28–33), and for identifying the risk of symptomatic infection by VOCs (34, 35). In VE trials, a CoR can be a CoP if the CoR reliably predicts VE against the clinical endpoint, thereby acting not just as an intrinsic susceptibility factor or marker of pathogen exposure. In this case, the CoR could be a surrogate of the endpoint and could be useful for licensure of new vaccines.
A CoR would likely comprise a measure of the immune component plus determinants that act to modify such a measure (a multi-component composite CoR). While there is no scientific evidence for an absolute humoral or cellular CoP against SARS-CoV-2, identification of a multi-component composite CoR might be useful to guide the use of vaccines or patient management. In general, the immune component of a composite CoR should be easily measured by widely available technologies that are amenable to automation, are scalable, cost-efficient, and have a rapid turn-around time. Given the relative complexity, cost and pre-analytic requirements for cellular immune response testing, the preferred candidate for the immune component of a CoR would be detection of humoral immune response(s) (i.e., antibody). This perspective evaluates the various elements that need to be accommodated in the development of an antibody-based composite CoR for reinfection with SARS-CoV-2 or severe COVID-19.
Viral load varies widely between infected individuals and over time (36), with viral emissions independent of symptom severity (37). Exposure to SARS-CoV-2 is tempered by the use of personal protective measures and, at the population level, adherence to public health measures that reduce exposure has been variable (38, 39), making assessment of exposure dose complex.
Controlled human infections to directly study the impact of viral inoculum and disease severity are controversial (40), and only one human challenge trial of SARS-CoV-2 using a single low inoculum dose has been reported to date (41). However, the initial infective dose of SARS-CoV-2 is thought to be associated with disease severity (42–44), since relationships between dose and severity exist for many other viral infections (44). Evidence from SARS-CoV-2 animal models suggests that the route of transmission similarly affects disease severity (45).
Risk reduction depends on the dominant variant in circulation. Continued evolution of SARS-CoV-2 can lead to significant changes in viral transmission and impact reinfection rates (46). Mechanistically, the receptor binding domain (RBD) within the viral spike (S) glycoprotein engages in initiation of infection via interaction with the angiotensin converting enzyme-2 (ACE2) receptor (47). The RBD is a target for many nAbs (47) and mutations are frequently located at the RBD–ACE2 interface (48). It is therefore not surprising that changes to the viral epitope can reduce antibody binding (48), helping to drive immune escape from anti-RBD nAbs (49), decreasing previously generated protective immunity (50–52), and leading to variant-specific risks of severe illness (53, 54).
Patient differences impact susceptibility to reinfection and disease severity. The immune response declines with increasing age (55, 56), and age is the strongest predictor of SARS-CoV-2 infection–fatality ratio (57). Older individuals have been shown to exhibit reduced binding antibody titers and neutralization following vaccination (58–60). Pregnant women are also at high risk of severe outcomes (61). Similarly, immunocompromised or immunosuppressed individuals, or those affected by cancer or human immunodeficiency virus (HIV), exhibit reduced immune responses to infection or an increased risk of hospitalization (62–66). Other co-morbidities are frequently observed in those with severe COVID-19 (67, 68).
COVID-19 vaccines include recombinant subunit, nucleic acid, viral vector and whole virus vaccines, among others, and some vaccines have been adapted for Omicron variants (69). The use of different vaccines, combinations, the number of boosters received, the interval between boosters, the occurrence of natural infection, and combinations thereof, trigger the immune system to varying degrees in depth, breadth or duration of response (35, 66, 70–83). Pre-existing heterotypic immunity, due to past infections with other coronaviruses, may also influence the immune response to SARS-CoV-2 (84, 85).
Following primary infection, severely ill patients exhibit higher binding and neutralizing antibody titers or activity compared with individuals with mild disease (86–91). Persistence of nAbs has also been associated with disease severity (92). In the event of reinfection, there is an implicit assumption that nAb titers ameliorate severe COVID-19 (93, 94). In brief, in infection-naïve individuals, post-vaccination antibody titers (anti-S IgG and nAbs) correlate with higher vaccine efficacy (71), and post-vaccination anti-RBD IgG and nAbs levels associate with protection against infection and symptomatic disease even during the Omicron era (95) or inversely correlate with risk of death (anti-S IgG below 20th percentile) (96). Generally, individuals with higher nAbs (levels or capacity) are considered increasingly protected from infection (97–99), symptomatic reinfection (99–101), severe disease (100), or death (102) compared with individuals with lower nAbs. There is evidence that neutralization capacity can be strain specific (103).
In summary, viral and host elements modify the risk of reinfection or development of severe COVID-19 in various manners (Figure 1).
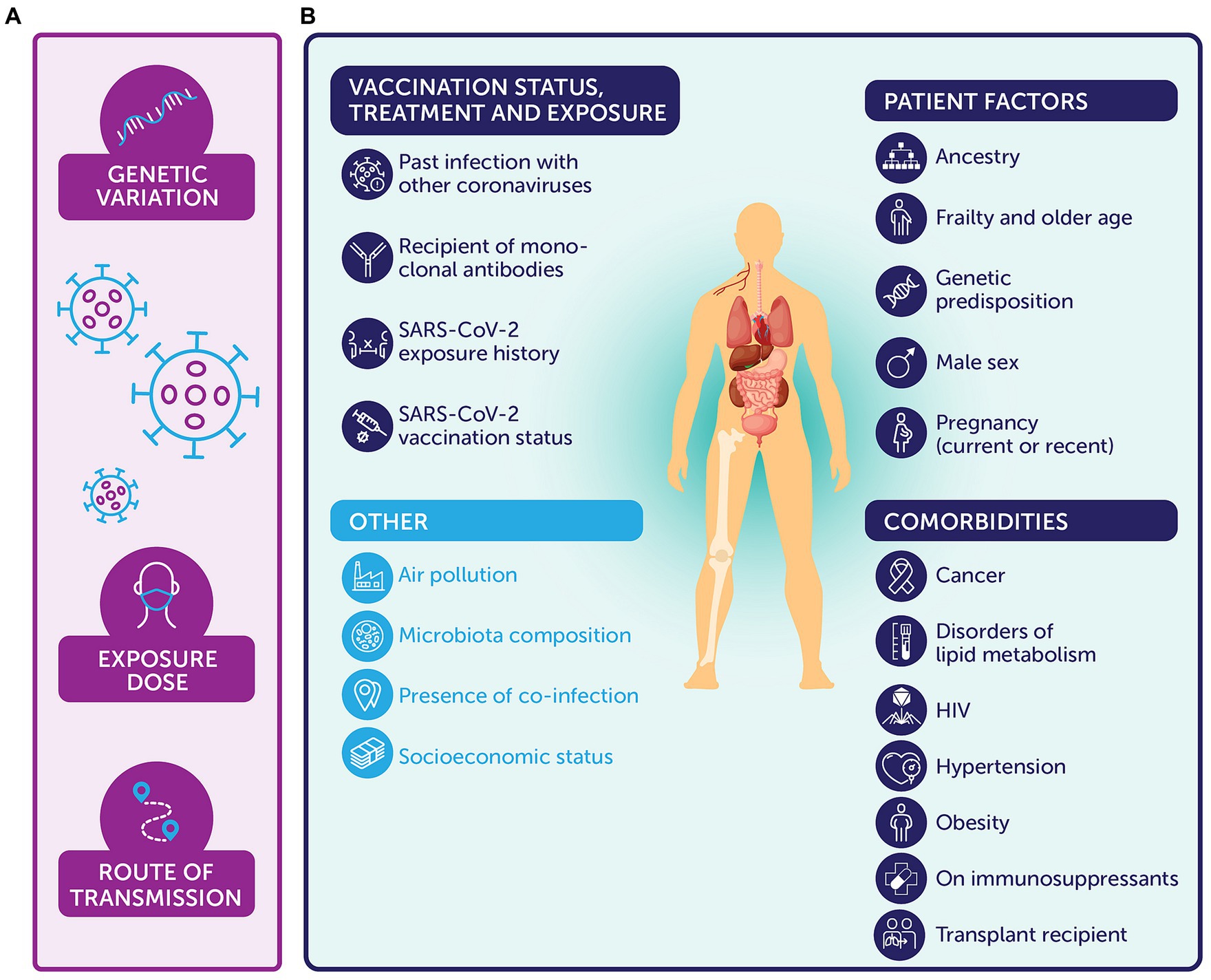
Figure 1. Summary of host and viral elements that can impact the immune response and response to SARS-CoV-2 (selected examples, not exhaustive, variables ordered alphabetically within figure [not according to importance]). (A) Viral factors include genetic variation (50–54), the exposure dose (42–44), and route of transmission (45). (B) Host factors include patient factors, such as: ancestry, for example, non-European ancestry (68); frailty (45) and older age (55–60, 68); genetic predisposition (68, 104–107), including gene variants at 3p21.31 (68, 107) and variants involved in immune signaling [e.g., TLR7 (105) and interferon (106)]; male sex (68); and current or recent pregnancy (61). Equally, past infection with other coronaviruses (84, 85), whether an individual has received monoclonal antibodies (108), and exposure history or vaccination status (type, provision of boosters, or intervals) (35, 66, 70–83) are also relevant. Comorbidities similarly affect the immune response, such as: whether an individual has a history of malignancy (66) or has received recent chemotherapy for cancer (63); has disorders of lipid metabolism (67); is a transplant recipient (62); has uncontrolled HIV (65); has hypertension (67); is obese (67, 68); or takes immunosuppressants (64). Other relevant variables include air pollution (45), microbiota composition (45), presence of co-infections (45), and socioeconomic status (45, 68).
The antibody component of a composite CoR should be developed under defined conditions. To provide insight into these conditions, an understanding of antibody dynamics is required.
Natural infection with SARS-CoV-2 elicits a diversity of antibodies including those targeting S and nucleocapsid (N) antigens (75, 109) and the development of anti-RBD IgG antibodies is associated with improved patient survival (110). A detailed systematic review of 66 studies investigated antibody responses (111). Collectively, the evidence supports the induction of IgM production in the acute phase of natural infection (peak prevalence: 20 days) followed by IgA (peak prevalence: 23 days), IgG (peak prevalence: 25 days), and nAbs (peak prevalence: 31 days) after symptom onset (111).
Serum IgG has the longest half-life compared with the relatively transient IgA or IgM (112). A longitudinal analysis of 4,558 individuals, measuring total anti-N antibodies, revealed that, while total antibodies begin to decline after 90–100 days, they may persist for over 500 days after natural infection (113). Specifically measuring nAb via plaque reduction neutralization test (PRNT) shows that infection yields a robust nAb response in most individuals (86). Some studies report that anti-S antibodies show greater persistence than anti-N antibodies (114, 115).
Dramatic inductions of anti-S or anti-RBD IgG antibodies is indicative of vaccination (75, 116, 117). Primary vaccination by some vaccines [but not all (118)], or boosters generates high nAb titers (117, 119, 120) or neutralizing responses (116). Notably, nAbs wane over time (35) with a half-life of 108 days (100)—although the level of decay may be assay or variant dependent (119) – and multiple clinical factors affect the duration of neutralization responses after primary vaccination (66) (see also Figure 1).
Numerous immunoassays for the detection of antibodies against SARS-CoV-2 are available, differing in the immunoglobulin class detected, target viral antigen, format, and output [qualitative, (semi)-quantitative] [reviewed in detail (121, 122)].
Head-to-head comparisons from the pre-Omicron era reveal variable levels of performance between the assays (123–127), caused by numerous technical factors including assay methodology, format and antibodies used, timing of testing, and the targeted viral antigen. Comparison studies show that sensitivity for detecting prior infection by different serologic assays changes over time (128). Commercial assays developed early during the pandemic are based on ancestral/wild-type antigens. Subsequently, there is potential for differential performance in the Omicron-era: in particular, S- and RBD-specific immunoassays have shown significantly reduced performance (129–131), and decreased comparability of quantitative results (132).
Most common commercial immunoassays detect both binding and nAbs without differentiating between them, however certain assays measuring IgG or total antibodies correlate well with neutralizing capacity (28, 97, 133–139), acting as surrogates of neutralization. Cell-based virus neutralization tests can be used to measure neutralizing capability, but these are typically not readily available in clinical laboratories due to inherent test performance challenges associated with their methodology (including the need for biosafety level 3 containment for live-virus neutralization assays), time and cost (140).
Mature immune responses are dominated by IgG. Serologic assays that measure IgG or total antibodies (if skewed toward IgG) that correlate with neutralizing activity and focus on anti-RBD should be used for the serologic component of a composite CoR; anti-N antibodies are unlikely to be neutralizing as the N protein is located within the viral envelope (75).
Assays should be adapted for accurate measurement of the modified antigen, if applicable. However, frequent adaptation of assays is unlikely if several variants are circulating in parallel and due to regulatory requirements for assays. Therefore, studies are needed to determine assay applicability in the present conditions, especially since RBD mutations frequently occur and recombinant versions of RBD or S are commonly used in immunoassays (122). Accordingly, the upper and lower thresholds of any CoR may need modification.
External ring trials show poor comparability of assays from different manufacturers (141, 142) and there are significant challenges with the current binding antibody units (BAU) standardization, due to multiple factors, including different assay methods, antibody class(es) detected and target antigen used. Of note, BAU reference materials were derived from UK convalescent individuals infected in 2020 (143) (pre-Omicron), and there are vastly different BAU standardized values (144). While new reference materials include VOCs, they still contain antibodies derived during the pre-Omicron era (145). Antibody measurements should be harmonized across assays from different manufacturers, irrespective of the different epitopes utilized, to reduce variability. To support this, there is an urgent need for external quality assessment, production of robust traceable certified reference materials, standards for different variants, and improved documentation of the methods on laboratory reports. Age-specific normalization of reference intervals in defined groups, by means of z-log transformation and documentation in antibody passes, may further improve the comparability of assays. Stakeholders should agree on minimum performance-based criteria to develop the gold standard for CoR, allowing validation of secondary assays.
Finally, systemic cellular assays could provide a comprehensive profile of the immune response, especially in immunocompromised and susceptible individuals who are not able to mount a robust antibody response. Currently, they lack scientific evidence and their use in clinical practice still remains uncertain.
Systemic anti-SARS-CoV-2 antibody testing can be performed on blood, plasma/serum, or dried blood spots (DBS) (122, 146–148). An advantage of whole blood or DBS collection is the ease in obtaining the sample. While many methodologies focus on systemic testing, infection with SARS-CoV-2 or vaccination against COVID-19 induces mucosal antibodies (149, 150), thus secretions such as saliva offer another possibility. Antibody dynamics will differ depending on the material in question (151), and sample types are subject to specific idiosyncrasies, such as additional pre-processing, that need to be accounted for (152). Of note, the collection protocol (passive drool versus swab-stimulated saliva, for instance) can influence the antibody yield (153). Currently secretion-based testing is less suitable for a composite CoR as performance is variable (154).
A composite CoR will likely be sample matrix-specific. Our preference is for plasma/serum, as this sample matrix has the largest evidence base, shows the least variability, experiences less interference than whole blood, and is consistent with CoRs established for other infectious diseases. DBS would be also possible, but variability is high, and few laboratories have an established workflow.
Formats include high-throughput automated enzyme immunoassay/ electrochemiluminescence immunoassay/enzyme-linked immunosorbent assay (certified and used in central laboratories and hospitals), point-of-care (POC) testing (used in emergencies and outpatients setting), and direct-to-consumer testing (at-home use with online services). POC testing is gaining in popularity, but methodological variation is higher (155) and any method that relies upon sampling from untrained individuals is less reliable for (semi)quantitative measurements (156).
We recommend automated assays that are approved by location-specific regulatory agencies and performed in certified and centralized laboratories. Home sampling/DBS would contribute to a reduction in clinician workload, particularly in high-density residential facilities, but methods are not yet sufficiently robust. Currently, there is no clear benefit in POC testing as urgent results are not critical.
Considering antibody dynamics, several important questions arise: what is the optimal time point for measurement; would the timing differ depending on the vaccine schedule, and/or the presence of previous infection of a specified severity; should antibody levels be measured once or serially? While single values can be plotted into modeled curves showing decrease rates over time, serial measurements could further refine the composite CoR. Only individuals with symptomatic disease or vaccination are known to stabilize the curve—infections that are sufficiently mild to lack detection will impact the composite CoR model.
As most individuals have experienced infection or vaccination, and titers are generally high and more stable than with single exposures, sampling should be performed annually or less. Serologic evaluation should be conducted more frequently in the older adult or immunocompromised than the general population (time interval to be defined), depending on any underlying disease and/or treatment.
A composite CoR would be helpful particularly for high-risk groups, such as solid organ transplant recipients (157), and those in occupations with high risk of exposure to SARS-CoV-2. However, whether a composite CoR would operate at the individual or population level is yet uncertain.
For health policymakers, a composite CoR could be useful for: (1) predicting the durability of protection, supporting serosurveys to determine the protection levels of individuals and populations; (2) aiding decision-making with regard to monitoring vaccination efficacy and identifying individuals who would benefit from booster vaccinations; (3) evaluating the need for extra protection of vulnerable communities in the face of new variants with low cross protection and less efficacious vaccines; (4) licensing new vaccines; and (5) developing clear immunologic vaccine trial endpoints.
A previous systematic review by Perry and colleagues found mixed evidence for a serologic CoP, with the lack of standardization between laboratory methodology, differing assay targets and sampling time points, and the lack of information on the SARS-CoV-2 variant confounding interpretation (158). We have highlighted various parameters that should be controlled for in any measure of risk, some of which will be challenging to obtain (such as host genetics). Comparing different protection studies is also difficult as infectious pressure in the observation time period is often uncertain as, in reality, community data are incomplete and the number of oligosymptomatic infections is unclear. Of course, individual responses to infection and vaccination with regards to antibody production will make long-term assessment difficult, intrinsic risk will vary by age and protection will not be linear (139, 159). To ensure an acceptable level of accuracy, it will also be important to assess the composite CoR in geographic settings where extrinsic environmental factors, host genetic backgrounds, and circulating variants contribute to the overall effect on the immune response. All the variables previously described need to be thought of in the general context of laboratory diagnostics, paying attention to sensitivity, specificity, positive/negative predictive value, reliability, precision, dilution, linearity, robustness, stability, preanalytics, scalability (automation), cost-efficiency, In Vitro Diagnostic Regulation certification, and the use of qualified standard and control materials. Laboratory quality is essential for meaningful follow-up of quantitative antibody levels.
While the development of a composite CoR is a sizeable undertaking, steps can be taken to address this need. Studies need to adapt to the requirements of new variants, controlling for patient settings (vaccination types, earlier infections), and levels of disease severity. The emergence of VOCs means that a CoR will undoubtedly be variant-specific and the timing of infections and vaccination, how variants impact disease severity, antibody kinetics, and assay reactivity, must be respected. Frequently revisiting the data would be helpful as overall epidemiology changes; since almost all epidemiologic population-based studies have ended, background data is increasingly difficult to acquire, and this must be reversed. While serologic testing has retreated from the political agenda and public interest, we have an obligation to broaden the scientific knowledge base, and collect data to inform public health authorities, given that COVID-19 still causes a significant number of deaths and there is a considerable population of those with post-acute sequelae of SARS-CoV-2 infection [long COVID; (160)].
A composite CoR will differ depending on the clinical endpoint (26). Definitions of symptomatic or severe disease are often not consistent across studies (100). Clinical outcomes must be precisely defined: an evaluation of the primary endpoints of 19 clinical trials for severe COVID-19 revealed the complexity of this task, reporting 12 different primary endpoints (161). In addition, the ideal timeframe for predictive ability is yet to be determined.
While we support the development of a composite CoR and serologic testing by high- quality controlled assays, viruses such as influenza have significant strain variation and similar disease severity, so the importance of a composite CoR for SARS-CoV-2 should be judged against other pathogens of interest. Assessment of cost-effectiveness will likely inform upon the need for a composite CoR.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
SH: Conceptualization, Writing – original draft, Writing – review & editing. CD: Conceptualization, Writing – review & editing. JI: Conceptualization, Writing – review & editing. ET: Conceptualization, Writing – review & editing. AW: Conceptualization, Writing – original draft, Writing – review & editing.
The authors declare that this study received funding from Roche Diagnostics International Ltd. All authors participated in a SARSCoV-2 expert panel convened between February and June 2022, for which they received an honorarium sponsored by this funder. The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article, or in the decision to submit it for publication, but was involved by providing review.
The authors thank Corrinne Segal of Elements Communications Limited (London, United Kingdom) for editorial assistance. This article has previously been submitted to the Authorea preprint server and can be found at https://doi.org/10.22541/au.169412008.83234734/v1.
SH has received grants from Roche Diagnostics, Sysmex and Volition, consulting fees from Instand e.V, EQAS, Merck KG, Roche Diagnostics and Thermo Fisher Scientific, speaker’s honoraria from BMS, Medica, Roche Diagnostics and Trillium, and has leadership roles in the International Society of Oncology and Biomarkers (board member and secretary), DGKL Competence Field Molecular Diagnostic (vice speaker) and Federal Medical Association, D5 Group (delegate of the DGKL). CD has received speaker’s honoraria from Abbott Diagnostics, Roche Diagnostics and Siemens Healthineers. JI has received grants from Roche Diagnostics. ET has received consulting fees from EUROIMMUN US, Serimmune and Roche Diagnostics, speaker’s honoraria from the American Society for Microbiology and EUROIMMUN US, and support for meetings from the American Society for Microbiology, the New York City Branch of the American Society of Microbiology and the Pan American Society for Clinical Virology. AW has received grants from numerous different public fundings, including the German Center for Infection Research, Fraunhofer Gesellschaft, and German Aif and Zim programs, royalties or licenses from Smart United GmbH, consulting fees from Roche Diagnostics and Roche Pharma, speaker’s honoraria from BÄMI and Roche Diagnostics, support for meetings from BÄMI and Roche Diagnostics, participated in advisory boards for Roche Diagnostics, declares stock or stock options in Smart United GmbH and Munich Innovative Biosolutions UG (haftungsbeschränkt), and has received reduced rates for materials and equipment from EUROIMMUN and Roche Diagnostics. SH is a founder of CEBIO and SFZ BioCoDE.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Rahman, MM, Masum, MHU, Wajed, S, and Talukder, A. A comprehensive review on COVID-19 vaccines: development, effectiveness, adverse effects, distribution and challenges. VirusDis. (2022) 33:1–22. doi: 10.1007/s13337-022-00755-1
2. Hale, T, Angrist, N, Goldszmidt, R, Kira, B, Petherick, A, Phillips, T, et al. A global panel database of pandemic policies (Oxford COVID-19 government response tracker). Nat Hum Behav. (2021) 5:529–38. doi: 10.1038/s41562-021-01079-8
3. Hale, T, Petherick, A, Anania, J, Andretti, B, Angrist, Noam, Barnes, Roy, et al. Variation in government responses to COVID-19. Version 14.1. Blavatnik School of Government Working Paper. (2022). Available at: www.bsg.ox.ac.uk/covidtracker (Accessed June 13, 2023).
4. Qi, H, Liu, B, Wang, X, and Zhang, L. The humoral response and antibodies against SARS-CoV-2 infection. Nat Immunol. (2022) 23:1008–20. doi: 10.1038/s41590-022-01248-5
5. Liu, M, Gan, H, Liang, Z, Liu, L, Liu, Q, Mai, Y, et al. Review of therapeutic mechanisms and applications based on SARS-CoV-2 neutralizing antibodies. Front Microbiol. (2023) 14:1122868. doi: 10.3389/fmicb.2023.1122868
6. Moss, P . The T cell immune response against SARS-CoV-2. Nat Immunol. (2022) 23:186–93. doi: 10.1038/s41590-021-01122-w
7. Hansen, CH, Michlmayr, D, Gubbels, SM, Mølbak, K, and Ethelberg, S. Assessment of protection against reinfection with SARS-CoV-2 among 4 million PCR-tested individuals in Denmark in 2020: a population-level observational study. Lancet. (2021) 397:1204–12. doi: 10.1016/s0140-6736(21)00575-4
8. Abu-Raddad, LJ, Chemaitelly, H, Coyle, P, Malek, JA, Ahmed, AA, Mohamoud, YA, et al. SARS-CoV-2 antibody-positivity protects against reinfection for at least seven months with 95% efficacy. EClinicalMedicine. (2021) 35:100861. doi: 10.1016/j.eclinm.2021.100861
9. Hall, VJ, Foulkes, S, Charlett, A, Atti, A, Monk, EJM, Simmons, R, et al. SARS-CoV-2 infection rates of antibody-positive compared with antibody-negative health-care workers in England: a large, multicentre, prospective cohort study (SIREN). Lancet. (2021) 397:1459–69. doi: 10.1016/s0140-6736(21)00675-9
10. Lumley, SF, Rodger, G, Constantinides, B, Sanderson, N, Chau, KK, Street, TL, et al. An observational cohort study on the incidence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and B.1.1.7 variant infection in healthcare workers by antibody and vaccination status. Clin Infect Dis. (2022) 74:1208–19. doi: 10.1093/cid/ciab608
11. Abu-Raddad, LJ, Chemaitelly, H, Ayoub, HH, Coyle, P, Malek, JA, Ahmed, AA, et al. Introduction and expansion of the SARS-CoV-2 B.1.1.7 variant and reinfections in Qatar: a nationally representative cohort study. PLoS Med. (2021) 18:e1003879. doi: 10.1371/journal.pmed.1003879
12. Chemaitelly, H, Bertollini, R, and Abu-Raddad, LJ. Efficacy of natural immunity against SARS-CoV-2 reinfection with the beta variant. N Engl J Med. (2021) 385:2585–6. doi: 10.1056/NEJMc2110300
13. Nordström, P, Ballin, M, and Nordström, A. Risk of SARS-CoV-2 reinfection and COVID-19 hospitalisation in individuals with natural and hybrid immunity: a retrospective, total population cohort study in Sweden. Lancet Infect Dis. (2022) 22:781–90. doi: 10.1016/s1473-3099(22)00143-8
14. Altarawneh, HN, Chemaitelly, H, Hasan, MR, Ayoub, HH, Qassim, S, AlMukdad, S, et al. Protection against the Omicron variant from previous SARS-CoV-2 infection. N Engl J Med. (2022) 386:1288–90. doi: 10.1056/NEJMc2200133
15. Pulliam, JRC, van Schalkwyk, C, Govender, N, von Gottberg, A, Cohen, C, Groome, MJ, et al. Increased risk of SARS-CoV-2 reinfection associated with emergence of Omicron in South Africa. Science. (2022) 376:eabn4947. doi: 10.1126/science.abn4947
16. Guedes, AR, Oliveira, MS, Tavares, BM, Luna-Muschi, A, Lazari, CDS, Montal, AC, et al. Reinfection rate in a cohort of healthcare workers over 2 years of the COVID-19 pandemic. Sci Rep. (2023) 13:712. doi: 10.1038/s41598-022-25908-6
17. Chemaitelly, H, Nagelkerke, N, Ayoub, HH, Coyle, P, Tang, P, Yassine, HM, et al. Duration of immune protection of SARS-CoV-2 natural infection against reinfection. J Travel Med. (2022) 29:taac109. doi: 10.1093/jtm/taac109
18. Bowe, B, Xie, Y, and Al-Aly, Z. Acute and postacute sequelae associated with SARS-CoV-2 reinfection. Nat Med. (2022) 28:2398–405. doi: 10.1038/s41591-022-02051-3
19. Yang, SL, Ripen, AM, Lee, JV, Koh, K, Yen, CH, Chand, AK, et al. Time from last immunity event against infection during Omicron-dominant period in Malaysia. Int J Infect Dis. (2023) 128:98–101. doi: 10.1016/j.ijid.2022.12.025
20. Stein, C, Nassereldine, H, Sorensen, RJD, Amlag, JO, Bisignano, C, Byrne, S, et al. Past SARS-CoV-2 infection protection against re-infection: a systematic review and meta-analysis. Lancet. (2023) 401:833–42. doi: 10.1016/S0140-6736(22)02465-5
21. Carabelli, AM, Peacock, TP, Thorne, LG, Harvey, WT, Hughes, J, Peacock, SJ, et al. SARS-CoV-2 variant biology: immune escape, transmission and fitness. Nat Rev Microbiol. (2023) 21:162–77. doi: 10.1038/s41579-022-00841-7
22. World Health Organization . Updated working definitions and primary actions for SARS-CoV-2 variants. (2023). Available at: https://www.who.int/publications/m/item/updated-working-definitions-and-primary-actions-for--sars-cov-2-variants (Accessed April 13, 2023).
23. Viana, R, Moyo, S, Amoako, DG, Tegally, H, Scheepers, C, Althaus, CL, et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature. (2022) 603:679–86. doi: 10.1038/s41586-022-04411-y
24. World Health Organization. One year since the emergence of COVID-19 virus variant Omicron. (2022). Available at: https://www.who.int/news-room/feature-stories/detail/one-year-since-the-emergence-of-Omicron (Accessed April 17, 2023).
25. Plotkin, SA, and Gilbert, PB. Nomenclature for immune correlates of protection after vaccination. Clin Infect Dis. (2012) 54:1615–7. doi: 10.1093/cid/cis238
26. Plotkin, SA . Correlates of protection induced by vaccination. Clin Vaccine Immunol. (2010) 17:1055–65. doi: 10.1128/CVI.00131-10
27. Gilbert, PB, Qin, L, and Self, SG. Evaluating a surrogate endpoint at three levels, with application to vaccine development. Stat Med. (2008) 27:4758–78. doi: 10.1002/sim.3122
28. Gilbert, PB, Montefiori, DC, McDermott, AB, Fong, Y, Benkeser, D, Deng, W, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. (2022) 375:43–50. doi: 10.1126/science.abm3425
29. Fong, Y, McDermott, AB, Benkeser, D, Roels, S, Stieh, DJ, Vandebosch, A, et al. Immune correlates analysis of the ENSEMBLE single Ad26.COV2.S dose vaccine efficacy clinical trial. Nat Microbiol. (2022) 7:1996–2010. doi: 10.1038/s41564-022-01262-1
30. Ramasamy, MN, Minassian, AM, Ewer, KJ, Flaxman, AL, Folegatti, PM, Owens, DR, et al. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. (2021) 396:1979–93. doi: 10.1016/s0140-6736(20)32466-1
31. Feng, S, Phillips, DJ, White, T, Sayal, H, Aley, PK, Bibi, S, et al. Correlates of protection against symptomatic and asymptomatic SARS-CoV-2 infection. Nat Med. (2021) 27:2032–40. doi: 10.1038/s41591-021-01540-1
32. Li, G, Cappuccini, F, Marchevsky, NG, Aley, PK, Aley, R, Anslow, R, et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 (AZD1222) vaccine in children aged 6-17 years: a preliminary report of COV006, a phase 2 single-blind, randomised, controlled trial. Lancet. (2022) 399:2212–25. doi: 10.1016/s0140-6736(22)00770-x
33. Cristiano, A, Nuccetelli, M, Pieri, M, Sarubbi, S, Pelagalli, M, Calugi, G, et al. Serological anti-SARS-CoV-2 neutralizing antibodies association to live virus neutralizing test titers in COVID-19 paucisymptomatic/symptomatic patients and vaccinated subjects. Int Immunopharmacol. (2021) 101:108215. doi: 10.1016/j.intimp.2021.108215
34. Nilles, EJ, Paulino, CT, de St Aubin, M, Duke, W, Jarolim, P, Sanchez, IM, et al. Tracking immune correlates of protection for emerging SARS-CoV-2 variants. Lancet Infect Dis. (2023) 23:153–4. doi: 10.1016/s1473-3099(23)00001-4
35. Cromer, D, Steain, M, Reynaldi, A, Schlub, TE, Wheatley, AK, Juno, JA, et al. Neutralising antibody titres as predictors of protection against SARS-CoV-2 variants and the impact of boosting: a meta-analysis. Lancet Microbe. (2022) 3:e52–61. doi: 10.1016/s2666-5247(21)00267-6
36. Jones, TC, Biele, G, Mühlemann, B, Veith, T, Schneider, J, Beheim-Schwarzbach, J, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. (2021) 373:eabi5273. doi: 10.1126/science.abi5273
37. Zhou, J, Singanayagam, A, Goonawardane, N, Moshe, M, Sweeney, FP, Sukhova, K, et al. Viral emissions into the air and environment after SARS-CoV-2 human challenge: a phase 1, open label, first-in-human study. Lancet Microbe. (2023) 4:e579–90. doi: 10.1016/s2666-5247(23)00101-5
38. Kale, D, Herbec, A, Beard, E, Gold, N, and Shahab, L. Patterns and predictors of adherence to health-protective measures during COVID-19 pandemic in the UK: cross-sectional and longitudinal findings from the HEBECO study. BMC Public Health. (2022) 22:2347. doi: 10.1186/s12889-022-14509-7
39. Hutchins, HJ, Wolff, B, Leeb, R, Ko, JY, Odom, E, Willey, J, et al. COVID-19 mitigation behaviors by age group - United States, April-June 2020. MMWR Morb Mortal Wkly Rep. (2020) 69:1584–90. doi: 10.15585/mmwr.mm6943e4
40. Jamrozik, E, and Selgelid, MJ. COVID-19 human challenge studies: ethical issues. Lancet Infect Dis. (2020) 20:e198–203. doi: 10.1016/s1473-3099(20)30438-2
41. Killingley, B, Mann, AJ, Kalinova, M, Boyers, A, Goonawardane, N, Zhou, J, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge in young adults. Nat Med. (2022) 28:1031–41. doi: 10.1038/s41591-022-01780-9
42. Khosroshahi, HT, and Mardomi, A. The initial infectious dose of SARS-CoV-2 and the severity of the disease: possible impact on the incubation period. Future Virol. (2021) 16:369–73. doi: 10.2217/fvl-2020-0330
43. Van Damme, W, Dahake, R, van de Pas, R, Vanham, G, and Assefa, Y. COVID-19: does the infectious inoculum dose-response relationship contribute to understanding heterogeneity in disease severity and transmission dynamics? Med Hypotheses. (2021) 146:110431. doi: 10.1016/j.mehy.2020.110431
44. Spinelli, MA, Glidden, DV, Gennatas, ED, Bielecki, M, Beyrer, C, Rutherford, G, et al. Importance of non-pharmaceutical interventions in lowering the viral inoculum to reduce susceptibility to infection by SARS-CoV-2 and potentially disease severity. Lancet Infect Dis. (2021) 21:e296–301. doi: 10.1016/s1473-3099(20)30982-8
45. Zsichla, L, and Müller, V. Risk factors of severe COVID-19: a review of host, viral and environmental factors. Viruses. (2023) 15:175. doi: 10.3390/v15010175
46. Markov, PV, Ghafari, M, Beer, M, Lythgoe, K, Simmonds, P, Stilianakis, NI, et al. The evolution of SARS-CoV-2. Nat Rev Microbiol. (2023) 21:361–79. doi: 10.1038/s41579-023-00878-2
47. Klasse, PJ, and Moore, JP. Antibodies to SARS-CoV-2 and their potential for therapeutic passive immunization. Elife. (2020) 9:e57877. doi: 10.7554/eLife.57877
48. Harvey, WT, Carabelli, AM, Jackson, B, Gupta, RK, Thomson, EC, Harrison, EM, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. (2021) 19:409–24. doi: 10.1038/s41579-021-00573-0
49. Souza, PFN, Mesquita, FP, Amaral, JL, Landim, PGC, Lima, KRP, Costa, MB, et al. The spike glycoprotein of SARS-CoV-2: a review of how mutations of spike glycoproteins have driven the emergence of variants with high transmissibility and immune escape. Int J Biol Macromol. (2022) 208:105–25. doi: 10.1016/j.ijbiomac.2022.03.058
50. Liu, C, Ginn, HM, Dejnirattisai, W, Supasa, P, Wang, B, Tuekprakhon, A, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cells. (2021) 184:4220–36.e13. doi: 10.1016/j.cell.2021.06.020
51. Migueres, M, Dimeglio, C, Trémeaux, P, Abravanel, F, Raymond, S, Lhomme, S, et al. Influence of immune escape and nasopharyngeal virus load on the spread of SARS-CoV-2 Omicron variant. J Infect. (2022) 84:e7–9. doi: 10.1016/j.jinf.2022.01.036
52. Wang, P, Nair, MS, Liu, L, Iketani, S, Luo, Y, Guo, Y, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. (2021) 593:130–5. doi: 10.1038/s41586-021-03398-2
53. Kow, CS, Ramachandram, DS, and Hasan, SS. Risk of severe illness in patients infected with SARS-CoV-2 of Delta variant: a systematic review and meta-analysis. Infect Dis. (2022) 54:614–7. doi: 10.1080/23744235.2022.2055787
54. Kow, CS, Ramachandram, DS, and Hasan, SS. The risk of mortality and severe illness in patients infected with the Omicron variant relative to delta variant of SARS-CoV-2: a systematic review and meta-analysis. Ir J Med Sci. (2023) 192:2897–904. doi: 10.1007/s11845-022-03266-6
55. Ciabattini, A, Nardini, C, Santoro, F, Garagnani, P, Franceschi, C, and Medaglini, D. Vaccination in the elderly: the challenge of immune changes with aging. Semin Immunol. (2018) 40:83–94. doi: 10.1016/j.smim.2018.10.010
56. Nian, Y, Minami, K, Maenesono, R, Iske, J, Yang, J, Azuma, H, et al. Changes of T-cell immunity over a lifetime. Transplantation. (2019) 103:2227–33. doi: 10.1097/tp.0000000000002786
57. COVID-19 Forecasting Team . Variation in the COVID-19 infection–fatality ratio by age, time, and geography during the pre-vaccine era: a systematic analysis. Lancet. (2022) 399:1469–88. doi: 10.1016/S0140-6736(21)02867-1
58. Collier, DA, Ferreira, I, Kotagiri, P, Datir, RP, Lim, EY, Touizer, E, et al. Age-related immune response heterogeneity to SARS-CoV-2 vaccine BNT162b2. Nature. (2021) 596:417–22. doi: 10.1038/s41586-021-03739-1
59. Bichara, CDA, Queiroz, MAF, da Silva Graça Amoras, E, Vaz, GL, Vallinoto, IMVC, Bichara, CNC, et al. Assessment of anti-SARS-CoV-2 antibodies post-Coronavac vaccination in the Amazon region of Brazil. Vaccines. (2021) 9:1169. doi: 10.3390/vaccines9101169
60. Walsh, EE, Frenck, RW Jr, Falsey, AR, Kitchin, N, Absalon, J, Gurtman, A, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med. (2020) 383:2439–50. doi: 10.1056/NEJMoa2027906
61. Allotey, J, Stallings, E, Bonet, M, Yap, M, Chatterjee, S, Kew, T, et al. Clinical manifestations, risk factors, and maternal and perinatal outcomes of coronavirus disease 2019 in pregnancy: living systematic review and meta-analysis. BMJ. (2020) 370:m3320. doi: 10.1136/bmj.m3320
62. Li, J, Ayada, I, Wang, Y, den Hoed, CM, Kamar, N, Peppelenbosch, MP, et al. Factors associated with COVID-19 vaccine response in transplant recipients: a systematic review and meta-analysis. Transplantation. (2022) 106:2068–75. doi: 10.1097/tp.0000000000004256
63. Wankhede, D, Grover, S, and Hofman, P. Determinants of humoral immune response to SARS-CoV-2 vaccines in solid cancer patients: a systematic review and meta-analysis. Vaccine. (2023) 41:1791–8. doi: 10.1016/j.vaccine.2023.01.072
64. Shen, C, Risk, M, Schiopu, E, Hayek, SS, Xie, T, Holevinski, L, et al. Efficacy of COVID-19 vaccines in patients taking immunosuppressants. Ann Rheum Dis. (2022) 81:875–80. doi: 10.1136/annrheumdis-2021-222045
65. Höft, MA, Burgers, WA, and Riou, C. The immune response to SARS-CoV-2 in people with HIV. Cell Mol Immunol. (2023). doi: 10.1038/s41423-023-01087-w
66. Zhao, M, Slotkin, R, Sheth, AH, Pischel, L, Kyriakides, TC, Emu, B, et al. Serum neutralizing antibody titers 12 months after coronavirus disease 2019 messenger RNA vaccination: correlation to clinical variables in an adult, US population. Clin Infect Dis. (2023) 76:e391–9. doi: 10.1093/cid/ciac416
67. Kompaniyets, L, Pennington, AF, Goodman, AB, Rosenblum, HG, Belay, B, Ko, JY, et al. Underlying medical conditions and severe illness among 540,667 adults hospitalized with COVID-19, March 2020-March 2021. Prev Chronic Dis. (2021) 18:E66. doi: 10.5888/pcd18.210123
68. Shelton, JF, Shastri, AJ, Ye, C, Weldon, CH, Filshtein-Sonmez, T, Coker, D, et al. Trans-ancestry analysis reveals genetic and nongenetic associations with COVID-19 susceptibility and severity. Nat Genet. (2021) 53:801–8. doi: 10.1038/s41588-021-00854-7
69. Yadav, T, Kumar, S, Mishra, G, and Saxena, SK. Tracking the COVID-19 vaccines: the global landscape. Hum Vaccin Immunother. (2023) 19:2191577. doi: 10.1080/21645515.2023.2191577
70. Lim, SY, Kim, JY, Jung, J, Yun, SC, and Kim, SH. Waning of humoral immunity depending on the types of COVID-19 vaccine. Infect Dis. (2023) 55:216–20. doi: 10.1080/23744235.2023.2165707
71. Earle, KA, Ambrosino, DM, Fiore-Gartland, A, Goldblatt, D, Gilbert, PB, Siber, GR, et al. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. (2021) 39:4423–8. doi: 10.1016/j.vaccine.2021.05.063
72. Nham, E, Ko, JH, Song, KH, Choi, JY, Kim, ES, Kim, HJ, et al. Kinetics of vaccine-induced neutralizing antibody titers and estimated protective immunity against wild-type SARS-CoV-2 and the Delta variant: a prospective nationwide cohort study comparing three COVID-19 vaccination protocols in South Korea. Front Immunol. (2022) 13:968105. doi: 10.3389/fimmu.2022.968105
73. Lafon, E, Jäger, M, Bauer, A, Reindl, M, Bellmann-Weiler, R, Wilflingseder, D, et al. Comparative analyses of IgG/IgA neutralizing effects induced by three COVID-19 vaccines against variants of concern. J Allergy Clin Immunol. (2022) 149:1242–52.e12. doi: 10.1016/j.jaci.2022.01.013
74. Greaney, AJ, Loes, AN, Gentles, LE, Crawford, KHD, Starr, TN, Malone, KD, et al. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci Transl Med. (2021) 13:eabi9915. doi: 10.1126/scitranslmed.abi9915
75. Meyers, J, Windau, A, Schmotzer, C, Saade, E, Noguez, J, Stempak, L, et al. SARS-CoV-2 antibody profile of naturally infected and vaccinated individuals detected using qualitative, semi-quantitative and multiplex immunoassays. Diagn Microbiol Infect Dis. (2022) 104:115803. doi: 10.1016/j.diagmicrobio.2022.115803
76. Townsend, JP, Hassler, HB, Sah, P, Galvani, AP, and Dornburg, A. The durability of natural infection and vaccine-induced immunity against future infection by SARS-CoV-2. Proc Natl Acad Sci U S A. (2022) 119:e2204336119. doi: 10.1073/pnas.2204336119
77. Yu, Y, Esposito, D, Kang, Z, Lu, J, Remaley, AT, De Giorgi, V, et al. mRNA vaccine-induced antibodies more effective than natural immunity in neutralizing SARS-CoV-2 and its high affinity variants. Sci Rep. (2022) 12:2628. doi: 10.1038/s41598-022-06629-2
78. Vietri, MT, D'Elia, G, Caliendo, G, Passariello, L, Albanese, L, Molinari, AM, et al. Antibody levels after BNT162b2 vaccine booster and SARS-CoV-2 Omicron infection. Vaccine. (2022) 40:5726–31. doi: 10.1016/j.vaccine.2022.08.045
79. Munro, APS, Janani, L, Cornelius, V, Aley, PK, Babbage, G, Baxter, D, et al. Safety and immunogenicity of seven COVID-19 vaccines as a third dose (booster) following two doses of ChAdOx1 nCov-19 or BNT162b2 in the UK (COV-BOOST): a blinded, multicentre, randomised, controlled, phase 2 trial. Lancet. (2021) 398:2258–76. doi: 10.1016/s0140-6736(21)02717-3
80. Andrews, N, Tessier, E, Stowe, J, Gower, C, Kirsebom, F, Simmons, R, et al. Duration of protection against mild and severe disease by Covid-19 vaccines. N Engl J Med. (2022) 386:340–50. doi: 10.1056/NEJMoa2115481
81. Cerqueira-Silva, T, Andrews, JR, Boaventura, VS, Ranzani, OT, de Araújo Oliveira, V, Paixão, ES, et al. Effectiveness of CoronaVac, ChAdOx1 nCoV-19, BNT162b2, and Ad26.COV2.S among individuals with previous SARS-CoV-2 infection in Brazil: a test-negative, case-control study. Lancet Infect Dis. (2022) 22:791–801. doi: 10.1016/s1473-3099(22)00140-2
82. Buckner, CM, Kardava, L, El Merhebi, O, Narpala, SR, Serebryannyy, L, Lin, BC, et al. Interval between prior SARS-CoV-2 infection and booster vaccination impacts magnitude and quality of antibody and B cell responses. Cell. (2022) 185:4333–46.e14. doi: 10.1016/j.cell.2022.09.032
83. Hornsby, H, Nicols, AR, Longet, S, Liu, C, Tomic, A, Angyal, A, et al. Omicron infection following vaccination enhances a broad spectrum of immune responses dependent on infection history. Nat Commun. (2023) 14:5065. doi: 10.1038/s41467-023-40592-4
84. Souris, M, Tshilolo, L, Parzy, D, Lobaloba Ingoba, L, Ntoumi, F, Kamgaing, R, et al. Pre-pandemic cross-reactive immunity against SARS-CoV-2 among Central and West African populations. Viruses. (2022) 14:2259. doi: 10.3390/v14102259
85. Devaux, CA, and Fantini, J. Unravelling antigenic cross-reactions toward the world of coronaviruses: extent of the stability of shared epitopes and SARS-CoV-2 anti-spike cross-neutralizing antibodies. Pathogens. (2023) 12:713. doi: 10.3390/pathogens12050713
86. Lau, EHY, Tsang, OTY, Hui, DSC, Kwan, MYW, Chan, WH, Chiu, SS, et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat Commun. (2021) 12:63. doi: 10.1038/s41467-020-20247-4
87. Vanshylla, K, Di Cristanziano, V, Kleipass, F, Dewald, F, Schommers, P, Gieselmann, L, et al. Kinetics and correlates of the neutralizing antibody response to SARS-CoV-2 infection in humans. Cell Host Microbe. (2021) 29:917–29.e4. doi: 10.1016/j.chom.2021.04.015
88. Chen, X, Pan, Z, Yue, S, Yu, F, Zhang, J, Yang, Y, et al. Disease severity dictates SARS-CoV-2-specific neutralizing antibody responses in COVID-19. Signal Transduct Target Ther. (2020) 5:180. doi: 10.1038/s41392-020-00301-9
89. Maciola, AK, La Raja, M, Pacenti, M, Salata, C, De Silvestro, G, Rosato, A, et al. Neutralizing antibody responses to SARS-CoV-2 in recovered COVID-19 patients are variable and correlate with disease severity and receptor-binding domain recognition. Front Immunol. (2022) 13:830710. doi: 10.3389/fimmu.2022.830710
90. Hansen, CB, Jarlhelt, I, Pérez-Alós, L, Hummelshøj Landsy, L, Loftager, M, Rosbjerg, A, et al. SARS-CoV-2 antibody responses are correlated to disease severity in COVID-19 convalescent individuals. J Immunol. (2021) 206:109–17. doi: 10.4049/jimmunol.2000898
91. Wang, P, Liu, L, Nair, MS, Yin, MT, Luo, Y, Wang, Q, et al. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect. (2020) 9:2091–3. doi: 10.1080/22221751.2020.1823890
92. Chia, WN, Zhu, F, Ong, SWX, Young, BE, Fong, SW, Le Bert, N, et al. Dynamics of SARS-CoV-2 neutralising antibody responses and duration of immunity: a longitudinal study. Lancet Microbe. (2021) 2:e240–9. doi: 10.1016/s2666-5247(21)00025-2
93. U.S Food and Drug Administration . EUA for convalescent plasma. (2021). Available at: https://www.fda.gov/media/141477/download (Accessed June 23, 2023).
94. U.S Food and Drug Administration . EUA for casirivimab and imdevimab. (2020). Available at: https://www.fda.gov/media/143891/download (Accessed June 23, 2023).
95. Gilboa, M, Gonen, T, Barda, N, Cohn, S, Indenbaum, V, Weiss-Ottolenghi, Y, et al. Factors associated with protection from SARS-CoV-2 Omicron variant infection and disease among vaccinated health care workers in Israel. JAMA Netw Open. (2023) 6:e2314757. doi: 10.1001/jamanetworkopen.2023.14757
96. Vikström, L, Fjällström, P, Gwon, YD, Sheward, DJ, Wigren-Byström, J, Evander, M, et al. Vaccine-induced correlate of protection against fatal COVID-19 in older and frail adults during waves of neutralization-resistant variants of concern: an observational study. Lancet Reg Health Eur. (2023) 30:100646. doi: 10.1016/j.lanepe.2023.100646
97. Dimeglio, C, Herin, F, Martin-Blondel, G, Miedougé, M, and Izopet, J. Antibody titers and protection against a SARS-CoV-2 infection. J Infect. (2022) 84:248–88. doi: 10.1016/j.jinf.2021.09.013
98. Bergwerk, M, Gonen, T, Lustig, Y, Amit, S, Lipsitch, M, Cohen, C, et al. Covid-19 breakthrough infections in vaccinated health care workers. N Engl J Med. (2021) 385:1474–84. doi: 10.1056/NEJMoa2109072
99. Lumley, SF, O'Donnell, D, Stoesser, NE, Matthews, PC, Howarth, A, Hatch, SB, et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. (2021) 384:533–40. doi: 10.1056/NEJMoa2034545
100. Khoury, DS, Cromer, D, Reynaldi, A, Schlub, TE, Wheatley, AK, Juno, JA, et al. Neutralizing antibody levels are highly predictive of immune protection from symptomatic SARS-CoV-2 infection. Nat Med. (2021) 27:1205–11. doi: 10.1038/s41591-021-01377-8
101. Gillot, C, Bayart, JL, Closset, M, Cabo, J, Maloteau, V, Dogné, JM, et al. Peri-infection titers of neutralizing and binding antibodies as a predictor of COVID-19 breakthrough infections in vaccinated healthcare professionals: importance of the timing. Clin Chem Lab Med. (2023) 61:1670–5. doi: 10.1515/cclm-2023-0134
102. Dispinseri, S, Secchi, M, Pirillo, MF, Tolazzi, M, Borghi, M, Brigatti, C, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun. (2021) 12:2670. doi: 10.1038/s41467-021-22958-8
103. Dimeglio, C, Herin, F, Da-Silva, I, Gernigon, C, Porcheron, M, Chapuy-Regaud, S, et al. Decreased efficiency of neutralizing antibodies from previously infected or vaccinated individuals against the B.1.617.2 (delta) SARS-CoV-2 variant. Microbiol Spectr. (2022) 10:e0270621. doi: 10.1128/spectrum.02706-21
104. Pairo-Castineira, E, Clohisey, S, Klaric, L, Bretherick, AD, Rawlik, K, Pasko, D, et al. Genetic mechanisms of critical illness in COVID-19. Nature. (2021) 591:92–8. doi: 10.1038/s41586-020-03065-y
105. Matuozzo, D, Talouarn, E, Marchal, A, Zhang, P, Manry, J, Seeleuthner, Y, et al. Rare predicted loss-of-function variants of type I IFN immunity genes are associated with life-threatening COVID-19. Genome Med. (2023) 15:22. doi: 10.1186/s13073-023-01173-8
106. Kousathanas, A, Pairo-Castineira, E, Rawlik, K, Stuckey, A, Odhams, CA, Walker, S, et al. Whole-genome sequencing reveals host factors underlying critical COVID-19. Nature. (2022) 607:97–103. doi: 10.1038/s41586-022-04576-6
107. COVID-19 Host Genetics Initiative . Mapping the human genetic architecture of COVID-19. Nature. (2021) 600:472–7. doi: 10.1038/s41586-021-03767-x
108. Kim, PS, Dimcheff, DE, Siler, A, Schildhouse, RJ, and Chensue, SW. Effect of monoclonal antibody therapy on the endogenous SARS-CoV-2 antibody response. Clin Immunol. (2022) 236:108959. doi: 10.1016/j.clim.2022.108959
109. Azak, E, Karadenizli, A, Uzuner, H, Karakaya, N, Canturk, NZ, and Hulagu, S. Comparison of an inactivated Covid19 vaccine-induced antibody response with concurrent natural Covid19 infection. Int J Infect Dis. (2021) 113:58–64. doi: 10.1016/j.ijid.2021.09.060
110. Secchi, M, Bazzigaluppi, E, Brigatti, C, Marzinotto, I, Tresoldi, C, Rovere-Querini, P, et al. COVID-19 survival associates with the immunoglobulin response to the SARS-CoV-2 spike receptor binding domain. J Clin Invest. (2020) 130:6366–78. doi: 10.1172/jci142804
111. Arkhipova-Jenkins, I, Helfand, M, Armstrong, C, Gean, E, Anderson, J, Paynter, RA, et al. Antibody response after SARS-CoV-2 infection and implications for immunity: a rapid living review. Ann Intern Med. (2021) 174:811–21. doi: 10.7326/m20-7547
112. Sterlin, D, Mathian, A, Miyara, M, Mohr, A, Anna, F, Claër, L, et al. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci Transl Med. (2021) 13:eabd2223. doi: 10.1126/scitranslmed.abd2223
113. Swartz, MD, DeSantis, SM, Yaseen, A, Brito, FA, Valerio-Shewmaker, MA, Messiah, SE, et al. Antibody duration after infection from SARS-CoV-2 in the Texas coronavirus antibody rresponse survey. J Infect Dis. (2023) 227:193–201. doi: 10.1093/infdis/jiac167
114. Abraha, I, Eusebi, P, Germani, A, Pasquarelli, E, Pascolini, S, Antonietti, R, et al. Temporal trends and differences of SARS-CoV-2-specific antibody responses in symptomatic and asymptomatic subjects: a longitudinal study from Umbria in Italy. BMJ Open. (2022) 12:e056370. doi: 10.1136/bmjopen-2021-056370
115. Ripperger, TJ, Uhrlaub, JL, Watanabe, M, Wong, R, Castaneda, Y, Pizzato, HA, et al. Orthogonal SARS-CoV-2 serological assays enable surveillance of low-prevalence communities and reveal durable humoral immunity. Immunity. (2020) 53:925–33.e4. doi: 10.1016/j.immuni.2020.10.004
116. Hvidt, AK, Baerends, EAM, Søgaard, OS, Stærke, NB, Raben, D, Reekie, J, et al. Comparison of vaccine-induced antibody neutralization against SARS-CoV-2 variants of concern following primary and booster doses of COVID-19 vaccines. Front Med. (2022) 9:994160. doi: 10.3389/fmed.2022.994160
117. Rosseto-Welter, EA, Rodrigues, SS, de Figueiredo, AB, França, CN, Oliveira, DBL, Bachi, ALL, et al. Cellular and humoral immune responses to vaccination for COVID-19 are negatively impacted by senescent T cells: a case report. Vaccines. (2023) 11:840. doi: 10.3390/vaccines11040840
118. Omran, EA, Habashy, RE, Ezz Elarab, LA, Hashish, MH, El-Barrawy, MA, Abdelwahab, IA, et al. Anti-spike and neutralizing antibodies after two doses of COVID-19 sinopharm/BIBP vaccine. Vaccine. (2022) 10:1340. doi: 10.3390/vaccines10081340
119. Lyke, KE, Atmar, RL, Islas, CD, Posavad, CM, Szydlo, D, Paul Chourdhury, R, et al. Rapid decline in vaccine-boosted neutralizing antibodies against SARS-CoV-2 Omicron variant. Cell Rep Med. (2022) 3:100679. doi: 10.1016/j.xcrm.2022.100679
120. Furukawa, K, Tjan, LH, Kurahashi, Y, Sutandhio, S, Nishimura, M, Arii, J, et al. Assessment of neutralizing antibody response against SARS-CoV-2 variants after 2 to 3 doses of the BNT162b2 mRNA COVID-19 vaccine. JAMA Netw Open. (2022) 5:e2210780. doi: 10.1001/jamanetworkopen.2022.10780
121. Fox, T, Geppert, J, Dinnes, J, Scandrett, K, Bigio, J, Sulis, G, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. (2022) 2022:CD013652. doi: 10.1002/14651858.CD013652.pub2
122. Theel, ES . Performance characteristics of high-throughput serologic assays for severe acute respiratory syndrome coronavirus 2 with Food and Drug Administration emergency use authorization: a review. Clin Lab Med. (2022) 42:15–29. doi: 10.1016/j.cll.2021.10.006
123. Olbrich, L, Castelletti, N, Schälte, Y, Garí, M, Pütz, P, Bakuli, A, et al. Head-to-head evaluation of seven different seroassays including direct viral neutralisation in a representative cohort for SARS-CoV-2. J Gen Virol. (2021) 102:001653. doi: 10.1099/jgv.0.001653
124. The National SARS-CoV-2 Serology Assay Evaluation Group . Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis. (2020) 20:1390–400. doi: 10.1016/s1473-3099(20)30634-4
125. Harritshøj, LH, Gybel-Brask, M, Afzal, S, Kamstrup, PR, Jørgensen, CS, Thomsen, MK, et al. Comparison of 16 serological SARS-CoV-2 immunoassays in 16 clinical laboratories. J Clin Microbiol. (2021) 59:e02596-20. doi: 10.1128/jcm.02596-20
126. Chiereghin, A, Zagari, RM, Galli, S, Moroni, A, Gabrielli, L, Venturoli, S, et al. Recent advances in the evaluation of serological assays for the diagnosis of SARS-CoV-2 infection and COVID-19. Front Public Health. (2020) 8:620222. doi: 10.3389/fpubh.2020.620222
127. Riester, E, Findeisen, P, Hegel, JK, Kabesch, M, Ambrosch, A, Rank, CM, et al. Performance evaluation of the Roche Elecsys anti-SARS-CoV-2 S immunoassay. J Virol Methods. (2021) 297:114271. doi: 10.1016/j.jviromet.2021.114271
128. Muecksch, F, Wise, H, Batchelor, B, Squires, M, Semple, E, Richardson, C, et al. Longitudinal serological analysis and neutralizing antibody levels in coronavirus disease 2019 convalescent patients. J Infect Dis. (2021) 223:389–98. doi: 10.1093/infdis/jiaa659
129. Migueres, M, Chapuy-Regaud, S, Miédougé, M, Jamme, T, Lougarre, C, Da Silva, I, et al. Current immunoassays and detection of antibodies elicited by Omicron SARS-CoV-2 infection. J Med Virol. (2023) 95:e28200. doi: 10.1002/jmv.28200
130. Springer, DN, Perkmann, T, Jani, CM, Mucher, P, Prüger, K, Marculescu, R, et al. Reduced sensitivity of commercial spike-specific antibody assays after primary infection with the SARS-CoV-2 Omicron variant. Microbiol Spectr. (2022) 10:e0212922. doi: 10.1128/spectrum.02129-22
131. Habermann, E, Frommert, LM, Ghannam, K, Nguyen My, L, Gieselmann, L, Tober-Lau, P, et al. Performance of commercial SARS-CoV-2 wild-type and Omicron BA.1 antibody assays compared with pseudovirus neutralization tests. J Clin Virol. (2023) 165:105518. doi: 10.1016/j.jcv.2023.105518
132. Rössler, A, Knabl, L, Raschbichler, LM, Peer, E, von Laer, D, Borena, W, et al. Reduced sensitivity of antibody tests after Omicron infection. Lancet Microbe. (2023) 4:e10–1. doi: 10.1016/s2666-5247(22)00222-1
133. Montesinos, I, Dahma, H, Wolff, F, Dauby, N, Delaunoy, S, Wuyts, M, et al. Neutralizing antibody responses following natural SARS-CoV-2 infection: dynamics and correlation with commercial serologic tests. J Clin Virol. (2021) 144:104988. doi: 10.1016/j.jcv.2021.104988
134. Theel, ES, Johnson, PW, Kunze, KL, Wu, L, Gorsh, AP, Granger, D, et al. SARS-CoV-2 serologic assays dependent on dual-antigen binding demonstrate diverging kinetics relative to other antibody detection methods. J Clin Microbiol. (2021) 59:e0123121. doi: 10.1128/jcm.01231-21
135. Chapuy-Regaud, S, Miédougé, M, Abravanel, F, Da Silva, I, Porcheron, M, Fillaux, J, et al. Evaluation of three quantitative anti-SARS-CoV-2 antibody immunoassays. Microbiol Spectr. (2021) 9:e0137621. doi: 10.1128/spectrum.01376-21
136. Lustig, Y, Sapir, E, Regev-Yochay, G, Cohen, C, Fluss, R, Olmer, L, et al. BNT162b2 COVID-19 vaccine and correlates of humoral immune responses and dynamics: a prospective, single-Centre, longitudinal cohort study in health-care workers. Lancet Respir Med. (2021) 9:999–1009. doi: 10.1016/s2213-2600(21)00220-4
137. Rubio-Acero, R, Castelletti, N, Fingerle, V, Olbrich, L, Bakuli, A, Wölfel, R, et al. In search of the SARS-CoV-2 protection correlate: head-to-head comparison of two quantitative S1 assays in pre-characterized oligo-/asymptomatic patients. Infect Dis Ther. (2021) 10:1505–18. doi: 10.1007/s40121-021-00475-x
138. Jochum, S, Kirste, I, Hortsch, S, Grunert, VP, Legault, H, Eichenlaub, U, et al. Clinical utility of Elecsys anti-SARS-CoV-2 S assay in COVID-19 vaccination: an exploratory analysis of the mRNA-1273 phase 1 trial. Front Immunol. (2021) 12:798117. doi: 10.3389/fimmu.2021.798117
139. Regev-Yochay, G, Lustig, Y, Joseph, G, Gilboa, M, Barda, N, Gens, I, et al. Correlates of protection against COVID-19 infection and intensity of symptomatic disease in vaccinated individuals exposed to SARS-CoV-2 in households in Israel (ICoFS): a prospective cohort study. Lancet Microbe. (2023) 4:e309–18. doi: 10.1016/s2666-5247(23)00012-5
140. Lu, Y, Wang, J, Li, Q, Hu, H, Lu, J, and Zeliang, C. Advances in neutralization assays for SARS-CoV-2. Scand J Immunol. (2021) 94:e13088. doi: 10.1111/sji.13088
141. Ast, V, Costina, V, Eichner, R, Bode, A, Aida, S, Gerhards, C, et al. Assessing the quality of serological testing in the COVID-19 pandemic: results of a European external quality assessment (EQA) scheme for anti-SARS-CoV-2 antibody detection. J Clin Microbiol. (2021) 59:e0055921. doi: 10.1128/JCM.00559-21
142. Perkmann, T, Mucher, P, Osze, D, Muller, A, Perkmann-Nagele, N, Koller, T, et al. Comparison of five anti-SARS-CoV-2 antibody assays across three doses of BNT162b2 reveals insufficient standardization of SARS-CoV-2 serology. J Clin Virol. (2023) 158:105345. doi: 10.1016/j.jcv.2022.105345
143. World Health Organization . Establishment of the WHO International Standard And Reference Panel for anti-SARS-CoV-2 antibody. (2020). WHO Expert Committee on Biological Standardization (WHO/BS/2020.2403). Available at: https://www.who.int/publications/m/item/WHO-BS-2020.2403 (Accessed June 23, 2023).
144. Kroidl, I, Winter, S, Rubio-Acero, R, Bakuli, A, Geldmacher, C, Eser, TM, et al. Studying temporal titre evolution of commercial SARS-CoV-2 assays reveals significant shortcomings of using BAU standardization for comparison. Virol J. (2023) 20:200. doi: 10.1186/s12985-023-02167-z
145. National Institute for Biological Standards and Control . WHO International Standard: 1st International Standard for antibodies to SARS-CoV-2 variants of concern (NIBSC code: 21/338, instructions for use, version 2.0) (2022). Available at: https://www.nibsc.org/documents/ifu/21-338.pdf (Accessed October 30, 2023).
146. Beyerl, J, Rubio-Acero, R, Castelletti, N, Paunovic, I, Kroidl, I, Khan, ZN, et al. A dried blood spot protocol for high throughput analysis of SARS-CoV-2 serology based on the Roche Elecsys anti-N assay. EBioMedicine. (2021) 70:103502. doi: 10.1016/j.ebiom.2021.103502
147. Decru, B, Van Elslande, J, Weemaes, M, Houben, E, Empsen, I, André, E, et al. Comparison of the diagnostic performance with whole blood and plasma of four rapid antibody tests for SARS-CoV-2. Clin Chem Lab Med. (2020) 58:e197–9. doi: 10.1515/cclm-2020-0817
148. Castelletti, N, Paunovic, I, Rubio-Acero, R, Beyerl, J, Plank, M, Reinkemeyer, C, et al. A dried blood spot protocol for high-throughput quantitative analysis of SARS-CoV-2 RBD serology based on the Roche Elecsys system. Microbiol Spectr. (2023) In press
149. Fröberg, J, and Diavatopoulos, DA. Mucosal immunity to severe acute respiratory syndrome coronavirus 2 infection. Curr Opin Infect Dis. (2021) 34:181–6. doi: 10.1097/qco.0000000000000724
150. Guerrieri, M, Francavilla, B, Fiorelli, D, Nuccetelli, M, Passali, FM, Coppeta, L, et al. Nasal and salivary mucosal humoral immune response elicited by mRNA BNT162b2 COVID-19 vaccine compared to SARS-CoV-2 natural infection. Vaccines. (2021) 9:1499. doi: 10.3390/vaccines9121499
151. Cervia, C, Nilsson, J, Zurbuchen, Y, Valaperti, A, Schreiner, J, Wolfensberger, A, et al. Systemic and mucosal antibody responses specific to SARS-CoV-2 during mild versus severe COVID-19. J Allergy Clin Immunol. (2021) 147:545–57.e9. doi: 10.1016/j.jaci.2020.10.040
152. Russell, MW, and Mestecky, J. Mucosal immunity: the missing link in comprehending SARS-CoV-2 infection and transmission. Front Immunol. (2022) 13:957107. doi: 10.3389/fimmu.2022.957107
153. Martínez-Subiela, S, Franco-Martínez, L, Rubio, CP, Muñoz-Prieto, A, Torres-Cantero, A, Tecles, F, et al. Measurement of anti SARS-CoV-2 RBD IgG in saliva: validation of a highly sensitive assay and effects of the sampling collection method and correction by protein. Clin Chem Lab Med. (2022) 60:1683–9. doi: 10.1515/cclm-2022-0418
154. Heaney, CD, Pisanic, N, Randad, PR, Kruczynski, K, Howard, T, Zhu, X, et al. Comparative performance of multiplex salivary and commercially available serologic assays to detect SARS-CoV-2 IgG and neutralization titers. J Clin Virol. (2021) 145:104997. doi: 10.1016/j.jcv.2021.104997
155. Conklin, SE, Martin, K, Manabe, YC, Schmidt, HA, Miller, J, Keruly, M, et al. Evaluation of serological SARS-CoV-2 lateral flow assays for rapid point-of-care testing. J Clin Microbiol. (2021) 59:e02020–14. doi: 10.1128/jcm.02020-20
156. Sims, MD, Podolsky, RH, Childers, KL, Higgins, B, Trueman, J, Homayouni, R, et al. Dried blood spots are a valid alternative to venipuncture for COVID-19 antibody testing. J Immunol Methods. (2023) 513:113420. doi: 10.1016/j.jim.2022.113420
157. Kaufman, HW, Meyer, WA, Clarke, NJ, Radcliff, J, Rank, CM, Freeman, J, et al. Assessing vulnerability to COVID-19 in high-risk populations: the role of SARS-CoV-2 spike-targeted serology. Popul Health Manag. (2023) 26:29–36. doi: 10.1089/pop.2022.0241
158. Perry, J, Osman, S, Wright, J, Richard-Greenblatt, M, Buchan, SA, Sadarangani, M, et al. Does a humoral correlate of protection exist for SARS-CoV-2? A systematic review. PLoS One. (2022) 17:e0266852. doi: 10.1371/journal.pone.0266852
159. Haveri, A, Solastie, A, Ekström, N, Österlund, P, Nohynek, H, Nieminen, T, et al. Neutralizing antibodies to SARS-CoV-2 Omicron variant after third mRNA vaccination in health care workers and elderly subjects. Eur J Immunol. (2022) 52:816–24. doi: 10.1002/eji.202149785
160. O'Mahoney, LL, Routen, A, Gillies, C, Ekezie, W, Welford, A, Zhang, A, et al. The prevalence and long-term health effects of Long Covid among hospitalised and non-hospitalised populations: a systematic review and meta-analysis. EClinicalMedicine. (2023) 55:101762. doi: 10.1016/j.eclinm.2022.101762
Keywords: SARS-CoV-2, immunity, antibodies, models and modeling, innate and adaptive immune response, patient-centered care, vaccines, clinical utility
Citation: Holdenrieder S, Dos Santos Ferreira CE, Izopet J, Theel ES and Wieser A (2023) Clinical and laboratory considerations: determining an antibody-based composite correlate of risk for reinfection with SARS-CoV-2 or severe COVID-19. Front. Public Health. 11:1290402. doi: 10.3389/fpubh.2023.1290402
Received: 07 September 2023; Accepted: 30 November 2023;
Published: 28 December 2023.
Edited by:
Ritthideach Yorsaeng, Chulalongkorn University, ThailandReviewed by:
Igor Stoma, Gomel State Medical University, BelarusCopyright © 2023 Holdenrieder, Dos Santos Ferreira, Izopet, Theel and Wieser. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefan Holdenrieder, cy5ob2xkZW5yaWVkZXJAdHVtLmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.