
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Public Health, 27 November 2023
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 11 - 2023 | https://doi.org/10.3389/fpubh.2023.1283113
 Diego Cantoni1
Diego Cantoni1 Martin Mayora-Neto1
Martin Mayora-Neto1 Mariliza Derveni2
Mariliza Derveni2 Kelly da Costa1
Kelly da Costa1 Joanne Del Rosario1
Joanne Del Rosario1 Veronica O. Ameh3,4
Veronica O. Ameh3,4 Claude T. Sabeta4,5
Claude T. Sabeta4,5 Bethany Auld2
Bethany Auld2 Arran Hamlet6
Arran Hamlet6 Ian M. Jones7
Ian M. Jones7 Edward Wright2
Edward Wright2 Simon D. Scott1
Simon D. Scott1 Efstathios S. Giotis8,9*
Efstathios S. Giotis8,9* Ashley C. Banyard10*
Ashley C. Banyard10* Nigel Temperton1*
Nigel Temperton1*Introduction: The Eidolon helvum fruit bat is one of the most widely distributed fruit bats in Africa and known to be a reservoir for several pathogenic viruses that can cause disease in animals and humans. To assess the risk of zoonotic spillover, we conducted a serological survey of 304 serum samples from E. helvum bats that were captured for human consumption in Makurdi, Nigeria.
Methods: Using pseudotyped viruses, we screened 304 serum samples for neutralizing antibodies against viruses from the Coronaviridae, Filoviridae, Orthomyxoviridae and Paramyxoviridae families.
Results: We report the presence of neutralizing antibodies against henipavirus lineage GH-M74a virus (odds ratio 6.23; p < 0.001), Nipah virus (odds ratio 4.04; p = 0.00031), bat influenza H17N10 virus (odds ratio 7.25; p < 0.001) and no significant association with Ebola virus (odds ratio 0.56; p = 0.375) in this bat cohort.
Conclusion: The data suggest a potential risk of zoonotic spillover including the possible circulation of highly pathogenic viruses in E. helvum populations. These findings highlight the importance of maintaining sero-surveillance of E. helvum, and the necessity for further, more comprehensive investigations to monitor changes in virus prevalence, distribution over time, and across different geographic locations.
Throughout the course of human history, viral zoonotic spillover events have been sporadic and sometimes catastrophic, resulting in several highly lethal pandemics (1). With approximately 60–75% of all human infectious diseases arising from zoonotic transmission (2), it is crucial to remain vigilant in identifying and monitoring potential sources of zoonotic spillover, such as wildlife reservoirs, in order to prevent future outbreaks. Over the past four decades, bats have been identified as a significant source of zoonotic events that have sparked major outbreaks of viruses with considerable implications for human health (3). As the second most diverse mammalian order, bats have been linked to the transmission of a range of viruses, including coronaviruses, filoviruses, lyssaviruses, and henipaviruses, among others (4, 5). With over 12,000 bat-derived virus sequences spanning 30 viral families, having relevance to both veterinary and medical sectors, the need for surveillance of bat populations is critical to assess and mitigate the risk of potential zoonotic spillover events.
Eidolon helvum, the straw-colored fruit bat, is one of the most widely distributed fruit bats in Africa (6). It has extensive migratory patterns of over 2,000 kilometers and is hypothesized to migrate based on availability of fruits to increase reproductive success (7, 8). According to the DBatVir database (9) (accessed on 25th August, 2023) 17 different viruses, including Coronaviridae and Paramyxoviridae, have been detected in E. helvum.
In several countries within Africa, bats, including E. helvum, are often hunted for either bushmeat (10, 11) or as a form of pest control to mitigate fruit farming losses (12). One of these countries is Nigeria, where E. helvum can be found in bushmeat markets to be sold as food (12–15) or as resources for traditional medicine (16). Though many of these practices have been limited by policies imposed due to the Ebola outbreak in 2014 (17), continued bat hunting for human consumption greatly increases the risk of zoonotic transmission, making surveillance vital for public health and wildlife conservation efforts in the region.
Although serological evidence of neutralizing antibodies in bat sera is not definitive proof of active virus infection in bats, it does suggest that bats have been exposed to the virus or closely related viruses, and have mounted an adaptive immune response. Screening for neutralizing antibodies in bats using highly pathogenic viruses requires use of high containment facilities, which can be avoided by using pseudotyped viruses (PVs) in neutralization assays as they are considered safe for handling under biosafety level 2 conditions. Pseudotyped virus neutralization assays (PVNA) have gained widespread usage for detecting neutralizing antibodies due to their high sensitivity and robust correlation with live virus neutralization assays (18).
This study aimed to preliminarily assess the potential presence of pathogenic viruses in bats hunted for bushmeat, to allow further, more comprehensive follow-up investigations focusing on the presence of animal and human pathogenic viruses in Nigeria. We screened 304 serum samples from E. helvum bats that were captured for human consumption in Makurdi, Nigeria using PVs expressing the viral glycoproteins of several viruses known to pose high public health risks (Table 1). Due to limited volumes of sera, we prioritized the order of screening based on the viruses’ potential risk to animal and human health (19–21). Our findings indicate the presence of neutralizing antibodies against important representatives of different virus families, which may suggest the circulation of several highly pathogenic viruses with pandemic potential, or closely related viruses, in colonies of E. helvum in Nigeria, and therefore, warrants further comprehensive research.
All sera were collected from terminally bled straw-colored fruit bats (Eidolon helvum) that were captured for human consumption in Makurdi, Benue State Nigeria (7o44′25.7″N 8o31′52.8″E). The bats in Makurdi were collected from roosts in trees in and around the Benue State Government House, and on trees in private residences close to the government house. Bat roost sites and seasons when bats roost in Makurdi were identified by collaborating and interacting with local bat hunters. Convenient sampling was done by collecting samples only on days when bat hunters set traps to capture bats. Bat roost sites were visited once every week over a period of 12–14 weeks. Personnel protective equipment with respirators were worn by those involved in the sampling of bats captured for human consumption. The bats were captured by setting mist nets in the evenings when they went out to feed. The bats were trapped in the nets as they return to their roost sites on trees during the early hours of the following morning. The bats were then carefully taken down alive from the nets by the hunters, identified morphologically using phenotypic properties and bled from the brachial vein, using a 5 mL syringe and a 21-gauge hypodermic needle by the investigator. Bats were also sampled from roosts on trees in private residences where permissions were gained. Sampling was carried out for two consecutive seasons (November 2017–March 2018 and November 2018–March 2019). The whole blood samples were placed in well-labeled 7 mL tubes. These were placed on ice in cooler boxes and transported to the Amadu Ali Centre for Public Health and Comparative Medicine Laboratory, Federal University of Agriculture Makurdi. Sera were harvested from whole blood by centrifuging at 3,000 revolutions per minute for 10 min. Serum samples were placed in labeled 2 mL screwcap cryovials, heat-inactivated at 56°C for 30 min and stored at −20°C, then later transported to the Animal and Plant Health Agency, Weybridge (United Kingdom) after an import license was approved (License Number: ITIMP19.1408). Ethical approval for this research study was granted by the Animal Ethics Committee and Research Ethics Committee of University of Pretoria (certificate numbers V092-18 and REC097-18). The Director/Chief Veterinary Officer of Nigeria, Department of Veterinary and Pest Control Services, Federal Ministry of Agriculture and Pest Control Services, Abuja Nigeria granted permission to sampling of bat populations (license number VDS/194/S.4/11/T/85).
Lentiviral (HIV) based pseudotypes used for this study were generated and characterized as described in detail, Influenza H17 (22), SARS-CoV-1 (23), SARS-CoV-2 (24), RaTG13 (25), EBOV and MARV (26), while NiV and GH-M74a were adapted from Khetawat et al. (27). Briefly, 1.0 μg of p8.91 plasmid encoding the HIV gag-pol was mixed with 1.5 μg of pCSFLW reporter gene and 1 μg of each surface viral glycoproteins as described in the studies cited in the previous sentence and in Table 2. After mixing the plasmids in 200 μL of Opti-MEM (ThermoFisher, Woolwich, UK), Fugene HD (Promega, Southampton, UK) transfection reagent was added at a 1:3 (plasmid:Fugene HD) ratio and incubated for 15 min prior to adding the transfection complexes to HEK293T/17 cells in T-75 cell culture flasks. Pseudotypes were harvested at 48- and 72-h post transfection, whereby culture media was removed from the flasks and filtered through a 0.45 μM cellulose acetate filter (Corning, Deeside, UK). Samples were aliquoted and frozen at −80°C for long term storage prior to use.
Pseudotyped viruses were titrated by serially diluting filtered pseudotypes with fresh DMEM starting from a 1:2 to a final 1:512 dilution using white 96-well flat bottom plates (ThermoFisher, Woolwich, UK). Then appropriate target cells that allow permissibility for infection were added (Table 2) and plates were returned to the incubator. After 48 h, cells were lysed with Bright-Glo reagent (Promega, Southampton, UK), and luciferase expression levels were assessed using a GloMax plate reader (Promega, Southampton, UK).
Sera were initially screened at single point dilution (1:100, except NiV and GH-M74a at 1:50). Each plate had wells containing PV only to determine maximum pseudotype entry. Samples where a 1 log decrease in RLU compared to the no-serum virus-only control was observed were then selected for cytotoxicity assay using Cell Titre Glo kit (Promega, Southampton, UK) and light microscopy to verify viability of cells prior to undertaking PVNA (data not shown). Neutralization assays were carried out by mixing bat sera, positive control sera (EBOV: WHO International Standard for Ebola virus Antibodies, 15/262, H17: mouse polyclonal antibodies raised to purified H17 antigen expressed using the baculovirus system as described in Loureiro et al. (28) and Shelton et al. (29), or positive monoclonal antibody NiV: m102.4 (30), with cell culture media at a starting input of 1:40 ratio (sera: cell culture media) and diluted either 4-fold (GH-M74a and NiV) to 1:5,000 or 8-fold (all other viruses) to 1:5120. PVs were added to the plates at a minimum RLU input of 105 per well, incubated for 1 h at 37°C, followed by addition of target cells at a density of 104 cells per well, except for NiV and GH-M74a where a cell density of 204 cells per well was used. Plates were incubated at 37°C and 5% CO2 for 48 h prior to lysis using Bright-Glo reagent according to manufacturer’s protocol and monitoring of luciferase expression using a GloMax plate reader. Regression curves were fitted using GraphPad Prism 8 software (San Diego, CA, United States) as described previously (31). Some PVNAs tested fewer bat serum samples, due to the limited volumes supplied for this study. Currently, no positive control reagent exists for GH-M74a.
Data were analyzed using STATA version 16 (StataCorp, College Station, TX, United States). Descriptive statistics were used to summarize the distribution of the variables. Bivariate logistic regression was used to determine the association between the presence or absence of neutralizing antibodies against each virus. Multivariable logistic regression was used to adjust for confounding factors. The data were coded as binary variables (1 = positive, 0 = negative). Logistic regression was used to determine the association between the presence or absence of neutralizing antibodies against each virus and the dependent variable was the presence or absence of neutralizing antibodies against each virus. Crude and adjusted odds ratios (OR) were estimated with 95% confidence intervals (CI) to measure the strength of association between each variable and the presence or absence of neutralizing antibodies.
Our PVNA screening revealed the presence of neutralizing antibodies against several of the pseudotyped viruses tested (Figures 1A,B; Table 3). However, no neutralization was observed against the Coronaviridae members SARS-CoV-2, SARS-CoV, and the bat coronavirus RaTG13. Although neutralizing antibodies against EBOV PVs were detected in a single sample (n = 1/278), no neutralization was observed against MARV. We also found several samples positive for neutralizing antibodies against influenza A virus H17N10 PVs (n = 26/304), indicating the presence of positive or cross-reactive H17 neutralizing antibodies. Additionally, positive samples were detected for Nipah virus (NiV) (n = 16/54) and GH-M74a (n = 36/54) from the Paramyxoviridae PVs.
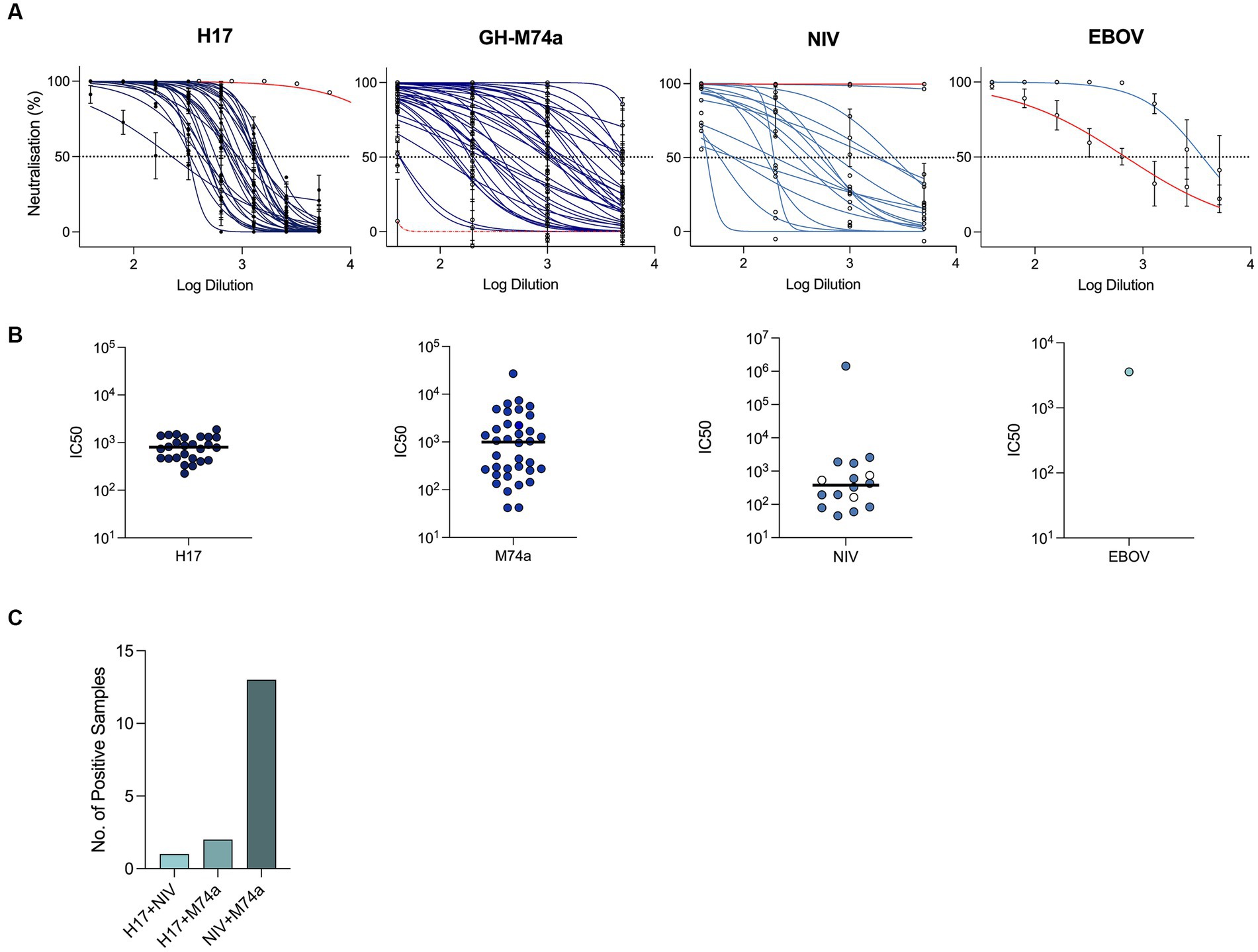
Figure 1. Positive detection of neutralizing antibodies in E. helvum by PVNA assay. Neutralization curves derived from pMN assays against positive samples that were selected from the initial screening, red curves denoting positive control reagents, with the exception of GH-M-74a which was not neutralized by NiV 102.4 mAb (red dotted line) (A). IC50s calculated by pMN assays against each PV (B). In the panel displaying samples positive for NiV, empty white circles denote samples that did not cross-neutralize GH-M74a. Number of samples positive for neutralizing antibodies against multiple viruses (C).
Our logistic regression analysis shows that the presence of neutralizing antibodies is significantly associated with virus type, as indicated by the p-values for the virus coefficients. Specifically, the odds of samples having neutralizing antibodies are significantly higher for H17 (odds ratio = 7.25, p < 0.001), NiV (odds ratio = 4.04, p = 0.00031), and GH-M74a (odds ratio = 6.23, p < 0.001) compared to the reference category of SARS-CoV-2. EBOV did not show a significant association with neutralizing antibody status (odds ratio = 0.56, p = 0.375).
We further analyzed the data to investigate the presence of samples positive for neutralizing antibodies against multiple viruses (Figure 1C). Our analysis revealed one sample positive for H17 and NiV, two samples positive for H17 and GH-M74a, and 13 samples positive for both NiV and GH-M74a. These findings suggest that multiple viruses may have circulated within the same bat or that antibodies to one of the three viruses may cross-neutralize against multiple viruses.
This study investigated the seroprevalence of neutralizing antibodies against several highly pathogenic viruses in E. helvum bats in Nigeria. We report a high level of seroprevalence against GH-M74a (66.7% of samples) and NiV (29.6%). The high level of seropositivity against GH-M74a could be the result of a recent resolved infection or the presence of multiple pathogens circulating with similar antigenicity. Given that the two viruses share a key conserved region in their attachment protein, and that GH-M74a antibodies have been shown to cross neutralize NiV (32), the significance of our study becomes evident when we consider the detection of three unique samples that, notably, exhibited neutralization exclusively against NiV but not GH-M74a. This suggests the potential existence of a closely related virus to NiV, or, though unlikely, the presence of NiV, which has yet to be identified in bats in Africa. In any case, the predictive value of a virus’s capability to utilize highly conserved receptors is significant when assessing the potential for viral emergence and cross-species transmission (33) Therefore, our results may indicate the presence of a closely related virus whose pathogenicity and virulence remain to be characterized. The henipavirus designated Ghana virus (GhV) remains the only African henipavirus that has been fully sequenced, isolated from E. helvum (34). However, Henipavirus and henipa-like virus antibodies have been detected in E. helvum (32, 35, 36) and other species, of which the literature has been thoroughly reviewed in Mbu’u et al. (37). Despite their detection, overall seroprevalence in bats remains low. Interestingly, Pernet et al. (32) detected a 3–4% seroprevalence rate from 497 human blood samples against Hendra virus, GH-74a and NiV, of which almost all positive samples were derived from individuals who reported butchering bats for bushmeat. This finding, combined with the outcomes of our study, underscores the ongoing elevated risk of exposure for individuals engaged in the bushmeat trade.
We also observed seropositivity toward the H17N10 PVs, a bat Influenza A-like virus that contains unconventional HA and NA proteins (38). The evolutionary distinct H17N10 along with H18N11 virus (not screened for in this study) were originally recovered from asymptomatic fruit bats of the Neotropic bat family Phyllostomidae (Sturnira lilium and Artibeus planirostris) in several countries of Central and South America (38–40). These viruses have attracted considerable attention following reports that their entry in host cells is mediated by the conserved trans-species MHC-II proteins, suggesting zoonotic potential (41, 42). So far, bats in Central and S. America, have been found seropositive for H17N10 and H18N11, but not in a study in Central Europe (43), and to our knowledge this is the first report of H17-neutralizing samples in non-Neotropical bat species. Considering the lack of a validated serological assay to screen E. helvum bat sera specifically for H17N10, we cannot exclude serological cross-reactivity with heterologous H17-like antigens. Nonetheless, the potential presence of undiscovered H17 or H17-like IAV species in E. helvum, such as the ones described in Phylostomidae is tantalizing, cannot be ruled out and warrants further examination by unbiased approaches, i.e., metagenomics in subsequent studies.
Although we only detected a single sample positive for neutralizing antibodies against EBOV, similar studies have also reported very low seroprevalence rates in E. helvum; 1 out of 262 samples from Ghana (44), and 19 out of 748 samples from Zambia (45). These low numbers could be due to the fact E. helvum bat cells have been shown to be refractory toward EBOV infection (46). On the other hand, a report detected much higher seroprevalence of EBOV antibodies in E. helvum from Cameroon, with 107 out of 817 positive samples (47). Again, it is vital to consider the possibility of cross-neutralization, not only with other pathogenic species of ebolaviruses but potentially closely related undiscovered filoviruses, which may not be pathogenic in humans such as the newly discovered Bombali virus in Africa, and shares cross-neutralization against EBOV antibodies (48).
There are several limitations to our study. Relative to the size of bat colonies, 304 samples is small and as a result may not accurately represent potential viruses in circulation within the colony. Secondly, the use of PVs alone, without nucleic acid testing does not definitively inform whether the antibodies we detected were generated by infection of the stated virus or a close relative, resulting in cross neutralizing antibodies, which we have detailed in the discussion. Nonetheless, the preliminary nature of our data in this study would suggest that it is worth carrying out a more comprehensive investigation of E. helvum bats in Makurdi to assess the potential risks posed to the human and animal population.
In summary, our serological screening of E. helvum sera obtained from bats has revealed the presence of antibodies that can neutralize PVs displaying the glycoproteins of several highly pathogenic viruses. The capture and preparation of these bats for human consumption suggests a potential for direct exposure to bat bodily fluids, thereby elevating the risk of cross-species transmission of the viruses and other pathogens. Given that human settlements are encroaching into areas known to harbor large bat colonies, especially in areas that are known for bat roosting, the risk of zoonotic spillover will continue to increase. Therefore, monitoring of bat viruses especially in the large bat populations in Sub-Saharan Africa is crucial in order to better understand the prevalence and transmission of viruses and to mitigate the risk of potential spillover events.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was approved by Approval to sample bat populations (VDS/194/S.4/11/T/85) was obtained from the Director/Chief Veterinary Officer of Nigeria, Department of Veterinary and Pest Control Services, Federal Ministry of Agriculture and Pest Control Services, Abuja Nigeria. The study was conducted in accordance with the local legislation and institutional requirements.
DC: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Validation, Visualization, Writing – original draft, Writing – review & editing. MM-N: Data curation, Formal analysis, Investigation, Methodology, Validation, Writing – review & editing. BA: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. KD: Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. JDR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – review & editing. VA: Data curation, Investigation, Methodology, Resources, Writing – review & editing. CS: Investigation, Resources, Validation, Writing – review & editing. MD: Conceptualization, Data curation, Formal analysis, Methodology, Writing – review & editing. AH: Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. EW: Conceptualization, Investigation, Methodology, Resources, Supervision, Writing – original draft, Writing – review & editing. SS: Investigation, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. EG: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – review & editing. AB: Investigation, Resources, Supervision, Validation, Writing – review & editing. NT: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Resources, Supervision, Validation, Visualization, Writing – review & editing. IJ: Resources, Writing - review and editing, Validation.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. AB was part funded by the UK Department for the Environment, Food and Rural Affairs (Defra) and the devolved Scottish and Welsh governments under grants SE2213 and SV3006. EG is supported by funding by the University of Essex COVID-19 Rapid and Agile and the Faculty of Science and Health Research Innovation and Support Funds.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer WM declared a shared affiliation with the authors VA and CS to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Smith, I, and Wang, L-F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr Opin Virol. (2013) 3:84–91. doi: 10.1016/j.coviro.2012.11.006
2. Ellwanger, JH, and Chies, JAB. Zoonotic spillover: understanding basic aspects for better prevention. Genet Mol Biol. (2021) 44:e20200355. doi: 10.1590/1678-4685-GMB-2020-0355
3. Wynne, JW, and Wang, L-F. Bats and viruses: friend or foe? PLoS Pathog. (2013) 9:e1003651. doi: 10.1371/journal.ppat.1003651
4. Ingala, MR, Simmons, NB, and Perkins, SL. Bats are an untapped system for understanding microbiome evolution in mammals. mSphere. (2018) 3:e00397–18. doi: 10.1128/mSphere.00397-18
5. Glennon, EE, Restif, O, Sbarbaro, SR, Garnier, R, Cunningham, AA, Suu-Ire, RD, et al. Domesticated animals as hosts of henipaviruses and filoviruses: a systematic review. Vet J. (2018) 233:25–34. doi: 10.1016/j.tvjl.2017.12.024
6. Peel, AJ, Sargan, DR, Baker, KS, Hayman, DTS, Barr, JA, Crameri, G, et al. Continent-wide panmixia of an African fruit bat facilitates transmission of potentially zoonotic viruses. Nat Commun. (2013) 4:2770. doi: 10.1038/ncomms3770
7. Ossa, G, Kramer-Schadt, S, Peel, AJ, Scharf, AK, and Voigt, CC. The movement ecology of the straw-colored fruit bat, Eidolon helvum, in sub-Saharan Africa assessed by stable isotope ratios. PLoS One. (2012) 7:e45729. doi: 10.1371/journal.pone.0045729
8. Markotter, W, Coertse, J, De Vries, L, Geldenhuys, M, and Mortlock, M. Bat-borne viruses in Africa: a critical review. J Zool. (1987) 311:77–98. doi: 10.1111/jzo.12769
9. Chen, L, Liu, B, Yang, J, and Jin, Q. DBatVir: the database of bat-associated viruses. Database (Oxford). (2014) 2014:bau021. doi: 10.1093/database/bau021
10. Richter, HV, and Cumming, GS. Food availability and annual migration of the straw-colored fruit bat (Eidolon helvum). J Zool. (2006) 268:35–44. doi: 10.1111/j.1469-7998.2005.00020.x
11. Kamins, AO, Restif, O, Ntiamoa-Baidu, Y, Suu-Ire, R, Hayman, DTS, Cunningham, AA, et al. Uncovering the fruit bat bushmeat commodity chain and the true extent of fruit bat hunting in Ghana. West Africa Biol Conserv. (2011) 144:3000–8. doi: 10.1016/j.biocon.2011.09.003
12. Mickleburgh, S, Waylen, K, and Racey, P. Bats as bushmeat: a global review. Oryx. (2009) 43:217–34. doi: 10.1017/S0030605308000938
13. Meseko, C, Shittu, I, and Adedeji, A. The bush meat trade thrives in Nigeria despite anxiety over coronavirus. Trans R Soc Trop Med Hyg. (2020) 114:639–41. doi: 10.1093/trstmh/traa060
14. Friant, S, Paige, SB, and Goldberg, TL. Drivers of bushmeat hunting and perceptions of zoonoses in Nigerian hunting communities. PLoS Negl Trop Dis. (2015) 9:e0003792. doi: 10.1371/journal.pntd.0003792
15. Funmilayo, O. Fruit bats for meat: are too many taken? Oryx. (1978) 14:377–8. doi: 10.1017/S0030605300016008
16. Adeyanju, TE, Alarape, AA, Musila, S, Adeyanju, AT, Omotoriogun, TC, Medina-Jerez, W, et al. Human–bat relationships in southwestern Nigerian communities. Anthrozoös. (2023) 36:407–25. doi: 10.1080/08927936.2023.2166715
17. Funk, SM, Fa, JE, Ajong, SN, Eniang, EA, Dendi, D, Di Vittorio, M, et al. Pre- and post-Ebola outbreak trends in wild meat trade in West Africa. Biol Conserv. (2021) 255:109024. doi: 10.1016/j.biocon.2021.109024
18. Bentley, EM, Mather, ST, and Temperton, NJ. The use of pseudotypes to study viruses, virus sero-epidemiology and vaccination. Vaccine. (2015) 33:2955–62. doi: 10.1016/j.vaccine.2015.04.071
19. Janik, E, Ceremuga, M, Niemcewicz, M, and Bijak, M. Dangerous pathogens as a potential problem for public health. Medicina. (2020) 56:591. doi: 10.3390/medicina56110591
20. Ching, PKG. de los Reyes VC, Sucaldito MN, Tayag E, Columna-Vingno AB, Malbas FF, et al. outbreak of Henipavirus infection, Philippines, 2014. Emerg Infect Dis. (2015) 21:328–31. doi: 10.3201/eid2102.141433
21. Chua, KB. Nipah virus outbreak in Malaysia. J Clin Virol. (2003) 26:265–75. doi: 10.1016/S1386-6532(02)00268-8
22. Del Rosario, JMM, da Costa, KAS, Asbach, B, Ferrara, F, Ferrari, M, Wells, DA, et al. Exploiting pan influenza a and pan influenza B Pseudotype libraries for efficient vaccine antigen selection. Vaccine. (2021) 9:741. doi: 10.3390/vaccines9070741
23. Carnell, G, Grehan, K, Ferrara, F, Molesti, E, and Temperton, N. An optimized method for the production using PEI, titration and Neutralizationof SARS-CoV spike luciferase Pseudotypes. Bio Protoc. (2017) 7:e2514. doi: 10.21769/BioProtoc.2514
24. Genova, CD, Sampson, A, Scott, S, Cantoni, D, Mayora-Neto, M, Bentley, E, et al. Production, titration, neutralisation, storage and lyophilisation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) lentiviral Pseudotypes. Bio-protocol. (2021) 11:e4236–6. doi: 10.21769/BioProtoc.4236
25. Cantoni, D, Mayora-Neto, M, Thakur, N, Elrefaey, AME, Newman, J, Vishwanath, S, et al. Pseudotyped bat coronavirus RaTG13 is efficiently neutralised by convalescent sera from SARS-CoV-2 infected patients. Commun Biol. (2022) 5:409–8. doi: 10.1038/s42003-022-03325-9
26. Neto, MM, Wright, E, Temperton, N, Soema, P, ten Have, R, Ploemen, I, et al. Application and comparison of lyophilisation protocols to enhance stable long-term storage of filovirus pseudotypes for use in antibody neutralisation tests. J Appl Microbiol. (2023) 134:lxac067. doi: 10.1093/jambio/lxac067
27. Khetawat, D, and Broder, CC. A functional Henipavirus envelope glycoprotein Pseudotyped lentivirus assay system. Virol J. (2010) 7:312. doi: 10.1186/1743-422X-7-312
28. Loureiro, S, Ren, J, Phapugrangkul, P, Colaco, CA, Bailey, CR, Shelton, H, et al. Adjuvant-free immunization with hemagglutinin-fc fusion proteins as an approach to influenza vaccines. J Virol. (2011) 85:3010–4. doi: 10.1128/jvi.01241-10
29. Shelton, H, Ayora-Talavera, G, Ren, J, Loureiro, S, Pickles, RJ, Barclay, WS, et al. Receptor binding profiles of avian influenza virus hemagglutinin subtypes on human cells as a predictor of pandemic potential. J Virol. (2011) 85:1875–80. doi: 10.1128/JVI.01822-10
30. Playford, EG, Munro, T, Mahler, SM, Elliott, S, Gerometta, M, Hoger, KL, et al. Safety, tolerability, pharmacokinetics, and immunogenicity of a human monoclonal antibody targeting the G glycoprotein of henipaviruses in healthy adults: a first-in-human, randomised, controlled, phase 1 study. Lancet Infect Dis. (2020) 20:445–54. doi: 10.1016/S1473-3099(19)30634-6
31. Ferrara, F, and Temperton, N. Pseudotype neutralization assays: from laboratory bench to data analysis. Methods Protocols. (2018) 1:8. doi: 10.3390/mps1010008
32. Pernet, O, Schneider, BS, Beaty, SM, LeBreton, M, Yun, TE, Park, A, et al. Evidence for henipavirus spillover into human populations in Africa. Nat Commun. (2014) 5:5342. doi: 10.1038/ncomms6342
33. Holmes, EC. What can we predict about viral evolution and emergence? Curr Opin Virol. (2013) 3:180–4. doi: 10.1016/j.coviro.2012.12.003
34. Drexler, JF, Corman, VM, Müller, MA, Maganga, GD, Vallo, P, Binger, T, et al. Bats host major mammalian paramyxoviruses. Nat Commun. (2012) 3:796. doi: 10.1038/ncomms1796
35. Muleya, W, Sasaki, M, Orba, Y, Ishii, A, Thomas, Y, Nakagawa, E, et al. Molecular epidemiology of paramyxoviruses in frugivorous Eidolon helvum bats in Zambia. J Vet Med Sci. (2014) 76:611–4. doi: 10.1292/jvms.13-0518
36. Drexler, JF, Corman, VM, Gloza-Rausch, F, Seebens, A, Annan, A, Ipsen, A, et al. Henipavirus RNA in African bats. PLoS One. (2009) 4:e6367. doi: 10.1371/journal.pone.0006367
37. Mbu’u, CM, Mbacham, WF, Gontao, P, Sado Kamdem, SL, AMN, N, Groschup, MH, et al. Henipaviruses at the Interface between bats, livestock and human population in Africa. Vector-Borne Zoon Dis. (2019) 19:455–65. doi: 10.1089/vbz.2018.2365
38. Tong, S, Li, Y, Rivailler, P, Conrardy, C, Castillo, DAA, Chen, L-M, et al. A distinct lineage of influenza a virus from bats. Proc Natl Acad Sci. (2012) 109:4269–74. doi: 10.1073/pnas.1116200109
39. Tong, S, Zhu, X, Li, Y, Shi, M, Zhang, J, Bourgeois, M, et al. New World Bats Harbor diverse influenza a viruses. PLoS Pathog. (2013) 9:e1003657. doi: 10.1371/journal.ppat.1003657
40. Campos, ACA, Góes, LGB, Moreira-Soto, A, de Carvalho, C, Ambar, G, Sander, A-L, et al. Bat Influenza A(HL18NL11) Virus in Fruit Bats. Brazil Emerg Infect Dis. (2019) 25:333–7. doi: 10.3201/eid2502.181246
41. Giotis, ES. Inferring the urban transmission potential of bat influenza viruses. Front Cell Infect Microbiol. (2020) 10:264. doi: 10.3389/fcimb.2020.00264
42. Giotis, ES, Carnell, G, Young, EF, Ghanny, S, Soteropoulos, P, Wang, L-F, et al. Entry of the bat influenza H17N10 virus into mammalian cells is enabled by the MHC class II HLA-DR receptor. Nat Microbiol. (2019) 4:2035–8. doi: 10.1038/s41564-019-0517-3
43. Fereidouni, S, Kwasnitschka, L, Balkema Buschmann, A, Müller, T, Freuling, C, Schatz, J, et al. No virological evidence for an influenza a - like virus in European bats. Zoonoses Public Health. (2015) 62:187–9. doi: 10.1111/zph.12131
44. Hayman, DTS, Emmerich, P, Yu, M, Wang, L-F, Suu-Ire, R, Fooks, AR, et al. Long-term survival of an urban fruit bat seropositive for Ebola and Lagos bat viruses. PLoS One. (2010) 5:e11978. doi: 10.1371/journal.pone.0011978
45. Ogawa, H, Miyamoto, H, Nakayama, E, Yoshida, R, Nakamura, I, Sawa, H, et al. Seroepidemiological prevalence of multiple species of filoviruses in fruit bats (Eidolon helvum) migrating in Africa. J Infect Dis. (2015) 212:S101–8. doi: 10.1093/infdis/jiv063
46. Ng, M, Ndungo, E, Kaczmarek, ME, Herbert, AS, Binger, T, Kuehne, AI, et al. Filovirus receptor NPC1 contributes to species-specific patterns of ebolavirus susceptibility in bats. elife. (2015) 4:e11785. doi: 10.7554/eLife.11785
47. Djomsi, DM, Mba Djonzo, FA, Ndong Bass, I, Champagne, M, Lacroix, A, Thaurignac, G, et al. Dynamics of antibodies to ebolaviruses in an Eidolon helvum bat Colony in Cameroon. Viruses. (2022) 14:560. doi: 10.3390/v14030560
48. Bentley, EM, Richardson, S, Derveni, M, Rijal, P, Townsend, AR, Heeney, JL, et al. Cross-neutralisation of novel Bombali virus by Ebola virus antibodies and convalescent plasma using an optimised Pseudotype-based neutralisation assay. Trop Med Infect Dis. (2021) 6:155. doi: 10.3390/tropicalmed6030155
Keywords: Eidolon helvum , pseudotypes, Ebola virus, Nipah virus, Marburg virus, henipavirus, H17N10, Ghana bat henipavirus
Citation: Cantoni D, Mayora-Neto M, Derveni M, da Costa K, Del Rosario J, Ameh VO, Sabeta CT, Auld B, Hamlet A, Jones IM, Wright E, Scott SD, Giotis ES, Banyard AC and Temperton N (2023) Serological evidence of virus infection in Eidolon helvum fruit bats: implications for bushmeat consumption in Nigeria. Front. Public Health. 11:1283113. doi: 10.3389/fpubh.2023.1283113
Received: 25 August 2023; Accepted: 02 November 2023;
Published: 27 November 2023.
Edited by:
Ahmed Mostafa, National Research Centre, EgyptReviewed by:
Wanda Markotter, University of Pretoria, South AfricaCopyright © 2023 Cantoni, Mayora-Neto, Derveni, da Costa, Del Rosario, Ameh, Sabeta, Auld, Hamlet, Jones, Wright, Scott, Giotis, Banyard and Temperton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Efstathios S. Giotis, ZS5naW90aXNAZXNzZXguYWMudWs=; Ashley C. Banyard, QXNobGV5LkJhbnlhcmRAYXBoYS5nb3YudWs=; Nigel Temperton, Ti5UZW1wZXJ0b25Aa2VudC5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.