
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health , 23 November 2023
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 11 - 2023 | https://doi.org/10.3389/fpubh.2023.1251609
Objective: We investigated the epidemiological surveillance of the intestinal colonization and nosocomial infection of carbapenem-resistant Enterobacteriales (CRE) isolates from inpatients, which can provide the basis for developing effective prevention.
Methods: A total of 96 CRE strains were collected from 1,487 fecal samples of hospitalized children between January 2016 and June 2017, which were defined as the “CRE colonization” group. In total, 70 CRE clinical isolates were also randomly selected for the comparison analysis and defined as the “CRE infection” group. The antimicrobial susceptibility of all strains was determined by the microdilution broth method. Polymerase chain reaction (PCR) was used to analyze carbapenemase genes, plasmid typing, and integrons. Multilocus sequence typing was further used to determine clonal relatedness.
Results: In the “CRE colonization” group, Klebsiella pneumoniae was mostly detected with a rate of 42.7% (41/96), followed by Escherichia coli (34.4%, 33/96) and Enterobacter cloacae (15.6%, 15/96). The ST11 KPC-2 producer, ST8 NDM-5 producer, and ST45 NDM-1 producer were commonly present in carbapenem-resistant K. pneumoniae (CRKPN), carbapenem-resistant E. coli (CRECO), and carbapenem-resistant E. cloacae (CRECL) isolates, respectively. In the “CRE infection” group, 70% (49/70) of strains were K. pneumoniae, with 21.4% E. cloacae (15/70) and 5.7% E. coli (4/70). The ST15 OXA-232 producer and ST48 NDM-5 producer were frequently observed in CRKPN isolates, while the majority of NDM-1-producing CRECL isolates were assigned as ST45. Phylogenetic analysis showed that partial CRE isolates from intestinal colonization and nosocomial infection were closely related, especially for ST11 KPC-2-producing CRKPN and ST45 NDM-1-producing CRECL. Furthermore, plasmid typing demonstrated that IncF and IncFIB were the most prevalent plasmids in KPC-2 producers, while IncX3/IncX2 and ColE were widely spread in NDM producer and OXA-232 producer, respectively. Then, class 1 integron intergrase intI1 was positive in 74.0% (71/96) of the “CRE colonization” group and 52.9% (37/70) of the “CRE infection” group.
Conclusion: This study revealed that CRE strains from intestinal colonization and nosocomial infection showed a partial correlation in the prevalence of CRE, especially for ST11 KPC-2-producing CRKPN and ST45 NDM-1-producing CRECL. Therefore, before admission, long-term active screening of rectal colonization of CRE isolates should be emphasized.
Enterobacteriales represent a large family of gram-negative bacteria, which cause a series of nosocomial infections including pneumonia, bloodstream infections, and urinary tract infections. Carbapenems including ertapenem, imipenem, and meropenem are commonly considered the last line of defense to treat multidrug-resistant bacterial infections, and the wide and unreasonable use of these antibiotics has consequently resulted in the increasing incidence of antimicrobial resistance. Carbapenem-resistant Enterobacteriales (CRE) are regarded as the top-ranked bacteria in the priority list of resistant pathogens of the World Health Organization and have become a severe threat to global public health (1, 2). In China, the overall incidence of CRE infection per 10,000 discharges was 4.0 and differed significantly by region (3). Production of carbapenemases including Klebsiella pneumoniae carbapenemase (KPC) and New Delhi metallo-β-lactamase (NDM) was the main mechanism of CRE. Then, the rapid spread of CRE severely limited the options for antibiotics and prolonged the length of hospital stay, ultimately resulting in high mortality.
The family Enterobacteriales naturally colonizes in the gastrointestinal tract, which is the most important reservoir for CRE strains. Previous studies reported that the prevalence of CRE colonization rates was less than or ~5%, but it would be higher in critical patients in the intensive care unit (4–7). Furthermore, fecal carriage of CRE isolates is regarded as a risk factor for developing CRE infections. A recent study showed that 50% of the patients with CRE rectal colonization could develop CRE-associated pneumonia and simultaneously were more likely to have a long hospital stay and high hospitalization expense than patients with non-colonized CRE (8). Therefore, early and rapid detection of CRE colonization among high-risk patients will be helpful to implement strategies to prevent CRE dissemination.
Many guidelines recommend that active screening of rectal colonization of CRE is an easy and important tactic to control CRE infection, and rectal swabs are considered as the preferred sample for screening on account of good patient compliance and fewer side effects (9, 10). Accordingly, we started the CRE screening for hospitalized children in 2016 and continuously monitored the change in CRE prevalence. However, the distribution of characteristics between the colonized CRE and clinical CRE isolates among children still needs to be summarized. In this study, we will investigate epidemiological surveillance of the colonization/infection of CRE isolates from inpatients, which can provide the basis for developing effective prevention measures for CRE infections.
To compare the colonization/infection of CRE isolates from inpatients, this study was designed in Shanghai Children's Hospital between January 2016 and June 2017. The inclusion criteria were as follows: Fecal sample was collected from hospitalized children aged 0 to 18 years who received the routine fecal culture testing, and only the first fecal sample of each inpatient was enrolled. The exclusion criteria were as follows: (1) patients older than 18 years; (2) patients from an outpatient clinic; and (3) patients with duplicate fecal samples. A total of 1,487 fecal samples from 1,487 inpatients were transported to clinical microbiology for fecal culture testing and were further screened by performing the meropenem disk screening method. Moreover, 96 CRE strains were detected with positive results, which were defined as the “CRE colonization” group. Meanwhile, we randomly selected 70 CRE clinical isolates from the 210 clinical CRE isolates using the random number generation function in Microsoft Office Excel 2010 (Microsoft Corporation, Redmond, WA, USA), and these CRE strains were isolated from different samples including sputum, blood, urine, and pus, which caused nosocomial infections. These clinical isolates were incorporated for the comparison analysis and defined as the “CRE infection” group. The clinical information of all enrolled inpatients including patient demographics, hospital ward, use of antibiotics, and treatment outcomes was also collected from electronic medical records. All CRE isolates were identified by the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) system using MALDI Biotyper (Bruker Daltonik GmbH, Bremen, Germany). These isolates were stored at −80°C in a 40% glycerol broth medium for further analysis.
The minimum inhibitory concentrations (MICs) of antimicrobial agents including cefazolin, cefuroxime, ceftazidime, ceftriaxone, cefotaxime, cefepime, ampicillin–sulbactam, piperacillin–tazobactam, ertapenem, imipenem, meropenem, aztreonam, ciprofloxacin, levofloxacin, amikacin, gentamicin, trimethoprim–sulfamethoxazole, ceftazidime–avibactam, tigecycline, and colistin were determined by the microdilution broth method. The results were interpreted by the breakpoints of Clinical and Laboratory Standards Institute (CLSI) 2019. The interpretive criterion for tigecycline was based on the breakpoints of the Food and Drug Administration (FDA). Escherichia coli ATCC 25922 and E. coli ATCC 35218 were used for quality control.
All the CRE isolates were screened for the presence of carbapenemase genes including blaNDM, blaKPC, blaOXA − 48, blaIMP, blaAIM, blaVIM, blaGIM, and blaSIM by polymerase chain reaction (PCR) as previously described (11) (Supplementary Table 1S). The amplicons were sequenced, and nucleotide sequences were further analyzed and compared with the sequences available at the National Center for Biotechnology Information (NCBI) website (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Genetic evolutionary relationships were identified for K. pneumoniae, E. coli, and Enterobacter cloacae using multilocus sequence typing (MLST) schemes. The housekeeping genes of K. pneumoniae (ropB, gapA, mdh, pgi, phoE, infB, and tonB) and E. coli (dinB, icdA, pabB, polB, putB, trpA, trpB, and uidA) were performed according to the protocol available on the website of Institute Pasteur MLST (https://bigsdb.pasteur.fr/). Meanwhile, amplification of housekeeping genes for E. cloacae was also conducted as described on the PubMLST website (https://pubmlst.org/). The STs were assigned by sequencing and submitting the sequences to the MLST database for alignment. MEGA 7.0 software was used for Phylogenetic analysis.
For all CRE isolates, the molecular typing of the plasmids was performed using the PCR-based replicon typing, and 17 pairs of primers (HI1, HI2, I1, L/M, N, FIA, FIB, W, Y, P, FIC, A/C, T, FIIAs, F, K, and B/O) were amplified as described previously. In addition, we used new primers typing IncX groups to IncX1-IncX4 and new primers for ColE-type plasmid (12–14). The amplified products were electrophoresed in 1.5% agarose gels, stained with Green Nucleic Acid Stain (Sangon, Shanghai, China), and visualized under ultraviolet light using the Gel Doc 2000 system (Bio-Rad).
The class 1, 2, and 3 integrons were screened by PCR amplification for intI1, intI2, and intI3 genes. Furthermore, amplification of the intI1 variable region was performed using the primers 5′-CS and 3′-CS under the conditions and annealing temperatures as described previously (15, 16) (Supplementary Table 1S). To identify different types of gene cassettes, same-sized amplicons were analyzed by restriction fragment length polymorphism (RFLP) with digested RsaI and Hinf I genes (TaKaRa Bio Inc., Tokyo, Japan), which were dependent on the species. Amplicons showing the same pattern were considered to be identical gene cassettes, and then, one representative product of each distinct RFLP was purified and sequenced by Sangon Biotech Co. Ltd (Shanghai, China). The sequencing results were analyzed using the BLAST program on the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
The statistical analysis was performed using SPSS 25.0 for Windows (version 25.0; SPSS Inc., Chicago, IL, USA). For continuous variables, data were described as median, which was compared using the Mann–Whitney U-test. Multivariate analysis was used to identify the independent predictors, and we calculated the odds ratios (ORs) and the 95% confidence intervals (CIs) for each variable. A value of P ≤ 0.05 was considered statistically significant.
A total of 96 non-duplicate samples were detected as CRE-positive with a carriage of 6.5% (96 out of 1,487). The clinical characteristics of enrolled patients between the “CRE colonization” group and the “CRE infection” group are presented in Table 1. The CRE strains between the two groups were predominantly isolated from the department of neonatology, with severe underlying diseases including pneumonia, sepsis, and gastrointestinal infections. More than 50% of patients in the “CRE colonization” group and the “CRE infection” group had a history of invasive procedures and antibiotic exposures such as carbapenems (P ≤ 0.05). It is worth noting that 14.6% of CRE carriers in the “CRE colonization” group had CRE infections after the detection of CRE. The death rates between the “CRE colonization” group and the “CRE infection” group were 2.1 and 2.9%, respectively. Multivariate analysis showed that the underlying diseases of gastrointestinal infections (OR = 0.044, 95% CI: 0.006–0.342, P = 0.003) and invasive procedures (OR = 180.081, 95% CI: 16.681–1,944.066, P < 0.001; Table 2) are independent risk factors for the colonization/infection of CRE isolates.
Among 96 isolates enrolled in the “CRE colonization” group, K. pneumoniae was mostly detected with a rate of 42.7% (41/96), followed by E. coli (34.4%, 33/96) and E. cloacae (15.6%, 15/96). Other species including four Klebsiella oxytoca, one Klebsiella aerogenes, one Proteus mirabilis, and one Citrobacter freundii were also observed in the intestinal tract of children, as shown in Figure 1A. Meanwhile, 70% (49/70) strains in the “CRE infection” group were K. pneumoniae, with 21.4% E. cloacae (15/70), 5.7% E. coli (4/70), 1.4% K. oxytoca (1/70), and 1.4% K. aerogenes (1/70; Figure 1B). K. pneumoniae is commonly isolated not only from children with CRE intestinal colonization but also from children with CRE nosocomial infection.
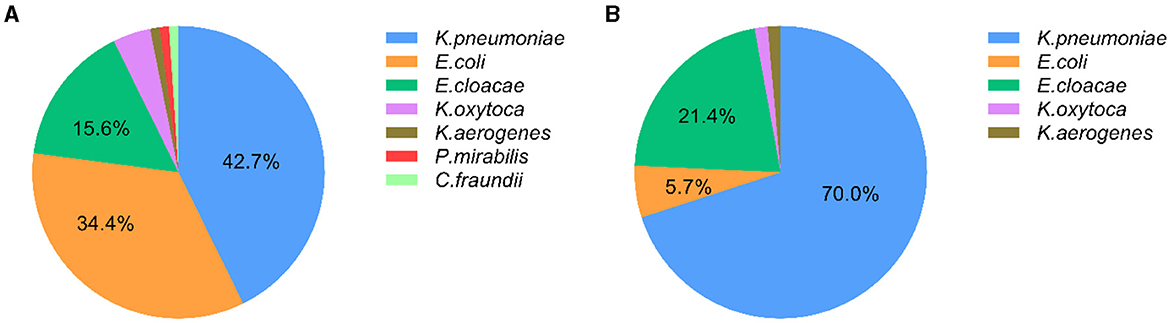
Figure 1. Distribution of carbapenem-resistant Enterobacteriales (CRE) in inpatients with CRE intestinal colonization and CRE infection. (A) The distribution of CRE strains from intestinal colonization; (B) The distribution of CRE strains from nosocomial infection.
In the “CRE colonization” group, 95.8% (92/96) of CRE isolates produced carbapenemases. NDM-1 was the major carbapenemase (43.8%), with KPC-2, NDM-5, and IMP being 22.9%, 19.8%, and 7.3%, respectively. KPC-2 and NDM-1 were observed in 53.7% (22/41) and 36.6% (15/41) of carbapenem-resistant K. pneumoniae (CRKPN) isolates, while NDM-5 and NDM-1 account for 48.5% (16/33) and 42.4% (13/33) of carbapenem-resistant E. coli (CRECO) strains, respectively. The NDM-1 was most frequently found in carbapenem-resistant E. cloacae (CRECL; Figure 2). However, in the “CRE infection” group, NDM-1, OXA-232, NDM-5, and KPC-2 account for 41.4%, 25.7%, 20%, and 12.9% of CRE isolates, respectively. The majority of OXA-232, NDM-5, NDM-1, and KPC-2 were detected in CRKPN strains, with four CRECO strains and fifteen CRECL strains containing NDM-1 carbapenemase (Figure 2).
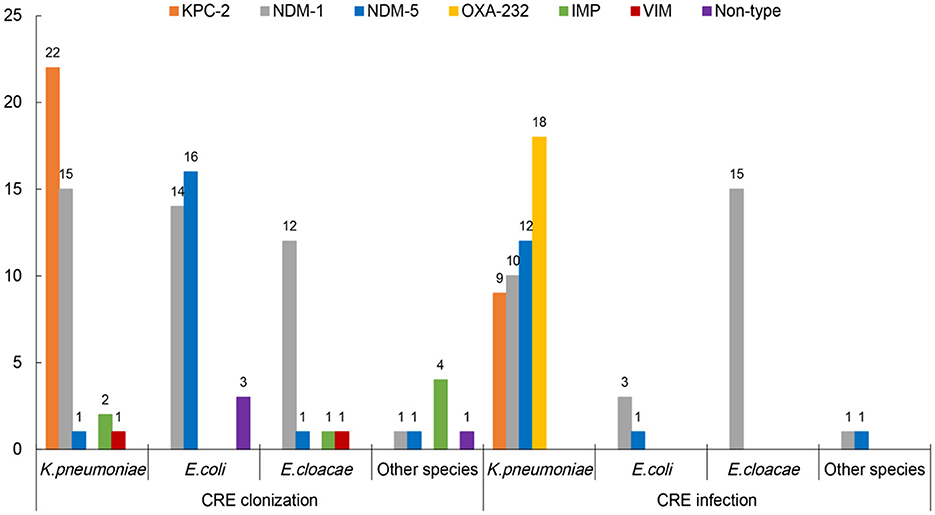
Figure 2. The carbapenemases of carbapenem-resistant Enterobacteriales (CRE) strains from intestinal colonization and nosocomial infection.
Antimicrobial susceptibility results demonstrated that all CRE isolates exhibited high resistance to cephalosporins and carbapenems, as well as ampicillin–sulbactam and piperacillin–tazobactam, while all strains were susceptible to tigecycline and colistin. However, the resistant pattern of different species from intestinal colonization and nosocomial infection varied, with CRKPN isolates being the most serious. As shown in Figure 3, CRKPN isolates in the “CRE infection” group showed higher resistance to amikacin and gentamicin than these species showed in the “CRE colonization” group. The in vitro activity of aztreonam, ciprofloxacin, levofloxacin, and trimethoprim–sulfamethoxazole against CRECO isolates in the “CRE infection” group was higher than these antimicrobial strains in the “CRE colonization” group. Of note, the resistance spectrum of KPC-2-producing isolates was different from the other type of carbapenemase, especially for ciprofloxacin, levofloxacin, amikacin, gentamicin, and trimethoprim–sulfamethoxazole (Figure 4).
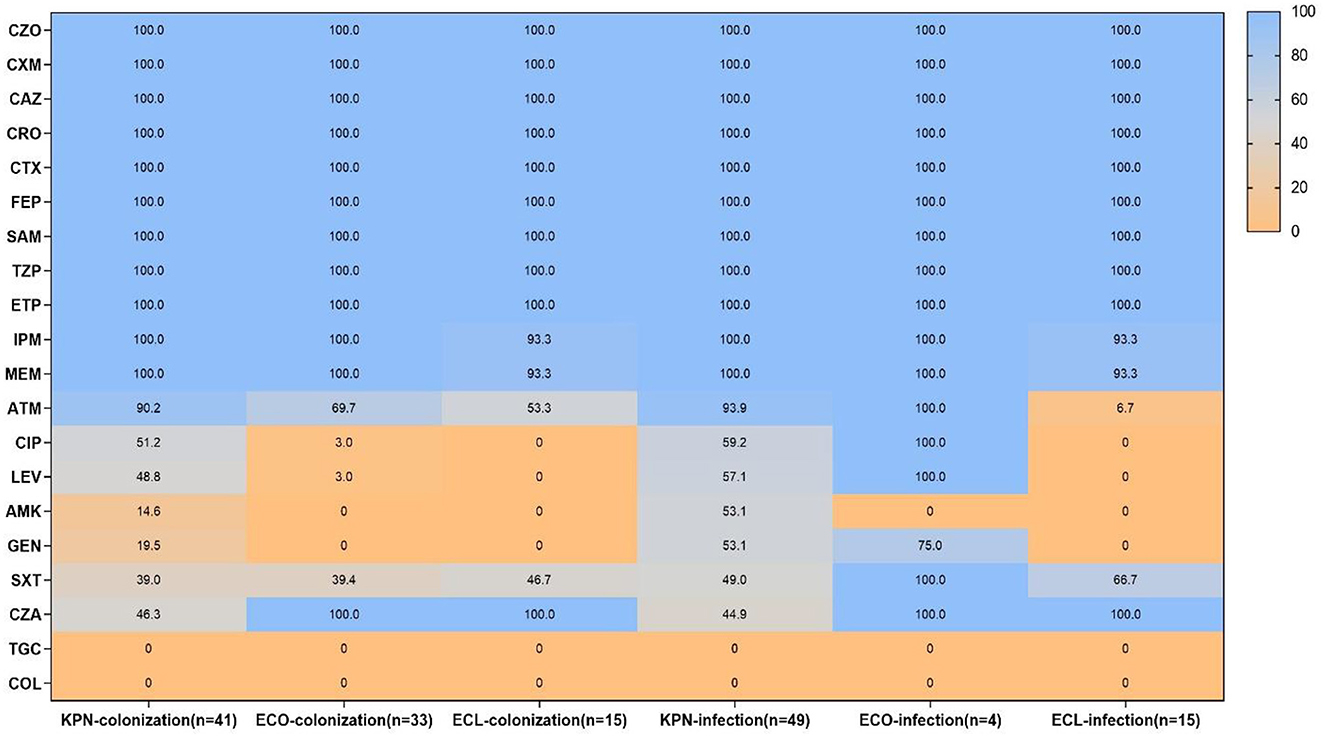
Figure 3. Antimicrobial resistance pattern of carbapenem-resistant Enterobacteriales (CRE) strains from intestinal colonization and nosocomial infection. KPN, Klebsiella pneumoniae; ECO, Escherichia coli; ECL, Enterobacter cloacae; CZO, cefazolin; CXM, cefuroxime; CAZ, ceftazidime; CRO, ceftriaxone; CTX, cefotaxime; FEP, cefepime; SAM, ampicillin-sulbactam; TZP, piperacillin–tazobactam; ETP, ertapenem; IPM, imipenem; MEM, meropenem; ATM, aztreonam; CIP, ciprofloxacin; LEV, levofloxacin; AMK, amikacin; GEN, gentamicin; SXT, trimethoprim-sulfamethoxazole; CZA, ceftazidime-avibactam; TGC, tigecycline; COL, colistin.
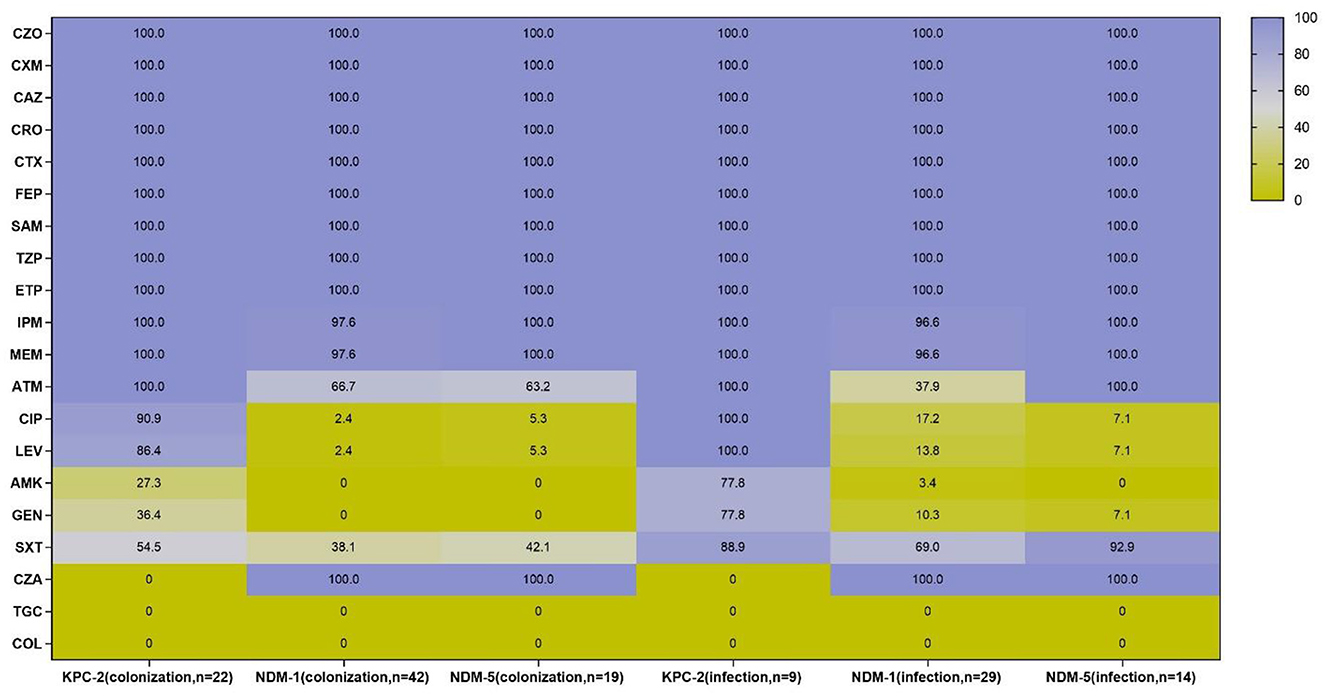
Figure 4. Antimicrobial resistance profiles of KPC-2, NDM-1, and NDM-5-producing strains from intestinal colonization and nosocomial infection. CZO, cefazolin, CXM, cefuroxime, CAZ, ceftazidime, CRO, ceftriaxone, CTX, cefotaxime, FEP, cefepime, SAM, ampicillin-sulbactam, TZP, piperacillin–tazobactam, ETP, ertapenem, IPM, imipenem, MEM, meropenem, ATM, aztreonam, CIP, ciprofloxacin, LEV, levofloxacin, AMK, amikacin, GEN, gentamicin, SXT, trimethoprim-sulfamethoxazole, CZA, ceftazidime-avibactam, TGC, tigecycline; COL, colistin.
According to the MLST analysis, CRE isolates were assigned to various ST types in different species (K. pneumoniae, 11 STs; E. coli, 11 STs; and E. cloacae, nine STs; Figure 5). Noticeably, ST11 (95.4%, 21/22) was the most predominant type in KPC-2-producing K. pneumoniae in the “CRE colonization” group, while ST15 and ST48 were frequently represented ST in OXA-232-producing K. pneumoniae and NDM-5-producing K. pneumoniae isolates in the “CRE infection” group, respectively. Moreover, the distribution of STs was relatively dispersive in carbapenemase-producing E. coli between the two groups, with 41.2% (7/17) of NDM-5 producers and 47.1% (8/17) of NDM-1 producers being typed as ST8 and ST1015, respectively. Additionally, the majority of CRECL strains harboring NDM-1 carbapenemase were assigned to ST45 in “CRE colonization” (83.3%, 10/12) and “CRE infection” groups (73.3%, 11/15). Phylogenetic analysis showed that partial CRE isolates from intestinal colonization and nosocomial infection were closely related, especially for ST11 KPC-2-producing CRKPN and ST45 NDM-1-producing CRECL.
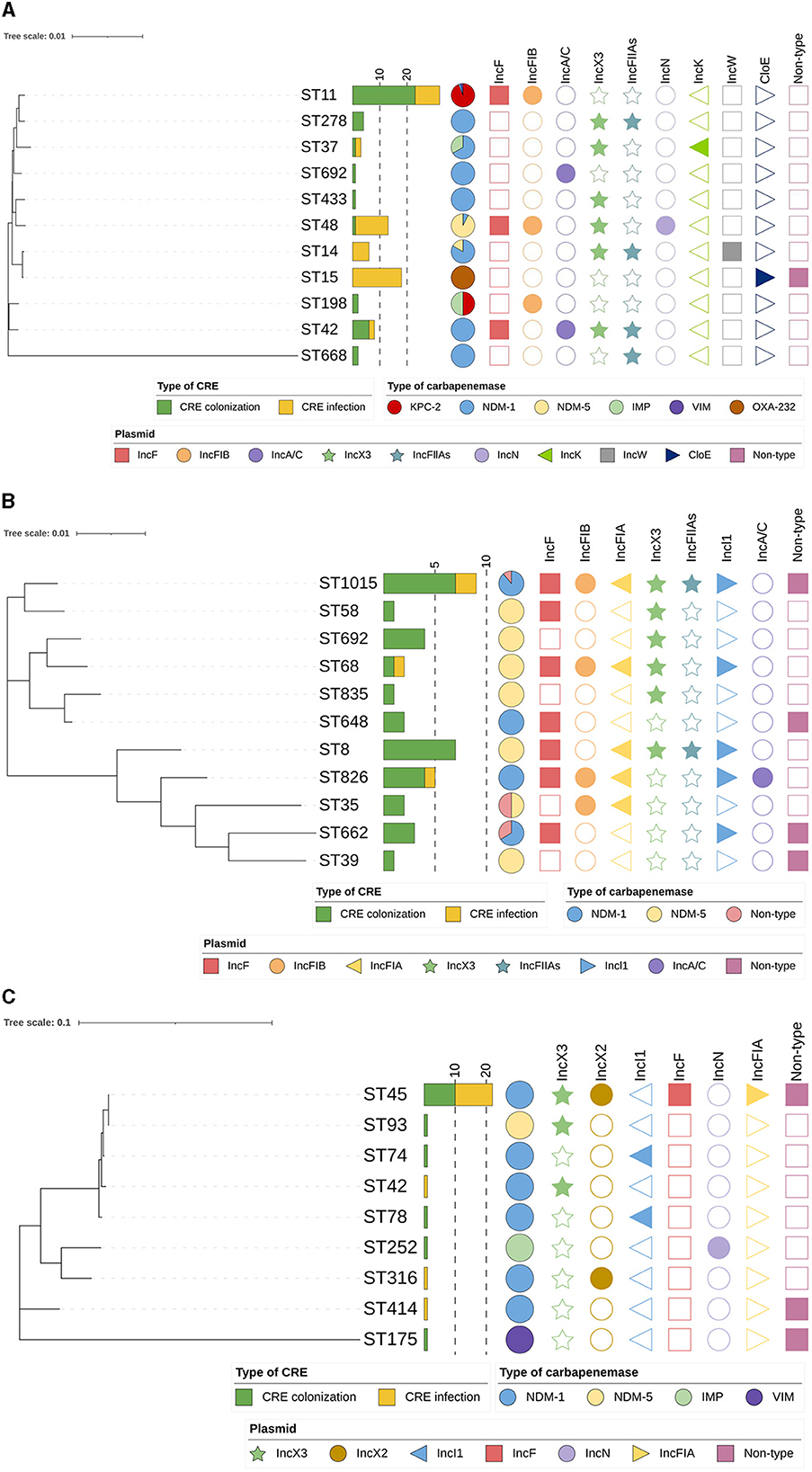
Figure 5. Phylogenetic tree of MLST data among CRE isolates analyzed by MEGA 7. (A) Phylogenetic tree of carbapenem-resistant Klebsiella pneumoniae; (B) Phylogenetic tree of carbapenem-resistant Escherichia coli; (C) Phylogenetic tree of carbapenem-resistant Enterobacter cloacae.
The plasmid replicon types of CRE isolates showed that isolates containing particular carbapenemase carried different types of plasmids (Figure 5). Of CRKPN isolates, most ST11 strains (81.3%, 26 out of 32) had plasmid replicon-type IncF together with IncFIB, which harbored the blaKPC − 2 gene. ST48 isolates (92.3%, 12 out of 13) and ST15 isolates (94.4%, 17 out of 18) carried IncX3 plasmid and ColE plasmid, respectively. Among CRECO isolates, IncX3 and IncA/C plasmids were relatively highly observed in several ST types. However, 54.5% of ST45 blaNDM − 1 carrying CRECL isolates possessed plasmid replicon-type IncX2.
In the “CRE colonization” group, class 1 integron intergrase intI1 was detected in 71 isolates (74.0%, 71 out of 96), with 6 different gene cassettes. The prevalence of complex class 1 integrons varied in different species. The most common gene cassette in CRKPN and CRECL isolates was dfrA12-orf-aadA2, while aar3-dfrA27 was the most detected gene among CRECO isolates. However, in the “CRE infection” group, 37 isolates (52.9%, 37/70) were observed positive with class 1 integron, and the gene cassettes dfrA12-orf-aadA2 and aadA1-aacA6 were common in CRKPN and CRECL isolates, respectively. No isolate was detected with class 2 integron intergrase intI2 and class 3 integron intergrase intI3.
Recently, the CRE isolates have been increasingly identified in children, and their horizontal spread among different species has contributed to a dilemma of no drugs available for clinical anti-infective therapy. Children are considered a special population, and once they are infected with CRE strains, it will greatly limit the choice of antimicrobial agents. Colonization of CRE strains in the gastrointestinal tract implies the potential for nosocomial transmission, and rapid active screening of CRE is essential for reducing and controlling CRE infections. Notably, the proportion of CRE colonization in hospitalized patients ranged from 15.5 to 52% and varied among different countries, different crowds, and different departments (17–20). This study revealed a carriage rate of 6.5%, which was lower than the previous literature. Furthermore, evaluating the special risk factors for the colonization/infection of CRE isolates is crucial for identifying high-risk patients. Consistent with other studies (21, 22), invasive procedures such as mechanical ventilation were identified as risk factors for colonization or infection with CRE. Additionally, long hospitalization stays and exposure to antibiotic use also increase the likelihood of cross-transmission of CRE isolates.
The diversity of CRE bacterial species was observed in this study, and the most common CRE isolates both in intestinal colonization and nosocomial infection was CRKPN, which is in agreement with the findings of previous studies (23, 24). Notably, the rapid increase of CRKPN in recent years is a critical threat to global health and causes high mortality (25). In addition, the proportion of CRECO in the “CRE colonization” group was higher than those in the “CRE infection” group, while the proportion of CRECL in the “CRE colonization” group was lower than those in the “CRE infection” group. It could be speculated that CRECO isolates were easier to detect in fecal samples, but it should be further verified by expanding the number of specimens. CRECL represents the third most common CRE followed by CREKPN and CRECL isolates, and the increased incidence of these isolates has constantly been reported worldwide, which requires more clinical attention (26, 27).
The prevalence of carbapenemase types among CRE strains has varied by regions: metallo-β-lactamases (MBLs) including NDM and VIM were dominant in isolates collected in Africa, the Middle East, and Asia/Pacific; KPC was mainly detected in Latin America and North America; and OXA-48-like carbapenemases were predominant in Europe (23). However, a study conducted in China displayed that the most prevalent carbapenemase gene was blaKPC − 2 among K. pneumoniae isolates from adult patients and blaNDM among E. coli isolates from children (28). In this study, among CRKPN isolates, KPC-2 producers were the most detected in the “CRE colonization” group, while OXA-232 producers were predominantly found in the “CRE infection” group. Notably, KPC-2-bearing ST11 CRKPN isolates were widely disseminated throughout China (28, 29) and had been reported to be colonized in the intestinal tract (30). In recent years, OXA-48-like carbapenemase, including OXA-162 and OXA-181, was increasingly prevalent in CRE strains (31, 32), and OXA-232 carbapenemase, a variant of OXA-181 with one amino acid substitution, was found in ST15 K. pneumoniae strains in Shanghai, China (33). Even though in this study, the OXA-48-like carbapenemase was not distributed in colonized CRKPN isolates, the clinicians should still be aware of the emergence of OXA-48-like carbapenemase in the gastrointestinal tract, which was predominantly distributed in fecal colonization (34). Interestingly, the frequency of NDM-carrying isolates has increased worldwide, and NDM-type has spread globally and is detected in various species. Among CRECO strains, NDM-type carbapenemase, including NDM-1 and NDM-5, was frequently detected in the “CRE colonization” group and the “CRE infection” group. Meanwhile, NDM-1-type carbapenemase was the most common among ST45 CRECL isolates both in the “CRE colonization” group and the “CRE infection” group, showing the high similarity of CRECL between colonization and infection. The trend of carbapenemase types in the colonization/infection of CRE isolates should be further concerned.
Mobile genetic elements containing integrons and plasmids highlight their role in the horizontal spread of carbapenemase genes among CRE isolates (35). Other resistant genes in mobile genetic elements also contribute to resistance to antibiotics and result in therapeutic failure of antibiotics. This study revealed that various plasmids were detected in CRE isolates, and no difference in the distribution of plasmids was found between the “CRE colonization” group and the “CRE infection” group. A high incidence of IncF-type plasmid in blaKPC − 2-carrying CRKPN isolates was observed, while IncX type, especially IncX3 and IncX2, was observed in most blaNDM-carrying CRE isolates. The presence of IncF and IncX3 plasmid-mediated the clonal dissemination of carbapenemase genes reported throughout the world (36, 37). Furthermore, integrons with classes 1–3 were known as multidrug-resistant integrons that may ultimately contribute to the wide spread of “superbugs” including CRE (38). A high frequency of class 1 integrons was identified in the “CRE colonization” group (74.0%) and the “CRE infection” group (52.9%), which is in agreement with an earlier report (16). Urgent surveillance of mobile genetic elements among CRE isolates should be conducted to control the spread of these pathogens.
Our study still has several limitations. First, this study was performed in one hospital, and the results were not suitable for other institutions. Therefore, multicenter design and comprehensive capture of the colonization/infection of CRE isolates should be further conducted. Second, even though we recommended CRE screening for all hospitalization patients, the active screening was still not frequently performed for all inpatients. The fecal sample was collected from hospitalized children who received the routine fecal culture testing. It might have led to the low CRE-positive rate. Finally, the sample size was relatively small and could not fully reflect the prevalence of intestinal colonization and nosocomial infection of CRE strains.
In conclusion, this study described the prevalence of intestinal colonization and nosocomial infection with CRE isolates in children, and KPC-2, NDM-1, NDM-5, and OXA-232 carbapenemases were the main types found in the “CRE colonization” group and the “CRE infection” group. Then, a few intestinal colonized CRE strains were probably linked to clinically infected strains, especially for ST11 KPC-2-producing CRKPN and ST45 NDM-1-producing CRECL. Therefore, in the case of the outbreak of these high-risk CRE clones from intestinal colonization, we should reinforce the surveillance of CRE from intestinal colonization and long-term active screening of rectal colonization of CRE isolates before admission should also be emphasized.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
This study was approved by the Ethics Committee of Shanghai Children's Hospital (Shanghai, China) [code: 2019R031-F01].
FP, WW, and HZ designed this study. FP, PC, YD, and FY conducted methodology, acquisition of data, and data analysis. FP wrote the first draft of the manuscript. WW and HZ critically revised and edited the manuscript. All authors read and approved the final manuscript.
This study was financially supported by the Shanghai Municipal Commission of Health and Family Planning (201940253), Shanghai Rising Stars of Medical Talents Youth Development Program (Youth Medical Talents—Clinical Laboratory Practitioner Program), research project of Shanghai Children's Hospital (2020YGZQ06), and the National Natural Science Foundation of China (Grant No. 32100716).
PC was employed by Suzhou Molarray Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2023.1251609/full#supplementary-material
CRE, carbapenem-resistant Enterobacteriales; KPC, Klebsiella pneumoniae carbapenemase; NDM, New Delhi metallo-β-lactamase; CRKPN, carbapenem-resistant K. pneumoniae; CRECO, carbapenem-resistant E. coli; CRECL, carbapenem-resistant E. cloacae; MLST, multilocus sequence typing.
1. World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics are Urgently Needed (2017). Available online at: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed February 2, 2023).
2. Salomao MC, Freire MP, Lazari CS, Cury AP, Rossi F, Segurado AAC, et al. Transmission of carbapenem-resistant enterobacterales in an overcrowded emergency department: controlling the spread to the hospital. Clin Infect Dis. (2023) 77:S46–52. doi: 10.1093/cid/ciad263
3. Zhang Y, Wang Q, Yin Y, Chen H, Jin L, Gu B, et al. Epidemiology of carbapenem-resistant enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother. (2018) 62:e01882-17. doi: 10.1128/AAC.01882-17
4. Zeng G, Pang Y, Zheng J, Zhuo C, Guo Y, Liang J, et al. Colonization with carbapenem-resistant Enterobacteriaceae contributes to unfavorable outcomes in end-stage liver disease patients. Antibiotics. (2022) 11:1667. doi: 10.3390/antibiotics11111667
5. Yee R, Fisher S, Bergman Y, Chambers KK, Tamma PD, Carroll KC, et al. Combined selective culture and molecular methods for the detection of carbapenem-resistant organisms from fecal specimens. Eur J Clin Microbiol Infect Dis. (2021) 40:2315–21. doi: 10.1007/s10096-021-04281-8
6. Mannathoko N, Mosepele M, Gross R, Smith RM, Alby K, Glaser L, et al. Colonization with extended-spectrum cephalosporin-resistant Enterobacterales (ESCrE) and carbapenem-resistant Enterobacterales (CRE) in healthcare and community settings in Botswana: an antibiotic resistance in communities and hospitals (ARCH) study. Int J Infect Dis. (2022) 122:313–20. doi: 10.1016/j.ijid.2022.06.004
7. Shu LB, Lu Q, Sun RH, Lin LQ, Sun QL, Hu J, et al. Prevalence and phenotypic characterization of carbapenem-resistant Klebsiella pneumoniae strains recovered from sputum and fecal samples of ICU patients in Zhejiang Province, China. Infect Drug Resist. (2019) 12:11–8. doi: 10.2147/IDR.S175823
8. Yuan W, Xu J, Guo L, Chen Y, Gu J, Zhang H, et al. Clinical risk factors and microbiological and intestinal characteristics of carbapenemase-producing enterobacteriaceae colonization and subsequent infection. Microbiol Spectr. (2022) 10:e0190621. doi: 10.1128/spectrum.01906-21
9. Magiorakos AP, Burns K, Rodriguez Bano J, Borg M, Daikos G, Dumpis U, et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant enterobacteriaceae into healthcare settings: guidance from the european centre for disease prevention and control. Antimicrob Resist Infect Control. (2017) 6:113. doi: 10.1186/s13756-017-0259-z
10. Zeng M, Xia J, Zong Z, Shi Y, Ni Y, Hu F, et al. Guidelines for the diagnosis, treatment, prevention and control of infections caused by carbapenem-resistant gram-negative Bacilli. J Microbiol Immunol Infect. (2023) 56:653–71. doi: 10.1016/j.jmii.2023.01.017
11. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. (2011) 70:119–23. doi: 10.1016/j.diagmicrobio.2010.12.002
12. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by Pcr-based replicon typing. J Microbiol Methods. (2005) 63:219–28. doi: 10.1016/j.mimet.2005.03.018
13. Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, et al. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. (2012) 68:43–50. doi: 10.1016/j.plasmid.2012.03.001
14. Potron A, Rondinaud E, Poirel L, Belmonte O, Boyer S, Camiade S, et al. Genetic and biochemical characterisation of Oxa-232, a carbapenem-hydrolysing class D beta-lactamase from Enterobacteriaceae. Int J Antimicrob Agents. (2013) 41:325–9. doi: 10.1016/j.ijantimicag.2012.11.007
15. Zheng F, Sun J, Cheng C, Rui Y. Molecular characteristics of carbapenem-resistant gram-negative bacteria in Southern China. Microb Drug Resist. (2015) 21:178–85. doi: 10.1089/mdr.2014.0085
16. Rui Y, Lu W, Li S, Cheng C, Sun J, Yang Q. Integrons and insertion sequence common region 1 (ISCR1) of carbapenem-non-susceptible gram-negative bacilli in fecal specimens from 5000 patients in southern China. Int J Antimicrob Agents. (2018) 52:571–6. doi: 10.1016/j.ijantimicag.2018.06.015
17. Wangchinda W, Thamlikitkul V, Watcharasuwanseree S, Tangkoskul T. Active surveillance for carbapenem-resistant Enterobacterales (CRE) colonization and clinical course of CRE colonization among hospitalized patients at a University Hospital in Thailand. Antibiotics. (2022) 11:1401. doi: 10.3390/antibiotics11101401
18. Tinelli M, Rossini A, Scudeller L, Zabzuni D, Errico G, Fogato E, et al. Dynamics of carbapenemase-producing enterobacterales intestinal colonisation in the elderly population after hospital discharge, Italy, 2018-2020. Int J Antimicrob Agents. (2022) 59:106594. doi: 10.1016/j.ijantimicag.2022.106594
19. Wu YL, Hu XQ, Wu DQ, Li RJ, Wang XP, Zhang J, et al. Prevalence and risk factors for colonisation and infection with carbapenem-resistant enterobacterales in intensive care units: a prospective multicentre study. Intensive Crit Care Nurs. (2023) 79:103491. doi: 10.1016/j.iccn.2023.103491
20. Tran DM, Larsson M, Olson L, Hoang NTB, Le NK, Khu DTK, et al. High prevalence of colonisation with carbapenem-resistant enterobacteriaceae among patients admitted to vietnamese hospitals: risk factors and burden of disease. J Infect. (2019) 79:115–22. doi: 10.1016/j.jinf.2019.05.013
21. Mijac V, Brkic S, Milic M, Siljic M, Cirkovic V, Perovic V, et al. Intestinal colonization of preterm neonates with carbapenem resistant Enterobacteria at hospital discharge. Antibiotics. (2023) 12:284. doi: 10.3390/antibiotics12020284
22. Chiotos K, Tamma PD, Flett KB, Naumann M, Karandikar MV, Bilker WB, et al. Multicenter study of the risk factors for colonization or infection with carbapenem-resistant Enterobacteriaceae in children. Antimicrob Agents Chemother. (2017) 61:e01440-17. doi: 10.1128/AAC.01440-17
23. Estabrook M, Muyldermans A, Sahm D, Pierard D, Stone G, Utt E. Epidemiology of resistance determinants identified in meropenem-nonsusceptible Enterobacterales collected as part of a global surveillance study, 2018 to 2019. Antimicrob Agents Chemother. (2023) 67:e0140622. doi: 10.1128/aac.01406-22
24. Pan F, Tian D, Wang B, Zhao W, Qin H, Zhang T, et al. Fecal carriage and molecular epidemiology of carbapenem-resistant enterobacteriaceae from outpatient children in Shanghai. BMC Infect Dis. (2019) 19:678. doi: 10.1186/s12879-019-4298-3
25. Pajand O, Rahimi H, Badmasti F, Gholami F, Alipour T, Darabi N, et al. Various arrangements of mobile genetic elements among Cc147 subpopulations of Klebsiella pneumoniae harboring Bla(Ndm-1): a comparative genomic analysis of carbapenem resistant strains. J Biomed Sci. (2023) 30:73. doi: 10.1186/s12929-023-00960-0
26. Zhu Z, Xie X, Yu H, Jia W, Shan B, Huang B, et al. Epidemiological characteristics and molecular features of carbapenem-resistant Enterobacter strains in China: a multicenter genomic study. Emerg Microbes Infect. (2023) 12:2148562. doi: 10.1080/22221751.2022.2148562
27. Gartzonika K, Politi L, Mavroidi A, Tsantes AG, Spanakis N, Priavali E, et al. High prevalence of clonally related St182 Ndm-1-producing Enterobacter cloacae complex clinical isolates in Greece. Int J Antimicrob Agents. (2023) 62:106837. doi: 10.1016/j.ijantimicag.2023.106837
28. Han R, Shi Q, Wu S, Yin D, Peng M, Dong D, et al. Dissemination of carbapenemases (Kpc, Ndm, Oxa-48, Imp, and Vim) among carbapenem-resistant enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. (2020) 10:314. doi: 10.3389/fcimb.2020.00314
29. Hu Y, Liu C, Shen Z, Zhou H, Cao J, Chen S, et al. Prevalence, Risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008-2018. Emerg Microbes Infect. (2020) 9:1771–9. doi: 10.1080/22221751.2020.1799721
30. Migliorini LB, Leaden L, de Sales RO, Correa NP, Marins MM, Koga PCM, et al. The gastrointestinal load of carbapenem-resistant enterobacteriacea is associated with the transition from colonization to infection by Klebsiella pneumoniae isolates harboring the Bla(Kpc) gene. Front Cell Infect Microbiol. (2022) 12:928578. doi: 10.3389/fcimb.2022.928578
31. Voulgari E, Poulou A, Dimitroulia E, Politi L, Ranellou K, Gennimata V, et al. Emergence of Oxa-162 carbapenemase- and Dha-1 Ampc cephalosporinase-producing sequence type 11 Klebsiella pneumoniae causing community-onset infection in Greece. Antimicrob Agents Chemother. (2015) 60:1862–4. doi: 10.1128/AAC.01514-15
32. Magobo RE, Ismail H, Lowe M, Strasheim W, Mogokotleng R, Perovic O, et al. Outbreak of Ndm-1- and Oxa-181-producing Klebsiella pneumoniae bloodstream infections in a neonatal unit, South Africa. Emerg Infect Dis. (2023) 29:1531–9. doi: 10.3201/eid2908.230484
33. Yin D, Dong D, Li K, Zhang L, Liang J, Yang Y, et al. Clonal dissemination of Oxa-232 carbapenemase-producing Klebsiella pneumoniae in neonates. Antimicrob Agents Chemother. (2017) 61:e00385-17. doi: 10.1128/AAC.00385-17
34. Liu C, Fang Y, Zeng Y, Lu J, Sun Q, Zhou H, et al. First report of oxa-181-producing Klebsiella pneumoniae in China. Infect Drug Resist. (2020) 13:995–8. doi: 10.2147/IDR.S237793
35. Shropshire WC, Konovalova A, McDaneld P, Gohel M, Strope B, Sahasrabhojane P, et al. Systematic analysis of mobile genetic elements mediating beta-lactamase gene amplification in noncarbapenemase-producing carbapenem-resistant enterobacterales bloodstream infections. mSystems. (2022) 7:e0047622. doi: 10.1128/msystems.00476-22
36. Zhou Y, Wu C, Wang B, Xu Y, Zhao H, Guo Y, et al. Characterization difference of typical Kl1, Kl2 and St11-Kl64 hypervirulent and carbapenem-resistant Klebsiella pneumoniae. Drug Resist Updat. (2023) 67:100918. doi: 10.1016/j.drup.2023.100918
37. Ariyoshi T, Aoki K, Kubota H, Sadamasu K, Ishii Y, Tateda K. Molecular characterization of Bla(Ndm)-carrying Incx3 plasmids: Bla(Ndm-16b) likely emerged from a mutation of Bla(Ndm-5) on Incx3 plasmid. Microbiol Spectr. (2022) 10:e0144922. doi: 10.1128/spectrum.01449-22
Keywords: carbapenem-resistant Enterobacteriales, intestinal colonization, nosocomial infection, KPC-2, NDM-1
Citation: Pan F, Chen P, Duan Y, Yu F, Weng W and Zhang H (2023) Prevalence of intestinal colonization and nosocomial infection with carbapenem-resistant Enterobacteriales in children: a retrospective study. Front. Public Health 11:1251609. doi: 10.3389/fpubh.2023.1251609
Received: 02 July 2023; Accepted: 09 October 2023;
Published: 23 November 2023.
Edited by:
J. Luis Espinoza, Kanazawa University, JapanReviewed by:
Mai Ha Thi Thao, Can Tho University of Medicine and Pharmacy, VietnamCopyright © 2023 Pan, Chen, Duan, Yu, Weng and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenhao Weng, d2VuZ3dlbmhhb0BzaGNoaWxkcmVuLmNvbS5jbg==; Hong Zhang, c2NoanlrMjAxNUAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.