- 1Fujian Provincial Center for Disease Control and Prevention, Fujian Provincial Key Laboratory of Zoonosis Research, Fuzhou, China
- 2Chinese Center for Disease Control and Prevention, Beijing, China
- 3Fuzhou Institute for Disease Control and Prevention of China Railway Nanchang Bureau Group Co., Ltd., Fuzhou, China
- 4National Health Commission Key Laboratory for Parasitic Disease Prevention and Control, Jiangsu Provincial Key Laboratory for Parasites and Vector Control Technology, Jiangsu Institute of Parasitic Diseases, Wuxi, China
- 5School of Public Health, Fujian Medical University, Fuzhou, China
Objective: HIV/AIDS remains a global public health problem, and understanding the structure of social networks of people living with HIV/AIDS is of great importance to unravel HIV transmission, propose precision control and reduce new infections. This study aimed to investigate the epidemiological characteristics of HIV transmission in Fujian province, southeastern China from 2015 to 2020 based on HIV molecular network.
Methods: Newly diagnosed, treatment-naive HIV/AIDS patients were randomly sampled from Fujian province in 2015 and 2020. Plasma was sampled for in-house genotyping resistance test, and HIV molecular network was created using the HIV-TRACE tool. Factors affecting the inclusion of variables in the HIV molecular network were identified using univariate and multivariate logistic regression analyses.
Results: A total of 1,714 eligible cases were finally recruited, including 806 cases in 2015 and 908 cases in 2020. The dominant HIV subtypes were CRF01_AE (41.7%) and CRF07_BC (38.3%) in 2015 and CRF07_BC (53. 3%) and CRF01_AE (29.1%) in 2020, and the prevalence of HIV drug resistance was 4.2% in 2015 and 5.3% in 2020. Sequences of CRF07_BC formed the largest HIV-1 transmission cluster at a genetic distance threshold of both 1.5 and 0.5%. Univariate and multivariate logistic regression analyses showed that ages of under 20 years and over 60 years, CRF07_BC subtype, Han ethnicity, sampling in 2015, absence of HIV drug resistance, married with spouse, sampling from three cities of Jinjiang, Nanping and Quanzhou resulted in higher proportions of sequences included in the HIV transmission molecular network at a genetic distance threshold of 1.5% (p < 0.05).
Conclusion: Our findings unravel the HIV molecular transmission network of newly diagnosed HIV/AIDS patients in Fujian province, southeastern China, which facilitates the understanding of HIV transmission patterns in the province.
Introduction
Following concerted efforts for over four decades, HIV remains a global public health problem (1). Currently, HIV transmission via blood, injection drug use or mother-to-child has been almost under effective control (2); however, there is no remarkable decline in the annual global number of HIV/AIDS cases (3). A recent study reported that the global age-standardized HIV/AIDS prevalence, death, and disability adjusted life years (DALYs) rate in 2019 increased by 307.26, 4.34, and 221.91 per 100,000 cases in relative to in 1990 (4). More importantly, resurgence of HIV/AIDS has been detected across the world (5, 6). Ending AIDS as a public health threat by 2030 is increasingly under a great challenge (7). Understanding the structure of social networks of people living with HIV/AIDS is of great importance to unravel HIV transmission, propose precision control and reduce new infections (8, 9).
Recently, characterization of HIV-1 transmission with the gene sequences from routine monitoring of HIV drug resistance based on molecular network analysis has been performed for identification of people at a high risk of transmission and implementation of immediate interventions (10–12). A recent molecular transmission network-based study identified young people outside of school as the main high-risk group for spreading HIV to students and the key group leading to the spread of HIV among young students in Guangxi, southern China (13). In addition, molecular transmission network analyses targeting older adults revealed that commercial sexual behaviors between older adult men and female sex workers led to the rapid spread of HIV in Zhejiang province, eastern China and Guangxi, southern China (14, 15). Based on molecular networks and spatial epidemiology, a network connectivity analysis showed that some geographically distant cities of Sichuan province had stronger transmission links than cities that were closer together, indicating that molecular monitoring is more effective to identify geographically dispersed propagation clusters (16). To investigate the epidemic characteristics of virus strains in various cities of Zhejiang province, a molecular network analysis was performed and Jiaxing City was found to have geographical clustering and a certain number of HIV/AIDS patients with high transmission risk (17). In the current study, a molecular transmission network analysis of HIV/AIDS was performed, based on monitoring of HIV drug resistance, aiming to rapid identification of individuals with high-risk transmission and at high risk of HIV infection in Fujian province, southeastern China, and to reduce new HIV infection with immediate targeted interventions.
Materials and methods
Study subjects
All newly diagnosed HIV/AIDS patients were sampled from Fujian province in 2015 and 2020, and all participants were naïve for antiretroviral therapy. Participants’ demographics, route of HIV infection, HIV subtype and clinical symptoms were collected from medical records and individual investigation forms.
In-house genotyping resistance test
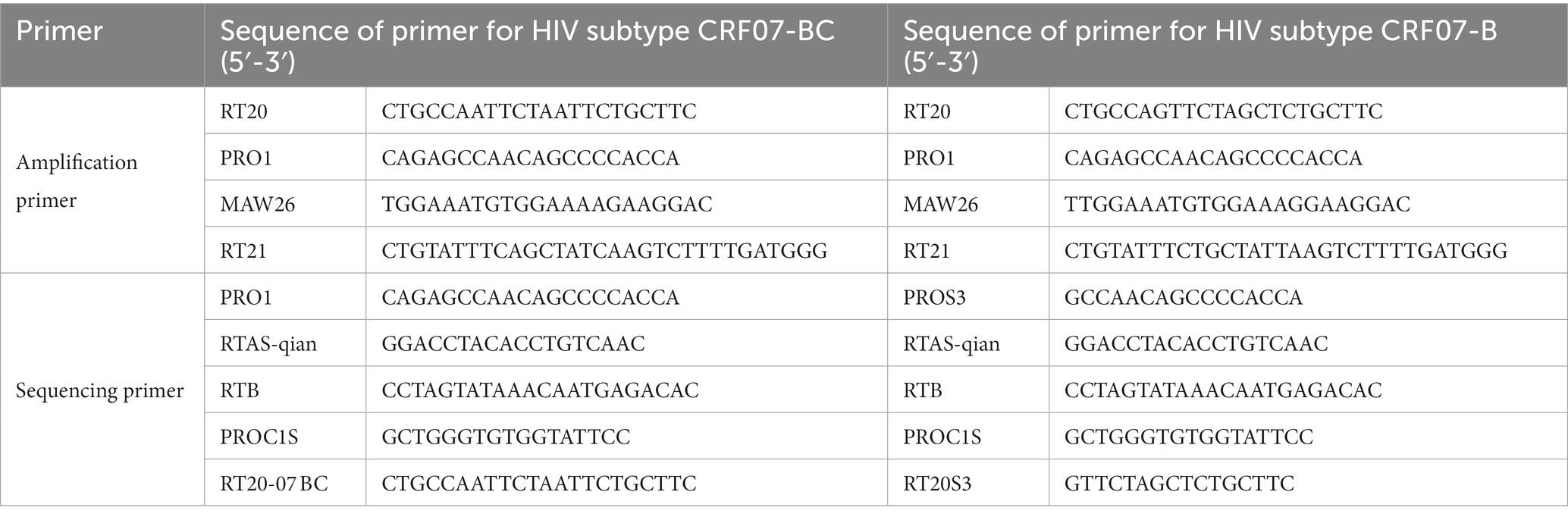
10 mL EDTA-anticoagulated blood was sampled from each participant, centrifuged within 6 h post-sampling, and the plasma was collected. Viral RNA was extracted from plasma using QIAamp Viral RNA Mini Kits (QIAGEN GmbH; Hilden, Germany), and subjected to reverse transcriptional amplification using in-house genotyping resistance test with the primers for implication of HIV subtypes CRF07-BC and B, which are effective to detect HIV-1 subtype A1, B, C, D, G, CRF01_AE, CRF02_AG, CRF06_cpx, CRF07_BC, CRF08_BC, CRF15_01B and other recombinant strains (18) (Table 1). The amplification products were purified and sequenced by Sangon Biotech (Shanghai, China). The fragments of amplified and sequenced target genes covered 4–99 amino acids in the protease region and 38 to 320 amino acids in the reverse transcriptase region.
Analysis of HIV drug resistance
The sequences were subjected to quality assessment, editing, assembly and mixed-base calling using the software ChromasPro version 2.4.1 and BioEdit version 7.2, and then uploaded to the Stanford HIV Drug Resistance Database (https://hivdb.stanford.edu/) for analysis of HIV drug resistance.
Generation of HIV molecular network
Due to the sampling time span of 5 years in this study, a molecular network was constructed using a 1.5% gene distance threshold, while a molecular network at a 0.5% gene distance threshold was created to highlight recent HIV infections and merge with the molecular network at a 1.5% gene distance threshold. All gene sequences with length of 1,000 bp and longer and mixed-base proportion of <5% were aligned and saved as.fas files, and all epidemiological data corresponding to each sequence were saved as.csv files. Pairwise genetic distances were estimated using the Tamura-Nei 93 (TN93) fast distance calculator, and HIV molecular network was created using the HIV-TRACE tool (19). All nodes in the HIV molecular network were assigned with epidemiological data, and molecular network maps were generated.
Statistical analysis
All data were entered into Microsoft Excel 2010 (Microsoft Corporation; Redmond, WA, United States). Differences of proportions were tested for statistical significance with chi-square test. Factors affecting the inclusion of variables in the HIV molecular network were identified using univariate and multivariate logistic regression models with the inclusion of sequence in the molecular cluster as a dependent variable and age, gender, molecular subtype, educational level, ethnicity, sampling time, route of HIV transmission, HIV drug resistance, marital status and sampling regions as independent variables. All statistical analyses were performed using the software SPSS version 22.0 (SPSS, Inc.; Chicago, IL, United States), and a p value of <0.05 was considered statistically significant.
Ethical statement
This study was approved by the Medical Ethics Review Committee of Fujian Provincial Center for Disease Control and Prevention (approval no. 2019–013). All procedures and performed following Declaration of Helsinki, as well as international and national laws, regulations and guidelines for human studies.
Results
Subject characteristics
A total of 1,858 treatment-naïve, newly diagnosed HIV/AIDS cases were enrolled, including 922 cases in 2015 and 936 cases in 2020. If samples with partial negative amplification and partial successful amplification with sequence length of 1,000 bp and shorter and/or mixed base of >5%, a total of 1,714 cases were included in the final analysis, including 806 cases in 2015 and 908 cases in 2020. Most cases were men (84.31%), and the highest number of cases was seen at ages of 20 to 29 years (28.35%). Sexual contact was the predominant route of HIV transmission (96.27%), and illiteracy and primary school was the main educational level (32.56%) (Table 2).
Trends in HIV subtype
Among totally 1,714 sequences, the predominant HIV subtypes were CRF07_BC (46.2%) and CRF01_AE (35.0%), and the dominant HIV subtypes were CRF01_AE (41.7%) and CRF07_BC (38.3%) in 2015 and CRF07_BC (53. 3%) and CRF01_AE (29.1%) in 2020. The proportion of the HIV CRF01_AE genotype reduced by 12.6% (χ2 = 5.5, p < 0.000,1) and the proportion of the HIV CRF07_BC genotype increased by 15% (χ2 = 23.9, p < 0.000,1) in 2020 relative to in 2015.
HIV drug resistance
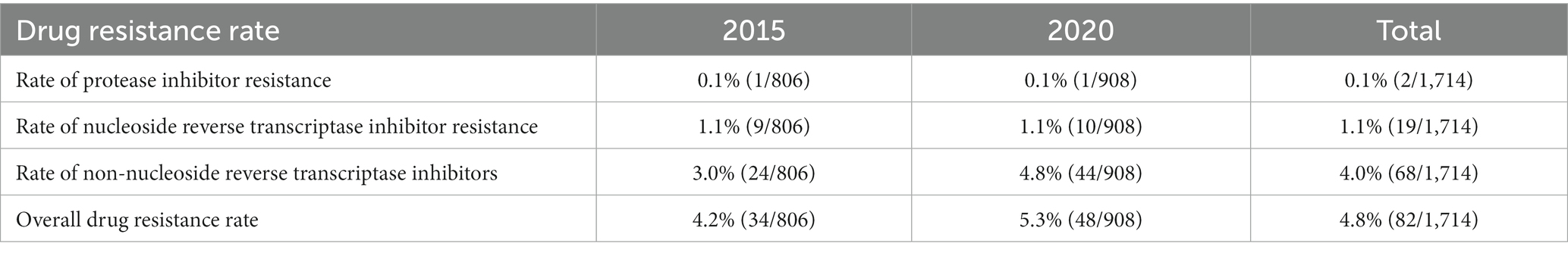
The prevalence of HIV drug resistance was 4.2% in 2015 and 5.3% in 2020, and a higher prevalence rate of non-nucleoside reverse transcriptase inhibitors (NNRTIs) resistance was seen than that of protease inhibitor and nucleoside reverse transcriptase inhibitor (NRTI) resistance in both 2015 and 2020 (Table 3).
HIV molecular network characteristics
A genetic distance threshold of 1.5% was used to create the HIV molecular network, and a HIV molecular network was generated at a genetic distance threshold of 0.5% for identifying recent HIV infections, which was merged with the HIV molecular network created at a genetic distance threshold of 1.5%. A total of 50.6% sequences were included in the HIV molecular network at a genetic distance threshold of 1.5%, which generated 162 HIV transmission clusters, and sequences of CRF07_BC formed the largest HIV-1 transmission cluster, which contained 272 nodes. Sequences of CRF01_AE formed the second largest HIV-1 transmission cluster, which contained 30 nodes, and there were 30 HIV transmission clusters that contained more than 4 nodes. At a genetic distance threshold of 0.5%, a total of 17.4% sequences were included in the HIV molecular network. Sequences of CRF07_BC formed the largest HIV-1 transmission cluster, which contained 11 nodes, and there were 12 HIV transmission clusters that contained more than 4 nodes (Figure 1).
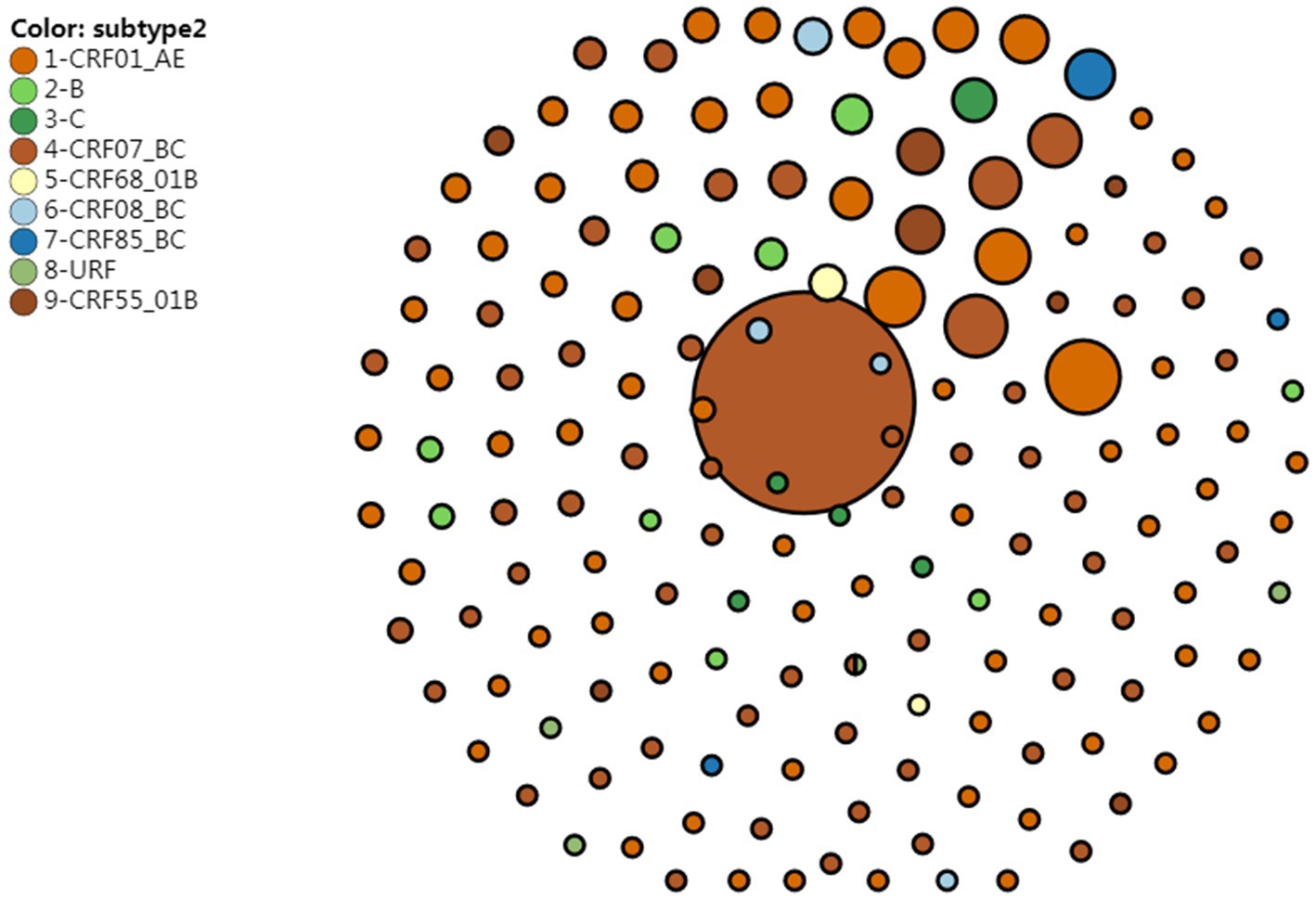
Figure 1. HIV molecular network reveals HIV transmission clusters at a genetic distance threshold of 0.5 and 1.5%.
Univariate analysis showed significant differences in the proportion of sequences included in the HIV transmission molecular network in terms of age groups, HIV subtype, emergence of HIV drug resistance and marital status at a 1.5% gene distance threshold (p < 0.05). Multivariate logistic regression analysis showed that ages of under 20 years and over 60 years, CRF07_BC subtype, Han ethnicity, sampling in 2015, absence of HIV drug resistance, married with spouse, sampling from JJ, NP, and QZ resulted in higher proportions of sequences included in the HIV transmission molecular network (p < 0.05) (Table 4).
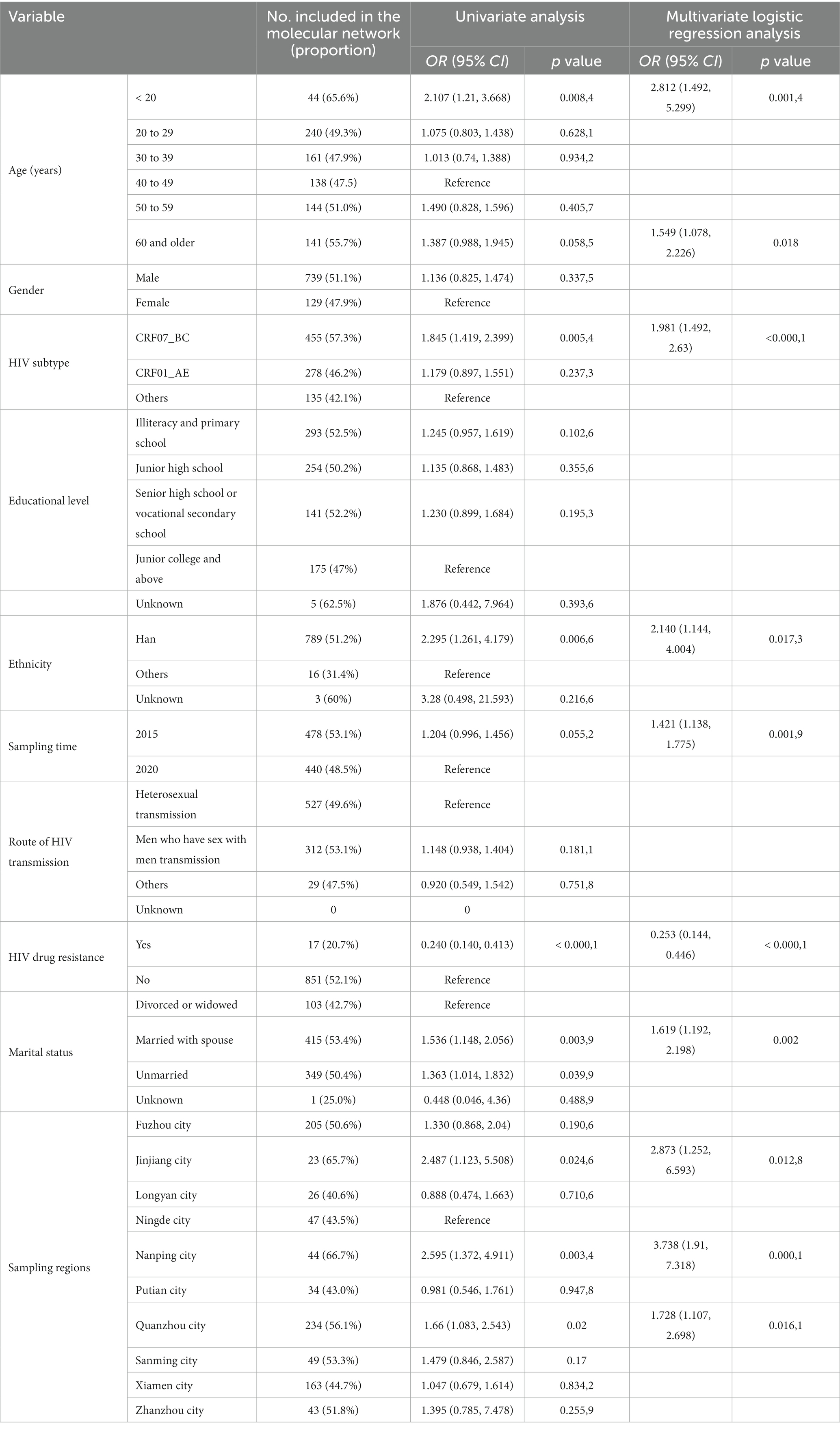
Table 4. Univariate and multivariate analyses of factors affecting the inclusion of sequences in the HIV molecular network.
There were three transmission clusters (11, 31, and 75) including sequences with DR mutations for HIV drug resistance, in the created HIV molecular network. These three clusters all carried E138G and V189E, which led to NNRTIs resistance. These three clusters, which contained 11, 4, and 2 nodes, were all formed by CRF55_01B subtype, and patients carrying these sequences were all men and predominantly single (Figure 2).
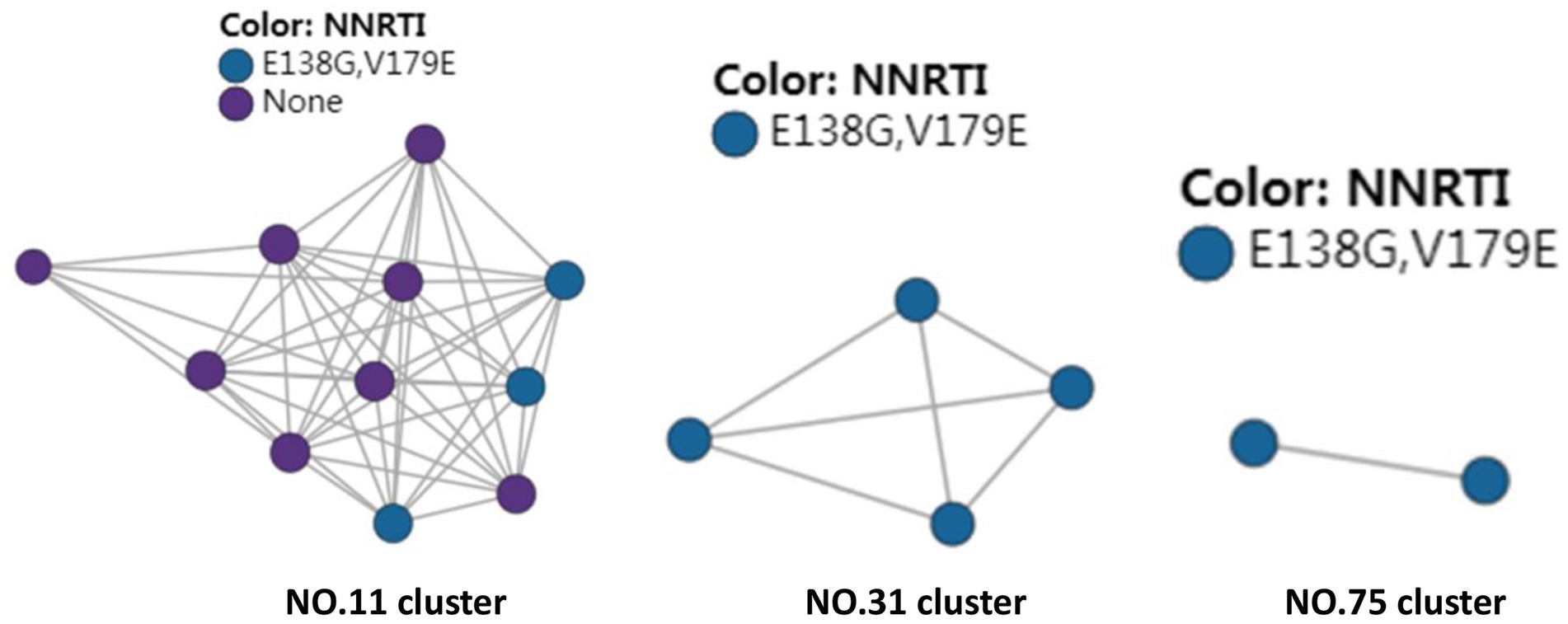
Figure 2. HIV molecular network identifies transmission clusters responsible for HIV drug resistance.
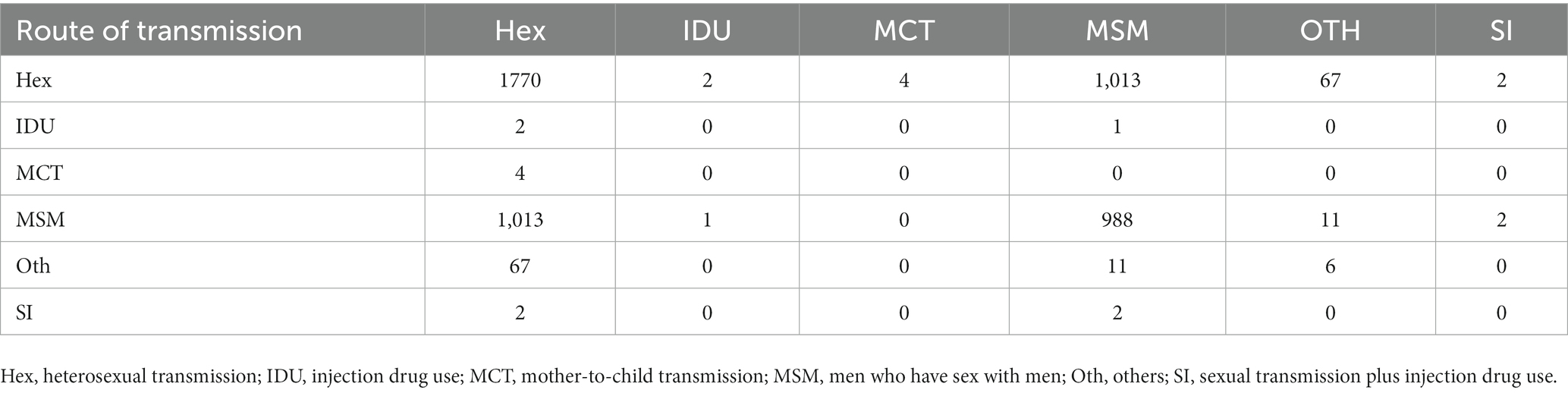
At a genetic distance at 1.5% gene distance threshold, there were 60.7% nodes with heterosexual transmission as the route of HIV transmission (527/868), including 402 males and 125 females and 35.9% with MSM as the route of HIV transmission (312/868) (Table 5), and there were 27% nodes with sampling sites in Quanzhou city (234/868), 23.6% in Fuzhou city (205/868) and 18.8% in Xiamen City (163/868) (Figure 3). A pie chart was created to display intraregional molecular networking, and the major connections in Nanping city (an inland region) came from local regions, suggesting that intraregional HIV transmission was predominant, while the HIV transmission in Longyan city (an inland region) was closely connected with three cities of Quanzhou, Fuzhou and Xiamen, and the HIV transmission in Sanming and Ningde cities was strongly connected with Fuzhou city. Molecular networking showed a high proportion of connections between Fuzhou city (capital of Fujian province) and cities along the coastal regions in Fujian province, which may be attributed to spatial proximity and convenient transportation. In addition, molecular networking revealed that the connections between Quanzhou city and other cities in Fujian province gradually reduced with the spatial distance, which may be attributed to spatial proximity (Figure 4).
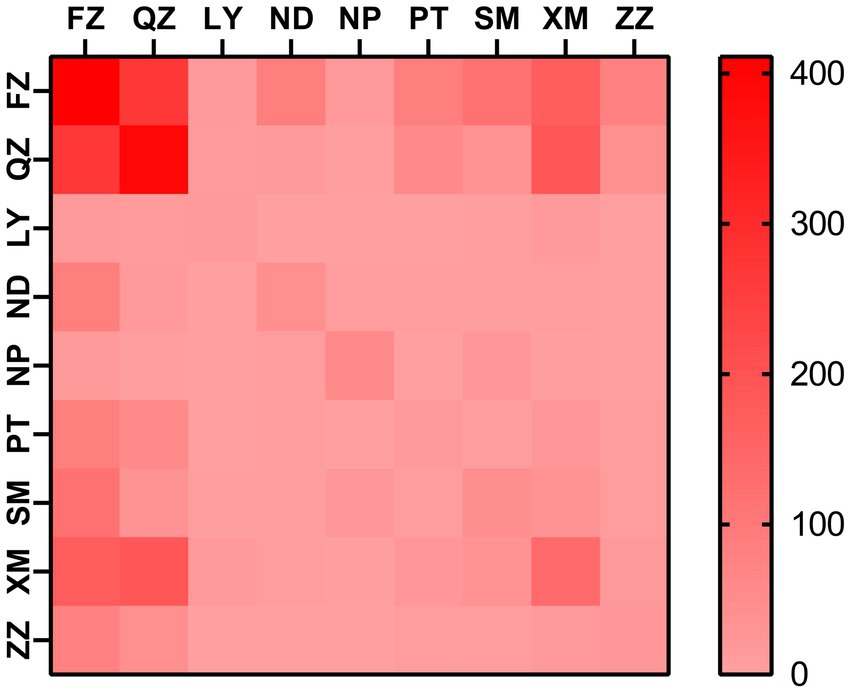
Figure 3. Heatmap for intraregional HIV molecular transmission network in Fujian province. FZ, Fuzhou city; LY, Longyan city; ND, Ningde city; NP, Nanping city; PT, Putian city; QZ, Quanzhou city; SM, Sanming city; XM, Xiamen city; ZZ, Zhangzhou city.
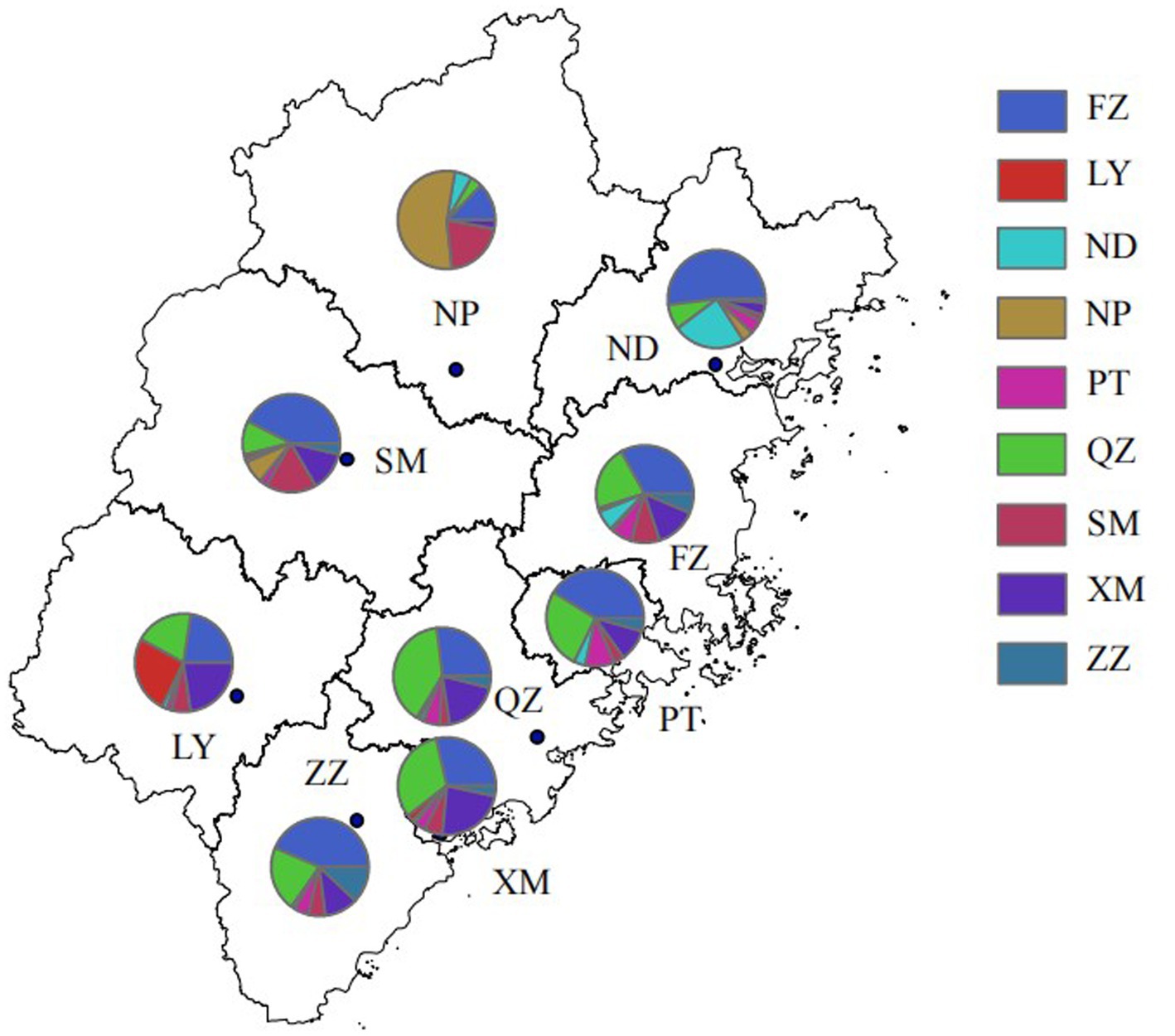
Figure 4. Intraregional HIV molecular network in Fujian province. FZ, Fuzhou city; LY, Longyan city; ND, Ningde city; NP, Nanping city; PT, Putian city; QZ, Quanzhou city; SM, Sanming city; XM, Xiamen city; ZZ, Zhangzhou city.
Discussion
Social network of HIV-infected individuals is of critical importance for HIV transmission (15–17). Therefore, understanding the social network of HIV-infected individuals is of great significance to unravel HIV transmission patterns, propose targeted and precision interventions and reduce new infections (20–22). Because of latent HIV infection (23), timing from HIV infection to diagnosis (24), a low possibility of acquiring HIV infection from a single high-risk behaviors (25), and difficulty in acquiring accurate sexual or drug use behaviors through traditional epidemiological surveys (26), conventional epidemiological approaches, such as history of infection contacts and traceability investigation, are difficult for analysis of HIV transmission networks, while molecular epidemiology is effective to supplement the shortcomings of conventional epidemiology and social network analysis (27, 28).
The development of molecular epidemiological approaches and highly efficient replication and low-fidelity reverse transcription of HIV-1 lead to high similarity but non-identity between progeny and parental viruses (29). Therefore, estimating the genetic distance between sequences from different infected individuals may be useful to identify the potential transmission pattern among HIV-infected cases that sequences corresponded to (30). It has been reported that HIV transmission network analysis using gene sequences from routine monitoring of HIV drug resistance based on molecular network is effective to guide the formulation of the AIDS control strategy (10–12), and HIV transmission network analysis has been included as one of the national control strategies for new HIV infections (31).
Currently, the common molecular network analysis approaches include pairwise genetic distance estimation, phylogenetic reconstruction alone and in combination, each approach has its advantages and disadvantages. The Cluster Picker tool is effective to identify the cluster that contains only two sequences, while the HIV-TRACE tool appears to identify large or small clusters. In this study, the genetic distance in the HIV molecular network was estimated with the TN93 fast distance calculator using the HIV-TRACE tool (32).
This is the first study to create the HIV molecular transmission network of newly diagnosed HIV/AIDS patients in Fujian province, southeastern China. At a genetic distance threshold of 1.5%, a total of 162 HIV transmission clusters were generated, with 30 transmission clusters containing more than 4 nodes, and sequences of CRF07_BC formed the largest HIV-1 transmission cluster, which contained 272 nodes. In addition, the transmission cluster derived from CRF07_BC contained 67 nodes at a genetic distance threshold of 0.5%, indicating the ongoing transmission of HIV CRF07_BC subtype. Univariate analysis showed significant differences in the proportion of sequences included in the HIV transmission molecular network in terms of age groups, HIV subtype, emergence of HIV drug resistance and marital status (p < 0.05). Multivariate logistic regression analysis showed that ages of under 20 years and over 60 years, CRF07_BC subtype, Han ethnicity, sampling in 2015, absence of HIV drug resistance, married with spouse, sampling from JJ, NP, and QZ resulted in higher proportions of sequences included in the HIV transmission molecular network (p < 0.05). Further studies to investigate the causes responsible for the high proportion of inclusion of sequences from HIV-infected individuals are needed for targeted interventions, so as to reduce HIV transmission and new infections.
There has been an increase in the number and proportion of newly reported HIV/AIDS cases among individuals at ages of 60 years and older in Fujian province from 2012 to 2022. Our findings showed the highest proportion of inclusion in the HIV molecular network among patients at ages of 60 years and older (55.7%), which is consistent with the epidemiological characteristics of HIV/AIDS in the province. Intensified epidemiological investigations and molecular network dynamic monitoring, timely identification of active transmission networks and high-risk carriers, and targeted interventions are required targeting these high-risk populations to reduce the HIV transmission risk and new infections.
The prevalence of HIV infection has been increasing in MSM over years (33). The prevalence of HIV-1 infection has been maintained at a high level among MSM in Fujian province. During the investigation and tracking of the transmission process of HIV-1 in MSM, conventional epidemiological tools based on questionnaire surveys and peer tracking may have various biases, resulting in low credibility of conclusions and subsequent unfavorable follow-up and behavioral interventions. Recently, HIV-1 molecular transmission networks using the viral gene sequences of infected individuals has gradually been employed to accurately identify potential transmission and determine active transmission networks, thus facilitating targeted intervention measures (34–38).
In this study, molecular network analysis showed more connections between the route of MSM and heterosexual transmission than the route of MSM, indicating the strong association between two routes of heterosexual transmission and MSM. in most of the transmission clusters, different routes of HIV transmission appear in the same network, and in different routes of HIV transmission, there is a “key person” who is a man who has sex with men but reports heterosexuality. During routine HIV/AIDS epidemiology surveys, data pertaining to sexual behaviors acquired may be inaccurate, which may affect the formulation of HIV/AIDS control measures. Therefore, by creating an HIV molecular network, we can find these errors and correct them through further investigations, so as to facilitate the understanding of the true epidemiological characteristics of HIV/AIDS in Fujian Province and provide insights into HIV/AIDS prevention and control.
In addition, the highest numbers of connections were observed between samples sites from Fuzhou city and Quanzhou city, indicating the important role of Fuzhou and Quanzhou cities in HIV transmission across Fujian province, which may be attributed to convenient transportation and geographical proximity.
The transmitted HIV drug resistance has shown a tendency toward a rise in Fujian province since 2008, and a more remarkable increase has been found since 2012. The overall prevalence of transmitted drug resistance was 4.4% among treatment-naive HIV-1-infected patients in Shaanxi province, northwestern China from 2003 to 2013, and the prevalence became stable between 2009 and 2013 (p = 0.982) (39). The overall prevalence of transmitted HIV drug resistance was 3.42% among newly diagnosed HIV-1 individuals in Jiangsu province, China during the period from 2009 through 2011 (40). In addition, the proportion of transmitted drug resistance was 4.4% among recently infected HIV-positive individuals in Hong Kong from 2007 to 2010 (41) and the prevalence of transmitted drug resistance of HIV-1 strains was 11.1% among individuals attending voluntary counseling and testing in Taiwan from 2006 to 2014 (42). In the current study, the overall prevalence of HIV drug resistance was 4.8%, and the prevalence was 4.2% in 2015 and 5.3% in 2020. In addition, molecular network analysis identified three transmission clusters (Cluster 11, 31 and 75), and these three clusters all carried E138G and V189E, which led to NNRTIs resistance. Epidemiological data showed that these three clusters were all formed by CRF55_01B subtype, and patients carrying these sequences were all men and predominantly single, indicating that the transmission of HIV drug resistance mainly occurs in MSM. Further studies to unravel the underlying mechanisms, screen high-risk population for HIV and implement targeted interventions are needed to avoid the spread of HIV drug resistance.
The current study has some limitations. First, HIV-uninfected individuals were not included. This study aimed to unravel the molecular transmission network of newly diagnosed HIV/AIDS patients in Fujian province, southeastern China. HIV molecular transmission network consists of a group of HIV-infected individuals with potential transmission relationships, and the HIV that individuals infect has a genetic similarity (43). Therefore, HIV-uninfected individuals at a high risk of infection were not recruited in the present study. Nevertheless, HIV-uninfected individuals at a potential risk of infection will be included in the creation of the HIV risk network based on field epidemiological surveys after construction of the HIV molecular network. Second, the molecular clusters generated from the HIV molecular network are only effective to identify the transmission relationship among individuals included in the molecular clusters, but fail to unravel the transmission of virus (44). Further field epidemiological surveys are required to identify potential HIV spreader.
In summary, we, for the first time, create the HIV molecular transmission network of newly diagnosed HIV/AIDS patients in Fujian province, southeastern China, which facilitates the understanding of HIV transmission patterns in the province. A comprehensive analysis of the transmission clusters in the HIV molecular network, demographics, epidemiological features and laboratory testing facilitates the formulation of targeted interventions for HIV/AIDS, so as to reduce new HIV infections.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The studies involving humans were approved by the Fujian Provincial Center for Disease Control and Prevention. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This study was supported by grants from the Fujian Provincial Health Technology Project (grant no. 2019-CX-9), the Pilot Project of Fujian Provincial Department of Science and Technology (grant no. 2019Y0053 and 2021Y0047).
Conflict of interest
LL was employed by Fuzhou Institute for Disease Control and Prevention of China Railway Nanchang Bureau Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Beyrer, C. A pandemic anniversary: 40 years of HIV/AIDS. Lancet. (2021) 397:2142–3. doi: 10.1016/S0140-6736(21)01167-3
2. De Cock, KM, Jaffe, HW, and Curran, JW. Reflections on 40 years of AIDS. Emerg Infect Dis. (2021) 27:1553–60. doi: 10.3201/eid2706.210284
3. WHO. Global progress report on HIV, viral hepatitis and sexually transmitted infections. Geneva: The World Health Organization (2021).
4. Tian, X, Chen, J, Wang, X, Xie, Y, Zhang, X, Han, D, et al. Global, regional, and national HIV/AIDS disease burden levels and trends in 1990-2019: a systematic analysis for the global burden of disease 2019 study. Front Pub Health. (2023) 11:1068664. doi: 10.3389/fpubh.2023.1068664
5. Govender, RD, Hashim, MJ, Khan, MA, Mustafa, H, and Khan, G. Global epidemiology of HIV/AIDS: a resurgence in North America and Europe. J Epidemiol Glob Health. (2021) 11:296–301. doi: 10.2991/jegh.k.210621.001
6. He, N. Research Progress in the epidemiology of HIV/AIDS in China. China CDC Wkly. (2021) 3:1022–30. doi: 10.46234/ccdcw2021.249
7. De Lay, PR, Benzaken, A, Karim, QA, Aliyu, S, Amole, C, Ayala, G, et al. Ending AIDS as a public health threat by 2030: time to reset targets for 2025. PLoS Med. (2021) 18:e1003649. doi: 10.1371/journal.pmed.1003649
8. Pagkas-Bather, J, Young, LE, Chen, YT, and Schneider, JA. Social network interventions for HIV transmission elimination. Curr HIV/AIDS Rep. (2020) 17:450–7. doi: 10.1007/s11904-020-00524-z
9. Jing, F, Zhang, Q, Tang, W, Wang, JZ, Lau, JT, and Li, X. Reconstructing the social network of HIV key populations from locally observed information. AIDS Care. (2021) 35:1243–50. doi: 10.1080/09540121.2021.1883514
10. Pines, HA, Wertheim, JO, Liu, L, Garfein, RS, Little, SJ, and Karris, MY. Concurrency and HIV transmission network characteristics among MSM with recent HIV infection. AIDS. (2016) 30:2875–83. doi: 10.1097/QAD.0000000000001256
11. Zhao, B, Song, W, Kang, M, Dong, X, Li, X, Wang, L, et al. Molecular network analysis reveals transmission of HIV-1 drug-resistant strains among newly diagnosed HIV-1 infections in a moderately HIV Endemic City in China. Front Microbiol. (2022) 12:797771. doi: 10.3389/fmicb.2021.797771
12. Dávila-Conn, V, García-Morales, C, Matías-Florentino, M, López-Ortiz, E, Paz-Juárez, HE, Beristain-Barreda, Á, et al. Characteristics and growth of the genetic HIV transmission network of Mexico City during 2020. J Int AIDS Soc. (2021) 24:e25836. doi: 10.1002/jia2.25836
13. Jiang, H, Lan, G, Zhu, Q, Liang, S, Li, J, Feng, Y, et al. Nonstudent young men put students at high risk of HIV acquisition in Guangxi, China: a phylogenetic analysis of surveillance data. Open Forum Infect Dis. (2022) 9:ofac042. doi: 10.1093/ofid/ofac042
14. Zhang, F, Yang, Y, Liang, N, Liang, H, Chen, Y, Lin, Z, et al. Transmission network and phylogenetic analysis reveal older male-centered transmission of CRF01_AE and CRF07_BC in Guangxi, China. Emerg Microbes Infect. (2023) 12:2147023. doi: 10.1080/22221751.2022.2147023
15. Jiang, J, Fan, Q, Zhang, J, Luo, M, Ding, X, Pan, X, et al. A geographic hotspot and emerging transmission cluster of the HIV-1 epidemic among older adults in a rural area of eastern China. AIDS Res Hum Retrovir. (2020) 36:712–20. doi: 10.1089/aid.2019.0293
16. Yuan, D, Yu, B, Liang, S, Fei, T, Tang, H, Kang, R, et al. HIV-1 genetic transmission networks among people living with HIV/AIDS in Sichuan, China: a genomic and spatial epidemiological analysis. Lancet Reg Health West Pac. (2021) 18:100318 doi: 10.1016/j.lanwpc.2021.100318
17. Ge, R, Zhu, GY, Pan, XH, Fan, Q, Chen, ZW, Zhang, JF, et al. Analysis on the HIV-1 molecular transmission characteristics of newly confirmed HIV/AIDS in Jiaxing city, 2017-2018. Chin J Epidemiol. (2021) 42:2118–24. doi: 10.3760/cma.j.cn112338-20210811-00631
18. Chen, JH, Wong, KH, Chan, K, Lam, HY, Lee, SS, Li, P, et al. Evaluation of an in-house genotyping resistance test for HIV-1 drug resistance interpretation and genotyping. J Clin Virol. (2007) 39:125–31. doi: 10.1016/j.jcv.2007.03.008
19. Kosakovsky Pond, SL, Weaver, S, Leigh Brown, AJ, and Wertheim, JO. HIV-TRACE (TRAnsmission cluster engine): a tool for large scale molecular epidemiology of HIV-1 and other rapidly evolving pathogens. Mol Biol Evol. (2018) 35:1812–9. doi: 10.1093/molbev/msy016
20. Wertheim, JO, Kosakovsky Pond, SL, Forgione, LA, Mehta, SR, Murrell, B, Shah, S, et al. Social and genetic networks of HIV-1 transmission in new York City. PLoS Pathog. (2017) 13:e1006000. doi: 10.1371/journal.ppat.1006000
21. Rothenberg, RB, Potterat, JJ, Woodhouse, DE, Muth, SQ, Darrow, WW, and Klovdahl, AS. Social network dynamics and HIV transmission. AIDS. (1998) 12:1529–36. doi: 10.1097/00002030-199812000-00016
22. Koku, E, and Felsher, M. The effect of social networks and social constructions on HIV risk perceptions. AIDS Behav. (2020) 24:206–21. doi: 10.1007/s10461-019-02637-y
23. Dufour, C, Gantner, P, Fromentin, R, and Chomont, N. The multifaceted nature of HIV latency. J Clin Invest. (2020) 130:3381–90. doi: 10.1172/JCI136227
24. Crepaz, N, Song, R, Lyss, SB, and Hall, HI. Estimated time from HIV infection to diagnosis and diagnosis to first viral suppression during 2014–2018. AIDS. (2021) 35:2181–90. doi: 10.1097/QAD.0000000000003008
25. Dosekun, O, and Fox, J. An overview of the relative risks of different sexual behaviours on HIV transmission. Curr Opin HIV AIDS. (2010) 5:291–7. doi: 10.1097/COH.0b013e32833a88a3
26. Anderson, JE, McCormick, L, and Fichtner, R. Factors associated with self-reported STDs: data from a national survey. Sex Transm Dis. (1994) 21:303–8. doi: 10.1097/00007435-199411000-00002
27. Vasylyeva, TI, Friedman, SR, Paraskevis, D, and Magiorkinis, G. Integrating molecular epidemiology and social network analysis to study infectious diseases: towards a socio-molecular era for public health. Infect Genet Evol. (2016) 46:248–55. doi: 10.1016/j.meegid.2016.05.042
28. Boccia, S, Pasquarella, C, Colotto, M, Barchitta, M, Quattrocchi, A, and Agodi, A. Public health genomics and GISIO working groups of the Italian Society of Hygiene, preventive medicine and public health (SItI). Molecular epidemiology tools in the management of healthcare-associated infections: towards the definition of recommendations. Epidemiol Prev. (2015) 39:21–6.
29. Rawson, JMO, Nikolaitchik, OA, Keele, BF, Pathak, VK, and Hu, WS. Recombination is required for efficient HIV-1 replication and the maintenance of viral genome integrity. Nucleic Acids Res. (2018) 46:10535–45. doi: 10.1093/nar/gky910
30. Tamura, K, and Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. (1993) 10:512–26. doi: 10.1093/oxfordjournals.molbev.a040023
31. Fauci, AS, Redfield, RR, Sigounas, G, Weahkee, MD, and Giroir, BP. Ending the HIV epidemic: a plan for the United States. JAMA. (2019) 321:844–5. doi: 10.1001/jama.2019.1343
32. Rose, SLK, Lamers, SL, Dollar, JJ, Grabowski, MK, Hodcroft, EB, Ragonnet-Cronin, M, et al. Identifying transmission clusters with cluster picker and HIV-TRACE. AIDS Res Hum Retrovir. (2017) 33:211–8. doi: 10.1089/aid.2016.0205
33. Dong, MJ, Peng, B, Liu, ZF, Ye, QN, Liu, H, Lu, XL, et al. The prevalence of HIV among MSM in China: a large-scale systematic analysis. BMC Infect Dis. (2019) 19:1000. doi: 10.1186/s12879-019-4559-1
34. Otani, M, Shiino, T, Kondo, M, Hachiya, A, Nishizawa, M, Kikuchi, T, et al. Japanese drug resistance HIV-1 surveillance network. Phylodynamic analysis reveals changing transmission dynamics of HIV-1 CRF01_AE in Japan from heterosexuals to men who have sex with men. Int J Infect Dis. (2021) 108:397–405. doi: 10.1016/j.ijid.2021.05.066
35. Tordoff, DM, Buskin, S, Lechtenberg, R, Golden, MR, Kerani, RP, and Herbeck, JT. Combining traditional and molecular epidemiology methods to quantify local HIV transmission among foreign-born residents. AIDS. (2021) 35:655–64. doi: 10.1097/QAD.0000000000002783
36. Chaillon, A, Essat, A, Frange, P, Smith, DM, Delaugerre, C, Barin, F, et al. Spatiotemporal dynamics of HIV-1 transmission in France (1999–2014) and impact of targeted prevention strategies. Retrovirology. (2017) 14:15. doi: 10.1186/s12977-017-0339-4
37. Oster, AM, France, AM, and Mermin, J. Molecular epidemiology and the transformation of HIV prevention. JAMA. (2018) 319:1657–8. doi: 10.1001/jama.2018.1513
38. Cao, DQ, Chen, JK, Tang, JL, He, TT, Lu, QL, and Yang, ZK. Characteristics of the molecular transmission network in newly confirmed human immunodeficiency virus type 1 infected cases from 2018 to 2019 in Shaoxing City, Zhejiang Province. Chin J Infect Dis. (2021) 39:157–62. doi: 10.3760/cma.j.cn311365-20200628-00683
39. Zhao, K, Kang, W, Liu, Q, Li, Y, Liu, Q, Jiang, W, et al. Genotypes and transmitted drug resistance among treatment-naive HIV-1-infected patients in a northwestern province, China: trends from 2003 to 2013. PLoS One. (2014) 9:e109821. doi: 10.1371/journal.pone.0109821
40. Guo, H, Xu, X, Hu, H, Zhou, Y, Yang, H, Qiu, T, et al. Low prevalence of the transmitted HIV-1 drug resistance among newly diagnosed HIV-1 individuals in Jiangsu Province, China during 2009-2011. BMC Public Health. (2015) 15:120. doi: 10.1186/s12889-015-1489-8
41. Jiamsakul, A, Sirivichayakul, S, Ditangco, R, Wong, KH, Li, PC, Praparattanapan, J, et al. TREAT Asia studies to evaluate resistance – surveillance study (TASER-S). Transmitted drug resistance in recently infected HIV-positive individuals from four urban locations across Asia (2007-2010) - TASER-S. AIDS Res Ther. (2015) 12:3. doi: 10.1186/s12981-015-0043-1
42. Lai, CC, Liu, WC, Fang, CT, Yang, JY, Chang, LH, Wu, PY, et al. Transmitted drug resistance of HIV-1 strains among individuals attending voluntary counselling and testing in Taiwan. J Antimicrob Chemother. (2016) 71:226–34. doi: 10.1093/jac/dkv284
43. Han, X, Zhao, B, An, M, Zhong, P, and Shang, H. Molecular network-based intervention brings us closer to ending the HIV pandemic. Front Med. (2020) 14:136–48. doi: 10.1007/s11684-020-0756-y
Keywords: HIV/AIDS, molecular network, epidemiological characteristic, southeastern China, drug resistance
Citation: Wang Z, Wang D, Lin L, Qiu Y, Zhang C, Xie M, Lu X, Lian Q, Yan P, Chen L, Feng Y, Xing H, Wang W and Wu S (2023) Epidemiological characteristics of HIV transmission in southeastern China from 2015 to 2020 based on HIV molecular network. Front. Public Health. 11:1225883. doi: 10.3389/fpubh.2023.1225883
Edited by:
Samuel R. Friedman, National Development and Research Institutes, United StatesReviewed by:
Benedetto Maurizio Celesia, UOC Infectious Diseases ARNAS Garibaldi, ItalyKledoaldo Lima, Universidade Federal de Pernambuco, Brazil
Copyright © 2023 Wang, Wang, Lin, Qiu, Zhang, Xie, Lu, Lian, Yan, Chen, Feng, Xing, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Wang, d2FuZ3dlaUBqaXBkLmNvbQ==; Shouli Wu, c2hvdWxpMTk3NUBzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Zhenghua Wang1†
Zhenghua Wang1† Dong Wang
Dong Wang Yi Feng
Yi Feng Hui Xing
Hui Xing Wei Wang
Wei Wang Shouli Wu
Shouli Wu