
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health, 23 August 2023
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 11 - 2023 | https://doi.org/10.3389/fpubh.2023.1216704
This article is part of the Research TopicHigh-level Antimicrobial Resistance or Hypervirulence in Emerging and Re-emerging “Super-Bug” Foodborne Pathogens: Detection, Mechanism, and Dissemination from Omics InsightsView all 16 articles
Background: Co-existence of colistin, β-lactam and carbapenem in multidrug-resistant Enterobacteriaceae isolates poses a serious threat to public health. In this study, we investigated and characterized the co-occurrence of blaCTX-M-65, blaOXA-1, and mcr-1.1 strain isolated from a clinical extensively-drug-resistant Escherichia coli ST744 in Shanghai.
Methods: Antimicrobial susceptibility test was carried out by agar dilution methods. Whole genome sequencing was conducted, and resistance genes, and sequence types of colistin in E. coli isolates were analyzed. Plasmid stability and amino acid mutations were assessed in E. coli isolates.
Results: A colistin resistant E. coli ST744, named ECPX221, was identified out of 145 fecal samples collected. The strain carries a 60,168 IncI2 plasmid with the mcr-1.1 gene. The strain also has blaCTX-M-65, blaOXA-1, dfrA14, qnrS1, cmlA5, arr2, ampC, aph(4)-Ia, sul1, and aadA5 resistance genes. The plasmid pECPX221 was capable of conjugation with an efficiency of 2.6 × 10−2. Notably, 45% of the transconjugants were determined as mcr-1.1-harboring in the colistin-free environment after 60 generation of passage. No mutations occurred in pmrB, mgrB, and phoPQ gene in the mcr-1.1-harboring transconjugants. Bioinformatic analysis indicated pECPX221 shared highly similar backbone with the previously reported mcr-1.1-harboring pAH62-1, pMFDS1339.1, pSCZE4, and p2018-10-2CC. Furthermore, sequencing and phylogenetic analyses revealed a similarity between other MCR-1-homolog proteins, indicating that ECPX221 was colistin resistant.
Conclusion: The stable transferable mcr-1.1-harboring plasmid found in the E. coli ST744 strain indicated the high risk to disseminate the extensively-drug-resistance phenotype among Enterobacteriaceae.
Multidrug resistance in Escherichia coli has become a concerning issue, displaying resistance to β-lactam antibiotics, particularly through the production of β-lactamases, including extended-spectrum β-lactamases (ESBLs) and carbapenemases (1). The increasing use of β-lactams and carbapenems over the past several decades has led to the increased use of colistin, which is now considered the last therapeutic option for treating infections caused by such organisms. However, the efficacy of colistin has been challenged by the emergence of plasmid-mediated mobile colistin resistance (mcr-1), which was found in Enterobacteriaceae in 2015 (2), and has since been disseminated in animals, meat products, humans (both fecal carriage and infections), and the environment in over 50 countries, covering six continents (3–5). In China, colistin has been adopted for treating carbapenem-resistant Enterobacteriaceae (CRE) infections since 2018 (6), thereby increasing the potential risk of the dissemination of mcr-1 (7). To date, mcr-1 positive plasmids have also been found in multidrug-resistant (MDR) and extensively drug-resistant (XDR) Enterobacteriaceae isolates carrying plasmid-borne carbapenemase and ESBL genes (8), which could generate resistance to multiple drugs and contribute to the spread of MDR bacteria in human populations (9).
Previous studies have reported mcr-1-harboring plasmids found in different Inc. types, while IncI2, IncHI2, and IncX4 appear to be the most common carriers of mcr-1 (3, 10–12). The mobility of mcr-1 is difficult to control because mcr-1-harboring plasmids often carry other antimicrobial resistance (AMR) genes, including those encoding resistance to β-lactams, fluoroquinolones, and tetracyclines (8, 13, 14). For example, Wang et al. reported E. coli strain QE11-421, which was isolated from a sputum sample of a 90-year-old male patient receiving treatment in the ICU, was an mcr-1-positive colistin-resistant isolate that co-harbored the blaKPC-2 gene conferring carbapenem resistance (15). Zhang et al. (16) reported the coexistence of the blaNDM-5, blaCTX-M-65, blaOXA-10, blaTEM-1 and mcr-1.1 genes detected in E. coli 20IR1127 strain belonging to ST156 lineage isolated from children in China. Lu reported three E. coli ST6775 coharboring tet(X4), mcr-1, and blaNDM-5 isolates from pigeons (17). In this study, we investigated the prevalence of colistin-resistant E. coli isolates from fecal samples in Shanghai and characterized the plasmid stability and persistence that contribute to colistin resistance in E. coli isolates.
Fresh fecal samples were collected from patients experiencing acute diarrhea within 3 days of symptom onset, and who had not received any antibiotics treatment from September 2021 to January 2022 from surveillance hospitals in Shanghai. The samples were collected using five sterile cotton swabs from multiple sites and placed into a 50 mL screw-cap-sealed centrifuge tube with C–B transport medium. For patients and infants who experienced difficulty in defecating, rectal swabs were used to collect the samples. The rectal cotton swab was soaked in normal saline and inserted 4–5 cm deep into the anus (2–3 cm for children), and gently rotated. At least two swabs were collected from the same patient and placed in C–B transport medium and 3 mL of preservation solution (containing 5% bovine serum cell maintenance solution). The samples were stored at 4°C and sent to the laboratory within 48 h after collection.
Each fecal sample was placed in sterile plastic bags containing 225 mL of Mueller–Hinton broth and incubated overnight at 37°C. The samples were then seeded on Nutrient broth plates (COMAGAL Microbial Technology, Shanghai) with 2 μg/mL colistin and incubated for 24 h at 37°C. To identify the isolate, a positive colony was selected by amplifying the mcr-1 gene via real-time PCR (RT-PCR). The strains were then identified by MALDI-TOF mass spectrometry using the VITEK MS system (BioMérieux Shanghai Co. Limited). Basic clinical data, including gender, age, and date of isolation, were collected for patients from whom the mcr-1-harboring strains were isolated. The mcr-1 gene screening was performed as following: briefly, the genomic DNA from each of the strains was extracted by boiling and freeze-thawing processes, and the resulting supernatant was used as the template. The specific primer used in this study was: mcr-1-RT-F: 5′-CGCGATGCTACTGATCACCA-3′, mcr-1-RT-R: 5′-GGTCGTATCATAGACCGTGCC-3′, and the mcr-1-probe: VIC-5′-TTATCATCGTATCGCTATGTGCTA-3′-MGB.
A total of 30 antimicrobial agents (Shanghai Fosun Biological Technology Co., Ltd.) were used for antimicrobial susceptibility testing via broth microdilution method, including ampicillin (AMP), ampicillin/sulbactam 2:1 ratio (AMS), tetracycline (TET), chloramphenicol (CHL), trimethoprim/sulfamethoxazole (SXT), cefazolin (CFZ), cefotaxime (CTX), ceftazidime (CAZ), cefoxitin (CFX), gentamicin (GEN), imipenem (IMP), nalidixic acid (NAL), azithromycin (AZI), tigecycline (TIG), ciprofloxacin (CIP), amoxicillin/clavulanic acid (AMC), cefotaxime/clavulanic acid (CTC), ceftazidime/clavulanic acid (CAC), colistin (CT), aztreonam (ATM), cefuroxime (CXM), amikacin (AMI), cefepime (CPM), meropenem (MEM), levofloxacin (LEV), ertapenem (ETP), ceftazidime/avibactam (CZA), streptomycin (STR), and norfloxacin (NOR). The double-disc synergy test and a modified carbapenem inactivation method were used to confirm the production of ESBL and carbapenemase, respectively, according to Clinical and Laboratory Standards Institute (CLSI) guidelines. E. coli ATCC®25922™ strain was used as quality control. The European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints defines colistin resistance as 2 μg/mL for Enterobacteriaceae (18).
To investigate whether the mcr-1 gene was present on a transferable plasmid, we performed a filter mating assay using rifamycin-resistant E. coli C600 as the recipient strain. Both the original isolates and recipient E. coli C600 were grown overnight in LB broth and adjusted to a 0.5 McFarland standard. The donor bacteria were mixed with recipient E. coli C600 at a ratio of 1:3 to 5 mL of fresh LB broth and then incubated at 37°C overnight. Transconjugants were selected on LB plates supplemented with rifampicin (40 μg/mL) and colistin (2 μg/mL). Putative transconjugants were confirmed using PCR and antimicrobial susceptibility tests. The mobilization efficiency were calculated by dividing the number of transconjugant colonies by the number of donor colonies (19).
The plasmid stability assay was conducted to determine the stability of the mcr-1-harboring plasmids in the absence of antibiotic selective pressure. The strains were grown in LB broth containing colistin (2 μg/mL) and then transferred to a fresh LB broth without antibiotics. The cultures were periodically passaged for 60 days, with serial passages performed every 24 h, resulting in approximately 600 generations of bacterial growth. Cultures from passages 2, 4, 6, 8, 10, 20, 30, 40, 50, and 60 were diluted and plated onto LB plates containing colistin and LB plates without antibiotics. The frequency of stable plasmids was calculated as the number of colonies grown on the LB plate containing colistin and the antibiotic-free LB plate, divided by the total number of colonies on both plates, multiplied by 100%. The distribution of all mcr-1-harboring plasmids from different antimicrobial environments and passages was monitored by RT-PCR and Sanger sequencing to identify any changes in the plasmid composition over time.
To assess the genetic mutation in the colistin-resistant E. coli isolates, the colistin resistance genes including mgrB, pmrAB, phoPQ were amplified by real-time quantitative PCR as previously described (20, 21). Mutations that occurred in colistin-resistant E. coli isolates were determined by comparing to their corresponding parental reference genomes.
The genomic DNA was extracted and sequencing libraries were generated using the TruSeq DNA Sample Preparation Kit (Illumina, USA). The genome sequencing was then performed with standard protocol and were sequenced with 150-bp paired-end strategy by using the Illumina Novaseq 6,000 (Sangon Biotech Company, Shanghai, China). Data assembly was carried out after adapter contamination removal and data filtering by using AdapterRemoval and SOAPec. Scaffold and contig construction were performed using SPAdes (version 3.12.0) and A5-miseq, respectively, with integration of all assembled results to obtain a complete sequence.
Subsequently, the VFDB (Virulence Factors of Pathogenic Bacteria)1 and CARD (The Comprehensive Antibiotic Resistance)2 database were used to retrieve the pathogenicity genes and antibiotic resistance genes, respectively. The mcr-1-carrying contigs generated by Illumina sequencing were examined for Inc. types by PlasmidFinder version 2.1.3 The set of close typing sequences was determined by PubMLST4 and then compared with the typing sequences of Escherichia spp. The linear comparison of complete plasmid sequences was created by EasyFig version 2.2.2.5 The plasmid construction map was generated by SnapGene 6.1.2 software (Insightful Science, United States). The genome-wide similarities was generated by FastANI version 1.33.6 Sequences were deposited to NCBI website under the Bioproject PRJNA929103.
The MCR-1 and MCR-1-like proteins’ homologous sequences were extracted from NCBI through BLASTp search (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 20 June 2023), with MCR-1 protein of ECPX221 in this study obtained from the sequencing data. Aligned sequences of MCR-1 obtained from ClustalW version 2.07 were used to construct a phylogenetic tree through the Maximum Likelihood Method of MEGA X (Mega Limited, Auckland, New Zealand). To confirm the results, 1,000 bootstrap repetitions were used.
This study was reviewed and approved by the ethical committee of the Shanghai Municipal Centre for Disease Control and Prevention.
Out of 145 fecal samples collected between September 2021 and January 2022, only one E. coli isolate, named ECPX221, was found to harbor the mcr-1.1 gene. Antimicrobial resistance testing was performed on this mcr-1.1-positive strain, and it was found to exhibit colistin resistance at 2 μg/mL (as shown in Table 1). Furthermore, ECPX221 was identified as an extended-spectrum β-lactamase producer, and was found to be resistant to CIP, AMP, AMS, CFZ, CTX, CXM, SXT, NAL, CHL, TET, ATM, LEV, and NOR. However, it was found to be susceptible to 15 other common antibiotics, including CFX, CPM, CZA, IMP, ETP, TIG, and others (as shown in Table 1). Apart from mcr-1.1, ECPX221 was also found to carry resistance genes for blaCTX-M-65, blaOXA-1, dfrA14, qnrS1, cmlA5, arr2, ampC, aph(4)-Ia, sul1, and aadA5.
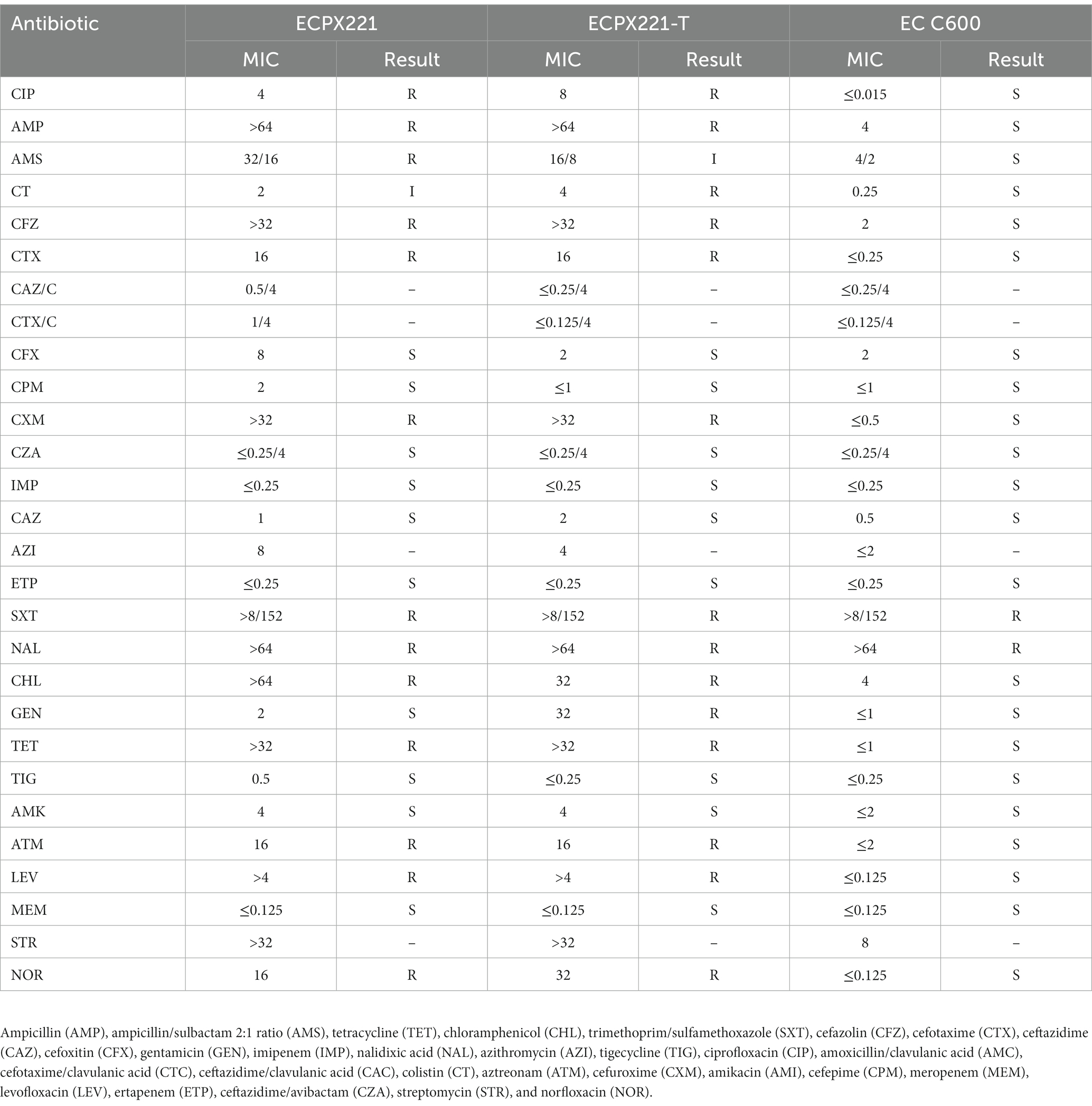
Table 1. Information for the mcr-1.1-harboring Escherichia coli strain ECPX221 identified in this study and the transconjugant isolate ECPX221-T. The wild type EC C600 was used as control.
The result indicated that the mcr-1.1-harboring plasmid was capable of successful transfer from the donor strain to the recipient strain (E. coli C600). The conjugation of ECPX221 to E. coli C600 via horizontal transfer was achieved with an average efficiency of 2.6 × 10−2. The transconjugant ECPX221-T, which was confirmed to harbor mcr-1.1 gene, exhibited a MIC value of 4 μg/mL to colistin, which represented a significant increase when compared to the wild type E. coli C600 (0.25 μg/mL). Therefore, it was speculated that the transconjugants acquired the colistin resistance gene from the donor strain.
In order to assess the plasmid stability, we analyzed the dynamics of the pECPX221 plasmid by passaging the ECPX221-T strain carrying the plasmid with colistin for 60 days. Next day, 97% of the transconjugants (29 positive colonies out of 30 colonies) were detected as mcr-1.1 positive in an antibiotic-free environment, while all transconjugants were positive in the colistin environment. On day 30, only 60% of the transconjugants (15 positive colonies out of 25 colonies) were detected as mcr-1.1-carrying isolates in the antibiotic-free environment, and this result decreased further to 45% on day 60, while the positive rate remained at 94% in the plate with colistin (Figure 1). To assess genetic mutations related to colistin resistance in the mcr-1-harboring plasmid in E. coli, we examined key genes such as mgrB, pmrAB, and phoPQ in the mcr-1.1-harboring and non-harboring transconjugants. However, our results showed that none of the aforementioned genes were mutated in the mcr-1.1-harboring transconjugants.
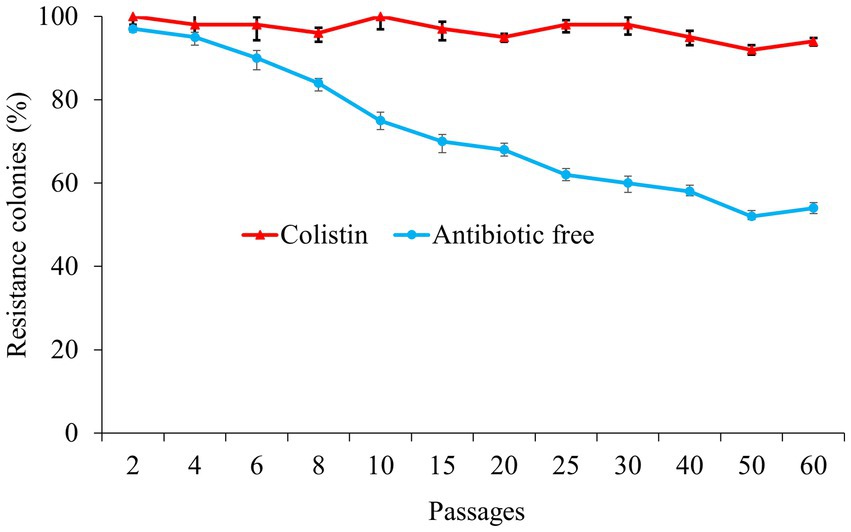
Figure 1. The resistance colonies that harboring mcr-1.1 gene in the colistin (red) and antibiotic free environment (blue).
We sequenced the genomes of the EXPX221 and the sequence data showed that it belonging to the replicon types IncI2. BLASTn analysis showed that the backbone of the plasmid pECPX221 (GenBank Accession No. GCA_028527545) was strikingly similar with (the query cover of 100% and the identities 99%) other previously sequenced mcr-1.1-harboring IncI2 plasmids, such as pAH62-1 from E. coli AH62 (GenBank Accession No. CP055260), pMFDS1339.1 from E. coli MFDS1339 (GenBank Accession No. MK852553), pSCZE4 from E. coli SCZE5 (GenBank Accession No. CP051226), and p2018-10-2CC from E. coli 2018–10-2CC (GenBank Accession No. LC511662). In all, the ANI heatmap showed that these IncI2 plasmids bearing mcr-1.1 showed very high architectural conservation (Figure 2). Furthermore, the BLAST comparison of pECPX221, pAH62-1, pMFDS1339.1, pSCZE4 and p2018-10-2CC revealed that their mcr-1.1 insertion sites differed (Figure 3). An approximately 2.5 kb mcr-1.1-pap2 element was identified in the above-mentioned plasmids. In addition, an integrase core domain protein as IS481-like element ISEc19 family transposase (WP_010723086) was identified in pECPX221, but only found in pMFDS1339.1. The putative conjugal transfer components of pECPX221 were also detected by using oriTfinder. The vir gene family encoding VirB1 to VirB11 were identified as T4SS belonging to Type IV secretion system was predicted on pECPX221 (Figure 4). The relaxase in pECPX221 from 45,033 to 49,346 nt was found to be 99% identity with the relaxase (WP_124777228.1) that obtained from E. coli. This evidence confirms that pECPX221 is a conjugative plasmid.
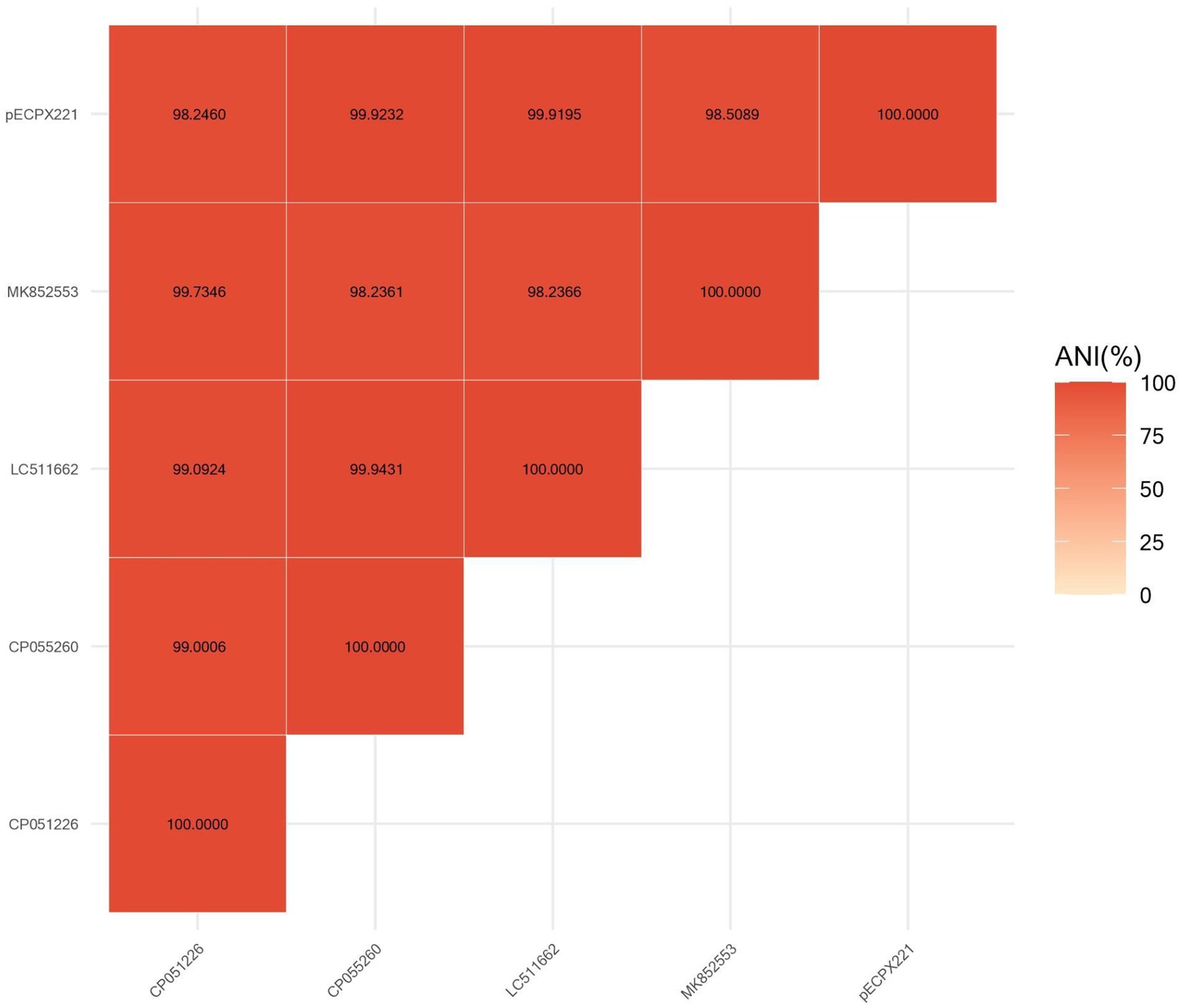
Figure 2. Comparison of pECPX221 and other mcr-1-harboring plasmids including pAH62-1, pMFDS1339.1, pSCZE4, and p2018-10-2CC by FastANI software.
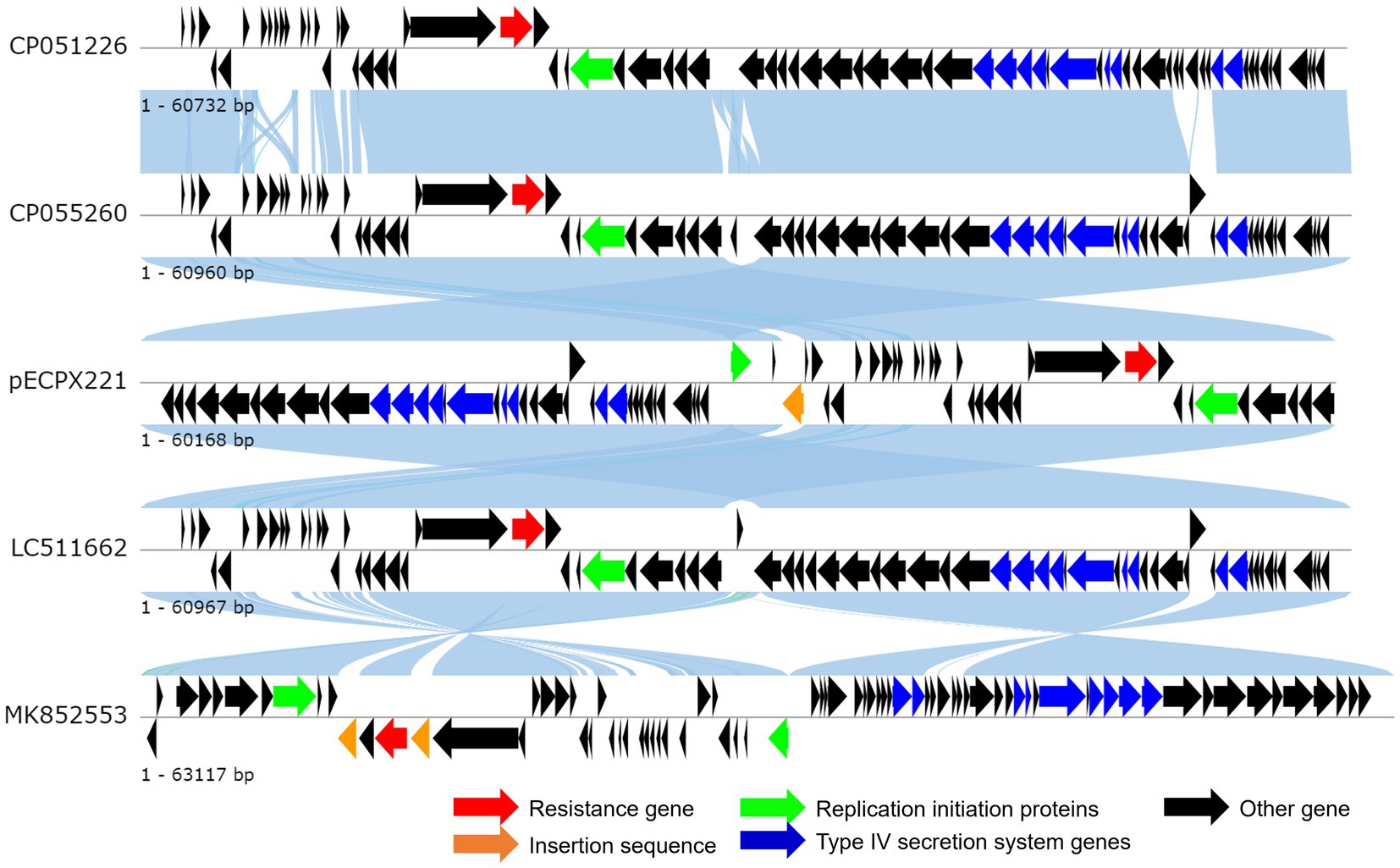
Figure 3. Linear comparison of complete plasmid sequences of plasmid Escherichia coli pECPX221 (this study), pAH62-1 from E. coli AH62 (GenBank Accession No. CP055260), pMFDS1339.1 from E. coli MFDS1339 (GenBank Accession No. MK852553), pSCZE4 from E. coli SCZE5 (GenBank Accession No. CP051226), and p2018-10-2CC from E. coli 2018–10-2CC (GenBank Accession No. LC511662).
A total of 54 MCR-1 proteins originated from E. coli (Supplementary File S1), and 26 proteins of MCR-1 gene obtained from bacteria other than E. coli (Supplementary File S2) were categorized for further analysis. In the case of protein acquisition from the NCBI database, above 50% of query coverage was set as the screening point. These two sets of proteins (Supplementary Files S1, S2), including ECPX221 strain harboring MCR-1 in this study, were used for phylogenetic analysis (Figure 5). The sequenced MCR-1 of ECPX221 in this study clearly showed its genomic confirmation as mcr-1 genes by highly aligning with mcr-1 genes of E. coli as well as other bacteria origins. In addition, the phylogenetic tree showed that ECPX221 strain in this study was mostly of Asian origin and that they were closely related.
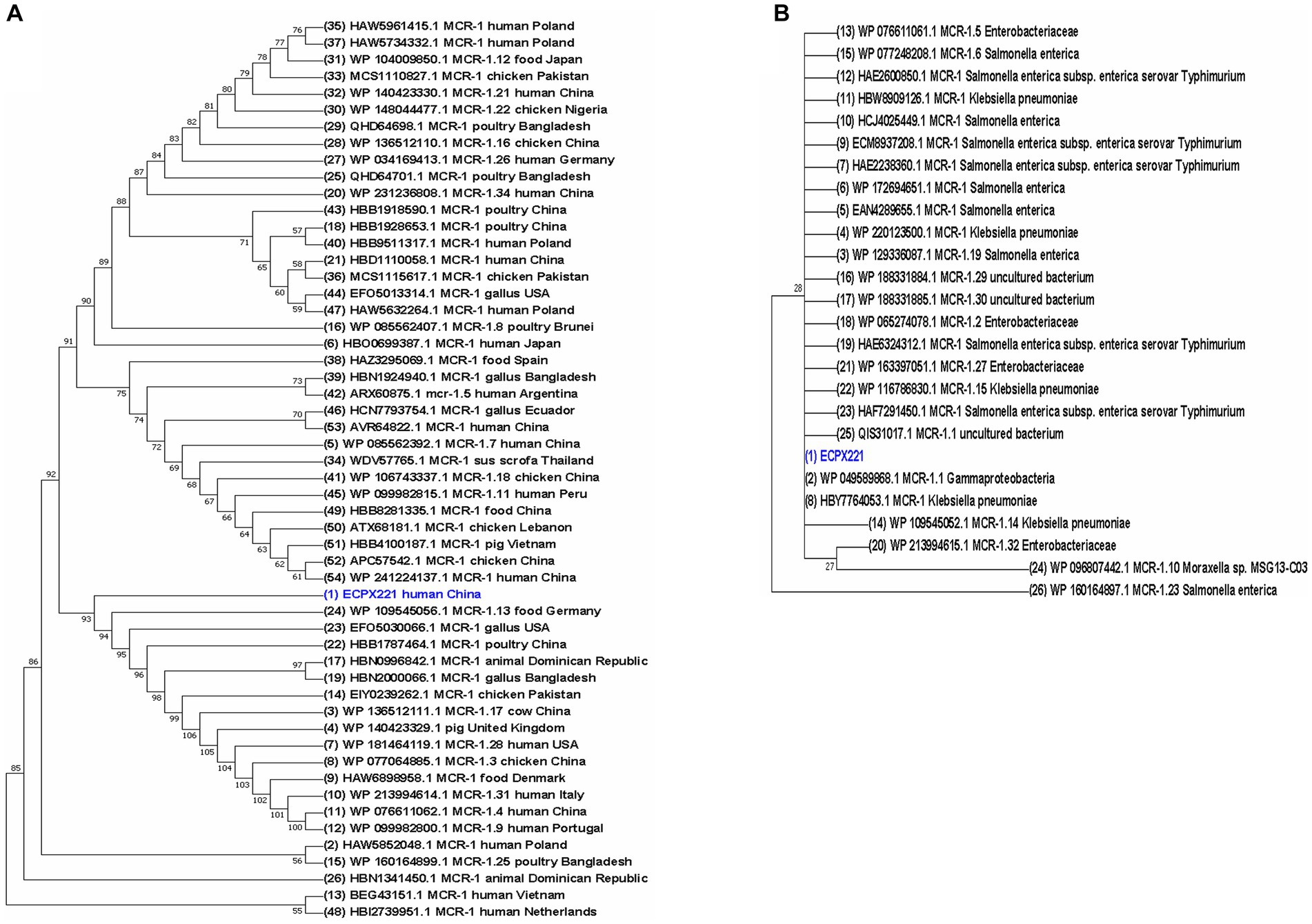
Figure 5. Phylogenetic study of MCR-1 and MCR-1-like proteins reveals ancestral origins and diversification. Using amino acid sequences from MCR-1 protein of ECPX221 in this study, the BLAST search tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi, Accessed on 20 June 2023) was used to retrieve related sequences of MCR-1 and MCR-1-like proteins from the NCBI database. MCR-1 and MCR-1-like proteins of E. coli, Salmonella, and strains containing LptA and others were among the sequences categorized. Using aligned MCR-1 sequences from CLUSTALW, the maximum likelihood method of MEGA X was used to create a phylogenetic tree. (A) Phylogeny of 54 MCR-1 proteins of E. coli origin retrieved from NCBI database including sequenced MCR-1 gene of ECPX221 from this study. (B) Phylogeny of 26 MCR-1 proteins retrieved from NCBI database except E. coli strains.
Colistin resistance has become a serious issue in food animals such as pigs and chickens, as it has been frequently used since the 1950s (22, 23). Livestock and poultry are known to be the main reservoir for colistin resistance, and the discovery of the stable plasmid-mediated mcr-1 gene in E. coli has helped us understand the potential transmission of colistin resistance between animals and humans (2). In this study, we have identified the co-occurrence of ESBL genes (blaCTX-M-65 and blaOXA-1) and mcr-1.1-producing E. coli ST744 isolates from clinical fecal samples in Shanghai. The plasmid pECPX221 contains four typical conjugal modules: a T4CP gene, an oriT-like region for transfer origin, a relaxase gene, and a gene cluster for the bacterial T4SS apparatus. Additionally, pECPX221 contains the mcr-1.1-pap2 cassette, which has been shown to be capable of horizontally transferring into various plasmid replicon types (24).
The prevalence of coexistence of mcr-1, ESBL genes carrying E. coli was found to be only 0.7% (1/145) in clinical fecal samples, which is similar as that one E. coli strain harboring mcr-1 and blaNDM-5 reported in companion animals that was collected from six different cities including Harbin, Yangzhou, Chongqing, Wuhan, Chengdu and Guangzhou (0.8%, 1/129) (25). Also, it was not that high when comparing with that has been reported in other regions in China such as in Shandong (3.5%) (26), Shanghai (3.9%) (27), Henan (7.7%) (28) and some other countries like in Pakistan (23.2%) (29), Lebanon (18%) (30), Japan (4.84%) (31) and Bolivia (38.3%) (32), but a little higher than that children patients with diarrhoea in Shanghai (0.28%) (33). This low prevalence could be attributed to the strict usage of colistin in Shanghai. According to the Shanghai Catalogue of Graded Management of Clinical Application of Antibacterial Drugs (2021 version) (34), colistin is only used for extensively resistant gram-negative infections such as Pseudomonas aeruginosa, Acinetobacter baumannii, and Enterobacteriaceae.
E. coli is the first and most prevalent species carrying the mcr-1 gene in Enterobacteriaceae and can be isolated from raw meat and fecal samples of animals and humans in China (2, 35). The co-existence of ESBL genes such as blaCTX-M-65 and mcr-1-harboring E. coli have been reported globally, particularly in animal source E. coli isolates in northern China (36, 37). In eastern China, Zhejiang Province identified the co-occurrence of blaNDM-5, blaCTX-M-65, blaOXA-10, blaTEM-1, and mcr-1.1 genes isolated from human E. coli ST156 in 2022 (16). Jiangsu Province reported an ESBL, carbapenemase- and mcr-1-producing E. coli ST648 strain isolated from a urine sample, which was found to have three transferable resistance plasmids (38). Apart from E. coli, one isolate named CFSA664 was found to co-harbor the mcr-1 gene and blaCTX-M-65 in Salmonella enterica serotype Indiana from retail chickens in Jiangsu (39). However, the co-existence of blaCTX-M-65 and mcr-1 was found in E. coli ST117 isolated from a veterinary hospital in Shanghai (40) and EC1CT136A isolated from broiler farms in Ecuador (41). Our study identified, for the first time, the co-existence of blaCTX-M-65, blaOXA-1, and mcr-1.1 from a human fecal isolate in E. coli ST744 in Shanghai. This finding indicates a high risk of disseminating this extensively drug-resistant E. coli, which poses a threat to public health.
WGS data indicated the presence of various determinants of antibiotic resistance, suggesting that carbapenem and colistin co-resistant strains may be selected with the use of any antibiotics. Colistin resistance typically arises from selective pressure resulting from the use of polymyxins (2). In China, colistin was widely used as a growth promoter in animal production until 2017, when it was banned due to the identification of plasmid-mediated colistin-resistant isolates in the country (42, 43). Furthermore, colistin therapy in humans was introduced in February 2017 (7). As a result, it is essential to note that the prevalence of colistin-resistant isolates may be underestimated, as colistin susceptibility is not routinely tested in clinical samples from outpatients.
Plasmid horizontal transfer of mcr-1 gene was widely reported in human, food, animals, and the environment in a lot of countries and regions worldwide (44). In this study, the plasmid replicon type of IncI2, the major type of plasmids spreading globally that promote E. coli resistance, was identified. The ISApl1, which is consistently associated with the mcr-1 gene and its related cassette that can be inserted into a variety of genetic loci in different plasmids, was missing in our study, this was similar to the previously reports on the absence of ISApl1 in E. coli strain and other Enterobacteriaceae such as E. fergusonii (45). The diversity of plasmids carrying the mcr-1 gene has been shown to increase with the use of colistin in clinical settings, suggesting that colistin administration could promote the dissemination of diverse resistance plasmids among E. coli isolates (46). The result of plasmid stability assay revealed that pECPX221 remained stable in the recipient bacterial strain, which was consistent with other E. coli strains (47, 48).
The phylogenetic analysis indicated that the sequenced mcr-1 genes of E. coli are homologous to previously reported mcr-1 genes from E. coli and other bacteria origins. The sequence of the mcr-1 identified in our study is identical to that of the mcr-1 previously identified from E. coli (GenBank Accession No: A0A0R6L508). The phylogenetic tree demonstrates a close relation among some studied mcr-1 strains, and an evolutionary relationship to other mcr-1 genes like strain WP109545056, EF05030066, and HBB1787464. Furthermore, the phylogenetic tree revealed that the mcr-1-positive ECPX221 in this study is related to many MCR-1 variants. For example, it is closely related to the mcr-1.13 strain isolated from meat, and its spread could result in widespread resistance to colistin.
There are two limitations to this study: Firstly, the E. coli isolates were collected solely from fecal samples in Shanghai province, China. To obtain more accurate results, it would be beneficial to include isolates from additional regions with prolonged monitoring. Secondly, the fitness cost of ECPX221 needs to be assessed in order to evaluate the potential plasmid loss imposed by pECPX221.
The study found that the prevalence of mcr-1.1-harboring E. coli among clinical fecal isolates in Shanghai is low. However, the strain ECPX221, which carries the mcr-1.1 gene, exhibited extensive antimicrobial resistance profiles and additional resistance genes. The results of the conjugation experiment confirmed the horizontal transfer of the mcr-1.1 gene, and the mcr-1.1-harboring plasmid pECPX221 was found to be stable in the recipient strain. Phylogenetic analysis showed an evolutionary linkage between MCR-1 and MCR-1 homolog proteins. Given the crucial role of colistin as a last-line treatment option against infections caused by multidrug-resistant Gram-negative bacteria, continuous surveillance is urgently needed to monitor the spread of the coexistence of the mcr-1.1 gene and other important resistance genes.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA929103.
The studies involving humans were approved by Shanghai Municipal Center for Disease Control and Prevention. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
JF wrote the draft, revised the manuscript, designed this study, and responsible for the whole experiment. HYW, YZ, JYL, YC, YTW, JYF, and QS participated in the whole experiment process. ZAY and MC managed the experiment and provided suggestions and revisions. All authors contributed to the article and approved the submitted version.
The work was supported by the Three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai (2023-2025; Grant no. GWVI-3).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2023.1216704/full#supplementary-material
1. ^http://www.mgc.ac.cn/VFs/main.htm
3. ^https://cge.cbs.dtu.dk/services/PlasmidFinder/
5. ^https://mjsull.github.io/Easyfig/
1. van Duin, D, and Doi, Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence. (2017) 8:460–9. doi: 10.1080/21505594.2016.1222343
2. Liu, YY, Wang, Y, Walsh, TR, Yi, LX, Zhang, R, Spencer, J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. (2016) 16:161–8. doi: 10.1016/S1473-3099(15)00424-7
3. Sun, J, Zhang, H, Liu, YH, and Feng, Y. Towards understanding MCR-like colistin resistance. Trends Microbiol. (2018) 26:794–808. doi: 10.1016/j.tim.2018.02.006
4. Wang, C, Feng, Y, Liu, L, Wei, L, Kang, M, and Zong, Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect. (2020a) 9:508–16. doi: 10.1080/22221751.2020.1732231
5. Wang, R, van Dorp, L, Shaw, LP, Bradley, P, Wang, Q, Wang, X, et al. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. (2018) 9:1179. doi: 10.1038/s41467-018-03205-z
6. Liang, Q, Huang, M, and Xu, Z. Early use of polymyxin B reduces the mortality of carbapenem-resistant Klebsiella pneumoniae bloodstream infection. Braz J Infect Dis. (2019) 23:60–5. doi: 10.1016/j.bjid.2018.12.004
7. Shen, Y, Zhou, H, Xu, J, Wang, Y, Zhang, Q, Walsh, TR, et al. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol. (2018) 3:1054–62. doi: 10.1038/s41564-018-0205-8
8. Poirel, L, Jayol, A, and Nordmann, P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. (2017) 30:557–96. doi: 10.1128/CMR.00064-16
9. Yang, YQ, Zhang, AY, Ma, SZ, Kong, LH, Li, YX, Liu, JX, et al. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J Antimicrob Chemother. (2016) 71:2336–8. doi: 10.1093/jac/dkw243
10. Kong-Ngoen, T, Santajit, S, Tunyong, W, Pumirat, P, Sookrung, N, Chaicumpa, W, et al. Antimicrobial resistance and virulence of non-typhoidal Salmonella from retail foods marketed in Bangkok, Thailand. Foods. (2022) 11:661. doi: 10.3390/foods11050661
11. Matamoros, S, van Hattem, JM, Arcilla, MS, Willemse, N, Melles, DC, Penders, J, et al. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. (2017) 7:15364. doi: 10.1038/s41598-017-15539-7
12. Hu, Y, Zhang, C, and Zhang, J. Antimicrobial Resistance in Non-typhoidal Salmonella from Retail Foods Collected in 2020 in China. Zoonoses (2023) 3. doi: 10.15212/ZOONOSES-2023-0001
13. Guo, L, Wang, J, Wang, S, Su, J, Wang, X, and Zhu, Y. Genome characterization of mcr-1-positive Escherichia coli isolated from pigs with postweaning Diarrhea in China. Front Vet Sci. (2020) 7:503. doi: 10.3389/fvets.2020.00503
14. Lu, X, Xiao, X, Liu, Y, Li, R, and Wang, Z. Emerging opportunity and Destiny of mcr-1- and tet(X4)-Coharboring plasmids in Escherichia coli. Microbiol Spectr. (2021) 9:e0152021. doi: 10.1128/Spectrum.01520-21
15. Wang, Y, Liu, H, Wang, Q, Du, X, Yu, Y, and Jiang, Y. Coexistence of Bla(KPC-2)-IncN and mcr-1-IncX4 plasmids in a ST48 Escherichia coli strain in China. J Glob Antimicrob Resist. (2020b) 23:149–53. doi: 10.1016/j.jgar.2020.08.023
16. Zhang, X, Fang, C, Zhang, J, Hua, W, He, R, and Zhou, M. Carbapenemase- and colistin resistant Escherichia coli strains from children in China: high genetic diversity and first report of Bla (NDM-5), Bla (CTX-M-65), Bla (OXA-10), Bla (TEM-1), and mcr-1.1 genes co-occurrence in E. coli ST156. Infect Drug Resist. (2022) 15:5315–20. doi: 10.2147/IDR.S378574
17. Lu, X, Du, Y, Peng, K, Zhang, W, Li, J, Wang, Z, et al. Coexistence of tet(X4), mcr-1, and Bla(NDM-5) in ST6775 Escherichia coli isolates of animal origin in China. Microbiol Spectr. (2022) 10:e0019622. doi: 10.1128/spectrum.00196-22
18. European Committee on Antimicrobial Susceptibility Testing. (2022). Clinical breakpoints-bacteria (v 12.0).
19. Wang, P, Xiong, Y, Lan, R, Ye, C, Wang, H, Ren, J, et al. pO157_Sal, a novel conjugative plasmid detected in outbreak isolates of Escherichia coli O157:H7. J Clin Microbiol. (2011) 49:1594–7. doi: 10.1128/JCM.02530-10
20. Cannatelli, A, Di Pilato, V, Giani, T, Arena, F, Ambretti, S, Gaibani, P, et al. In vivo evolution to colistin resistance by PmrB sensor kinase mutation in KPC-producing Klebsiella pneumoniae is associated with low-dosage colistin treatment. Antimicrob Agents Chemother. (2014) 58:4399–403. doi: 10.1128/AAC.02555-14
21. Haeili, M, Javani, A, Moradi, J, Jafari, Z, Feizabadi, MM, and Babaei, E. MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front Microbiol. (2017) 8:2470. doi: 10.3389/fmicb.2017.02470
22. Rhouma, M, Beaudry, F, and Letellier, A. Resistance to colistin: what is the fate for this antibiotic in pig production? Int J Antimicrob Agents. (2016) 48:119–26. doi: 10.1016/j.ijantimicag.2016.04.008
23. Feng, J, Xu, Z, Zhuang, Y, Liu, MX, Luo, JY, Wu, YT, et al. Research progress on detection technology of colistin resistance (in Chinese). J Pathogen Biol. (2023) 18:854–9. doi: 10.13350/j.cjpb.230722
24. Li, A, Yang, Y, Miao, M, Chavda, KD, Mediavilla, JR, Xie, X, et al. Complete sequences of mcr-1-harboring plasmids from extended-Spectrum-beta-lactamase- and Carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother. (2016) 60:4351–4. doi: 10.1128/AAC.00550-16
25. Kuang, X, Yang, R, Ye, X, Sun, J, Liao, X, Liu, Y, et al. NDM-5-producing Escherichia coli co-Harboring mcr-1 gene in companion animals in China. Animals (Basel). (2022) 12:1310. doi: 10.3390/ani12101310
26. Bi, Z, Berglund, B, Sun, Q, Nilsson, M, Chen, B, Tarnberg, M, et al. Prevalence of the mcr-1 colistin resistance gene in extended-spectrum beta-lactamase-producing Escherichia coli from human faecal samples collected in 2012 in rural villages in Shandong Province, China. Int J Antimicrob Agents. (2017) 49:493–7. doi: 10.1016/j.ijantimicag.2016.12.018
27. Xie, J, Liang, B, Xu, X, Yang, L, Li, H, Li, P, et al. Identification of mcr-1-positive multidrug-resistant Escherichia coli isolates from clinical samples in Shanghai, China. J Glob Antimicrob Resist. (2022) 29:88–96. doi: 10.1016/j.jgar.2022.02.008
28. Lu, X, Zhang, P, Du, P, Zhang, X, Wang, J, Yang, Y, et al. Prevalence and genomic characteristics of mcr-positive Escherichia coli strains isolated from humans, pigs, and foods in China. Microbiol Spectr. (2023) 11:e0456922. doi: 10.1128/spectrum.04569-22
29. Hameed, F, Khan, MA, Bilal, H, Muhammad, H, and Tayyab Ur, R. Detection of MCR-1 gene in multiple drug resistant Escherichia coli and Klebsiella pneumoniae in human clinical samples from Peshawar, Pakistan. Comb Chem High Throughput Screen. (2021) 24:737–42. doi: 10.2174/1386207323666200914100119
30. Hassan, J, Mann, D, Li, S, Deng, X, and Kassem, II. Emergence of the Mobile colistin resistance gene, mcr-1, in multidrug-Resistant E. coli isolated from the Fecal matter of toddlers in a community. Antimicrob Agents Chemother. (2021) 65:e00243-21. doi: 10.1128/AAC.00243-21
31. Nakano, A, Nakano, R, Nishisouzu, R, Suzuki, Y, Horiuchi, S, Kikuchi-Ueda, T, et al. Prevalence and relatedness of mcr-1-mediated colistin-resistant Escherichia coli isolated from livestock and farmers in Japan. Front Microbiol. (2021) 12:664931. doi: 10.3389/fmicb.2021.664931
32. Giani, T, Sennati, S, Antonelli, A, Di Pilato, V, di Maggio, T, Mantella, A, et al. High prevalence of carriage of mcr-1-positive enteric bacteria among healthy children from rural communities in the Chaco region, Bolivia, September to October 2016. Euro Surveill. (2018) 23:1800115. doi: 10.2807/1560-7917.ES.2018.23.45.1800115
33. Feng, J, Zhuang, Y, Luo, J, Xiao, Q, Wu, T, Chen, Y, et al. Prevalence of colistin-resistant mcr-1-positive Escherichia coli isolated from children patients with diarrhoea in Shanghai, 2016-2021. J Glob Antimicrob Resist. (2023) 34:166–175. doi: 10.1016/j.jgar.2023.06.006
34. Shanghai Municipal Health Commission (2021). "Shanghai catalogue of graded management of clinical application of antibacterial drugs (2021 version)", Shanghai Municipal Health Commission Shanghai.
35. Zhang, S, Huang, Y, Yang, G, Lei, T, Chen, M, Ye, Q, et al. High prevalence of multidrug-resistant Escherichia coli and first detection of IncHI2/IncX4-plasmid carrying mcr-1 E. coli in retail ready-to-eat foods in China. Int J Food Microbiol. (2021) 355:109349. doi: 10.1016/j.ijfoodmicro.2021.109349
36. Chen, Y, Liu, Z, Zhang, Y, Zhang, Z, Lei, L, and Xia, Z. Increasing prevalence of ESBL-producing multidrug resistance Escherichia coli from diseased pets in Beijing, China from 2012 to 2017. Front Microbiol. (2019) 10:2852. doi: 10.3389/fmicb.2019.02852
37. Wu, C, Wang, Y, Shi, X, Wang, S, Ren, H, Shen, Z, et al. Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008-2014. Emerg Microbes Infect. (2018) 7:30. doi: 10.1038/s41426-018-0033-1
38. Lu, Y, Kang, H, and Fan, J. A novel Bla (CTX-M-65)-Harboring IncHI2 plasmid pE648CTX-M-65 isolated from a clinical extensively-drug-resistant Escherichia coli ST648. Infect Drug Resist. (2020) 13:3383–91. doi: 10.2147/IDR.S269766
39. Hu, Y, He, Y, Nguyen, SV, Liu, C, Liu, C, Gan, X, et al. Antimicrobial resistance of Salmonella Indiana from retail chickens in China and emergence of an mcr-1-harboring isolate with concurrent resistance to ciprofloxacin, cefotaxime, and colistin. Front Microbiol. (2022) 13:955827. doi: 10.3389/fmicb.2022.955827
40. Lin, H, Chen, W, Zhou, R, Yang, J, Wu, Y, Zheng, J, et al. Characteristics of the plasmid-mediated colistin-resistance gene mcr-1 in Escherichia coli isolated from a veterinary hospital in Shanghai. Front Microbiol. (2022a) 13:1002827. doi: 10.3389/fmicb.2022.1002827
41. Vinueza-Burgos, C, Ortega-Paredes, D, Narvaez, C, De Zutter, L, and Zurita, J. Characterization of cefotaxime resistant Escherichia coli isolated from broiler farms in Ecuador. PLoS One. (2019) 14:e0207567. doi: 10.1371/journal.pone.0207567
42. Walsh, TR, and Wu, Y. China bans colistin as a feed additive for animals. Lancet Infect Dis. (2016) 16:1102–3. doi: 10.1016/S1473-3099(16)30329-2
43. Wang, Y, Xu, C, Zhang, R, Chen, Y, Shen, Y, Hu, F, et al. Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect Dis. (2020c) 20:1161–71. doi: 10.1016/S1473-3099(20)30149-3
44. Touati, A, and Mairi, A. Plasmid-determined colistin resistance in the north African countries: a systematic review. Microb Drug Resist. (2021) 27:121–33. doi: 10.1089/mdr.2019.0471
45. Lin, J, Tang, B, Zheng, X, Chang, J, Ma, J, He, Y, et al. Emergence of Incl2 plasmid-mediated colistin resistance in avian Escherichia fergusonii. FEMS Microbiol Lett. (2022b) 369:fnac016. doi: 10.1093/femsle/fnac016
46. Huang, H, Dong, N, Shu, L, Lu, J, Sun, Q, Chan, EW, et al. Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China, 2014-2019. Emerg Microbes Infect. (2020) 9:237–45. doi: 10.1080/22221751.2020.1717380
47. Cheng, P, Yang, Y, Cao, S, Liu, H, Li, X, Sun, J, et al. Prevalence and characteristic of swine-origin mcr-1-positive Escherichia coli in Northeastern China. Front Microbiol. (2021) 12:712707. doi: 10.3389/fmicb.2021.712707
Keywords: MCR-1, Escherichia coli STstrain-744 (ST744), colistin, stability, plasmid
Citation: Feng J, Wu H, Zhuang Y, Luo J, Chen Y, Wu Y, Fei J, Shen Q, Yuan Z and Chen M (2023) Stability and genetic insights of the co-existence of blaCTX-M-65, blaOXA-1, and mcr-1.1 harboring conjugative IncI2 plasmid isolated from a clinical extensively-drug resistant Escherichia coli ST744 in Shanghai. Front. Public Health. 11:1216704. doi: 10.3389/fpubh.2023.1216704
Received: 04 May 2023; Accepted: 03 August 2023;
Published: 23 August 2023.
Edited by:
Scott Van Nguyen, American Type Culture Collection, United StatesReviewed by:
Amjad Islam Aqib, Cholistan University of Veterinary and Animal Sciences, PakistanCopyright © 2023 Feng, Wu, Zhuang, Luo, Chen, Wu, Fei, Shen, Yuan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhengan Yuan, eXVhbnpoZW5nYW5Ac2NkYy5zaC5jbg==; Min Chen, Y2hlbm1pbkBzY2RjLnNoLmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.