- 1EcoHealth Alliance, New York, NY, United States
- 2Centre for Integrative Ecology, School of Life and Environmental Sciences, Deakin University, Geelong, VIC, Australia
- 3One Health Laboratory, International Centre for Diarrheal Diseases Research, Bangladesh (icddr,b), Dhaka, Bangladesh
- 4Institute of Epidemiology, Disease Control and Research (IEDCR), Dhaka, Bangladesh
- 5Queensland Alliance for One Health Sciences, School of Veterinary Science, University of Queensland, Brisbane, QLD, Australia
Waterfowl are considered to be natural reservoirs of the avian influenza virus (AIV). However, the dynamics of transmission and evolutionary patterns of AIV and its subtypes within duck farms in Bangladesh remain poorly documented. Hence, a cross-sectional study was conducted in nine districts of Bangladesh between 2019 and 2021, to determine the prevalence of AIV and its subtypes H5 and H9, as well as to identify risk factors and the phylodynamics of H5N1 clades circulating in domestic duck farms. The oropharyngeal and cloacal swab samples were tested for the AIV Matrix gene (M-gene) followed by H5, H7, and H9 subtypes using rRT-PCR. The exploratory analysis was performed to estimate AIV and its subtype prevalence in different production systems, and multivariable logistic regression model was used to identify the risk factors that influence AIV infection in ducks. Bayesian phylogenetic analysis was conducted to generate a maximum clade credibility (MCC) tree and the maximum likelihood method to determine the phylogenetic relationships of the H5N1 viruses isolated from ducks. AIV was detected in 40% (95% CI: 33.0–48.1) of the duck farms. The prevalence of AIV was highest in nomadic ducks (39.8%; 95% CI: 32.9–47.1), followed by commercial ducks (24.6%; 95% CI: 14.5–37.3) and backyard ducks (14.4%; 95% CI: 10.5–19.2). The H5 prevalence was also highest in nomadic ducks (19.4%; 95% CI: 14.0–25.7). The multivariable logistic regression model revealed that ducks from nomadic farms (AOR: 2.4; 95% CI: 1.45–3.93), juvenile (AOR: 2.2; 95% CI: 1.37–3.61), and sick ducks (AOR: 11.59; 95% CI: 4.82–32.44) had a higher risk of AIV. Similarly, the likelihood of H5 detection was higher in sick ducks (AOR: 40.8; 95% CI: 16.3–115.3). Bayesian phylogenetic analysis revealed that H5N1 viruses in ducks belong to two distinct clades, 2.3.2.1a, and 2.3.4.4b. The clade 2.3.2.1a (reassorted) has been evolving silently since 2015 and forming at least nine subgroups based on >90% posterior probability. Notably, clade 2.3.4.4b was introduced in ducks in Bangladesh by the end of the year 2020, which was genetically similar to viruses detected in wild birds in Japan, China, and Africa, indicating migration-associated transmission of an emerging panzootic clade. We recommend continuing AIV surveillance in the duck production system and preventing the intermingling of domestic ducks with migratory waterfowl in wetlands.
1. Introduction
The avian influenza virus (AIV) has garnered increased attention recently because of its impact on productivity, commerce, and human health. The highly pathogenic avian influenza (HPAI) H5N1 virus has been linked to poultry epidemics and occasional human infections worldwide (1). In Bangladesh, the epidemic of H5N1 in poultry was reported for the first time in 2007. Since then, the disease has spread throughout the country, with 585 H5N1 outbreaks reported until the end of 2020 (2–4). In contrast, the first human case of H5N1 was detected through exposure to slaughtered poultry in Bangladesh on May 22, 2008 (5). AIV is now considered to be endemic and some recent research has identified a high percentage of AIV in birds from farms and live bird markets (LBM) including peri-urban and rural settings (6–9). Waterfowl from the order of Anseriformes (including ducks, geese, and swans), are distributed worldwide due to aquatic habitats and are considered one of the major natural reservoirs for AIV (10, 11). Other than domestic duck species, migratory waterfowl stopover for a few days to several weeks to rest at foraging areas (wetlands and lakes) along their migratory routes (10, 12). The AIV can spread to and from domestic duck populations due to the length of stay and wetland of both domestic and migratory duck populations, and the asymptomatic nature of infected individuals increases the likelihood that the virus will spread to other species (13). When an infected duck defecates in a specific wetland or waterbody, the AIV enters the environment and infects other ducks easily while they access the same areas. Although AIV has been replicated in the respiratory tract, we cannot overlook the fecal shedding of the AIV (14). Consequently, wetlands and water bodies can become contaminated with AIV through the defecation of infected birds, therefore, transmission of the virus is more likely when a significant number of birds roost on a small wetland (15). This evidence can be corroborated by another study in which the authors recovered the virus from the lake surface, where many different duck species graze (16). So, the high AIV titer in feces, the stability of the virus in the water, and the higher number of positive cloacal than tracheal samples suggest the virus persists in duck populations through fecal-oral transmission (17). Therefore, the present study is conducted to estimate the prevalence and risk factors of AIV in domestic ducks under different rearing systems and landscapes.
Bangladesh is an agriculture-based country where the total livestock population comprises around 311.8 million chickens and 63.85 million ducks throughout the country (18), which are housed in over 53 thousand commercial broiler farms, 18 thousand commercial layer farms, and 6.5 thousand commercial duck farms, whereas in rural settings on an average, each household rears 6.8 chickens and ducks in backyard systems for their consumption or even commercial activity (19, 20). Furthermore, Bangladesh is also known as a riverine country due to its numerous transboundary rivers, suitable habitats, and wetlands that attract millions of migratory birds of 244 species each winter (October to March) and allow them to intermingle with resident aquatic wild birds and domestic ducks (21, 22). Ducks are typically raised for household and commercial production in Bangladesh using nomadic or semi-scavenging systems. Consequently, domestic ducks have frequent access to wetlands and interact closely with various migratory bird species, which may facilitate the evolution and emergence of novel strains of AIV and eventually lead to widespread outbreaks of the virus. The reservoir duck species are able to shed and transmit the virus from the respiratory and intestinal tracts, showing few or no symptoms of the disease. Therefore, understanding the epidemiology of the origin and circulation pattern of H5N1 in the duck population in Bangladesh is deemed a priority.
The AIV RNA prevalence in domestic ducks in parts of Bangladesh has been previously documented as 0.9–89% (23–25), whereas the dominant AIV subtypes were H5 and H9 in ducks (26). Furthermore, since the first detection of HPAI H5N1 viruses, various clades, including 2.2.2, 2.3.2, 2.3.4.2 (27, 28), 2.3.2.1a (29, 30), and 2.3.4.4 (31) clade of H5N6, have been identified in Bangladesh. Besides, the novel reassortant H5N1 clade 2.3.2.1a has already been isolated from the LBMs in Bangladesh, having a close relatedness to the virus isolated from birds sampled in one of the four regions of this country (32). Furthermore, during the last 3 years, clade 2.3.4.4b of the H5N1 virus has recently spread to domestic poultry and wild birds widely in Europe, Africa, Asia, and America, leading to the loss of over 33 million domestic birds (33). On the other hand, northwest Spain encountered an outbreak of 2.3.4.4b H5N1 in Minks (34). Also, this clade of H5N1 was also detected in mammals like harbor porpoises in Sweden (35) and dolphins, Sea lions, Sanderlings, Pelicans, and Cormorants in Peru (36). There have been 893 sporadic human A(H5N1) cases reported from 21 countries since 1997, and eight of those cases have been caused by clade 2.3.4.4b since 2022, which raises the possibility of a pandemic (37, 38). Both nomadic and backyard ducks are reared in a free scavenging system in Bangladesh, sharing open wetlands with large numbers of migratory waterfowl, and other wild birds and transmission of HPAI H5N1 may occur easily where the migratory birds are considered one of the potential routes for introducing new clades of HPAI H5N1 in Bangladesh (39). The surveillance of AIV in ducks from different production systems and patterns of AIV and subtype circulation within these systems are not well documented. Molecular characterization and evolutionary dynamics of HPAI H5N1 in the duck population are crucial. Therefore, we conducted this study to know the prevalence of AIV and their subtypes H5 and H9, risk factors, and phylodynamics of H5N1 clades circulating in domestic ducks in the different production systems in Bangladesh.
2. Methodology
2.1. Ethical approval
The study protocol was approved by the ethics committee of Chattogram Veterinary and Animal Science University (CVASU) bearing the number CVASU/Dir(R&E) EC/2019/126(1) and CVASU/Dir(R&E)EC/2020/191/7.
2.2. Study design and site selection
Bangladesh currently has three duck production systems: nomadic farms, backyard farms, and commercial farms. Nomadic duck farming is a traditional approach to duck production where the ducks are kept in a free-range habitat and are allowed to roam and feed in various regions (40). In the backyard farming system, household ducks are kept overnight near or within the farmer’s house and travel only over a short distance for scavenging (27), and in the commercial farming system, ducks are kept in total confinement (41). Considering the duck farming patterns, we conducted a cross-sectional study and purposive sampling to find out AIV, H5, and H9 subtype prevalence as well as risk factors among ducks in different production systems from 2019 to 2021 in Bangladesh. The study sites were selected based on duck density, the presence of wetlands, and migrating waterfowl. Data on the distribution of migratory bird staging areas in Bangladesh was obtained from the literature (42, 43). Additionally, the duck density data were gathered from the Bangladesh agriculture census 2019 (44). Figure 1 depicts the nine selected districts of Bangladesh, namely Dhaka, Faridpur, Cumilla, Kushtia, Meherpur, Moulovibazar, Sylhet, Sirajganj, and Rajshahi, which represent the wide spectrum of duck-rearing practices across the country. The sampling of nomadic ducks from Sylhet and Moulovibazar represented the wetland habitats of Haor basin (45), while Kushtia, Meherpur, and Sirajganj were considered as Jamuna floodplains (46). In wetlands, domestic ducks and migratory birds share foraging habits and intermingle. Consequently, backyard ducks were also sampled in wetlands areas. In addition, backyard ducks were sampled from Dhaka, Cumilla, and Faridpur. The samples of commercial duck farms were collected from Cumilla, Dhaka, Kushtia, Meherpur, Rajshahi, and Sylhet (Figure 1).
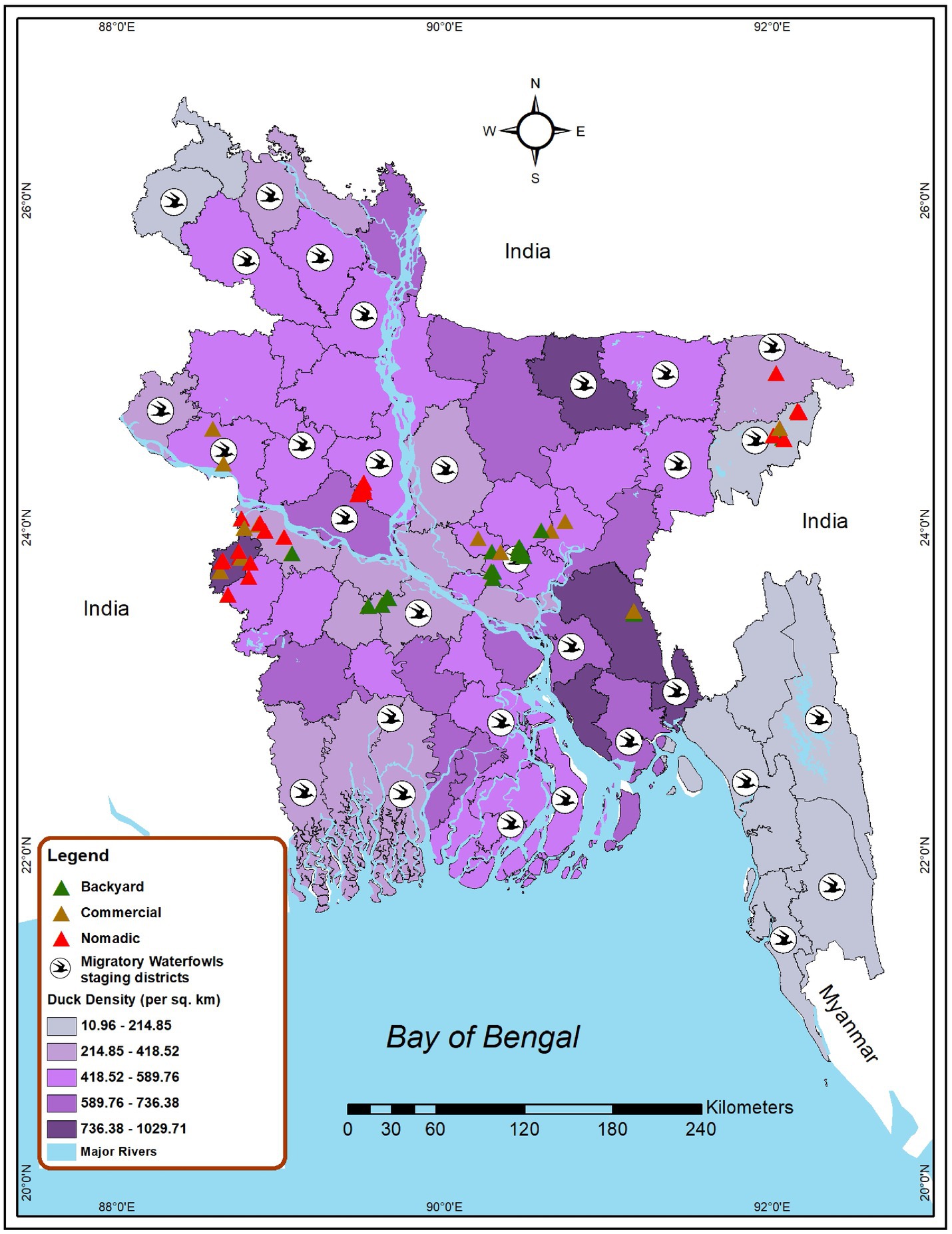
Figure 1. Map locating the site selected for investigating AIV risk analysis among duck farms in Bangladesh (2019–2021). Green triangles represent backyard duck farms, brown triangles represent commercial duck farms, and red triangles represent nomadic duck farms chosen for sampling in this study. Bird symbols denote districts that have migratory bird staging areas. The intensity of the color gradient shows the density of ducks in a district.
2.3. Sample and data collection procedure
We sampled a total of 522 ducks from 171 farms, with 270 ducks coming from 127 backyard farms, 61 ducks from 11 commercial farms, and 191 from 33 nomadic farms. The samples were collected from both sick and healthy ducks, and common signs observed in sick ducks were torticollis, lack of coordination, leg paralysis, and sudden death, which have also been associated with H5N1 symptoms in previous studies (24, 47). Pooled oropharyngeal with cloacal swabs were collected from each duck by an experienced field veterinarian while causing the birds as little distress as possible. The biological specimens were collected by wearing appropriate personal protective equipment like coveralls, gloves, and other safety equipment. Immediately after sampling, the swabs sticks were placed into a 1.8 ml cryovial containing 1 mL viral transport medium (VTM). Each vial was marked using a unique identification number and placed in the portable dry shipper before transport to the laboratory. In the lab, all the samples were stored at –80°C freezer until further laboratory evaluation. A pre-tested questionnaire and face-to-face interview were used to collect all biosecurity-related data, that could potentially be a risk factor.
2.4. Virological testing
The viral RNA was extracted from the pooled swab samples (oropharyngeal and cloacal) using a KingFisher Flex 96-well robot (Thermo Scientific, Waltham, MA) and the MagMAX 96 AI/ND Viral RNA Isolation Kit (Ambion, Inc. Austin, TX) in accordance with the manufacturer’s instructions. Real-time reverse transcriptase PCR (rRT-PCR) was used in conjunction with reference primers and probes to detect the presence of the AIV (InfluA) Matrix (M) gene in viral RNA, as described by the CDC and Spackman (48, 49). Then, InfluA (M-gene) positive samples were examined with specific subtypes primers of H5, H7, and H9 as previously described (49, 50). The samples were considered as AIV positive for the M-gene if the cycle threshold (Ct) was less than 40 and as H5, H7, and H9 positive if Ct < 37 (51). Samples that tested positive for the M gene but negative for H5, H7, and H9 were classified as A/untyped.
2.5. H5N1 sequencing
The viral RNA was extracted using QIAamp viral RNA minikit (Qiagen). The influenza segments were amplified following the protocol described by Zhou et al. (52). After amplification, PCR amplicons were visualized by agarose gel electrophoresis, followed by purification in an AMPure XP Bead. Subsequent nanopore sequencing libraries were prepared using Ligation Sequencing Kit (SQK-LSK109) and the Native barcoding approach. In 2019, the Sanger sequencing was deployed to amplify and subjected to partial sequencing of HA and NA genes of the 2 H5N1 virus described by Hoffmann (53). In 2021, the final library was quantified in the Qubit 1× dsDNA High Sensitivity Assay Kit (Invitrogen) with a Qubit 4 fluorometer (Invitrogen) and loaded onto the FLO-MIN106D flow cell on an Oxford Nanopore MinION MK 1C platform. Raw fast5 reads were base called by real-time base-calling with Guppy 4.3.4, released with MinKNOW software with the fast base-calling mode, and subsequent analyses were performed in the appropriate bioinformatics tools. The HA and NA segments of H5N1 sequences were submitted to GenBank under the accession numbers from OQ430759 to OQ430762 and OQ423229 to OQ423237.
2.6. Statistical analysis
The frequency, percentage, and univariate value of p were computed at the socio-demographic level of the duck farmer, along with duck-rearing practices in different production systems and landscapes. A descriptive analysis was computed to determine the prevalence of AIV, H5, and H9 according to the different factors at the individual bird and flock levels. The cross-tabulation and chi-square tests were performed to identify the risk factors between AIV and H5N1 with different bird-level risk factors. Furthermore, the risk factors that were determined as significant at univariate analysis were forwarded to multivariable logistic regression. The likelihood ratio (Wald test) with a value of p of ≤0.05 was used to identify the primary risk factor. The results were presented as Adjusted Odds Ratios (AOR), 95% confidence intervals, and values of p The data generated from this study were stored in MS Excel 2021 and checked the data integrity in MS Excel. We used RStudio version 4.1.2 for statistical analysis. We used “lme4” and “tidyverse” packages for the analysis in R software. The ArcGIS1 software was used to create a duck density map and to visualize the spatial distribution of migratory waterfowl staging areas and duck farming sites of studied districts (Figure 1). The district-level administrative shape file was retrieved from freely available DIVA-GIS2 (54).
2.7. Bayesian phylogenetic analysis of H5N1 viruses
To identify the clade diversity of H5N1 viruses circulating among ducks in Bangladesh, On January 1, 2023, all accessible HA gene sequences of A/H5N1 HPAIs found in Bangladesh from ducks with full-length HA sequences were retrieved from the GISAID Epiflu database (55). The HA sequences of H5N1 from 2007 to 2022, were retrieved from GISAID and NCBI and then the artifacts sequence were removed. A Maximum Clade Credibility (MCC) tree using the Bayesian Markov Chain Monte Carlo approach was generated using the temporal information of the sequence data to estimate the evolution of H5N1 viruses in Bangladesh (BEAST 1.10.4) (56). The uncorrelated lognormal clock model with the Bayesian Skyline tree prior was used with 10 million generations (57). The gamma-distributed rate variation among sites with four rate categories (HKYþG) (58) was used. The sampling frequency was 1,000. We visualized the MCC trees in FigTree v1.4.4.3 To identify the phylogenetic relationships of the seven H5N1 viruses sequenced in this study, the maximum likelihood method was used. For each gene segment of HA and NA, we used BLAST best matches to select the relevant sequences. TIM + F + G4 model for HA segments and K3Pu + F + G4 for NA segments was chosen by minimum BIC values using IQ-Tree (59). For each tree, we used 1,000 bootstrap replicates for generating the trees. The maximum likelihood tree was also visualized using Figtree v1.4.4.
3. Results
3.1. Prevalence of AIV, H5, H9, and A/Untyped in ducks and farming types
AIV prevalence for the overall sampled duck was 24.9% (130/522) (95% CI: 21.3–28.9) (Figure 2). Across the farming system, AIV prevalence was highest in the nomadic duck (76/191) (39.8%; 95% CI: 32.9–47.1) followed by commercial (15/61) (24.6%; 95% CI: 14.5–37.3) and backyard duck (39/270) (14.4%; 95% CI: 10.5–19.2). The H5 prevalence was prominent in nomadic ducks (37/191) (19.4%; 95% CI: 14.0–25.7). There was no H9 subtype found in commercial and nomadic ducks but in two backyard ducks (1.6, 95% CI: 0–8.8) (Figure 2). None of the sample was positive for H7.
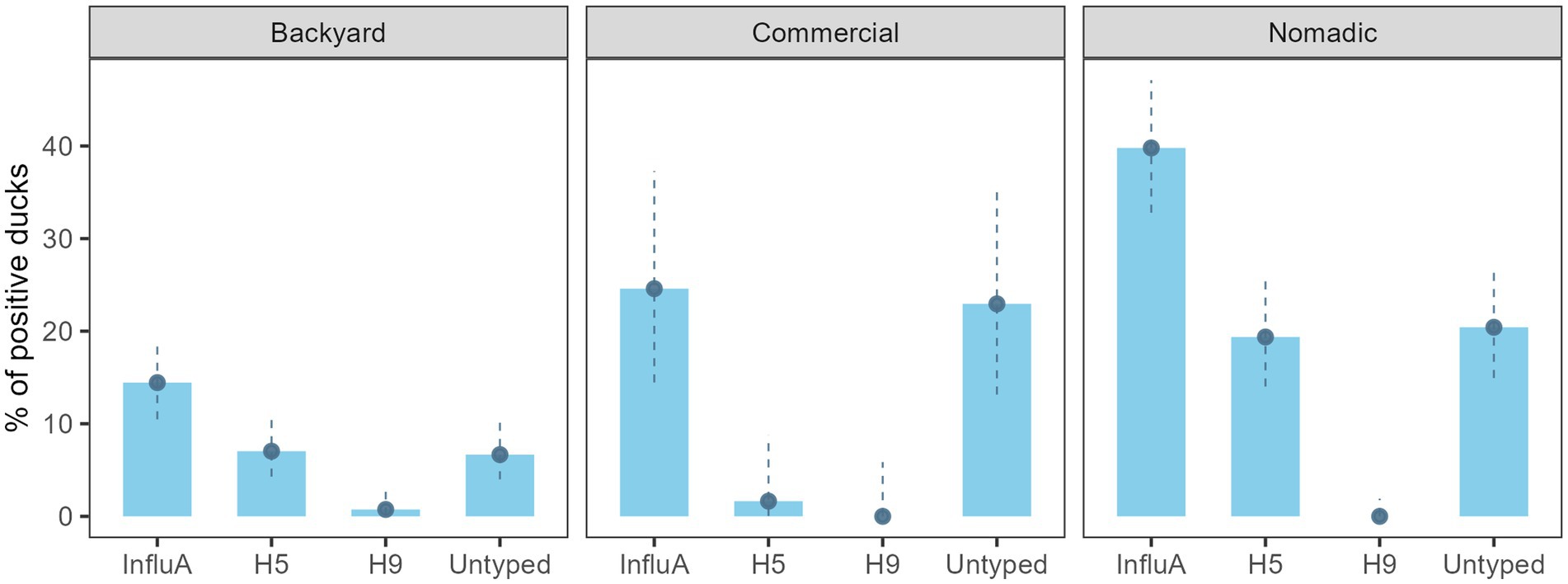
Figure 2. Bangladesh duck level InfluA (M gene), H5, H9, and A/Untyped prevalence with 95% confidence interval during 2019–2021.
On the other hand, AIV prevalence for the overall duck farm was 40.4% (69/171) (95% CI: 33.0–48.1%), backyard farm was 29.1% (37/127) (95% CI: 21.4–37.9), the commercial farm was 63.6% (7/11) (95% CI: 30.8–89.1), and the nomadic farm was 75.8% (25/33) (95% CI: 57.7–88.9) (Figure 3). The H5 subtype was higher (14/33) (42.4%; 95% CI: 25.5–60.8) in the nomadic farming system. In backyard farming, the prevalence of H5 and A/Untyped subtypes on farms were similar (18/127) (14.2%; 95% CI: 8.6–21.5) (Figure 3).
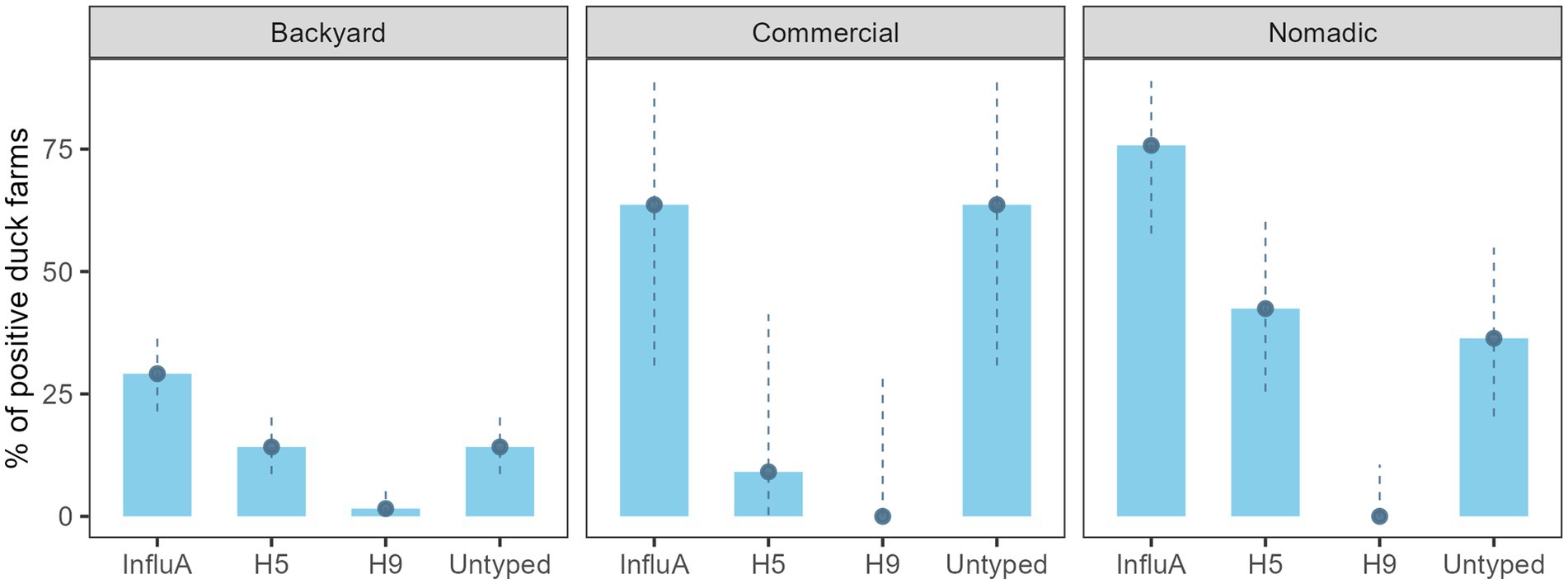
Figure 3. Duck farm level InfluA (M gene), H5, H9, and A/Untyed prevalence with 95% confidence interval in Bangladesh (2019–2021).
3.2. Association of AIV and its subtypes with migratory waterfowl interface
We found evidence of an association between AIV subtypes and migratory waterfowl present in that area. A/H5 and A/Untyped were significantly associated with the presence of migratory waterfowl (Figure 4).
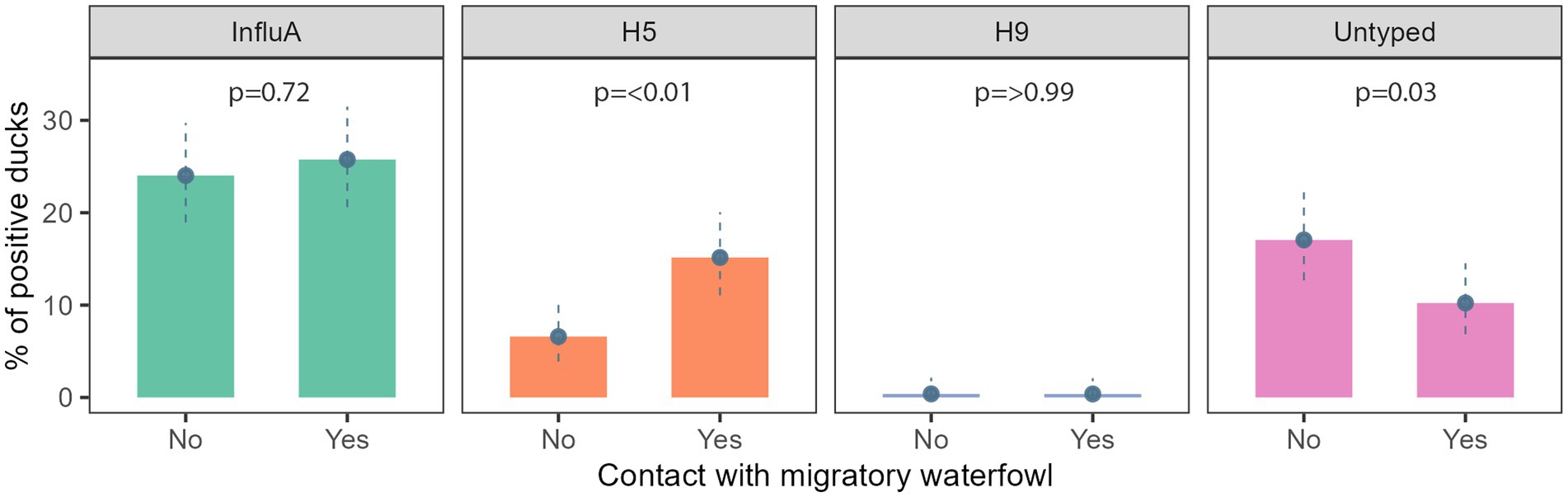
Figure 4. Prevalence of InfluA and its subtypes with migratory waterfowl contact in Bangladesh during 2019–2021.
3.3. Risk factor for the circulation of AIV in ducks
We had four variables to check for association with AIV and H5. The farming system, age, and health condition were significantly associated with AIV and H5. Among the farming system, the nomadic system had a higher prevalence for AIV (39.8%; 95% CI: 32.8–47.1%) and H5 (19.4%; 95% CI: 32.8–47.1), whereas backyard and commercial were less positive. Juvenile age group birds were significantly more positive than adults, and sick birds were the most affected by AIV (83.3%; 95% CI: 67.2–93.7) and H5 (80.6%; 95% CI: 64.0–91.8) (Table 1). The sick ducks developed neurological signs including uncoordinated gait circling and torticollis at the terminal stage, digestive symptoms (whitish feces, fecal attached to the plumage and cloaca) and respiratory distress, dilated pupils and followed by death.
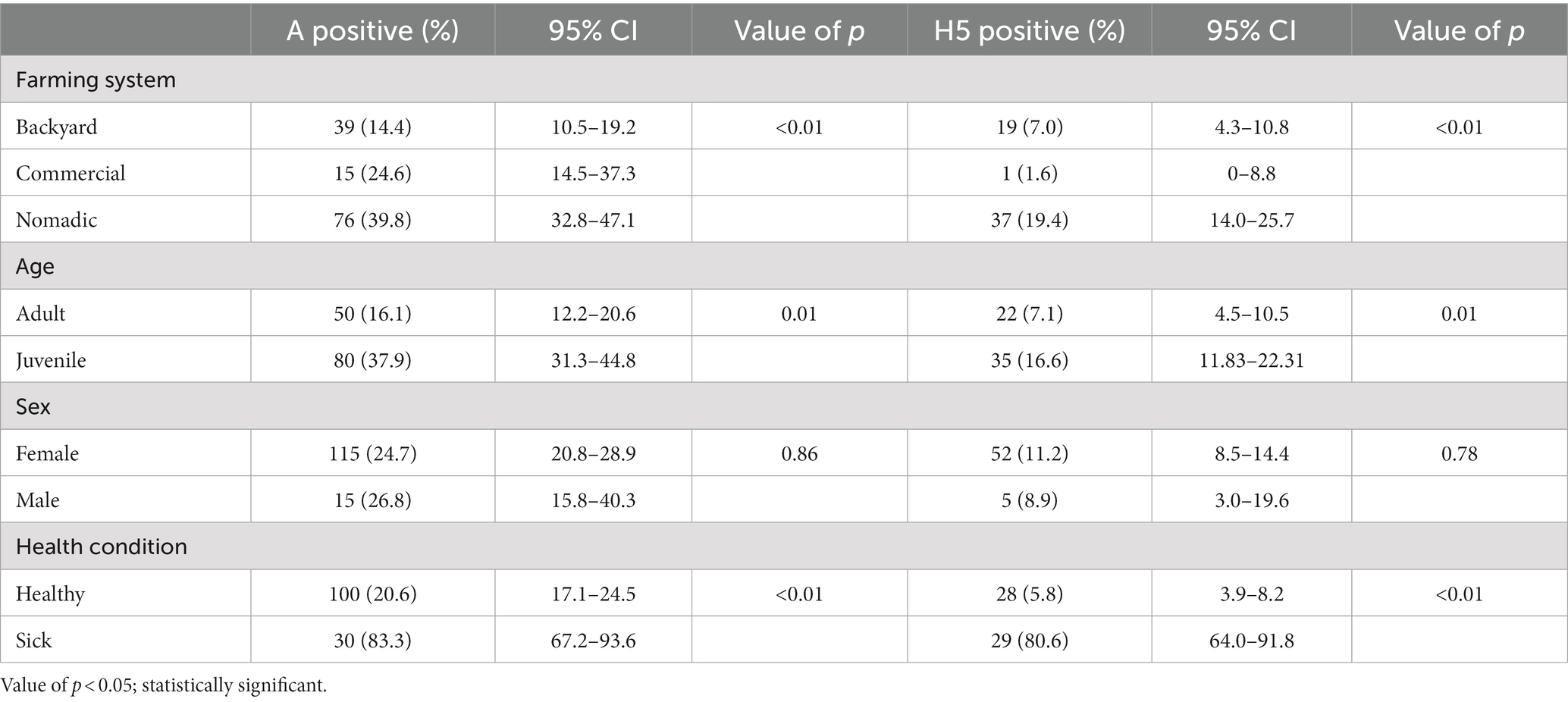
Table 1. Cross table with chi-square analysis between AIV and bird-level factors of duck Bangladesh isolates (2019–2021).
In the multivariable logistic regression model, we found three variables as significant risk factors for AIV and one risk factor for A/H5. The nomadic farming system had 2.39 times (95% CI: 1.45–3.93) higher odds of affecting AIV than backyard farming (p = 0.01). Compared to adults, juvenile ducks had 2.22 times (95% CI: 1.37–3.61) odds of having AIV (p = 0.01). The AIV detection in sick ducks (the ducks displayed dilated pupils and white feces remained on the plumage surrounding the cloaca and neurologic symptoms include an uncoordinated gait, tremors, and torticollis) was 11.59 times (95% CI: 4.82–32.44) more likely (p < 0.01) than healthy birds (Table 2). We found health conditions to be a significant risk factor for H5. The odds of H5 detection in sick birds were 46.5 times (95% CI: 18.7–130.3) more likely than healthy ones (p < 0.01) (Table 2).
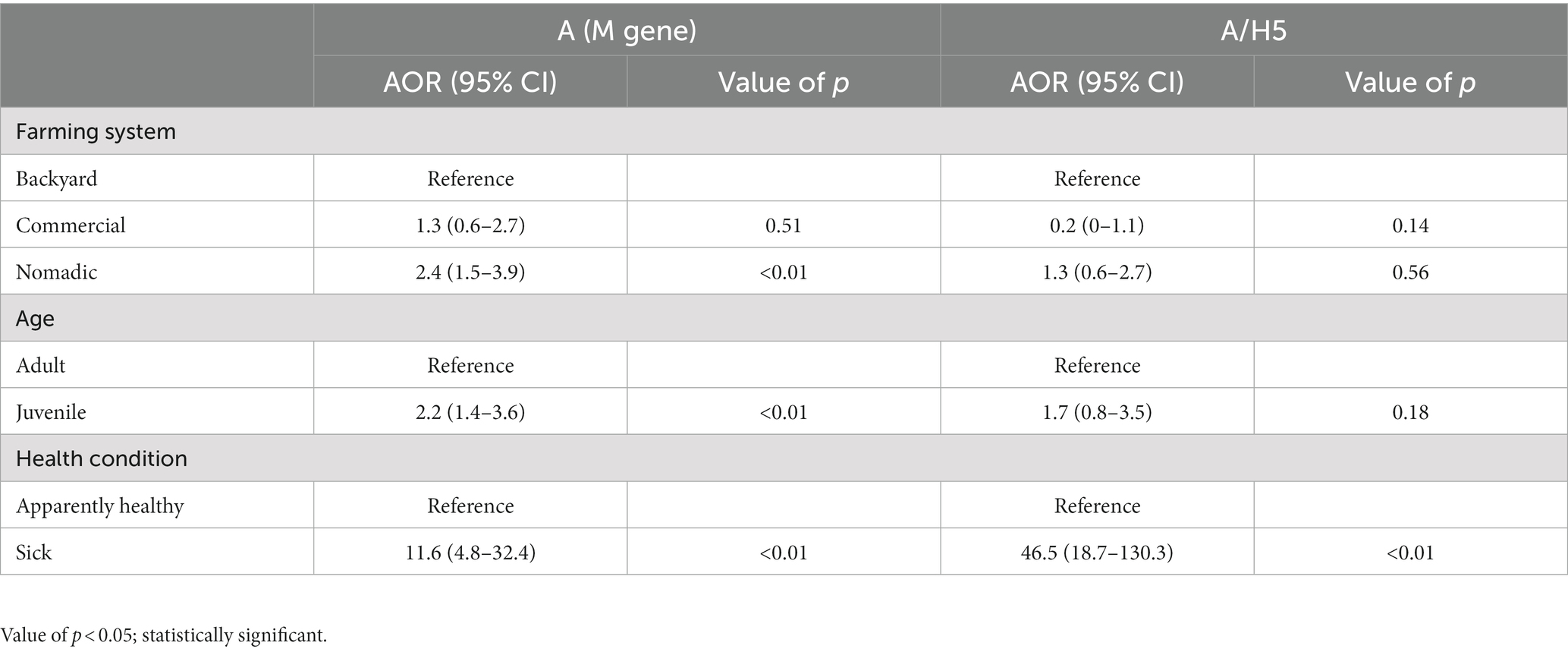
Table 2. Risk factors of AIV and A/H5 circulation in individual ducks from the different production systems in Bangladesh (2019–2021).
3.4. Bayesian phylogenetic analysis of the evolution of H5N1 clades in Bangladeshi ducks
The Bayesian phylogenetic tree (Figure 5) indicates that clade 2.3.2.1a has been circulating in ducks in Bangladesh since 2011. In 2015, the novel reassortant of the clade 2.3.2.1a H5N1 virus was discovered in ducks (Figure 5). The majority of H5N1 viruses detected in waterfowl are novel reassortant of clade 2.3.2.1a. Seven H5N1 sequences were identified as belonging to the emerging panzootic clade 2.3.4.4b (Figure 5). Sequences from this emerging clade clustered with white-tailed eagles from Japan (Hokkaido), geese from China (Hunan), and chickens and ducks from Africa (Nigeria and Benin). These sequences share a similarity of between 98.65 and 98.97% with H5N1 viruses of clade 2.3.4.4b from Japan and a similarity of 99.30% with viruses from China. This new clade may have been introduced to Bangladesh by the end of 2020 (Figure 5). Figure 6 shows clade 2.3.2.1a has been silently evolving among ducks, and based on posterior probability >90%, and that the clade has formed at least nine subgroups among ducks in Bangladesh. Currently, only subclade R9 of clade 2.3.2.1a is circulating in ducks in Bangladesh.

Figure 5. Bayesian phylogenetic tree of H5 HA viruses of diverse clades in Duck in Bangladesh. Taxon labels with blue color indicate the viruses found in ducks under this study, and taxon labels with green color indicate those reassorted H5N1 viruses detected as clade 2.3.4.4b.
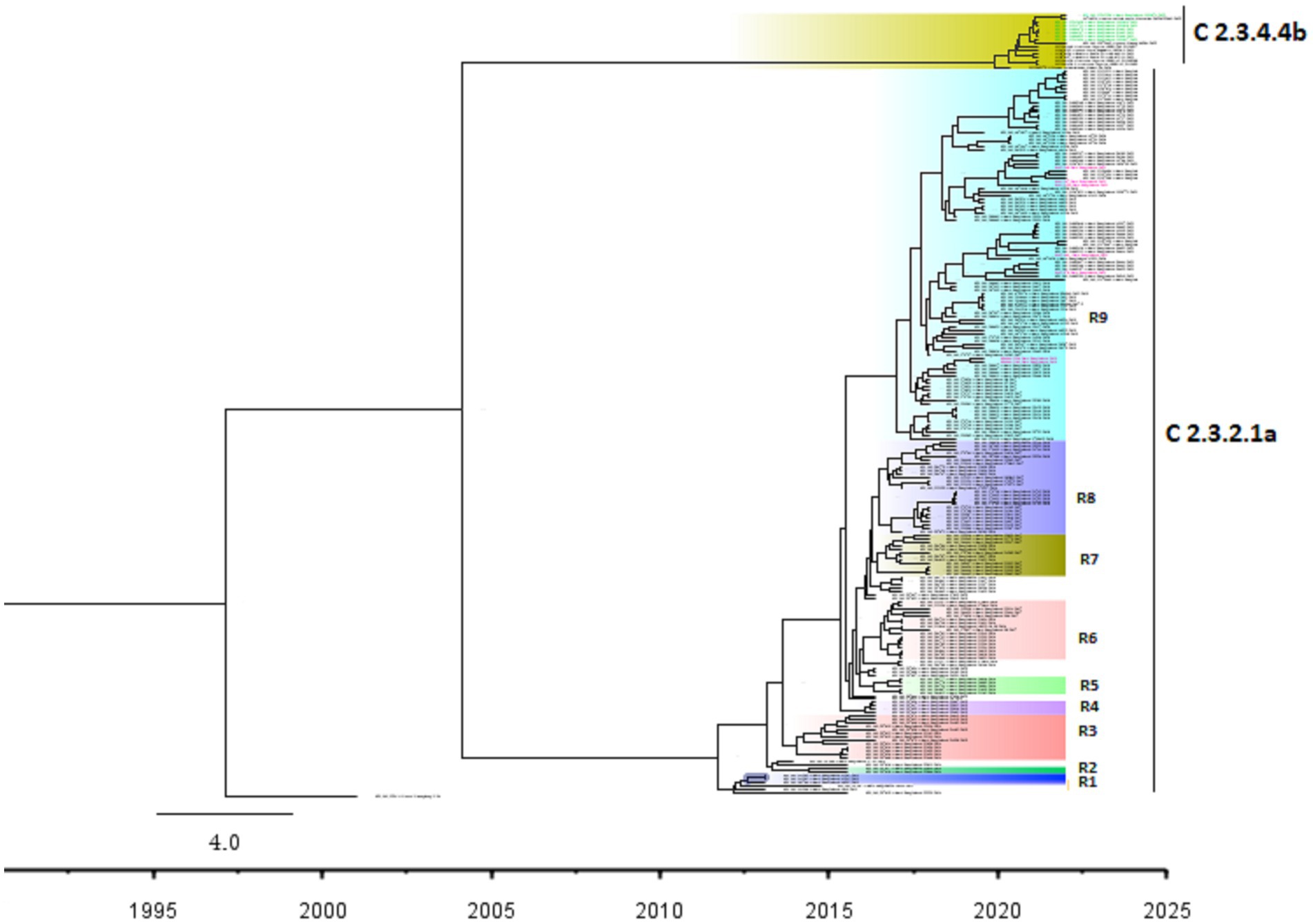
Figure 6. Bayesian phylogenetic tree of H5 HA viruses of diverse clades along with subgroups based on >90% posterior probability in Duck in Bangladesh. Taxon labels with red color indicate the viruses found in ducks under this study, and taxon labels with green color indicate those reassorted H5N1 viruses detected as clade 2.3.4.4b. Each colored box indicates a subgroup.
3.5. Maximum likelihood phylogenetic analysis of HA and NA sequences of H5N1 viruses isolated in ducks in Bangladesh
Figures 7, 8 present the maximum likelihood phylogenetic trees of HA and NA gene segments of H5N1 viruses sequenced in this study. Five H5N1 viruses were detected in 2021 and the two viruses in 2019 belonged to the newly reassorted clade 2.3.2.1a. However, they clustered in different groups within this clade. The two virus sequences from 2019 have clustered together. The maximum likelihood tree of HA also shows that the viruses we detected in duck hosts are similar to those found in chickens. BAIV-570 and BAIV-404 have clustered within a group with virus sequences obtained from chicken (Bootstrap value>95%).
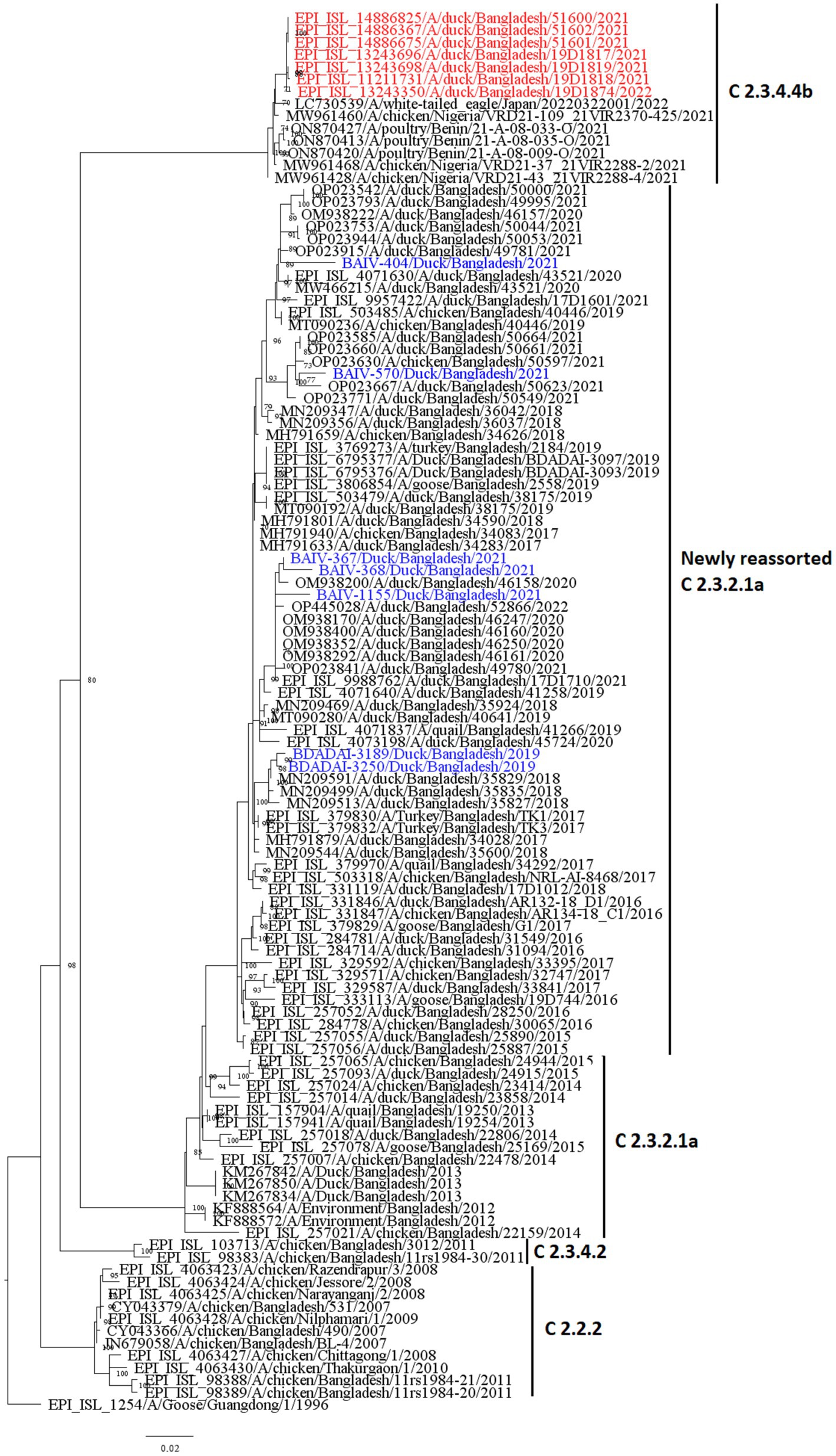
Figure 7. Maximum likelihood tree of HA sequences of H5N1 viruses in Bangladesh. Taxon labels with blue color indicate the viruses found in ducks under this study, and taxon labels with red color indicate those reassorted H5N1 viruses detected as clade 2.3.4.4b.

Figure 8. Maximum likelihood tree of 6 NA sequences of H5N1 viruses. Taxon labels with blue color indicate the viruses found in ducks under this study, and taxon labels with red color indicate those reassorted H5N1 viruses detected as HA clade 2.3.4.4b.
4. Discussion
4.1. Prevalence and risk factors of AIV and subtypes in the different duck production systems
The high prevalence of AIV with the H5N1 subtype in nomadic ducks compared to backyard and commercial ducks was consistent with the other study conducted by Khatun et al. (60), reported a higher prevalence of AIV in ducks reared in the hoar (wetland) region where the nomadic system is prevalent. This is because of the higher density of migratory birds in the hoar area, with a possible most increased interaction between the native duck and migratory bird species (61). Previous studies in Bangladesh detected AIV with H5N1 in both domestic and migratory ducks in wetland areas where domestic ducks and migratory birds shared the same feeding habitats in wetlands (62, 63).
Furthermore, the farm-level prevalence of AIV was also higher in nomadic ducks, supported by Hasan et al. (61) because the grazing land ecosystem is a critical factor for the circulation and spread of AIV. Concerning risk factors, there is a significant association among different farming systems, which is also supported by Henning et al. (64) reported that the birds that used to scavenge are most frequently affected. Juvenile ducks were mostly affected by both AIV M-gene and H5 subtype, which was supported by Strurm-Ramirez et al. (65). Our study revealed that the farming system significantly impacts the presence of AIV in ducks. The odds of AIV have been observed to be greater in ducks from nomadic farms than in backyard ducks. As low-lying areas with vast bodies of water are a favorable environment for raising nomadic ducks, and they have more interaction with migrating waterfowl than a backyard or commercial ducks, previous studies have shown that nomadic ducks are more susceptible to the AIV (66, 67). Our study also showed that juvenile ducks are more likely to be infected by AIV than adult ducks. A study in Canada also reported a higher detection rate of AIV in juvenile ducks than in adults (68). Adult birds presumably have acquired immunity or an enhanced immune response, but juvenile birds are immunologically more naive, rendering them more vulnerable to viral infection than adult birds (69).
Our study also showed that detecting AIV and A/H5 is higher in sick ducks than in apparently healthy ducks. Previous studies in Bangladesh and other countries have reported similar results for ducks and poultry birds (2, 70–73). Even though ducks can secrete large quantities of a deadly virus without manifesting any outward signs of disease, H5N1 can cause the birds to have breathing difficulties such as gaping (mouth breathing), nasal snicking (coughing sound), sneezing, gurgling, or rattling. Since AIV causes bird sickness, the detection rate of AIV and H5 is higher in sick ducks (74–76).
4.2. Phylodynamic of multiple clades of H5N1 viruses in duck farms
According to our study, multiple H5N1 virus clades are spreading in Bangladesh. Our study shows that two clades of H5N1 viruses are now circulating among ducks in Bangladesh. These two clades, 2.3.2.1a and 2.3.4.4b, of H5N1 viruses in ducks, have also been detected in nearby countries such as India and China (77, 78). Our study shows that clade 2.3.2.1a has been detected in ducks since 2011 and has become endemic in ducks in Bangladesh. Similar to our findings, other studies reported that this clade reassorted, resulting in a new subclade in 2015 (79). According to Barman et al. (32), this novel reassortant clade 2.3.2.1a virus emerged in Bangladesh via reassortment with LPAI viruses transmitted by migrating birds. Despite vaccination of commercial chicken farms in Bangladesh, Clade 2.3.2.1a HPAI has caused ongoing outbreaks in Bangladesh since 2011. Our study also shows that viruses of clade 2.3.2.1a have created at least nine subgroups within ducks based on >90% posterior probability. This suggests that the virus of this clade is silently evolving, and ducks may play an important role in the emergence of new clades in Bangladesh. Prior studies have also shown that clade 2.3.2.1a HPAIs are circulating in LBMs and domestic ducks in Bangladesh, where they play an important role in the maintenance and development of new reassortant viruses (25, 80).
The findings of the phylogenetic study also revealed that clade 2.3.4.4b was introduced to Bangladesh by the end of 2020. Cui et al. (33) also reported that one H5N1 virus from Bangladesh clustered with Chinese viruses within the 2.3.4.4b clade. Since its emergence in the Netherlands in October 2020, H5N1 viruses with the clade 2.3.4.4b HA gene have spread to several countries in Europe, Africa, Asia, and North and South America (81, 82). On the other hand, in China, Between September 2021 and March 2022, H5N1 viruses bearing the HA clade 2.3.4.4b were discovered in wild birds and domestic poultry (33). Our study shows that seven viruses clustering within the 2.3.4.4b clade have similarities with viruses from Japan and China. So, it might be possible that migratory birds of the Central Asian flyway may influence the transmission of this novel clade in Bangladesh. Though the H5N1 viruses with clade 2.3.4.4b have only been detected in Ducks in Bangladesh, this clade has been detected in wild birds and domestic Anseriformes and Galliformes in other countries (83–85). On the other hand, this clade has also caused outbreaks in minks in Spain (86). More than 50 thousand mink were killed and their carcasses destroyed, and it was assumed that wild birds may have played a major role in the transmission of the virus (34). This virus has also been detected in harbor porpoises in Sweden (35) and dolphins, sea lions, sanderlings, pelicans, and cormorants in Peru (36), along with 8 human cases since 2022 (37). It is extremely alarming because the H5N1 virus is known to spread poorly among mammals; humans almost exclusively contract it from infected birds. However, it has since been established that the 2.3.4.4b outbreak in minks spread throughout a tightly-knit mammalian population (87). Given that the virus has already been introduced to Bangladesh, it is likely that this clade may also spread to chickens and other poultry through ducks and wild birds. As a result, there is a danger of transmission among humans as well as the possibility of a pandemic. We recommend carrying out a thorough risk analysis so that decision-makers may fully comprehend the risks connected to AIV and H5N1 outbreaks, the possible effects of the epidemic, and the steps that can be done to prevent or mitigate the disease’s transmission.
5. Conclusion
This study demonstrates that H5N1 circulating in all three duck farming production systems and nomadic farms poses a higher risk of AIV infection than those from residential or commercial farms. Age and health of ducks influence the risk of AIV and H5N1 infection in populations of ducks. Clades 2.3.2.1a and 2.3.4.4b of H5N1 are circulating in Bangladeshi waterfowl. The duck farmer should receive appropriate training to enhance farm biosecurity practices in order to prevent the spread of AIV. Enhanced AIV surveillance is necessary for both domestic and migratory waterfowl, with a focus on Anseriformes production systems, to analyze the genetic diversity of H5N1 viruses and to determine the evolution of the virus at high-risk interfaces between domestic ducks and migratory birds.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The studies involving human participants were reviewed and approved by the Ethics Committee (EC) (Protocol: CVASU/Dir(R&E) EC/2020/191/7) of the Chattogram Veterinary and Animal Sciences University. The participants provided their written informed consent to participate in this study. The animal study was reviewed and approved by the Chattogram Veterinary and Animal Sciences University-Animal Experimentation Ethics Committee (protocol: CVASU/Dir(R&E) EC/2020/191/7). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
AI: conceptualization. AI, MdH, and SI: methodology. AI, MdH, SI, and MAS: field investigation. AI, MAS, and MI: validation.AI, EA, and MI: software and formal analysis. AI and EA:visualization and writing—original draft. AI, EA, MMH, and MI: data curation. MEH, MM, and MZR: laboratory analysis.AI, MAS, MI, and MMH: writing—review, and editing. TS and MZR: supervision. MMH, TS, and MZR: project administration and funding acquisition. All authors contributed to the article and approved the submitted version. All authors have read and approved the final version of the manuscript.
Funding
The sample collection was supported by the University Grant Commission (UGC) of Bangladesh through Chattogram Veterinary and Animal Sciences University (CVASU), grant number UGC/CVASU#06, and the United States Agency for International Development (USAID) Emerging Pandemic Threats PREDICT project (cooperative agreement number: AID-OAA-A-14-00102) through EcoHealth Alliance. We thank the governments of Bangladesh, Canada, Sweden, and the United Kingdom for providing core/unrestricted support to icddr, b. The team was partially supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID) National Institutes of Health (NIH) (U01AI153420).
Acknowledgments
We acknowledge the Institute of Epidemiology, Disease Control and Research (IEDCR) Bangladesh and EcoHealth Alliance, “The Conservation, Food and Health Foundation(CFHF)” and the Centre for Integrative Ecology at Deakin University, Australia, for their support in conducting this research. We acknowledge the use of sequences and metadata from the EpiFlu database of the Global Initiative on Sharing Avian Influenza Data (GISAID) and the animal surveillance database of the Influenza Research Database.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Sun, Y, Zhao, X, Qian, J, Jiang, M, Jia, M, Xu, Y, et al. High activity levels of avian influenza upwards 2018–2022: a global epidemiological overview of fowl and human infections. One Health. (2023) 16:100511. doi: 10.1016/j.onehlt.2023.100511
2. Islam, A, Amin, E, Hasan, R, Hassan, MM, Miah, M, Samad, MA, et al. Patterns and risk factors of avian influenza a (H5) and a (H9) virus infection in pigeons and quail at live bird markets in Bangladesh, 2017–2021. Front Veter Sci. (2022) 9:9. doi: 10.3389/fvets.2022.1016970
3. FAO. EMPRES global animal disease information system (EMPRES-i). Available at: https://empres-i.apps.fao.org/ (accessed on April 20, 2023). (2023)
4. Islam, A, Islam, S, Amin, E, Hasan, R, Hassan, MM, Miah, M, et al. Annual trading patterns and risk factors of avian influenza a/H5 and a/H9 virus circulation in Turkey birds (Meleagris gallopavo) at live bird markets in Dhaka city, Bangladesh. Front Veter Sci. (2023) 9:684. doi: 10.3389/fvets.2022.1016970
5. IEDCR. First human infection with influenza a H5N1 confirmed in Bangladesh. Available at: https://www.iedcr.org/index.php?option=com_content&view=article&id=61&Itemid=115 (Accessed March 27, 2023) (2012)
6. Rahman, MA, Belgrad, JP, Sayeed, MA, Abdullah, MS, Barua, S, Chisty, NN, et al. Prevalence and risk factors for avian influenza virus (H5 and H9) contamination in peri-urban and rural live bird markets in Bangladesh. Front Public Health. (2023) 11:1349.doi: 10.3389/fpubh.2023.1148994
7. Islam, A, Islam, S, Islam, M, Hossain, ME, Munro, S, Samad, MA, et al. Prevalence and risk factors of avian influenza viruses among household ducks in Chattogram, Bangladesh. Vet Res Commun. (2022) 46:471–80.doi: 10.1007/s11259-021-09874-4
8. Chowdhury, M, Islam, S, Hossain, ME, Rahman, MZ, Zulkar Nine, HSM, Sadik, AS, et al. Detection of influenza a and adenovirus in captive wild birds in Bangladesh. Int J Infect Dis. (2020) 101:229. doi: 10.1016/j.ijid.2020.11.034
9. Islam, A, Islam, S, Rahman, MK, Hossain, ME, Samad, MA, Rostral, MK, et al. Circulation of avian influenza H5N1 and H9N2 virus in dhamrai and savar upazila, Bangladesh. Int J Infect Diseases. (2023) 130:S75. doi: 10.1016/j.ijid.2023.04.186
10. Olsen, B, Munster, VJ, Wallensten, A, Waldenström, J, Osterhaus, ADME, and Fouchier, RAM. Global patterns of influenza a virus in wild birds. Science. (2006) 312:384–8. doi: 10.1126/science.1122438
11. Hassan, MM, Hoque, MA, Debnath, NC, Yamage, M, and Klaassen, M. Are poultry or wild birds the main reservoirs for avian influenza in Bangladesh? EcoHealth. (2017) 14:490–500. doi: 10.1007/s10393-017-1257-6
12. Torrontegi, O, Alvarez, V, Acevedo, P, Gerrikagoitia, X, Höfle, U, and Barral, M. Long-term avian influenza virus epidemiology in a small Spanish wetland ecosystem is driven by the breeding Anseriformes community. Vet Res. (2019) 50:4. doi: 10.1186/s13567-019-0623-5
13. Mateus-Anzola, J, Martínez-López, B, Espinosa-García, AC, and Ojeda-Flores, R. Global subtype diversity, spatial distribution patterns, and phylogenetic analysis of avian influenza virus in water. Transbound Emerg Dis. (2022) 69:e344–55. doi: 10.1111/tbed.14307
14. Zhang, H, Xu, B, Chen, Q, Chen, J, and Chen, Z. Characterization of an H10N8 influenza virus isolated from Dongting lake wetland. Virol J. (2011) 8:1–9. doi: 10.1186/1743-422X-8-42
15. Blagodatski, A, Trutneva, K, Glazova, O, Mityaeva, O, Shevkova, L, Kegeles, E, et al. Avian influenza in wild birds and poultry: dissemination pathways, monitoring methods, and virus ecology. Pathogens. (2021) 10:630. doi: 10.3390/pathogens10050630
16. Ahrens, AK, Selinka, HC, Wylezich, C, Wonnemann, H, Sindt, O, Hellmer, HH, et al. Investigating environmental matrices for use in avian influenza virus surveillance—surface water, sediments, and avian fecal samples. Microbiol Spectrum. (2023) 11:e02664–22. doi: 10.1128/spectrum.02664-22
17. Kim, JK, Negovetich, NJ, Forrest, HL, and Webster, RG. Ducks: the “Trojan horses” of H5N1 influenza. Influenza Other Respir Viruses. (2009) 3:121–8. doi: 10.1111/j.1750-2659.2009.00084.x
18. DLS. Livestock economy at a glance. Available at: http://dls.portal.gov.bd/sites/default/files/files/dls.portal.gov.bd/page/ee5f4621_fa3a_40ac_8bd9_898fb8ee4700/2022-07-18-03-43-37d18965a6458cda3c542ab146480962.pdf (Accessed November 27, 2022). (2022)
19. Banglapedia. Livestock. Available at https://en.banglapedia.org/index.php/Livestock#:~:text=Livestock%20population%20in%20Bangladesh%20is,chicken%20and%2034.1%20million20ducks (Accessed November 27, 2022). (2021).
20. Karmoker, Y. Self sufficiency in protein: poultry industry in Bangladesh. Available at: https://businessinspection.com.bd/poultry-industry-in-bangladesh/ (Accessed November 27, 2022).(2022).
21. Mahmud, I., Bangladesh saw 100,000 more migratory birds in 2019. Prothom Alo. Available at: https://en.prothomalo.com/bangladesh/Bangladesh-saw-100-000-more-migratory-birds-in (Accessed November 27, 2022). (2019).
22. Hoque, MA, Hassan, MM, Haque, E, Shaikat, AH, Khan, SA, Alim, A, et al. A survey of gastro-intestinal parasitic infection in domestic and wild birds in Chittagong and greater Sylhet. Bang Prevent Veter Med. (2014) 117:305–12. doi: 10.1016/j.prevetmed.2014.07.012
23. Ansari, WK, Parvej, MS, el Zowalaty, ME, Jackson, S, Bustin, SA, Ibrahim, AK, et al. Surveillance, epidemiological, and virological detection of highly pathogenic H5N1 avian influenza viruses in duck and poultry from Bangladesh. Vet Microbiol. (2016) 193:49–59. doi: 10.1016/j.vetmic.2016.07.025
24. Haider, N, Sturm-Ramirez, K, Khan, SU, Rahman, MZ, Sarkar, S, Poh, MK, et al. Unusually high mortality in waterfowl caused by highly pathogenic avian influenza a (H5N1) in Bangladesh. Transbound Emerg Dis. (2017) 64:144–56. doi: 10.1111/tbed.12354
25. Khan, SU, Gurley, ES, Gerloff, N, Rahman, MZ, Simpson, N, Rahman, M, et al. Avian influenza surveillance in domestic waterfowl and environment of live bird markets in Bangladesh, 2007-2012. Sci Rep. (2018) 8:9396. doi: 10.1038/s41598-018-27515-w
26. Islam, A, Qayum, MO, Hossain, ME, Islam, S, Islam, K, Alam, HMS, et al. Epidemiological investigation of H9N2 virus circulation in backyard poultry farms and humans in a rural community. BANG Int J Infect Diseases. (2023) 130:S72. doi: 10.1016/j.ijid.2023.04.179
27. Dutta, P, Islam, A, Sayeed, MA, Rahman, MA, Abdullah, MS, Saha, O, et al. Epidemiology and molecular characterization of avian influenza virus in backyard poultry of Chattogram, Bangladesh. Infect Genet Evol. (2022) 105:105377. doi: 10.1016/j.meegid.2022.105377
28. Hoque, MA, Tun, HM, Hassan, MM, Khan, SA, Islam, SKMA, Islam, MN, et al. Molecular epidemiology of influenza a (H5N1) viruses, Bangladesh, 2007–2011. Prev Vet Med. (2013) 111:314–8. doi: 10.1016/j.prevetmed.2013.06.003
29. Islam, A, Islam, S, Samad, MA, Hossain, ME, Hassan, MM, Alexandersen, S, et al. Epidemiology and molecular characterization of multiple avian influenza A/H5 subtypes circulating in house crow (Corvus splendens) and poultry in Bangladesh. Int J Infect Dis. (2022) 116:S92–3. doi: 10.1016/j.ijid.2021.12.218
30. Islam, A, Islam, S, Hossain, ME, Samad, MA, Billah, MM, Hassan, MM, et al. One health investigation of house crow (Corvus splendens) mortality event linked to the potential circulation of H5N1 virus at live bird Markets in Northwestern Bangladesh. Int J Infect Dis. (2022) 116:S112. doi: 10.1016/j.ijid.2021.12.265
31. Islam, A, Hossain, ME, Islam, S, Samad, MA, Rahman, MK, Chowdhury, MGA, et al. Detection and genetic characterization of avian influenza a (H5N6) virus clade 2.3. 4.4 in isolates from house crow and poultry in Bangladesh, 2017. Int J Infect Dis. (2020) 101:339–40. doi: 10.1016/j.ijid.2020.09.894
32. Barman, S, Marinova-Petkova, A, Hasan, MK, Akhtar, S, el-Shesheny, R, Turner, JCM, et al. Role of domestic ducks in the emergence of a new genotype of highly pathogenic H5N1 avian influenza a viruses in Bangladesh. Emerg Microbes Infect. (2017) 6:1–13. doi: 10.1038/emi.2017.60
33. Cui, P, Shi, J, Wang, C, Zhang, Y, Xing, X, Kong, H, et al. Global dissemination of H5N1 influenza viruses bearing the clade 2.3. 4.4 b HA gene and biologic analysis of the ones detected in China. Emerg Microbes Infect. (2022) 11:1693–704. doi: 10.1080/22221751.2022.2088407
34. Agüero, M, Monne, I, Sánchez, A, Zecchin, B, Fusaro, A, Ruano, MJ, et al. Highly pathogenic avian influenza a (H5N1) virus infection in farmed minks, Spain, October 2022. Eur Secur. (2023) 28:2300001. doi: 10.2807/1560-7917.ES.2023.28.3.2300001
35. Thorsson, E, Zohari, S, Roos, A, Banihashem, F, Bröjer, C, and Neimanis, A. Highly pathogenic avian influenza a (H5N1) virus in a harbor porpoise, Sweden. Emerg Infect Dis. (2023) 29:852–5. doi: 10.3201/eid2904.221426
36. Leguia, M, Garcia-Glaessner, A, Munoz-Saavedra, B, Juarez, D, Barrera, P, Calvo-Mac, C, et al. Highly pathogenic avian influenza a (H5N1) in marine mammals and seabirds in Peru. bioRxiv. (2023). doi: 10.1101/2023.03.03.531008
37. CDC, Technical report: highly pathogenic avian influenza A(H5N1) viruses. Available at: https://www.cdc.gov/flu/avianflu/spotlights/2022-2023/h5n1-technical-report.htm#infections-among-mammals (Accessed May 3, 2023). (2023).
38. WHO, Avian influenza weekly update number 891. Available at: https://www.who.int/docs/default-source/wpro---documents/emergency/surveillance/avian-influenza/ai_20230414.pdf?sfvrsn=5f006f99_113 (Accessed May 3, 2023). (2023).
39. Parvin, R, Kamal, AH, Haque, ME, Chowdhury, EH, Giasuddin, M, Islam, MR, et al. Genetic characterization of highly pathogenic H5N1 avian influenza virus from live migratory birds in Bangladesh. Virus Genes. (2014) 49:438–48. doi: 10.1007/s11262-014-1118-0
40. Henning, J, Henning, KA, Long, NT, Ha, NT, Vu, LT, and Meers, J. Characteristics of two duck farming systems in the Mekong Delta of Viet Nam: stationary flocks and moving flocks, and their potential relevance to the spread of highly pathogenic avian influenza. Trop Anim Health Prod. (2013) 45:837–48. doi: 10.1007/s11250-012-0296-9
41. das, SC, Chowdhury, SD, Khatun, MA, Nishibori, M, Isobe, N, and Yoshimura, Y. Poultry production profile and expected future projection in Bangladesh. Worlds Poult Sci J. (2008) 64:99–118. doi: 10.1017/S0043933907001754
42. Muzaffar, SB, Takekawa, JY, Prosser, DJ, Douglas, DC, Yan, B, and Xing, Z. Ecological determinants of highly pathogenic avian influenza (H5N1) outbreaks in Bangladesh. PLoS One. (2012) 7:e33938. doi: 10.1371/journal.pone.0033938
43. Hassan, MM, el Zowalaty, ME, Islam, A, Khan, SA, Rahman, MK, Järhult, JD, et al. Seasonal movements and migration of Pallas’s gulls Larus ichthyaetus from Qinghai Lake. China Forktail. (2008) 2008:100–7.
45. Khan, KA, Saha, S, Hossain, M, Haque, M, Haq, M, and Islam, M. Epidemiological investigation of recurrent outbreaks of duck plague in selected Haor (wetland) areas of Bangladesh. J Adv Veter Anim Res. (2018) 5:131–9. doi: 10.5455/javar.2018.e256
46. Hossain, M, Hoque, M, Fournie, G, Das, G, Henning, J, Järhult, JD, et al.Management and health of stationary and nomadic ducks in the coastal and Haor areas of Bangladesh. (2019)
47. Khan, SU, Berman, LS, Haider, N, Gerloff, N, Rahman, MZ, Shu, B, et al. Investigating a crow die-off in January–February 2011 during the introduction of a new clade of highly pathogenic avian influenza virus H5N1 into Bangladesh. Arch Virol. (2014) 159:509–18. doi: 10.1007/s00705-013-1842-0
48. Spackman, E. Avian influenza virus detection and quantitation by real-time RT-PCR. Anim Influenza Virus. (2014):105–18. doi: 10.1007/978-1-4939-0758-8_10
49. CDC laboratory support for influenza surveillance (CLSIS). Centers for Disease Control and Prevention. Atlanta, GA, USA (2013).
50. Ali, MZ, Hasan, M, and Giasuddin, M. Potential risk factors of avian influenza virus infection in asymptomatic commercial chicken flocks in selected areas of Bangladesh during 2019. J Adv Vet Ani Res. (2021) 8:51–7. doi: 10.5455/javar.2021.h484
51. Kim, Y, Biswas, PK, Giasuddin, M, Hasan, M, Mahmud, R, Chang, YM, et al. Prevalence of avian influenza a (H5) and a (H9) viruses in live bird markets, Bangladesh. Emerg Infect Dis. (2018) 24:2309–16. doi: 10.3201/eid2412.180879
52. Zhou, B, Donnelly, ME, Scholes, DT, St. George, K, Hatta, M, Kawaoka, Y, et al. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and swine origin human influenza a viruses. J Virol. (2009) 83:10309–13. doi: 10.1128/JVI.01109-09
53. Hoffmann, E, Stech, J, Guan, Y, Webster, RG, and Perez, DR. Universal primer set for the full-length amplification of all influenza a viruses. Arch Virol. (2001) 146:2275–89. doi: 10.1007/s007050170002
54. Hijmans, RJ, Guarino, L, Bussink, C, Mathur, P, Cruz, M, Barrentes, I, et al.Diva-gis. (2004), Vsn.
55. Elbe, S, and Buckland-Merrett, G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Global Chall. (2017) 1:33–46. doi: 10.1002/gch2.1018
56. Drummond, AJ, and Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. (2007) 7:214–8. doi: 10.1186/1471-2148-7-214
57. Baele, G, Li, WL, Drummond, AJ, Suchard, MA, and Lemey, P. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol. (2012) 30:239–43. doi: 10.1093/molbev/mss243
58. Hasegawa, M, Kishino, H, and Yano, T-A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. (1985) 22:160–74. doi: 10.1007/BF02101694
59. Kalyaanamoorthy, S, Minh, BQ, Wong, TKF, von Haeseler, A, and Jermiin, LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. (2017) 14:587–9. doi: 10.1038/nmeth.4285
60. Khatun, A, Giasuddin, M, Islam, KM, Khanom, S, Samad, MA, Islam, MR, et al. Surveillance of avian influenza virus type a in semi-scavenging ducks in Bangladesh. BMC Vet Res. (2013) 9:1–8. doi: 10.1186/1746-6148-9-196
61. Hassan, MM, el Zowalaty, ME, Islam, A, Khan, SA, Rahman, MK, Järhult, JD, et al. Prevalence and diversity of avian influenza virus hemagglutinin sero-subtypes in poultry and wild birds in Bangladesh. Veter Sci. (2020) 7:73. doi: 10.3390/vetsci7020073
62. Islam, A, Hill, N, Mikolon, A, Sturm-Ramirez, K, Rahman, M, Paul, S, et al. Avian influenza a viruses in wild birds and domestic ducks in Bangladesh, in options for the control of avian influenza VIII, Cape Town, South Africa (2013) 5–10:2013.
63. Islam, A, Mikolon, A, Khan, M, Rahman, A, Paul, S, Islam, A, et al.A survey of avian influenza in wild birds and domestic ducks in Bangladesh, in international conference on emerging infectious diseases 2012. (2012)
64. Henning, KA, Henning, J, Morton, J, Long, NT, Ha, NT, and Meers, J. Farm-and flock-level risk factors associated with highly pathogenic avian influenza outbreaks on small holder duck and chicken farms in the Mekong Delta of Viet Nam. Prev Vet Med. (2009) 91:179–88. doi: 10.1016/j.prevetmed.2009.05.027
65. Sturm-Ramirez, K, Hulse-Post, DJ, Govorkova, EA, Humberd, J, Seiler, P, Puthavathana, P, et al. Are ducks contributing to the endemicity of highly pathogenic H5N1 influenza virus in Asia? J Virol. (2005) 79:11269–79. doi: 10.1128/JVI.79.17.11269-11279.2005
66. Sarkar, S, Khan, SU, Mikolon, A, Rahman, MZ, Abedin, J, Zeidner, N, et al. An epidemiological study of avian influenza a (H5) virus in nomadic ducks and their raising practices in northeastern Bangladesh, 2011-2012. Influenza Other Respir Viruses. (2017) 11:275–82. doi: 10.1111/irv.12438
67. Hidayat, M, Dewi, AM, Schoonman, L, Wibawa, H, Lubis, EP, Lockhart, C, et al. Investigating the endemic presence and persistence of HPAI H5N1 virus on Java. Indonesia: Authorea Preprints (2020).
68. Huang, Y, Wille, M, Dobbin, A, Robertson, GJ, Ryan, P, Ojkic, D, et al. A 4-year study of avian influenza virus prevalence and subtype diversity in ducks of Newfoundland. Canada Canadian J Microbiol. (2013) 59:701–8. doi: 10.1139/cjm-2013-0507
69. Wahlgren, J. Influenza a viruses: an ecology review. Infect Ecol Epidemiol. (2011) 1:6004. doi: 10.3402/iee.v1i0.6004
70. Islam, A, Islam, S, Amin, E, Shano, S, Samad, MA, Shirin, T, et al. Assessment of poultry rearing practices and risk factors of H5N1 and H9N2 virus circulating among backyard chickens and ducks in rural communities. PLoS One. (2022) 17:e0275852. doi: 10.1371/journal.pone.0275852
71. Kayali, G, Kandeil, A, el-Shesheny, R, Kayed, AS, Gomaa, MM, Maatouq, AM, et al. Active surveillance for avian influenza virus, Egypt, 2010–2012. Emerg Infect Dis. (2014) 20:542–51. doi: 10.3201/eid2004.131295
72. Scheibner, D, Ulrich, R, Fatola, OI, Graaf, A, Gischke, M, Salaheldin, AH, et al. Variable impact of the hemagglutinin polybasic cleavage site on virulence and pathogenesis of avian influenza H7N7 virus in chickens, turkeys and ducks. Sci Rep. (2019) 9:1–13. doi: 10.1038/s41598-019-47938-3
73. Islam, A, Islam, S, Flora, MS, Amin, E, Woodard, K, Webb, A, et al. Epidemiology and molecular characterization of avian influenza a viruses H5N1 and H3N8 subtypes in poultry farms and live bird markets in Bangladesh. Sci Rep. (2023) 13:7912. doi: 10.1038/s41598-023-33814-8
74. Ligon, BL. Avian influenza virus H5N1: a review of its history and information regarding its potential to cause the next pandemic In:. Seminars in pediatric infectious diseases, vol. 16: Elsevier (2005). 326–35.
75. Cardona, CJ, Xing, Z, Sandrock, CE, and Davis, CE. Avian influenza in birds and mammals. Comp Immunol Microbiol Infect Dis. (2009) 32:255–73. doi: 10.1016/j.cimid.2008.01.001
76. Young, SG, Carrel, M, Malanson, G, Ali, M, and Kayali, G. Predicting avian influenza co-infection with H5N1 and H9N2 in northern Egypt. Int J Environ Res Public Health. (2016) 13:886. doi: 10.3390/ijerph13090886
77. Li, Y, Li, X, Lv, X, Xu, Q, Zhao, Z, Qin, S, et al. Highly pathogenic avian influenza A (H5Nx) virus of Clade 2.3. 4.4 b emerging in Tibet, China, 10. Microbiol Spectr. (2022)10:e00643–22. doi: 10.1128/spectrum.00643-22
78. Nagarajan, S, Tosh, C, Smith, DK, Peiris, JSM, Murugkar, HV, Sridevi, R, et al. Avian influenza (H5N1) virus of clade 2.3. 2 in domestic poultry in India. PLoS One. (2012) 7:e31844. doi: 10.1371/journal.pone.0031844
79. Islam, A, Klaassen, M, Hossain, ME, Samad, MA, Hassan, MM, Amin, E, et al. Risk assessment, detection, and molecular characterization of H5N1 virus in a backyard chicken flock and a commercial FARM in Bangladesh. Int J Infect Dis. (2023) 130:S79–80. doi: 10.1016/j.ijid.2023.04.196
80. Biswas, P, Giasuddin, M, Nath, BK, Islam, MZ, Debnath, NC, and Yamage, M. Biosecurity and circulation of influenza a (H5N1) virus in live-bird markets in Bangladesh, 2012. Transbound Emerg Dis. (2017) 64:883–91. doi: 10.1111/tbed.12454
81. Bevins, SN, Shriner, SA, Cumbee JC Jr,, Dilione, KE, Douglass, KE, Ellis, JW, et al. Intercontinental movement of highly pathogenic avian influenza a (H5N1) clade 2.3. 4.4 virus to the United States, 2021. Emerg Infect Dis. (2022) 28:1006–11. doi: 10.3201/eid2805.220318
82. European Food Safety Authority, European Centre for Disease Prevention, Control, European Union Reference Laboratory for Avian InfluenzaAdlhoch, C, Fusaro, A, Gonzales, JL, Kuiken, T, Marangon, S, et al. Control, European Union reference Laboratory for Avian Influenza, et al., avian influenza overview December 2021–march 2022. EFSA J. (2022) 20:e07289. doi: 10.2903/j.efsa.2022.7289,
83. Mosaad, Z, Elhusseiny, MH, Zanaty, A, Fathy, MM, Hagag, NM, Mady, WH, et al. Emergence of highly pathogenic avian influenza a virus (H5N1) of clade 2.3. 4.4 b in Egypt, 2021–2022. Pathogens. (2023) 12:90. doi: 10.3390/pathogens12010090
84. Sanogo, IN, Djegui, F, Akpo, Y, Gnanvi, C, Dupré, G, Rubrum, A, et al. Highly pathogenic avian influenza a (H5N1) clade 2.3. 4.4 b virus in poultry, Benin, Emerging. Infect Diseases. (2021) 28:2534–7. doi: 10.3201/eid2812.221020
85. Ouoba, LB, Habibata-Zerbo, L, Zecchin, B, Barbierato, G, Hamidou-Ouandaogo, S, Palumbo, E, et al. Emergence of a reassortant 2.3. 4.4 b highly pathogenic H5N1 avian influenza virus containing H9N2 PA gene in Burkina Faso, West Africa, in 2021. Viruses. (2022) 14:1901
86. De Vries, E, and De Haan, CA. Highly pathogenic influenza a (H5N1) viruses in farmed mink outbreak contain a disrupted second sialic acid binding site in neuraminidase, similar to human influenza a viruses. Eur Secur. (2023) 28:2300085. doi: 10.2807/1560-7917.ES.2023.28.7.2300085
Keywords: avian influenza, HPAI H5N1, waterfowl, risk factors, phylogeny, 2.3.2.1a, 2.3.4.4b, zoonotic
Citation: Islam A, Hossain ME, Amin E, Islam S, Islam M, Sayeed MA, Hasan MM, Miah M, Hassan MM, Rahman MZ and Shirin T (2023) Epidemiology and phylodynamics of multiple clades of H5N1 circulating in domestic duck farms in different production systems in Bangladesh. Front. Public Health. 11:1168613. doi: 10.3389/fpubh.2023.1168613
Edited by:
Xinjian Zhang, Centers for Disease Control and Prevention (CDC), United StatesReviewed by:
Yassmin Moatasim, National Research Centre, EgyptAmal Anis Mahdi Eid, Zagazig University, Egypt
Copyright © 2023 Islam, Hossain, Amin, Islam, Islam, Sayeed, Hasan, Miah, Hassan, Rahman and Shirin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ariful Islam, YXJpZkBlY29oZWFsdGhhbGxpYW5jZS5vcmc=
 Ariful Islam
Ariful Islam Mohammad Enayet Hossain
Mohammad Enayet Hossain Emama Amin
Emama Amin Shariful Islam
Shariful Islam Monjurul Islam
Monjurul Islam Md Abu Sayeed4
Md Abu Sayeed4 Md Mehedi Hasan
Md Mehedi Hasan Mojnu Miah
Mojnu Miah Mohammad Mahmudul Hassan
Mohammad Mahmudul Hassan Mohammed Ziaur Rahman
Mohammed Ziaur Rahman