
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Public Health, 20 February 2023
Sec. Occupational Health and Safety
Volume 11 - 2023 | https://doi.org/10.3389/fpubh.2023.1073658
This article is part of the Research TopicBiomonitoring as an exposure assessment tool in occupational health - challenges and ways forwardView all 5 articles
Introduction: Epigenetic marks have been proposed as early changes, at the subcellular level, in disease development. To find more specific biomarkers of effect in occupational exposures to toxicants, DNA methylation studies in peripheral blood cells have been performed. The goal of this review is to summarize and contrast findings about DNA methylation in blood cells from workers exposed to toxicants.
Methods: A literature search was performed using PubMed and Web of Science. After first screening, we discarded all studies performed in vitro and in experimental animals, as well as those performed in other cell types other than peripheral blood cells. Results: 116 original research papers met the established criteria, published from 2007 to 2022. The most frequent investigated exposures/labor group were for benzene (18.9%) polycyclic aromatic hydrocarbons (15.5%), particulate matter (10.3%), lead (8.6%), pesticides (7.7%), radiation (4.3%), volatile organic compound mixtures (4.3%), welding fumes (3.4%) chromium (2.5%), toluene (2.5%), firefighters (2.5%), coal (1.7%), hairdressers (1.7%), nanoparticles (1.7%), vinyl chloride (1.7%), and others. Few longitudinal studies have been performed, as well as few of them have explored mitochondrial DNA methylation. Methylation platforms have evolved from analysis in repetitive elements (global methylation), gene-specific promoter methylation, to epigenome-wide studies. The most reported observations were global hypomethylation as well as promoter hypermethylation in exposed groups compared to controls, while methylation at DNA repair/oncogenes genes were the most studied; studies from genome-wide studies detect differentially methylated regions, which could be either hypo or hypermethylated.
Discussion: Some evidence from longitudinal studies suggest that modifications observed in cross-sectional designs may be transitory; then, we cannot say that DNA methylation changes are predictive of disease development due to those exposures.
Conclusion: Due to the heterogeneity in the genes studied, and scarcity of longitudinal studies, we are far away from considering DNA methylation changes as biomarkers of effect in occupational exposures, and nor can we establish a clear functional or pathological correlate for those epigenetic modifications associated with the studied exposures.
In 2011, Szyf (1), in a very illustrative and synthetic work, coined the term “toxicomethylomics.” He discussed the correlation of DNA methylation and gene expression in different genomic regions, including so-called “regulatory sequences” as well as intragenic or gene body sequences, informing future studies of human populations exposed to chemicals with mutagenic or genotoxic properties. At the time of its publication, around five research papers had been published exploring DNA methylation changes in occupationally exposed populations. In the subsequent decade, there were significant advancements in the technologies used to study DNA methylation, progressing work in the field from those analyzing methylation of specific genes to epigenome-wide analysis using microarray technology and high-throughput sequencing platforms that interrogate DNA methylation at single-nucleotide resolution. Here, we narratively review publications on DNA methylation in peripheral blood cells from people occupationally exposed to toxic agents to evaluate the insight that has been gained and the direction of the field.
A literature search was performed using PubMed and Web of Science. We established two searches: one with the keywords “DNA methylation” and “occupational exposure” and another search with key words “DNA methylation” and “workers.” The searches were performed between August 2022 and October 2022. After first screening (yielding 150 research papers), we read every abstract to determine if the work was performed in blood cells from healthy workers, discarding all in vitro and experimental animal studies as well as those performed in other human cell types; we also discarded duplicated papers. From the selected abstracts, we read the whole paper, identifying the DNA methylation technique utilized in the manuscript, the genomic target(s) (including mitochondrial DNA) and the main findings. Studies matching our inclusion criteria were entered into a database and summarized. Typically, DNA methylation research is performed describing the exposed participants, the disease/consequences of that exposure, the analysis platform for studying DNA methylation, and other endpoints such as impact upon gene expression (Figure 1). We categorized the included publications according to the compound studied. For those studies where a diverse range of compounds were analyzed, the papers were categorized according to the task/job (i.e., hairdressers, firefighters).
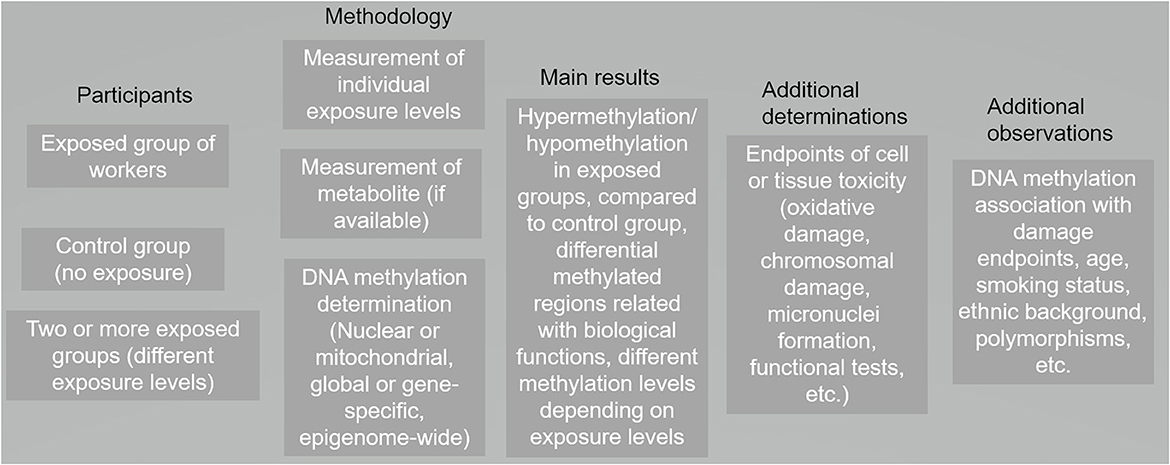
Figure 1. Panorama of the general design and results in toxicomethylomics studies in blood cells from occupationally–exposed individuals published from 2007 to 2022 (116 research papers).
One hundred and sixteen research papers met the aforementioned criteria, which were each published between 2007 and 2022. These studies addressed a total of 30 different compounds and occupations. A summary of the studies characteristics is shown in Table 1. In the following section, we review their main findings according to the exposure type, describing the results according to the publication date (from the oldest to the most current). Appearance of results try to follow an order according to the chemical similarity of the compounds (organic compounds, material obtained from combustion, metals etc.). Before the section “miscellaneous” which groups studies where only one research report has been performed for each toxicant, we grouped studies performed in occupationally-exposed groups; such studies did not mention a specific exposure, but, since they were performed in peripheral blood in a group of workers, they accomplished our inclusion criteria (Summary is presented in Table 2).
The first paper regarding epigenetic effects in workers exposed to a carcinogenic compound was in people exposed to this very well-known leukemogen. In 2007, Bollati et al. (2) used pyrosequencing and reported hypomethylation of the repetitive elements Alu (taking its name from the Arthrobacter luteus restriction endonuclease) and the long interspersed nuclear element-1 (LINE-1), as a surrogate for global DNA methylation. Their work also examined methylation at the promoter region of two genes, revealing loss of methylation for the Melanoma-associated antigen 1 gene (MAGE-1) and hypermethylation of p15, a tumor repressor gene. This first report of global hypomethylation (LINE-1) in response to exposure to a carcinogenic compound has since been confirmed by a number of other studies, as has the observation of p15 hypermethylation. Moreover, participants in this study were considered to be exposed to “low-levels,” which suggests DNA methylation as an early biological response marker even to low exposures. Five years later, Xing et al. (3) measured promoter methylation of the p16 (also a tumor repressor gene) promoter region in benzene exposed workers. In addition, they measured expression of p16 at the mRNA level. They reported promoter hypermethylation in the exposed group, and methylation levels negatively correlated with p16 expression; the first evidence that promoter methylation in response to benzene exposure can silence gene expression. Although examined in only a few individuals (eleven exposed vs. eight controls), those results were supported by similar observations in cell lines exposed to benzene metabolites. In the same year, Seow et al. (4) showed an interesting negative correlation between urinary S-phenyl-mercapturic acid (SPMA, a main metabolite of benzene) with methylation levels of Alu elements and the p15 gene promoter, which represents a link between xenobiotic biotransformation enzyme activity and DNA methylation. Also in 2012, Carugno et al. (5) using data from the same population studied by Bollati et al. (2), described an association between mitochondrial DNA copy number and both LINE-1 hypomethylation and p15 hypermethylation, providing evidence for the relationship between nuclear epigenetic marks and the function of mitochondria; organelles which are highly affected by toxic chemicals via redox imbalance.
Further, gene polymorphisms are known to influence the body's response to toxic chemicals. Hence, Xing et al. (6) investigated the relationship between single nucleotide polymorphisms (SNPs) within four genes encoding metabolic enzymes and promoter methylation of eleven genes associated with hematotoxicity in benzene-exposed workers. They observed that ERCC excision repair 3 gene (ERCC3) promoter methylation was increased in the exposed group, and also that individuals carrying a genetic variant mapping to Epoxide Hydrolase 1 gene (EPHX1) had decreased ERCC3 methylation. In 2014, Yang et al. (7) employed the widely-used Illumina Infinium 450K methylation microarray platform for epigenome-wide analysis, which revealed three hypermethylated genes: Protein kinase, cGMP-dependent, Regulatory, Type I (PRKG1K), Par-3 Family Cell Polarity Regulator (PARD3), and Ephrin type-A receptor 8 (EPHA8) and two hypomethylated: Signal Transducer And Activator Of Transcription 3 (STAT3) as well as Interferon Gamma Receptor 1 (IFNGR1) in four benzene-exposed individuals compared to controls. Importantly, hypermethylation was shown to decrease gene expression, while hypomethylated genes showed higher expression. They concluded that aberrant hypomethylation of STAT3 might be a potential biomarker of chronic benzene poisoning.
Other authors have tried to relate DNA damage markers with methylation, such as Li et al. (8), who described that DNA damage, as well as SPMA urinary levels, were negatively associated with O-6-Methylguanine-DNA Methyltransferase gene (MGMT) methylation. MGMT methylation was also significantly decreased in the benzene-exposed group. In the same year, Zhang et al. in an effort to elucidate the underlying mechanisms of global hypomethylation caused by benzene exposure, investigated the relationship between global methylation and several endpoints related with DNA damage (including mutations) and polymorphisms in DNA Methyltransferase 3 Alpha gene (DNMT3A) and DNA Methyltransferase 3 Beta (DNMT3B). They described the association between SNPS within both the DNMT3A and DNMT3B genes and global DNA methylation. In that same year, Zheng et al. (9), analyzed ERCC3 promoter methylation in benzene-exposed workers and its possible relationship with hematotoxicity markers. Methylation at 2 CpGs were correlated negatively with neutrophil abundance. Ren et al. (10), analyzed global DNA methylation and its relationship with polymorphisms within the methylenetetrahydrofolate reductase (MTHFR) gene, which encodes a key enzyme required for the production of methyl donors for DNMTs. Individuals homozygous for the TT allele at rs1801133 displayed lower methylation levels compared with those homozygous for the wild type allele. These findings illustrate the interaction between genetic variants and the epigenome, including variants within genes encoding the epigenetic machinery of the cell and also enzymes that may mediate response to environmental exposures. In the same year, Jamebozorgi et al. (11), described higher promoter methylation for p15INK4b, a cyclin-dependent kinase inhibitor gene, in workers exposed to benzene compared to controls. Looking for possible methylation changes in repetitive elements subfamilies, Rota et al. (12) reported that only L1-Pa5 was hypomethylated in petrol station workers (89 workers) compared to controls, with Alu subfamilies showing no such effect, in contrast to previous findings. Costa-Amaral et al. (13), did not find differences in gene-specific methylation for the Cyclin Dependent Kinase Inhibitor 2B (CDKN2B) and Kruppel Like Factor 6 (KLF6) genes in Brazilian workers exposed to benzene. In a very interesting study using a longitudinal approach, Ren et al. (14) followed benzene-exposed workers from 2009 to 2013, analyzing DNA methylation once in 2009 and again in 2013, measuring methylation of LINE-1, MGMT, and Human MutL homolog 1 gene (hMLH1). LINE-1 methylation was higher in 2013 compared to 2009, while promoter methylation in hMLH1 was lower and corresponded with the reduction in benzene exposure. Zhang et al. (15), studying the base excision repair (BER) pathway and their polymorphisms, in benzene-exposed workers, found that the rs1130409 variant allele (GG+GT) was associated with low global DNA methylation. Wang et al. (16) studied mitochondrial DNA methylation, specifically at the Mitochondrially Encoded Cytochrome c Oxidase I (MT-COI) gene, and reported two interesting results: mitochondrial methylation was lower in the exposed group; and methylation of MT-COI was correlated with white blood cell count and also with platelet levels. Epigenome-wide analysis was performed on a large scale for the first time by Ren et al. (17), using the Infinium 450K methylation microarray. They reported 442 sites in benzene-exposed participants were hypermethylated, corresponding to 253 genes, and 237 sites were hypomethylated, corresponding to 130 genes. Correlation analyses showed that promoter methylation of Colony Stimulating Factor 3 Receptor (CSF3R) and Coagulation Factor II Thrombin Receptor (F2R) genes were both highly correlated with gene expression. The CSF3R gene is of particular interest as it encodes a protein involved in the differentiation of granulocytes. In persons exposed to the highest benzene levels, in the poisoning cases, the proportion of neutrophils in the blood was significantly different by CSF3R methylation levels, demonstrating a potential function role for this gene in the toxicity associated with benzene exposure. By their side, Ji et al. (18) reported lower global DNA methylation in benzene-exposed workers compared to controls, with mitochondrial DNA copy number negatively correlated with global DNA methylation; and also Pan et al. (19), using methylation-specific PCR to analyze the Urothelial Cancer Associated 1 gene (UCA1), found that methylation at the promoter region of this gene is negatively correlated with benzene exposure time. Together, there is a wealth of data to demonstrate both global hypomethylation and gene-specific changes in response to benzene exposure, with CSF3R a particularly interesting candidate. In 2022 we have data from three research works: van der Laan et al. (20) using the 450K microarray, found that occupational exposure to benzene accelerated epigenetic aging in the blood and was also associated with shorter estimated telomere length. Zhang et al. (21), studying promoter methylation of the Long intergenic non-protein coding RNA 173 (LINC00173) vs. the expression of this RNA in benzene exposed workers, found that LINC00173 promoter methylation was negatively correlated with its expression. Very recently, Phillips et al. (22), in a cross-sectional epigenome-wide association study, using the Human Methylation 450 Bead Chips, investigated differences in blood cell DNA methylation among 50 benzene-exposed subjects and 48 controls, and analyzed CpG-level and regional-level. They report genome-wide significant alterations in methylation at 340 CpG sites, and also in mean methylation levels at a large genomic region; also, Pathway analysis of genes sites revealed an impact on the AMPK signaling pathway.
VOCs are chemical compounds present in a myriad of labor settings. The most widely-studied of these is benzene, which has been discussed in the previous section. However, other VOCs have been described as contributing to asthma (toluene), have been classified as carcinogenic in animals (ethylbenzene) or are reprotoxic and toxic to the nervous system (such as n-hexane). These mixtures are frequently found in organic solvents. Epigenetic effects in workers exposed to these kind of mixtures were first evaluated in 2012 by Godderis et al. (24), who evaluated DNA global methylation in workers exposed to solvents. They observed global hypermethylation in response to exposure, which was negatively associated with exposure levels. Furthermore, SNPs within the Glutathione S-Transferase Pi gene (GSTP1), encoding an enzyme involved in detoxification processes, were found to be significantly associated with this global DNA hypomethylation. In 2017, Jiménez-Garza et al. (25), studying groups of Mexican workers exposed to solvents via work at a shoe factory or gas station in comparison to office-based controls, evaluated promoter methylation of the Interleukin-6 (IL6), Cytochrome P450 2E1 (CYP2E1), and Inducible Nitric oxide synthase (iNOS) genes. IL6 promoter methylation was higher in gas station attendants (the group with the highest exposure levels to benzene), while CYP2E1 promoter methylation was negatively correlated with benzene exposure levels. Furthermore, positive correlations were observed between CYP2E1 promoter methylation and both iNOS promoter methylation and urinary levels of SPMA. A year later, the same group of researchers (26) studied promoter methylation for a panel of genes including Cyclooxygenase II (COX-2), GSTP1, Heme Oxygenase 1 (HMOX-1), iNOS, Superoxide Dismutase 1 (SOD1), Tumor necrosis alpha (TNF-α), and Topoisomerase II alpha (TOP2A). This time they added another group for comparison: shoe workers exposed to very high solvent levels, excepting benzene. Hypermethylation of TOP2A, SOD1, TNF-α was observed among these shoe workers. Further to their previous observations of correlations between epigenetic changes of different genes, significant correlations were observed between GSTP1 promoter methylation and methylation of both iNOS and COX-2. Ethylbenzene exposure levels were significantly correlated with TOP2A methylation. Interestingly, a “cumulated time of exposure” showed significant negative correlation with methylation of both TNF-α and iNOS. In a subset of workers taken from the two latter mentioned studies, Jiménez-Garza et al. (27) and now studying methylation mapping to the 5′UTR region of genes, analyzed methylation of the CYP2E1, IL-6, SOD1, and TNF-α genes. They reported lower methylation levels for IL-6 and higher methylation levels for TNF-α for the mixtures-exposed group compared to controls. Recently, Yu et al. (28), studied genome-wide DNA methylation in blood from Korean workers exposed to a binary mixture (toluene and xylene) and compared results with workers exposed to a tertiary mixture (toluene, xylene, and ethylbenzene group). Authors observed the highest amount of differentially methylated regions in the group exposed to the tertiary mixture, which underlines the complexity of studying the effects of a mixture of different compounds and making appropriate inferences.
Toluene is a VOC that is frequently used in many industries. Toluene chronic exposure has been related with respiratory affections and, when combined with other VOCs, may increase their hematotoxic effects. To date, only three studies have examined DNA methylation in response to exposure to this compound. In 2015, Jiménez-Garza et al. (29) analyzed promoter methylation of COX-2, CYP2E1, GSTP1, HMOX-1, IL-6, iNOS, SOD1, TNF-α, and TOP2A in Mexican workers from a tannery who are exposed to toluene. While they did not report statistically significant differences between exposed individuals and controls, positive correlations were observed between toluene airborne levels and both CYP2E1 and IL6 promoter methylation. CYP2E1 promoter methylation levels were higher in toluene-exposed smokers compared to non-smokers, and were also correlated with GSTP1 and SOD1 promoter methylation. One year later, Hong et al. (30) studied DNA methylation in Korean workers exposed to toluene. They reported 26 genes that were upregulated and hypomethylated, as well as 32 genes downregulated and hypermethylated in the exposed group. In 2020, studying a subset of the toluene-exposed workers from their 2015 publication, Jiménez-Garza et al. examined 5'UTR methylation of five genes: CYP2E1, IL-6, SOD1, and TNF-α. They observed hypomethylation in the 5′UTR region of the CYP2E1 gene in toluene-exposed workers compared to controls, further underlining the apparent importance of this gene in our understanding of the effects of toluene exposure.
There are different labor activities where workers are exposed to PAHs. Many of these compounds have carcinogenic properties, with benzo[a]pyrene specifically causing lung cancer, while other PAHs such as naphthalene, acenaphthylene, acenaphthene, fluorene, anthracene, phenanthrene, fluoranthene, pyrene, chrysene, benz[a] anthracene, benzo[b]fluoranthene, benzo[k] fluoranthene, B[a]P, indeno[1,2,3-cd]pyrene, benzo[g,h,i]-perylene, and dibenz[a,h]anthracene have also shown carcinogenic potential. The first study we identified was that of Pavanello et al. (31), who investigated coke-oven workers exposed to PAHs. They studied methylation of the p53, p16, Hypermethylated in cancer 1 protein (HIC1), and IL-6 genes as well as of Alu and LINE-1 repetitive elements. They observed Alu and LINE-1 hypermethylation in the exposed workers, while gene-specific IL-6 methylation was significantly and positively correlated with PAH exposure levels. The next year, the same research group (32), studied the same population of coke-oven workers and reported TP53 promoter hypermethylation in response to exposure, with TP53 hypomethylation also demonstrated to be one of the principal determinants of shorter telomere length. In 2012, Yang et al. (33) examined 69 workers exposed to PAHs, and specifically focused upon p16 (INK4α) promoter methylation. They reported p16 (INK4α) hypermethylation in exposed workers, and they also observed that hypermethylated CpG sites were positively correlated with levels of urinary 1 hydroxypyrene (1-OHP) and with the frequency of cytokinesis block micronucleus. That same year, Ouyang et al. (34), studied promoter methylation of the GSTP1, Interferon Gamma (IFN-g), RAD21 cohesin complex component (RAD21), and Dual Specificity Phosphatase 22 (DUSP22) genes in firefighters exposed to PAHs. Firefighters had a higher prevalence of DUSP22 promoter hypomethylation in the blood, and the extent of hypomethylation correlated with duration of firefighting service. One year later, Duan et al. (35), examined global methylation (using LINE-1 as a surrogate marker) and promoter-specific methylation of the MGMT gene in 82 PAH-exposed workers. They reported global hypomethylation as well as hypomethylation of MGMT. In addition, LINE-1 methylation was inversely associated with DNA damage as determined by Comet assay, and a significant increase of micronuclei in the low MGMT methylation group. Pavanello et al. (36), studying TP53 methylation in Polish workers exposed to PAHs, found hypomethylation of the TP53 promoter, contrary to their previous study of 2010. That same year, Alegría-Torres et al. (37), studied promoter methylation of five genes: TP53, TNF-a, IFN-c, IL-6, and Interleukin 12 (IL-12) in Mexican brickmakers. They did not find any difference in promoter methylation of these genes, but they reported negative correlations between promoter DNA methylation of two genes (IL-12 and TP53) with urinary 1-OHP concentration; a metabolite from PAH exposure.
In 2015, Zhang et al. (38) examined methylation of the p14(ARK), p15(INK4b), and p16(INK4a) genes in coke oven workers. The three genes studied were hypermethylated, with two of these correlated with years worked and also with urinary 1-OHP levels. The next year, Zhang et al. (39) examined promoter methylation of the p16, Ras Association Domain Family Member 1 (RASSF1A) and MGMT genes, as well as LINE-1, in workers exposed to diesel exhaust. The exposed group showed lower methylation levels for all three genes compared to controls, with methylation levels negatively correlated with markers of cytogenetic damage. In 2017, He et al. (40) found that Tripartite Motif Containing 36 gene (TRIM36) promoter hypermethylation was positively correlated with the level of urinary 1-OHP in 151 exposed workers. One year later, Li et al. (41) analyzed epigenome-wide methylation in Chinese participants exposed to different levels of PAHs (including an occupationally exposed group from coke-oven emissions) using the 450 k microarray. They found hypermethylated CpGs mapping to the Four And A Half LIM Domains 2 (FHL2) gene, as well as gene expression positively associated with methylation. In the same year, (47) investigated global and AHRR promoter methylation in coke oven workers. They observed that the workers with high urinary 1-OHP have hypomethylation of LINE-1. Heavy smoking was associated with a significantly increased risk of hypomethylation of Aryl-Hydrocarbon Receptor Repressor (AHRR) gene, in line with numerous other reports of AHRR serving as an effective biomarker of tobacco smoke exposure. Alhamdow et al. (42) studied F2R like thrombin or trypsin receptor 3 (F2RL3) and AHRR (exon 1 region) in chimney sweeps from Sweden. In these two genes that are known to be associated with tobacco smoke exposure, they found hypomethylation of both in the exposed group, and that non-smoking chimney sweeps had lower average F2RL3 methylation levels. Wahlberg et al. (43) similarly studied AHRR and F2RL3 in chimney sweeps and again reported AHRR hypomethylation, but they also found that variants within the Filaggrin (FLG) gene were associated with ~2.5% higher methylation of F2RL3. Liu et al. (44) examined methylation at the promoter region of Cytochrome P450 1A1 (CYP1A1) gene in Chinese coke-oven workers, with hypomethylation observed in workers highly exposed to PAHs and a significant relationship between CYP1A1 hypomethylation and high 8-hydroxydeoxyguanosine (8-OHdG); a specific marker of oxidative stress. In a third publication from that year, Fu et al. (45), also studying Chinese coke oven workers, examined 8-oxoguanine DNA glycosylase (OGG1) gene promoter methylation and described its positive correlation with urinary 1-OHP levels. In the two most recent papers, both from 2020, Zhao et al. (46) reported IFN-γ hypermethylation and IL-10 hypomethylation in coke-oven workers, with IL-10 methylation inversely correlated with urine 1-OHP concentration. Finally, Alhamdow et al. investigating exposure to high-molecular-weight PAHs in chimney sweeps, confirmed hypomethylation of AHRR and F2RL3 in exons 1 and 2, respectively.
Exposure to PM is associated with increased risk of asthma and cardiovascular disease. Although PM is present in high levels in the ambient air of polluted cities, some workers are continuously exposed to these particles in the occupational setting. Most of the studies investigating epigenetic changes in occupational cohorts exposed to PM come from the same research groups, evaluating different endpoints and contrasting the results obtained in Italian steel workers and Chinese truck drivers. Two studies were published in 2011 where Italian steel workers were studied. Dioni et al. (49) measured Human Telomerase Reverse Transcriptase (hTERT) methylation in peripheral blood, comparing pre-shift methylation levels with the post-shift changes in the same workday (with no control group), but no significant differences were observed. The other report from that year is the one from Hou et al. (53) who investigated, also in steel workers, promoter methylation levels of four tumor suppressor genes: Adenomatous polyposis coli (APC); p16: TP53; and RASSF1A. They also compared methylation before and after shifts, with mean promoter methylation levels of APC and p16 significantly higher in post-exposure samples compared to baseline samples. Furthermore, APC methylation was positively associated with PM10 exposure. Conversely, the mean levels of TP53 and RASSF1A promoter methylation were significantly decreased in post-exposure samples.
Two years later, Byun et al. (59) studied methylation in 10 repetitive element subfamilies by their evolutionary origins in three different groups of workers occupationally exposed to PM: steel workers (the aforementioned Italian steel workers); gas station attendants; and Chinese truck drivers (120 workers in total). LINE-1 elements showed a differential effect by evolutionary age, with older elements showing greater hypomethylation in response to PM exposure. That same year, Byun et al. (60) examined mitochondrial DNA methylation and reported that participants with high metal-rich PM10 exposure showed higher Mitochondrially Encoded tRNA Phenylalanine (MT-TF) and Mitochondrially Encoded 12S RRNA (MT-RNR1) methylation in comparison to control subjects, with MT-RNR1 methylation positively correlated with mtDNA copy number. In the same year, Tarantini et al. (50) studied promoter methylation of the Nitric Oxide Synthase 3 (NOS3) and Endothelin 1 (EDN1) genes in steel workers and reported an inverse correlation between NOS3 methylation exposure to PM10, while EDN1 methylation showed a similar negative correlation with zinc exposure. Kile et al. (51), studying boilermaker welders, examined iNOS promoter methylation, assessing pre- and post-shift changes. They reported that PM2.5 exposure was associated with increased methylation of this gene, with a positive association between levels and the number of years worked as a boilermaker. In 2014, Fan et al. (52) measured methylation of repetitive elements in the genome in boilermakers exposed to high levels of PM2.5. They reported no differences in pre-shift and post-shift methylation of tandem repeats but did observe a positive correlation between PM2.5 exposure and LINE-1 methylation. In another study of truck drivers in Beijing, Hou et al. (53) measured methylation of three tandem repeat elements in relation to exposure to different components of PM. NBL2 methylation (a DNA repeated sequence) was positively associated with concentrations of silicon and calcium, while in office workers SATα methylation (Satellite alpha, a repetitive element) was positively associated with sulfur. From a similar study performed in the same population, Guo et al. (54) reported the effects of short-term exposure to inhalable particulate matter, and again focused upon tandem repeat sequences within the genome. Both personal PM2.5 and ambient PM10 levels showed an effect upon SATα methylation that was stronger among higher-exposed truck drivers than office workers, while ambient PM10 was negatively correlated with NBL2 methylation. In 2017, Mercorio et al. (55) studied 63 male workers from an electric furnace steel plant and examined methylation of the Wp promoter of the Epstein-Barr Virus (EBV-Wp) and the promoter of the human-endogenous-retrovirus w elements (HERV-w). They reported EBV-Wp hypomethylation following exposure, yet PM and metal components were shown to exhibit a positive effect upon EBV-Wp methylation. We detected two important studies in 2022: first, Honkova et al. (56) studied policeman exposed to different levels of PM 2.5, benzo[a]pyrene and NO2 from three different cities; DNA methylation was analyzed by a genome-wide microarray method. They describe 13,643 differentially methylated CpG loci between policeman from two different cities; they also found two hypomethylated loci annotated to the DNA repair gene XRCC5; groups of differentially methylated loci annotated to the same gene were linked to diabetes mellitus, respiratory diseases, the dopaminergic system of the brain and neurodegenerative diseases. Finally, Guo et al. (57), investigated mitochondrial DNA (mtDNA) methylation in workers from a coke-oven plant exposed concurrently to PAH's and PM 2.5 mtDNA methylation was measured by bisulfite pyrosequencing of two genes of ATP synthase (MT-ATP6 and MT-ATP8). They observed a significant, negative association between methylation of platelet mitochondrial gene MT-ATP6 and PM exposure.
There are a wide variety of chemical compounds that function as pesticides. Acute health effects are common in highly exposed people, but chronic exposures are also known to have serious detrimental effects upon human health. In 2016, the first report on farm workers exposed to pesticides associated with DNA methylation appeared: Howard et al., using the 450k microarray platform, identified three CpG sites that are hypermethylated in exposed farmworkers compared to non-farmworkers following Bonferroni correction, suggestive of a general decrease in gene expression of the regulated genes. Pathway analysis, incorporated 33 other CpG sites that showed significant changes between the exposure groups that did not survive Bonferroni correction, particularly revealed enrichment in genes implicated in immune function, as well as growth hormone signaling and DNA damage response. One year later, Rusiecki et al. (63) analyzed methylation at the promoter region of three genes: Cadherin 1 (CDH1), GSTP1, MGMT, and LINE-1 (as a surrogate marker of global methylation) in pesticide applicators from the USA. They categorized results according to having a high pesticide exposure event (HPEEs), with those individuals who have ever experienced an HPEE (n = 142), associated with hypermethylation of the GSTP1 promoter and loss of methylation at the MGMT promoter, with greater effects observed in applicators of more advanced age and with lower plasma folate levels. In 2018, Benedetti et al. (64) published their findings in 137 farmers exposed to a pesticide mixture, which revealed global hypermethylation in the exposed group, as determined by high performance liquid chromatography rather than the more widely-used LINE-1 assay. In direct contrast to this, Benitez-Trinidad et al. (65) observed LINE-1 hypomethylation in exposed workers (urban sprayers; n = 190). The latter is supported by Kahl et al. (62), who similarly reported global hypomethylation, in addition to p16 promoter hypermethylation, in pesticide-exposed tobacco farmers. Importantly, they reported that global DNA methylation levels were associated with elevated oxidative stress, which may help to bring mechanistic insight into the relationship between exposure, the epigenome and human health. In a similar population, Kahl et al. (61) examined global DNA methylation in relation to nutritional intake, but they did not identify significant associations. The fourth study published in 2018 came from van der Plaat et al. (66) who, using Illumina 450K microarrays, described methylation changes associated with high pesticide exposure at 31 CpGs mapping to 29 genes. Significant impacts upon gene expression were only identified for seven of these, which may suggest a broader impact upon the epigenome than locus-specific modulation of gene expression. In 2019, Herrera-Moreno et al. (67) looked for methylation changes within the 5′UTR region of the CDKN2B gene and the promoter of Cyclin Dependent Kinase Inhibitor 2A (CDKN2A) in Mexican workers exposed to pesticides. CDKN2B hypomethylation was observed in exposed individuals, with a signification association between methylation and exposure levels, while hypermethylation of the CDKN2A promoter was observed in moderately-exposed individuals. Finally, Paredes-Céspedes et al. (68), determined methylation profiles of the CDKN2B and CDKN2A genes in a genetically conserved population exposed to pesticides. They observed that the farmer group presented a higher methylation percentage of CDKN2B than the non-farmer group, but no differences in CDKN2A were observed between groups. A positive correlation between methylation of CpG site 3 of CDKN2B and time working in the field was observed in the farmer group.
Lead is a toxic metal with adverse health effects, particularly to the nervous system. Indeed, according to the World Health Organization, lead exposure accounted for more than one-million deaths in 2017. The first report of epigenetic changes in occupationally-exposed individuals is from 2010, when Kovatsi et al. (70) analyzed p16 promoter methylation in nine exposed workers. Although they used a qualitative approach to exposure, p16 promoter hypermethylation was apparent among exposed workers. Li et al. (71) studied methylation of the Aminolevulinate Dehydratase (ALAD) gene promoter in 103 workers from a battery plant. This gene encodes an enzyme whose activity is inhibited by lead and can thereby result in anemia. The study revealed ALAD hypermethylation in exposed workers and a concurrent decrease in gene expression, with ALAD methylation associated with increased risk of lead poisoning. Together, these findings have brought mechanistic insight into the inhibition of normal ALAD activity through lead exposure. In 2013, Li et al. (72) examined LINE-1 methylation in 53 exposed battery plant workers and 57 age-matched controls and reported hypomethylation in exposed participants, with a significant associated between methylation levels and blood lead levels. One year later (101), the same group performed further analysis within the same cohort to examine GSTP1 promoter methylation, but no statistically significant differences were present. In 2017, Devóz et al. (73) analyzed global methylation in Brazilian workers employed in automotive battery factories, which further confirmed a significant correlation between DNA methylation and blood/plasma lead concentrations. A year later, Yu et al. (74) analyzed the methylation of six human tumor suppressor genes, four of which were hypermethylated (hMLH1, p14, p15, and p16) in the exposed group, while two (GSTP1 and MGMT) showed no significant changes. In 2019, epigenome-wide analysis through the expanded Illumina Infinium EPIC (850K) microarray platform was performed by Zhang et al. (15) who identified 180 differentially methylated genes between high- and low-exposure groups. Through pathway analysis, 57 significant gene ontology enrichment terms were identified, including glutathione derivative biosynthetic process and nervous system development; thereby potentially bringing mechanistic insight into the toxic effects of lead exposure. The most recent study of lead exposure upon the epigenome is that of Devóz et al. (75), who examined promoter methylation of four genes related to glutathione activity: Glutamate-Cysteine Ligase Catalytic Subunit (GCLC); Glutathione Peroxidase 1 (GPX1); Glutathione-Disulfide Reductase (GSR); and GSTP1. In workers exposed to lead, high blood lead levels were associated with reduced GCLC and GSTP1 methylation, potentially supporting the findings of Zhang et al. and indicating that this pathway may be crucial to understanding biological response to lead exposure. The two most recent research works about lead exposure and DNA methlyation, are the one from Issah et al. (102), where they studied global DNA methylation (using LINE-1 as an indicator) in electronic waste workers exposed to lead; they did not find statistically significant differences between exposed group (n = 100) vs. control group (n = 51). Finally, Meng et al. (76), in an effort of relating DNA damage induced by lead in exposed workers, analyzed methylation using the 850K Beadchip platform, comparing methylation in workers with an already established DNA-damage biomarker, with workers without such damage as well as controls; first, differentially methylated positions (DMPs) between the controls and the exposed workers were identified, In addition, DMPs were identified between the DNA-undamaged and DNA-damaged workers. Methylation levels of four candidate genes were measured by pyrosequencing in an independent sample set; the result of comparisons between the controls and the lead-exposed workers show that DMPs were significantly enriched in genes related to nerve conduction and cell cycle. Between DNA-damaged group and DNA-undamaged group, differentially methylated genes were enriched in the pathways related to cell cycle and DNA integrity checkpoints.
Chromium VI (Cr VI) is carcinogenic and targets the respiratory system, kidneys, liver, skin, and eyes. The first study of DNA methylation and chromium exposure is from 2012, when Wang et al. recruited 115 workers from chromate producing facilities and examined global DNA methylation patterns. They observed global hypomethylation in exposed workers, which they hypothesized to be the product of depleted serum folate levels that were also observed with exposure. In 2016, Yang et al. (77) performed the first study of mitochondrial DNA methylation in relation to chromium exposure, which revealed MT-TF and MT-RNR1 hypomethylation and lower levels of methylation of these genes in workers with highest blood chromium levels. Most recently, in 2018 Hu et al. (78) examined promoter methylation of genes implicated in DNA damage response and repair. They observed OGG1, MGMT, and RAD51 hypermethylation in exposed workers, with a positive correlation observed between exposure level and methylation, and a negative correlation between methylation and expression. These findings may suggest that chromium exposure leads to down-regulation of tumor-suppressor genes, thereby providing mechanistic insight into the carcinogenic processes associated with exposure.
Prolonged exposure to welding fumes may cause lung damage and various types of cancer, including that of the lung, larynx and urinary tract. Health effects from certain fumes may include metal fume fever, stomach ulcers, kidney damage and nervous system damage. Three studies have been performed that have examined DNA methylation changes in workers exposed to welding fumes. In the first of these, from 2015, Li et al. (80) studied welders from Sweden and assessed methylation in a non-specified regions of tumor-suppressor genes such as Homeobox A9 (HOXA9), Short Stature Homeobox 2 (SHOX2), CDKN2A, MGMT, and APC. They found APC higher methylation in welders. This was also associated with wood burning at home and has been shown elsewhere to be associated with tobacco smoke (103), suggesting that different products of combustion may have a similar effect upon this gene. That same year, Hossain et al. investigated methylation of four CpG sites mapping to exon 2 of the F2RL3 gene in 101 welders and 127 controls. They found hypomethylation at one CpG site, while the number of years of welding work was associated with hypomethylation of another. As the F2RL3 gene has been most widely studied in relation to tobacco smoke exposure, this study provides further evidence of overlap between the epigenetic changes associated with different types of combustion. Also in 2017, Xu et al. (81) investigated the impact upon the mitochondrial epigenome. They reported hypomethylation of the regulatory D-Loop region and of the MT-TF gene, with their methylation associated with particle-containing welding fumes exposure. Very recently, Dauter et al. (82) in a longitudinal study where 78 welders and 96 controls were examined on two occasions 6 years apart, measured DNA methylation of CpG sites within the cancer-related genes AHRR, F2RL3, and B3GNTL1 by pyrosequencing. They report that, compared with controls, welders showed a significant decrease over time in DNA methylation of B3GNTL1 CpG1 and CpG4. In addition, exposure to respirable dust and cumulative exposure was associated with a decrease in methylation of F2RL3 CpG2 among all welders.
Many different disorders have been associated with exposure to coal, including pneumoconiosis, fibrosis, asbestosis, silicosis, emphysema, loss of lung function, and cancer. To date, there have been two studies of the effect of coal upon the human epigenome. The first, by de Souza et al. (84), examined global DNA methylation in coal miners, which revealed global hypermethylation in exposed workers compared with controls. In 2020, the same group of researchers investigated possible relationships between global methylation and polymorphisms in xenobiotic-metabolizing enzymes. Interestingly, they reported that variants within the CYP1A1 gene were associated with lower global methylation levels in coal-exposed individuals, but no significant impact was observed for variants in glutathione-associated genes or others.
Radiation is a well-known cause of cancer. For most individuals, exposure predominantly comes from natural sources, but some occupations can result in significantly higher exposure levels. In 2015, Lee et al. (86), studied methylation levels at the repetitive elements LINE-1 and Sat-2 in nuclear power plant workers. Global DNA methylation levels were lower in radiation-exposed workers than in controls, with LINE-1 methylation negatively correlated with total cumulative radiation dose and with the presence of chromosomal abnormalities. One year later, Kuzmina et al. (87) examined RASSF1A, CDKN2A, and GSTP1 promoter methylation in individuals with a history of exposure to ionizing radiation (including those involved with clean-up of the Chernobyl nuclear plant) in comparison to controls. They observed that GSTP1 and p16 hypermethylation were correlated with exposure. Expanding on their work in relation to the long-term effects of radiation exposure, Kuzmina et al. (88) performed a validation study in a separate cohort of 49 workers from another nuclear plant. They again reported associations with methylation of the GSTP1 and p16 genes, and they additionally observed hypermethylation of the TP53 and SOD3 gene promoters in comparison to non-exposed individuals.
In 2019, Cho et al. (89), examined global methylation in 40 radiographers compared to 28 controls via both LINE-1 assay and luminometric methylation assay (LUMA). They observed global hypomethylation in the exposed group, with methylation of LINE-1 elements associated with cumulative radiation dosage and also associated with chromosomal abnormalities. In the same year, Chen et al. (90) studied 117 physicians exposed to low doses of ionizing radiation. Although marginal, they observed global hypomethylation, suggesting an impact of even comparatively low dosages upon the epigenome.
The impact of nanoparticles upon the epigenome is a field that is only now beginning to develop. The first publication on this topic, published in 2020 by Rossnerova et al. (92), used the Illumina EPIC (850 K) microarray to profile DNA methylation in a group of 20 long-term exposed research workers and 20 controls. They reported hypomethylation at 341 CpG sites and hypermethylation of 364, with leading hits mapping to genes involved in pathways such as immune function and xenobiotic detoxification. More recently, in 2021 Rossnerova et al. (91) reported their findings from a longitudinal study where they analyzed DNA methylation in leukocytes from 10 participants occupationally exposed to nanoparticles and 4 non-exposed controls over the course of 4 years. While they observed methylation changes within both groups, these were more pronounced in the exposed subjects, which they hypothesized to be part of adaptation to chronic nanoparticle exposure.
To date, there have been only two studies of the effects of formaldehyde upon the epigenome. In 2019 Barbosa et al. (104) analyzed global methylation in 49 beauty salon workers, which revealed hypermethylation in exposed workers. More recently, van der Laan et al. (20) measured epigenetic age acceleration in 31 Chinese workers occupationally exposed to formaldehyde in comparison to 39 non-exposed controls, but they did not observe statistically significant differences.
Phillips et al. (93) performed epigenome-wide analysis of DNA methylation using the 450 k microarray in 67 workers exposed to TCE (30 low exposure and 37 high exposure) compared to 73 non-exposed control subjects. Interestingly, they reported increased variation in methylation patterns among exposed individuals, and perhaps on account of this only 25 CpG sites were determined to show significant differences in comparison to controls. Notably, the Tripartite Motif Containing 68 (TRIM68) gene promoter was hypomethylated with TCE exposure, although the implications of this finding are not currently clear. The aforementioned study by van der Laan et al. (20) of epigenetic age in response to environmental exposures also examined the impact of TCE, and in contrast to formaldehyde, it was demonstrated that exposure to TCE increased age acceleration. Together, these studies may suggest an impact of TCE upon the epigenome, but that this appears to be broad and variable, as opposed to inducing gene-specific changes.
Chronic exposure to smoke is one of the main risks for firefighters, and one study of the impact of their exposure to PAHs was described in a previous section. Here, we include two studies that focus on their profession, rather than exposure to a specific compound(s). The first of these, from Zhou et al. (94), examined differences between newly recruited firefighters and incumbents, using the Illumina Infinium EPIC microarray. They identified four CpGs, including three mapping to the Yip1 Domain Family Member 6 (YIPF6), Mercaptopyruvate Sulfurtransferase (MPST) and PC-esterase domain-containing protein 1B (PCED1B) genes, with significant changes in methylation, with three showing higher methylation in incumbents. Global methylation assessment was also able to discriminate between incumbents (including by years of service) and new recruits. The other study performed in firefighters is the very recent publication by Goodrich et al. (95). In this work, once again the Illumina EPIC microarray was used, this time to compare 31 Hispanic white and 163 non-Hispanic white firefighters. Five xenobiotic metabolizing genes were reported to be differentially methylated, including three CpG sites within the promoter region of the biotransformation gene Sulfotransferase C2 (SULT1C2) having lower methylation in Hispanic compared to non-Hispanic firefighters. In 2022, the same group of scientists, performed a repeated measures methylation study in firefighters, using a platform that interrogates up to 850,000 methylation sites, measuring methylation levels prior to live fire training and 20–37 months later. They report 680 CpG sites with altered methylation, including 60 with at least a 5% methylation difference at follow-up; genes with differentially methylated CpG sites were enriched in biological pathways related to cancers, neurological function, cell signaling and transcription regulation.
Working as a hairdresser has been associated with increased risk for cancer, particularly bladder cancer. We found two publications performed in these workers that analyzed DNA methylation. Li et al. (97) recruited 295 hairdressers and analyzed DNA methylation of the following genes: APC; CDKN2A; Death Associated Protein Kinase 1 (DAPK1); GSTP1; MGMT; POU domain, class 4, transcription factor 2 (POU4F2); RASSF1A; RUNX Family Transcription Factor 3 (RUNX3); and Twist Family BHLH Transcription Factor 1 (TWIST1). They did not observe statistically significant differences between groups, but they reported that “hair waving” was associated with less frequent CDKN2A methylation. In 2019, Liljedahl et al. (98) performed gene-specific analysis of DNA methylation in the same cohort of workers studied in the former work. This time they analyzed a greater number of genes: APC; CDKN2A; DAPK1; Docking Protein 1 (DOK1); GSTP1; MGMT; POU4F2; RASSF1A; RUNX3; SHOX2; and TWIST1. They confirmed no significant differences between groups, but now described that carriers of genetic variants within the FLG gene showed higher methylation of CDKN2A.
Wu et al. (99) studied methylation of the MGMT and hMLH1 gene promoters in 101 workers exposed to this compound. MGMT was demonstrated to be hypomethylated, which was only observed in individuals showing chromosomal abnormalities. In 2022, Zhao et al. (100) performing a whole-genome bisulfite sequencing in peripheral leukocytes from three workers exposed to vynil chloride and their respective controls, found that exposed workers showed A total of 9,534 differentially methylated regions (DMRs), of which 4,816 were hypomethylated and 4,718 were hypermethylated. After that first approach, they also selected 50 participants from each group and then methylation of differentially methylated genes (DMGs) found were verified by methylation-specific PCR (MSP) and agarose gel electrophoresis: the coincidence rate was 60–100%. The most involved pathways from the differentially methylated regions observed, corresponded to cancer, neuroactive ligand-receptor interaction, and axon guidance.
In this section, we review all those research works for chemical compounds that comprise the sole study of each.
In 2014, Nylander-French et al. (105) studied 20 automotive spray-painters and performed epigenome-wide analysis by 450K microarray. They reported that methylation at 114 CpG loci were significantly associated with the exposure and urine biomarker levels, with Latrophilin 3 (LPHN3) and Scavenger Receptor Class A Member 5 (SCARA5) appearing to be of particular interest.
Yang et al. (106) measured DNA global methylation in aluminum-exposed pot room workers, without inclusion of a control group. They observed significant decreases in global DNA methylation with increasing levels of exposure.
Janasik et al. (107) recruited 61 occupationally-exposed men and assessed global methylation as well as promoter methylation of the Nuclear Factor Erythroid 2–related Factor 2 (NRF2) and Kelch-like ECH-associated protein 1 (KEAP1) genes. They observed both global and gene-specific hypermethylation in the exposed group, with levels of arsenic exposure positively correlated with global DNA methylation.
Yu et al. (108) analyzed 47 healthy subjects with exposure to asbestos and compared them to both a control group and to 52 individuals diagnosed with benign asbestos-related disorders. They observed that the groups with asbestos exposure and with benign disorders both showed decreased global DNA methylation levels compared with controls, and with no significant difference between asbestos-exposed individuals with and without benign disorders.
Ghosh et al. (109) studied multi-wall carbon nanotubes exposed workers, analyzing both global methylation and promoter methylation for the following genes: DNA methyltransferase 1 (DNMT1); Histone Deacetylase 4 (HDAC4); Nuclear Protein, Coactivator Of Histone Transcription (NPAT/ATM); and Shikimate Kinase 1 (SKI). They did not observe an impact upon global methylation, but the DNMT1 promoter was hypermethylated while HDAC and NPATM were hypermethylated too. Interestingly, SKI showed both gains and losses of methylation at different CpG sites.
Wang et al. (110) recruited 180 workers exposed to coal tar pitch and examined methylation of the p16, Fragile Histidine Triad Diadenosine Triphosphatase (FHIT) and RASSF1A gene promoters. They did not find significant differences between groups, although they observed that p16 methylation was higher in exposed individuals when analysis was restricted to younger individuals (<40).
Silva et al. (111) studied CDKN2A, MLH1, and APC promoter methylation and LINE-1 as a surrogate marker of global methylation in 59 workers exposed to the construction environment and in 49 unexposed workers. They observed higher average levels of methylation for all three genes and hypomethylation of the LINE-1.
Shen et al. (112) analyzed promoter methylation of p16, RASSF1A, and MGMT in workers with occupational exposure to diesel engine exhaust. All three were reported to be hypomethylated and showed with negative correlations with the presence of DNA adducts.
Ouyang et al. (113), studied promoter methylation in 131 workers, with hypermethylation of GSTM1, DUSP22, IFN-γ, and IL-4. IFN-γ observed in the exposed group.
In 2019, van der Plaat et al. (114) analyzed, by means of the Illumina 450K microarray platform, differentially methylated regions in exposed workers. Exposure to gasses/fumes and to mineral dust were associated with 14 and 7 differentially methylated regions (DMRs), respectively. Three of these showed changes in responses to both dust and to gasses and fumes, which mapped to the 60S Ribosomal Protein L1 (RPL1, one DMR) and Long Intergenic Non-Protein Coding RNA 2169 (LINC02169, two DMRs) genes. Many of the observed changes were demonstrated to impact upon gene expression, thereby potentially providing mechanistic insight into the impact of this exposure.
In 2015, Searles Nielsen et al. (115) assessed DNA methylation at three CpG sites in first exon of the NOS2 gene in blood taken from 201 welders, which revealed hypomethylation in exposed workers.
Yang et al. (116) studied gene-specific methylation of the p15 and p16 genes in nickel exposed workers. The p15 gene was observed to be hypermethylated, with workers having higher urinary nickel at increased risk of hypermethylation.
Wang et al. (117), using an epigenome-wide approach, investigated methylation profiles in male employees in an e-waste dismantling area from China. Exposure was associated with 391 hypomethylation sites and 553 hypermethylation sites, with gene expression demonstrated to be affected for ten genes.
In this review, we have summarized current knowledge on the impact of toxic agents upon the epigenome (DNA methylation specifically) in the blood of occupationally-exposed workers. Occupational exposures are particularly different, compared with incidental exposure in the general population, on account of the frequency, duration and dosage of exposure, with individuals often continuously exposed for 8 h or more for 5 or 6 days at a constant level. These unique characteristics of occupational exposures make such individuals an almost controllable model of exposure, compared to incidental (and often non-continuous) exposures of varying degrees in general population. Subsequently, the study of occupational exposures, as in these collated 116 research papers, can provide strong evidence and great insight into epigenetic mechanisms of toxicity. As summarized in Table 1, most of the chemicals studied are related to carcinogenesis, with benzene exposure the most widely studied. When taking into consideration that our review has exclusively focused upon studies of apparently healthy individuals and not those who have already developed disease, all statistically significant results in exposed groups may be considered as early modifications related to disease onset. While epigenome-wide analysis is increasingly common, the most widely-used platform to date has been bisulfite pyrosequencing, which has typically been used to assess global DNA methylation via LINE-1 or Alu assay, but also to focus upon genes related with DNA repair and other tumor-suppressor genes. We regret that few studies have examined the mitochondrial epigenome, and this is clearly an area that warrants further study. Almost 90% of the papers reported significant differences in an exposed group compared with controls, with global hypomethylation observed almost invariably with a wide range of exposures, and promoter hypermethylation also being commonly reported. Only three papers reported concurrent hypomethylation and hypermethylation of different CpG sites within the same gene or locus, of which two reported hypermethylation in the promoter region and hypomethylation within the 5′UTR of the same gene. As methylation within the 5′UTR is very frequently associated with gene transcription, such observations require further work to understand the potential functional consequences. At this point, results from epigenome-wide studies cannot be readily compared due to the complexity of analytical approaches and the comparatively small number of studies. An important question within the field is the persistence of the observed changes (or lack thereof), and therefore it is worth to highlight and to contrast the results from the only two longitudinal studies where authors performed repeated methylation measures. Ren et al. (14) monitored 35 workers exposed to benzene, comparing their methylation levels in 2009 and later in 2013. During this time their exposure levels decreased, and subsequently they reported that LINE-1 methylation levels increased in 2013 compared to 2009 while hMLH1 methylation levels decreased. On the other hand, Rossnerova et al. (91) reported what they call a “higher stability” in five differentially methylated CpG sites evaluated four times annually in 10 workers exposed to nanoparticles, suggesting an enforcement of epigenetic changes through continuous exposure. As these results suggest dynamic changes in DNA methylation in response to exposure levels, which are therefore likely to fluctuate over the lifecourse, they raise some important questions: how transitory are the observed global hypomethylation and gene-specific hypermethylation within exposed individuals? If they are more dynamic, what would measurement of DNA methylation at a single timepoint tell us about the real risk to develop disease? It should be noted that the observations from the two aforementioned studies were in genes from very different cellular pathways, in addition to the toxicant of exposure being different. Another important contribution are those results showing correlation of methylation levels with an already established clinical or diagnostic endpoint, such as neutrophil abundance in the blood or cell/tissue damage endpoints such as cytokinesis-block micronucleus.
At light of all considerations mentioned in the former paragraph, regarding the few longitudinal studies, and also that the epigenetic modifications observed in a certain window of time may be transitory, we think that, with the results obtained in the 116 papers reviewed, we cannot give an insight about the possible consequences of such DNA alterations, since this would be speculative. Based also in the former commentary, we think that results obtained in the several studies do not allow as to claim that observed changes in DNA methylation may be considered as functional or pathological correlates. We consider that, the interest for investigating global and/or promoter hypermethylation as a result of toxicant exposures, mainly for those toxicants considered carcinogenic in humans, comes from the observations in tumors and/or already cancerous cells, where these two modifications were published first in the early eighties and confirmed in the nineties by several studies. However, two considerations are important to interpret these results obtained in occupationally-exposed workers: (1) they are healthy people, and (2) DNA methylation was performed in peripheral blood cells; then, we cannot generalize in the sense that all epigenetic modifications found in blood cells reflects the changes related with carcinogenesis that may occur in other target cells. For instance, in this review we have found that, occupational exposure to arsenic (a known carcinogenic agent) caused global hypermethylation in blood cells, while trichloroethylene and diesel exhaust exposure (also related with cancer development in different tissues) caused gene promoter hypomethylation; on the contrary, 95% of the studies in workers exposed to benzene report hypomethylation of Alu and LINE-1, as surrogate markers of global methylation; however, benzene, contrary to arsenic, trichloroethylene and diesel exhaust, does affect cells from the hematopoietic system. This tells us about the lack of homogeneity for the different results, and therefore, so far, we cannot say that modifications observed in DNA methylation are predictive of certain disease development.
Over the course of 15 years, many and varied results have been published in the toxicomethylomics field, specifically in blood cells from occupational-exposed individuals. It is difficult to generalize the observations those studies have described, mostly because of the cross-sectional nature of the existing publications and their heterogeneous nature. We are still far from considering methylation changes (either global or gene-specific) as a biomarker of effect for most chemical exposures. Even though global hypomethylation and gene-specific promoter hypermethylation (albeit for different genes) may be considered as consistent observations for different exposures, to date there has been no establishment of functional or pathological correlates for those epigenetic modifications. We suggest to create a global network of experts for co-ordinated examination of genes and pathways for the same chemical exposure, with particular focus on longitudinal studies, in order to help bring consensus for how epigenetic changes may be implicated in the mechanisms of toxicity for disease development. Furthermore, we also propose that more studies should examine mitochondrial DNA, since this organelle is very susceptible to early damage, mostly from reactive oxygen species, which is a common toxicological alteration related with all mechanisms of toxicity.
OJ-G: concept design, paper writing, and bibliographic research. LG: supervision and writing. MG: writing and concept design. TB: style correction and concept design. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Szyf M. The implications of DNA methylation for toxicology: toward toxicomethylomics, the toxicology of DNA methylation. Toxicol Sci. (2011) 120:235–55. doi: 10.1093/toxsci/kfr024
2. Bollati V, Baccarelli A, Hou L, Bonzini M, Fustinoni S, Cavallo D, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. (2007) 67:876–80. doi: 10.1158/0008-5472.CAN-06-2995
3. Xing C, Wang Q, Tian QH, Li B, Ni Y, Yin S, et al. Hypermethylation and downregulation of tumor suppressor gene p16 in benzene poisoning. Wei Sheng Yan Jiu. (2012) 41:247–50.
4. Seow WJ, Pesatori AC, Dimont E, Farmer PB, Albetti A, Ettinger AS, et al. Urinary benzene biomarkers and DNA methylation in Bulgarian petrochemical workers: study findings and comparison of linear and beta regression models. PLoS ONE. (2012) 7:e50471doi: 10.1371/journal.pone.0050471
5. Carugno M, Pesatori AC, Dioni L, Hoxha M, Bollati V, Albetti B, et al. Increased mitochondrial DNA copy number in occupations associated with low-dose benzene exposure. Environ Health Perspect. (2012) 120:210–5. doi: 10.1289/ehp.1103979
6. Xing C, Chen Q, Li G, Zhang L, Zheng M, Zou Z, et al. Microsomal epoxide hydrolase (EPHX1) polymorphisms are associated with aberrant promoter methylation of ERCC3 and hematotoxicity in benzene-exposed workers. Environ Mol Mutagen. (2013) 54:397–405. doi: 10.1002/em.21786
7. Yang J, Bai W, Niu P, Tian L, Gao A. Aberrant hypomethylated STAT3 was identified as a biomarker of chronic benzene poisoning through integrating DNA methylation and mRNA expression data. Exp Mol Pathol. (2014) 96:346–53. doi: 10.1016/j.yexmp.2014.02.013
8. Li J, Zhang X, He Z, Sun Q, Qin F, Huang Z, et al. MGMT hypomethylation is associated with DNA damage in workers exposed to low-dose benzene. Biomarkers. (2017) 22:470–5. doi: 10.1080/1354750X.2016.1274335
9. Zheng M, Lin F, Hou F, Li G, Zhu C, Xu P, et al. Association between promoter methylation of gene ERCC3 and benzene hematotoxicity. Int J Environ Res Public Health. (2017) 14:921. doi: 10.3390/ijerph14080921
10. Ren JC, Wu YX, Wu Z, Zhang GH, Wang H, Liu H, et al. MTHFR gene polymorphism is associated with DNA hypomethylation and genetic damage among benzene-exposed workers in southeast China. J Occup Environ Med. (2018) 60:e188–92. doi: 10.1097/JOM.0000000000001288
11. Jamebozorgi I, Majidizadeh T, Pouryagoub G, Mahjoubi F. Aberrant DNA methylation oftwo tumor suppressor genes, p14ARF and p15INK4b, after chronic occupational exposure to low level of benzene. Int J Occup Environ Med. (2018) 9:145–51. doi: 10.15171/ijoem.2018.1317
12. Rota F, Conti A, Campo L, Favero C, Cantone L, Motta V, et al. Epigenetic and Transcriptional modifications in repetitive elements in petrol station workers exposed to benzene and MTBE. Int J Environ Res Public Health. (2018) 15:735. doi: 10.3390/ijerph15040735
13. Costa-Amaral IC, Carvalho LVB, Santos MVC, Valente D, Pereira AC, Figueiredo VO, et al. Environmental assessment and evaluation of oxidative stress and genotoxicity biomarkers related to chronic occupational exposure to benzene. Int J Environ Res Public Health. (2019) 16:2240. doi: 10.3390/ijerph16122240
14. Ren J, Cui JP, Luo M, Liu H, Hao P, Wang X, et al. The prevalence and persistence of aberrant promoter DNA methylation in benzene-exposed Chinese workers. PLoS ONE. (2019) 14:e0220500. doi: 10.1371/journal.pone.0220500
15. Zhang XX, He Z, Feng B, Shao H. An epigenome-wide DNA methylation study of workers with an occupational exposure to lead. J Appl Toxicol. (2019) 39:1311–9. doi: 10.1002/jat.3816
16. Wang DP, Cai DY, Yang XL, Lu X, Lin DF, Li PM, et al. Study of methylation of mitochondrial MT-COI of benzene poisoning. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. (2020) 38:664–8. doi: 10.3760/cma.j.cn121094-20200409-00174
17. Ren JC, Wang T, Wu H, Zhang GH, Sun D, Guo K, et al. Promoter hypermethylation in CSF3R induces peripheral neutrophil reduction in benzene-exposure poisoning. Environ Mol Mutagen. (2020) 61:786–96. doi: 10.1002/em.22382
18. Ji B, Xiao LY, Ren JC, Zhang GH, Wang Y, Dong T, et al. Gene-Environment interactions between environmental response genes polymorphisms and mitochondrial DNA copy numbers among benzene workers. J Occup Environ Med. (2021) 63:e408–15. doi: 10.1097/JOM.0000000000002225
19. Pan Z, Zhong B, Ling X, Zhang H, Tan Q, Huang D, et al. The DNMT1-associated lncRNA UCA1 was upregulated in TK6 cells transformed by long-term exposure to hydroquinone and benzene-exposed workers via DNA hypomethylation. J Biochem Mol Toxicol. (2021) 35:e22920. doi: 10.1002/jbt.22920
20. van der Laan L, Cardenas A, Vermeulen R, Fadadu RP, Hubbard AE, Phillips RV, et al. Epigenetic aging biomarkers and occupational exposure to benzene, trichloroethylene and formaldehyde. Environ Int. (2022) 158:106871. doi: 10.1016/j.envint.2021.106871
21. Zhang H, Pan Z, Ling X, Tan Q, Yuan Q, Qin J, et al. LINC00173 interacts with DNMT1 to regulate LINC00173 expression via promoter methylation in hydroquinone-induced malignantly transformed TK6 cells and benzene-exposed workers. Toxicol Sci. (2022) 187:311–24. doi: 10.1093/toxsci/kfac004
22. Phillips RV, Wei L, Cardenas A, Hubbard AE, McHale CM, Vermeulen R, et al. Epigenome-wide association studies of occupational exposure to benzene and formaldehyde. Epigenetics. (2022) 25:1–19. doi: 10.1080/15592294.2022.2115604
23. Zhang GH, Lu Y, Ji BQ, Ren JC, Sun P, Ding S, et al. Do mutations in DNMT3A/3B affect global DNA hypomethylation among benzene-exposed workers in Southeast China?: Effects of mutations in DNMT3A/3B on global DNA hypomethylation. Environ Mol Mutagen. (2017) 58:678–87. doi: 10.1002/em.22136
24. Godderis L, De Raedt K, Tabish AM, Poels K, Maertens N, De Ruyck K, et al. Epigenetic changes in lymphocytes of solvent-exposed individuals. Epigenomics. (2012) 4:269–77. doi: 10.2217/epi.12.23
25. Jiménez-Garza O, Guo L, Byun HM, Carrieri M, Bartolucci GB, Zhong J, et al. Promoter methylation status in genes related with inflammation, nitrosative stress and xenobiotic metabolism in low-level benzene exposure: searching for biomarkers of oncogenesis. Food Chem Toxicol. (2017) 109 (Pt 1):669–76. doi: 10.1016/j.fct.2017.08.019
26. Jiménez-Garza O, Guo L, Byun HM, Carrieri M, Bartolucci GB, Barrón-Vivanco BS, et al. Aberrant promoter methylation in genes related to hematopoietic malignancy in workers exposed to a VOC mixture. Toxicol Appl Pharmacol. (2018) 339:65–72. doi: 10.1016/j.taap.2017.12.002
27. Jiménez-Garza O, Linares-Segovia B, Ruiz-García L, Monroy-Torres R, Hernández-Luna MA. 5'UTR methylation in different genes from workers exposed to volatile organic compounds: a new insight for considering an epigenetic mark as a functional correlate. Toxicol Lett. (2020) 330:59–64. doi: 10.1016/j.toxlet.2020.05.001
28. Yu SY, Koh EJ, Kim SH, Lee SY, Lee JS, Son SW, et al. Integrated analysis of multi-omics data on epigenetic changes caused by combined exposure to environmental hazards. Environ Toxicol. (2021) 36:1001–10. doi: 10.1002/tox.23099
29. Jiménez-Garza O, Baccarelli AA, Byun HM, Márquez-Gamiño S, Barrón-Vivanco BS, Albores A. CYP2E1 epigenetic regulation in chronic, low-level toluene exposure: relationship with oxidative stress and smoking habit. Toxicol Appl Pharmacol. (2015) 286:207–15. doi: 10.1016/j.taap.2015.04.016
30. Hong JY, Yu SY, Kim SY, Ahn JJ, Kim Y, Kim GW, et al. Association analysis of toluene exposure time with high-throughput mRNA expressions and methylation patterns using in vivo samples. Environ Res. (2016) 146:59–64. doi: 10.1016/j.envres.2015.12.014
31. Pavanello S, Bollati V, Pesatori AC, Kapka L, Bolognesi C, Bertazzi PA, et al. Global and gene-specific promoter methylation changes are related to anti-B[a]PDE-DNA adduct levels and influence micronuclei levels in polycyclic aromatic hydrocarbon-exposed individuals. Int J Cancer. (2009) 125:1692–7. doi: 10.1002/ijc.24492
32. Pavanello S, Pesatori AC, Dioni L, Hoxha M, Bollati V, Siwinska E, et al. Shorter telomere length in peripheral blood lymphocytes of workers exposed to polycyclic aromatic hydrocarbons. Carcinogenesis. (2010) 31:216–21. doi: 10.1093/carcin/bgp278
33. Yang P, Ma J, Zhang B, Duan H, He Z, Zeng J, et al. CpG site-specific hypermethylation of p16INK4α in peripheral blood lymphocytes of PAH-exposed workers. Cancer Epidemiol Biomarkers Prev. (2012) 21:182–90. doi: 10.1158/1055-9965.EPI-11-0784
34. Ouyang B, Baxter CS, Lam HM, Yeramaneni S, Levin L, Haynes E, et al. Hypomethylation of dual specificity phosphatase 22 promoter correlates with duration of service in firefighters and is inducible by low-dose benzo[a]pyrene. J Occup Environ Med. (2012) 54:774–80. doi: 10.1097/JOM.0b013e31825296bc
35. Duan H, He Z, Ma J, Zhang B, Sheng Z, Bin P, et al. Global and MGMT promoter hypomethylation independently associated with genomic instability of lymphocytes in subjects exposed to high-dose polycyclic aromatic hydrocarbon. Arch Toxicol. (2013) 87:2013–22. doi: 10.1007/s00204-013-1046-0
36. Pavanello S, Dioni L, Hoxha M, Fedeli U, Mielzynska-Svach D, Baccarelli AA. Mitochondrial DNA copy number and exposure to polycyclic aromatic hydrocarbons. Cancer Epidemiol Biomarkers Prev. (2013) 22:1722–9. doi: 10.1158/1055-9965.EPI-13-0118
37. Alegría-Torres JA, Barretta F, Batres-Esquivel LE, Carrizales-Yáñez L, Pérez-Maldonado IN, Baccarelli A, et al. Epigenetic markers of exposure to polycyclic aromatic hydrocarbons in Mexican brickmakers: a pilot study. Chemosphere. (2013) 91:475–80. doi: 10.1016/j.chemosphere.2012.11.077
38. Zhang H, Li X, Ge L, Yang J, Sun J, Niu Q. Methylation of CpG island of p14: p15(INK4b) and p16(INK4a) genes in coke oven workers. Hum Exp Toxicol. (2015) 34:191–7. doi: 10.1177/0960327114533576
39. Zhang X, Li J, He Z, Duan H, Gao W, Wang H, et al. Associations between DNA methylation in DNA damage response-related genes and cytokinesis-block micronucleus cytome index in diesel engine exhaust-exposed workers. Arch Toxicol. (2016) 90:1997–2008. doi: 10.1007/s00204-015-1598-2
40. He Z, Li D, Ma J, Chen L, Duan H, Zhang B, et al. TRIM36 hypermethylation is involved in polycyclic aromatic hydrocarbons-induced cell transformation. Environ Pollut. (2017) 225:93–103. doi: 10.1016/j.envpol.2017.03.001
41. Li J, Zhu X, Yu K, Jiang H, Zhang Y, Wang B, et al. Exposure to polycyclic aromatic hydrocarbons and accelerated DNA methylation aging. Environ Health Perspect. (2018) 126:067005. doi: 10.1289/EHP2773
42. Alhamdow A, Lindh C, Hagberg J, Graff P, Westberg H, Krais AM, et al. DNA methylation of the cancer-related genes F2RL3 and AHRR is associated with occupational exposure to polycyclic aromatic hydrocarbons. Carcinogenesis. (2018) 39:869–78. doi: 10.1093/carcin/bgy059
43. Wahlberg K, Liljedahl ER, Alhamdow A, Lindh C, Lidén C, Albin M, et al. Filaggrin variations are associated with PAH metabolites in urine and DNA alterations in blood. Environ Res. (2019) 177:108600. doi: 10.1016/j.envres.2019.108600
44. Liu Y, Li X, Zhang B, Fu Y, Yang A, Zhang H, et al. CYP1A1 methylation mediates the effect of smoking and occupational polycyclic aromatic hydrocarbons co-exposure on oxidative DNA damage among Chinese coke-oven workers. Environ Health. (2019) 18:69. doi: 10.1186/s12940-019-0508-0
45. Fu Y, Niu Y, Pan B, Liu Y, Zhang B, Li X, et al. OGG1 methylation mediated the effects of cell cycle and oxidative DNA damage related to PAHs exposure in Chinese coke oven workers. Chemosphere. (2019) 224:48–57. doi: 10.1016/j.chemosphere.2019.02.114
46. Zhao H, Gao Y, Song T, Jia Y. TH1/TH2 type cytokine expression and DNA aberrant methylation analysis in peripheral blood of coke oven workers. Wei Sheng Yan Jiu. (2020) 49:242–8. doi: 10.19813/j.cnki.weishengyanjiu.2020.02.013
47. Yang J, Liu Y, Zhang H, Zhang H, Wang W, Fan Y. Urinary 1-hydroxypyrene and smoking are determinants of LINE-1 and AhRR promoter methylation in coke oven workers. Mutat Res Genet Toxicol Environ Mutagen. (2018) 826:33–40. doi: 10.1016/j.mrgentox.2018.01.001
48. Alhamdow A, Essig YJ, Krais AM, Gustavsson P, Tinnerberg H, Lindh CH, et al. Fluorene exposure among PAH-exposed workers is associated with epigenetic markers related to lung cancer. Occup Environ Med. (2020) 77:488–95. doi: 10.1136/oemed-2020-106413
49. Dioni L, Hoxha M, Nordio F, Bonzini M, Tarantini L, Albetti B, et al. Effects of short-term exposure to inhalable particulate matter on telomere length, telomerase expression, and telomerase methylation in steel workers. Environ Health Perspect. (2011) 119:622–7. doi: 10.1289/ehp.1002486
50. Tarantini L, Bonzini M, Tripodi A, Angelici L, Nordio F, Cantone L, et al. Blood hypomethylation of inflammatory genes mediates the effects of metal-rich airborne pollutants on blood coagulation. Occup Environ Med. (2013) 70:418–25. doi: 10.1136/oemed-2012-101079
51. Kile ML, Fang S, Baccarelli AA, Tarantini L, Cavallari J, Christiani DC. A panel study of occupational exposure to fine particulate matter and changes in DNA methylation over a single workday and years worked in boilermaker welders. Environ Health. (2013) 12:47. doi: 10.1186/1476-069X-12-47
52. Fan T, Fang SC, Cavallari JM, Barnett IJ, Wang Z, Su L, et al. Heart rate variability and DNA methylation levels are altered after short-term metal fume exposure among occupational welders: a repeated-measures panel study. BMC Public Health. (2014) 14:1279. doi: 10.1186/1471-2458-14-1279
53. Hou L, Zhang X, Zheng Y, Wang S, Dou C, Guo L, et al. Altered methylation in tandem repeat element and elemental component levels in inhalable air particles. Environ Mol Mutagen. (2014) 55:256–65. doi: 10.1002/em.21829
54. Guo L, Byun HM, Zhong J, Motta V, Barupal J, Zheng Y, et al. Effects of short-term exposure to inhalable particulate matter on DNA methylation of tandem repeats. Environ Mol Mutagen. (2014) 55:322–35. doi: 10.1002/em.21838
55. Mercorio R, Bonzini M, Angelici L, Iodice S, Delbue S, Mariani J, et al. Effects of metal-rich particulate matter exposure on exogenous and endogenous viral sequence methylation in healthy steel-workers. Environ Res. (2017) 159:452–7. doi: 10.1016/j.envres.2017.08.042
56. Honkova K, Rossnerova A, Chvojkova I, Milcova A, Margaryan H, Pastorkova A, et al. Genome-Wide DNA methylation in policemen working in cities differing by major sources of air pollution. Int J Mol Sci. (2022) 23:1666. doi: 10.3390/ijms23031666
57. Guo L, Wang Y, Yang X, Wang T, Yin J, Zhao L, et al. Aberrant mitochondrial DNA methylation and declined pulmonary function in a population with polycyclic aromatic hydrocarbon composition in particulate matter. Environ Res. (2022) 214 (Pt 1):113797. doi: 10.1016/j.envres.2022.113797
58. Hou L, Zhang X, Tarantini L, Nordio F, Bonzini M, Angelici L, et al. Ambient PM exposure and DNA methylation in tumor suppressor genes: a cross-sectional study. Part Fibre Toxicol. (2011) 8:25. doi: 10.1186/1743-8977-8-25
59. Byun HM, Panni T, Motta V, Hou L, Nordio F, Apostoli P, et al. Effects of airborne pollutants on mitochondrial DNA methylation. Part Fibre Toxicol. (2013) 10:18. doi: 10.1186/1743-8977-10-18
60. Byun HM, Motta V, Panni T, Bertazzi PA, Apostoli P, Hou L, et al. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol. (2013) 10:28. doi: 10.1186/1743-8977-10-28
61. Kahl VFS, Dhillon V, Fenech M, de Souza MR, da Silva FN, Marroni NAP, et al. Occupational exposure to pesticides in tobacco fields: the integrated evaluation of nutritional intake and susceptibility on genomic and epigenetic instability. Oxid Med Cell Longev. (2018) 2018:7017423. doi: 10.1155/2018/7017423
62. Kahl VFS, Dhillon VS, Simon D, da Silva FR, Salvador M, Branco CDS, et al. Chronic occupational exposure endured by tobacco farmers from Brazil and association with DNA damage. Mutagenesis. (2018) 33:119–28. doi: 10.1093/mutage/gex045
63. Rusiecki JA, Beane Freeman LE, Bonner MR, Alexander M, Chen L, Andreotti G, et al. High pesticide exposure events and DNA methylation among pesticide applicators in the agricultural health study. Environ Mol Mutagen. (2017) 58:19–29. doi: 10.1002/em.22067
64. Benedetti D, Lopes Alderete B, de Souza CT, Ferraz Dias J, Niekraszewicz L, Cappetta M, et al. DNA damage and epigenetic alteration in soybean farmers exposed to complex mixture of pesticides. Mutagenesis. (2018) 33:87–95. doi: 10.1093/mutage/gex035
65. Benitez-Trinidad AB, Medina-Díaz IM, Bernal-Hernández YY, Barrón-Vivanco BS, González-Arias CA, Herrera-Moreno JF, et al. Relationship between LINE-1 methylation pattern and pesticide exposure in urban sprayers. Food Chem Toxicol. (2018) 113:125–33. doi: 10.1016/j.fct.2018.01.035
66. van der Plaat DA, de Jong K, de Vries M, van Diemen CC, Nedeljković I, Amin N, et al. Occupational exposure to pesticides is associated with differential DNA methylation. Occup Environ Med. (2018) 75:427–35. doi: 10.1136/oemed-2017-104787
67. Herrera-Moreno JF, Medina-Díaz IM, Bernal-Hernández YY, Ramos KS, Alvarado-Cruz I, Quintanilla-Vega B, et al. Modified CDKN2B (p15) and CDKN2A (p16) DNA methylation profiles in urban pesticide applicators. Environ Sci Pollut Res Int. (2019) 26:15124–35. doi: 10.1007/s11356-019-04658-5
68. Paredes-Céspedes DM, Bernal-Hernández YY, Herrera-Moreno JF, Rojas-García AE, Medina-Díaz IM, González-Arias CA, et al. Methylation patterns of the CDKN2B and CDKN2A genes in an indigenous population exposed to pesticides. Hum Exp Toxicol. (2022) 41:9603271211063161. doi: 10.1177/09603271211063161
69. Howard TD, Hsu FC, Chen H, Quandt SA, Talton JW, Summers P, et al. Changes in DNA methylation over the growing season differ between North Carolina farmworkers and non-farmworkers. Int Arch Occup Environ Health. (2016) 89:1103–10. doi: 10.1007/s00420-016-1148-0
70. Kovatsi L, Georgiou E, Ioannou A, Haitoglou C, Tzimagiorgis G, Tsoukali H, et al. p16 promoter methylation in Pb2+ -exposed individuals. Clin Toxicol. (2010) 48:124–8. doi: 10.3109/15563650903567091
71. Li C, Xu M, Wang S, Yang X, Zhou S, Zhang J, et al. Lead exposure suppressed ALAD transcription by increasing methylation level of the promoter CpG islands. Toxicol Lett. (2011) 203:48–53. doi: 10.1016/j.toxlet.2011.03.002
72. Li C, Yang X, Xu M, Zhang J, Sun N. Epigenetic marker (LINE-1 promoter) methylation level was associated with occupational lead exposure. Clin Toxicol (Phila). (2013) 51:225–9. doi: 10.3109/15563650.2013.782410
73. Devóz PP, Gomes WR, De Araújo ML, Ribeiro DL, Pedron T, Greggi Antunes LM, et al. Lead (Pb) exposure induces disturbances in epigenetic status in workers exposed to this metal. J Toxicol Environ Health A. (2017) 80:1098–105. doi: 10.1080/15287394.2017.1357364
74. Yu LB, Tu YT, Huang JW, Zhang YN, Zheng GQ, Xu XW, et al. Hypermethylation of CpG islands is associated with increasing chromosomal damage in Chinese lead-exposed workers. Environ Mol Mutagen. (2018) 59:549–56. doi: 10.1002/em.22194
75. Devóz PP, Reis MBD, Gomes WR, Maraslis FT, Ribeiro DL, Antunes LMG, et al. Adaptive epigenetic response of glutathione (GSH)-related genes against lead (Pb)-induced toxicity, in individuals chronically exposed to the metal. Chemosphere. (2021) 269:128758. doi: 10.1016/j.chemosphere.2020.128758
76. Meng Y, Zhou M, Wang T, Zhang G, Tu Y, Gong S, et al. Occupational lead exposure on genome-wide DNA methylation and DNA damage. Environ Pollut. (2022) 304:119252. doi: 10.1016/j.envpol.2022.119252
77. Yang L, Xia B, Yang X, Ding H, Wu D, Zhang H, et al. Mitochondrial DNA hypomethylation in chrome plating workers. Toxicol Lett. (2016) 243:1–6. doi: 10.1016/j.toxlet.2015.11.031
78. Hu G, Li P, Cui X, Li Y, Zhang J, Zhai X, et al. Cr(VI)-induced methylation and down-regulation of DNA repair genes and its association with markers of genetic damage in workers and 16HBE cells. Environ Pollut. (2018) 238:833–43. doi: 10.1016/j.envpol.2018.03.046
79. Wang TC, Song YS, Wang H, Zhang J, Yu SF, Gu YE, et al. Oxidative DNA damage and global DNA hypomethylation are related to folate deficiency in chromate manufacturing workers. J Hazard Mater. (2012) 213–4:440–6. doi: 10.1016/j.jhazmat.2012.02.024
80. Li H, Hedmer M, Wojdacz T, Hossain MB, Lindh CH, Tinnerberg H, et al. Oxidative stress, telomere shortening, and DNA methylation in relation to low-to-moderate occupational exposure to welding fumes. Environ Mol Mutagen. (2015) 56:684–93. doi: 10.1002/em.21958
81. Xu Y, Li H, Hedmer M, Hossain MB, Tinnerberg H, Broberg K, et al. Occupational exposure to particles and mitochondrial DNA—relevance for blood pressure. Environ Health. (2017) 16:22. doi: 10.1186/s12940-017-0234-4
82. Dauter UM, Alhamdow A, Cediel-Ulloa A, Gliga AR, Albin M, Broberg K. Cancer-related changes and low-to-moderate exposure to welding fumes: a longitudinal study. Scand J Work Environ Health. (2022) 48:21–30. doi: 10.5271/sjweh.3988
83. Hossain MB, Li H, Hedmer M, Tinnerberg H, Albin M, Broberg K. Exposure to welding fumes is associated with hypomethylation of the F2RL3 gene: a cardiovascular disease marker. Occup Environ Med. (2015) 72:845–51. doi: 10.1136/oemed-2015-102884
84. de Souza MR, Kahl VFS, Rohr P, Kvitko K, Cappetta M, Lopes WM, et al. Shorter telomere length and DNA hypermethylation in peripheral blood cells of coal workers. Mutat Res Genet Toxicol Environ Mutagen. (2018) 836 (Pt. B):36–41. doi: 10.1016/j.mrgentox.2018.03.009
85. de Souza MR, Rohr P, Kahl VFS, Kvitko K, Cappetta M, Lopes WM, et al. The influence of polymorphisms of xenobiotic-metabolizing and DNA repair genes in DNA damage, telomere length and global DNA methylation evaluated in open-cast coal mining workers. Ecotoxicol Environ Saf. (2020) 189:109975. doi: 10.1016/j.ecoenv.2019.109975
86. Lee Y, Kim YJ, Choi YJ, Lee JW, Lee S, Cho YH, et al. Radiation-induced changes in DNA methylation and their relationship to chromosome aberrations in nuclear power plant workers. Int J Radiat Biol. (2015) 91:142–9. doi: 10.3109/09553002.2015.969847
87. Kuzmina NS, Lapteva N, Rubanovich AV. Hypermethylation of gene promoters in peripheral blood leukocytes in humans long term after radiation exposure. Environ Res. (2016) 146:10–7. doi: 10.1016/j.envres.2015.12.008
88. Kuzmina NS, Lapteva NS, Rusinova GG, Azizova TV, Vyazovskaya NS, Rubanovich AV. Gene hypermethylation in blood leukocytes in humans long term after radiation exposure - validation set. Environ Pollut. (2018) 234:935–42. doi: 10.1016/j.envpol.2017.12.039
89. Cho YH, Jang Y, Woo HD, Kim YJ, Kim SY, Christensen S, et al. LINE-1 hypomethylation is associated with radiation-induced genomic instability in industrial radiographers. Environ Mol Mutagen. (2019) 60:174–84. doi: 10.1002/em.22237
90. Chen B, Dai Q, Zhang Q, Yan P, Wang A, Qu L, et al. The relationship among occupational irradiation, DNA methylation status, and oxidative damage in interventional physicians. Medicine. (2019) 98:e17373. doi: 10.1097/MD.0000000000017373
91. Rossnerova A, Honkova K, Chvojkova I, Pelclova D, Zdimal V, Hubacek JA, et al. Individual DNA methylation pattern shifts in nanoparticles-exposed workers analyzed in four consecutive years. Int J Mol Sci. (2021) 22:7834. doi: 10.3390/ijms22157834
92. Rossnerova A, Honkova K, Pelclova D, Zdimal V, Hubacek JA, Chvojkova I, et al. DNA methylation profiles in a group of workers occupationally exposed to nanoparticles. Int J Mol Sci. (2020) 21:2420. doi: 10.3390/ijms21072420
93. Phillips RV, Rieswijk L, Hubbard AE, Vermeulen R, Zhang J, Hu W, et al. Human exposure to trichloroethylene is associated with increased variability of blood DNA methylation that is enriched in genes and pathways related to autoimmune disease and cancer. Epigenetics. (2019) 14:1112–24. doi: 10.1080/15592294.2019.1633866
94. Zhou J, Jenkins TG, Jung AM, Jeong KS, Zhai J, Jacobs ET, et al. DNA methylation among firefighters. PLoS ONE. (2019) 14:e0214282. doi: 10.1371/journal.pone.0214282
95. Goodrich JM, Furlong MA, Caban-Martinez AJ, Jung AM, Batai K, Jenkins T, et al. Differential DNA methylation by hispanic ethnicity among firefighters in the united states. Epigenet Insights. (2021) 14:25168657211006159. doi: 10.1177/25168657211006159
96. Goodrich JM, Jung AM, Furlong MA, Beitel S, Littau S, Gulotta J, et al. Repeat measures of DNA methylation in an inception cohort of firefighters. Occup Environ Med. (2022) 79:656–63. doi: 10.1136/oemed-2021-108153
97. Li H, Åkerman G, Lidén C, Alhamdow A, Wojdacz TK, Broberg K, Albin M. Alterations of telomere length and DNA methylation in hairdressers: a cross-sectional study. Environ Mol Mutagen. (2016) 57:159–67. doi: 10.1002/em.21991
98. Liljedahl ER, Wahlberg K, Lidén C, Albin M, Broberg K. Genetic variants of filaggrin are associated with occupational dermal exposure and blood DNA alterations in hairdressers. Sci Total Environ. (2019) 653:45–54. doi: 10.1016/j.scitotenv.2018.10.328
99. Wu F, Liu J, Qiu YL, Wang W, Zhu SM, Sun P, et al. Correlation of chromosome damage and promoter methylation status of the DNA repair genes MGMT and hMLH1 in Chinese vinyl chloride monomer (VCM)-exposed workers. Int J Occup Med Environ Health. (2013) 26:173–82. doi: 10.2478/s13382-013-0079-1
100. Zhao X, Hao Y, Wang Q, Shen Y, Cheng Y, Li B, et al. Novel deoxyribonucleic acid methylation perturbations in workers exposed to vinyl chloride. Toxicol Ind Health. (2022) 38:377–88. doi: 10.1177/07482337221098600
101. Li C, Yang X, Xu M, Zhang J, Sun N. Association between GSTP1 CpG methylation and the early phase of lead exposure. Toxicol Mech Methods. (2014) 24:111–5. doi: 10.3109/15376516.2013.859195
102. Issah I, Arko-Mensah J, Rozek LS, Zarins KR, Dwomoh D, Agyekum TP, et al. Association between toxic and essential metals in blood and global DNA methylation among electronic waste workers in Agbogbloshie, Ghana. Environ Sci Pollut Res Int. (2022) 29:72946–56. doi: 10.1007/s11356-022-20954-z
103. Barrow TM, Klett H, Toth R, Böhm J, Gigic B, Habermann N, et al. Smoking is associated with hypermethylation of the APC 1A promoter in colorectal cancer: the ColoCare Study. J Pathol. (2017) 243:366–75. doi: 10.1002/path.4955
104. Barbosa E, Dos Santos ALA, Peteffi GP, Schneider A, Müller D, Rovaris D, et al. Increase of global DNA methylation patterns in beauty salon workers exposed to low levels of formaldehyde. Environ Sci Pollut Res Int. (2019) 26:1304–14. doi: 10.1007/s11356-018-3674-7
105. Nylander-French LA, Wu MC, French JE, Boyer JC, Smeester L, Sanders AP, et al. DNA methylation modifies urine biomarker levels in 1,6-hexamethylene diisocyanate exposed workers: a pilot study. Toxicol Lett. (2014) 231:217–26. doi: 10.1016/j.toxlet.2014.10.024
106. Yang X, Yuan Y, Lu X, Yang J, Wang L, Song J, et al. The relationship between cognitive impairment and global DNA methylation decrease among aluminum potroom workers. J Occup Environ Med. (2015) 57:713–7. doi: 10.1097/JOM.0000000000000474
107. Janasik B, Reszka E, Stanislawska M, Jablonska E, Kuras R, Wieczorek E, et al. Effect of arsenic exposure on NRF2-KEAP1 pathway and epigenetic modification. Biol Trace Elem Res. (2018) 185:11–9. doi: 10.1007/s12011-017-1219-4
108. Yu M, Lou J, Xia H, Zhang M, Zhang Y, Chen J, et al. Global DNA hypomethylation has no impact on lung function or serum inflammatory and fibrosis cytokines in asbestos-exposed population. Int Arch Occup Environ Health. (2017) 90:265–74. doi: 10.1007/s00420-017-1195-1
109. Ghosh M, Öner D, Poels K, Tabish AM, Vlaanderen J, Pronk A, et al. Changes in DNA methylation induced by multi-walled carbon nanotube exposure in the workplace. Nanotoxicology. (2017) 11:1195–210. doi: 10.1080/17435390.2017.1406169
110. Wang Y, Duan X, Zhang Y, Wang S, Yao W, Wang S, et al. [DNA methylation and telomere damage in occupational people exposed to coal tar pitch]. Zhonghua Lao Dong Wei Sheng Zhi Ye Bing Za Zhi. (2015) 33:507–11.
111. Silva IR, Ramos MCAS, Arantes LMRB, Lengert AVH, Oliveira MA, et al. Evaluation of DNA methylation changes and micronuclei in workers exposed to a construction environment. Int J Environ Res Public Health. (2019) 16:902. doi: 10.3390/ijerph16060902
112. Shen ML, He ZN, Zhang X, Duan HW, Niu Y, Bin P, et al. Association of etheno-DNA adduct and DNA methylation level among workers exposed to diesel engine exhaust. Zhonghua Yu Fang Yi Xue Za Zhi. (2017) 51:556–61. doi: 10.3760/cma.j.issn.0253-9624.2017.06.019
113. Ouyang B, Bernstein DI, Lummus ZL, Ying J, Boulet LP, Cartier A, et al. Interferon-γ promoter is hypermethylated in blood DNA from workers with confirmed diisocyanate asthma. Toxicol Sci. (2013) 133:218–24. doi: 10.1093/toxsci/kft079
114. van der Plaat DA, Vonk JM, Terzikhan N, de Jong K, de Vries M, La Bastide-van Gemert S, et al. Occupational exposure to gases/fumes and mineral dust affect DNA methylation levels of genes regulating expression. Hum Mol Genet. (2019) 28:2477–85. doi: 10.1093/hmg/ddz067
115. Searles Nielsen S, Checkoway H, Criswell SR, Farin FM, Stapleton PL, Sheppard L, et al. Inducible nitric oxide synthase gene methylation and parkinsonism in manganese-exposed welders. Parkinsonism Relat Disord. (2015) 21:355–60. doi: 10.1016/j.parkreldis.2015.01.007
116. Yang J, Chen W, Li X, Sun J, Guo Q, Wang Z. Relationship between urinary nickel and methylation of p15, p16 in workers exposed to nickel. J Occup Environ Med. (2014) 56:489–92. doi: 10.1097/JOM.0000000000000168
Keywords: epigenetics, occupational toxicology, DNA methylation, biomarker of effect, mitochondrial DNA methylation, DNA repair genes, peripheral blood
Citation: Jiménez-Garza O, Ghosh M, Barrow TM and Godderis L (2023) Toxicomethylomics revisited: A state-of-the-science review about DNA methylation modifications in blood cells from workers exposed to toxic agents. Front. Public Health 11:1073658. doi: 10.3389/fpubh.2023.1073658
Received: 18 October 2022; Accepted: 25 January 2023;
Published: 20 February 2023.
Edited by:
Sophie Ndaw, Institut National de Recherche et de Sécurité pour la prévention des maladies professionnelles et des accidents du travail (INRS), FranceReviewed by:
Juliana Jalaludin, Universiti Putra Malaysia, MalaysiaCopyright © 2023 Jiménez-Garza, Ghosh, Barrow and Godderis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lode Godderis, 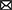 bG9kZS5kb2RkZXJpc0BrdWxldXZlbi5iZQ==
bG9kZS5kb2RkZXJpc0BrdWxldXZlbi5iZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.