
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health, 06 July 2022
Sec. Digital Public Health
Volume 10 - 2022 | https://doi.org/10.3389/fpubh.2022.922510
This article is part of the Research TopicBig Data Analytics for Smart Healthcare applicationsView all 109 articles
 Wen-Cai Liu1,2†
Wen-Cai Liu1,2† Ming-Xuan Li2†
Ming-Xuan Li2† Shi-Nan Wu2†
Shi-Nan Wu2† Wei-Lai Tong1,3
Wei-Lai Tong1,3 An-An Li1,3
An-An Li1,3 Bo-Lin Sun1,3
Bo-Lin Sun1,3 Zhi-Li Liu1,3
Zhi-Li Liu1,3 Jia-Ming Liu1,3*
Jia-Ming Liu1,3*Breast cancer (BC) was the most common malignant tumor in women, and breast infiltrating ductal carcinoma (IDC) accounted for about 80% of all BC cases. BC patients who had bone metastases (BM) were more likely to have poor prognosis and bad quality of life, and earlier attention to patients at a high risk of BM was important. This study aimed to develop a predictive model based on machine learning to predict risk of BM in patients with IDC. Six different machine learning algorithms, including Logistic regression (LR), Naive Bayes classifiers (NBC), Decision tree (DT), Random Forest (RF), Gradient Boosting Machine (GBM), and Extreme gradient boosting (XGB), were used to build prediction models. The XGB model offered the best predictive performance among these 6 models in internal and external validation sets (AUC: 0.888, accuracy: 0.803, sensitivity: 0.801, and specificity: 0.837). Finally, an XGB model-based web predictor was developed to predict risk of BM in IDC patients, which may help physicians make personalized clinical decisions and treatment plans for IDC patients.
As one of the most common malignancies, breast cancer (BC) accounted for 30% of all cancers in women (1). The incidence of BC continued to increase at a rate of ~0.5% per year, which was attributed at least in part to the continued decline in fertility and increased body weight (2). In the cancer statistical report, the number of BC patients exceeded 2.1 million, and infiltrating ductal carcinoma (IDC) was the most common one among different types of BC (3, 4). Early diagnosis provides a favorable prognosis and better overall survival for BC patients. In North America, early screening for BC significantly increased the 5-year survival rate to over 80% for BC patients (5). In recent years, BC patients still had a high incidence of distant metastatic recurrence, which was an important indicator for poor prognosis. Recent studies have suggested that age, menopausal status, T, N stages, histological grade and HR/HER2 status were risk factors for BM in BC patients (6–8). For distant metastases, bone was the most common site, and more than 60% of BC patients developed bone metastases (BM) (9). Another study indicated over 20% of patients developed BM within an average follow-up duration of 8 years. And further survival prognostic modeling showed a 40-month median survival time for patients with BM (10). In addition, BC patients with BM often developed secondary clinical complexities that took up significant medical resources (11). Early diagnosis of BM from BC will help in the timely prevention and treatment of complications and increase the quality of life for BC patients.
Currently, precision medicine has preceded four concepts: predictive, personalized, preventive and participatory (12). The technology of big data analytics is becoming a clinical imperative (13). This means that we need to use advanced technology to analyze large amounts of medical data to provide recommendations for individualized treatment. Many studies have used machine learning (ML) techniques to study clinical risk factors associated with cancer metastases for early detection (14–16). It is known that the most common type of pathology in BC is IDC (4). But few studies focused on incorporating machine learning to predict the risk of BM from IDC patients.
In this study, we attempted to develop a predictive model to predict the BM risk in IDC patients based on machine learning, and to assist clinicians in implementing more rational clinical decisions as well as to enable patients to receive earlier treatment. Our contribution includes:
• Machine learning algorithms were used instead of traditional statistical regression methods to process data on clinical characteristics of IDC patients to identify patients at high risk for BM.
• This paper compared various algorithms that could be used to process patient data and identified XGB as the best method for processing the data. Further, the hyperparameters of the XGB algorithm were fine-tuned using a random search method to improve performance.
• We performed importance analysis of the features included in the model using the permutation importance method, and these features were further analyzed and understood from a clinical medicine perspective in the discussion section.
• This study proposed a machine learning based solution that could assist clinicians in making individualized diagnoses of BM for IDC patients.
The rest of the paper was organized as follows: the materials and methods section provided a detailed documentation of the materials and methods used, as well as the dataset description, statistical analyses, data pre-processing, feature engineering, classification algorithms used and evaluation metrics. Results of the experiment are discussed in Section Results and then further discussed in Section Discussion. Conclusions, limitations and future work section concluded the results and provided limitations and the future direction of the current work.
The training and internal test set data was derived from the Surveillance, Epidemiology, and End Results (SEER) database, and the external test set data was derived from the First Affiliated Hospital of Nanchang University in China. A data set of 311,408 IDC patients was included from the SEER database (2010–2017) and sliced into a training set and an internal test set randomly in a ratio of 7:3. An external validation set included data from 1,243 IDC patients of our hospital (2010–2017). The exclusion criteria for clinical data were as follows: (1) unknown information of T, N stage, race, laterality, breast subtype, grade and marital status. (2) Other cases with unknown primary tumor and metastatic status. The detailed exclusion criteria were shown in Figure 1. Information of all variables was complete for these patients.
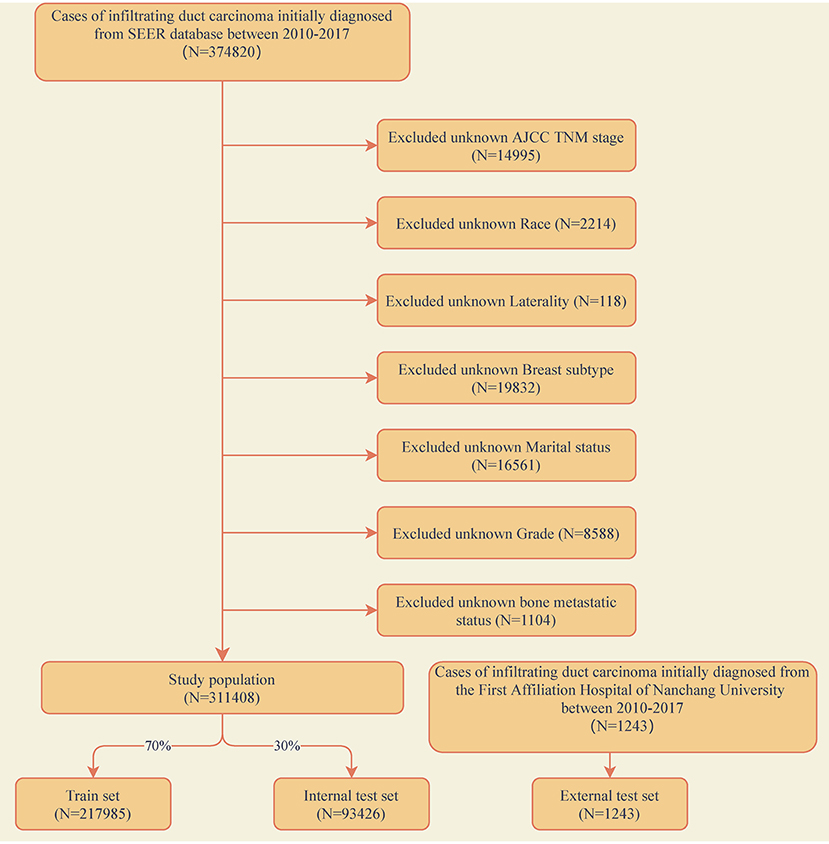
Figure 1. Flow diagram of the study population selected from the Surveillance, Epidemiology, and End Results (SEER) database and the First Affiliated Hospital of Nanchang University. According to the inclusion and exclusion criteria, a total of 311,408 patients of SEER were included in this study, and they were randomly cut into the training and internal test sets in a 7:3 ratio. Data from the First Affiliated Hospital of Nanchang University (n = 1,243) as an external test set.
We selected nine variables from the SEER database and our hospital that may affect BM in patients with IDC, including age at diagnosis, race, sex, grade, T, N stage, breast subtype, laterality and marital status. All cases included in this study were staged using the 7th edition of the AJCC TNM staging system and the relevant guidelines of the SEER project.
The statistical analyses in this study were all performed by Python (version 3.8, Python Software Foundation) and SPSS (version 26, IBM, USA). Data from SEER were randomly sliced into training and internal test sets in a ratio of 7:3 using python. The training set was used to construct the models, and the internal test set and external test set were used for model validation and evaluation. A heat map was drawn to determine the association among the variables. A univariate analysis was performed to compare variables between patients with and without BM. For categorical data, the chi-square test was used, and for continuous non-normally distributed data, the Wilcoxon rank-sum test was used. Variables with a P < 0.05 in univariate analysis were enclosed within the construction of machine learning models and multivariate logistic regression was performed to identify the risk factors of BM from IDC patients.
Category variables such as T and N stages were processed using label encoding methods. The univariate analysis was used to screen for meaningful combinations of features for predicting the risk of IDC patients with BM. Correlation analysis was used to analyze the correlation among the selected features. Feature importance analysis was performed on the variables based on the Permutation Importance principle (17, 18).
The purpose of this study was to accurately predict the clinical outcome of a specific patient based on multiple variables. So the predictive power and accuracy of the model were important. Thus, to evaluate the model, we considered the area under the receiver operating characteristic curve (AUC), accuracy, sensitivity (recall rate) and specificity score in the study. The following terms were used in the equations: TP (True Positive); TN (True Negative); FP (False Positive); and FN (False Negative).
All the algorithmic models were built based on scikit-learn (version 0.24.2). The random oversampling method in imbalanced-learn (version 0.9.1) was used to deal with the imbalance of data distribution.
In this study, we used six different machine learning algorithms: Logistic regression (LR), Naive Bayes classifiers (NBC), Decision tree (DT), Random Forest (RF), Gradient Boosting Machine (GBM) and Extreme gradient boosting (XGB) (19–24). The ML algorithms were trained and adjusted to predict the BM in IDC patients. Random search method in scikit-learn was used to adjust the hyperparameters of the model. Then, the predictive performance of the ML models was evaluated in internal 10-fold cross-validation of the train set, internal and external test sets and the AUC, accuracy, sensitivity (recall rate) and specificity score were evaluated. Then, we selected the best-performing model to build a web predictor.
A total of 311,408 cases who were first diagnosed with IDC in the SEER database from 2010 to 2017 were included. Of these cases, 7,949 (2.55%) complicated with BM and 303,459 (97.45%) without BM. All demographic and clinicopathological characteristics of these patients were demonstrated in detail in Table 1. All cases were randomly divided into a training set (n = 217,985) and an internal test set (n = 93,426) in a ratio of 7:3. Data for the external test set were derived from IDC patients firstly diagnosed in our hospital from 2010 to 2017 (N = 1,243). The details of the training and test sets were shown in Table 2.
According to the univariate analysis, patients' age, gender, race, grade, T, N stage, breast subtype and marital status were significantly associated with BM in patients with IDC (P < 0.05; Table 3). Variables with a P < 0.05 in the univariate analysis were selected for multivariate logistic regression analysis, in order to identify the risk factors of BM in IDC patients. According to these results, age, race, grade, T, N stage, breast subtype, and marital status were found to be independent factors for BM (Table 3).
To identify the effect of these features on prediction, correlation tests between each other were performed. Correlation analysis among dataset features provides information about the degree of interaction among features. The heat map showed the relevance of these features on the prediction of ML algorithm (Figure 2). The figure showed a positive correlation among T-stage, N-stage and pathological grade. This was consistent with clinical experience that poorly differentiated tumor tissue was poorly demarcated from surrounding normal tissue, which meant it was more aggressive.
The feature importance of each ML model for predicting BM was illustrated in Figure 3. Permutation importance principle was used to analyze the relative feature importance on the variables in each ML model. The basic idea of the principle was: (1) Train the model. (2) Disrupt the data in one of the columns and use that dataset for prediction, evaluating the decrease in prediction accuracy to reflect the importance of that feature variable. (3) Restore the validation dataset and repeat the second step to analyze the other feature variables. Although the relative feature importance in different ML models varied slightly, T, N stage, breast subtype, grade, and marital status were the top-ranked variables in most models. In contrast, the race was the last one in most models. But it also contributed to the model. In the XGB model, the relative feature importance was sorted in descending order by T, N stage, breast subtype, grade, marital status, laterality, age, sex, and race.
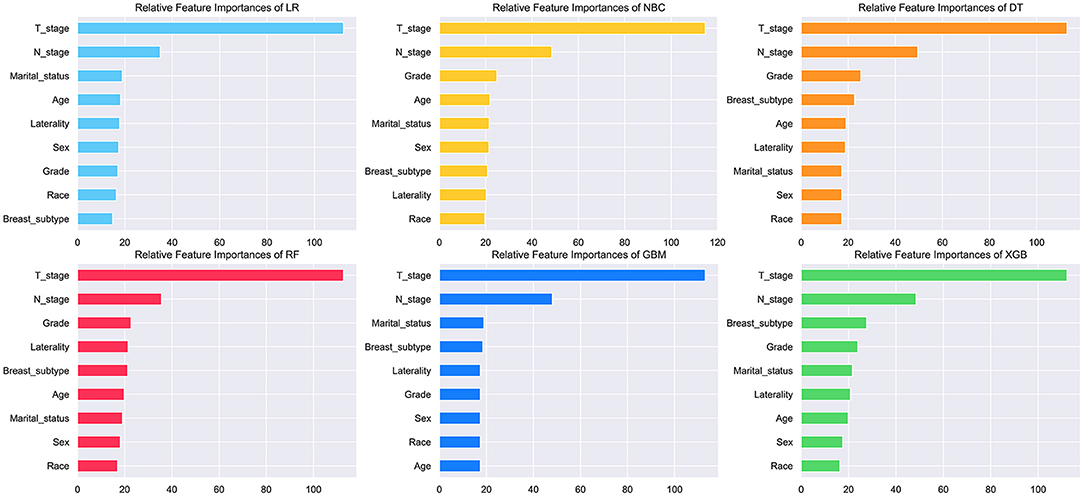
Figure 3. Relative feature importance of different models. The plot showed the ranking of the relative importance of features in all models.
The prediction performance of all models was compared with the internal 10-fold cross-validation of the training set, internal and external test sets, as shown in Figures 4–6 and Table 4. In the internal test set, the XGB model was the best performer with an AUC of 0.857, an accuracy of 0.787, a sensitivity of 0.787 and a specificity of 0.791. In the external test set, the XGB model showed a relatively better performance with an AUC of 0.888, an accuracy of 0.803, a sensitivity of 0.801 and a specificity of 0.837 (Table 4). Finally, we used the highest-performing model to create a web predictor.

Figure 4. Ten-fold cross-validation results of different machine learning models in the training set. DT, Decision tree; LR, Logistic regression; GBM, Gradient Boosting Machine; NBC, Naive Bayes classification; RF, Random Forest; XGB, Extreme gradient boosting.
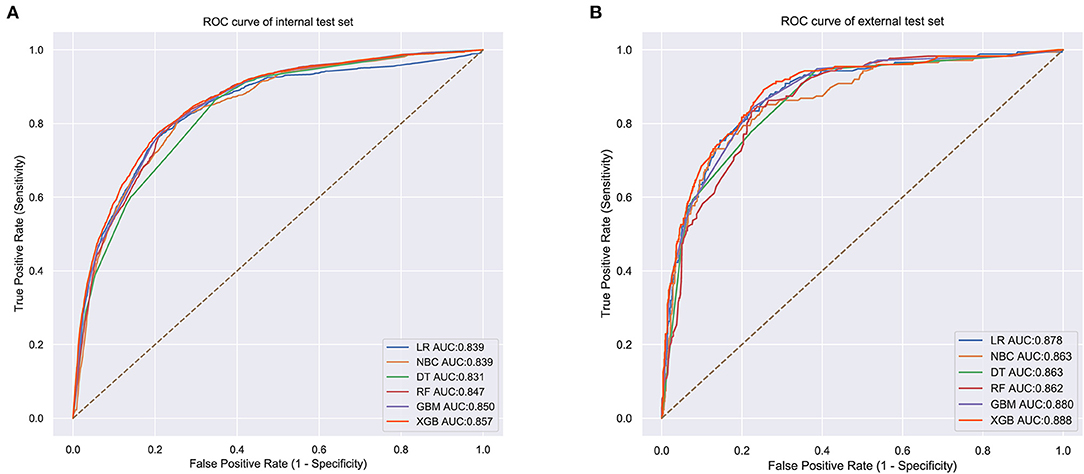
Figure 5. (A) Internal test ROC curve of different machine learning models. (B) External test ROC curve of different machine learning models.
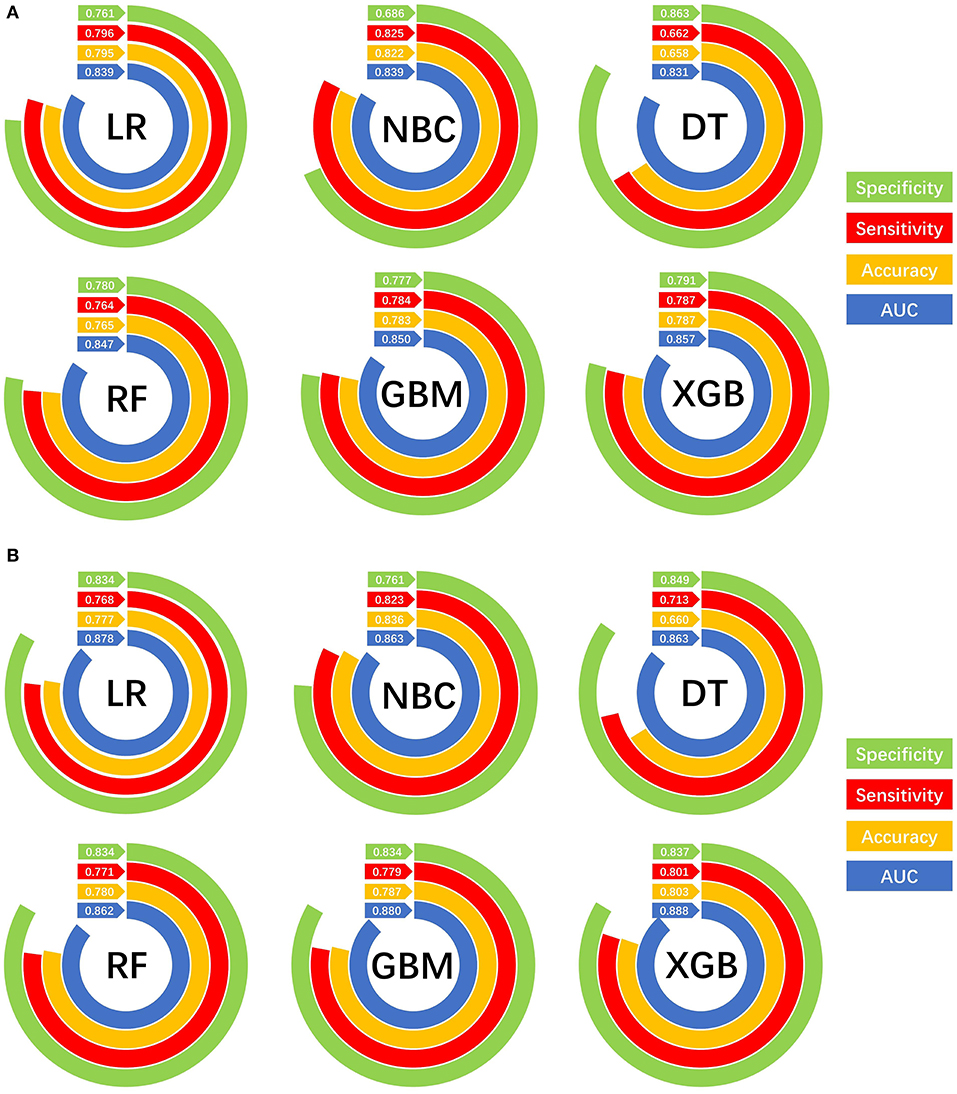
Figure 6. Prediction performances of different machine learning models. (A) Internal validation of different machine learning models. (B) External validation of different machine learning models.
Based on the XGB model, a ML algorithm for optimal predictive performance, the web predictor mentioned above was developed to predict the risk of BM in IDC patients. We can predict the BM risk in IDC patients simply by setting variables in the sidebar of the website (Figure 7) (https://share.streamlit.io/liuwencaincu/breast-cancer/main/breast.py).
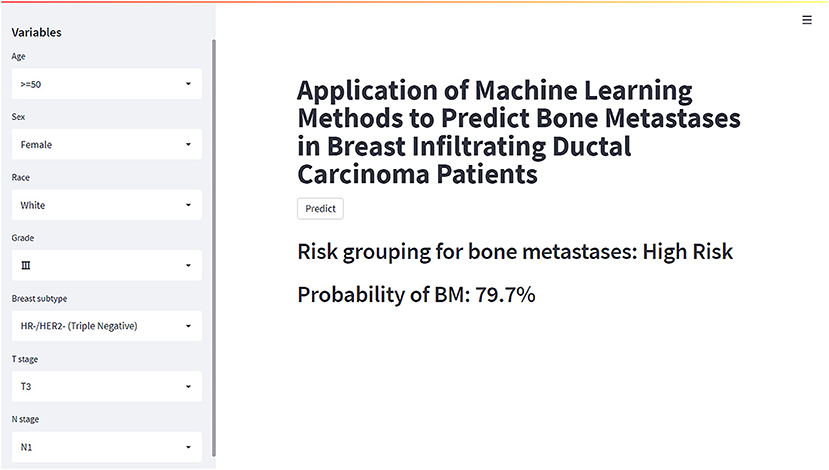
Figure 7. The web calculator for predicting bone metastases in breast infiltrating duct carcinoma patients.
Breast cancer was the most common malignant tumor in women, and IDC accounted for about 80% of all BC cases (3). BC patients who had BM were more likely to have poor prognosis and bad quality of life (25), and earlier attention to patients at a high risk of BM was important. However, metastatic BC was a highly heterogeneous disease, and bone was the most common site of distance metastasis from BC (3, 5, 26). A decline in quality of life was common among patients with BC who suffered from BM. And the risk of skeletal-related events significantly increased following BM in BC patients, such as pain, fracture and hypercalcemia (27, 28). The life expectancy of patients with BC benefitted from early detection and systemic therapy. Therefore, the prognosis of patients with late detection was often poor against a background of increasing incidence of BM.
Bone scintigraphy is often used to distinguish BC patients likely to develop BM. However, this method may not be suitable for early screening due to the expense and radiation damage associated with bone imaging. To help address these potential problems, we built a predictive model using ML technologies to predict BM in IDC patients and identify patients at a high risk of BM.
Presently, new techniques in ML and artificial intelligence (AI) help us succeed in translational research in many fields (29–31). Given the success of ML techniques in other fields, coupled with the large amount of data available in healthcare, ML techniques will have a promising future in the medical field (32–34). In particular, the emergence of electronic medical records (EMR) has generated a large accumulation of clinical data sets, including clinical diagnoses and laboratory data. ML in the medical field can lead to accurate diagnosis and personalized patient care (35). Since the outbreak of the COVID-19, scientists have used ML techniques to develop a variety of predictive and diagnostic models based on big clinical data from patients to help health care professionals work together to address the pandemic (36–38). Statistical and comprehensive reviews of ML in medical diagnosis by Bhavsar et al. (35, 39) suggested that ML techniques can help medical professionals reduce diagnostic errors, improve healthcare services, and cut treatment costs. And in the cancer metastases field, some clinical prediction models in predicting the risk of BM based on ML algorithms have been developed to assist clinicians in personalizing patient diagnosis (40, 41).
This study was novel in using ML algorithms to predict the risk of BM in IDC patients. To our knowledge, researchers in the medical field have only used traditional linear statistical models to predict the risk of BM in IDC patients, and few studies applied ML techniques to them (42). A ML model based on XGB algorithm was developed to accurately predict the risk of BM, outperforming other models developed in this study. The model established in this study had great discrimination and showed satisfactory specificity and sensitivity.
In this study, several models were constructed and validated to predict BM risk in patients with IDC using current ML methods, and logistic regression analysis demonstrated that age, race, grade, T, N stage, breast subtype, and marital status were independent risk factors for BM. After analyzing the performance of six ML algorithms, we found that the XGB method performed the best (AUC-0.888). The XGB algorithm added a regular term to the objective function to control the complexity of the model and avoid overfitting, while supporting column sampling to enhance the stability of the model (19). This may be part of the reason why it achieved the best performance in our study. To increase the practicality of this model implementation, we created an online web calculator for calculating individual BM probability in IDC patients.
Previous study indicated that age and race were investigated broadly as risk factors for BC metastasis (43). It was reported that white women were less likely to have BM from BC than black women (44). In our study, blacks also had a higher incidence of BM than those of Whites, American Indians and Asia-Pacific people. Chen et al. and Wang et al. found that elderly women were more likely to develop BM from BC (45, 46). In the present study, we found that advanced age was a risk factor for BM in patients with IDC.
Tumor size was positively correlated with the chance of BM. Yazdani et al. found that tumor size was a risk factor of BM from BC and a larger tumor size increased the likelihood of BM (47). In this study, patients with N3 stage were significantly more likely to develop BM than patients with other stages. And the average N stage in patients with BM was higher than that of the non-metastatic population. Yazdani et al. reported that more cancerous axillary lymph nodes increase the risk of BM (47). Colleoni et al. also found that there was the highest frequency of BM in patients who had four or more cancerous axillary lymph nodes (48). In addition, patients with higher-grade IDC had a higher risk of BM than those with grade I (Well differentiated) in this study. One reason for this may be that poorly differentiated tumor tissue was poorly demarcated from surrounding normal tissue, which meant it was more aggressive.
Additionally, it has been found that different breast subtypes showed different trends of BM. Previous study indicated that the incidence of BM was higher in patients with HR+/HER2- (Luminal A) and HR+/HER2+ (Luminal B) (49). In the current study, we found patients with HR+/HER2+ (Luminal B) were more likely to develop BM than those with other subtypes of BC.
Another risk factor for BM in our model was marital status. Unmarried patients had a higher risk of BM than those who were married. Zhao et al. and Gao et al. reported that marital status was a prognostic factor influencing the survival of metastatic BC patients (50, 51). Thus, we believed that the lifestyle habits and psychological factors of unmarried women may influence the distance metastasis of IDC patients.
As far as we know, this study was the first attempt to use ML algorithms to predict the risk of BM for IDC patients. Although some previous prediction models for BC based on the SEER database have been developed, it verified just from the SEER database and whether it can be used in different regions was not clear (26, 45). In addition, these studies only used nomogram as a visualization tool and did not provide a web predictor. External test of the model was important to validate the stability of the model for different regional populations. Therefore, in this study, we used data from the SEER database to build a prediction model based on the XGB algorithm and validated it with a cohort from China. Through our predictive model, we can predict the risk of BM in patients with IDC in the early stage, indicate the related risks before the progression to the late stage, and accept the corresponding treatment regimen as soon as possible, which can significantly improve the prognosis of IDC patients. Furthermore, we developed a web predictor based on the model to predict the risk of BM in IDC patients. Clinicians can easily enter information about the patient's relevant variables into the model on the web page, and the model will calculate the patient's risk of developing BM. Nowadays, precision medicine has preceded four concepts: predictive, personalized, preventive and participatory. Due to the ML model we built, we can predict the risk of BM to a particular patient. The rapidity and accuracy of the prediction output allowed clinicians to make personalized decisions for their patients and could be used as a basis for clinicians to explain their decisions to patients and involved them in their treatment choices. From the clinician's point of view, substantial advances in ML had potential implications in clinical practice, including diagnosis, risk stratification and prognosis, treatment planning, and advances in precision medical methods (52). Of course, they always had the final decision when it came to interpretation based on their domain expertise.
Overall, this study used ML algorithms to construct and validate a clinical prediction model for predicting the risk of BM in patients with IDC based on large samples. Among all these algorithms, XGB performed the best. And we built an easy-to-use web calculator based on the XGB model, which can help physicians to individualize the diagnosis and treatment of BM in IDC patients.
Although our model achieved good results in prediction, there were still some limitations in it. First, it was a retrospective study, which needed to be further verified by prospective study. Second, only one external validation set was used to validate the model, and further efforts were required to validate the performance of the model in a more diverse population. Third, the SEER database just recorded the initial diagnosis of a patient, which meant that further information was lack and we were unable to access this information for further analysis.
For future work, we will focus on prospective and diverse population validation of the models to verify the performance and stability. These models are then expected to be integrated into applications that assist clinicians in medical decision-making. This can be a step toward a semi-autonomous diagnostic system that can assist clinicians in making individualized diagnoses of BM for IDC patients.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://seer.cancer.gov/.
We received permission to access the research data file in the SEER program from the National Cancer Institute, US. Approval was waived by the Local Ethics Committee, as SEER data is publicly available and de-identified. This study was approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University, and cases from the First Affiliated Hospital of Nanchang University signed a written informed consent form.
W-CL and J-ML designed the study. W-CL, S-NW, W-LT, A-AL, and B-LS performed analysis and generated the figures and tables. W-CL, M-XL, and S-NW wrote the manuscript. Z-LL and J-ML critically reviewed the manuscript. All authors have read and approved the manuscript.
This work was supported by the Department of Science and Technology Program of Jiangxi Province, China (No. 20203BBG73045).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Mr. Wenxing Qian of Department of Computer Science, Beijing Jiaotong University for his assistance in algorithm and Ms. Yixin Lai of School of Pharmacy, Tongji Medical College, Huazhong University of Science and Technology for her assistance in literature search.
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics (2021. CA Cancer J Clin. (2021) 71:7–33. doi: 10.3322/caac.21654
2. Pfeiffer RM, Webb-Vargas Y, Wheeler W, Gail MH. Proportion of US trends in breast cancer incidence attributable to long-term changes in risk factor distributions cancer. Epidemiol Biomarkers Prev. (2018) 27:1214–22. doi: 10.1158/1055-9965.EPI-18-0098
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. (2018) 68:394–424. doi: 10.3322/caac.21492
4. Molland J, Donnellan M, Janu N, Carmalt H, Kennedy C, Gillett D. Infiltrating lobular carcinoma—a comparison of diagnosis, management and outcome with infiltrating duct carcinoma. Breast. (2004) 13:389–96. doi: 10.1016/j.breast.2004.03.004
5. DeSantis CE, Fedewa SA, Goding Sauer A, Kramer JL, Smith RA, Jemal A. Breast cancer statistics, 2015: Convergence of incidence rates between black and white women. CA Cancer J Clin. (2016) 66:31–42. doi: 10.3322/caac.21320
6. Zhang H, Zhu W, Biskup E, Yang W, Yang Z, Wang H, et al. Incidence, risk factors and prognostic characteristics of bone metastases and skeletal-related events (SREs) in breast cancer patients: a systematic review of the real world data. J Bone Oncol. (2018) 11:38–50. doi: 10.1016/j.jbo.2018.01.004
7. Chen WZ, Shen JF, Zhou Y, Chen XY, Liu JM, Liu ZL. Clinical characteristics and risk factors for developing bone metastases in patients with breast cancer. Sci Rep. (2017) 7:11325. doi: 10.1038/s41598-017-11700-4
8. Yamashiro H, Takada M, Nakatani E, Imai S, Yamauchi A, Tsuyuki S, et al. Prevalence and risk factors of bone metastasis and skeletal related events in patients with primary breast cancer in Japan. Int J Clin Oncol. (2014) 19:852–62. doi: 10.1007/s10147-013-0643-5
9. Kennecke H, Yerushalmi R, Woods R, Cheang MCU, Voduc D, Speers CH, et al. Metastatic behavior of breast cancer subtypes. J Clin Oncol. (2010) 28:3271–7. doi: 10.1200/JCO.2009.25.9820
10. Kuchuk I, Hutton B, Moretto P, Ng T, Addison CL, Clemons M. Incidence, consequences and treatment of bone metastases in breast cancer patients-experience from a single cancer centre. J Bone Oncol. (2013) 2:137–44. doi: 10.1016/j.jbo.2013.09.001
11. Lüftner D, Lorusso V, Duran I, Hechmati G, Garzon-Rodriguez C, Ashcroft J, et al. Health resource utilization associated with skeletal-related events in patients with advanced breast cancer: results from a prospective, multinational observational study. Springerplus. (2014) 3:328. doi: 10.1186/2193-1801-3-328
12. Alonso-Betanzos A, Bolón-Canedo V. Big-data analysis, cluster analysis, and machine-learning approaches. Adv Exp Med Biol. (2018) 1065:607–26. doi: 10.1007/978-3-319-77932-4_37
13. Baştanlar Y, Ozuysal M. Introduction to machine learning. Methods Mol Biol. (2014) 1107:105–28. doi: 10.1007/978-1-62703-748-8_7
14. Li W, Hong T, Liu W, Dong S, Wang H, Tang ZR, et al. Development of a machine learning-based predictive model for lung metastasis in patients with ewing sarcoma. Front Med (. (2022) 9:807382. doi: 10.3389/fmed.2022.807382
15. Li W, Liu W, Hussain Memon F, Wang B, Xu C, Dong S, et al. An external-validated prediction model to predict lung metastasis among osteosarcoma: a multicenter analysis based on machine learning. Comput Intell Neurosci. (2022) 2022:2220527. doi: 10.1155/2022/2220527
16. Li W, Yafeng L, Liu W, Tang Z, Dong S, Lei M, et al. Machine learning-based prediction of lymph node metastasis among osteosarcoma patients. Front Oncol. (2022) 12:1418. doi: 10.3389/fonc.2022.797103
17. Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics (. (2010) 26:1340–7. doi: 10.1093/bioinformatics/btq134
19. Chen T, Guestrin C. Xgboost: A scalable tree boosting system. In: Proceedings of the 22nd ACM Sigkdd International Conference on Knowledge Discovery and Data Mining. (2016). p. 785–94.
21. Tang J, Deng C, Huang G-B. Extreme learning machine for multilayer perceptron. IEEE Trans Neural Netw Learn Syst. (2015) 27:809–21. doi: 10.1109/TNNLS.2015.2424995
22. Sperandei S. Understanding logistic regression analysis. Biochem Med. (2014) 24:12–8. doi: 10.11613/BM.2014.003
23. Myles AJ, Feudale RN, Liu Y, Woody NA, Brown SD. An introduction to decision tree modeling. J Chem Soc. (2004) 18:275–85. doi: 10.1002/cem.873
24. Rish I. An empirical study of the naive Bayes classifier. In: IJCAI 2001 Workshop on Empirical Methods in Artificial Intelligence, Vol. 3. (2001). p. 41–6.
25. Costa L, Badia X, Chow E, Lipton A, Wardley A. Impact of skeletal complications on patients' quality of life, mobility, and functional independence. Support Care Cancer. (2008) 16:879–89. doi: 10.1007/s00520-008-0418-0
26. Wu Q, Li J, Zhu S, Wu J, Chen C, Liu Q, et al. Breast cancer subtypes predict the preferential site of distant metastases: a SEER based study. Oncotarget. (2017) 8:27990–6. doi: 10.18632/oncotarget.15856
27. Fornetti J, Welm AL, Stewart SA. Understanding the bone in cancer metastasis. J Bone Miner Res. (2018) 33:2099–113. doi: 10.1002/jbmr.3618
28. Metzger-Filho O, Sun Z, Viale G, Price KN, Crivellari D, Snyder RD, et al. Patterns of Recurrence and outcome according to breast cancer subtypes in lymph node-negative disease: results from international breast cancer study group trials VIII and IX. J Clin Oncol. (2013) 31:3083–90. doi: 10.1200/JCO.2012.46.1574
29. Khan MU, Lee SUJ, Abbas S, Abbas A, Bashir AK. Detecting wake lock leaks in android apps using machine learning. IEEE Access. (2021) 9:125753–67. doi: 10.1109/ACCESS.2021.3110244
30. Akbar A, Ibrar M, Jan MA, Bashir AK, Wang L. SDN-enabled adaptive and reliable communication in IoT-Fog environment using machine learning and multiobjective optimization. IEEE Intern Thin J. (2021) 8:3057–65. doi: 10.1109/JIOT.2020.3038768
31. Triantafyllidis AK, Tsanas A. Applications of machine learning in real-life digital health interventions. Rev Literat. (2019) 21:e12286. doi: 10.2196/12286
32. Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. (2018) 284:603–19. doi: 10.1111/joim.12822
33. Toh TS, Dondelinger F, Wang D. Looking beyond the hype: Applied AI and machine learning in translational medicine. EBioMedicine. (2019) 47:607–15. doi: 10.1016/j.ebiom.2019.08.027
34. Deo RC. Machine learning in medicine. Circulation. (2015) 132:1920–30. doi: 10.1161/CIRCULATIONAHA.115.001593
35. Bhavsar KA, Abugabah A, Singla J, AlZubi AA, Bashir AK. A comprehensive review on medical diagnosis using machine learning. Comput Mater Continua. (2021) 67:1997. doi: 10.32604/cmc.2021.014943
36. Mohan S, Abugabah A, Kumar Singh S, Kashif Bashir A, Sanzogni L. An approach to forecast impact of Covid-19 using supervised machine learning model Software. Pract Exp. (2022) 52:824–40. doi: 10.1002/spe.2969
37. Iwendi C, Bashir AK, Peshkar A, Sujatha R, Chatterjee JM, Pasupuleti S, et al. COVID-19 patient health prediction using boosted random forest algorithm. Front Public Health. (2020) 8:357. doi: 10.3389/fpubh.2020.00357
38. Ayoub A, Mahboob K, Javed A-R, Rizwan M, Gadekallu T-R, Abidi M-H, et al. Classification and categorization of COVID-19 outbreak in Pakistan. Comput Materi Continua. (2021) 69 (:1253–69. doi: 10.32604/cmc.2021.015655
39. Bhavsar KA, Singla J, Al-Otaibi YD, Song O-Y, Zikria YB, Bashir AK. Medical diagnosis using machine learning: a statistical review. Comput Mater Continua. (2021) 67:107–25. doi: 10.32604/cmc.2021.014604
40. Liu WC Li ZQ, Luo ZW, Liao WJ, Liu ZL, Liu JM. Machine learning for the prediction of bone metastasis in patients with newly diagnosed thyroid cancer. Cancer Med. (2021) 10:2802–11. doi: 10.1002/cam4.3776
41. Liu W-C, Li M-X, Qian W-X, Luo Z-W, Liao W-J, Liu Z-L, et al. Application of machine learning techniques to predict bone metastasis in patients with prostate cancer. Cancer Manag Res. (2021) 13:8723. doi: 10.2147/CMAR.S330591
42. Huang Z, Hu C, Liu K, Yuan L, Li Y, Zhao C, et al. Risk factors, prognostic factors, and nomograms for bone metastasis in patients with newly diagnosed infiltrating duct carcinoma of the breast: a population-based study. BMC Cancer. (2020) 20:1–17. doi: 10.1186/s12885-020-07635-1
43. Weigelt B, Peterse JL, van 't Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. (2005) 5:591–602. doi: 10.1038/nrc1670
44. DeSantis C, Jemal A, Ward E. Disparities in breast cancer prognostic factors by race, insurance status, and education. Cancer Causes Control. (2010) 21:1445–50. doi: 10.1007/s10552-010-9572-z
45. Chen MT, Sun HF, Zhao Y, Fu WY, Yang LP, Gao SP, et al. Comparison of patterns and prognosis among distant metastatic breast cancer patients by age groups: a SEER population-based analysis. Sci Rep. (2017) 7:9254. doi: 10.1038/s41598-017-10166-8
46. Wang R, Zhu Y, Liu X, Liao X, He J, Niu L. The Clinicopathological features and survival outcomes of patients with different metastatic sites in stage IV breast cancer. BMC Cancer. (2019) 19:1091. doi: 10.1186/s12885-019-6311-z
47. Yazdani A, Dorri S, Atashi A, Shirafkan H, Zabolinezhad H. Bone metastasis prognostic factors in breast cancer. Breast Cancer. (2019) 13:1178223419830978. doi: 10.1177/1178223419830978
48. Colleoni M, O'Neill A, Goldhirsch A, Gelber RD, Bonetti M, Thürlimann B, et al. Identifying breast cancer patients at high risk for bone metastases. J Clin Oncol. (2000) 18:3925–35. doi: 10.1200/JCO.2000.18.23.3925
49. Gong Y, Zhang J, Ji P, Ling H, Hu X, Shao ZM. Incidence proportions and prognosis of breast cancer patients with bone metastases at initial diagnosis. Cancer Med. (2018) 7:4156–69. doi: 10.1002/cam4.1668
50. Zhao W, Wu L, Zhao A, Zhang M, Tian Q, Shen Y, et al. A nomogram for predicting survival in patients with de novo metastatic breast cancer: a population-based study. BMC Cancer. (2020) 20:982. doi: 10.1186/s12885-020-07449-1
51. Gao T, Shao F. Risk factors and prognostic factors for inflammatory breast cancer with bone metastasis: a population-based study. J Orthop Surg. (2021) 29:23094990211000144. doi: 10.1177/23094990211000144
Keywords: breast cancer, infiltrating ductal carcinoma, bone metastases, machine learning, prediction
Citation: Liu W-C, Li M-X, Wu S-N, Tong W-L, Li A-A, Sun B-L, Liu Z-L and Liu J-M (2022) Using Machine Learning Methods to Predict Bone Metastases in Breast Infiltrating Ductal Carcinoma Patients. Front. Public Health 10:922510. doi: 10.3389/fpubh.2022.922510
Received: 18 April 2022; Accepted: 16 June 2022;
Published: 06 July 2022.
Edited by:
Ali Kashif Bashir, Manchester Metropolitan University, United KingdomReviewed by:
Chuan-Gui Song, Affiliated Union Hospital of Fujian Medical University, ChinaCopyright © 2022 Liu, Li, Wu, Tong, Li, Sun, Liu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia-Ming Liu, bGl1amlhbWluZ2RyQGhvdG1haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.