- 1Emergency Department, Zigong Fourth People's Hospital, Zigong, China
- 2Artificial Intelligence Key Laboratory of Sichuan Province, Zigong, China
- 3Institute of Medical Big Data, Zigong Academy of Artificial Intelligence and Big Data for Medical Science, Sichuan, China
- 4Department of Critical Care Medicine, Affiliated Jinhua Hospital, Zhejiang University School of Medicine, Jinhua, China
- 5Key Laboratory of Emergency and Trauma of Ministry of Education, Hainan Provincial Key Laboratory for Tropical Cardiovascular Diseases Research, The First Affiliated Hospital of Hainan Medical University, Research Unit of Island Emergency Medicine of Chinese Academy of Medical Sciences, Hainan Medical University, Haikou, China
- 6Department of Health Management Center, Zigong Fourth People's Hospital, Zigong, China
- 7Department of Gynecology, Fushun County Maternal and Child Health Hospital, Fushun, China
- 8Department of Obstetrics, Fushun County Maternal and Child Health Hospital, Fushun, China
- 9Department of Emergency Medicine, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, China
- 10Key Laboratory of Precision Medicine in Diagnosis and Monitoring Research of Zhejiang Province, Hangzhou, China
Patients treated in the intensive care unit (ICU) are closely monitored and receive intensive treatment. Such aggressive monitoring and treatment will generate high-granularity data from both electronic healthcare records and nursing charts. These data not only provide infrastructure for daily clinical practice but also can help to inform clinical studies. It is technically challenging to integrate and cleanse medical data from a variety of sources. Although there are several open-access critical care databases from western countries, there is a lack of this kind of database for Chinese adult patients. We established a critical care database involving patients with infection. A large proportion of these patients have sepsis and/or septic shock. High-granularity data comprising laboratory findings, baseline characteristics, medications, international statistical classification of diseases (ICD) code, nursing charts, and follow-up results were integrated to generate a comprehensive database. The database can be utilized for a variety of clinical studies. The dataset is fully accessible at PhysioNet(https://physionet.org/content/icu-infection-zigong-fourth/1.0/).
Background and Summary
Infection is common in the intensive care unit (1, 2). There are two categories of infections for patients in the intensive care unit (ICU) due to the place where the infection was acquired. One type of infection is the infection present on ICU admission, and most of such patients are transferred to ICU due to the development of sepsis and/or septic shock (3, 4). The other type of infection is the infection acquired after ICU admission, which is also termed the nosocomial infection (1). Critically ill patients are at increased risk of infection because of compromised immunity, use of intravascular catheters, and endotracheal intubation (5, 6). Irrespective of the places where the infection is acquired, the infection can cause systematic inflammatory response (SIRS), sepsis, and septic shock. These complications are associated with a significantly increased risk of mortality (7, 8). Although sepsis has been widely investigated in the literature (4, 9, 10), the raw data are typically not publicly available due to confidential or legal issues. The restricted data usage created a barrier to reproducing and verifying the results.
Although several open-access critical care databases from western countries have been created to promote data sharing and reuse for the scientific community (11–15), there is a lack of such database comprising Chinese adult patients. Since the Chinese population is the largest in the world, exploring infection/sepsis in the Chinese population is the key to achieving the goal proposed by the surviving sepsis campaign (16). Furthermore, a dataset, especially those generated from electronic healthcare records is large in volume. Secondary analysis of such dataset can generate novel insights into the diseases of interest (13, 17–19). Thus, creating a critical care database relating to patients with infection can help to promote collaborative research across the globe to reveal more insights into the infections in critically ill patients.
The rationales to include all critical patients with infection are 2 folds. First, such a database allows the capturing of longitudinal characteristics before and after infection in critically ill patients. This feature can be explored by restricting patients who acquired infection during ICU stay. A typical example is the subjects with intracranial hemorrhage who developed aspiration pneumonia in ICU. Risk factors for the development of infection can be analyzed. Second, for patients who had the infection before ICU admission, the severity spectrum ranging from infection, systematic inflammatory response syndrome, sepsis, severe sepsis, and septic shock can be captured. Third, the diagnosis of sepsis with international statistical classification of diseases (ICD) code is not accurate because there are many versions of sepsis definition. To include all infection patients allows the exploration of the agreement between these definitions. Clinical studies to develop the sepsis early warning system required the whole spectrum of diseases to be included in the database (20, 21). The critical care database comprises high granularity data including laboratory findings, baseline characteristics, medications, ICD-10 code, and nursing charts, and follow-up results were integrated to make a comprehensive database. The database can be utilized for a variety of clinical study purposes, such as epidemiology of risk factors, predictive analytics, natural language processing, and subphenotype identification.
Methods
Study Setting and Population
The study was conducted in Zigong Fourth People's Hospital, Sichuan, China from January 2019 to December 2020, and was approved by the Ethics Committee of Zigong Fourth People's Hospital (Approval Number: 2021-014). Informed consent was waived due to the retrospective design of the study. The study complies with the Declaration of Helsinki.
All patients who transferred to any type of ICU in the hospital from January 2019 to December 2020 were potentially eligible to be included in the database. Electronic healthcare records of consecutive ICU patients with the diagnosis of infection, irrespective of the place where the infection was acquired, were included in the database. Infection was defined according to the diagnosis descriptions that contained keywords such as “infection”, “pneumonia” and “-itis”. Because the original diagnosis description was recorded in simplified Chinese, in which the above keywords were extracted via “Ganran” and “Yan”. Some autoimmune or connective tissue diseases such as systemic lupus erythematosis (SLE), multiple sclerosis, rheumatoid arthritis, and Sjögren's syndrome were excluded manually.
Database Development
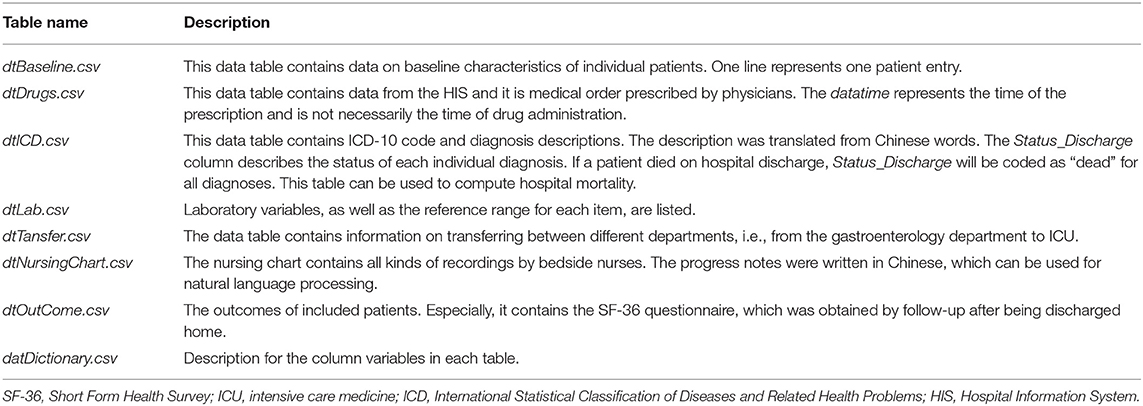
The critical care database was populated with data that have been acquired during routine clinical practice. Thus, the establishment of the database did not interfere with the clinical practice and was not associated with increased burden on healthcare providers, as well as risks for patients. Data were exported from several information systems including electronic healthcare records (EHR), hospital information system (HIS), laboratory information system (LIS), and critical care nursing chart system. The database was finally organized into seven tables in “.csv” format (Table 1). These data tables can be related to each other by patient ID (i.e., INP_NO or PATIENT_ID).
The core table dtBaseline contained baseline demographics of included patients, and it can be linked to other tables by either INP_NO or PATIENT_ID. PATIENT_ID was used to identify unique patients and INP_NO was used to identify unique hospital admission.
The dtOutCome table was generated by manual entry during follow-up. The Death_Date was recorded as hours from admission. The Short-Form Health Survey (SF-36) questionnaire was applied to evaluate the functional outcome of those who survived the critical illness. The Short Form Health Survey is a 36-item, patient-reported survey of patient health, which taps eight health concepts: bodily pain, physical functioning, role limitations due to physical health problems, role limitations due to personal or emotional problems, social functioning, energy/fatigue, emotional well-being, and general health perceptions. It also includes a single item that provides an indication of perceived change in health (22). The long-term mortality followed at 1 to 2 years after discharge was added if the patients' family members were willing to provide such information. In case a patient died after hospital discharge, the date was recorded.
Different from previous similar databases such as MIMIC-III which only contain laboratory values measured during ICU stay, we included all laboratory values during the index hospitalization including those measured outside ICU (11). We believe this can help to capture the full trajectory of pathophysiological changes before and after critical illness. For example, the identification of patients with acute kidney injury (AKI) is usually challenging if baseline serum creatinine (measured before the onset of the critical illness) is not available (23). Some stamp time points of laboratory measurements are earlier than the hospital admission time because these were measured in the emergency room or outpatient visit before hospital admission.
Deidentification
The data were deidentified before incorporating into the critical care database. The Health Insurance Portability and Accountability Act (HIPAA) protected health information identifiers including patient name, cell phone/telephone numbers, address, and any other variables that could uniquely identify the individual in structured data sources. The key variables PATIENT_ID and INP_NO were randomly assigned a unique number and the original patient ID and hospital ID were removed. Event time points were replaced with an offset value measured in hours from the hospital admission time (i.e., hospital admission time was the zero point). The original time points were removed from the dataset. Patients older than 89 years were assigned a random number from 90 to 120 for the age variable.
Data Records
The study generated a relational database consisting of seven tables (Table 2). The database integrated comprehensive information of 2,790 patients in ICU with infection from January 2019 to December 2020. Tables are linked by identifiers such as INP_NO or PATIENT_ID. INP_NO refers to unique hospital admission and PATIENT_ID refers to a unique subject.
High-granularity charted events such as progress notes, fluid intake, consciousness, vital signs, mechanical ventilator parameters, Richmond Agitation-Sedation Scale (RASS), and critical-care pain observation tool (CPOT) scores are recorded in the nursing chart table. Information from different sources might be inconsistent. For example, a drug may be prescribed by the physician as recorded in the dtDrugs table. However, the drug is actually not administered and thus will not be found in the dtNursingChart table. Our approach is to keep these tables independent for clarity because these tables reflect different sources of information and contain information for prognostic or predictive analytics. For example, the physician may prescribe analgesics for a patient on admission, but this patient actually does not experience pain or agitation and the analgesics are not actually administered. However, the presence of medical order reflects the physician's expectation and thus may contain prognostic information. The dataset is available at PhysioNet (https://physionet.org/content/icu-infection-zigong-fourth/1.0/).
Technical Validation
Data were retrospectively extracted from the information systems in the Zigong Fourth People's Hospital. Firstly, the required data were exported from an electronic healthcare database with the assistance of an information technology technician (Zhou). The exported data were then reviewed by three expert critical care physicians (PX, LC, and ZZ). Most variables recorded in Chinese such as diagnosis description, laboratory item, and department name were translated into English. However, the progress notes from the nursing chart remained in Chinese because such information can be used for natural language processing. Some embedding features might be lost or modified when they are translated into other languges (24). In the meantime, some impossible date entries (follow-up date earlier than the discharge date), impossible values from the nursing chart (i.e., respiratory rate = 2), and outliers (i.e., tidal volume = 30) were either removed or updated after a manual check. Data were finalized and fully anonymized on August 20, 2021.
Usage Notes
Data Access
The critical care database is provided as a collection of comma-separated value (CSV) files. Such files can be easily processed with popular languages scripts such as PostreSQL, MySQL, R (version 4.01, The R Foundation for Statistical Computing), and MonetDB. In particular, the relational database can be easily managed with the tidyverse pipeline. In tidyverse pipeline, all packages can be fit together seamlessly and users do not need to worry about compatibility issues between different functions from different sources, and tidyverse scripts are easier to write, read, and understand than base R code (25). Users are required to formally request access to the database.
Baseline Characteristics of Included Patients
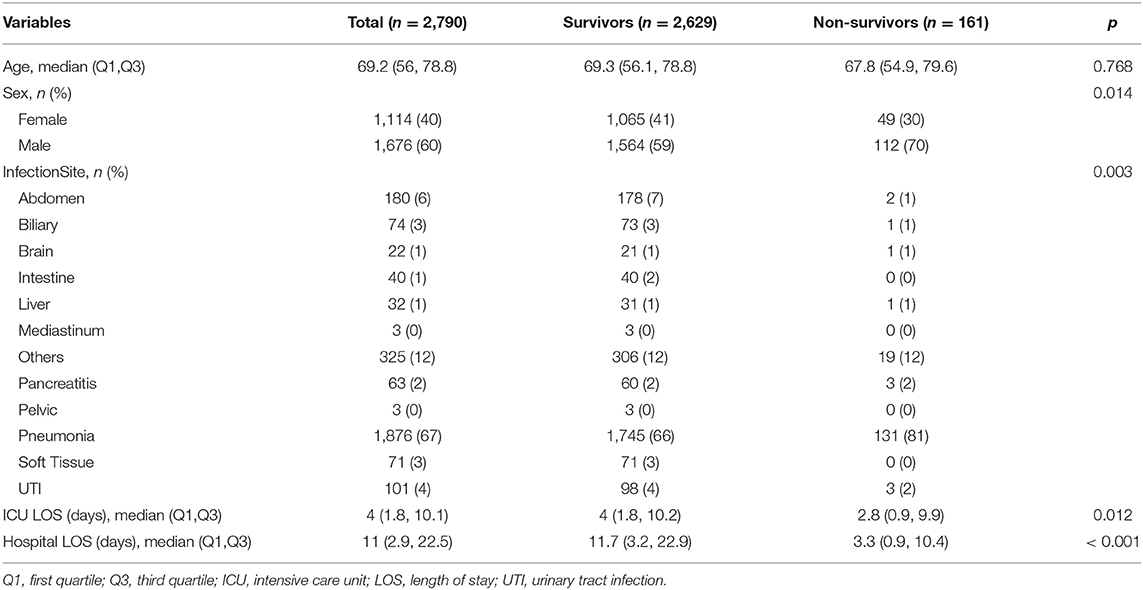
The overall mortality rate at hospital discharge was 5.8% (161/2,790). The proportion of men was higher in non-survivors than that in survivors (70 vs. 59%; p = 0.014). Patients with pneumonia were more likely to die than other sites of infection. However, non-survivors showed a shorter length of stay in both hospital and ICU, which was attributable to the fact that many severely ill patients chose to withdraw life-support interventions and died shortly after a few days of treatment.
Sample data for a single patient stay in the ICU are shown in Figure 1. The patient was transferred to ICU and experienced septic shock. Norepinephrine was used to maintain blood pressure. Organ failures including acute kidney injury, respiratory failure, and circulatory shock occurred sequentially during the disease course. Supportive treatments such as continuous renal replacement therapy (CRRT), mechanical ventilation (MV), and vasopressor were used. However, the clinical conditions deteriorate and suffered from sudden cardiac arrest (Figure 1).

Figure 1. Sample data for a single patient stay in the ICU. (A) Chart events extracted from free texts recorded in the nursing chart; (B) Structured numeric data extracted from the nursing chart. CRRT, continuous renal replacement therapy; MV, mechanical ventilation; CPR, cardiopulmonary resuscitation; SBP, systolic blood pressure; DBP, diastolic blood pressure.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://physionet.org/content/icu-infection-zigong-fourth/1.0/.
Ethics Statement
The studies involving human participants were reviewed and approved by Ethics Committee of Zigong Fourth People's Hospital. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author Contributions
ZZ, LC, and PX conceived the idea. YZ and SY curated data. RC and WH checked the accuracy of the data. FW performed a patient follow-up. All authors contributed to the article and approved the submitted version.
Funding
PX received funding from the RUIYI emergency medical research fund (202013), Open Foundation of Artificial Intelligence Key Laboratory of Sichuan Province (2020RYY03), and a Research project of the Health and Family Planning Commission of Sichuan Province (17PJ136). ZZ received funding from Yilu Gexin-Fluid Therapy Research Fund Project (YLGX-ZZ-2020005), Health Science and Technology Plan of Zhejiang Province (2021KY745), the Key Laboratory of Tropical Cardiovascular Diseases Research of Hainan Province (Grant No. KLTCDR-202001), and the Key Laboratory of Emergency and Trauma (Hainan Medical University), Ministry of Education (Grant No. KLET-202017). LC received funding from the Key Laboratory of Emergency and Trauma (Hainan Medical University), Ministry of Education (Grant No. KLET-202118).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dawit TC, Mengesha RE, Ebrahim MM, Tequare MH, Abraha HE. Nosocomial sepsis and drug susceptibility pattern among patients admitted to adult intensive care unit of Ayder Comprehensive Specialized Hospital, Northern Ethiopia. BMC Infect Dis. (2021) 21:824. doi: 10.1186/s12879-021-06527-4
2. Sang L, Xi Y, Lin Z, Pan Y, Song B, Li CA, et al. Secondary infection in severe and critical COVID-19 patients in China: a multicenter retrospective study. Ann Palliat Med. (2021) 10:8557–70. doi: 10.21037/apm-21-833
3. Abe T, Yamakawa K, Ogura H, Kushimoto S, Saitoh D, Fujishima S, et al. Epidemiology of sepsis and septic shock in intensive care units between sepsis-2 and sepsis-3 populations: sepsis prognostication in intensive care unit and emergency room (SPICE-ICU). J Intensive Care. (2020) 8:44. doi: 10.1186/s40560-020-00465-0
4. Zhang Z, Bokhari F, Guo Y, Goyal H. Prolonged length of stay in the emergency department and increased risk of hospital mortality in patients with sepsis requiring ICU admission. Emerg Med J. (2019) 36:82–7. doi: 10.1136/emermed-2018-208032
5. Nasa P, Juneja D, Singh O, Dang R, Singh A. An observational study on bloodstream extended-spectrum beta-lactamase infection in critical care unit: incidence, risk factors and its impact on outcome. Eur J Intern Med. (2012) 23:192–5. doi: 10.1016/j.ejim.2011.06.016
6. Patterson L, McMullan R, Harrison DA. Individual risk factors and critical care unit effects on Invasive Candida Infection occurring in critical care units in the UK: a multilevel model. Mycoses. (2019) 62:790–5. doi: 10.1111/myc.12956
7. Markwart R, Saito H, Harder T, Tomczyk S, Cassini A, Fleischmann-Struzek C, et al. Epidemiology and burden of sepsis acquired in hospitals and intensive care units: a systematic review and meta-analysis. Intensive Care Med. (2020) 46:1536–51. doi: 10.1007/s00134-020-06106-2
8. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (sepsis-3). JAMA. (2016) 315:801–10. doi: 10.1001/jama.2016.0287
9. Sakr Y, Jaschinski U, Wittebole X, Szakmany T, Lipman J, Ñamendys-Silva SA, et al. Sepsis in intensive care unit patients: worldwide data from the intensive care over nations audit. Open Forum Infect Dis. (2018) 5:ofy313. doi: 10.1093/ofid/ofy313
10. Walkey AJ, Lagu T, Lindenauer PK. Trends in sepsis and infection sources in the United States. A population-based study. Ann Am Thorac Soc. (2015) 12:216–20. doi: 10.1513/AnnalsATS.201411-498BC
11. Johnson AEW, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. (2016) 3:160035. doi: 10.1038/sdata.2016.35
12. Pollard TJ, Johnson AEW, Raffa JD, Celi LA, Mark RG, Badawi O, et al. The eICU Collaborative Research Database, a freely available multi-center database for critical care research. Sci Data. (2018) 5:180178. doi: 10.1038/sdata.2018.178
13. Schenck EJ, Hoffman KL, Cusick M, Kabariti J, Sholle ET, Campion TR Jr. Critical carE database for advanced research (CEDAR): an automated method to support intensive care units with electronic health record data. J Biomed Inform. (2021) 118:103789. doi: 10.1016/j.jbi.2021.103789
14. Thoral PJ, Kompanje EJO, Kaplan L, Peppink JM, Driessen RH, Sijbrands EJG, et al. Sharing ICU patient data responsibly under the society of critical care medicine/European society of intensive care medicine joint data science collaboration: the Amsterdam University Medical Centers Database (AmsterdamUMCdb) Example. Crit Care Med. (2021) 49:e563–77. doi: 10.1097/CCM.0000000000004916
15. Fleuren LM, Dam TA, Tonutti M, de Bruin DP, Lalisang RCA, Gommers D, et al. The Dutch Data Warehouse, a multicenter and full-admission electronic health records database for critically ill COVID-19 patients. Critical Care. (2021) 25:304. doi: 10.1186/s13054-021-03733-z
16. Nunnally ME, Ferrer R, Martin GS, Martin-Loeches I, Machado FR, De Backer D, et al. The Surviving Sepsis Campaign: research priorities for the administration, epidemiology, scoring and identification of sepsis. Intensive Care Med Exp. (2021) 9:34. doi: 10.1186/s40635-021-00400-z
17. Zhang Z, Mo L, Ho KM, Hong Y. Association between the use of sodium bicarbonate and mortality in acute kidney injury using marginal structural cox model. Crit Care Med. (2019) 47:1402–8. doi: 10.1097/CCM.0000000000003927
18. Zhang Z, Zhu C, Mo L, Hong Y. Effectiveness of sodium bicarbonate infusion on mortality in septic patients with metabolic acidosis. Intensive Care Med. (2018) 44:1888–95. doi: 10.1007/s00134-018-5379-2
19. Zhang Z, Chen L, Xu P, Hong Y. Predictive analytics with ensemble modeling in laparoscopic surgery: a technical note. Laparoscopic, Endoscopic and Robotic Surgery. (2022). doi: 10.1016/j.lers.2021.12.003
20. Sabir L, Ramlakhan S, Goodacre S. Comparison of qSOFA and hospital early warning scores for prognosis in suspected sepsis in emergency department patients: a systematic review. Emerg Med J. (2021). doi: 10.1136/emermed-2020-210416
21. Tarabichi Y, Cheng A, Bar-Shain D, McCrate BM, Reese LH, Emerman C, et al. Improving timeliness of antibiotic administration using a provider and pharmacist facing sepsis early warning system in the emergency department setting: a randomized controlled quality improvement initiative. Crit Care Med. (2021) 50:418–27. doi: 10.1097/CCM.0000000000005267
22. Brazier J, Roberts J, Deverill M. The estimation of a preference-based measure of health from the SF-36. J Health Econ. (2002) 21:271–92. doi: 10.1016/S0167-6296(01)00130-8
23. Kashani KB. Automated acute kidney injury alerts. Kidney Int. (2018) 94:484–90. doi: 10.1016/j.kint.2018.02.014
24. Newman-Griffis D, Lehman JF, Rosé C, Hochheiser H. Translational NLP: a new paradigm and general principles for natural language processing research. Proc Conf. (2021) 2021:4125–38. doi: 10.18653/v1/2021.naacl-main.325
Keywords: critical care, database, open access, infections, big data and analytics
Citation: Xu P, Chen L, Zhu Y, Yu S, Chen R, Huang W, Wu F and Zhang Z (2022) Critical Care Database Comprising Patients With Infection. Front. Public Health 10:852410. doi: 10.3389/fpubh.2022.852410
Received: 11 January 2022; Accepted: 14 February 2022;
Published: 17 March 2022.
Edited by:
Yong Ming Yao, First Affiliated Hospital of Chinese PLA General Hospital, ChinaReviewed by:
Zhi Mao, People's Liberation Army General Hospital, ChinaQilin Yang, The Second Affiliated Hospital of Guangzhou Medical University, China
Copyright © 2022 Xu, Chen, Zhu, Yu, Chen, Huang, Wu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongheng Zhang, emhfemhhbmcxOTg0QHpqdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Ping Xu
Ping Xu Lin Chen
Lin Chen Yuanfang Zhu6†
Yuanfang Zhu6† Zhongheng Zhang
Zhongheng Zhang