- 1Department of Information Technology, Faculty of Computing and Information Technology, King Abdulaziz University, Jeddah, Saudi Arabia
- 2Centre of Artificial Intelligence for Precision Medicines, King Abdulaziz University, Jeddah, Saudi Arabia
- 3Mathematics Department, Faculty of Science, Al-Azhar University, Naser City, Egypt
- 4Department of Pharmacy Practice, Faculty of Pharmacy, King Abdulaziz University, Jeddah, Saudi Arabia
- 5Department of Computer Science, Faculty of Computers and Information Canal Suez University, Ismalila, Egypt
- 6Department of Pharmaceutics, Faculty of Pharmacy, King Abdulaziz University, Jeddah, Saudi Arabia
- 7Center of Excellence for Drug Research and Pharmaceutical Industries, King Abdulaziz University, Jeddah, Saudi Arabia
- 8Department of Diagnostic Radiology, Faculty of Applied Medical Sciences, King Abdulaziz University, Jeddah, Saudi Arabia
- 9Imaging Unit, King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia
- 10Department of Mathematics, Faculty of Science, Taif University, Taif, Saudi Arabia
- 11Department of Mathematics, Faculty of Science, Sohag University, Sohag, Egypt
Diagnosis is a crucial precautionary step in research studies of the coronavirus disease, which shows indications similar to those of various pneumonia types. The COVID-19 pandemic has caused a significant outbreak in more than 150 nations and has significantly affected the wellness and lives of many individuals globally. Particularly, discovering the patients infected with COVID-19 early and providing them with treatment is an important way of fighting the pandemic. Radiography and radiology could be the fastest techniques for recognizing infected individuals. Artificial intelligence strategies have the potential to overcome this difficulty. Particularly, transfer learning MobileNetV2 is a convolutional neural network architecture that can perform well on mobile devices. In this study, we used MobileNetV2 with transfer learning and augmentation data techniques as a classifier to recognize the coronavirus disease. Two datasets were used: the first consisted of 309 chest X-ray images (102 with COVID-19 and 207 were normal), and the second consisted of 516 chest X-ray images (102 with COVID-19 and 414 were normal). We assessed the model based on its sensitivity rate, specificity rate, confusion matrix, and F1-measure. Additionally, we present a receiver operating characteristic curve. The numerical simulation reveals that the model accuracy is 95.8% and 100% at dropouts of 0.3 and 0.4, respectively. The model was implemented using Keras and Python programming.
Introduction
The coronavirus disease (COVID-19) has endangered social lives and created disturbing financial costs. Many studies have attempted to find a way to manage its spread and the resulting death. Moreover, many research propositions have been made to evaluate the existence and seriousness of pneumonia triggered by COVID-19 (1–4). Compared to CT, although radiography is easily accessible in hospitals worldwide, X-ray images are considered less delicate for examining patients with COVID-19. Primary analysis is crucial for instant seclusion of the infected individuals. Moreover, it decreases the rate of infection in a healthy population because of the accessibility of sufficient therapy or vaccination for the virus.
Chest radiography is an essential method for identifying pneumonia. This is easy to accomplish with rapid medical identification. Upper body CT has a high level of susceptibility for the identification of COVID-19 (5). The X-ray image reveals aesthetic keys associated with the virus (6, 7). They considered the tremendous rate of suspected individuals and a minimal variety of trained radiologists. These automated identification methods with refined abnormalities help diagnose diseases and raise early medical identification quality with high accuracy.
A therapeutic image in the form of a chest X-ray is crucial for the automatic clinical identification of COVID-19. A clinical approach for identifying COVID-19 based on artificial intelligence (AI) can be as precise as a human, conserve the radiotherapist period, and perform clinical identification less expensively and faster than standard techniques. AI explanations can be influential approaches for dealing with such problems. A machine learning structure was used to predict COVID-19 from a chest X-ray image. In contrast to classic medical image category approaches, which stick to a two-phase treatment (handmade function abstraction plus identification), we used an end-to-end deep determining structure that directly discovers COVID-19 from raw pictures without any pre-processing (8–14). Deep learning (DL)-based variations, specifically convolutional neural networks (CNNs), have been shown to perform better than timeless AI approaches. Recently, various processing system ideas and clinical image evaluation tasks have been utilized in several issues, including categorization, dissection, and face identification (15–19).
Convolutional neural network (CNN) is considered among the finest therapeutic imaging applications (20), particularly for categorization. CNN is better suited for large databases and requires computational assets (storage and memory). In most cases (as in this research), the database is inadequate; therefore, it is insufficient for building and training a CNN. Thus, obtaining the benefit of the CNN's power and transfer learning can reduce the computational cost (21, 22). A proficient CNN on a large and varied image database can perform specific classification tasks (23). Hence, many pre-trained designs such as VGG-Net, (24), Resnet (25), NAS-Net (26), Mobile-Net (27), Inceptions (28), and Xception (29) have won many of the world image classification competitions.
Recently, Bhattachary et al. in (30, 31) presented a survey of most of the DL models in the last five years, used to identify COVID-19 with different data sets. Kermany et al. (32) used the DL framework with transfer learning, Rajaraman et al. (33) used a customized CNN model- VGG16, Wang, et al. (34) proposed AI and DL based Frameworks, Shan et al. (35) presented Human-In-the-Loop Strategy and DL Based Segmentation Network VBNet, Ghoshal and Tucker (36) used Drop weights-based Bayesian Convolutional Network, Apostolopoulos and Mpesiana (37) presented Transfer Learning based on CNN, Esmail et al. (38) used CNN Architecture. Khan et al. (39) presented COVIDX-Net comprising Deep CNN Models. Sahiner et al. (40) proposed Deep CNN Models – ResNet50, InceptionV3, and Inception- ResNetV2.
How can the researchers in poor and developing countries quickly and appropriately contribute to stopping the spread of the virus and thus contributing to the economy's growth? Using AI and machine learning models with transfer learning can contribute to answering this question. This article, which we contribute to, is considered to prevent the spread of the virus.
Because of the absence of a public image database of patients with COVID-19, numerous research studies reporting on the choices for the adaptive discovery of COVID-19 from X-ray images were not easy. This is because they are difficult to train, and many images are required to discover these networks without overfitting. In this work, data augmentation of the training/testing data set was applied.
In this study, we present a COVID-19 X-ray classification technique based on transfer learning with Mobile NetV2, present in the appendix, which is a pre-trained design (without segmentation). This technique addresses the COVID-19 image deficiency issue. As an alternative to training the CNN from the beginning, our method fine tunes the last layer of the design pre-trained variation on Image-NET (presented in Section The Proposed Architecture net and Strategies for Using the Transfer Learning). The above methods assisted in training the versions with conveniently not easily offered images and they attained excellent efficiency.
The MobileNet model proved to be highly efficient in fruit classification with 102 varieties (41, 42) and the classification of systemic sclerosis skin (43). After reviewing the latest modern studies, the latest (30, 31) studies, and to the best of my knowledge, the MobileNet was not used to identify the coronavirus.
The main contribution of this paper is to investigate and evaluate the performance of MobileNetv2 as a lightweight convolutional neural with transfer-learning and augmentation data for the early detection of coronavirus, to propose and adopt a new model structure for MobileNetv2 for binary classification, and to investigate the COVID-Computer-aided diagnostic (CCAD) tool based on the MobilenetV2 developed.
This paper is organized as follows: The methods and materials are described as the general projected framework in Section Materials and Methods. The results and discussions in Section Experimental Results and Discussion describe the database, experimental research studies, and measurement tools. Finally, the conclusions and future work are presented in Section Conclusion and Future Work.
Materials and Methods
We utilized a CNN to extricate highlights from COVID-19 X-ray images. We took on an extraordinary CNN called a pre-trained show wherever the arrangement is already prepared on the Image-NET database, and it comprised numerous images (creature, plants, …on 1,000 courses). Transfer learning was utilized by exchanging weights obtained and booked right into a pre-trained adaptation system Mobile NetV2.
Transfer Learning
We had few images in the present task to conduct the training, particularly in the COVID-19 category. Thus, transfer learning is essential. Transfer learning enhances learning in a new job by moving expertise from previous works that have already been reviewed. That is, we used techniques that have been pre-trained on image classification tasks rather than learning a new design. The advantages of transfer learning are as follows:
1. Higher begin: The first ability (before fine adjustment of the creation) on the resource design is more significant.
2. Higher slope: The rate of ability enhancement throughout the training of the source design is steeper.
3. Higher asymptote: The converged skill of the skilled version is much better. When transfer learning is applied to the design, higher precision degrees are quicker.
Description of Deep CNN
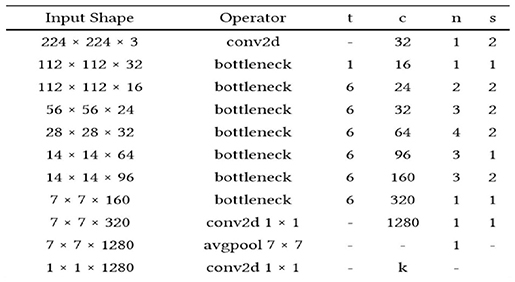
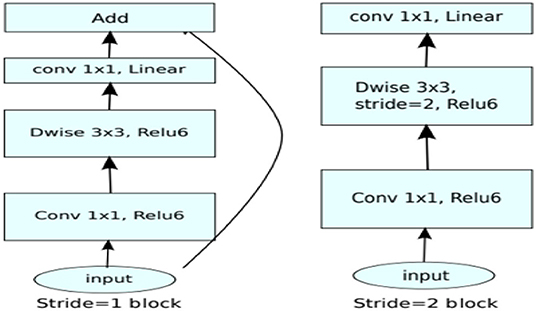
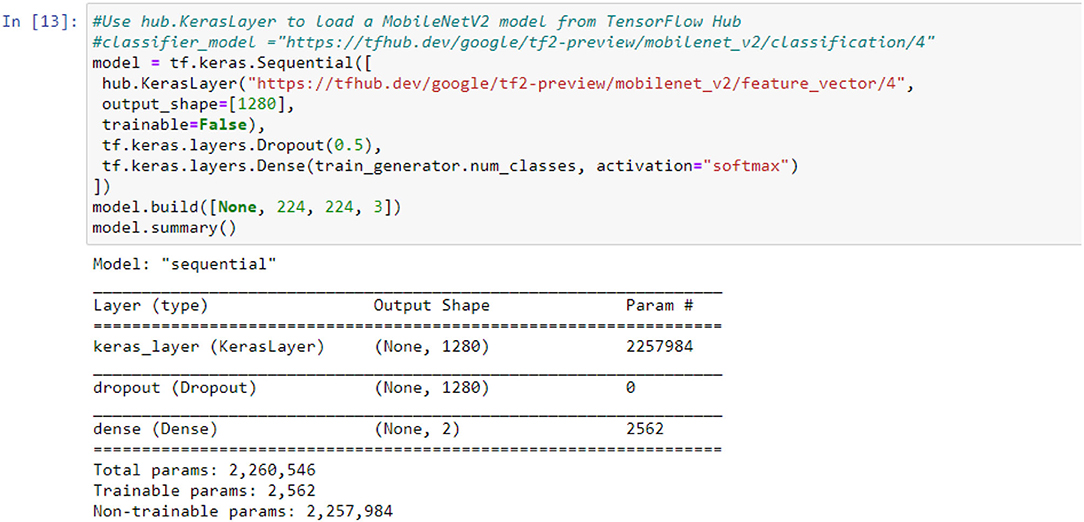
The application system is illustrated in Tables 1, 2, changing the highlights from the M to N channels, with walk s and advancement perspective t. This blockage incorporates a 1 × 1 convolution layer and sometimes the depth-wise convolutional layer. In addition, in requiring the advantage of serial actuation rather than the non-linear actuation included after the pointwise convolutional layer, it fulfills its objective down-sampling by setting basis s within the depth-wise convolutional layer. The entire organized Mobile-NetV2 system (Figure 1) is summarized in Table 2. The two-dimensional (2D) convolution (conv2d) layer is the essential complication, avg-pool is the average pooling, c is the number of the result channels, and n is the copied numbers. CNNs have 19 layers; the center layers are utilized for extricating capacities and the final layers are utilized for categorization. Based on the exchange learning guideline, we utilized Mobile-NetV2 pre-trained by Image-NET as a settled separator.
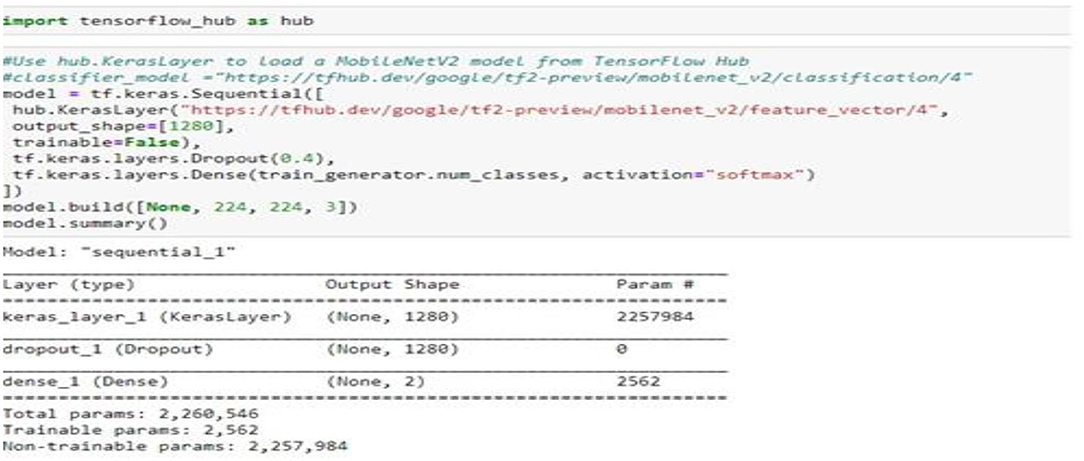
The mobile-NetV2 network is a base model without top classification layers, a deal for attribute abstraction. A conv2d layer with a dropout and soft-max categorizer layer is added to the basic design in Figure 2.
The design of the proposed model is summarized in Figure 2; we notice that the total number of coefficients is 2,260,546 for the new model. Only 2,562 coefficients are trainable.
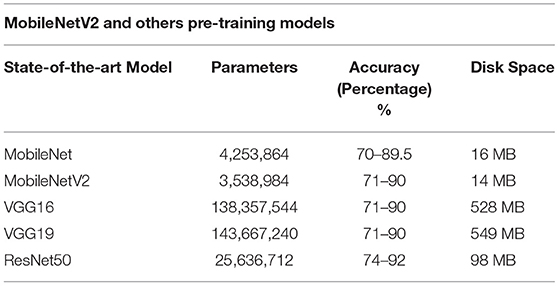
Table 3 shows the differences between the MobileNetV2 model and the other models regarding requirements and efficiency.
Experimental Results and Discussion
Database Description
The first dataset includes 309 chest X-ray images (102 were infected with COVID-19 and 207 were normal) see Figure 3. The second dataset includes 516 chest X-rays images (102 were infected with COVID-19 and 414 were normal). All Data sets are organized in two folders, one for training and the second for testing /validation. In each folder of the two (training and testing), two open folders equal to the number of classes, then in each class, the images examples of this class are put. The dataset was obtained from two different open-source databases:
1. GitHub database [https://github (44)].
2. https://kaggle.com/c/rsna-pneumonia-detection-challenge.
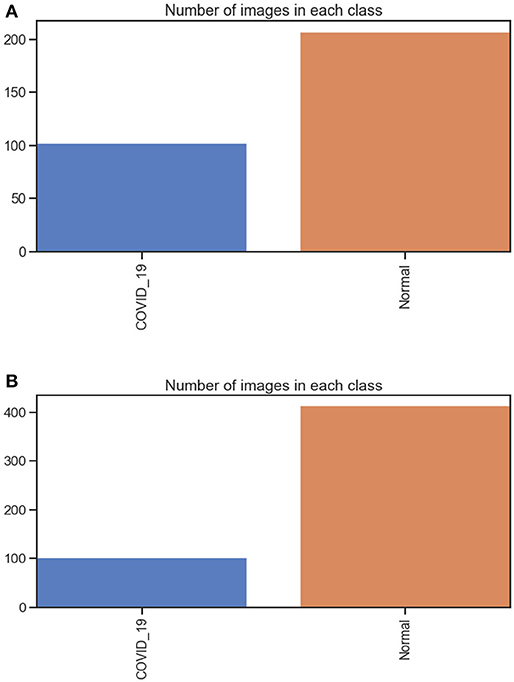
Figure 3. (A) number of images in each class dataset A. (B) number of images in each class dataset B.
They include 8,851 normal samples and 6,012 samples infected with pneumonia, and the chest X-ray image dimension is adjusted to 224 × 224 to satisfy the input conditions of the Mobile-NetV2 model.
Open-source CNN versions are readily available for training from scratch and transfer learning (customizing parts of an extant design for a new job). These versions are frequently tested on the ImageNet dataset of more than 1 million classifieds. The following section presents the proposed structure based on the MobileNetV2.
The Proposed Architecture Net and Strategies for Using the Transfer Learning
1. Building the generator of the training data.
The batches of tensor image data were generated using the class ImageDataGenerator(); also, this class is used as real-time data augmentation with different parameters. With the flow_from_directory() method, which receives two arguments, the directory(from which the images are loaded for creating the batches) and target size (we Adapt the: Target size of the loaded image, which is (256, 256) by default. To (128, 128). In addition, the method receives 3 other arguments (color_mode: RGB, class_mode:' “categorical”, batch_size:32).
2. Building the validation data generator (is like building the generator of the training data).
3. Loading the MobileNetv2.
Load the MobileNetv2 from the TensorFlow.Keras.Applications and store in the COVID19Model. Then, remove the last Final conect (FC) layers from the original model. The number of classes is 1,000, and the number of parameters is 4,231,976 trainable parameters in the network. To make this model suitable for working with our data set (Covid19) in two classes: change the size image from (224, 224, 3) to (128, 128, 3). we set in the MobileNet() function argument input_shape = (128, 128), and include_top = False (meaning load the model without FC layers) then make the model return just a vector of length equal to the number of classes. Also, the trainable argument of the loaded model is set to False.
4. Adding new FC Layers to the modified model.
We do not need to build a new architecture but use the architecture in the modified model stored in the COVID 19 model. Using the Sequential class in Keras, two layers were added at the top of the modified Mobile Net architecture, which are the: a) Average pooling layer b) FC layer with two neurons using the Dense class. (Note that an FC layer is also called a dense layer, and therefore the class used for building the FC layer is called Dense.) Adapting two layers is better than adapting one layer. Adapting three layers is better than just adapting 1 or 2 layers.
5. Compiling the new model.
Use the compile () method with the arguments optimizer, loss and metrics as: optimizer=tf.keras.optimizers.RMSprop (lr=0.0001), loss =“binary_crossentropy” and metrics=[“accuracy”]).
6. Finally, MobileNet transfer learning over the COVID-19 dataset.
The fit–_generator() method is used. With the argument:
generator: Train generator. steps_per_epoch: set to ceil (number of training samples/batch size). epochs: Number of training epochs. validation_data: Validation generator. validation_steps: calculated as ceil (number of validation samples/batch size.
7. Finally, save the model with extension.h5, which is a format for saving structured data.
Data Augmentation
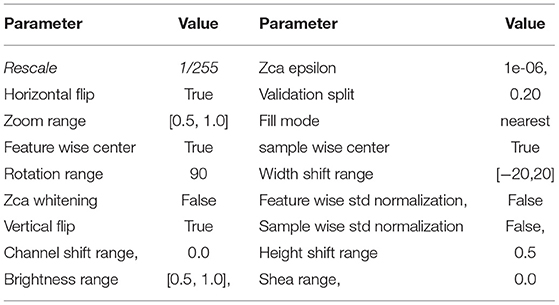
In machine learning (ML) and deep learning (DL) applications, the availability of large-scale, high-quality datasets plays a major role in the accuracy of the results. Keras ImageDataGenerator class provides a fast and simple method to increase your images. It offers various augmentation strategies like standardization, rotation, shifts, turns, brightness modification, etc. Data augmentation of the training/testing data set was applied to each image. For this, we use used the Keras ImageDataGenerator () class with the flowing set of constructor parameters: Table 4.
Experimental Environment
To survey the execution of Mobile-NetV2 with exchange learning, TensorFlow 2 (Tensorflow free) was introduced on Windows-10, a 64-bit working framework. Mobile-NetV2 was run using Python (Python free), and it was pre-trained using the Image-NET database. The exploratory framework is as follows: Intel R Center i7-8250U CPU @1.70 GHz, 1900 MHz 7 Core(s). 9 consistent Pr. and the memory was 16 GB. We used the Keras guide (45) and TensorFlow backend (46) in the implementation model.
Evaluation Metrics
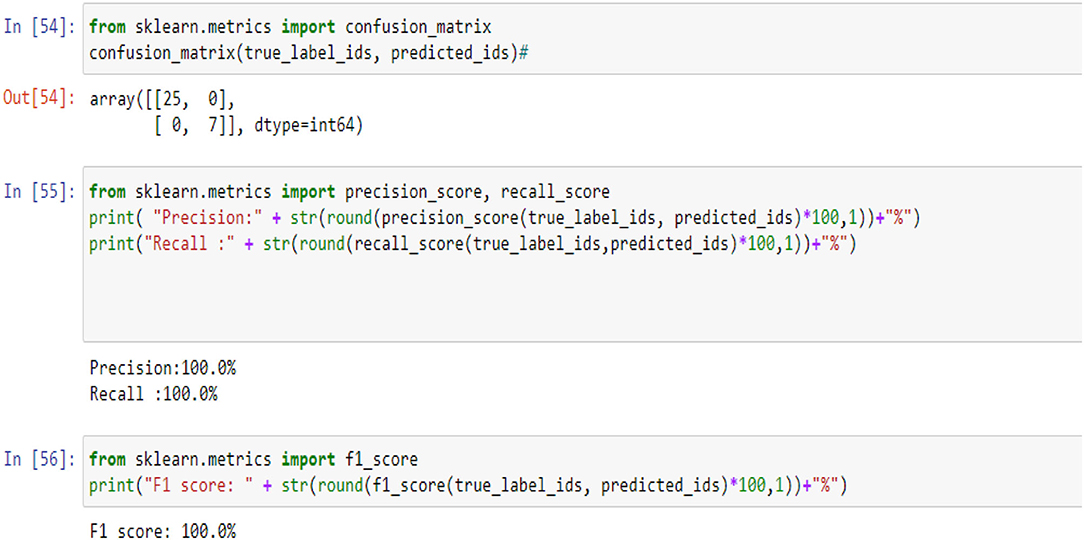
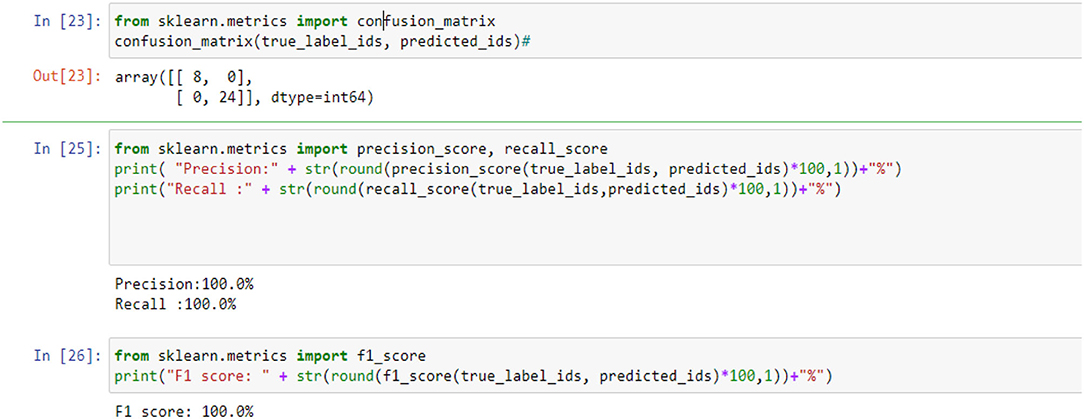
In the classification Models, most researchers used only accuracy as a performance metric. But the accuracy in some cases is not enough because of the imbalance problem in the data. To measure the model's performance and avoid the problem of data imbalance in the classes, we used more than one performance metric, such as the confusion matrix, F1, Precision, Recall, and Area Under Curve (AUC), which is one of the highest used metrics for evaluation. It is used for binary classification problems. The performance of the classification models can be assessed using various metrics, such as classification precision, sensitivity, specificity, F1-score, and accuracy. Sensitivity and specificity can be calculated using the following equations (45–47): Precision: measure the true patterns correctly predicted from the total predicted patterns in a true class. Accuracy: measure the ratio of correct predictions over the total number of examples evaluated, Recall: used to measure the fraction of positive examples that are correctly classified, and F1: a measure that provides the absolute average between accuracy and recall.
where true positive (TP) is the correctly categorized number and false positive (FP) is the incorrectly recognized number. False-negative (FN) is the image of a category that is found as an additional class and true negative (TN) is an image that does not belong to any category and is not categorized as any class.
Evaluation Results
We prepared the experiment with our database utilizing the Adam optimizer and a batch measure of 30. We performed our exploration by selecting a dropout rate (0.4, 0.5). Additionally, we utilized a learning rate rise to 1e-3 for the Adam optimizer and prepared for ten epochs within the pre-training organization; in any case, we select a much lower preparation rate e−5 for 31 epochs during the fine-adjustment phase.
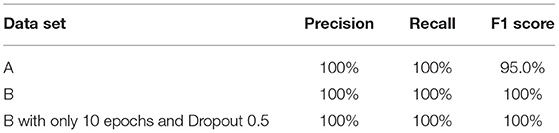
Case 1 (Dataset A)
Database B
Figures 4A,B, 8A,B, 13A,B, and Table 5 represents the accuracy and loss (training and validation) for data set A, Data set B and Dataset B with only 10 epochs and a dropout of 0.5, respectively.
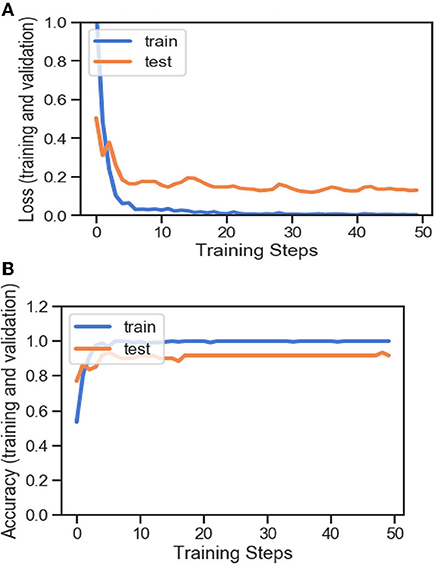
Figure 4. (A) Accuracy and loss (training and validation). (B) Accuracy and loss (training and validation).
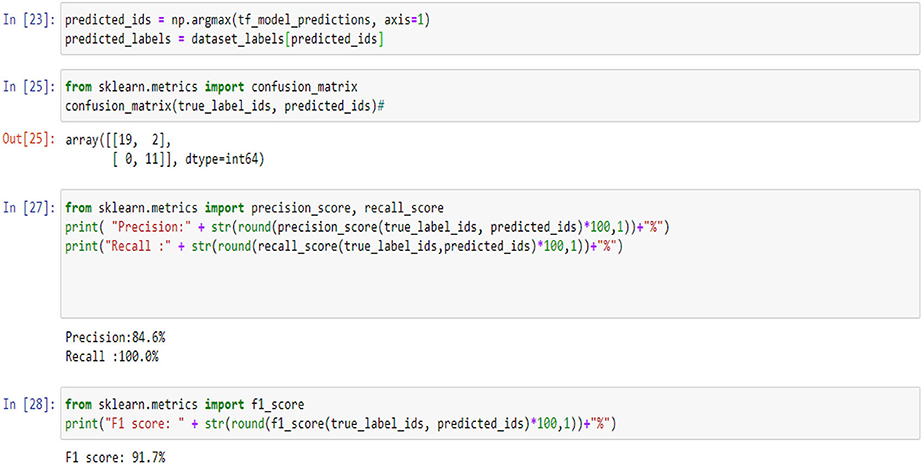
Figures 5, 9, 14 and Tables 6, 7 show the Confusion matrix, precision, recall, and F1 measurements for data set A, Data set B, and Dataset B with only 10 epochs and a dropout of 0.5, respectively.
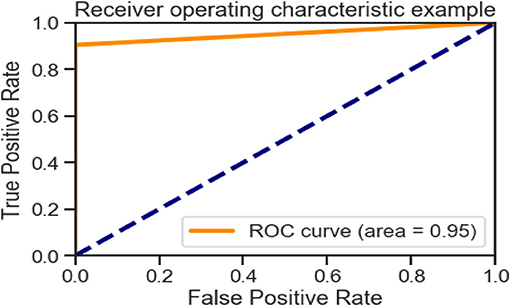
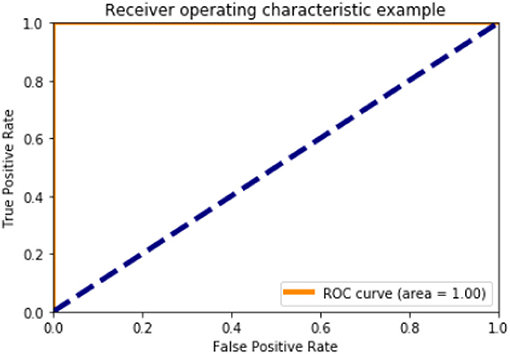
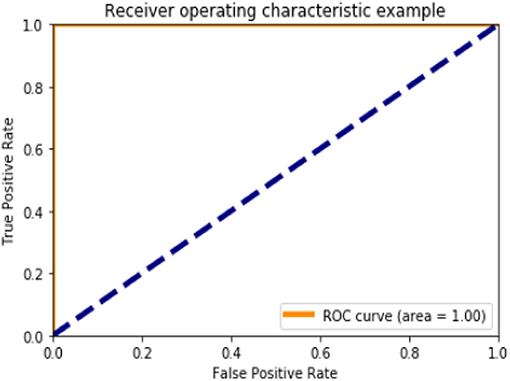
Figures 6, 10, 15 and Table 8 represents the receiver operating characteristic (ROC) Curve for data set A, Data set B, and Dataset B with only 10 epochs and a dropout of 0.5, respectively (see Figure 12).
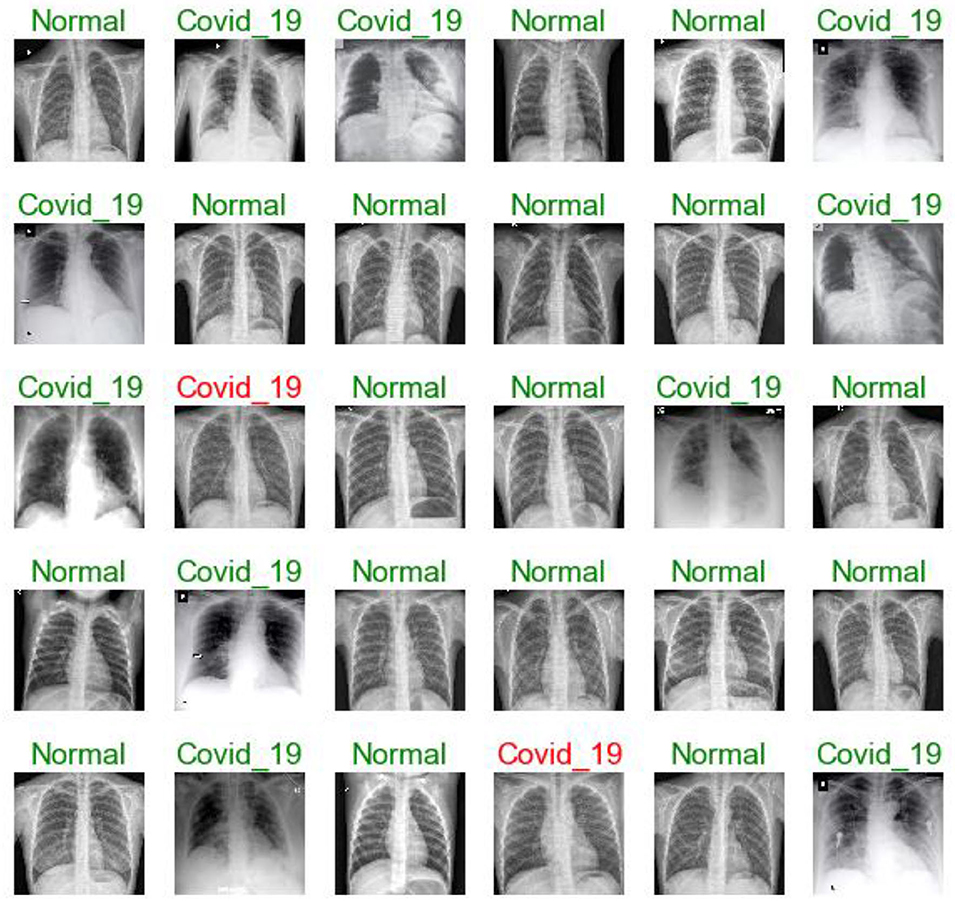
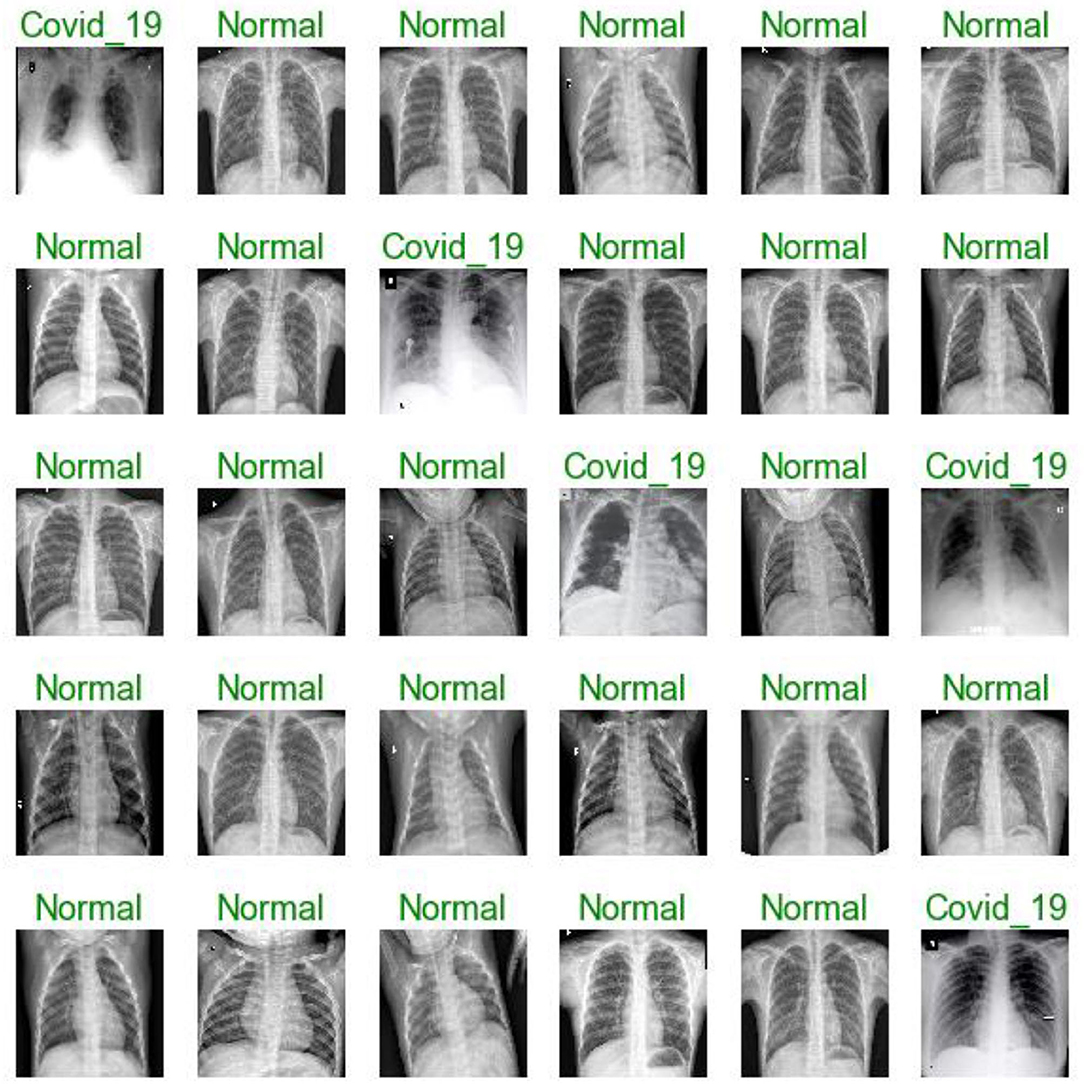
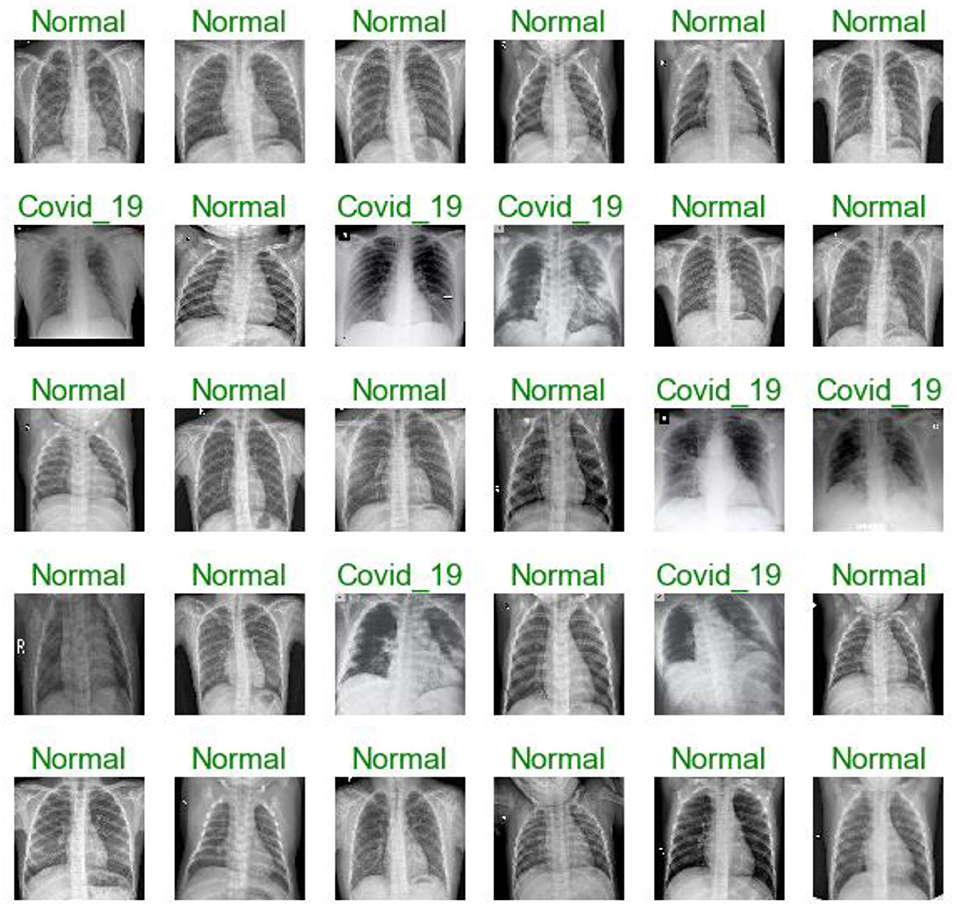
Figures 7, 11, 16 are present the model predictions results (green: correct, and red: incorrect) for data set A, Data set B, and Dataset B with only 10 epochs and a dropout of 0.5, respectively.
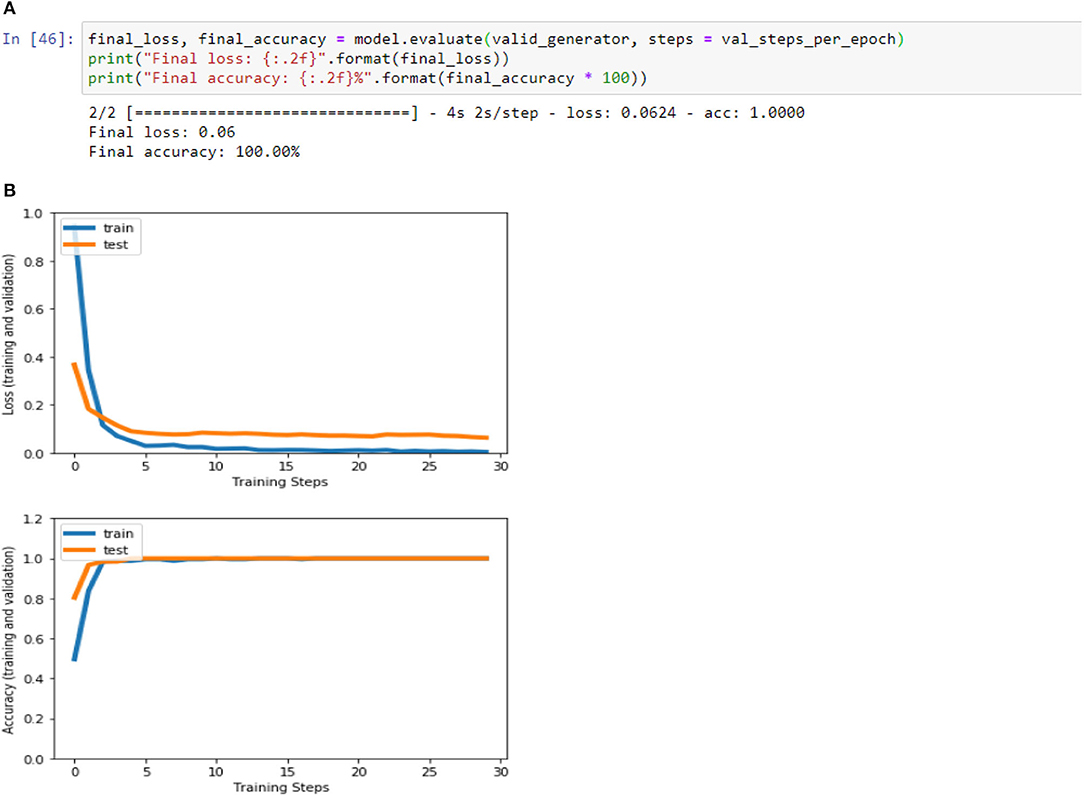
Figure 8. (A) Accuracy and loss (training and validation)–Database B. (B) Accuracy and loss (training and validation)–Database B.
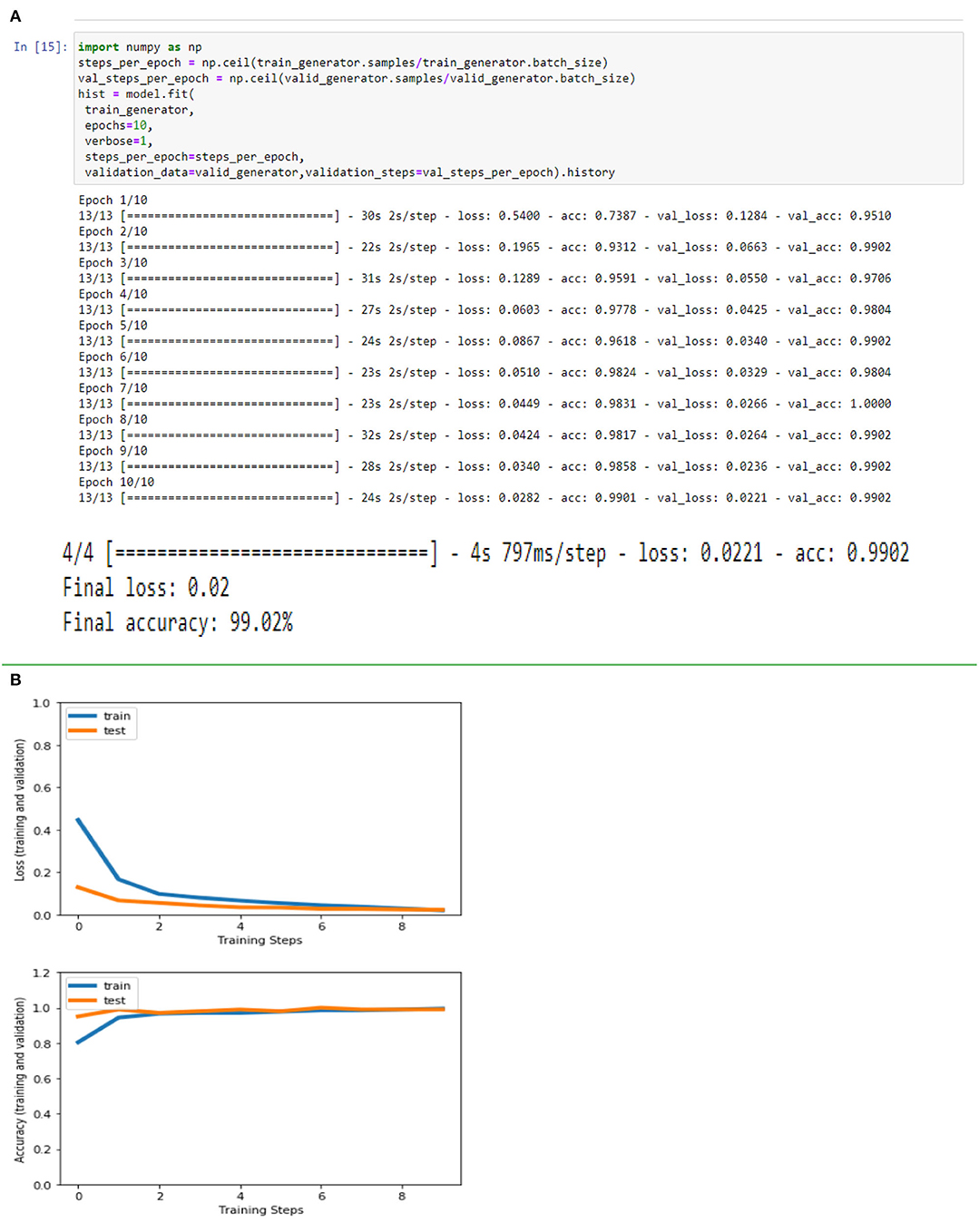
Figure 13. (A) Accuracy and loss (training and validation)–Database B with 10 epochs. (B) Accuracy and loss (training and validation)–Database B with 10 epochs.
Based on the results (all figures and tables), it is shown that the proposed classification strategy DL MobileNetV2 might considerably influence the automated detection and extraction of essential features from X-ray images associated with the medical diagnosis of COVID-19.
The preparation of a deep CNN (D-CNN) takes days and hours, and it requires a large database to induce significant precision. Most assignments or information were related. The model coefficients learned can be utilized within the modern demonstration through transfer learning to accelerate and maximize the learning execution of the modern model. We observed from our tests that preparing D-CNNs, particularly for Mobile-NetV2, with a transfer learning strategy could hasten the preparation method.
Moreover, distinct profundity convolutions and modified remaining direct blockages can diminish the number of coefficients. Thus, Mobile-NetV2 can be rapidly sent in versatile and inserted apparatus. Because the database utilized in this study is small, we trained our model utilizing dropout, which is a proficient strategy for minimizing overfitting in neural systems by dodging sophisticated adaptations on preparing information.
DL Challenge in the Diagnosis of COVID19 Chest Xray
Some limitations of the study research can be overcome in future research, specifically, a more in-depth analysis that requires much more patient data. Establishing models to identify COVID-19 cases from similar viral cases, such as severe acute respiratory syndrome (SARS), and a higher range of common pneumonia is needed. Nonetheless, this work contributes to the possibility of an affordable, fast, and automated diagnosis of the Coronavirus disease.
In the last two years, much DL research on X-rays has been conducted to promote and propose a primary diagnostic tool for COVID-19. These studies have shown good performance results and the best DL algorithm models. These studies have proven that DL algorithms can classify or differentiate between normal and COVID19 chest X-rays. Hence, to keep the study more reliable and to be part of the primary diagnosis tool, perhaps, now is the time to move the study to classify the severity level of COVID-19 where the levels of COVID-19 severity infection are classified into five stages. They are beginning from stage 1 up to stage 5. Stage 1 is defined as the early stage of infection, where these levels classify based on SpO2 level respiratory frequency and difficulty of breathing symptoms. Table 10 shows the severity level and its common symptoms. To certify this future works, deep learning researchers and medical doctors must work together to recognize or build the actual image samples for the algorithm.
Conclusion and Future Work
This study presents a recognizable verification framework for COVID-19 based on a light neural network, Mobile-NetV2, with a transfer-learning procedure and chest X-ray images. We utilized the pre-trained neural system, Mobile-NetV2, which was prepared using the Image-NET database. The conventional convolution layer was used as the beat layer of a recent study. To reduce overfitting, we connected the recession to the recently included conv2d layer. Mobile-NetV2 demonstrates employment to extricate highlights, and the soft-max categorizer is employed to categorize highlights.
The performance rates indicate that computer-aided diagnostic models based on the convolutional neural MobilenetV2 networks may be employed to diagnose COVID-19. The proposed strategy achieved a distinguishing proof accuracy of 99.9 and 100% in our chest X-ray pictures database, with 309 and 512 images (102 infected with COVID-19, 207 normal; 102 infected with COVID 19, 414 normal, respectively) (see Table 9). The proposed framework can be employed in limited computing and low power devices such as smartphones because Mobile-NetV2 is a lightweight neural arrangement.
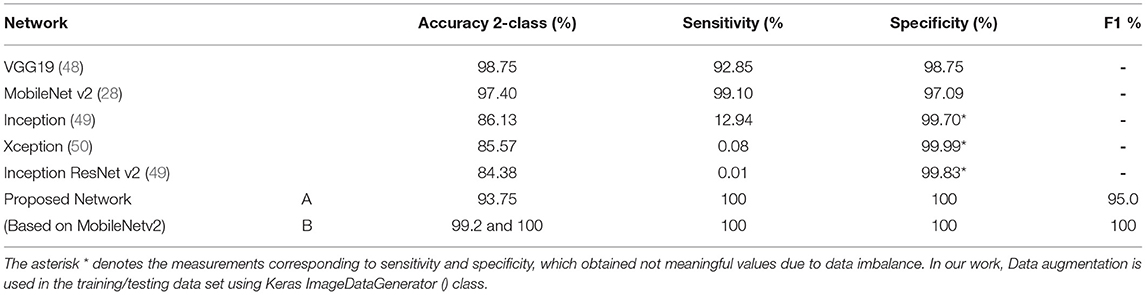
Table 9. Compare the results obtained using the proposed network with other networks (37).
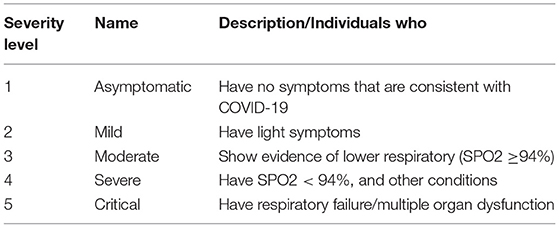
Table 10. Severity level of COVID-19 (51).
In the future, we plan to promote a hybrid system based on MobileNetV2 as a feature extractor with the Whale Optimization Algorithm (WOA) for efficient feature selection and other classifiers such as sport vector machine or Extra Randomized Tree (ERT) algorithm (52).
Data Availability Statement
The datasets analysed in this study are available at: GitHub database [https://github (30)] and https://kaggle.com/c/rsna-pneumonia-detection-challenge. Further inquiries can be directed to the corresponding author/s.
Ethics Statement
Ethics approval and written informed consent were not required for this study in accordance with national guidelines and local legislation.
Author Contributions
MR supervised the project. GA, HA, and SA designed the model, the computational framework, and analysed the data. AN and MR carried out the implementation. GA and HA performed the calculations. GA and SA wrote the manuscript with input from all authors. All authors provided critical feedback and helped shape the research, analysis and manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported financially by Institution Fund projects under Grant No. (IFPRC-215-249-2020).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We extend their appreciation to the Deputyship for Research and Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IFPRC-215-249-2020). and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
References
1. Lai CC, Shih TP, Ko WC, Tang HJ. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int J Antimicrobial Agents. (2020) 55:105924. doi: 10.1016/j.ijantimicag.2020.105924
2. Chen W, Peter WH, Frederick GH, George FGA. novel coronavirus outbreak of global health concern. Lancet. (2020) 395:470–3. doi: 10.1016/S0140-6736(20)30185-9
3. Wong HY, Lam HY, Fong AH, Leung ST, Chin TW, Lo CS, et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. (2019) 296:E72–8. doi: 10.1148/radiol.2020201160
4. Cho S, Lim S, Kim C, Wi S, Kwon T, Youn WS. Enhancement of soft-tissue contrast in cone-beam Computed tomography (CT) using an anti-scatter grid with a sparse sampling approach. Phys Medica. (2020) 70:1–9. doi: 10.1016/j.ejmp.2020.01.004
5. Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W. Correlation of chest Computed tomography (CT) and RT-PCR testing in coronavirus disease 2019/2020 (COVID- 19) in China: a report of 1014 case. Radiology. (2020) 296. doi: 10.1148/radiol.2020200642
6. Kanne JP, Little BP, Chung JH, Elicker BM. Essentials for radiologists on COVID-19: an update - radiology scientific expert panel. Radiology. (2020) 296:E113–4. doi: 10.1148/radiol.2020200527
7. Wang L, Wong A. Covid-net, A tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images. arXiv:2003.09871. (2020). doi: 10.1038/s41598-020-76550-z
8. Montagnon E, Cerny M, Cadrin-Chênevert A, Hamilton V, Derennes T, Ilinca A. Deep learning work?ow in radiology: a primer. Insights into Imag. (2020) 11:22. doi: 10.1186/s13244-019-0832-5
9. Ibrahim A, Mohammed S, Ali HA, Hussein SE. Breast cancer segmentation from thermal images based on chaotic Salp swarm algorithm. IEEE Access. (2020) 8:122121–34. doi: 10.1109/ACCESS.2020.3007336
10. Han J, Zhang D, Cheng G, Liu N, Xu D. Advanced deep learning techniques for salient and category-specific object detection: a survey. IEEE Signal Process Mag. (2018) 35:84–100. doi: 10.1109/MSP.2017.2749125
11. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556. (2014). Available online at: http://arxiv.org/abs/1409.1556
12. Al-Dhamari A, Sudirman R, Humaimi Mahmood NH. and Transfer deep learning along with binary support vector machine for abnormal behavior detection. IEEE Access. (2020) 8:61085–95. doi: 10.1109/ACCESS.2020.2982906
13. Yu S, Xie L, Liu L, Xia D. Learning long-term temporal features with deep neural networks for human action recognition. IEEE Access. (2020) 8:1840–50. doi: 10.1109/ACCESS.2019.2962284
14. Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Into Imag. (2018) 9:611–29. doi: 10.1007/s13244-018-0639-9
15. LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE. (1998) 86:2278–324. doi: 10.1109/5.726791
16. Arayanan VB, Kendall A, Cipolla R. SegNet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intel. (2017) 39:2481–95. doi: 10.1109/TPAMI.2016.2644615
17. Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. Adv Neural Inform Process Syst. (2015) 28:91–9. Available online at: https://arxiv.org/abs/1506.01497.
18. Dong C, Loy CC, He K, Tang X. Learning a deep convolutional network for image super-resolution. In: European Conference on Computer Vision. Springer, Cham. (2014). doi: 10.1007/978-3-319-10593-2_13
19. Minaee S, Abdolrashidi A, Su H, Bennamoun M, Zhang D. Biometric recognition using deep learning: a survey. arXiv:1912.00271.(2019).
20. Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M. A survey on deep learning in medical image analysis. Med Image Anal. (2017) 42:60–88. doi: 10.1016/j.media.2017.07.005
21. Sharif Razavian A, Azizpour H, Sullivan J, Carlsson S. Cnn features off-the-shelf: an astounding baseline for recognition. In: Proceedings of the IEEE Conference on computer vision and pattern recognition workshops. (2014). p. 806–813 doi: 10.1109/CVPRW.2014.131
22. Donahue J, Jia Y, Vinyals O, Hoffman J, Zhang N, Tzeng E, et al. DeCAF: a deep convolutional activation feature for generic visual recognition. In: International Conference on Machine Learning. (2014). p. 647–655.
23. Nguyen L, Lin D, Lin Z, Cao J. Deep cnns for microscopic image categorization by exploiting transfer learning and feature concatenation. In: 2018 IEEE International Symposium on Circuits and Systems (ISCAS). (2018). p. 1–5. doi: 10.1109/ISCAS.2018.8351550
24. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.(2014).
25. He K, Zhang XR. Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2016). p. 770–778. doi: 10.1109/CVPR.2016.90
26. Zoph B, Vasudevan V, Shlens J, Quoc Le Q. Automl for large scale image categorization and object detection. Available online at: https://kaggle.com/c/rsna-pneumonia-detection-challenge, Blog. (2017).
27. Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T. Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv preprint arXiv:1704.04861. (2017).
28. Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D. Going deeper with convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2015). p. 1–9. doi: 10.1109/CVPR.2015.7298594
29. Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence. (2017).
30. Bhattacharya S, Maddikunta PKR, Pham QV, Gadekallu TR, Chowdhary CL, Alazab M. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: a survey. Sustain Cities Soc. (2021) 65:102589. doi: 10.1016/j.scs.2020.102589
31. Manoj M, Srivastava G, Somayaji SRK, Gadekallu TR, Maddikunta PKR, Bhattacharya S. An Incentive Based Approach for COVID-19 using Blockchain Technology. In: IEEE Globecom Workshops. (2020). doi: 10.1109/GCWkshps50303.2020.9367469
32. Kermany DS, Goldbaum M, Cai W, Valentim CC, Liang H, Baxter SL. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 1725. (2018) 1122–31. doi: 10.1016/j.cell.2018.02.010
33. Rajaraman S, Candemir S, Kim I, Thoma G, Antani S. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Appl Sci. (2018) 8:10. doi: 10.3390/app8101715
34. Wang S, Kang B, Ma J, Zeng X, Xiao M, Guo J. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). Eur Radiol. (2021) 31:6096–104. doi: 10.1007/s00330-021-07715-1
35. Shan F, Gao Y, Wang J, Shi W, Shi N, Han M. Lung infection quantification of COVID-19 in CT images with deep learning. arXiv preprint arXiv:2003.04655. (2020)
36. Ghoshal B, Tucker A. Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection. arXiv preprint arXiv:2003.10769. (2020)
37. Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. (2020) 43:635–40. doi: 10.1007/s13246-020-00865-4
38. Esmail K, Hemdan ME, Shouman MA. Cascaded deep learning classifiers for computer-aided diagnosis of COVID-19 and pneumonia diseases in X-ray scans. Compl Intell Syst. (2021) 7:235–47. doi: 10.1007/s40747-020-00199-4
39. Khan AI, Shah JL, Bhat MM. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput Methods Programs Biomed. (2020) 196:10558. doi: 10.1016/j.cmpb.2020.105581
40. Sahiner A, Hameed AA, Jamil A. Identifying Pneumonia in SARS-CoV-2 Disease from Images using Deep Learning. Manchester J Arti Intell Appl Sci. (2021) 2. Available online at: https://mjaias.co.uk/mj-en.
41. Liu J, Wang X. Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant Methods. (2020) 16:83. doi: 10.1186/s13007-020-00624-2
42. Xiang Q, Wang X, Li R, Zhang G, Lai J, Hu Q. Fruit image classification based on Mobilenetv2 with transfer learning technique. In: Proceedings of the 3rd International Conference on Computer Science and Application Engineering. (2019). doi: 10.1145/3331453.3361658
43. Akay M, Du Y, Sershen CL, Wu M, Chen TY, Assassi S, et al. Deep learning classification of systemic sclerosis skin using the MobileNetV2 model. IEEE Eng Med Biol. (2021) 2:104–10. doi: 10.1109/OJEMB.2021.3066097
44. Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M. Covid-19 image data collection: Prospective predictions are the future. arXiv preprint arXiv:2006.11988. (2020)
45. Chollet FK. A Python Deep Learning Library. Available online at: https://keras.io (2015). p. 48.
46. Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. Tensorflow: Large-scale machine learning on heterogeneous systems. arXiv preprint arXiv:1603.04467. (2015). Available online at: https://www.tensorflow.org/api_docs/python/tf/keras/backend
47. Abu-Zinadah H, Abdel Azim G. Segmentation of epithelial human type 2 cells images for the indirect immune fluorescence based on modified quantum entropy. EURASIP J Image Video Process. (2021). 2021:1–9. doi: 10.1186/s13640-021-00554-6
48. Ragab M, Eljaaly K, Alhakamy NA, Alhadrami HA, Bahaddad AA, Abo-Dahab SM, et al. Deep ensemble model for COVID-19 diagnosis and classification using chest CT images. Biology. (2022) 11:43. doi: 10.3390/biology11010043
49. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556. (2015)
50. Chollet FXD. learning with depthwise separable convolutions. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. (2017). p. 1251–1258. doi: 10.1109/CVPR.2017.195
51. Markom MA, Taha SM, Adom AH, Sukor AA, Nasir AA, Yazid H. A review: deep learning classification performance of normal and COVID-19 chest X-ray images. J Phys. (2021) 2071:012003. doi: 10.1088/1742-6596/2071/1/012003
Keywords: machine learning, convolution neural networks, transfer learning, MobileNetV2, COVID-19
Citation: Ragab M, Alshehri S, Azim GA, Aldawsari HM, Noor A, Alyami J and Abdel-khalek S (2022) COVID-19 Identification System Using Transfer Learning Technique With Mobile-NetV2 and Chest X-Ray Images. Front. Public Health 10:819156. doi: 10.3389/fpubh.2022.819156
Received: 20 November 2021; Accepted: 12 January 2022;
Published: 03 March 2022.
Edited by:
Thippa Reddy Gadekallu, VIT University, IndiaCopyright © 2022 Ragab, Alshehri, Azim, Aldawsari, Noor, Alyami and Abdel-khalek. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mahmoud Ragab, bXJhZ2FiQGthdS5lZHUuc2E=
 Mahmoud Ragab
Mahmoud Ragab Samah Alshehri
Samah Alshehri Gamil Abdel Azim
Gamil Abdel Azim Hibah M. Aldawsari6,7
Hibah M. Aldawsari6,7 Adeeb Noor
Adeeb Noor S. Abdel-khalek
S. Abdel-khalek