- 1Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, China
- 2Department of Laboratory, Yongzhou Central Hospital, Yongzhou, China
- 3Department of Medical Lab Science, School of Laboratory Medicine and Life Science, Wenzhou Medical University, Wenzhou, China
Carbapenem-resistant Klebsiella pneumonia (CRKP) infections has become a concerning threat. However, knowledge regarding the characteristics of intestinal CRKP isolates is limited. This study aimed to investigate and compare the clinical, virulence and molecular epidemiological characteristics of intestinal colonization and extraintestinal infections CRKP strains. The clinical characteristics were investigated retrospectively. Polymerase chain reaction was used to investigate the capsular serotype, virulence genes and carbapenemase genes. Capsular polysaccharide quantification assay, serum resistance assay, biofilm formation assay, and infection model of Galleria mellonella larvae were performed to compare the virulence and pathogenicity. Besides, multilocus-sequence-typing (MLST) and pulsed-field-gel-electrophoresis (PFGE) were conducted to explore the homology of intestinal CRKP isolates. A total of 54 intestinal CRKP isolates were included. The main capsular serotypes were K14, K64, and K19. C-reactive protein and the proportion of ICU isolation of the infection group were significantly higher than that of the colonization group (P < 0.05). The carrier rates of various virulence genes of CRKP in the infection group were mostly higher than those in the colonization group, wherein the carrier rates of peg-344 and rmpA were significantly different (P < 0.05). There was no significant difference in capsular polysaccharides, antiserum ability, biofilm formation ability between the two group (P > 0.05), but the lethality of the infection group to Galleria mellonella was significantly higher than that of the colonization group (P < 0.05). The MLST categorized the 54 isolates into 13 different sequence types. PFGE revealed that homology among the 54 CRKP strains was <80%. This study suggested that the CRKP strains in the infection group had higher virulence than those in the colonization group. The development of CRKP isolates colonizing in the intestine should be addressed in future clinical surveillance.
Introduction
With the widespread use of carbapenems, the emergence of carbapenem-resistant Klebsiella pneumoniae (CRKP) has been increasingly reported worldwide (1–3). Numerous reports indicated that CRKP was considered as a serious threat to global health (4, 5).
The intestinal tract of hospitalized patients is an enormous repository of antibiotic-resistant bacteria (6). More seriously, antibiotic-resistant bacteria can not only colonize in the intestine tract, but also transfer to other tissues through intestinal metastasis, leading to serious extraintestinal infections (7), which will be an important risk factor for systemic infection in-hospital death of patients. However, studies have shown that not all Klebsiella pneumoniae (K. pneumoniae) colonized in the intestine could cause further infection (8), and the majority of patients with colonized pathogens did not suffer from infection for several years or even decades (9).
The highly pathogenic bacterium whose strong virulence has been attributed to its ability to resist the host's innate immunity and further infect the host invasively (10, 11). Studies have shown that high-virulence isolates usually have higher capsule content, antiserum ability, and carry a variety of virulence factors, including high-viscosity phenotypes, capsular serotypes, virulence genes, and high-virulence-related clone types, etc. (12, 13). The capsular polysaccharide around bacteria is a crucial virulence factor of K. pneumoniae, participating in the resistance to phagocytosis and serum bactericidal activity (14). Capsular polysaccharides are also closely related to the hyperviscosity phenotype, K1/K2 capsular serotype, and mucus phenotype regulatory gene A (rmpA) (7, 15). In addition, the adhesion of fimbriae is another vital virulence factor of K. pneumoniae. The gene fimH and mrkD promote bacterial adhesion to host tissues and organs by encoding type 1 and type 3 fimbriae of K. pneumoniae, leading to bacterial colonization and pathogenicity. Among them, mrkD can also promote the development of biofilms, which improves the bacteria's resistance to host defense and antibiotics (16).
This study retrospectively investigated the clinical and microbiological characteristics of patients with intestinal CRKP and compared the patients characteristics with CRKP asymptomatic intestinal colonization and extraintestinal infections. The virulence genes (fimH, mrkD, peg-344, rmpA, rmpA2, icuA) and virulence phenotype (capsular polysaccharides, antiserum ability, biofilm formation ability and the lethality of Galleria mellonella larvae) were studied to understand the virulence characteristics of the intestinal CRKP strains. Finally, the molecular epidemiological characteristics of the intestinal CRKP strains were analyzed by MLST and PFGE to provide a theoretical basis for the reasonable clinical management of the intestinal CRKP strains.
Materials and Methods
Bacterial Isolates
A total of 54 intestinal CRKP isolates were included in 2019, in The First Affiliated Hospital of Wenzhou Medical University in Wenzhou, China. All isolates were identified by the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) system. These isolates were stored at −80°C for further research. The intestinal CRKP isolates were divided into two groups by clinical data and homology analysis (MLST and PFGE). One group was the CRKP that caused only asymptomatic intestinal colonization (colonization group); the another group was the CRKP that caused extraintestinal infections after intestinal colonization (infection group). If the MLST and PFGE of the extraintestinal infection isolates were consistent with those isolated from the intestine, and the infection occurred later than the colonization of the intestinal strain, we thought it was caused by the colonization of the intestinal strain. The information on the strains isolated from extraintestinal infection sites was shown in Supplementary Table S1.
Clinical Data Collection
The medical records were reviewed to integrally collect the data of the patients with intestinal CRKP strain during the study period. The study data included the following variables: demographic characteristics (gender and age), underlying or concomitant conditions (diabetes, malignant tumors, cardiovascular diseases, liver insufficiency, renal insufficiency, trauma, etc.), admission temperature, invasive procedures, use of hormones or immunosuppressants, inflammation indicators [white blood cell (WBC) count, C-reactive protein (CRP), procalcitonin (PCT)], admission to intensive care unit (ICU), and prognosis, etc.
Antimicrobial Susceptibility Testing
The MICs of antimicrobial agents, including fosfomycin (FOS) and amikacin (AMK) were determined using the agar dilution method, and the MICs of tigecycline (TGC), colistin (COL) and ceftazidime/avibactam (CZA) were measured via the broth microdilution method (BMD). The results were interpreted by the latest guidelines published by the Clinical and Laboratory Standards Institute (CLSI; Pittsburgh, PA, USA) 2020 (17) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST 2020) (http://www.eucast.org).
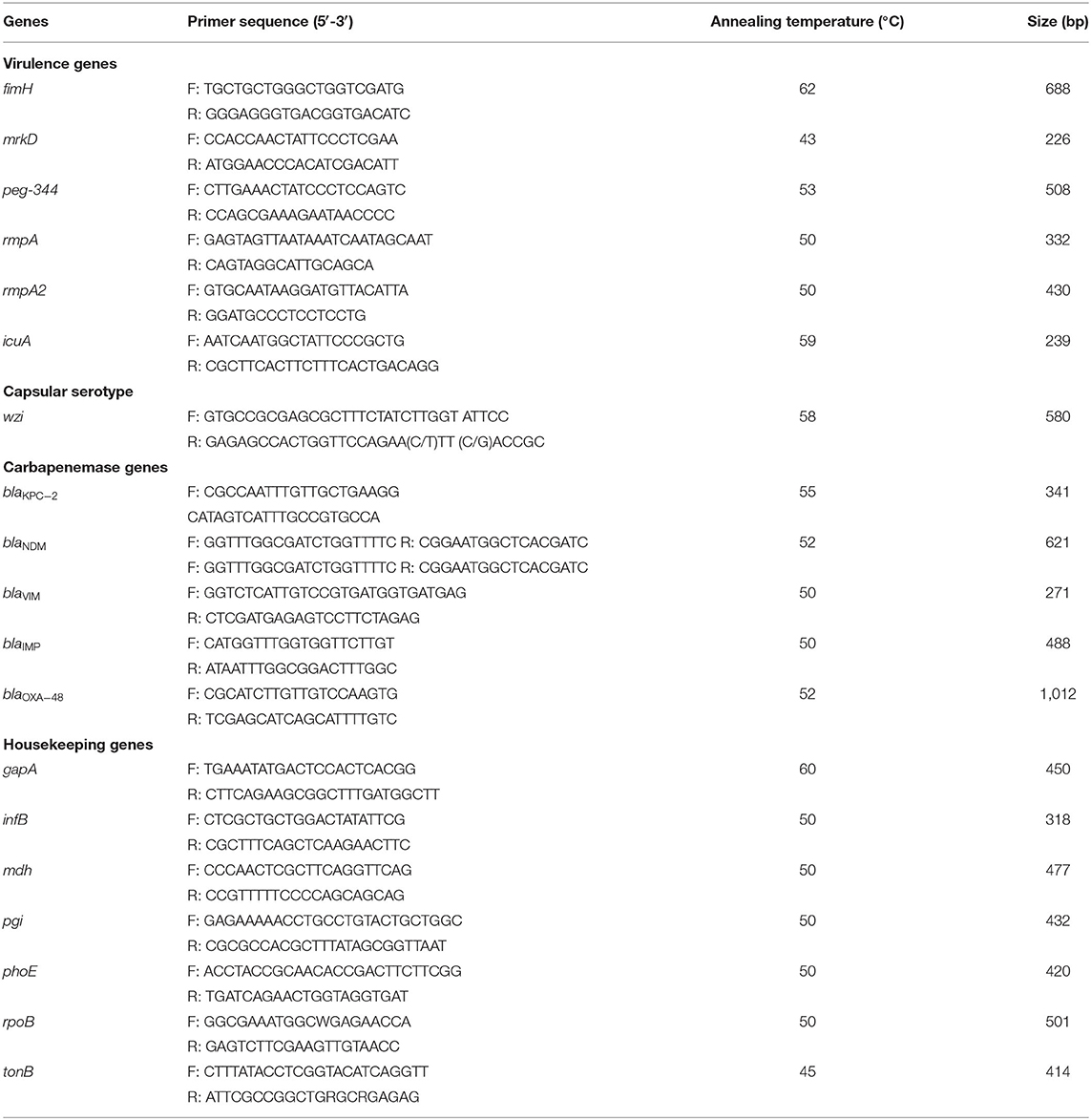
Polymerase Chain Reaction (PCR) for Capsular Serotypes, Virulence Genes, and Carbapenemase Genes
DNA was extracted from CRKP strains. Subsequently, capsular serotype-specific gene (wzi), virulence genes (e.g., fimH, mrkD, peg-344, rmpA, rmpA2 and icuA) and carbapenemase genes were amplified by PCR using specific primers as previously described (1, 18–20) (Table 1). The positive PCR products were verified by sequencing at Beijing Genomics Institute Technology Co. Ltd. (Shanghai, China). Nucleotide sequences were compared using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Capsular Polysaccharide Quantification Assay
Capsular polysaccharide quantification assay was performed as described previously (21, 22), with minor modifications. Briefly, 500 μL of cultured bacteria suspensions were resuspended in phosphate-buffered saline (PBS) and adjusted to 108 CFU/mL, and a 1.2 mL sodium tetraborate in sulfuric acid was added in the resuspensions that were later placed in the ice bath and incubated for 5 min at 100°C, and then place on ice for 10 min. A 20 μL volume of 1.5 mg/mL m-hydroxyldiphenyl was then added and mixed. After a 5 min incubation at room temperature, the absorbance at 590 nm was measured. The glucuronic acid content was determined from a standard curve of glucuronic acid and expressed as μg/108 CFU. Results were presented as the mean and standard deviation (Mean ± SD) of the data of three independent experiments.
Serum Resistance Assay
Serum resistance assay was performed as described previously (23), with minor modifications. Briefly, overnight bacterial cultures were diluted 1:100 into 10 mL of fresh Luria-Bertani (LB) medium and incubated until the bacterial suspension equaled to an OD600 of 0.5. Then, a 1 mL aliquot of the culture was washed with PBS and resuspended in 1 mL of PBS. Next, 100 μL of the bacterial suspension was mixed with 300 μL of normal human serum (NHS) and PBS. Bacteria-PBS mix was regarded as control. After mixing, the serum-bacteria and PBS-bacteria suspensions were incubated at 37°C for 3 h. To calculate the serum bactericidal effect, a 100 μL aliquot was taken from each suspension and 10-fold serial dilutions were plated onto LB plates. The serum bactericidal effect was expressed as the ratio of the CFUs in the serum-bacteria suspension to the CFUs in the PBS-bacteria suspension. All experiments were performed in triplicate, and results were expressed as survival percentage.
Biofilm Formation Assay
The biofilm formation assay was measured using the method of Wilksch et al. (24). Briefly, clinical isolates were grown to logarithmic phase in LB broth and diluted 1:100 into fresh LB broth. A total of 200 μL of each dilution were added to a 96-well polystyrene microtiter plate and blank controls were set at the same time, and each strain set three duplicate wells. Then, the plate was incubated at 37°C for 24 h. Planktonic cells were removed, and the wells were washed three times with sterile water and then stained with 200 μL 0.1% crystal violet for 10 min and rinsed three times with sterile water. Stained biofilms were solubilized with 95% (V/V) ethanol and quantified by measuring the OD595. Each sample was measured in triplicates and averages of absorbance values were used for analysis.
Infection Model of Galleria mellonella Larvae
The virulence of K. pneumoniae was evaluated by the G. mellonella larvae infection model, as previously described (25). Briefly, serial dilutions of K. pneumoniae (104, 105, 106, 107 CFU/mL) were prepared in PBS. Subsequently, a 10 μL of each dilution was injected into the last left proleg. Larvae injected with 10 μL PBS were used as the control group. Ten larvae weighing between 200 and 250 mg were randomly selected for each dilution. The larvae were incubated at 37°C in the dark and observed at 24 h intervals in a week. Larvae were considered dead when they repeatedly failed to respond to physical stimuli. The primary outcome for the G. mellonella model was rapidity and extent of mortality of larvae assessed with Kaplan-Meier analysis and log-rank test.
Multi-Locus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE)
In this study, seven housekeeping genes of K. pneumoniae (rpoB, gapA, phoE, mdh, tonB, pgi, and infB) were amplified by polymerase chain reaction (PCR) and sequenced. The genotypes of all isolates were identified according to the protocol provided on the designated website. The alleles and STs were assigned according to the Pasteur Institute multilocus sequence typing online database for K. pneumoniae (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html).
PFGE was performed on our strains based on our previously described method (1), with only minor modifications. Genomic DNA was extracted from the K. pneumoniae strains, and digested with Xba I (Takara 1093A, Japan) for 2 h. PFGE was performed using a CHEF-Mapper XA PFGE system (Bio-Rad, USA) for 18 h with a molecular weight of 30–600 kb and a switch time of 6–36 s. DNA fingerprints were then revealed by Ethidium-bromide staining. The GelDoc XR gel imaging system (Bio-Rad, USA) was used to visualize stripe patterns.
Statistical Analysis
All statistical analyses were performed using SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). The Chi-Square test and Fisher's exact test were used to compare categorical variables. Student's t-test and Mann-Whitney U test were used to compare continuous variables with and without normal distribution, respectively. P-value <0.05 was considered statistically significant. All tests were two-tailed.
Results
Clinical Characteristics
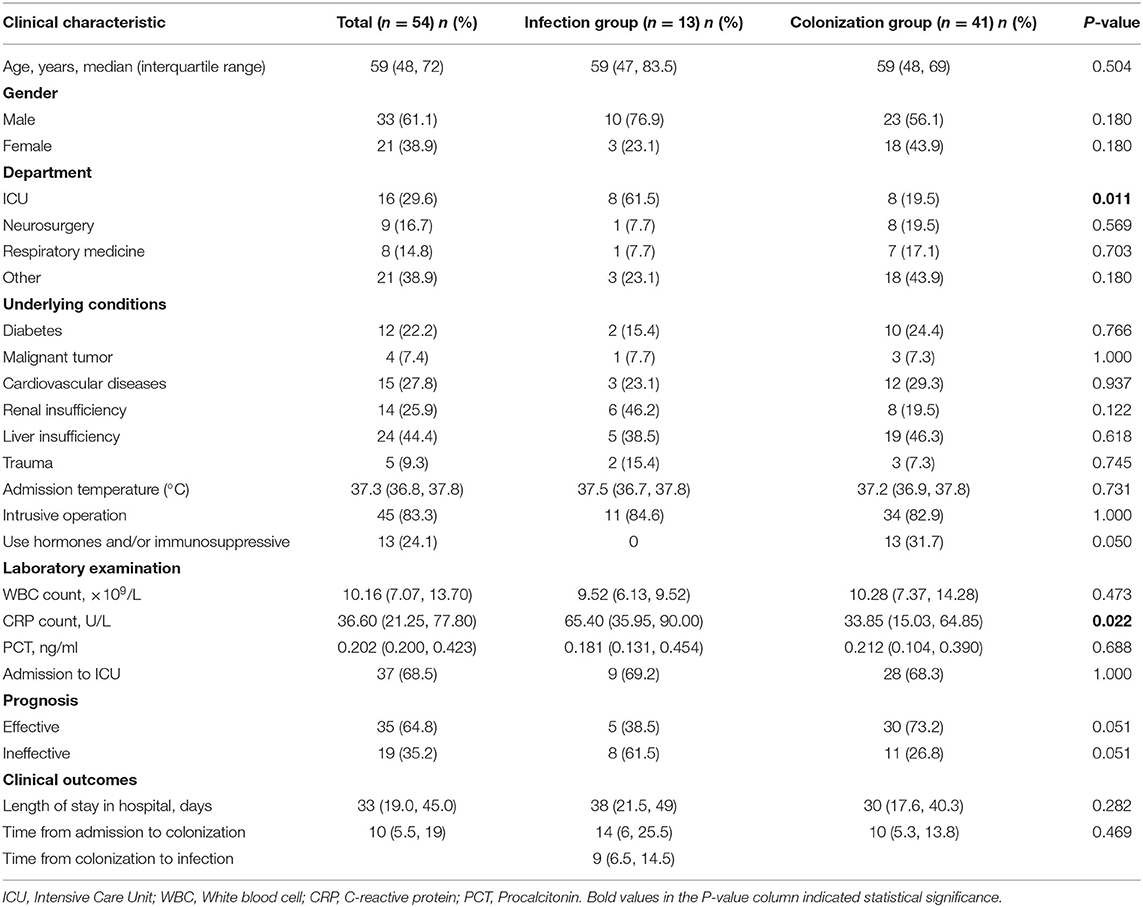
The results showed that the median age of patients with intestinal CRKP was 59-year-old, and males accounted for 61.1%. 51 cases (94.4%) suffered from concomitant diseases, including 12 (22.2%) diabetes, 15 (27.8%) cardiovascular and cerebrovascular diseases, 14 (25.9%) renal insufficiency, 24 (44.4%) liver insufficiency, 5 (9.3%) trauma and so on. Inflammatory indicators include leukocytosis, C-reactive protein, and procalcitonin, all of which were increased. 45 (83.3%) have received invasive procedures, and 37 (68.5%) have been admitted to the ICU. Intestinal CRKP strains were mainly from ICU (29.6%), neurosurgery (16.7%) and respiratory medicine (14.8%). Among the 54 patients with intestinal CRKP strains, 13 caused extraintestinal infections after intestinal colonization (defined as the infection group), and 41 were only intestinal asymptomatic colonization (defined as the colonization group). C-reactive protein and the proportion of ICU isolates in the patients of the infected group were significantly higher than those in the colonization group (P < 0.05). The clinical data of 54 patients were shown in Table 2.
Antimicrobial Susceptibility Testing
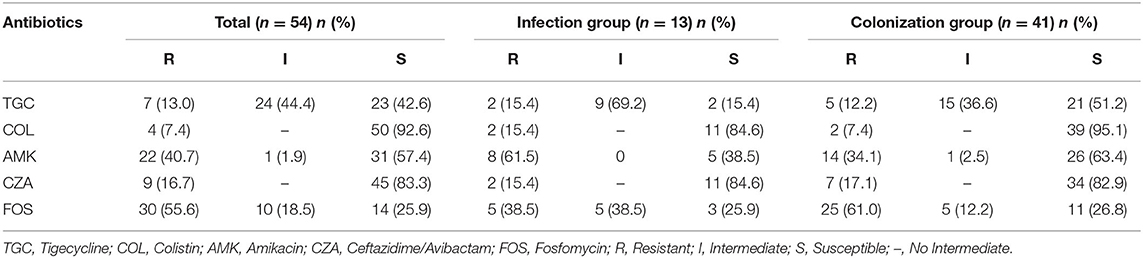
The resistance rates of 54 CRKP strains to TGC, COL, CAZ/AVI, AMK, and FOS were 13, 7.4, 16.7, 40.7, and 55.6%, respectively. The susceptible rates of 54 CRKP strains to TGC, COL, CAZ/AVI, AMK, and FOS were 42.6, 92.6, 83.3, 57.4, and 25.9%, respectively. There were higher susceptible rates to colistin and ceftazidime/avibactam, and a higher intermediate rates (69.2%) to tigecycline (Table 3).
Carbapenemase Genes
83.3% (45/54) of CRKP strains carried the blaKPC−2 gene, and blaKPC−2 was the dominant carbapenemase gene no matter in the infection group (69.2%, 9/13) or the colonization group (87.8%, 36/41). In addition, a strain carrying blaNDM was also detected in the colonization group strains. blaVIM, blaIMP and blaOXA−48 were not detected in all strains. The results of carbapenemase genes of 54 CRKP strains were shown in Figure 1.
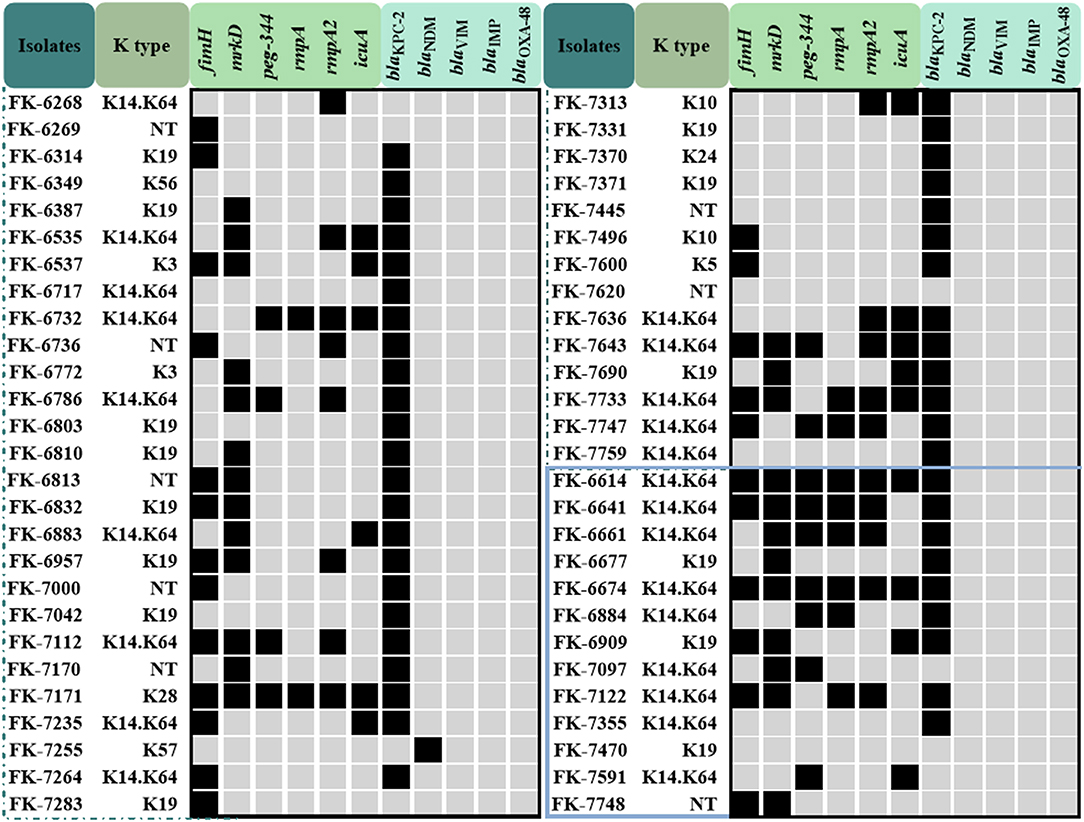
Figure 1. The capsular serotype, virulence genes and carbapenemase genes map of the colonization group and the infection group CRKP strains. The strains in the blue box represent the infection group strains, the others are the colonization group strains.
Capsular Serotypes and Virulence Genes
In the infection group, K14 and K64 serotypes accounted for 69.2% (9/13), followed by K19 serotypes accounting for 23.1% (3/13), and one strain could not be typed; in the colonization group strains, K14 and K64 serotypes accounted for 42.6% (23/41), K19 serotype accounted for 25.9% (14/41), and 8 strains could not be typed. The carrier rates of fimH, mrkD, peg-344, rmpA, rmpA2, and icuA of 54 CRKP strains were 44.4, 88.9, 24.1, 20.4, 33.3, and 27.8%, respectively. The carrier rate of virulence genes in the infection group was higher than that of the colonization group except for mrkD, and the difference of peg-344 and rmpA was significant (P < 0.05). The results of the capsular serotype and virulence genes of 54 CRKP strains were shown in Figure 1 and Table 4.
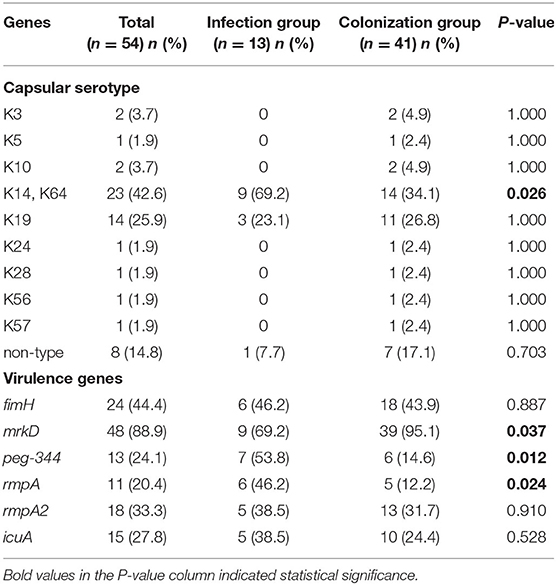
Table 4. The capsular serotype and virulence genes of the infection group and the colonization group.
The Virulence Phenotype
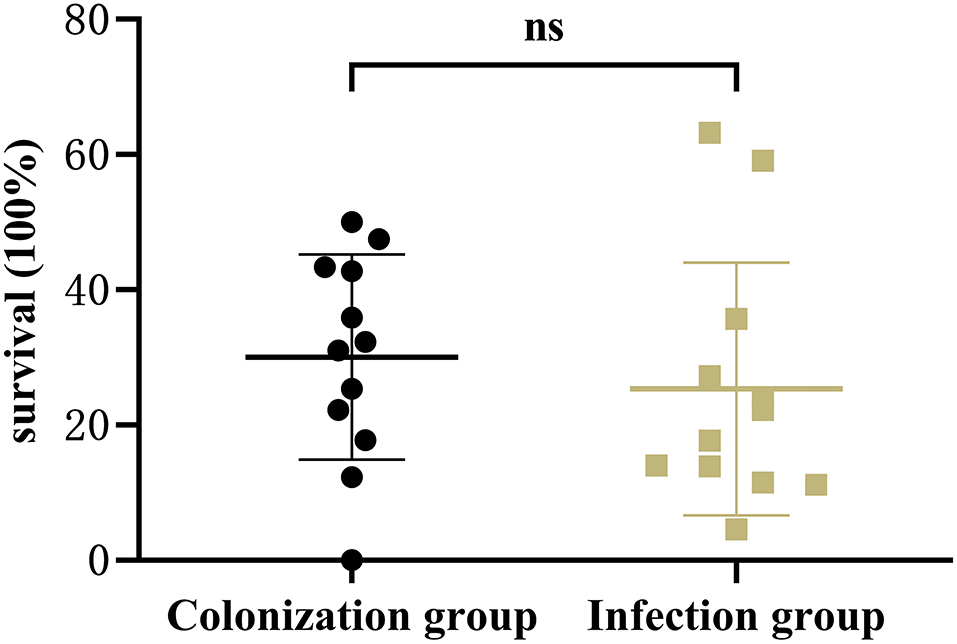
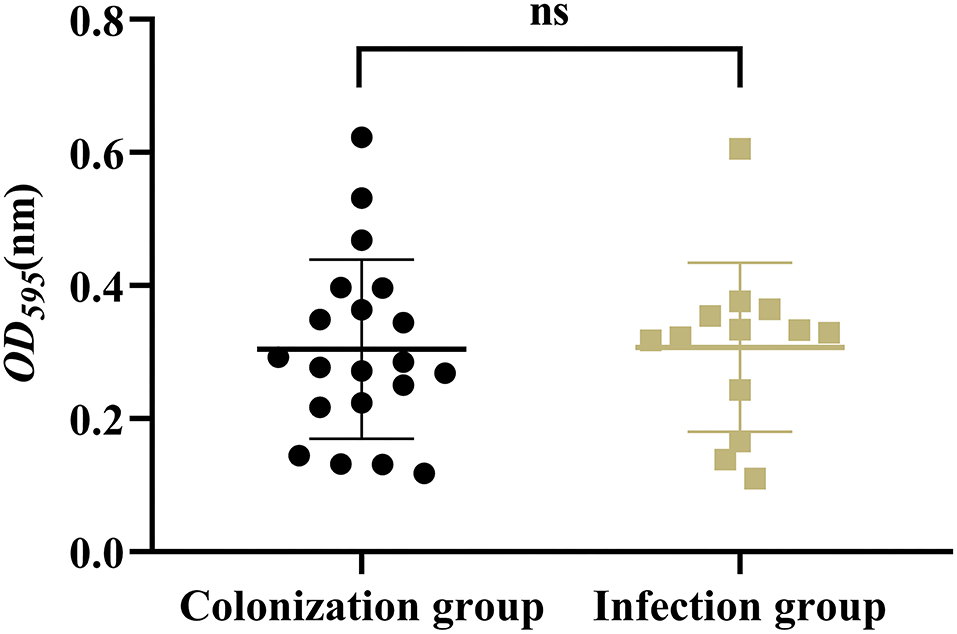
Results showed that there was no significant difference in the capsular polysaccharide content, antiserum effect and biofilm formation between colonization group and infection group strains (P > 0.05) (Figures 2–4); The survival situations of G. mellonella larvae infected with CRKP strain were shown in Figure 5. The results showed that the lethality rate of G. mellonella larvae in the infection group was significantly higher than that of the colonization group (P < 0.05). The lethality rate of the two groups was significantly higher than that of the PBS control group (P < 0.05).
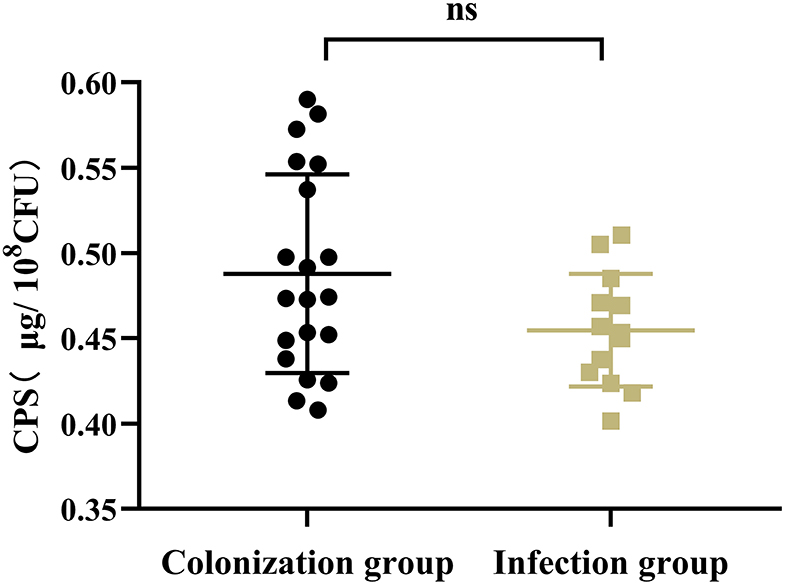
Figure 2. Capsular polysaccharide content of intestinal CRKP strains. ns, no significant difference.
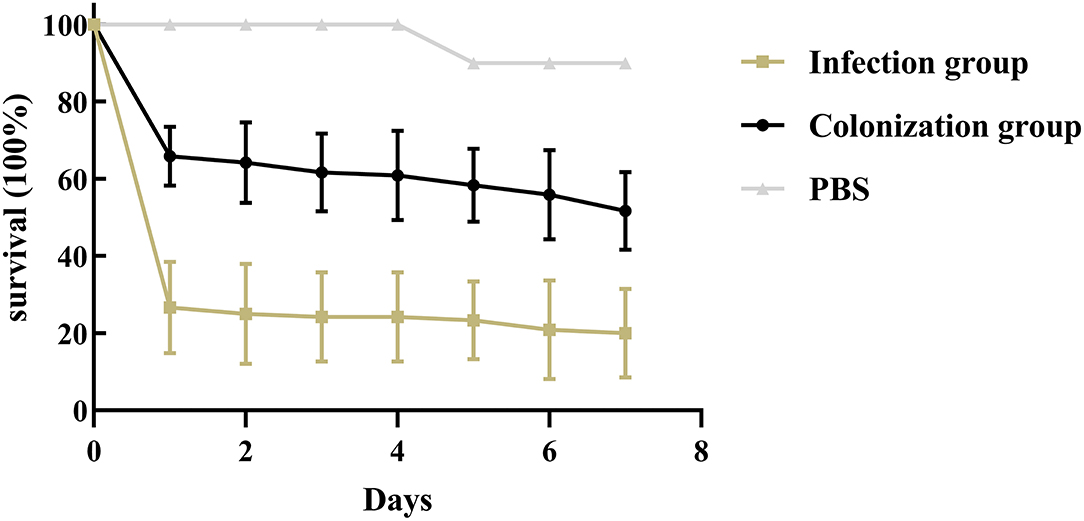
Figure 5. Infection model of Galleria mellonella larvae. Survival of Galleria mellonella larvae infected by intestinal CRKP strains.
Multi-Locus Sequence Typing (MLST) and Pulsed-Field Gel Electrophoresis (PFGE)
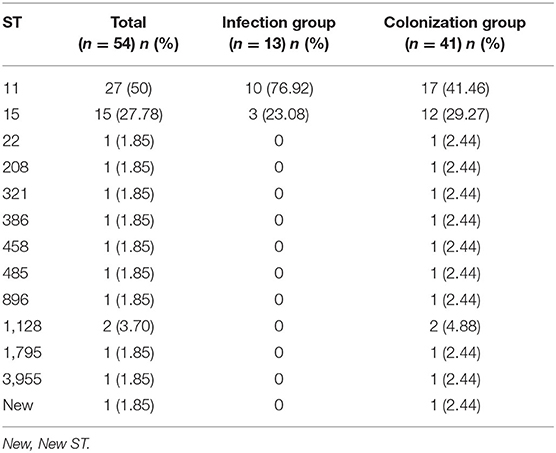
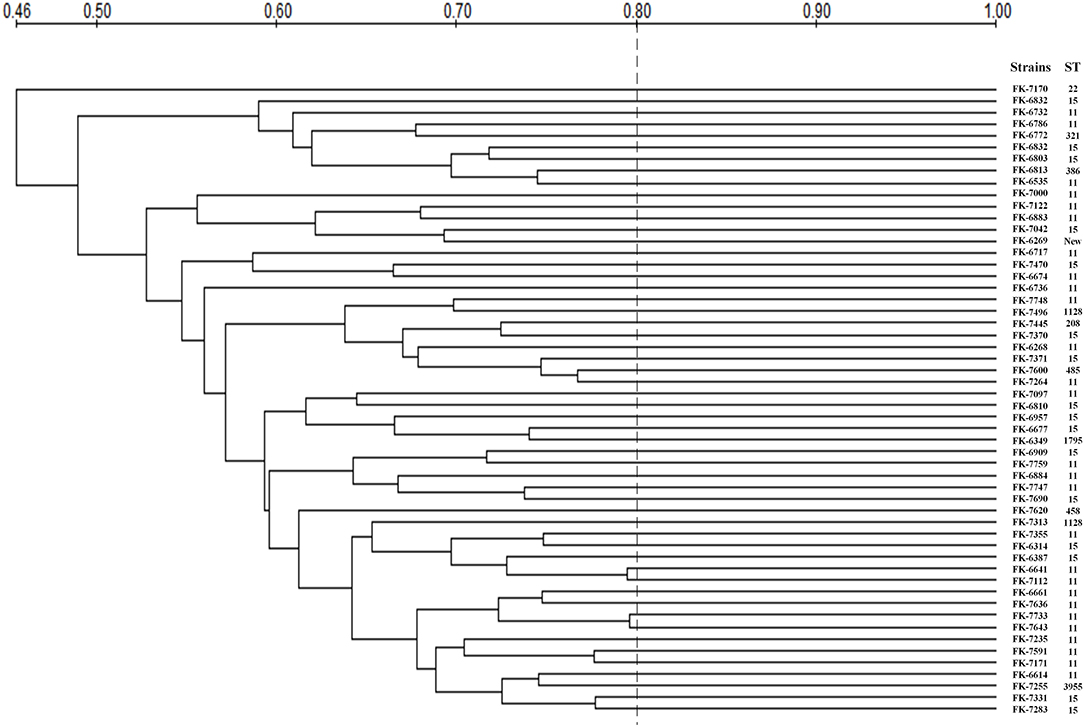
The MLST and PFGE results of 54 strains were shown in Table 5 and Figure 6. MLST showed that the 54 CRKP strains belonged to 13 different ST, among which ST11 and ST15 were the most concentrated, accounting for 27 (50%) and 15 (27.78%), respectively. In infection group, there were only two ST types: ST11 and ST15. However, in the colonization group, there were a few other rare ST types besides ST11 and ST15, such as ST22, ST208, ST321, ST386, ST458, ST485, ST896, ST1128, ST1795, ST395. In addition, a new kind of ST was observed in infection group. PFGE showed that 54 CRKP strains belonged to different pulse types, and their similarities were all <80%. The strains displayed low phylogeny in both the colonization group and infection group, indicating that no clonal transmission has been found.
Discussion
In recent years, the detection rate of CRKP strains has been reported to be increasing (26), which has brought great challenges to clinical treatment. In order to understand the difference between intestinal colonization and causing-extraintestinal infections CRKP strains, we investigated and compared the clinical and microbiological characteristics of intestinal CRKP isolates.
In this study, patients with intestinal CRKP were mainly middle-aged and elderly men, consistent with the conclusion of Chen et al. (27), most of which were accompanied by diabetes, liver and kidney insufficiency, and cardiovascular diseases. The CRP of patients in the infection group was significantly higher than that of the colonization group (P < 0.05), which suggested that the CRP could be used as an early warning factor for secondary infection after colonization. Among patients with intestinal colonized CRKP, 45 patients (83.3%) have received invasive procedures, and 37 patients (68.5%) have been admitted to ICU, which suggested that invasive procedures and ICU admission may be the cause of CRKP colonization, which is consistent with the conclusions of previous reports (27, 28). The number of patients in ICU, neurosurgery, and respiratory was much more, and the isolation rate of the infection group strains in the ICU was significantly higher than that in the colonization group (P < 0.05), suggesting that for patients in ICU, neurosurgery and respiratory should undergo routine CRKP screening when they are admitted to the hospital to prevent and control CRKP infection. The results of clinical data showed that the median time from admission to intestinal CRKP colonization of patients was 10-day, and the median time from intestinal CRKP colonization to infection was 9-day. It is necessary to take preventive and control measures as early as possible for the prevention and treatment of infection.
Our study showed that the most prevalent carbapenemase gene was blaKPC−2 among CRKP isolates, which is consistent with previous studies (29, 30). There were 8 strains that have not been detected in any of blaKPC−2, blaNDM, blaVIM, blaIMP, or blaOXA−48, which may be caused by other resistance mechanisms, such as extended-spectrum β-lactamase, efflux pumps and porin mutations (31). Due to the production of carbapenemase and extended-spectrum β-lactamase (32), CRKP strains are resistant to general antibacterial drugs such as carbapenem and cephalosporins. At present, only a few drugs, such as colistin, tigecycline, some aminoglycoside drugs, and the recently approved ceftazidime/avibactam, still have good antibacterial activity against CRKP (33–35). The antimicrobial susceptibility testing showed that the intestinal CRKP strains had a high resistance rate to amikacin and fosfomycin, and the resistance rate to tigecycline and ceftazidime/avibactam were relatively low, accounting for 13 and 16.7%, respectively, which are lower than the results of Park and Zhou et al. (36, 37). Among all antibiotics tested, the most susceptible on CRKP strains was colistin, which was lower than the resistance rate reported by Rojas et al. (38). Colistin is still a good choice for the treatment of CRKP infection. However, the side effects such as nephrotoxicity and neurotoxicity should also be taken seriously. In severe CRKP infection, colistin is often recommended to combined with other antibiotics (such as tigecycline, meropenem, gentamicin, or fosfomycin, etc.) (39).
The virulence level of K. pneumoniae is closely related to the invasion and defense. High virulence helps the bacteria resist the host's innate immunity and invasively infect the host (10, 11). High viscosity is one of the most extrusive characteristics of highly virulent K. pneumoniae. Although the high viscosity of the bacteria is related to high virulence, it is not a necessary condition that high-viscosity K. pneumoniae is not equal to high-virulent K. pneumoniae. Multilocus sequence typing (MLST) and capsular antigen serotypes have also been used to identify high-virulent K. pneumoniae, even some special STs (such as ST23, ST65, and ST86) and capsular serotypes (such as K1 and K2) are closely related to high-virulent K. pneumoniae, but these genotypes and serotypes can also exist in classic K. pneumoniae. In 54 researched strains, no matter in the colonization group or the infection group, no K1, K2 capsular serotypes and ST23, ST65, and ST86 strains were found, but this cannot completely rule out high-virulence K. pneumoniae. Research (40) reported that there were also high-virulent strains of K. pneumoniae in ST11, and the intestinal CRKP strains in this study are mainly concentrated in ST11, especially in the infection group, ST11 strains account for a larger proportion.
Studies (18) have reported that peg-344, iucA, rmpA, and rmpA2 and other virulence genes can be used as candidate biomarkers for identifying K. pneumonia with high virulence, which showed the accuracy was >0.95. These specific genes mediate the increasing production of siderophores and capsular polysaccharides, thereby enhancing the high viscosity and virulence of K. pneumoniae. Although such laboratory indicators have high specificity, they are not only caused by high-virulent K. pneumoniae, but also exists in classic K. pneumoniae. Our results showed that the carrier rate of virulence genes in the infection group was higher than that in the colonization group, and the difference of peg-344 and rmpA was significant (P < 0.05). Nevertheless, there was no significant difference in the production of capsular polysaccharides and antiserum killing ability of strains in infection group and colonization group. This may be closely related to the content of capsular polysaccharides in the strains. Studies have shown that capsular polysaccharides can hinder the reaction of bacteria with serum antibodies and other components (13, 21). Fimbriae adhesion is another prominent virulence factor for the increased virulence of K. pneumoniae. fimH and mrkD mediate adhesion by encoding K. pneumoniae type 1 and type 3 fimbriae, respectively, and promote bacterial adhesion to host tissues and organs, leading to bacterial colonization and pathogenicity. In addition, mrkD can also promote the development of biofilms, the physical barrier formed can hinder the attack of phagocytic killer cells and enzymes on bacteria, and increase the invasion of bacteria to the host and lead to antibiotics-resistance (16). However, our results showed that the carrier rate of mrkD in the infection group was significantly lower than that in the colonization group, but there was no significant difference in their biofilm formation ability. In order to explore the in vivo virulence and pathogenicity of the intestinal CRKP strains, we established the CRKP infection model of G. mellonella larvae. The results showed that the lethality rate of the infection group was significantly higher than that of the colonization group (P < 0.05), and the lethality rate of the two groups of strains was significantly higher than that of the PBS control group. It proved that the CRKP strains of infection group have higher virulence and stronger pathogenicity. Clinicians need to pay attention to patients with intestinal colonized CRKP, and take preventive and inspection measures in time to minimize the occurrence of highly virulent K. pneumoniae infections.
In this study, 54 intestinal CRKP strains belonged to different pulse types, suggesting that the strains had low homology. MLST showed that the intestinal colonized CRKP strains belonged to 13 ST types, among which ST11 and ST15 were the most concentrated, accounting for 27 (44.4%) and 15 (27.8%) respectively, and all the infection group strains were ST11 and ST15. The most common CRKP strains are ST11 and ST258 in the world, as well ST11 is the main ST in China, and ST258 is the most common in the United States (41, 42). ST11 is a single locus variant (tonB) of ST258, suggesting a close relationship between them. According to reports (42), ST11 is the main ST type of KPC-producing K. pneumoniae. Therefore, it can be speculated that K. pneumoniae of some STs (such as ST11 and ST258) are good carriers of drug-resistant plasmids and promote the spread of drug resistance. Some studies believed that (43) the ICEKp258.2 element of ST258 and ST11 strains can encode one or more factors, which may contribute to the persistence and spread of the strain. Specifically, ICEKp258.2 can encode IV fimbriae, which enables the strain to adhere to the surface of a variety of materials and/or improve its ability to exchange genetic information, thereby contributing to the persistence of the strain and stability (43). Therefore, we need to pay attention to the spread of ST11 K. pneumoniae between patients. In addition, ST15 is a single locus variant of ST14 (infB), which may represent the spread in the midwest of the United States, but it has now spread in China (42). ST15 and ST14 may belong to another potential clonal complex that may spread in China. According to reports (44), most of the isolates carrying blaOXA−232 belonged to ST15, therefore, attention should be paid to the prevalence of ST15 in the intestinal CRKP. The dominant clone group is relatively easy to develop carbapenem resistance, so molecular epidemiological monitoring can effectively control the occurrence and spread of drug-resistant bacterial infections in hospitalized patients.
The limitation of this study is the limited number of strains. This study showed that the CRKP strains in the infection group had higher virulence than those in the colonization group, which was mainly manifested in the carrier rates of various virulence genes and the mortality to Galleria mellonella, except capsular polysaccharide, antiserum ability and biofilm forming ability, which needs to be expanded in the future for further research.
Conclusions
Our results suggested that the CRKP strains in the infection group had higher virulence than those in the colonization group. The development of CRKP isolates colonizing in the intestine should be addressed in future clinical surveillance.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
All the investigation protocols in this study were approved by the Ethics Committee of The First Affiliated Hospital of Wenzhou Medical University. Informed consent was waived because this study with an observational approach had mainly focused on bacteria and involved no interventions to the patients.
Author Contributions
WL conducted the experiments, analyzed the data, and wrote the manuscript. YZ and TC participated in experiments. WZ and LC participated in writing. NH participated in the analysis of results. JC and TZ helped design the study. YS designed the study and corrected the manuscript. HW guided the revision of the manuscript. All authors have read and approved the manuscript.
Funding
We thank the National Natural Science Foundation of China (no. 81971986), the Health Department of Zhejiang Province of the People's Republic of China (no. 2019KY098), and the Planned Science and Technology Project of Wenzhou (no. Y2020974) for providing financial funding.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2021.783124/full#supplementary-material
References
1. Zhao Y, Zhang X, Torres VVL, Liu H, Rocker A, Zhang Y, et al. An outbreak of carbapenem-resistant and hypervirulent Klebsiella pneumoniae in an intensive care unit of a major teaching hospital in Wenzhou, China. Front Public Health. (2019) 7:229. doi: 10.3389/fpubh.2019.00229
2. Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. (2018) 18:37–46. doi: 10.1016/S1473-3099(17)30489-9
3. Veeraraghavan B, Shankar C, Karunasree S, Kumari S, Ravi R, Ralph R. Carbapenem resistant Klebsiella pneumoniae isolated from bloodstream infection: Indian experience. Pathog Glob Health. (2017) 111:240–6. doi: 10.1080/20477724.2017.1340128
4. Karampatakis T, Tsergouli K, Iosifidis E, Antachopoulos C, Karapanagiotou A, Karyoti A, et al. Impact of active surveillance and infection control measures on carbapenem-resistant Gram-negative bacterial colonization and infections in intensive care. J Hosp Infect. (2018) 99:396–404. doi: 10.1016/j.jhin.2018.05.010
5. Lu MC, Tang HL, Chiou CS, Wang YC, Chiang MK, Lai YC. Clonal dissemination of carbapenemase-producing Klebsiella pneumoniae: two distinct sub-lineages of Sequence Type 11 carrying bla(KPC-2) and bla(OXA-48). Int J Antimicrob Agents. (2018) 52:658–62. doi: 10.1016/j.ijantimicag.2018.04.023
6. Freedberg DE, Zhou MJ, Cohen ME, Annavajhala MK, Khan S, Moscoso DI, et al. Pathogen colonization of the gastrointestinal microbiome at intensive care unit admission and risk for subsequent death or infection. Intensive Care Med. (2018) 44:1203–11. doi: 10.1007/s00134-018-5268-8
7. Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. (2016) 80:629–61. doi: 10.1128/MMBR.00078-15
8. Zhang X, Zhao Y, Wu Q, Lin J, Fang R, Bi W, et al. Zebrafish and Galleria mellonella: models to identify the subsequent infection and evaluate the immunological differences in different Klebsiella pneumoniae intestinal colonization strains. Front Microbiol. (2019) 10:2750. doi: 10.3389/fmicb.2019.02750
9. Harada S, Tateda K, Mitsui H, Hattori Y, Okubo M, Kimura S, et al. Familial spread of a virulent clone of Klebsiella pneumoniae causing primary liver abscess. J Clin Microbiol. (2011) 49:2354–6. doi: 10.1128/JCM.00034-11
10. Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. (2019) 32:e00001-19. doi: 10.1128/CMR.00001-19
11. Liu Y, Liu PP, Wang LH, Wei DD, Wan LG, Zhang W. Capsular polysaccharide types and virulence-related traits of epidemic KPC-producing Klebsiella pneumoniae isolates in a Chinese University Hospital. Microb Drug Resist. (2017) 23:901–7. doi: 10.1089/mdr.2016.0222
12. Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, et al. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol. (2016) 6:165. doi: 10.3389/fcimb.2016.00165
13. Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, et al. Antimicrobial resistance of hypervirulent Klebsiella pneumoniae: epidemiology, hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. (2017) 7:483. doi: 10.3389/fcimb.2017.00483
14. Fung CP, Chang FY, Lin JC, Ho DM, Chen CT, Chen JH, et al. Immune response and pathophysiological features of Klebsiella pneumoniae liver abscesses in an animal model. Lab Invest. (2011) 91:1029–39. doi: 10.1038/labinvest.2011.52
15. Yu WL, Ko WC, Cheng KC, Lee CC, Lai CC, Chuang YC. Comparison of prevalence of virulence factors for Klebsiella pneumoniae liver abscesses between isolates with capsular K1/K2 and non-K1/K2 serotypes. Diagn Microbiol Infect Dis. (2008) 62:1–6. doi: 10.1016/j.diagmicrobio.2008.04.007
16. Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun. (2009) 77:5016–24. doi: 10.1128/IAI.00585-09
17. M100 Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Wayne, PA: Clinical and Laboratory Standards Institute (2020). p. 1–294.
18. Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. (2018) 56:e00776-18. doi: 10.1128/JCM.00776-18
19. Aboulela A, El-Sherbini E, Abu-Sheasha G, El-Raouf HA, El-Ghazzawi E, Gaballah A. Molecular strain typing of multidrug-resistant Klebsiella pneumoniae: capsular wzi gene sequencing versus multiple locus variable number tandem repeat analysis. Diagn Microbiol Infect Dis. (2020) 98:115139. doi: 10.1016/j.diagmicrobio.2020.115139
20. Tian XB, Zheng XK, Sun Y, Fang RC, Zhang SQ, Zhang XC, et al. Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from chinese patients during 2002–2017. Infect Drug Resist. (2020) 13:501–12. doi: 10.2147/IDR.S232010
21. Yang X, Wai-Chi Chan E, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. (2019) 4:2039–43. doi: 10.1038/s41564-019-0566-7
22. Filisetti-Cozzi TM, Carpita NC. Measurement of uronic acids without interference from neutral sugars. Anal Biochem. (1991) 197:157–62. doi: 10.1016/0003-2697(91)90372-Z
23. Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, Koh TH, et al. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol. (2011) 49:3761–5. doi: 10.1128/JCM.00977-11
24. Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. (2011) 7:e1002204. doi: 10.1371/journal.ppat.1002204
25. Insua JL, Llobet E, Moranta D, Pérez-Gutiérrez C, Tomás A, Garmendia J, et al. Modeling Klebsiella pneumoniae pathogenesis by infection of the wax moth Galleria mellonella. Infect Immun. (2013) 81:3552–65. doi: 10.1128/IAI.00391-13
26. Gao L, Lv Y, Li Y. Analysis of the drug resistance of carbapenem-resistant Klebsiella pneumoniae in the China antimicrobial resistance surveillance trial program, 2007–2018. Microb Drug Resist. (2020) 26:944–50. doi: 10.1089/mdr.2019.0299
27. Chen X, Liu Q, Liu WE, Yan Q. Risk factors for subsequential carbapenem-resistant Klebsiella pneumoniae clinical infection among rectal carriers with carbapenem-resistant Klebsiella pneumoniae. Infect Drug Resist. (2020) 13:1299–305. doi: 10.2147/IDR.S247101
28. Qin X, Wu S, Hao M, Zhu J, Ding B, Yang Y, et al. The colonization of carbapenem-resistant Klebsiella pneumoniae: epidemiology, resistance mechanisms, and risk factors in patients admitted to intensive care units in China. J Infect Dis. (2020) 221(Suppl. 2):S206–14. doi: 10.1093/infdis/jiz622
29. Hu YY, Liu CC, Shen ZQ, Zhou HW, Cao JM, Chen S, et al. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect. (2020) 9: 1771–9. doi: 10.1080/22221751.2020.1799721
30. Han RR, Shi QY, Wu S, Yin DD, Peng MJ, Dong D, et al. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. (2020) 10:314. doi: 10.3389/fcimb.2020.00314
31. Suay-García B, Pérez-Gracia MT. Present and future of carbapenem-resistant Enterobacteriaceae (CRE) infections. Antibiotics. (2019) 8:122. doi: 10.3390/antibiotics8030122
32. Di Domenico EG, Cavallo I, Sivori F, Marchesi F, Prignano G, Pimpinelli F, et al. Biofilm production by carbapenem-resistant Klebsiella pneumoniae significantly increases the risk of death in oncological patients. Front Cell Infect Microbiol. (2020) 10:561741. doi: 10.3389/fcimb.2020.561741
33. Zhang J, Yu L, Fu Y, Zhao Y, Wang Y, Zhao J, et al. Tigecycline in combination with other antibiotics against clinical isolates of carbapenem-resistant Klebsiella pneumoniae in vitro. Ann Palliat Med. (2019) 8:622–31. doi: 10.21037/apm.2019.09.11
34. Morici P, Florio W, Rizzato C, Ghelardi E, Tavanti A, Rossolini GM, et al. Synergistic activity of synthetic N-terminal peptide of human lactoferrin in combination with various antibiotics against carbapenem-resistant Klebsiella pneumoniae strains. Eur J Clin Microbiol Infect Dis. (2017) 36:1739–48. doi: 10.1007/s10096-017-2987-7
35. Shields RK, Nguyen MH, Chen L, Press EG, Potoski BA, Marini RV, et al. Ceftazidime-avibactam is superior to other treatment regimens against carbapenem-resistant Klebsiella pneumoniae bacteremia. Antimicrob Agents Chemother. (2017) 61:e00883-17. doi: 10.1128/AAC.00883-17
36. Park Y, Choi Q, Kwon GC, Koo SH. Molecular epidemiology and mechanisms of tigecycline resistance in carbapenem-resistant Klebsiella pneumoniae isolates. J Clin Lab Anal. (2020) 34:e23506. doi: 10.1002/jcla.23506
37. Zhou J, Yang J, Hu F, Gao K, Sun J, Yang J. Clinical and molecular epidemiologic characteristics of ceftazidime/avibactam-resistant carbapenem-resistant Klebsiella pneumoniae in a neonatal intensive care unit in China. Infect Drug Resist. (2020) 13:2571–8. doi: 10.2147/IDR.S256922
38. Rojas LJ, Salim M, Cober E, Richter SS, Perez F, Salata RA, et al. Colistin resistance in carbapenem-resistant Klebsiella pneumoniae: laboratory detection and impact on mortality. Clin Infect Dis. (2017) 64:711–8. doi: 10.1093/cid/ciw805
39. Tumbarello M, Viale P, Viscoli C, Trecarichi EM, Tumietto F, Marchese A, et al. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae: importance of combination therapy. Clin Infect Dis. (2012) 55:943–50. doi: 10.1093/cid/cis588
40. Wei DD, Wan LG, Deng Q, Liu Y. Emergence of KPC-producing Klebsiella pneumoniae hypervirulent clone of capsular serotype K1 that belongs to sequence type 11 in Mainland China. Diagn Microbiol Infect Dis. (2016) 85:192–4. doi: 10.1016/j.diagmicrobio.2015.03.012
41. Andrade LN, Curiao T, Ferreira JC, Longo JM, Clímaco EC, Martinez R, et al. Dissemination of blaKPC-2 by the spread of Klebsiella pneumoniae clonal complex 258 clones (ST258, ST11, ST437) and plasmids (IncFII, IncN, IncL/M) among Enterobacteriaceae species in Brazil. Antimicrob Agents Chemother. (2011) 55:3579–83. doi: 10.1128/AAC.01783-10
42. Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. (2011) 66:307–12. doi: 10.1093/jac/dkq431
43. Adler A, Khabra E, Chmelnitsky I, Giakkoupi P, Vatopoulos A, Mathers AJ, et al. Development and validation of a multiplex PCR assay for identification of the epidemic ST-258/512 KPC-producing Klebsiella pneumoniae clone. Diagn Microbiol Infect Dis. (2014) 78:12–5. doi: 10.1016/j.diagmicrobio.2013.10.003
Keywords: carbapenem-resistant, Klebsiella pneumonia, intestinal colonization, infection, virulence
Citation: Liao W, Huang N, Zhang Y, Sun Y, Chen T, Zeng W, Chen L, Wen H, Cao J and Zhou T (2021) Comparison of Carbapenem-Resistant Klebsiella pneumoniae Strains Causing Intestinal Colonization and Extraintestinal Infections: Clinical, Virulence, and Molecular Epidemiological Characteristics. Front. Public Health 9:783124. doi: 10.3389/fpubh.2021.783124
Received: 25 September 2021; Accepted: 12 November 2021;
Published: 03 December 2021.
Edited by:
Jian Wu, Zhejiang University, ChinaReviewed by:
Fupin Hu, Huashan Hospital, Fudan University, ChinaAhmad Al Atrouni, Lebanese University, Lebanon
Copyright © 2021 Liao, Huang, Zhang, Sun, Chen, Zeng, Chen, Wen, Cao and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianming Cao, d3pjam1pbmdAMTYzLmNvbQ==; Tieli Zhou, d3l6dGxpQDE2My5jb20=
 Wenli Liao1,2
Wenli Liao1,2 Yao Sun
Yao Sun Jianming Cao
Jianming Cao Tieli Zhou
Tieli Zhou