
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Public Health , 25 November 2021
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 9 - 2021 | https://doi.org/10.3389/fpubh.2021.766871
This article is part of the Research Topic "Source-tracking”, molecular epidemiology and antigenic diversity of SARS-CoV-2 infections causing coronavirus disease 2019, COVID-19. View all 37 articles
 Dhanasekaran Sakthivel1†
Dhanasekaran Sakthivel1† David Delgado-Diaz1†
David Delgado-Diaz1† Laura McArthur2
Laura McArthur2 William Hopper1
William Hopper1 Jack S. Richards1,3,4
Jack S. Richards1,3,4 Charles A. Narh1,3,4*
Charles A. Narh1,3,4*Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) is a recently emerged and highly contagious virus that causes coronavirus disease 2019 (COVID-19). As of August 24, 2021, there were more than 212 million confirmed COVID-19 cases and nearly 4.4 million deaths reported globally. Early diagnosis and isolation of infected individuals remains one of the most effective public health interventions to control SARS-CoV-2 spread and for effective clinical management of COVID-19 cases. Currently, SARS-CoV-2 infection is diagnosed presumptively based on clinical symptoms and confirmed by detecting the viral RNA in respiratory samples using reverse transcription polymerase chain reaction (RT-PCR). Standard RT-PCR protocols are time consuming, expensive, and technically demanding, which makes them a poor choice for large scale and point-of-care screening in resource-poor settings. Recently developed isothermal nucleic acid amplification tests (iNAAT), antigen and/or serological tests are cost-effective to scale COVID-19 testing at the point-of-care (PoC) and for surveillance activities. This review discusses the development of rapid PoC molecular tools for the detection and surveillance of SARS-CoV-2 infections.
The diagnosis of active SARS-CoV-2 infection is critical in epidemiological surveillance, infection control and contact tracing, clinical management, and for monitoring the impact of interventions against the spread of the virus. Current diagnostic tests fall into three main categories: molecular tests that detect the SARS-CoV-2 RNA, antigen tests that detect the presence of specific viral antigens, and serological tests that detect anti-SARS-CoV-2 immunoglobulins (Ig). COVID-19 diagnosis criteria vary among countries (1) but in every case, detection of SARS-CoV-2 RNA by reverse transcriptase PCR (RT-PCR) is considered a confirmatory diagnosis (2). However, RT-PCR is expensive and laborious; requiring viral RNA isolation, purification and reverse transcription to complementary DNA (cDNA) before amplification using PCR. Hence, it requires skilled personnel and dedicated laboratory space, thus limiting its use in resource-limited settings. More recently, serology and antigen-based test, and isothermal nucleic acid amplification test (iNAAT) have become available for the diagnosis of COVID-19 (Table 1). These tests have acceptable sensitivity and do not require sophisticated equipment, offer rapid turnaround time within an hour and can be performed at the point-of-care (PoC). To date, several RT-PCR diagnostic kits and, to a lesser extent antigen/antibody-based detection tests, isothermal amplification tests, clustered regularly interspaced short palindromic repeats (CRISPR-) and sequencing-based detection tools have been approved by the Food and Drug Administration (FDA) via Emergency Use Authorization (EUA) for the diagnosis of COVID-19. This review discusses the molecular, serological, and antigen diagnostic tools for detecting SARS-CoV-2 infections, their potential use for PoC diagnosis of COVID-19 (Table 1).
Our understanding of immune response against SARS-CoV-2 infections has rapidly unfolded as millions of individuals have been infected. Seroconversion in infected individuals has been observed between 1 and 2 weeks post-symptom onset (3–9). Studies on the immune responses of SARS-CoV-2-infected patients have shown increased presence of follicular helper T cells, activated CD4+ and CD8+ T cells with the detection of Immunoglobulin A (IgA), IgM, and IgG against the SARS-CoV-2 spike (S), nucleocapsid (N) and envelop (E) proteins (6, 10–13). Antibodies against SARS-CoV-2 have been shown to persist at least 12 months post-infection in most individuals (14–18). The majority of SARS-CoV-2 rapid diagnostic tests detect the presence of anti-SARS-CoV-2 antibodies (IgG, and/or IgM) in plasma or serum of infected individuals (FDA.gov).
Immunoassays for COVID-19 diagnosis target the most immunogenic proteins—N and S (6, 10, 11, 19–21). In serum and plasma specimens, anti-SARS-CoV-2 antibodies could be detected as early as 2 weeks post-symptom onset (21). However, infected individuals show different antibody profiles over the course of the disease (12, 22). SARS-CoV-2-specific IgA and IgM antibodies have been detected 5 days post-symptom onset while IgG was detected 14 days post-symptom onset (12, 22), indicating early and evolving infections, respectively. The majority of the immunoassays in use are based on enzyme linked immunosorbent assay (ELISA), immunochromatography (lateral-flow) and antigen microarray (6, 10, 20–23). ELISA offers high-throughput but requires experienced technicians, a laboratory space, and several other instruments, and thus, it is not feasible for PoC diagnosis. In contrast, lateral-flow-based assays are easy to use, do not require instruments, and have been developed and deployed as PoC tools for serological and antigen-based diagnosis of COVID-19 (24, 25).
In spite of their advantages (Table 1), serological tests are limited in the diagnosis of SARS-CoV-2 infections due to their poor sensitivity to detect mild and asymptomatic infections (26). In addition, reports of individuals who remain PCR-positive after seroconversion suggest that they may still be shedding viral RNA during the convalescent stage. However, this may not necessarily indicate the presence of viable virus (3, 4, 27–30). Therefore, serological tests may be limited to identification of past but not active infections. Considering their relatively lower cost and ease of use in comparison with RT-PCR, they could be used to initially screen vulnerable populations to estimate seropositivity rates.
In contrast to antibody tests, antigen tests detect the presence of specific SARS-CoV-2 antigens in respiratory samples. Oropharyngeal, nasal and nasopharyngeal specimens are the most compatible specimen types for the majority of COVID-19 antigen and NAAT methods (2, 31). Antigen tests are relatively more affordable than RT-PCR, and considerably sensitive when used during the infectious period of the disease (32, 33). They are recommended for routine testing among at risk populations (34, 35). To date, over 20 SARS-CoV-2 antigen tests have received FDA emergency use authorization, reporting analytical sensitivities down to 30 TCID50/mL and specificities of up to 99% (36).
Several NAAT tools have been developed to detect SARS-CoV-2 infections by amplifying the viral RNA from a wide range of sample types including nasal swabs and saliva samples. While RT-PCR is currently the gold standard for the detection of SARS-CoV-2 RNA due to its high sensitivity, other methods including recombinase polymerase amplification (RPA) and loop mediated isothermal amplification (LAMP, see Figure 1) have also been used for COVID-19 diagnosis (37–39). RT-PCR is expensive, laborious and requires skilled personnel, making it unsuitable for PoC diagnosis (37–39). The accuracy and sensitivity of RT-PCR is affected by the purity of the sample and/or extracted RNA. The global shortage of RNA extraction kits has had adverse impacts on COVID-19 diagnosis and control worldwide. Though extraction-free RT-qPCR protocols have been considered as alternatives for the standard SARS-CoV-2 RT-PCR method, laboratory optimizations are often required to minimize false-negative rates (40).
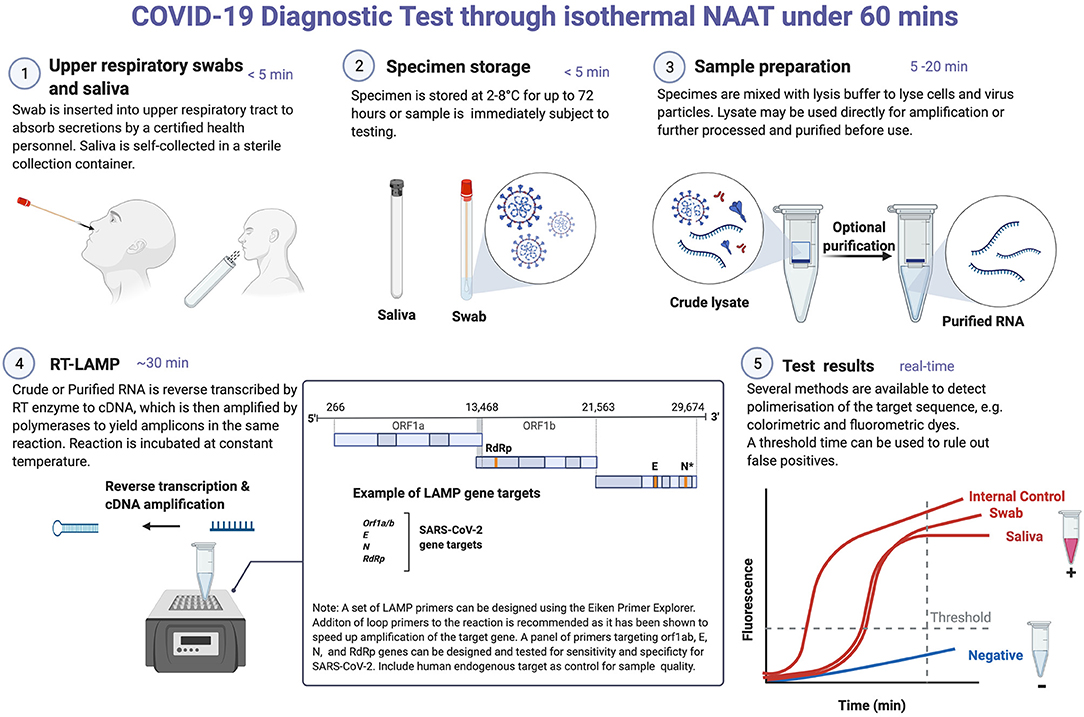
Figure 1. COVID-19 diagnostic testing through isothermal NAAT. Reverse transcriptase LAMP (RT-LAMP) detection of SARS-CoV-2 RNA in nasal swab and saliva samples. These samples can be stored in the refrigerator for 3 days prior to NAAT testing. Where testing can be done immediately, sample preparation and/or RNA extraction is performed, which may take between 5 and 20 min. RNA purification is often required for RT-PCR-based testing. This is done because contaminants in crude cell lysates could potentially reduce the polymerase activity of the reverse transcriptase and DNA polymerases used in RT-PCR. In contrast, the Bacillus stearothermophilus (Bst) DNA polymerase used in LAMP is more tolerant to inhibitors. For RT-LAMP testing, a set of four to six primers targeting any of the viral genes can be designed using online programs, e.g., Eiken Primer design software. It is recommended that a primer set targeting a human endogenous gene is included as a control for sample preparation/RNA extraction and amplification efficiency. In RT-LAMP, both cDNA synthesis (reverse transcriptase) and amplification (Bst) occur simultaneously and in the same reaction tube at a constant temperature (60–65°C). A double-stranded DNA intercalating dye can be added to detect amplicons either by colorimetry (show color change), fluorescent (for real-time detection) or both. RT-LAMP is prone to false-positive amplifications and as such any assay developed using this technique needs to be standardized for each test type. *In a NAAT-based assay, two targets on N gene were included in a single reaction to increase the test sensitivity. Created with Biorender.com.
Several reports suggest that RT-PCR positivity does not necessarily correlate with clinical infectivity since respiratory samples may contain non-viable virus, which could persist in the body for several weeks during the convalescent stage of the disease (41–44). Viable virus can be cultured from samples with low RT-PCR cycle threshold (Ct) values while samples with high Ct values are less likely to contain culturable virus (3, 45–47), suggesting that individuals with high Ct values (usually those at later stages of the disease) are less likely to spread the virus than those with low Ct values (usually those in the acute phase of the disease).
The performance of NAATs on the detection of SARS-CoV-2 RNA has been extensively reviewed elsewhere (48–52). Due to the lack of standardization in the NAAT testing algorithms including sample types and target genes, it is difficult to compare the analytical performance of the various test types (Table 1). A study by Vogels and colleagues found that test sensitivity was comparable among most of the primer-probe sets with the exception of primer sets targeting the RNA dependent RNA polymerase (RdRp-SARSr) gene segment, which resulted in lower sensitivity (53). Significant difference in sensitivity has been observed with commercial RT-PCR kits. For instance, Igloi and colleagues evaluated 13 commercial kits and reported analytical sensitivities that ranged from 3.3 to 330 viral RNA copies in the RT-PCR assays evaluated (54).
Published gene targets for the detection of SARS-CoV-2 have comparable specificity. While RT-PCR based commercial kits used by the China National Institute for Viral Disease Control and Prevention (CCDC) predominantly target the Open Reading Frame 1ab (orf1ab) and N genes (55), other commercially available RT-PCR kits target the RNA-dependent RNA polymerase (RdRP) and/or E genes (37). In other assays, multiple targets on the same gene are included in order to increase the test sensitivity, e.g., two targets on the N gene (56). The sensitivity of PCR-based detection has been improved with double strand excision of the target using the CRISPR gene-editing technique (38, 39, 57). The rising number of mutations in the viral genome, particularly in the S and orf1ab genes have raised concerns about the sensitivity of NAAT tools to detect SARS-CoV-2 including emerged variants—alpha (B.1.1.7), beta (B1.3.51), gamma (P.1), delta (B.1.617.2) and epsilon (B.1.427/B.1.429), which have been associated with high transmissibility and disease severity in many geographical regions (58, 59). It is important that NAAT diagnostic tools are routinely quality-checked to ensure that they detect all variants in circulation and meet international regulatory test performance criteria (60).
The sensitivity and performance of NAATs for accurate detection of SARS-CoV-2 relies on the specimen type and quality, and the method used for processing the sample (37, 61–65). According to the WHO guidelines, testing for SARS-CoV-2 viral RNA requires respiratory samples. Upper respiratory specimens (nasopharyngeal, nasal, and/or oropharyngeal swabs) are most suited for testing early-stage infections, especially in asymptomatic or mild cases, while lower respiratory specimens (sputum and/or endotracheal aspirate or bronchoalveolar lavage) are recommended if for patients in the post-symptomatic phase of the disease and those with severe disease (2). In addition to respiratory samples, detection of viral RNA in serum and fecal samples collected from infected patients has also been reported, in particular where respiratory specimen gave a negative test result (55, 63, 64). However, these samples provide no clear utility for accurate detection of active SARS-CoV-2 infection (66, 67).
Specimens collected from infected individuals at the pre-symptomatic phase through to the hyperinflammatory phase of COVID-19 have resulted in variable positive rates. Studies have shown that a few days prior to and during the symptomatic phase, sputum and nasopharyngeal swab samples gave higher PCR positivity compared to fecal samples. However, the opposite has been observed during the recovery phase (63, 68), demonstrating the potential utility of fecal samples for monitoring viral clearance during the recovery phase. Although a few studies have been able to recover viable virus from fecal samples and anal swabs of convalescent patients (69–71), it is important to note that the presence of viral RNA in feces may not be an indication of active infection but an indication of residual viral RNA being cleared from the body via shedding of infected epithelial cells.
Recent evidence has demonstrated the utility of sputum and saliva as specimens for detection of SARS-CoV-2 (72–76) (Figure 1). For instance, a comparison of sample positivity using quantitative RT-PCR showed that sputum samples had higher positive rates than throat and nasal swabs collected from the same patient (65). Other studies have also reported differences in test sensitivity comparing saliva and nasopharyngeal swabs (73, 76–81). Saliva has been recommended for COVID-19 diagnosis, in particular for surveillance activities. Saliva sampling is non-invasive and suitable for COVID-19 screening in vulnerable populations and in settings where swabs are in limited supply (79, 82, 83). Sputum offers comparable sensitivity to other respiratory samples for the detection of SARS-CoV-2 RNA (74, 84) but its use is limited in situations where patients are unable to expectorate enough sputum for testing (72, 74). Unless collected properly, sputum sampling poses a high risk of viral transmission. Therefore, nasal swabs are preferred over sputum for the detection of SARS-CoV-2 RNA by NAAT methods (Figure 1).
During the early stages of the pandemic, detection of SARS-CoV-2 infection was severely impacted due to the shortage of RNA extraction kits (85, 86). In certain circumstances, these shortages led to delays in diagnosis, which hampered public health control efforts. To increase accessibility to molecular diagnostic tools for COVID-19, several research laboratories developed and optimized NAAT protocols to simplify and obviate the need for RNA purification (87, 88). More cost-effective molecular tools will be needed for SARS-CoV-2 surveillance during and post-pandemic.
The global spread of SARS-CoV-2 and its associated morbidity and mortality requires cost-effective laboratory equipment and PoC diagnostic tools for screening at-risk populations. PoC tests are easy to use and could be readily deployed at healthcare centers, schools and airports, and among vulnerable populations in aged care centers. RT-PCR is generally performed in centralized Biosafety level 2 (BSL2) laboratories and require regulatory approval to undertake COVID-19 testing (37–39). The complexities and the long wait times (≥2 h) for RT-PCR test results makes it a less attractive tool for PoC diagnosis of COVID-19.
Development of a NAAT assay combining RNA extraction, cDNA synthesis and amplification in a single reaction tube, and without the need for sophisticated instruments offers huge prospects for COVID-19 diagnosis at the point-of-care (89, 90) (Figure 1). iNAATs including RPA and LAMP do not require expensive PCR equipment, tolerate crude lysates as input for amplification and can be integrated into portable isothermal instruments for PoC COVID-19 testing (91) (Figure 1). LAMP and RPA applications for SARS-CoV-2 detection have been reviewed elsewhere (89). Although iNAATs pose a higher risk of cross-contamination when compared to PCR-based diagnostic tools (Table 1), certain strategies have been shown to mitigate this problem (92–94). For instance, a pre-optimized closed-tube isothermal amplification coupled with quality control checks to eliminate false-positive and carry-over contamination would be optimum for SARS-CoV-2 testing (Figure 1). As countries scale-up efforts from control to elimination of SARS-CoV-2, cost-effective molecular tools including iNAAT that require minimal sample processing and can be integrated into portable isothermal devices for use at the point-of-care or in the field will be crucial to elimination efforts.
Laboratory testing for COVID-19 has been integral to public health efforts to control the spread of SARS-CoV-2 globally. However, the high cost and centralization of RT-PCR testing, and the long testing times from sample collection to receipt of test results could hamper SARS-CoV-2 control efforts. RT-LAMP-based testing methods overcome most of the limitations of RT-PCR and can be developed for PoC diagnosis of COVID-19. Since they are compatible with most sample types for detecting active SARS-CoV-2 infections. Thus, they could complement other low-cost diagnostic tools including RDTs and lateral-flow tests for monitoring SARS-CoV-2 transmission locally and globally. With the emergence of SARS-CoV-2 variants, routine quality control checks of NAAT diagnostic tools will be needed to ensure that they meet regulatory and test performance requirements.
CN and JR conceived and designed the review. CN, DS, and DD-D conducted the literature review and wrote majority of the manuscript. LM and JR contributed to literature review and writing. JR and WH critically revised the manuscript. All authors have read and approved the manuscript for publication.
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia (APP1161076 to JR). Burnet Institute received funding from the NHMRC Independent Research Institutes Infrastructure Support Scheme, and the Victorian State Government Operational Infrastructure Support Scheme. CN was partly supported by a COVID-19 grant (Ref. No. BSAC-COVID-64) from the British Society for Antimicrobial and Chemotherapy. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
ZiP Diagnostics is commercializing a COVID-19 point of care test. CN, DS, DD-D, WH, and JR have employment at ZiP.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Benitez MA, Velasco C, Sequeira AR, Henriquez J, Menezes FM, Paolucci F. Responses to COVID-19 in five Latin American countries. Health Policy and Technology. (2020) 9:525–59. doi: 10.1016/j.hlpt.2020.08.014
2. WHO. Diagnostic Testing for SARS-CoV-2: Interim Guidance. Available online at: https://apps.who.int/iris/bitstream/handle/10665/334254/WHO-2019-nCoV-laboratory-2020.6-eng.pdf?sequence=1&isAllowed=y (accessed April 20, 2021).
3. Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. (2020) 581:465–9. doi: 10.1038/s41586-020-2196-x
4. Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARS-CoV-2 in patients with novel coronavirus disease 2019. Clin Infect Dis. (2020) 71:2027–34. doi: 10.1093/cid/ciaa344
5. Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Mueller MA, et al. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. medRxiv [preprint]. (2020).
6. Okba NMA, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerging Infect Dis. (2020) 26:1478–88. doi: 10.3201/eid2607.200841
7. Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. (2020) 58:e00461–20. doi: 10.1128/JCM.00461-20
8. Long QX, Liu BZ, Deng HJ, Wu GC, Deng K, Chen YK, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. (2020) 26:845–8. doi: 10.1038/s41591-020-0897-1
9. Wan WY, Lim SH, Seng EH. Cross-reaction of sera from COVID-19 patients with SARS-CoV assays. Ann Acad Med Sing. (2020) 49:523–6. doi: 10.47102/annals-acadmedsg.2020120
10. Haveri A, Smura T, Kuivanen S, Osterlund P, Hepojoki J, Ikonen N, et al. Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020. Eurosurveillance. (2020) 25. doi: 10.2807/1560-7917.ES.2020.25.11.2000266
11. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2951-z
12. Guo L, Ren L, Yang S, Xiao M, Chang, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin Infect Dis. (2020) 71:778–85. doi: 10.1093/cid/ciaa310
13. Thevarajan I, Nguyen THO, Koutsakos M, Druce J, Caly L, van de Sandt CE, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. (2020) 26:453–5. doi: 10.1038/s41591-020-0819-2
14. Haveri A, Ekstrom N, Solastie A, Virta C, Osterlund P, Isosaari E, et al. Persistence of neutralizing antibodies a year after SARS-CoV-2 infection in humans. Eur J Immunol. (2021) 1–12. doi: 10.1002/eji.202149535
15. Alfano G, Fontana F, Morisi N, Giaroni F, Mori G, Guaraldi G, et al. One-year persistence of neutralizing anti-SARS-CoV-2 antibodies in dialysis patients recovered from COVID-19. Hemod Int. (2021) 25:E53–6. doi: 10.1111/hdi.12963
16. She Q, Xiao K, Yang H, Liu B, Pang X, Du J, et al. Antibodies can last for more than one year after SARS-CoV-2 infection: a follow-up study from survivors of COVID-19. Front Med. (2021) 8:967. doi: 10.3389/fmed.2021.684864
17. Wang Z, Muecksch F, Schaefer-Babajew D, Finkin S, Viant C, Gaebler C, et al. Naturally enhanced neutralizing breadth against SARS-CoV-2 one year after infection. Nature. (2021) 595:426–31. doi: 10.1038/s41586-021-03696-9
18. Sherina N, Piralla A, Du L, Wan H, Kumagai-Braesch M, Andrell J, et al. Persistence of SARS-CoV-2-specific B and T cell responses in convalescent COVID-19 patients 6-8 months after the infection. Med. (2021) 2:281–95 e4. doi: 10.1016/j.medj.2021.02.001
19. Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. (2020) 181:281–92 e6. doi: 10.1016/j.cell.2020.11.032
20. Diao B, Wen K, Chen J, Liu Y, Yuan Z, Han C, et al. Diagnosis of acute respiratory syndrome coronavirus 2 infection by detection of nucleocapsid protein. medRxiv [preprint]. (2020). doi: 10.1101/2020.03.07.20032524
21. Amanat F, Stadlbauer D, Strohmeier S, Nguyen TH, Chromikova V, McMahon M, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. (2020) 26:1033–6. doi: 10.1038/s41591-020-0913-5
22. Zhang P, Gao Q, Wang T, Ke Y, Mo F, Jia R, et al. Evaluation of recombinant nucleocapsid and spike proteins for serological diagnosis of novel coronavirus disease 2019 (COVID-19). medRxiv. (2020) 2020:20036954. doi: 10.1101/2020.03.17.20036954
23. Khan S, Nakajima R, Jain A, de Assis RR, Jasinskas A, Obiero JM, et al. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. bioRxiv [preprint]. (2020). doi: 10.1101/2020.03.24.006544
24. Wu JL, Tseng WP, Lin CH, Lee TF, Chung MY, Huang CH, et al. Four point-of-care lateral flow immunoassays for diagnosis of COVID-19 and for assessing dynamics of antibody responses to SARS-CoV-2. J Infect. (2020) 81:435–42. doi: 10.1016/j.jinf.2020.06.023
25. Michel M, Bouam A, Edouard S, Fenollar F, Di Pinto F, Mege JL, et al. Evaluating ELISA, immunofluorescence, and lateral flow assay for SARS-CoV-2 serologic assays. Front Microbiol. (2020) 11:597529. doi: 10.3389/fmicb.2020.597529
26. Liu ZL, Liu Y, Wan LG, Xiang TX, Le AP, Liu P, et al. Antibody profiles in mild and severe cases of COVID-19. Clin Chem. (2020) 66:1102–4. doi: 10.1093/clinchem/hvaa137
27. Liu WD, Chang SY, Wang JT, Tsai MJ, Hung CC, Hsu CL, et al. Prolonged virus shedding even after seroconversion in a patient with COVID-19. J Infect. (2020) 81:318–56. doi: 10.1016/j.jinf.2020.03.063
28. Jin Y, Wang M, Zuo Z, Fan C, Ye F, Cai Z, et al. Diagnostic value and dynamic variance of serum antibody in coronavirus disease 2019. Int J Infect Dis. (2020) 94:49–52. doi: 10.1016/j.ijid.2020.03.065
29. Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z, et al. Viral kinetics and antibody responses in patients with COVID-19. medRxiv [preprint]. (2020). doi: 10.1101/2020.03.24.20042382
30. Agarwal V, Venkatakrishnan A, Puranik A, Kirkup C, Lopez-Marquez A, Challener DW, et al. Long-term SARS-CoV-2 RNA shedding and its temporal association to IgG seropositivity. Cell Death Disc. (2020) 6:1–9. doi: 10.1038/s41420-020-00375-y
31. CDC. Interim Guidance for Antigen Testing for SARS-CoV-2. (2020). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html#previous (accessed April 23, 2021).
32. Nordgren J, Sharma S, Olsson H, Jamtberg M, Falkeborn T, Svensson L, et al. SARS-CoV-2 rapid antigen test: high sensitivity to detect infectious virus. J Clin Virol. (2021) 140:104846. doi: 10.1016/j.jcv.2021.104846
33. Pickering S, Batra R, Merrick B, Snell LB, Nebbia G, Douthwaite S, et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: a single-centre laboratory evaluation study. Lancet Microbe. (2021) 2:e461–71. doi: 10.1016/S2666-5247(21)00143-9
34. Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. (2021) 7:eabd5393. doi: 10.1126/sciadv.abd5393
35. Mina MJ, Parker R, Larremore DB. Rethinking Covid-19 test sensitivity - a strategy for containment. N Engl J Med. (2020) 383:e120. doi: 10.1056/NEJMp2025631
36. FDA. In Vitro Diagnostics EUAs. (2021). Available online at: https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas (accessed January 15, 2021).
37. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. (2020) 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045
38. Metsky HC, Freije CA, Kosoko-Thoroddsen T-SF, Sabeti PC, Myhrvold C. CRISPR-based COVID-19 surveillance using a genomically-comprehensive machine learning approach. bioRxiv. (2020) 2020:967026. doi: 10.1101/2020.02.26.967026
39. Yu L, Wu S, Hao X, Li X, Liu X, Ye S, et al. Rapid colorimetric detection of COVID-19 coronavirus using a reverse tran-scriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic plat-form: iLACO. medRxiv. (2020) 2020:20025874. doi: 10.1101/2020.02.20.20025874
40. Domnich A, De Pace V, Pennati BM, Caligiuri P, Varesano S, Bruzzone B, et al. Evaluation of extraction-free RT-qPCR methods for SARS-CoV-2 diagnostics. Arch Virol. (2021) 166:2825–8. doi: 10.1007/s00705-021-05165-0
41. La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. (2020) 39:1059. doi: 10.1007/s10096-020-03913-9
42. Rhee C, Kanjilal S, Baker M, Klompas M. Duration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infectivity: when is it safe to discontinue isolation? Clin Infect Dis. (2020) 72:1467–74. doi: 10.1093/cid/ciaa1249
43. van Kampen JJA, van de Vijver D, Fraaij PLA, Haagmans BL, Lamers MM, Okba N, et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19). Nat Commun. (2021) 12:267. doi: 10.1038/s41467-020-20568-4
44. Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. (2021) 2:e13–22. doi: 10.1016/S2666-5247(20)30172-5
45. Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. (2020) 71:2663–6. doi: 10.1093/cid/ciaa638
46. Young BE, Ong SWX, Ng LF, Anderson DE, Chia WN, Chia PY, et al. Viral dynamics and immune correlates of coronavirus disease 2019 (COVID-19) severity. Clin Infect Dis. (2021) 73:e2932–e2942. doi: 10.1093/cid/ciaa1280
47. Gniazdowski V, Morris CP, Wohl S, Mehoke T, Ramakrishnan S, Thielen P, et al. Repeat COVID-19 molecular testing: correlation of SARS-CoV-2 culture with molecular assays and cycle thresholds. Clin Infect Dis. (2020) 73:e860–9. doi: 10.1101/2020.08.05.20168963
48. Wang M, Chen D, Wu W, Tang H, Kan L, Zong Z, et al. Analytical performance evaluation of five RT-PCR kits for severe acute respiratory syndrome coronavirus 2. J Clin Lab Anal. (2021) 35:e23643. doi: 10.1002/jcla.23643
49. van Kasteren PB, van der Veer B, van den Brink S, Wijsman L, de Jonge J, van den Brandt A, et al. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol. (2020) 128:104412. doi: 10.1016/j.jcv.2020.104412
50. Krismawati H, Mahendra C, Hartanto A, Rokhmad MF, Sandi S, Irwanto A. Comparison study of commercial COVID-19 RT-PCR kits propose an approach to evaluate their performances. medRxiv [preprint]. (2021). doi: 10.1101/2021.01.20.21250143
51. FIND. Find Evaluation Update: Sars-Cov-2 Molecular Diagnostics. (2020). Available online at: https://www.finddx.org/covid-19-old/sarscov2-eval-molecular/ (accessed January 31, 2021).
52. Dinnes J, Deeks JJ, Berhane S, Taylor M, Adriano A, Davenport C, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. (2021) CD013705. doi: 10.1002/14651858.CD013705.pub2
53. Vogels CBF, Brito AF, Wyllie AL, Fauver JR, Ott IM, Kalinich CC, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. (2020) 5:1299–305. doi: 10.1038/s41564-020-0761-6
54. Igloi Z, Leven M, Abdel-Karem Abou-Nouar Z, Weller B, Matheeussen V, Coppens J, et al. Comparison of commercial realtime reverse transcription PCR assays for the detection of SARS-CoV-2. J Clin Virol. (2020) 129:104510. doi: 10.1016/j.jcv.2020.104510
55. Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. (2020) 323:1061–9. doi: 10.1001/jama.2020.1585
56. CDC. CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Diagnostic Panel. (2020). Available online at: https://www.fda.gov/media/134922/download (accessed April 29, 2021).
57. Guo L, Sun X, Wang X, Liang C, Jiang H, Gao Q, et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discovery. (2020) 6:1–4. doi: 10.1038/s41421-020-0174-y
58. McCallum M, Bassi J, De Marco A, Chen A, Walls AC, Di Iulio J, et al. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. (2021) 373:648–54. doi: 10.1126/science.abi7994
59. Singh J, Rahman SA, Ehtesham NZ, Hira S, Hasnain SE. SARS-CoV-2 variants of concern are emerging in India. Nat Med. (2021) 27:1131–3. doi: 10.1038/s41591-021-01397-4
60. FDA. Genetic Variants of SARS-CoV-2 May Lead to False Negative Results With Molecular Tests for Detection of SARS-CoV-2-Letter to Clinical Laboratory Staff and Health Care Providers. (2021). Available online at: https://www.fda.gov/medical-devices/letters-health-care-providers/genetic-variants-sars-cov-2-may-lead-false-negative-results-molecular-tests-detection-sars-cov-2 (accessed August 05, 2021).
61. Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, Chung YS, et al. Viral load kinetics of SARS-CoV-2 infection in first two patients in Korea. J Korean Med Sci. (2020) 35:e86. doi: 10.3346/jkms.2020.35.e86
62. Qiu H, Wu J, Hong L, Luo Y, Song Q, Chen D. Clinical and epidemiological features of 36 children with coronavirus disease 2019 (COVID-19) in Zhejiang, China: an observational cohort study. Lancet Infect Dis. (2020) 20:689–96. doi: 10.1016/S1473-3099(20)30198-5
63. Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. (2020) 5:434–5. doi: 10.1016/S2468-1253(20)30083-2
64. Xie C, Jiang L, Huang G, Pu H, Gong B, Lin H, et al. Comparison of different samples for 2019 novel coronavirus detection by nucleic acid amplification tests. Int J Infect Dis. (2020) 93:264–7. doi: 10.1016/j.ijid.2020.02.050
65. Yu F, Yan L, Wang N, Yang S, Wang L, Tang Y, et al. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. (2020) 71:793–8. doi: 10.1093/cid/ciaa345
66. Mesoraca A, Margiotti K, Viola A, Cima A, Sparacino D, Giorlandino C. Evaluation of SARS-CoV-2 viral RNA in fecal samples. Virol J. (2020) 17:86. doi: 10.1186/s12985-020-01359-1
67. Szymczak WA, Goldstein DY, Orner EP, Fecher RA, Yokoda RT, Skalina KA, et al. Utility of stool PCR for the diagnosis of COVID-19: comparison of two commercial platforms. J Clin Microbiol. (2020) 58:e01369–20. doi: 10.1128/JCM.01369-20
68. Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. (2020) 26:502–5. doi: 10.1038/s41591-020-0817-4
69. Xiao F, Sun J, Xu Y, Li F, Huang X, Li H, et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. (2020) 26:1920–2. doi: 10.3201/eid2608.200681
70. Yuan C, Zhu H, Yang Y, Cai X, Xiang F, Wu H, et al. Viral loads in throat and anal swabs in children infected with SARS-CoV-2. Emerg Microbes Infect. (2020) 9:1233–7. doi: 10.1080/22221751.2020.1771219
71. Yao H, Lu X, Chen Q, Xu K, Chen Y, Cheng M, et al. Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Dis. (2020) 6:76. doi: 10.1038/s41421-020-00226-1
72. Han H, Luo Q, Mo F, Long L, Zheng W. SARS-CoV-2 RNA more readily detected in induced sputum than in throat swabs of convalescent COVID-19 patients. Lancet Infect Dis. (2020) 20:655–6. doi: 10.1016/S1473-3099(20)30174-2
73. Mohammadi A, Esmaeilzadeh E, Li Y, Bosch R, Li J. SARS-CoV-2 detection in different respiratory sites: a systematic review and meta-analysis. EBio Med. (2020) 59:102903. doi: 10.1016/j.ebiom.2020.102903
74. Wang K, Zhang X, Sun J, Ye J, Wang F, Hua J, et al. Differences of severe acute respiratory syndrome coronavirus 2 shedding duration in sputum and nasopharyngeal swab specimens among adult inpatients with coronavirus disease 2019. Chest. (2020) 158:1876–84. doi: 10.1016/j.chest.2020.06.015
75. Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. (2020) 323:1843–4. doi: 10.1001/jama.2020.3786
76. Czumbel LM, Kiss S, Farkas N, Mandel I, Hegyi A, Nagy A, et al. Saliva as a candidate for COVID-19 diagnostic testing: a meta-analysis. Front Med. (2020) 7:465. doi: 10.3389/fmed.2020.00465
77. Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N Engl J Med. (2020) 383:1283–6. doi: 10.1056/NEJMc2016359
78. Becker D, Sandoval E, Amin A, De Hoff P, Leonetti N, Lim YW, et al. Saliva is less sensitive than nasopharyngeal swabs for COVID-19 detection in the community setting. medRxiv [preprint]. (2020). doi: 10.1101/2020.05.11.20092338
79. Williams E, Bond K, Zhang B, Putland M, Williamson DA. Saliva as a non-invasive specimen for detection of SARS-CoV-2. J Clin Microbiol. (2020) 58:e00776–20. doi: 10.1128/JCM.00776-20
80. Byrne RL, Kay GA, Kontogianni K, Aljayyoussi G, Brown L, Collins AM, et al. Saliva alternative to upper respiratory swabs for SARS-CoV-2 diagnosis. Emerg Infect Dis. (2020) 26:2770–1. doi: 10.3201/eid2611.203283
81. Jamal AJ, Mohammad M, Coomes E, Powis J, Li A, Paterson A, et al. Sensitivity of Nasopharyngeal Swabs and Saliva for the Detection of Severe Acute Respiratory Syndrome Coronavirus 2. Clin Infect Dis. (2021) 72:1064–6. doi: 10.1093/cid/ciaa848
82. Patriquin G, Davis I, Heinstein C, MacDonald J, Hatchette TF, LeBlanc JJ. Exploring alternative swabs for use in SARS-CoV-2 detection from the oropharynx and anterior nares. J Virol Meth. (2020) 285:113948. doi: 10.1016/j.jviromet.2020.113948
83. Hanson KE, Barker AP, Hillyard DR, Gilmore N, Barrett JW, Orlandi RR, et al. Self-collected anterior nasal and saliva specimens versus health care worker-collected nasopharyngeal swabs for the molecular detection of SARS-CoV-2. J Clin Microbiol. (2020) 58:e01824–20. doi: 10.1128/JCM.01824-20
84. Levican-Asenjo JE, Almonacid LI, Valenzuela G, Garcia T, Rojas L, Serrano E, et al. Viral shedding dynamics reveals sputum as a reliable and cost-saving specimen for SARS-CoV-2 diagnosis within the first 10 days since symptom onset: a prospective cohort study. medRxiv [preprint]. (2020). doi: 10.1101/2020.08.30.20183889
85. Fomsgaard AS, Rosenstierne MW. An alternative workflow for molecular detection of SARS-CoV-2 - escape from the NA extraction kit-shortage, copenhagen, denmark, March 2020. Eurosurveillance. (2020) 25:2000398. doi: 10.2807/1560-7917.ES.2020.25.14.2000398
86. Chu AW, Chan WM, Ip JD, Yip CC, Chan JF, Yuen KY, et al. Evaluation of simple nucleic acid extraction methods for the detection of SARS-CoV-2 in nasopharyngeal and saliva specimens during global shortage of extraction kits. J Clin Virol. (2020) 129:104519. doi: 10.1016/j.jcv.2020.104519
87. Mancini F, Barbanti F, Scaturro M, Errico G, Iacobino A, Bella A, et al. Laboratory management for SARS-CoV-2 detection: a user-friendly combination of the heat treatment approach and rt-Real-time PCR testing. Emerg Microbes Infect. (2020) 9:1393–6. doi: 10.1080/22221751.2020.1775500
88. Ulloa S, Bravo C, Parra B, Ramirez E, Acevedo A, Fasce R, et al. A simple method for SARS-CoV-2 detection by rRT-PCR without the use of a commercial RNA extraction kit. J Virol Meth. (2020) 285:113960. doi: 10.1016/j.jviromet.2020.113960
89. James AS, Alawneh JI. COVID-19 infection diagnosis: potential impact of isothermal amplification technology to reduce community transmission of SARS-CoV-2. Diagnostics. (2020) 10:399. doi: 10.3390/diagnostics10060399
90. Collier DA, Assennato SM, Warne B, Sithole N, Sharrocks K, Ritchie A, et al. Point of care nucleic acid testing for SARS-CoV-2 in hospitalized patients: a clinical validation trial and implementation study. Cell Rep Med. (2020) 1:100062. doi: 10.1016/j.xcrm.2020.100062
91. Österdahl MF, Lee KA, Lochlainn MN, Wilson S, Douthwaite S, Horsfall R, et al. Detecting SARS-CoV-2 at point of care: preliminary data comparing loop-mediated isothermal amplification (LAMP) to polymerase chain reaction (PCR). BMC Infect Dis. (2020) 20:1–8. doi: 10.1186/s12879-020-05484-8
92. Karthik K, Rathore R, Thomas P, Arun TR, Viswas KN, Dhama K, et al. New closed tube loop mediated isothermal amplification assay for prevention of product cross-contamination. MethodsX. (2014) 1:137–43. doi: 10.1016/j.mex.2014.08.009
93. Bao Y, Jiang Y, Xiong E, Tian T, Zhang Z, Lv J, et al. CUT-LAMP: contamination-free loop-mediated isothermal amplification based on the CRISPR/Cas9 cleavage. ACS Sensors. (2020) 5:1082–91. doi: 10.1021/acssensors.0c00034
94. Kellner MJ, Ross JJ, Schnabl J, Dekens MPS, Heinen R, Grishkovskaya I, et al. A rapid, highly sensitive and open-access SARS-CoV-2 detection assay for laboratory and home testing. bioRxiv. (2020) 2020:166397. doi: 10.1101/2020.06.23.166397
95. Lalli MA, Langmade JS, Chen X, Fronick CC, Sawyer CS, Burcea LC, et al. Rapid and extraction-free detection of SARS-CoV-2 from saliva by colorimetric reverse-transcription loop-mediated isothermal amplification. Clin Chem. (2020) 67:415–24. doi: 10.1093/clinchem/hvaa267
96. Brown JR, Atkinson L, Shah D, Harris K. Validation of an extraction-free RT-PCR protocol for detection of SARS-CoV2 RNA. medRxiv [preprint]. (2020). doi: 10.1101/2020.04.29.20085910
97. Aminian A, Safari S, Razeghian-Jahromi A, Ghorbani M, Delaney CP. COVID-19 outbreak and surgical practice: unexpected fatality in perioperative period. Ann Surgery. (2020) 272:e27–e9. doi: 10.1097/SLA.0000000000003925
98. Merindol N, Pépin G, Marchand C, Rheault M, Peterson C, Poirier A, et al. SARS-CoV-2 detection by direct rRT-PCR without RNA extraction. J Clin Virol. (2020) 128:104423. doi: 10.1016/j.jcv.2020.104423
99. Wee SK, Sivalingam SP, Yap EPH. Rapid direct nucleic acid amplification test without RNA extraction for SARS-CoV-2 using a portable PCR thermocycler. Genes. (2020) 11:664. doi: 10.3390/genes11060664
100. Rödel J, Egerer R, Suleyman A, Sommer-Schmid B, Baier M, Henke A, et al. Use of the variplex™ SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J Clin Virol. (2020) 132:104616. doi: 10.1016/j.jcv.2020.104616
101. Rabe BA, Cepko C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc Natl Acad Sci USA. (2020) 117:24450–8. doi: 10.1073/pnas.2011221117
102. Srivatsan S, Heidl S, Pfau B, Martin BK, Han PD, Zhong W, et al. SwabExpress: An end-to-end protocol for extraction-free covid-19 testing. Clin Chem. (2021). doi: 10.1093/clinchem/hvab132. [Epub ahead of print].
103. Wei S, Kohl E, Djandji A, Morgan S, Whittier S, Mansukhani M, et al. Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction. Sci Rep. (2021) 11:1–6. doi: 10.1038/s41598-021-81487-y
104. Grant PR, Turner MA, Shin GY, Nastouli E, Levett LJ. Extraction-free COVID-19 (SARS-CoV-2) diagnosis by RT-PCR to increase capacity for national testing programmes during a pandemic. bioRxiv [preprint]. (2020). doi: 10.1101/2020.04.06.028316
105. Smyrlaki I, Ekman M, Lentini A, de Sousa NR, Papanicolaou N, Vondracek M, et al. Massive and rapid COVID-19 testing is feasible by extraction-free SARS-CoV-2 RT-PCR. Nat Commun. (2020) 11:1–12. doi: 10.1038/s41467-020-18611-5
106. Lim B, Ratcliff J, Nawrot DA, Yu Y, Sanghani HR, Hsu CC, et al. Clinical validation of optimised RT-LAMP for the diagnosis of SARS-CoV-2 infection. Sci Rep. (2021) 11:16193. doi: 10.1038/s41598-021-95607-1
107. Graham TGW, Dugast-Darzacq C, Dailey GM, Nguyenla XH, Van Dis E, Esbin MN, et al. Open-source RNA extraction and RT-qPCR methods for SARS-CoV-2 detection. PLoS ONE. (2021) 16:e0246647. doi: 10.1371/journal.pone.0246647
108. Thi VLD, Herbst K, Boerner K, Meurer M, Kremer LP, Kirrmaier D, et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci Transl Med. (2020) 12:eabc7075. doi: 10.1126/scitranslmed.abc7075
109. Kriegova E, Fillerova R, Kvapil P. Direct-RT-qPCR detection of SARS-CoV-2 without RNA extraction as part of a COVID-19 testing strategy: from sample to result in one hour. Diagnostics. (2020) 10:605. doi: 10.3390/diagnostics10080605
110. Ganguli A, Mostafa A, Berger J, Lim J, Araud E, Baek J, et al. Reverse transcription loop-mediated isothermal amplification assay for ultrasensitive detection of SARS-CoV-2 in saliva and viral transport medium clinical samples. Anal Chem. (2021) 93:7797–807. doi: 10.1021/acs.analchem.0c05170
111. EC. European Commission COVID-19 in vitro Diagnostic Devices and Test Methods Database. (2021). Available online at: https://covid-19-diagnostics.jrc.ec.europa.eu/ (accessed October 06, 2021).
112. Lee JYH, Best N, McAuley J, Porter JL, Seemann T, Schultz MB, et al. Validation of a single-step, single-tube reverse transcription loop-mediated isothermal amplification assay for rapid detection of SARS-CoV-2 RNA. J Med Microbiol. (2020) 69:1169–78. doi: 10.1099/jmm.0.001238
113. Ganguli A, Mostafa A, Berger J, Aydin MY, Sun F, de Ramirez SAS, et al. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci USA. (2020) 117:22727–35. doi: 10.1073/pnas.2014739117
114. Chen X, Zhou Q, Li S, Yan H, Chang B, Wang Y, et al. Rapid and visual detection of SARS-CoV-2 using multiplex reverse transcription loop-mediated isothermal amplification linked with gold nanoparticle-based lateral flow biosensor. Front Cell Infect Microbiol. (2021) 656:581239. doi: 10.3389/fcimb.2021.581239
115. Karthik K, Aravindh Babu RP, Dhama K, Chitra MA, Kalaiselvi G, Alagesan Senthilkumar TM, et al. Biosafety concerns during the collection, transportation, and processing of COVID-19 samples for diagnosis. Arch Med Res. (2020) 51:623–30. doi: 10.1016/j.arcmed.2020.08.007
Keywords: COVID-19, SARS-CoV-2, point-of-care, diagnostics, isothermal amplification (LAMP), sample types, surveillance
Citation: Sakthivel D, Delgado-Diaz D, McArthur L, Hopper W, Richards JS and Narh CA (2021) Point-of-Care Diagnostic Tools for Surveillance of SARS-CoV-2 Infections. Front. Public Health 9:766871. doi: 10.3389/fpubh.2021.766871
Received: 30 August 2021; Accepted: 29 October 2021;
Published: 25 November 2021.
Edited by:
Zhongheng Zhang, Sir Run Run Shaw Hospital, ChinaReviewed by:
José Eduardo Levi, University of São Paulo, BrazilCopyright © 2021 Sakthivel, Delgado-Diaz, McArthur, Hopper, Richards and Narh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charles A. Narh, Y2hhcmxlcy5uYXJoQGJ1cm5ldC5lZHUuYXU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.