- Department of Biology, Stanford University, Stanford, CA, United States
Coronaviruses cause respiratory and digestive diseases in vertebrates. The recent pandemic, caused by the novel severe acute respiratory syndrome (SARS) coronavirus 2, is taking a heavy toll on society and planetary health, and illustrates the threat emerging coronaviruses can pose to the well-being of humans and other animals. Coronaviruses are constantly evolving, crossing host species barriers, and expanding their host range. In the last few decades, several novel coronaviruses have emerged in humans and domestic animals. Novel coronaviruses have also been discovered in captive wildlife or wild populations, raising conservation concerns. The evolution and emergence of novel viruses is enabled by frequent cross-species transmission. It is thus crucial to determine emerging coronaviruses' potential for infecting different host species, and to identify the circumstances under which cross-species transmission occurs in order to mitigate the rate of disease emergence. Here, I review (broadly across several mammalian host species) up-to-date knowledge of host range and circumstances concerning reported cross-species transmission events of emerging coronaviruses in humans and common domestic mammals. All of these coronaviruses had similar host ranges, were closely related (indicative of rapid diversification and spread), and their emergence was likely associated with high-host-density environments facilitating multi-species interactions (e.g., shelters, farms, and markets) and the health or well-being of animals as end-and/or intermediate spillover hosts. Further research is needed to identify mechanisms of the cross-species transmission events that have ultimately led to a surge of emerging coronaviruses in multiple species in a relatively short period of time in a world undergoing rapid environmental change.
Introduction
Coronaviruses (CoVs) cause respiratory and digestive diseases in humans and other animals, and are responsible for several emerging diseases. The severe acute respiratory syndrome (SARS) outbreak in 2002–2003 resulted in 8,422 human cases and 916 deaths in 33 countries (1). In 2012, Middle East respiratory syndrome (MERS) emerged, and over time has resulted in over 2,500 human cases and 866 deaths in 27 countries (2, 3). As of mid-2021, the novel coronavirus disease 2019 (COVID-19) pandemic has resulted in 4.2 million human deaths and 196.2 million cases in 221 countries and territories (4). Other animals have also been affected by these and other emerging coronaviruses, all of which resulted from cross-species transmission, and demonstrate the serious threat coronaviruses can pose to humans and other animals globally.
Named after their crown-shaped spike surface proteins, coronaviruses are enveloped, positive-sense single-stranded RNA viruses that belong to the family Coronaviridae, subfamily Orthocoronavirinae (5, 6). They split into four genera: Alphacoronavirus, Betacoronavirus, Deltacoronavirus, and Gammacoronavirus (5). The first two genera infect mammals primarily, whereas Gammacoronaviruses infect birds, and Deltacoronaviruses infect both mammals and birds (7). Coronaviruses further split into species; however, they exist as quasispecies due to the rapid evolution driven by their high mutation rates and homologous RNA recombination (8). Coronaviruses have the largest genomes (26.4–31.7 kb) of all known RNA viruses; thus, their genomes are especially prone to accumulation of mutations and recombined segments over time, which contributes to their diverse host range and potential for disease emergence (9).
Bats are considered reservoirs for most Alpha- and Betacoronaviruses, while wild birds are probable reservoirs for Gamma- and Deltacoronaviruses (10). Coronavirus spillover from reservoirs to other species, and subsequent cross-species transmission, is primarily mediated by recombination in the receptor-binding domain (RBD) of the spike protein (S) gene (11). The receptor-binding domain enables coronaviruses to infect hosts by binding to a host receptor, e.g., angiotensin-converting enzyme 2 (ACE2) in the case of SARS coronaviruses, for cell entry (7, 12, 13). Although research has revealed reservoirs and molecular mechanisms enabling cross-species transmission, and that viral evolution is facilitated by frequent cross-species transmission events (14), less is known about the environments favoring emerging coronavirus evolution in non-reservoir hosts.
Agriculture and industrialization expanded the global abundance of humans and domestic mammals (i.e., livestock and pets). Today, their combined biomass makes up 96% of all mammalian biomass on Earth (15). This may be the primary reason for disease emergence in humans and other animals (16). To help curb coronavirus disease emergence, it is important to identify current host ranges of existing coronaviruses in humans and domestic animals, and the circumstances associated with their cross-species transmission.
This review provides an updated succinct summary of known host ranges and cross-species transmissions of recently emerged coronaviruses in humans and domestic mammals. Moreover, I discuss commonalities among the ecological circumstances related to spillover and emergence of several coronaviruses in various mammalian hosts, and how these may inform One Health interventions for preventing disease emergence.
Emerging Human Coronaviruses
There are seven known human coronaviruses: the Betacoronaviruses SARS-CoV-1, MERS-CoV, and SARS-CoV-2, which caused SARS, MERS, and COVID-19, respectively, and the Alphacoronaviruses NL63 and 229E and Betacoronaviruses OC43 and HKU1, which cause the common cold in humans (17). The latter four may not be labeled as recently emerging coronaviruses, although they have spilled over at some point in the past. Bats are considered reservoirs for NL63 and 229E, whereas rodents are putative reservoirs for OC43 and HKU1 (17–19). NL63 possibly emerged several hundred years ago from recombination between ancestors to 229E in hipposiderid bats and coronaviruses circulating in African trident bats (19, 20). Based on phylogenetic analyses, cattle and camelids have been identified as probable intermediate spillover hosts for OC43 and 229E emergence one and two centuries ago, respectively (17, 18, 20). The bovine-to-human spillover that led to OC43 emergence likely coincided with a pandemic in 1890 (17, 21, 22). Indeed, OC43 and bovine coronavirus share 96% global nucleotide identity (23). Finally, extant lineages of HKU1 trace their most recent common ancestor to the 1950s, when it possibly spilled over from rodents (20).
Next, this section covers plausible spillover events—from reservoirs to humans via potential intermediate host species—that generated the recent SARS-CoV-1, MERS-CoV, and SARS-CoV-2, and their cross-species transmission potential.
SARS-CoV-1
Severe acute respiratory syndrome emerged in Guangdong, China, and caused the devastating 2000–2003 outbreak in several countries (1). Successful efforts curbed the epidemic: only a few cases occurred in late 2003 and early 2004 (24). There have been no known SARS-CoV-1-related cases since.
Based on genetic and epidemiologic investigations, the first SARS-CoV-1-infected individuals likely contracted the virus from masked palm civets or other wildlife in wet markets (24–27). Civet isolates revealed ongoing adaptation, suggesting that they were not reservoir hosts, but intermediate spillover hosts that contracted the virus from horseshoe bats (26–30). Substantial evidence confirms bats as SARS reservoirs (26, 28, 29, 31, 32).
Wildlife samples from a market in Shenzhen revealed that SARS-CoV-1 shared 99.8% nucleotide identity with isolates from civets and a raccoon dog, and that a ferret badger had seroconverted against SARS-CoV-1 (24, 26). Initial human cases reported direct or indirect contact with these animals via handling, killing, meat serving, or residing near wet markets (33). Surveys showed that animal (especially civet) traders, although asymptomatic, had disproportionately high seroconversion against SARS-CoV-1, suggesting they have been exposed to SARS-CoV-related viruses for several years before the SARS epidemic (24, 26). Intermediate spillover hosts were not necessarily required for the evolution of SARS-CoV-1, since a bat SARS-like coronavirus is able to bind to ACE2 in humans and civets for cell entry (34). Nonetheless, civets may have amplified the virus and brought it closer to humans (35).
Additional mammals are susceptible to SARS-CoV-1 infection. Cats, ferrets, guinea pigs, golden hamsters, common marmosets, grivets, and cynomolgus and rhesus macaques can be infected under experimental inoculation, seroconvert, and display similar pathological signs as humans, and the monkeys and guinea pigs usually display mild clinical signs, while cats and golden hamsters show no clinical signs (36–44). In two studies, inoculated ferrets only exhibited signs of lethargy (36, 37). Furthermore, cats and ferrets can shed SARS-CoV-1 and transmit the virus within each species (36). Cats have also been naturally infected by SARS-CoV-1 in an apartment block where residents had SARS, suggesting possible human-to-cat transmission (36). Although swine are susceptible to SARS-CoV-1 both experimentally and naturally, viral replication in (and shedding from) swine is poor (45–47). Mice and poultry are not susceptible to SARS-CoV-1 infection (45, 48, 49). Thus, SARS-CoV-1 was not uniquely adapted to humans, yet likely restricted to mammals.
MERS-CoV
Middle East respiratory syndrome cases are still being reported since it became endemic in the Arabian peninsula. Middle East respiratory syndrome does sporadically spread to other parts of the world, although with limited human-to-human transmission (50, 51). Most outbreaks originate from independent spillover events.
Bats are putative reservoirs for MERS, while dromedary camels and other camelids are intermediate spillover hosts (52–54). Although rare, camel-to-human transmission does occur (51, 55). Infected camels shed MERS-CoV via bodily fluids, especially nasal secretions, and exhibit sneezing, coughing, fever, and loss of appetite (56, 57). Camel care-takers or consumers of camel products are at risk of contracting MERS-CoV (51). People in direct or indirect contact with camels have disproportionately high seroconversion against MERS-CoV (58). Surveys from 2010 to 2013 in Saudi Arabia show that 90% of 310 and 74% of 203 camels were MERS-CoV seropositive (59, 60). Historical seropositive samples and phylogenetic analyses suggest that MERS-like coronaviruses have been circulating in camels for at least a few decades before MERS recently emerged in humans (52, 60–63). Camel markets with both live and dead animals are believed to serve as hotspots for MERS-CoV transmission (64).
MERS-CoV may infect additional species. Rhesus macaques, common marmosets, swine, llamas, rabbits, and alpacas have been infected experimentally, and the monkeys developed mild-to-moderate and moderate-to-severe disease, respectively, swine and llamas displayed rhinorrhea, while rabbits and alpacas showed no clinical signs, although alpacas shed MERS-CoV and transmitted it within its species (65–68). A virological survey found MERS-CoV in sheep, goats, donkeys, and a cow, but not in buffaloes, mules, or horses (69). A serological study confirms that equids might not be susceptible to MERS-CoV infection, although in vitro inoculation suggests otherwise (70). However, in an experimental inoculation study, sheep and horses did not show evidence of viral replication or seroconversion (68). Mice, golden hamsters, ferrets, and poultry are not considered susceptible to MERS-CoV infection, mainly because of their low host receptor homology with that of the MERS-CoV-susceptible species (67, 71).
SARS-CoV-2
The current COVID-19 pandemic was initially reported in Wuhan, China in 2019 (72, 73), although the origin of its pathogen, SARS-CoV-2, is still unclear. Its ancestor probably originated in bats, since SARS-CoV-2 is most closely related to the 2013 and 2019 isolates from horseshoe bats in Yunnan, China at the genome level, although not at the RBD level, suggesting neither might bind to human ACE2, and are thus not immediate ancestors of SARS-CoV-2 (72, 74, 75).
Conversely, isolates (pangolin-CoVs) from smuggled and diseased pangolins in Guangdong (2018–2019) are closely related to SARS-CoV-2 in the RBD region (76–80). Molecular binding simulations show that S proteins of SARS-CoV-2 and pangolin-CoVs can potentially recognize ACE2 in both humans and pangolins, suggesting possible pangolin-to-human spillover (76, 77). However, because pangolin-CoVs (including strains from Guangxi) are not the closest relatives to SARS-CoV-2 at the genome level, they are likely not direct ancestors of SARS-CoV-2 (76, 78, 79). Nevertheless, a 2019 pangolin-CoV isolate from Guangdong displayed high genome-wide similarity with both SARS-CoV-2 and SARS-CoV-2's closest relative (from bats), suggesting SARS-CoV-2 may have originated from recombination among coronaviruses present in bats and other wildlife (76, 77, 79, 81).
Like SARS-CoV-1, SARS-CoV-2 infects species with high ACE2 homology. Cats, ferrets, golden hamsters, tree shrews, common marmosets, grivets, and cynomolgus and rhesus macaques have been infected with SARS-CoV-2 experimentally, shed the virus, and displayed similar or milder clinical and pathological signs as humans, although cats may not show signs of disease (82–91). Conversely, dogs have low susceptibility to SARS-CoV-2, and show lack of clinical signs or dog-to-dog transmission, possibly due to their low levels of ACE2 in the respiratory tract (82, 91–93). Yet, cat-to-cat, ferret-to-ferret, hamster-to-hamster, and bat-to-bat transmission of SARS-CoV-2 have been confirmed experimentally (82, 90, 91, 94). However, mice, swine, and poultry are not susceptible to SARS-CoV-2 infection (49, 71, 82).
Accumulating evidence supports naturally occurring human-to-cat SARS-CoV-2 transmission, such as multiple reports worldwide of SARS-CoV-2-positive cats from confirmed or suspected SARS-CoV-2-positive owners (95). Natural human-to-dog transmission may be possible, as was confirmed by seroconversion and SARS-CoV-2 presence in two out of 15 dogs in close contact with COVID-19 patients, where the viral sequences from each dog-and-owner pair were identical (92). Serological and virological surveys, conducted several months after the pandemic started, indicate that SARS-CoV-2 prevalence is much lower in pet and street cats and dogs than in humans, even if pet owners had suspected or confirmed SARS-CoV-2 infection (96–100). Thus, cats and dogs can get infected under natural conditions, but rarely. However, certain environments might amplify natural infections and cross-species transmission. Human-to-mink, mink-to-mink, and mink-to-human transmission of SARS-CoV-2 have occurred on fur farms in several countries (95, 101–104). SARS-CoV-2 has also been transmitted to tigers, lions, and gorillas in zoos, raising concern for wildlife conservation (105).
Apart from the mink farm outbreaks, evidence so far suggests limited SARS-CoV-2 maintenance in domestic mammals or risk for secondary zoonoses (104). However, the panzootic potential of SARS-CoV-2 necessitates expanding veterinary surveillance (104, 106), especially if domestic and/or wild animals were to maintain SARS-CoV-2 as the human population undergoes vaccination, making COVID-19 control more difficult.
Emerging Coronaviruses in Domestic Mammals
Since the advent of agriculture (~8,000 BC), several spillover events have led to the emergence of novel pathogens in humans and domesticated animals (16). Genetic analyses place the common ancestor to all known coronaviruses at around 8,000 BC, and those of each genus at around 2,400–3,300 BC (10). Like humans, domestic mammals have been experiencing an increasing rate of novel coronavirus emergence, especially within the last century.
Bovine coronavirus (BCoV) likely emerged from rodent-CoVs around 1400 AD (17, 107). Bovine coronavirus is transmitted via the fecal–oral route, causing bloody diarrhea and respiratory infections in cattle (108–110). Bovine coronavirus-like viruses have also been detected in other domestic and wild ruminants (108). Bovine coronavirus can infect dogs experimentally, although subclinically (111). Turkeys show clinical signs of enteritis when infected with BCoV experimentally, but chickens are not susceptible (112). Equine-CoV, discovered in 1999, plausibly also descended from BCoV and causes enteritis in horses (113–115).
There are two dog coronaviruses: an Alphacoronavirus called canine enteric coronavirus (CCoV), transmitted fecal-orally, with serotypes CCoV-I and CCoV-II, and a Betacoronavirus called canine respiratory coronavirus (CRCoV), which causes kennel cough (116). Canine respiratory coronavirus was discovered in 2003 from a kennel outbreak (117). It was later also detected in samples from 1996 (118). It is closely related to BCoV and OC43, and genetic analyses suggest that CRCoV arose from a recent host-species shift of BCoV from bovine to canine hosts (117, 119).
Canine enteric coronavirus was first isolated from an outbreak in military dogs in 1971 (116). Initially, CCoV infections were believed to be restricted to the enteric tract causing mild diarrheal disease (120), but an increasing number of lethal pantropic infections suggests that CCoV is responsible for an emerging infectious disease in canines (116). There are three proposed subtypes of CCoV-II: original CCoV-IIa, recombinant CCoV-IIb, and CCoV-IIc (116). The two biotypes of CCoV-IIa have different tissue tropism and pathogenicity: “classical” CCoV-IIa is restricted to the small intestine causing enteritis, but the emerging “pantropic” CCoV-IIa causes leukopenia and is often fatal (116, 121). In 2019, an Asian pantropic CCoV-IIa strain was also isolated from a wolf in Italy (122), suggesting spillover to wildlife of imported strains (123). Cats and swine are also susceptible to CCoV (124–126).
There are six porcine coronaviruses: four Alphacoronaviruses, transmissible gastroenteritis virus (TGEV), porcine respiratory coronavirus (PRCoV), porcine epidemic diarrhea virus (PEDV), and swine acute diarrhea syndrome coronavirus (SADS-CoV), one Betacoronavirus, porcine haemagglutinating encephalomyelitis virus (PHEV), and one Deltacoronavirus, porcine deltacoronavirus (PDCoV) (127). Transmissible gastroenteritis virus, PEDV, SADS-CoV, and PDCoV cause severe enteritis that are fatal in piglets, PHEV causes digestive and/or neurological disease, and PRCoV causes mild respiratory disease (127).
Transmissible gastroenteritis virus, discovered in 1946 (128), likely emerged from CCoV-II (129), and its less virulent descendent PRCoV was identified in 1984 (130). Porcine haemagglutinating encephalomyelitis virus, first described in 1957, likely descended from BCoV (127). Porcine epidemic diarrhea virus emerged in the 1970s in Europe and Asia, likely from bat-CoVs, and was introduced in North America in 2013 after a new PEDV strain emerged in China in 2010 (131–134). A serological study indicates that PEDV subsequently spilled over from domestic to feral swine populations in the US (135). Porcine deltacoronavirus was first detected in swine samples from 2009 in Hong Kong (10, 132). In 2014, PDCoV caused the first-reported outbreaks in USA and South Korea (136, 137). It was proposed that the virus' ancestor originated from recombination between sparrow-CoV and bulbul-CoV (138). Porcine deltacoronavirus is most closely related to Deltacoronaviruses sampled from Asian leopard cats and ferret badgers in Guangdong and Guangxi markets (the first documented cases of Deltacoronaviruses in mammals) (139), suggesting that these species could have acted as intermediates for interspecies PDCoV spillover (140). In 2016, SADS outbreaks emerged in Guangdong with evidence strongly suggesting bat-to-swine spillover origin (141).
There is one coronavirus that primarily infects cats: feline coronavirus (FCoV). This Alphacoronavirus exists in two serotypes: FCoV-I and FCoV-II (142). Both cause digestive diseases and are transmitted fecal-orally. FCoV-I is the most common type, but less virulent than FCoV-II (143, 144). Comparative sequence studies indicate FCoV-I is genetically similar to CCoV-I, and FCoV-II emerged from recombination between FCoV-I and CCoV-II (121, 142, 145, 146). Conceivably, FCoV-I and CCoV-I evolved from a common ancestor, while CCoV-II and FCoV-II arose as more virulent recombinants (129). For each serotype, there are two biotypes with different pathogenicity: feline enteric coronavirus (FECV) and feline infectious peritonitis virus (FIPV). Feline enteric coronavirus usually causes mild diarrhea, whereas feline infectious peritonitis (FIP) is lethal. Feline infectious peritonitis virus evolves from FECV via within-host mutations in the S gene that alter cell tropism, and emerges during persistent infection of FECV (142, 147). However, a novel FIPV strain may have been transmitted horizontally (144). In 2004, a disease resembling FIP was also discovered in ferrets caused by an emerging ferret systemic coronavirus, a decade after the first and less virulent ferret coronavirus (enteric) was discovered (148). Feline infectious peritonitis likely emerged in the late 1950s, within a decade after the first TGE cases in swine in USA (128, 149). Thus, FCoV is closely related to TGEV and CCoV, and recombinants among all three have emerged (150–152), probably because all three can cross-infect cats, swine, and dogs (125, 151, 153–155).
Discussion
Coronaviruses in humans and domestic animals are closely related (Figure 1), and have emerged recently and at an increasing rate. The circumstances associated with their emergence are high-animal-density environments that favor interspecies interactions, such as kennels, shelters, farms, and markets (Table 1), which increase disease prevalence and promote cross-species transmission. Indeed, studies show that seroprevalence of CCoV is higher in kennels compared to the rest of the dog population, and shelters co-housing dogs with cats harbor recombinant canine-feline coronaviruses (116, 151, 153, 159). Further, commercial agriculture has led to large numbers of domestic animals living in close proximity to humans, possibly driving the emergence of OC43 from cattle, and 229E and MERS from camelids.
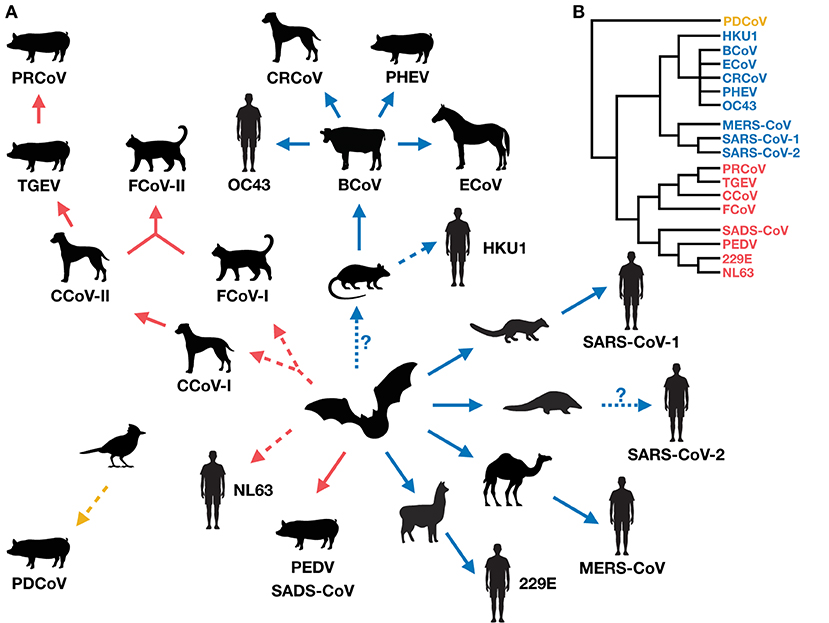
Figure 1. (A) The evolution and radiation of coronaviruses in humans and domestic mammals (via potential wild intermediate spillover host species). The radiation suggests there could be a vicious cycle of coronavirus emergence, whereby newly emerged viruses in new hosts increase the likelihood of producing more new recombinants. Red, blue and yellow arrows indicate the direction of spillover of coronavirus emergence for Alphacoronaviruses, Betacoronaviruses, and Deltacoronaviruses, respectively. Solid arrows represent direct (confirmed or suspected) coronavirus transmission between host species (although indirect transmission via an unidentified intermediate host is not excluded), and dashed arrows represent suspected indirect transmission via an unidentified intermediate host (although direct transmission is not excluded) (10, 17, 104, 127, 139, 141, 156). Dotted arrows with a question mark indicate uncertain spillover events. (B) A simplified phylogeny of the coronaviruses covered in this review, drawn from published findings (5, 129, 157).
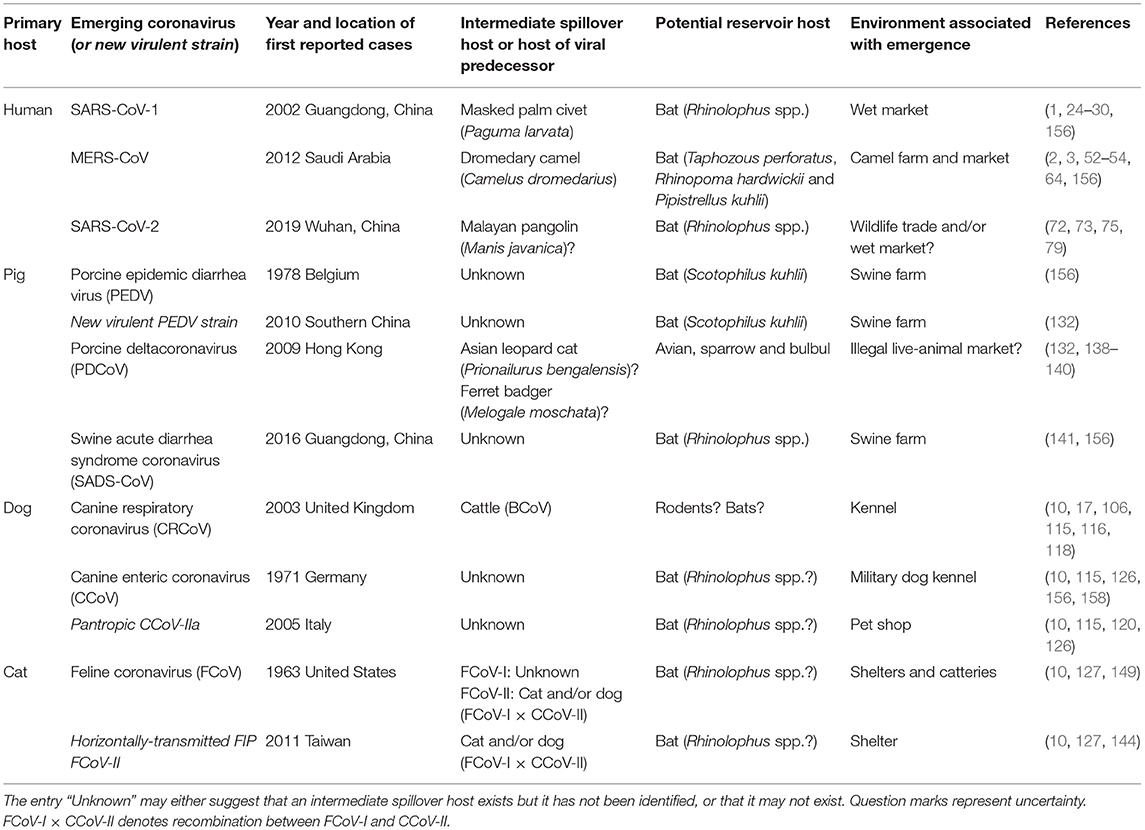
Table 1. First reported outbreaks and probable host species involved in the cross-species transmission events of recently emerging coronaviruses (or new virulent strains of re-emerging coronaviruses) in humans and domestic mammals covered in this review.
Additionally, animals kept under poor conditions or exposed to stress (e.g., during transport) suffer from poor health and suppressed immune systems, rendering them more susceptible to infections (64, 160). For example, mink fur farms, where animals are usually kept in small, unhygienic enclosures, generated new strains of SARS-CoV-2 causing secondary zoonoses (95, 101–103). The wildlife trade and wet markets are conducive to disease emergence as well, since animals are transported and kept in small, unhygienic cages next to many different animal species (160). Indeed, a study showed that civets in markets were disproportionately positive for SARS-CoV-1 compared to civets on the supplying farms (30). Further, SARS-CoV-1 isolates from a civet and a racoon dog at the same market, but from different regions of China, had an identical S-gene sequence, which differed from that of the other civet isolates, indicating the occurrence of cross-species transmission at the market (26). Accordingly, the concept of One Health is important for suppressing coronavirus emergence.
Little is still known about host ranges and cross-species transmissions of coronaviruses. Most studies on this topic have been motivated by finding appropriate animal models for vaccine development, or identifying potential host species enabling viral persistence. However, future studies should expand their surveys beyond domestic, captive, or common laboratory animals for a fuller comprehension of coronavirus emergence and the extent of its radiation (Figure 1A). Surveillance efforts of coronaviruses in the wild are underway (e.g., PREDICT, Global Virome Genome) (161, 162), which are important for identifying new coronaviruses with zoonotic potential [reviewed in (163)], tracking spillover pathways, and potentially filling in the host range gaps of known coronaviruses in humans and domestic mammals.
Concurrently with the global expansion of humans and domestic mammals, various coronaviruses have emerged as a result of cross-species transmission among humans, and domestic and wild animals. Conceivably, the human and domestic mammal population increase yielded a large enough susceptible population to maintain coronavirus circulation, provided more opportunities for novel coronavirus emergence via spillover among different species, and brought humans and domestic animals in closer contact with wild reservoirs (164–166). The mechanisms governing the surge and radiation of these recently emerged coronaviruses require further investigation. Actions reducing people's dependency on domestic animals and animal products, while improving the health of the animals remaining in captivity, may mitigate coronavirus emergence.
Author Contributions
NN performed the literature review and wrote the manuscript.
Funding
NN was supported by the Philanthropic Educational Organization (P.E.O.) Scholar Award from the International Chapter of the P.E.O. Sisterhood, the Environmental Venture Project Grant from the Stanford Woods Institute for the Environment, the Stanford Data Science Scholars program, and the Department of Biology at Stanford University.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The author thanks Tejas Athni, Alexander Becker, Marissa Childs, Lisa Couper, Isabel Delwel, Johannah Farner, Caroline Glidden, Mallory Harris, Isabel Jones, Morgan Kain, Devin Kirk, Christopher LeBoa, Andrea Lund, Erin Mordecai, Maike Morrison, Eloise Skinner, Susanne Sokolow, and Krti Tallam for helpful feedback and discussion on this topic.
References
1. da Silva PG, Mesquita JR, de São José Nascimento M, Ferreira VAM. Viral, host and environmental factors that favor anthropozoonotic spillover of coronaviruses: an opinionated review, focusing on SARS-CoV, MERS-CoV and SARS-CoV-2. Sci Total Environ. (2020) 750:141483. doi: 10.1016/j.scitotenv.2020.141483
2. Zaki AM, Van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. (2012) 367:1814–20. doi: 10.1056/NEJMoa1211721
3. de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the coronavirus study group. J Virol. (2013) 87:7790–2. doi: 10.1128/jvi.01244-13
4. CNN Health. World Covid-19 Tracker: Latest Cases and Deaths by Country. (2021). Available online at: https://www.cnn.com/interactive/2020/health/coronavirus-maps-and-cases/ (accessed January 10, 2021).
5. de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, et al. Family Coronaviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus Taxonomy Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press (2012). p. 806–28. doi: 10.1016/B978-0-12-384684-6.00068-9
6. Helmy YA, Fawzy M, Elaswad A, Sobieh A, Kenney SP, Shehata AA. The COVID-19 pandemic: a comprehensive review of taxonomy, genetics, epidemiology, diagnosis, treatment, and control. J Clin Med. (2020) 9:1225. doi: 10.3390/jcm9041225
7. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. (2016) 3:237–61. doi: 10.1146/annurev-virology-110615-042301
8. Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. (2011) 8:270–9. doi: 10.4161/rna.8.2.15013
9. Woo PCY, Huang Y, Lau SKP, Yuen KY. Coronavirus genomics and bioinformatics analysis. Viruses. (2010) 2:1805–20. doi: 10.3390/v2081803
10. Woo PCY, Lau SKP, Lam CSF, Lau CCY, Tsang AKL, Lau JHN, et al. Discovery of seven novel mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. (2012) 86:3995–4008. doi: 10.1128/jvi.06540-11
11. Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. (2016) 24:490–502. doi: 10.1016/j.tim.2016.03.003
12. Li W, Moore MJ, Vasllieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. (2003) 426:450–4. doi: 10.1038/nature02145
13. Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. (2020) 181:271.e8–80.e8. doi: 10.1016/j.cell.2020.02.052
14. Geoghegan JL, Duchêne S, Holmes EC. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. (2017) 13:e1006215. doi: 10.1371/journal.ppat.1006215
15. Bar-On YM, Phillips R, Milo R. The biomass distribution on Earth. Proc Natl Acad Sci USA. (2018) 115:6506–11. doi: 10.1073/pnas.1711842115
16. Harper KN, Armelagos GJ. Genomics, the origins of agriculture, and our changing microbe-scape: time to revisit some old tales and tell some new ones. Am J Phys Anthropol. (2013) 152(Suppl):135–52. doi: 10.1002/ajpa.22396
17. Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. (2018) 100:163–188. doi: 10.1016/bs.aivir.2018.01.001
18. Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, et al. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol. (2015) 89:11858–70. doi: 10.1128/jvi.01755-15
19. Tao Y, Shi M, Chommanard C, Queen K, Zhang J, Markotter W, et al. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol. (2017) 91:e01953–16. doi: 10.1128/jvi.01953-16
20. Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. (2017) 25:35–48. doi: 10.1016/j.tim.2016.09.001
21. Vijgen L, Keyaerts E, Lemey P, Maes P, Van Reeth K, Nauwynck H, et al. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. (2006) 80:7270–4. doi: 10.1128/jvi.02675-05
22. Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. (2005) 79:1595–604. doi: 10.1128/jvi.79.3.1595-1604.2005
23. Kin N, Miszczak F, Diancourt L, Caro V, Moutou F, Vabret A, et al. Comparative molecular epidemiology of two closely related coronaviruses, bovine coronavirus (BCoV) and human coronavirus OC43 (HCoV-OC43), reveals a different evolutionary pattern. Infect Genet Evol. (2016) 40:186–91. doi: 10.1016/j.meegid.2016.03.006
24. Wang LF, Eaton BT. Bats, civets and the emergence of SARS. Curr Top Microbiol Immunol. (2007) 315:325–44. doi: 10.1007/978-3-540-70962-6_13
25. Wang M, Yan M, Xu H, Liang W, Kan B, Zheng B, et al. SARS-CoV infection in a restaurant from palm civet. Emerg Infect Dis. (2005) 11:1860–5. doi: 10.3201/eid1112.041293
26. Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. (2003) 302:276–8. doi: 10.1126/science.1087139
27. Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci USA. (2005) 102:2430–5. doi: 10.1073/pnas.0409608102
28. Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, et al. Bats are natural reservoirs of SARS-like coronaviruses. Science. (2005) 310:676–9. doi: 10.1126/science.1118391
29. Yuan J, Hon CC, Li Y, Wang D, Xu G, Zhang H, Zhou P, et al. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J Gen Virol. (2010) 91:1058–62. doi: 10.1099/vir.0.016378-0
30. Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, et al. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol. (2005) 79:11892–900. doi: 10.1128/jvi.79.18.11892-11900.2005
31. Lau SKP, Li KSM, Huang Y, Shek C-T, Tse H, Wang M, Choi GKY, et al. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol. (2010) 84:2808–19. doi: 10.1128/jvi.02219-09
32. Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi HW, Wong BHL, Wong SSY, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. (2005) 102:14040–5. doi: 10.1073/pnas.0506735102
33. Xu RH, He JF, Evans MR, Peng GW, Field HE, Yu DW, Lee CK, et al. Epidemiologic clues to SARS origin in China. Emerg Infect Dis. (2004) 10:1030–7. doi: 10.3201/eid1006.030852
34. Ge XY, Li JL, Yang XL, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. (2013) 503:535–8. doi: 10.1038/nature12711
35. Plowright RK, Eby P, Hudson PJ, Smith IL, Westcott D, Bryden WL, et al. Ecological dynamics of emerging bat virus spillover. Proc R Soc B Biol Sci. (2014) 282:20142124. doi: 10.1098/rspb.2014.2124
36. Martina BEE, Haagmans BL, Kuiken T, Fouchier RAM, Rimmelzwaan GF, Van Amerongen G, et al. SARS virus infection of cats and ferrets. Nature. (2003) 425:915. doi: 10.1038/425915a
37. Van den Brand JM, Haagmans BL, Leijten L, van Riel D, Martina BE, Osterhaus AD, et al. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet Pathol. (2008) 45:551–62. doi: 10.1354/vp.45-4-551
38. Fouchier RAM, Kuiken T, Schutten M, Van Amerongen G, Van Doornum GJJ, Van Den Hoogen BG, et al. Koch's postulates fulfilled for SARS virus. Nature. (2003) 423:240. doi: 10.1038/423240a
39. Liang L, He C, Lei M, Li S, Hao Y, Zhu H, et al. Pathology of guinea pigs experimentally infected with a novel reovirus and coronavirus isolated from SARS patients. DNA Cell Biol. (2005) 24:485–90. doi: 10.1089/dna.2005.24.485
40. Roberts A, Vogel L, Guarner J, Hayes N, Murphy B, Zaki S, et al. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J Virol. (2005) 79:503–11. doi: 10.1128/jvi.79.1.503-511.2005
41. Lawler J V, Endy TP, Hensley LE, Garrison A, Fritz EA, Lesar M, et al. Cynomolgus macaque as an animal model for severe acute respiratory syndrome. PLoS Med. (2006) 3:677–86. doi: 10.1371/journal.pmed.0030149
42. Greenough TC, Carville A, Coderre J, Somasundaran M, Sullivan JL, Luzuriaga K, et al. pneumonitis and multi-organ system disease in common marmosets (Callithrix jacchus) infected with the severe acute respiratory syndrome-associated coronavirus. Am J Pathol. (2005) 167:455–63. doi: 10.1016/S0002-9440(10)62989-6
43. Rowe T, Gao G, Hogan RJ, Crystal RG, Voss TG, Grant RL, et al. Macaque model for severe acute respiratory syndrome. J Virol. (2004) 78:11401–4. doi: 10.1128/jvi.78.20.11401-11404.2004
44. McAuliffe J, Vogel L, Roberts A, Fahle G, Fischer S, Shieh WJ, et al. Replication of SARS coronavirus administered into the respiratory tract of African Green, rhesus and cynomolgus monkeys. Virology. (2004) 330:8–15. doi: 10.1016/j.virol.2004.09.030
45. Weingart HM, Copps J, Drebot MA, Marszal P, Smith G, Gren J, et al. Susceptibility of pigs and chickens to SARS coronavirus. Emerg Infect Dis. (2004) 10:179–84. doi: 10.3201/eid1002.030677
46. Wang M, Jing HQ, Xu HF, Jiang XG, Kan B, Liu QY, et al. Surveillance on severe acute respiratory syndrome associated coronavirus in animals at a live animal market of Guangzhou in 2004. Zhonghua Liu Xing Bing Xue Za Zhi. 26:84–7 (2005).
47. Chen W, Yan M, Yang L, Ding B, He B, Wang Y, et al. SARS-associated coronavirus transmitted from human to pig. Emerg Infect Dis. (2005) 11:446–8. doi: 10.3201/eid1103.040824
48. Swayne DE, Suarez DL, Spackman E, Tumpey TM, Beck JR, Erdman D, et al. Domestic poultry and SARS coronavirus, Southern China. Emerg Infect Dis. (2004) 10:914–6. doi: 10.3201/eid1005.030827
49. Yuan L, Tang Q, Cheng T, Xia N. Animal models for emerging coronavirus: progress and new insights. Emerg Microbes Infect. (2020) 9:949–61. doi: 10.1080/22221751.2020.1764871
50. Gao H, Yao H, Yang S, Li L. From SARS to MERS: evidence and speculation. Front Med. (2016) 10:377–82. doi: 10.1007/s11684-016-0466-7
51. Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. (2014) 370:2499–505. doi: 10.1056/NEJMoa1401505
52. Müller MA, Corman VM, Jores J, Meyer B, Younan M, Liljander A, et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg Infect Dis. (2014) 20:2093–5. doi: 10.3201/eid2012.141026
53. Chu DKW, Poon LLM, Gomaa MM, Shehata MM, Perera RAPM, Zeid DA, et al. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis. (2014) 20:1049–53. doi: 10.3201/eid2006.140299
54. Peck KM, Burch CL, Heise MT, Baric RS. Coronavirus host range expansion and Middle East respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu Rev Virol. (2015) 2:95–117. doi: 10.1146/annurev-virology-100114-055029
55. Hemida MG, Al-Naeem A, Perera RAPM, Chin AWH, Poon LLM, Peiris M. Lack of Middle East respiratory syndrome coronavirus transmission from infected camels. Emerg Infect Dis. (2015) 21:699–701. doi: 10.3201/eid2104.141949
56. Adney DR, van Doremalen N, Brown VR, Bushmaker T, Scott D, de Wit E, et al. Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels. Emerg Infect Dis. (2014) 20:1999–2005. doi: 10.3201/eid2012.141280
57. Hemida MG, Chu DKW, Poon LLM, Perera RAPM, Alhammadi MA, Ng HY, et al. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis. (2014) 20:1231–4. doi: 10.3201/eid2007.140571
58. Skariyachan S, Challapilli SB, Packirisamy S, Kumargowda ST, Sridhar VS. Recent aspects on the pathogenesis mechanism, animal models and novel therapeutic interventions for Middle East respiratory syndrome coronavirus infections. Front Microbiol. (2019) 10:569. doi: 10.3389/fmicb.2019.00569
59. Hemida MG, Perera RA, Wang P, Alhammadi MA, Siu LY, Li M, Poon LL, et al. Middle East respiratory syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Eurosurveillance. (2013) 18:20659. doi: 10.2807/1560-7917.ES2013.18.50.20659
60. Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, de Wit E, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. (2014) 5:e00884–14. doi: 10.1128/mBio.00884-14
61. Sabir JSM, Lam TTY, Ahmed MMM, Li L, Shen Y, Abo-Aba SEM, Qureshi MI, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. (2016) 351:81–4. doi: 10.1126/science.aac8608
62. Corman VM, Jores J, Meyer B, Younan M, Liljander A, Said MY, et al. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992-2013. Emerg Infect Dis. (2014) 20:1319–22. doi: 10.3201/eid2008.140596
63. Lau SKP, Wong ACP, Lau TCK, Woo PCY. Molecular evolution of MERS coronavirus: Dromedaries as a recent intermediate host or long-time animal reservoir? Int J Mol Sci. (2017) 18:2138. doi: 10.3390/ijms18102138
64. Hemida MG, Alnaeem A. Some One Health based control strategies for the Middle East respiratory syndrome coronavirus. One Heal. (2019) 8:100102. doi: 10.1016/j.onehlt.2019.100102
65. Adney DR, Bielefeldt-Ohmann H, Hartwig AE, Bowen RA. Infection, replication, and transmission of Middle East respiratory syndrome coronavirus in alpacas. Emerg Infect Dis. (2016) 22:1031–7. doi: 10.3201/eid2206.160192
66. Crameri G, Durr PA, Klein R, Foord A, Yu M, Riddell S, et al. Experimental infection and response to rechallenge of alpacas with Middle East respiratory syndrome coronavirus. Emerg Infect Dis. (2016) 22:1071–4. doi: 10.3201/eid2206.160007
67. Van Doremalen N, Munster VJ. Animal models of Middle East respiratory syndrome coronavirus infection. Antiviral Res. (2015) 122:28–38. doi: 10.1016/j.antiviral.2015.07.005
68. Vergara-Alert J, van den Brand JMA, Widagdo W, Muñoz M, Raj VS, Schipper D, et al. Livestock susceptibility to infection with Middle East respiratory syndrome coronavirus. Emerg Infect Dis. (2017) 23:232–40. doi: 10.3201/eid2302.161239
69. Kandeil A, Gomaa M, Shehata M, El-Taweel A, Kayed AE, Abiadh A, et al. Middle East respiratory syndrome coronavirus infection in non-camelid domestic mammals. Emerg Microbes Infect. (2019) 8:103–8. doi: 10.1080/22221751.2018.1560235
70. Meyer B, García-Bocanegra I, Wernery U, Wernery R, Sieberg A, Müller MA, et al. Serologic assessment of possibility for MERS-CoV infection in equids. Emerg Infect Dis. (2015) 21:181–2. doi: 10.3201/eid2101.141342
71. Suarez DL, Pantin-Jackwood MJ, Swayne DE, Lee SA, DeBlois SM, Spackman E. Lack of susceptibility to SARS-CoV-2 and MERS-CoV in poultry. Emerg Infect Dis. (2020) 26:3074–6. doi: 10.3201/EID2612.202989
72. Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. (2020) 579:270–3. doi: 10.1038/s41586-020-2012-7
73. Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. (2020) 579:265–9. doi: 10.1038/s41586-020-2008-3
74. Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, et al. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol. (2020) 30:2196.e3–203.e3. doi: 10.1016/j.cub.2020.05.023
75. Latinne A, Hu B, Olival KJ, Zhu G, Zhang L, Li H, et al. Origin and cross-species transmission of bat coronaviruses in China. Nat Commun. (2020) 11:1–15. doi: 10.1038/s41467-020-17687-3
76. Liu P, Jiang J-Z, Wan X-F, Hua Y, Li L, Zhou J, et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. (2020) 16:e1008421. doi: 10.1101/2020.02.18.954628
77. Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, Li N, et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature. (2020) 583:286–9. doi: 10.1038/s41586-020-2313-x
78. Lam TTY, Jia N, Zhang YW, Shum MHH, Jiang JF, Zhu HC, et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. (2020) 583:282–5. doi: 10.1038/s41586-020-2169-0
79. Zhang T, Wu Q, Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. (2020) 30:1346.e2–1351.e2. doi: 10.1016/j.cub.2020.03.022
80. Sironi M, Hasnain SE, Rosenthal B, Phan T, Luciani F, Shaw MA, et al. SARS-CoV-2 and COVID-19: a genetic, epidemiological, and evolutionary perspective. Infect Genet Evol. (2020) 84:104384. doi: 10.1016/j.meegid.2020.104384
81. Flores-Alanis A, Sandner-Miranda L, Delgado G, Cravioto A, Morales-Espinosa R. The receptor binding domain of SARS-CoV-2 spike protein is the result of an ancestral recombination between the bat-CoV RaTG13 and the pangolin-CoV MP789. BMC Res Notes. (2020) 13:398. doi: 10.1186/s13104-020-05242-8
82. Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. (2020) 368:1016–20. doi: 10.1126/science.abb7015
83. Rockx B, Kuiken T, Herfst S, Bestebroer T, Lamers MM, Munnink BBO, et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. (2020) 368:1012–5. doi: 10.1126/science.abb7314
84. Munster VJ, Feldmann F, Williamson BN, van Doremalen N, Pérez-Pérez L, Schulz J, et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. (2020) 585:268–72. doi: 10.1038/s41586-020-2324-7
85. Woolsey C, Borisevich V, Prasad AN, Agans KN, Deer DJ, Dobias NS, et al. Establishment of an African green monkey model for COVID-19 and protection against re-infection. Nat Immunol. (2021) 22:86–98. doi: 10.1038/s41590-020-00835-8
86. Sia SF, Yan LM, Chin AWH, Fung K, Choy KT, Wong AYL, et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature. (2020) 583:834–8. doi: 10.1038/s41586-020-2342-5
87. Chan JFW, Zhang AJ, Yuan S, Poon VKM, Chan CCS, Lee ACY, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. (2020) 71:2428–46. doi: 10.1093/cid/ciaa325
88. Zhao Y, Wang J, Kuang D, Xu J, Yang M, Ma C, et al. Susceptibility of tree shrew to SARS-CoV-2 infection. Sci Rep. (2020) 10:16007. doi: 10.1038/s41598-020-72563-w
89. Lu S, Zhao Y, Yu W, Yang Y, Gao J, Wang J, et al. Comparison of nonhuman primates identified the suitable model for COVID-19. Signal Transduct Target Ther. (2020) 5:1–9. doi: 10.1038/s41392-020-00269-6
90. Schlottau K, Rissmann M, Graaf A, Schön J, Sehl J, Wylezich C, et al. SARS-CoV-2 in fruit bats, ferrets, pigs, and chickens: an experimental transmission study. Lancet Microbe. (2020) 1:e218–25. doi: 10.1016/s2666-5247(20)30089-6
91. Bosco-Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, et al. Experimental infection of domestic dogs and cats with SARS-CoV-2: pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci USA. (2020) 117:26382–8. doi: 10.1073/pnas.2013102117
92. Sit THC, Brackman CJ, Ip SM, Tam KWS, Law PYT, To EMW, Yu VYT, et al. Infection of dogs with SARS-CoV-2. Nature. (2020) 586:776–8. doi: 10.1038/s41586-020-2334-5
93. Zhai X, Sun J, Yan Z, Zhang J, Zhao J, Zhao Z, et al. Comparison of severe acute respiratory syndrome coronavirus 2 spike protein binding to ACE2 receptors from human, pets, farm animals, and putative intermediate hosts. J Virol. (2020) 94:e00831–20. doi: 10.1128/jvi.00831-20
94. Richard M, Kok A, de Meulder D, Bestebroer TM, Lamers MM, Okba NMA, et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat Commun. (2020) 11:1–6. doi: 10.1038/s41467-020-17367-2
95. Hosie MJ, Hofmann-Lehmann R, Hartmann K, Egberink H, Truyen U, Addie DD, et al. Anthropogenic infection of cats during the 2020 COVID-19 pandemic. Viruses. (2021) 13:185. doi: 10.3390/v13020185
96. Sailleau C, Dumarest M, Vanhomwegen J, Delaplace M, Caro V, Kwasiborski A, et al. First detection and genome sequencing of SARS-CoV-2 in an infected cat in France. Transbound Emerg Dis. (2020) 67:2324–8. doi: 10.1111/tbed.13659
97. Temmam S, Barbarino A, Maso D, Behillil S, Enouf V, Huon C, et al. Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Heal. (2020) 10:100164. doi: 10.1016/j.onehlt.2020.100164
98. Chen J, Huang C, Zhang Y, Zhang S, Jin M. Severe acute respiratory syndrome coronavirus 2-specific antibodies in pets in Wuhan, China. J Clean Prod. (2020) 81:e68–9. doi: 10.1016/j.jinf.2020.06.045
99. Zhang Q, Zhang H, Gao J, Huang K, Yang Y, Hui X, et al. A serological survey of SARS-CoV-2 in cat in Wuhan. Emerg Microbes Infect. (2020) 9:2013–9. doi: 10.1080/22221751.2020.1817796
100. Deng J, Jin YY, Liu Y, Sun J, Hao L, Bai J, et al. Serological survey of SARS-CoV-2 for experimental, domestic, companion and wild animals excludes intermediate hosts of 35 different species of animals. Transbound Emerg Dis. (2020) 67:1745–9. doi: 10.1111/tbed.13577
101. Munnink BBO, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. (2021) 371:172–7. doi: 10.1126/science.abe5901
102. Costagliola A, Liguori G, Angelo D, Costa C, Ciani F, Giordano A. Do animals play a role in the transmission of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2)? A commentary. (2021) 11:16. doi: 10.3390/ani11010016
103. Oreshkova N, Molenaar RJ, Vreman S, Harders F, Oude Munnink BB, Van Der Honing RWH, et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance. (2020) 25:2001005. doi: 10.2807/1560-7917.ES.2020.25.23.2001005
104. Bonilauri P, Rugna G. Animal coronaviruses and SARS-CoV-2 in animals, what do we actually know? Life. (2021) 11:1–17. doi: 10.3390/life11020123
105. Gibbons A. Captive gorillas test positive for coronavirus. Science. (2021). doi: 10.1126/science.abg5458
106. Gollakner R, Capua I. Is covid-19 the first pandemic that evolves into a panzootic? Vet Ital. (2020) 56:11–2. doi: 10.12834/VetIt.2246.12523.1
107. Lau SKP, Woo PCY, Li KSM, Tsang AKL, Fan RYY, Luk HKH, et al. Discovery of a novel coronavirus, China Rattus coronavirus HKU24, from Norway rats supports the murine origin of Betacoronavirus 1 and has implications for the ancestor of Betacoronavirus lineage A. J Virol. (2015) 89:3076–92. doi: 10.1128/jvi.02420-14
108. Amer HM. Bovine-like coronaviruses in domestic and wild ruminants. Anim Heal Res Rev. (2019) 19:113–24. doi: 10.1017/S1466252318000117
109. Saif LJ. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract. (2010) 26:349–64. doi: 10.1016/j.cvfa.2010.04.005
110. Saif LJ, Redman DR, Moorhead PD, Theil KW. Experimentally induced coronavirus infections in calves: viral replication in the respiratory and intestinal tracts. Am J Vet Res. (1986) 47:1426–32.
111. Kaneshima T, Hohdatsu T, Hagino R, Hosoya S, Nojiri Y, Murata M, et al. The infectivity and pathogenicity of a group 2 bovine coronavirus in pups. J Vet Med Sci. (2007) 69:301–3. doi: 10.1292/jvms.69.301
112. Ismail MM, Cho KO, Ward LA, Saif LJ, Saif YM. Experimental bovine coronavirus in turkey poults and young chickens. Avian Dis. (2001) 45:157–63. doi: 10.2307/1593023
113. Pusterla N, Vin R, Leutenegger CM, Mittel LD, Divers TJ. Enteric coronavirus infection in adult horses. Vet J. (2018) 231:13–8. doi: 10.1016/j.tvjl.2017.11.004
114. Sanz MG, Kwon SY, Pusterla N, Gold JR, Bain F, Evermann J. Evaluation of equine coronavirus fecal shedding among hospitalized horses. J Vet Intern Med. (2019) 33:918–22. doi: 10.1111/jvim.15449
115. Guy JS, Breslin JJ, Breuhaus B, Vivrette S, Smith LG. Characterization of a coronavirus isolated from a diarrheic foal. J Clin Microbiol. (2000) 38:4523–6. doi: 10.1128/jcm.38.12.4523-4526.2000
116. Licitra BN, Duhamel GE, Whittaker GR. Canine enteric coronaviruses: Emerging viral pathogens with distinct recombinant spike proteins. Viruses. (2014) 6:3363–76. doi: 10.3390/v6083363
117. Erles K, Toomey C, Brooks HW, Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. (2003) 310:216–23. doi: 10.1016/S0042-6822(03)00160-0
118. Ellis JA, McLean N, Hupaelo R, Haines DM. Detection of coronavirus in cases of tracheobronchitis in dogs: a retrospective study from 1971 to 2003. Can Vet J. (2005) 46:447–8.
119. Erles K, Shiu KB, Brownlie J. Isolation and sequence analysis of canine respiratory coronavirus. Virus Res. (2007) 124:78–87. doi: 10.1016/j.virusres.2006.10.004
120. Tennant BJ, Gaskell RM, Kelly DF, Carter SD, Gaskell CJ. Canine coronavirus infection in the dog following oronasal inoculation. Res Vet Sci. (1991) 51:11–8. doi: 10.1016/0034-5288(91)90023-H
121. Buonavoglia C, Decaro N, Martella V, Elia G, Campolo M, Desario C, et al. Canine coronavirus highly pathogenic for dogs. Emerg Infect Dis. (2006) 12:492–4. doi: 10.3201/eid1203.050839
122. Alfano F, Dowgier G, Valentino MP, Galiero G, Tinelli A, Decaro N, et al. Identification of pantropic canine coronavirus in a wolf (Canis lupus italicus) in Italy. J Wildl Dis. (2019) 55:504–8. doi: 10.7589/2018-07-182
123. Alfano F, Fusco G, Mari V, Occhiogrosso L, Miletti G, Brunetti R, et al. Circulation of pantropic canine coronavirus in autochthonous and imported dogs, Italy. Transbound Emerg Dis. (2020) 67:1991–9. doi: 10.1111/tbed.13542
124. McArdle F, Bennett M, Gaskell RM, Tennant B, Kelly DF, Gaskell CJ. Induction and enhancement of feline infectious peritonitis by canine coronavirus. Am J Vet Res. (1992) 53:1500–6.
125. Woods RD, Cheville NF, Gallagher JE. Lesions in the small intestine of newborn pigs inoculated with porcine, feline, and canine coronaviruses. Am J Vet Res. (1981) 42:1163–9.
126. Woods RD, Wesley RD. Seroconversion of pigs in contact with dogs exposed to canine coronavirus. Can J Vet Res. (1992) 56:78–80.
127. Decaro N, Lorusso A. Novel human coronavirus (SARS-CoV-2): a lesson from animal coronaviruses. Vet Microbiol. (2020) 244:108693. doi: 10.1016/j.vetmic.2020.108693
128. Doyle LP, Hutchings LM. A transmissible gastroenteritis in pigs. J Am Vet Med Assoc. (1946) 108:257–9.
129. Lorusso A, Decaro N, Schellen P, Rottier PJM, Buonavoglia C, Haijema B-J, et al. Gain, preservation, and loss of a group 1a coronavirus accessory glycoprotein. J Virol. (2008) 82:10312–7. doi: 10.1128/jvi.01031-08
130. Pensaert M, Callebaut P, Vergote J. Isolation of a porcine respiratory, non-enteric coronavirus related to transmissible gastroenteritis. Vet Q. (1986) 8:257–61. doi: 10.1080/01652176.1986.9694050
131. Decaro N, Martella V, Saif LJ, Buonavoglia C. COVID-19 from veterinary medicine and one health perspectives: what animal coronaviruses have taught us. Res Vet Sci. (2020) 131:21–3. doi: 10.1016/j.rvsc.2020.04.009
132. Wang Q, Vlasova AN, Kenney SP, Saif LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. (2019) 34:39–49. doi: 10.1016/j.coviro.2018.12.001
133. Huang YW, Dickerman AW, Piñeyro P, Li L, Fang L, Kiehne R, et al. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. MBio. (2013) 4:737–50. doi: 10.1128/mBio.00737-13
134. Sun RQ, Cai RJ, Chen YQ, Liang PS, Chen DK, Song CX. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. (2012) 18:161–3. doi: 10.3201/eid1801.111259
135. Bevins SN, Lutman M, Pedersen K, Barrett N, Gidlewski T, Deliberto TJ, et al. Spillover of swine coronaviruses, United States. Emerg Infect Dis. (2018) 24:1390–2. doi: 10.3201/eid2407.172077
136. Wang L, Byrum B, Zhang Y. Detection and genetic characterization of Deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis. (2014) 20:1227–30. doi: 10.3201/eid2007.140296
137. Lee S, Lee C. Complete genome characterization of Korean porcine deltacoronavirus strain KOR/KNU14-04/2014. Genome Announc. (2014) 2:e01191–14. doi: 10.1128/genomeA.01191-14
138. Lau SKP, Wong EYM, Tsang C-C, Ahmed SS, Au-Yeung RKH, Yuen K-Y, et al. Discovery and sequence analysis of four deltacoronaviruses from birds in the Middle East reveal interspecies jumping with recombination as a potential mechanism for avian-to-avian and avian-to-mammalian transmission. J Virol. (2018) 92:e00265–18. doi: 10.1128/jvi.00265-18
139. Dong BQ, Liu W, Fan XH, Vijaykrishna D, Tang XC, Gao F, et al. Detection of a novel and highly divergent coronavirus from Asian leopard cats and Chinese ferret badgers in Southern China. J Virol. (2007) 81:6920–6. doi: 10.1128/jvi.00299-07
140. Ma Y, Zhang Y, Liang X, Lou F, Oglesbee M, Krakowka S, et al. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. MBio. (2015) 6:e00064. doi: 10.1128/mBio.00064-15
141. Zhou P, Fan H, Lan T, Yang XL, Shi WF, Zhang W, et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature. (2018) 556:255–9. doi: 10.1038/s41586-018-0010-9
142. Tekes G, Thiel HJ. Feline coronaviruses: pathogenesis of feline infectious peritonitis. Adv Virus Res. (2016) 96:193–218. doi: 10.1016/bs.aivir.2016.08.002
143. Lin CN, Chang RY, Su BL, Chueh LL. Full genome analysis of a novel type II feline coronavirus NTU156. Virus Genes. (2013) 46:316–22. doi: 10.1007/s11262-012-0864-0
144. Wang YT, Su BL, Hsieh LE, Chueh LL. An outbreak of feline infectious peritonitis in a Taiwanese shelter: epidemiologic and molecular evidence for horizontal transmission of a novel type II feline coronavirus. Vet Res. (2013) 44:1–9. doi: 10.1186/1297-9716-44-57
145. Pratelli A, Martella V, Decaro N, Tinelli A, Camero M, Cirone F, et al. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J Virol Methods. (2003) 110:9–17. doi: 10.1016/S0166-0934(03)00081-8
146. Pratelli A, Martella V, Pistello M, Elia G, Decaro N, Buonavoglia D, et al. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J Virol Methods. (2003) 107:213–22. doi: 10.1016/S0166-0934(02)00246-X
147. Rottier PJM, Nakamura K, Schellen P, Volders H, Haijema BJ. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J Virol. (2005) 79:14122–30. doi: 10.1128/jvi.79.22.14122-14130.2005
148. Murray J, Kiupel M, Maes RK. Ferret coronavirus-associated diseases. Vet Clin North Am - Exot Anim Pract. (2010) 13:543–60. doi: 10.1016/j.cvex.2010.05.010
149. Pedersen NC. A review of feline infectious peritonitis virus infection: 1963-2008. J Feline Med Surg. (2009) 11:225–58. doi: 10.1016/j.jfms.2008.09.008
150. Herrewegh AAPM, Smeenk I, Horzinek MC, Rottier PJM, de Groot RJ. Feline coronavirus type II strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J Virol. (1998) 72:4508–14. doi: 10.1128/jvi.72.5.4508-4514.1998
151. Benetka V, Kolodziejek J, Walk K, Rennhofer M, Möstl K. M gene analysis of atypical strains of feline and canine coronavirus circulating in an Austrian animal shelter. Vet Rec. (2006) 159:170–5. doi: 10.1136/vr.159.6.170
152. Wesley RD. The S gene of canine coronavirus, strain UCD-1, is more closely related to the S gene of transmissible gastroenteritis virus than to that of feline infectious peritonitis virus. Virus Res. (1999) 61:145–52. doi: 10.1016/S0168-1702(99)00032-5
153. Rennhofer M, Benetka V, Sommerfeld-Stur I, Möstl K. Epidemiological investigations on coronavirus infections in dogs and cats in an animal shelter. Wien Tierarz Monatss. (2005) 92:21–7.
154. Laber KE, Whary MT, Bingel SA, Goodrich JA, Smith AC, Swindle MM. Biology and diseases of swine. In: Fox J, Anderson L, Loew F, Quimby F, editors. Laboratory Animal Medicine. Burlington: Academic Press. (2002). p. 615–73. doi: 10.1016/b978-012263951-7/50018-1
155. Saif L, Wesley R. Transmissible gastroenteritis virus. In: Leman AD, Straw BE, Mengeling WL, D'Allaire S, Taylor DJ, editors. Diseases of Swine. Ames, IA: Iowa State University Press. (1992). p. 362–86.
156. Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. Bats and coronaviruses. Viruses. (2019) 11:41. doi: 10.3390/v11010041
157. Nakagawa S, Miyazawa T. Genome evolution of SARS-CoV-2 and its virological characteristics. BioMed Central. (2020). doi: 10.1186/s41232-020-00126-7
158. Pratelli A. Genetic evolution of canine coronavirus and recent advances in prophylaxis. Vet Res. (2006) 37:191–200. doi: 10.1051/vetres:2005053
159. Naylor MJ, Monckton RP, Lehrbach PR, Deane EM. Canine coronavirus in Australian dogs. Aust Vet J. (2001) 79:116–9. doi: 10.1111/j.1751-0813.2001.tb10718.x
160. Baker SE, Cain R, van Kesteren F, Zommers ZA, D'Cruze N, MacDonald DW. Rough trade: animal welfare in the global wildlife trade. Bioscience. (2013) 63:928–38. doi: 10.1525/bio.2013.63.12.6
161. Carlson CJ. From PREDICT to prevention, one pandemic later. Lancet Microbe. (2020) 1:e6–7. doi: 10.1016/s2666-5247(20)30002-1
162. Our History — Global Virome Project. Available online at: http://www.globalviromeproject.org/ (accessed May 30, 2021).
163. Jelinek HF, Mousa M, Alefishat E, Osman W, Spence I, Bu D, et al. Evolution, ecology, and zoonotic transmission of betacoronaviruses: a review. Front Vet Sci. (2021) 8:380. doi: 10.3389/fvets.2021.644414
164. Lorusso A, Calistri P, Petrini A, Savini G, Decaro N. Novel coronavirus (COVID-19) epidemic: a veterinary perspective. Vet Ital. (2020) 56:5–10. doi: 10.12834/VetIt.2173.11599.1
165. Roche B, Garchitorena A, Guégan JF, Arnal A, Roiz D, Morand S, et al. Was the COVID-19 pandemic avoidable? A call for a “solution-oriented” approach in pathogen evolutionary ecology to prevent future outbreaks. Ecol Lett. (2020) 23:1557–60. doi: 10.1111/ele.13586
Keywords: coronavirus, COVID-19, cross-species transmission, host range, MERS, One Health, SARS, spillover
Citation: Nova N (2021) Cross-Species Transmission of Coronaviruses in Humans and Domestic Mammals, What Are the Ecological Mechanisms Driving Transmission, Spillover, and Disease Emergence? Front. Public Health 9:717941. doi: 10.3389/fpubh.2021.717941
Received: 31 May 2021; Accepted: 24 August 2021;
Published: 30 September 2021.
Edited by:
Kris A. Murray, Medical Research Council the Gambia Unit (MRC), GambiaReviewed by:
Serge Morand, Centre National de la Recherche Scientifique (CNRS), FranceYan-wei Cheng, Henan Provincial People's Hospital, China
Rachel Graham, University of North Carolina at Chapel Hill, United States
Paolo Bonilauri, Istituto Zooprofilattico Sperimentale Lombardia ed Emilia Romagna (IZSLER), Italy
Copyright © 2021 Nova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nicole Nova, bmljb2xlLm5vdmFAc3RhbmZvcmQuZWR1
 Nicole Nova
Nicole Nova