Corrigendum: Identification of Variable Importance for Predictions of Mortality From COVID-19 Using AI Models for Ontario, Canada
- 1School of Engineering, University of Guelph, Guelph, ON, Canada
- 2Adastra Corporation, Toronto, ON, Canada
The Severe Acute Respiratory Syndrome Coronavirus 2 pandemic has challenged medical systems to the brink of collapse around the globe. In this paper, logistic regression and three other artificial intelligence models (XGBoost, Artificial Neural Network and Random Forest) are described and used to predict mortality risk of individual patients. The database is based on census data for the designated area and co-morbidities obtained using data from the Ontario Health Data Platform. The dataset consisted of more than 280,000 COVID-19 cases in Ontario for a wide-range of age groups; 0–9, 10–19, 20–29, 30–39, 40–49, 50–59, 60–69, 70–79, 80–89, and 90+. Findings resulting from using logistic regression, XGBoost, Artificial Neural Network and Random Forest, all demonstrate excellent discrimination (area under the curve for all models exceeded 0.948 with the best performance being 0.956 for an XGBoost model). Based on SHapley Additive exPlanations values, the importance of 24 variables are identified, and the findings indicated the highest importance variables are, in order of importance, age, date of test, sex, and presence/absence of chronic dementia. The findings from this study allow the identification of out-patients who are likely to deteriorate into severe cases, allowing medical professionals to make decisions on timely treatments. Furthermore, the methodology and results may be extended to other public health regions.
Introduction
Since the outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), also known as COVID-19, in Wuhan, China in December 2019, the virus has caused chaos, extreme and widespread illness and mortalities, and shutdowns of country-wide economies around the globe. As a measure of the sheer magnitude of the impacts, more than 11.9% of the US population have tested positive for the virus, as of January 25, 2021 (1). In Ontario, COVID-19 has infected 282,511 people and resulted in 6,614 deaths as of February 13, 2021 (2).
In response to the day-to-day challenges of managing the COVID-19 pandemic, massive efforts continue to be made to protect the integrity of hospital systems to allow both treatment of COVID-19 cases as needed, as well as to avoid having to delay elective surgeries. However, the growth in caseloads continues to be an ongoing major concern, needing improved predictions to use for capacity planning. To better understand and assess the capacity of health care systems to respond to the medical needs arising from COVID-19, enormous efforts are being expended involving the use of epidemiological and machine learning modeling, to understand the projection of caseloads of the virus. An example is an artificial neural network (ANN) model developed by Abdulaal et al. to predict a patient-specific, point-of-admission mortality risk to inform clinical management decisions (3). Their modeling analyzed patient features including demographics, co-morbidities, smoking history, and presenting symptoms of 398 patients and achieved an area under the curve (AUC) of 86% (3). However, while epidemiological modeling has significant value for certain questions, the parameters of these models are typically resolved through some estimation procedure and future case counts through different classes are identified. As an example, the SEIR model approach assigned individuals to the susceptible (S), exposed (E), infected (I), and recovered (R) classes (1, 2, 4). However, the potential to predict how positive tests for COVID-19 at health centers will translate to ICU cases and mortality are limited. This information will be critical as health systems in North America are rapidly approaching capacity.
With issues of the second wave of the pandemic ongoing at the time of submission of this paper, interest continues to grow in obtaining more comprehensive caseload and patient data and predictions. Given there are now more accurate data available from the first and second waves, inclusion of these data has considerable merit. Until now, data security and privacy issues have to-date limited the accessibility of alternate and detailed data sources. Through obtaining detailed COVID-19 pandemic-related data, more powerful artificial intelligence (AI) techniques can be feasibly applied, offering the ability to uncover and understand the value of alternative interpretation procedures.
This paper describes results arising from use of AI models based on data from Ontario Health Data Platform (22 February 2020–20 October 2020) with the aim to improve prediction of caseloads and mortality risks in patients throughout the province of Ontario, with a population of more than 14 million. The dataset includes extensive information from the first wave and the beginning of the second wave of COVID-19 cases including Canadian census information by designated area. The census location information is based on a size of approximately three blocks and hence is able to capture representation of ethnicity, income level, and other social differences, and can therefore be considered robust. The results from this paper may be extended further to other public health regions throughout the world. The paper is organized as follows: related works are discussed in section Related Works. Section Materials and Methods describes the materials and methods. Results and discussion are found in section Results and Discussion and then the paper is concluded in section Conclusions.
Related Works
AI models can be employed in the prediction of patient-specific, point-of-admission mortality risks helping to inform clinical management decisions. Thus, using AI models, healthcare practitioners have ways to accurately predict the potential for an individual ending up in the Intensive Care Unit (ICU). As well, due to the very high AUC, AI models have the capability to consider a broad range of patient data, including demographics, co-morbidities, and much more, to predict patient-specific mortality risk following their hospital admission.
The emergence of “big data” in the early 2000s has proven very beneficial for public health investigations. The capabilities of AI modeling have been able to accurately predict numerous public health trends such as in-hospital mortality and detection of emerging epidemics (4–6). With the emergence of the COVID-19 global epidemic, big data and machine learning models have enormous potential to understand who, and where, people are most at risk.
Since March 2020, COVID-19 has spread around the globe, affecting every country and causing more than 2.1 million deaths worldwide (7). As a result, researchers have focused on COVID-19, to understand individuals most at risk as well as how to best distribute scarce resources. Identification of co-morbidities and creation of models to predict COVID-19 mortality using big data has significant potential.
Predictive analytics for assessing the in-hospital risk of patients and patient mortality have traditionally been limited to clinical decision rules (rules to reduce uncertainties in medical decision-making), but these approaches are not easily updated with new developments or data. Additionally, clinical decision rules may take years to identify and test and have been criticized for their lack of generally applicability to entire populations (4, 5). Hence, in the current situation, with the rapid spread of COVID-19, clinical decision support systems are still being developed for this particular virus (8).
Both Hernesneimi et al. and Taylor et al. note that one of the biggest challenges in developing models to predict mortality is assembling quality data (4, 5). However, this is outweighed by the benefits of a machine learning model, which provides improved performance and ability to be generalized to entire populations, or readily adapted to new populations (9). As well, AI models are updateable over time to recognize changing trends in medical care, all of which are attainable dimensions, if provided the large datasets (4, 5).
Rodriguez-Nava et al. used a random forest algorithm that predicted ICU admissions with an AUC of 0.82 and mortality with an AUC of 0.70 (10). Similarly, Jimenez-Solemm et al. used a random forest machine learning model using a Danish dataset with 3,944 COVID-19 patients that predicted ICU admissions with an AUC of 0.820, mortality with an AUC of 0.902, hospital admission with an AUC of 0.820 and ventilator treatment with an AUC of 0.815 (11). They found age and BMI the most important features for predicting hospital admission and ventilator treatment (11). Among all models, the parameters that were determined to have the highest severity of COVID-19 were heightened if male, had hypertension, and presence of dementia (11).
A study by Schöning et al. aiming to distinguish between severe and non-severe COVID-19 also found hypertension, along with diabetes mellitus (Type 2) and renal impairment to be prognostic of severe disease (12). Schöning et al. used a variety of machine learning models which were trained using data from the first wave in Switzerland and externally validated using data from the second wave (confirming findings to be generalizable) and achieved an accuracy of AUC values ranging from 0.86 (decision tree induction) to 0.96 (support vector machine) (12).
Kim et al. used Korean National Health Insurance data to identify co-morbidities and factors that increase mortality using multivariate logistic regression analysis with a confidence interval of 95% (13). A total of 9,148 COVID-19 patients, including 130 fatalities, were linked to their national health insurance record data, considering 298 medical conditions as co-morbidities (13). Williamson et al. performed a similar analysis on behalf of NHS England, with a sample size of 10,926 COVID-19 related deaths (14). Using a multivariable Cox model, Williamson et al. were able to estimate hazard ratios for characteristics such as smoking status, obesity, sex, age, deprivation, heart disease, etc. with a 95% confidence interval (14). Both studies identified age, deprivation, diabetes, bronchitis and severe asthma as top risk factors for COVID-19 (12, 13). Additionally, Kim et al. found that dental disorders were associated with high co-morbidity risk (13).
Machine learning has been found to predict mortality more accurately than non-machine learning models (4, 5). For example, mortality due to acute coronary syndrome is reliably predicted using the GRACE score, based on patients' variables in admission to the hospital. A machine learning model developed by Hernesneimi et al. achieved greatly improved results compared to traditional GRACE score (4). The machine learning model was developed using XGBoost and included phenotype data about the patient as well as hospital admission variables (4). The machine learning model outdid the traditional GRACE score method with an AUC of 0.904 compared to an AUC of 0.802 for GRACE score (4). Another example is a machine learning model developed by Taylor et al. to predict in-hospital mortality of patients with sepsis (5). This model considered 500 clinical variables including demographic information such as age, sex, insurance and employment status as well as health status and medical test results. Random Forest was reported to be a relatively interpretable option and produced the best results when compared with XGBoost, logistic regression or CART models (5).
A similar study from Zheng et al. used machine learning models to evaluate patients and predict rapid deterioration of in-hospital COVID-19 patients to determine triage priority (15). The model developed by Zheng et al. used the results of blood tests from 601 COVID-19 patients to predict deterioration up to 12 days before death occurred and reported that decreased lymphocyte count and increased lactate dehydrogenase levels, c-reactive protein levels and neutrophil counts, all corresponded with an increased likelihood for rapid deterioration (15). The best results were found using XGBoost and achieved an AUC of 0.953 for a small dataset (15). In addition, the illness trajectory (moderate, severe, critical – states as defined by Israeli Ministry of Health) of COVID-19 patients was predicted by Roimi et al. with an AUC of 0.88 using only patients' age, sex and day-by-day clinical state using a multistate Cox regression-based model (16).
Estiri et al. used nested generalized linear models to predict mortality using electronic health records (EHRs) of 16,709 COVID-19 age-separated cohorts from Mass General Brigham (a Boston-based non-profit hospital) (17). The data included were from the beginning of the EHR up to 14 days prior to the positive polymerase chain reaction (PCR) test to ensure COVID-related medications were not included in the model as risk factors (17). The model which included patients under the age of 45 had an AUC of 0.898, 0.789 in the 45–65 cohort and 0.753 in the 65–85 cohort, thus able to provide relatively high accuracy by relying only on data already stored in EHRs (17).
A prognostic machine learning model created by researchers from the Department of Decision Medicine at the University of Maastricht in The Netherlands reported the severity of COVID-19 in a patient (8). The model is based on the data from 299 patients' blood-test data and age and is readily available as an application (8). A similar application developed by ClosedLoop.ai determines a “vulnerability score” for an individual's susceptibility to severe COVID-19 illness (18).
Machine learning models have also been trained to predict rapid deterioration due to COVID-19 and even detect COVID-19 from lung X-Rays (19, 20). Zhu et al. used deep transfer learning to determine COVID-19 severity from X-rays and Elgendi et al. used deep neural networks to differentiate COVID-19 patient X-rays from other types of pneumonias (19, 20). Both methods used convolutional neural networks to a high degree of accuracy, reducing the need for radiologists (who may be in short supply in low-resource clinics) to read x-rays (19).
Mehta et al. used caseload and mortality data from early in the COVID-19 pandemic (March 2020), to determine the vulnerability of a county in the US to a large COVID-19 outbreak with high mortality (21). Mehta et al. used XGBoost to train a model that would predict which counties in the US were most at-risk using county-level population statistics such as age, gender, and density as well as CDC data for the health within a county (21). They reported that population size of a county was by far the most important variable, with population density, longitude, hypertension prevalence, chronic respiratory mortality rate, cancer crude rate, and diabetes prevalence, also playing large roles (21). While Mehta et al. do not state which counties are found to be the most at-risk, COVID-19 has reached virtually all parts of the U.S., including both rural and urban areas (21, 22).
As widely apparent from the numerous information venues, the magnitude of the impacts to people and the economies around the world are staggering. Hence, there is enormous interest in using available data, and in the development of models, that can combine available information in any manner to improve predictions of caseloads, supporting alternative strategies for predicting patient-specific mortality risk.
Materials and Methods
Dataset Description
For this research, to ensure adherence to strict security protocols, extensive data for 57,390 individual cases from Ontario Health Data Platform where data related to positive COVID-19 tests were collected between 22 February and 20 October 2020 were obtained for use in machine learning modeling. The dataset contained epidemiological and demographic information, recovery/mortality outcome information and co-morbidities of individuals residing in Ontario at census level. The attributes which proved most useful in the machine learning and statistical models are indicated in Table 1. Co-morbidities and age were collected from patient health records as of January 1, 2020; hence diagnoses of other medical conditions after this date were excluded. Of the 57,390 cases included in the dataset, 2,822 patients died of COVID-19 and the remaining 54,568 either recovered from COVID-19 or are still hospitalized.
Several input variables were derived using 2016 census data for the designated area of the individual patients. Census data in Canada are collected at postal code level and hence, represent approximately at three city block intervals. Census data relied upon included: ethnic concentration (of residential area), commuter concentration, median income and household size (these values are unlikely to change significantly between date of census and start of pandemic). These values were converted into quintiles (division of the population into 5 equal groups according to the distribution of input variables) with “1” being the lowest quintile, and “5” being the highest. Long-term care residents (LTC) did not include census-designated area information and therefore were represented with a zero value in the AI modeling. As well, for case-specific data for which substantial data were not available, those cases were removed from the modeling prior to undertaking the modeling (decreasing the 280,000 data to 57,390 cases actually used in the modeling) since the remaining cases represented a very substantial dataset for analyses. Individuals with missing data were not included in the analyses, however, for LTC residents' variables derived based on postal code, such as income, ethnic concentration, household size, and commuter percentage, were coded as zero since postal code information was not available for these individuals.
Model Development
This study compared three machine learning models and one statistical model. The following models were employed: artificial neural network (ANN), Random Forest (RF), extreme gradient boosting decision tree (XGBoost) all of which are tree structure machine learning models, and logistic regression (23–25). These models were adopted due to their high accuracy in binary classification problems as well as their prevalence/adoption in previous literature. The hardware used for developing the models included: a virtual server from OHDP specifically to run the code, 8 virtual CPUs and 128 GB of RAM, all running on Centos 7 Operating System. The software used included: R for coding and Rstudio as the integrated development environment (IDE).
Data Processing
Prior to model calibration, the dataset was randomly split into two segments, namely an 80% training dataset as an operational characterization and a 20% testing dataset where each model was calibrated using the training dataset and assessed for accuracy using the testing dataset. A grid search approach was used to adjust the hyperparameters of the models using a 10-fold cross-validation technique, repeated three times per model and optimized to produce the maximum area under the receiver operating characteristic curve (Area Under Curve, or AUC).
The logistic regression's input variables were chosen by a stepwise Akaike Information Criterion (AIC) function (23). The computer programming language R was used to develop and analyze all models (26). The final predicted outcome, recovered or died, was determined by the probability of mortality for each recorded case outcome generated by the models, based on a 50% threshold value ensuring highest total accuracy.
Results and Discussion
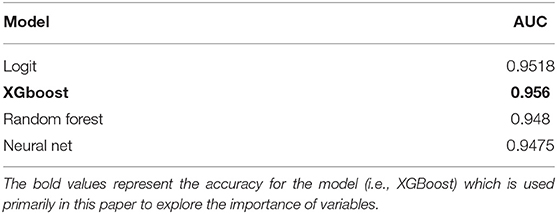
As a measure of accuracy, the Receiver Operating Characteristic curve (ROC) was calculated for each model. The Area Under the ROC curve was used to provide the basis of comparison between each model. Since an AUC value approaching “1” indicates high model accuracy while a value of 0.5 represents a model that is no better than random change, Table 2 demonstrates that all models utilized show strong predictive values.
All models developed have very high AUC values (>0.94). The most accurate model is XGBoost which has an AUC of 0.956. To the best of the authors' knowledge, this is the highest accuracy reported for COVID-19 mortality prediction models published to-date using Canadian data.
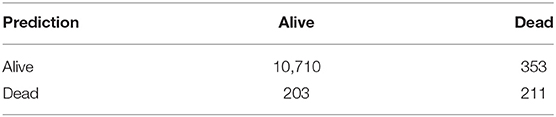
Although the AUC is a useful metric in understanding the overall accuracy of a binary prediction model, that metric does not provide specific accuracy for each class (accuracy in predicting survival vs. accuracy in predicting mortality). To highlight the accuracy of both predictions, a confusion matrix was developed. The confusion matrix as developed and highlighted in Table 3, compares the prediction accuracy of the XGBoost model with the reference (or actual outcome) of COVID-19 patients within the test dataset, assuming any patient with a predicted risk score below 0.5 will survive.
The confusion matrix indicates the model is extremely accurate in predicting which patients will survive, with 97% of the alive predictions actually surviving. The mortality prediction accuracy is substantially lower, with 49% of predicted deaths actually occurring. Even at 49%, the mortality prediction still provides a strong indicator on whether a COVID-19 patient will die and is helpful in strategic planning of possible medical caseloads, in terms of potential overload of the medical system.
It is noted that the mortality prediction accuracy of the modeling could be improved by increasing the risk score threshold, but this would also increase false negative predictions. Various methods have been created in an attempt to quantify the variable impact but many of them are flawed when using tree structure machine learning models (which includes XGBoost, Random Forests, decision trees, etc.). While the values could be adjusted to make a new set point, the models are quite accurate and are best represented by the AUC, meaning it is a good indicator of the actual risk of mortality. Overall, the results indicate the risk score predicted by the XGBoost model provides very strong insights regarding the outcome for Ontario COVID-19 patients, including the ability to plan for possible medical caseloads.
Variable Importance
Since the XGBoost model is the most accurate mortality prediction model developed for COVID-19 patients in Ontario, there is strong merit in investigating which variables are the most important and quantify how these variables contribute to the final predictions. To explore the impact of each variable on the final prediction, SHapley Additive exPlanations (SHAP) values have been used.
SHAP values determine the importance of a feature by comparing a model prediction with, and without, the feature for each observation within the training data. The SHAP values are calculated using SHAPforXGBoost R package and present the variable contribution on a log-odds scale (logarithm of the ratio of high mortality risk to low mortality risk) (27).
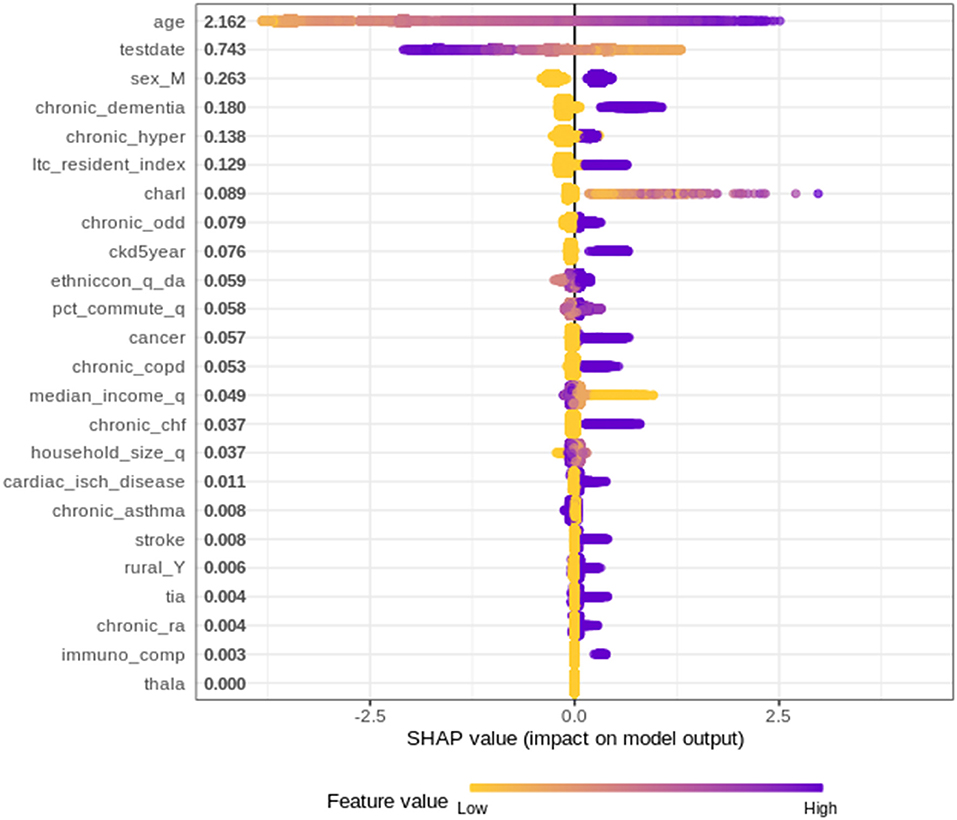
Figure 1 plots the SHAP value for each individual patient within the training dataset by input variable. The input variables, as listed on the y-axis, are ranked from most important (at the top) to least important (at the bottom) with their mean absolute SHAP value indicated next to the name. The X axis represents the SHAP value associated with each variable and patient within the training dataset (i.e., there is a plotted point for each case based on the influence that variable has on the prediction of that case). The color indicates whether the individual patients' input variable value was high (purple) or low (yellow). For example, in Figure 1 a “high” age has a high and “positive” impact on predicting mortality. The “high” comes from the purple color and the “positive” impact is shown in the X axis. Note, a range of SHAP values can exist per input variable value based on the SHAP values calculated for each observation and how they independently contribute to the machine learning model's predictions.
Overall, 24 variables were identified/ranked, with age as being unquestionably the most important variable for the XGBoost model. As a patient's age increases (approaches purple) the SHAP value impact increases, with a very high age being associated with an additional 2.5 increase in log-odds. The date of when someone tested positive for COVID-19 also demonstrated a strong impact on overall mortality risk, indicating as the positive test date increases (i.e., later in the pandemic) the risk of mortality decreases. The importance of the date when someone tested positive is very likely due to improved understanding of treatment options by the medical systems. Since the start of the pandemic in March 2020, health care workers and researchers worked fast to try to find the most effective ways to treat and prevent COVID-19 (e.g., including the use of antivirals and immune modulators, such as remdesivir, hydroxychloroquine, and dexamethasone) (28, 29). This finding described herein is supported by the European Medicines Agency indicated in June 2020 that it was discussing 132 potential treatments with developers, showing the rapid speed with which drugs and therapies against the virus are being developed (29, 30).
Other variables of high importance in terms of predicting the outcomes included “sex,” and chronic dementia, etc. on through the list of 24 variables, as depicted in Figure 1.
Conclusions
The impact of COVID-19 has been felt throughout populations around the world. In Ontario, COVID-19 has infected over 280,000 people with over 6,000 deaths (2). As the people of Ontario experience the second wave of the pandemic, the number of deaths associated with the pandemic will continue to increase. Understanding which Ontarians are most at risk will be important in determining how the medical system can most effectively deal with the pandemic, implement effective intervention strategies, and refine vaccination priorities.
This paper described a model with an accurate mortality prediction that can be used to assist Ontario's medical system in combating this pandemic and further highlight the risks faced by individual patients. By including patient demographics, co-morbidities, geographic, and census-based attributes, the XGBoost model developed surpassed all other Ontario mortality predictions published to date, with an AUC of 0.956; all four modeling approaches provided high AUCs where findings resulting from using logistic regression, XGBoost, ANN and RF, all demonstrate excellent discrimination (area under the curve for all four modeling approaches exceeded 0.948).
Findings related to the importance of 24 variables in characterizing mortality based on SHAP values, the importance of the variables found the most important parameters in order of importance were age, date of test, sex and chronic dementia, providing guidance to the medical professionals to identify the highest risks for out-patients who are likely to deteriorate into severe cases with the features indicating the risk of mortality.
Data Availability Statement
The data analyzed in this study is subject to the following licenses/restrictions: the dataset obtained from the Ontario Health Data Platform limits access to these data sets to trustworthy researchers, who are bound by law, ethics, the terms of their employment, and their research grants to protect the privacy and confidentiality of the data they work with. Requests to access these datasets should be directed to https://ohdp.ca/pre-application-form/.
Author Contributions
EAM, BS, JY, and SAG: conceptualization. BS: methodology and software. BS, EAM, JY, SAG, and BP: validation. BS, EAM, JY, SAG, and BP: formal analysis. EM: investigation. BS and EAM: data curation. BS, EAM, and BP: writing—original draft preparation. BS, EAM, JY, SAG, and BP: writing—review and editing. EAM: supervision, project administration, and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Natural Sciences and Engineering Research Council of Canada Alliance (Grant Number 401636) and University of Guelph Research Leadership Chair Professor funding.
Conflict of Interest
JY is employed by the company Adastra Corporation however Adastra has no financial or other conflicts of interest in this work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was supported by ICES, which is funded by an annual grant from the Ontario Ministry of Health (MOH) and the Ministry of Long-Term Care (MLTC). This study was supported by the Ontario Health Data Platform (OHDP), a Province of Ontario initiative to support Ontario's ongoing response to COVID-19 and its related impacts. The analyses, opinions, results, and conclusions reported in this paper are those of the authors and are independent from the funding sources. No endorsement by ICES, the OHDP, its partners, or the Province of Ontario is intended or should be inferred. Parts of this material are based on data and/or information compiled and provided by CIHI. However, the analyses, conclusions, opinions, and statements expressed in the material are those of the author(s), and not necessarily those of CIHI.
These datasets were linked using unique encoded identifiers and analyzed at ICES.
REB: The use of the data in this project is authorized under section 45 of Ontario's Personal Health Information Protection Act (PHIPA) and does not require review by a Research Ethics Board.
Access to datasets: The dataset from this study is held securely in coded form at ICES. While legal data sharing agreements between ICES and data providers (e.g., healthcare organizations and government) prohibit ICES from making the dataset publicly available, access may be granted to those who meet pre-specified criteria for confidential access, available at www.ices.on.ca/DAS (email: ZGFzQGljZXMub24uY2E=). The full dataset creation plan and underlying analytic code are available from the authors upon request, understanding that the computer programs may rely upon coding templates or macros that are unique to ICES and are therefore either inaccessible or may require modification.
OCR: Parts of this material are based on data and information provided by Cancer Care Ontario (CCO). The opinions, results, view, and conclusions reported in this paper are those of the authors and do not necessarily reflect those of CCO. No endorsement by CCO is intended or should be inferred.
ODB: We thank IQVIA Solutions Canada Inc. for use of their Drug Information Database.
Ontario Community Health Profiles Partnership (OCHPP): OCHPP created Ontario Marginalization Index (ON-Marg) which is a source for this paper as ON-Marg is used to understand inequalities in health and other social problems related to health among either population groups or geographic areas across Ontario.
Statistics Canada: Postal Code Conversion File and census data were adapted from Statistics Canada. This does not constitute an endorsement by Statistics Canada of this product.
References
1. Centres for Disease Control and Prevention. COVID Data Tracker Weekly Review. CDC (2020). Available online at: https://www.cdc.gov/coronavirus/2019-ncov/covid-data/covidview/index.html (accessed January 25, 2021).
2. Ontario Agency for Health Protection and Promotion (Public Health Ontario). Epidemiologic Summary: COVID-19 in Ontario – January 15, 2020 to February 15, 2021. Toronto, ON: Queen's Printer for Ontario (2021).
3. Abdulaal A, Patel A, Charani E, Denny S, Mughal N, Moore L. Prognostic modeling of COVID-19 using artificial intelligence in the United Kingdom: model development and validation. J Med Internet Res. (2020) 22:e20259. doi: 10.2196/20259
4. Scheele BC, Pasmans F, Skerratt LF, Berger L, Martel AN, Beukema W, et al. Extensive phenotype data and machine learning in prediction of mortalityin acute coronary syndrome–the MADDEC study. Ann Med. (2019) 51:156–63. doi: 10.1080/07853890.2019.1596302
5. Taylor RA, Pare JR, Venkatesh AK, Mowafi H, Melnick ER, Fleischman W, et al. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data–driven, machine learning approach. Acad Emerg Med. (2016) 23:269–78. doi: 10.1111/acem.12876
6. Poirier C, Lavenu A, Bertaud V, Campillo-Gimenez B, Chazard E, Cuggia M, et al. Real time influenza monitoring using hospital big data in combination with machine learning methods: comparison study. JMIL Public Health Surveill. (2018) 4:e11361. doi: 10.2196/11361
7. Johns Hopkins University and Medicine. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). Johns Hopkins University and Medicine (2020). Available online at: https://coronavirus.jhu.edu/map.html (accessed February 10, 2021).
8. Department of Precision Medicine. COVID-19 Risk. Department of Precision Medicine (2020). Available online at: https://covid19risk.ai/ (accessed November 18, 2020).
9. Tuite AR, Fisman DN, Greer AL. Mathematical modelling of COVID-19 transmission and mitigation strategies in the population of Ontario, Canada. CMAJ Open. (2020) 192:E497–505. doi: 10.1503/cmaj.200476
10. Rodriguez-Nava G, Trelles-Garcia DP, Yanez-Bello MA, Chung CW, Chaudry S, Khan A, et al. Using machine learning for prediction of poor clinical outcomes in adult patients hospitalized with COVID-19. Open Forum Infect Dis. (2020) 7:S162–3. doi: 10.1093/ofid/ofaa439.371
11. Jimenez-Solem E, Petersen TS, Hansen C, Hansen C, Lioma C, Igel C, et al. Developing and validating COVID-19 adverse outcome risk prediction models from a bi-national European cohort of 5594 patients. Sci Rep. (2021) 11:1–12. doi: 10.1038/s41598-021-81844-x
12. Schöning V, Liakoni E, Baumgartner C, Exadaktylos AK, Hautz WE, Atkinson A, et al. Development and validation of a prognostic COVID-19 severity assessment (COSA) score and machine learning models for patient triage at a tertiary hospital. J Transl Med. (2021) 19:1–11. doi: 10.1186/s12967-021-02720-w
13. Kim DW, Byeon KH, Kim J, Cho KD, Lee N. The correlation of comorbidities on the mortality in patients with COVID-19: an observational study based on the Korean National Health Insurance Big Data. J Korean Med Sci. (2020) 35:e243. doi: 10.3346/jkms.2020.35.e243
14. Williamson EJ, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. (2020) 584:430–36. doi: 10.1038/s41586-020-2521-4
15. Zheng Y, Zhu Y, Ji M, Wang R, Liu X, Zhang M, et al. A learning-based model to evaluate hospitalization priority in COVID-19 pandemics. Patterns. (2020) 1:100092. doi: 10.1016/j.patter.2020.100173
16. Roimi M, Gutman R, Somer J, Arie AB, Calman I, Bar-Lavie Y, et al. Development and validation of a machine learning model predicting illness trajectory and hospital utilization of COVID-19 patients—a nationwide study. JAMIA Open. (2021) ocab005. doi: 10.1093/jamia/ocab005
17. Estiri H, Strasser ZH, Klann JG, Naseri P, Wagholikar KB, Murphy SN. Predicting COVID-19 mortality with electronic medical records. NPJ Digit Med. (2021) 4:15. doi: 10.1038/s41746-021-00383-x
18. ClosedLoop. C-19 Index. (2020). Available online at: https://closedloop.ai/c19index/ (accessed November 18, 2020)
19. Zhu J, Shen B, Abbasi A, Hoshmand-Kochi M, Li H, Duong TQ. Deep transfer learning artificial intelligence accurately stages COVID-19 lung disease severity on portable chest radiographs. PLoS ONE. (2020) 15:e0236621. doi: 10.1371/journal.pone.0236621
20. Elgendi M, Nasir MU, Tang Q, Fletcher RR, Howard N, Menon C, et al. The performance of deep neural networks in differentiating chest x-Rays of COVID-19 patients from other bacterial and viral pneumonias. Front Med. (2020) 7:550. doi: 10.3389/fmed.2020.00550
21. Mehta M, Julaiti J, Griffin P, Kumara S. Early stage machine learning–based prediction of US county vulnerability to the COVID-19 pandemic: machine learning approach. JMIR Public Health Surveill. (2020) 6:e19446. doi: 10.2196/19446
22. John Hopkins University & Medicine. COVID-19 United States Cases by County. John Hopkins University & Medicine (2020). Avaialble online at: https://coronavirus.jhu.edu/us-map (accessed November 17, 2020).
24. Wiens J, Shenoy ES. Machine learning for healthcare: on the verge of a major shift in healthcare epidemiology. Clin Infect Dis. (2018) 66:149–53. doi: 10.1093/cid/cix731
25. Chen T, He T, Benesty M, Khotilovich V, Tang Y, Cho H, et al. xgboost: Extreme gradient boosting Version 1.0.0.2. (2021). Available online at: https://CRAN.R-project.org/package=xgboost (accessed January 26, 2021).
26. R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Core Team (2018).
27. Liu Y, Just A. SHAPforxgboost: SHAP Plots for 'XGBoost'. R Package Version 0.1.0. (2020). Available online at: https://github.com/liuyanguu/SHAPforxgboost/ (accessed January 26, 2021)
28. Greshko M. These promising coronavirus treatments are saving lives. Natl Geogr Res. (2020). Available online at: https://www.nationalgeographic.com/science/article/these-promising-coronavirus-treatments-are-saving-lives-cvdNatl (accessed April 28, 2021).
29. Robinson J. Everything you need to know about the COVID-19 therapy trials. Pharm J. (2021). Available online at: https://pharmaceutical-journal.com/article/feature/everything-you-need-to-know-about-the-covid-19-therapy-trials (accessed April 28, 2021).
30. Treatments and Vaccines for COVID-19. European Medicines Agency (2020). https://www.ema.europa.eu/en/human-regulatory/overview/public-health-threats/coronavirus-disease-covid-19/treatments-vaccines-covid-19 (accessed April 28, 2021).
Keywords: artificial intelligence, XGBoost, SHapley, COVID-19, mortality
Citation: Snider B, McBean EA, Yawney J, Gadsden SA and Patel B (2021) Identification of Variable Importance for Predictions of Mortality From COVID-19 Using AI Models for Ontario, Canada. Front. Public Health 9:675766. doi: 10.3389/fpubh.2021.675766
Received: 03 March 2021; Accepted: 24 May 2021;
Published: 21 June 2021.
Edited by:
Babak A. Ardekani, Nathan Kline Institute for Psychiatric Research, United StatesReviewed by:
Dejan Baskic, University of Kragujevac, SerbiaNa Hong, Digital China Health Technologies Co. Ltd., China
Copyright © 2021 Snider, McBean, Yawney, Gadsden and Patel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brett Snider, YnNuaWRlMDFAdW9ndWVscGguY2E=
 Brett Snider
Brett Snider Edward A. McBean1
Edward A. McBean1 Bhumi Patel
Bhumi Patel