- 1Department of Infectious Diseases, School of Medicine, Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, China
- 2Key Laboratory of Microbial Technology and Bioinformatics of Zhejiang Province, Hangzhou, China
- 3State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, Collaborative Innovation Centre for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital of Medicine School, Zhejiang University, Hangzhou, China
- 4Department of Hospital Epidemiology and Infection Control, School of Medicine, Sir Run Run Shaw Hospital, Zhejiang University, Hangzhou, China
Currently, the mechanism of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) transmission mechanism is unclear; however, it must be considered in conjunction with asymptomatic S. aureus strains colonization dynamics. This epidemiological study aimed to determine the role of the household in CA-MRSA transmission in China. Five patients with culture-confirmed CA-MRSA infection and five control patients were recruited from the Sir Run Run Shaw Hospital in Zhejiang, China, between December 2019 and January 2020. The household members of the patients, their pets, and environmental surfaces were sampled and screened for MRSA colonization. Mass spectrometry identification and antimicrobial susceptibility testing were performed on the MRSA isolates. Whole-genome sequencing and core genome multilocus sequence typing (cgMLST) were performed to determine the origin and transmission of the MRSA isolates in the households. Overall, 14 S. aureus-positive specimens (14.1%, 14/99) were obtained from the five households of patients with CA-MRSA infections, of which 12 (85.7%) were MRSA. The overall positivity of MRSA was 12.1% (12/99) among the samples from the CA-MRSA households, while no MRSA isolates were detected in the five control households. Most MRSA isolates belonged to epidemic CA-MRSA clones, such as ST59 (15/35, 42.9%) and ST508 (15/35, 42.9%). The cgMLST results confirmed that MRSA was transmitted among patients, contacts, and pets in the households and was present on environmental surfaces in the CA-MRSA patients' households. In conclusion, the study revealed that the home environment was an important MRSA reservoir. Therefore, focusing on MRSA decolonization in patients alone is not sufficient for infection control of CA-MRSA.
Introduction
Staphylococcus aureus is the main cause of infections in hospitals and communities and contributes significantly to the healthcare burden (1). S. aureus can cause various infections, from asymptomatic to invasive infections and from mild skin and soft tissue infections to life-threatening bacteremia. Since the mid-1990s, an increase in methicillin-resistant S. aureus (MRSA) infections has been reported among populations without exposure risk within the healthcare system, caused by community-associated MRSA (CA-MRSA) strains (2). However, the CA-MRSA dissemination and transmission mechanisms remain unclear.
Individuals who are asymptomatic carriers of S. aureus are the primary natural reservoir (3). Studies of CA-MRSA among family contacts of patients with CA-MRSA infection have found MRSA colonization rates of 8.7–37% (4). Unrecognized colonization of household contacts and contamination of the home environment may maintain colonization among individuals with MRSA. In some case reports, individuals who underwent appropriate antibiotic treatment for several months were cured only after disinfecting contaminated household surfaces (5, 6). In addition, methicillin-sensitive S. aureus (MSSA) and MRSA have been recovered from environmental samples collected in homes in which none of the inhabitants had signs of apparent infection (7, 8). There are still many unanswered questions about CA-MRSA colonization in the family environment. Some specific MRSA clones, such as ST-59 and ST-508, are prevalent in China. However, the specific transmission mechanism of CA-MRSA is unclear (9).
Thus, we conducted a pilot epidemiological study of the households of patients with CA-MRSA infection in China. We aimed to determine the potential role of other household members and the household environment in CA-MRSA transmission.
Materials and Methods
Participant Recruitment
Five patients diagnosed with CA-MRSA infection, including three with skin and soft tissue infections, one with septic arthritis, and one with mastoiditis, were enrolled in the study between December 2019 and January 2020. According to the United States Centers for Disease Control and Prevention definition, CA-MRSA infection is defined as any MRSA infection diagnosed in an outpatient or within 48 h of hospital admission in the absence of healthcare-associated MRSA risk factors. MRSA risk factors include hemodialysis, surgery, residence in a long-term care facility, hospitalization during the previous year, presence of an indwelling catheter or percutaneous device at culture time, or previous MRSA isolation from the patient (2). Five healthy subjects without Staphylococcus infection and their households were enrolled as the control group for comparison. These healthy subjects were all adult volunteers who lived in Zhejiang province without the following exclusion criteria: a history of S. aureus infection or colonization, chronic diseases and residence in a long-term care facility or hospitalization during the previous year, and medical workers. The five patients' households were labeled as A, B, C, D, and E, and the five control households were labeled F, G, H, I, and J.
Data and Specimen Collection
One visit was conducted at the home of each case patient. Samples were collected from four sources: the home environment, household members, pets, and poultry (10). Environmental surfaces presumed to be frequently touched (handles of doors, bathroom basin and kitchen sink, sofas, and computer keyboard and mouse) and used by most household members or presumed to play a role in transmission were sampled. We also collected samples from the nostrils of each family member and hairs on the backs of pets, such as dogs and cats, and livestock (Figure 1).
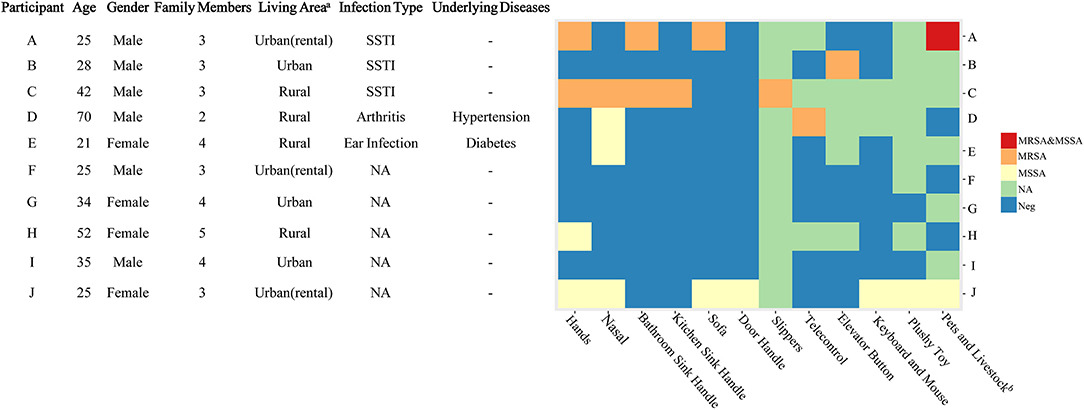
Figure 1. Characteristics of the participants and the methicillin-resistant Staphylococcus aureus isolates in their households. A, B, C, D, and E were the patients with CA-MRSA infection, and F, G, H, I, and J were the healthy subjects. CA, community-acquired; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible Staphylococcus aureus; NA, not available; Neg, negative; SSTI, skin and soft tissue infection. aUrban areas are densely populated with populations of ≥800,000. bDogs, cats, and chickens were included.
Samples were obtained from each environmental surface using a pre-wet sponge bar with Neutralizing Buffer (3M, Saint Paul, MN) using standard procedures (10). All patients and healthy subjects were required to refrain from any additional cleaning measures before sampling.
Culture, Identification, and Antimicrobial Susceptibility Testing
The samples were transported to the laboratory immediately after sampling, and each sample was inoculated into 2 mL of trypsin soy broth and incubated at 37°C overnight. The next day, 50 μL of the medium was streaked on CHROMagar Staph aureus plates (CHROMagar, Paris, France) and incubated at 37°C for 24 h. On the second day, we selected up to three pink or mauve colonies per sample and incubated them at 37°C for 24 h for further identification. Identification was performed based on mass spectrometry analysis using a microTyper MS (Skyray, Jiangsu, China) analyzer. Antimicrobial susceptibility testing was performed using agar dilution, and a D-zone test was performed according to the 2020 Clinical and Laboratory Standards Institute (CLSI) guidelines (cefoxitin, ciprofloxacin, levofloxacin, moxifloxacin, gentamicin, tetracycline, linezolid, vancomycin, trimethoprim-sulfamethoxazole, daptomycin, rifampicin, erythromycin, and clindamycin.) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (fosfomycin) (11). S. aureus ATCC 29213 was used as the control strain for antimicrobial susceptibility testing (12). All the isolates, MSSA and MRSA identified from the CHROMagar plates, were stored at −80°C for further analyses.
Whole-Genome Sequencing and Core Genome Multilocus Sequence Typing
Genomic DNA was extracted from S. aureus cultures using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions. All MRSA isolates recovered from the index patients and households were sequenced on an Illumina HiSeq X-10 platform using the 2 × 150-base-pair paired-end mode (Illumina, San Diego, CA) (11). We selected one isolate from each MSSA-positive sample for sequencing. The derived short reads were assembled into contigs using CLC Genomics Workbench software (version 12.0.0; CLC bio, Aarhus, Denmark). Genome assemblies were imported into SeqSphere+ software (version 6.0.0; Ridom) as FASTA files for multilocus sequence typing (MLST), core genome multilocus sequence typing (cgMLST) analysis, and staphylococcal protein A typing. The thresholds for interpreting clonality with cgMLST were as follows: ≤8 allelic differences were considered related, 9–29 allelic differences were considered possibly related, and ≥30 allelic differences were considered unrelated (13). For each household sample, S. aureus isolates with identical sequence types (ST) were considered to be redundant isolates.
Statistical Analysis
Fisher's exact test was used to determine the significance of differences in the MRSA isolation rates between the households of the patients with CA-MRSA and the control households. P-values of <0.05 were considered statistically significant.
Ethics
This study was approved by the local ethics committees of Sir Run Run Shaw Hospital (approval no. 20191201-1). Written informed consent was obtained from all participants or from a guardian if the participant was a minor.
Results
Study Participants
The mean age of the five CA-MRSA case-patients was 37 years (range: 21–70 years), and four out of five were male (Figure 1). The average time from the day of MRSA isolation to study enrolment was 19 days (range: 6–38 days). The average distance between the patients' houses and the Sir Run Run Shaw Hospital was 43 km (range: 6–88 km). Two patients lived in urban areas (>800,000 individuals in the census), while the other three lived in rural areas. Patient A had two cats and patient D had one dog in their households. The characteristics of the households of the control subjects are shown in Figure 1.
Prevalence of MRSA in the Household Contacts, Environment, and Pets
A total of 99 and 104 samples were collected from the households of the five CA-MRSA infection patients and healthy subjects, respectively. Up to three colonies were taken from each sample for identification and susceptibility testing as described in the methods. In total, 35 S. aureus isolates, including 32 MRSA and three MSSA, were identified from the samples of CA-MRSA patients' households, whereas only nine S. aureus isolates were identified from the samples of the healthy control households. None of the nine S. aureus isolates from the control households were MRSA strains. Thirty-five S. aureus isolates from patient households (32 MRSA and three MSSA), five MRSA isolates from index patients, and nine MSSA isolates from healthy control households were sequenced and analyzed (Supplementary Table 1). After removing the redundant S. aureus isolates, the total positive rate of S. aureus samples was 14.1% (14/99) in the case households and 8.7% (9/104) in the control households (P = 0.27). Of the 14 S. aureus-positive samples from the case households, 12 (85.7%) were MRSA strains (Table 1).
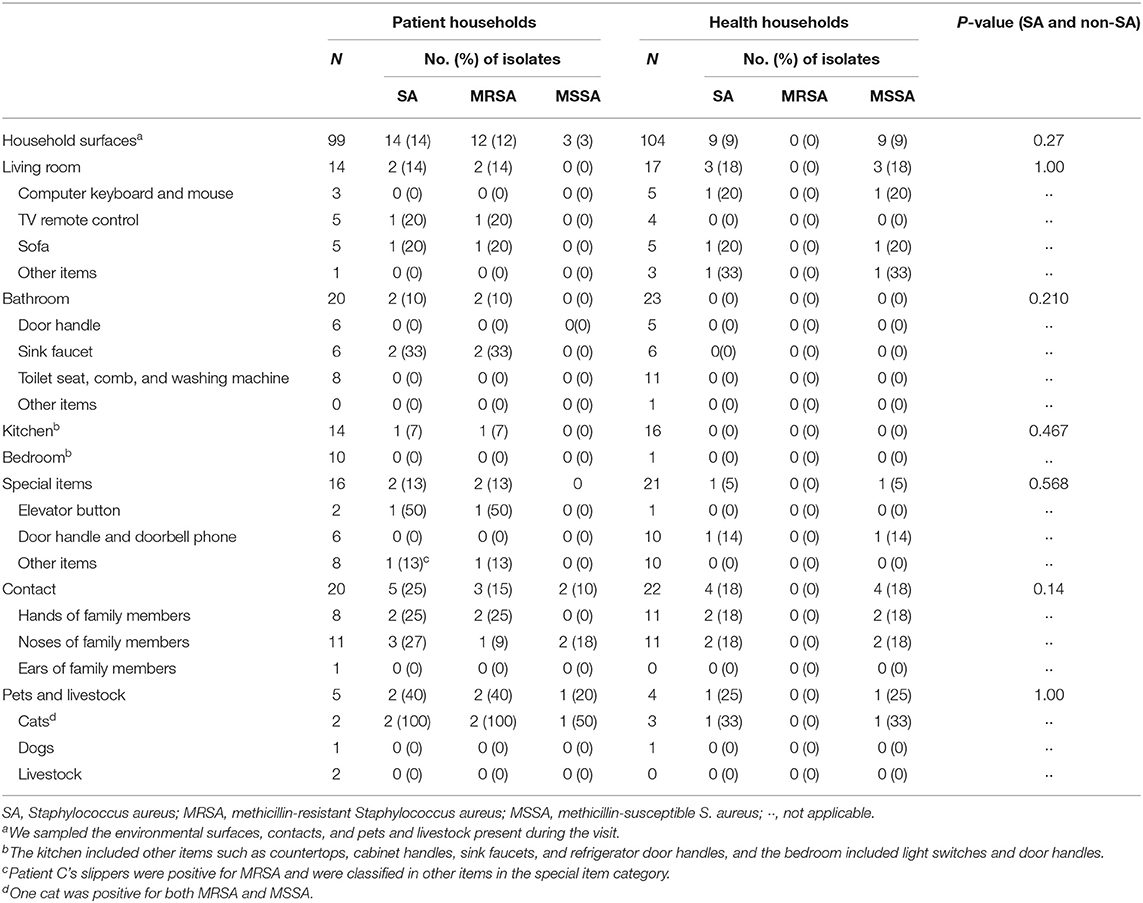
Table 1. Prevalence of Staphylococcus aureus on domestic environmental surfaces, contacts, and pets and poultry.
Among the five CA-MRSA patients' households, four had MRSA-contaminated environments, with a median number of two positive environmental samples (range: 0–5) per household. In the households of patients A, C, and D, MRSA was isolated from the handles of the bathroom basin, handles of kitchen sinks, slippers, sofa, and television remote control. However, from patient B's household, the only MRSA-positive environmental sample was obtained from the elevator button outside the patient's apartment. In patient E's household, none of the environmental surface samples tested positive for MRSA. In the control households, only MSSA isolates were found in the environmental samples from participant J's and H's households (Figure 1). We identified MRSA carriage in two cats of patient A, and one of the cats carried both MRSA and MSSA. No MRSA was isolated from the sample taken from patient D's dog.
Antimicrobial Susceptibility Testing and Molecular Typing
The minimum inhibitory antibiotic concentrations of 12 MRSA isolates from households and five MRSA isolates from patients are shown in Table 2. Fosfomycin results were interpreted in accordance with the EUCAST while others were according to the CLSI. As can be seen from Table 2, these strains were all resistant to cefoxitin, and none were resistant to gentamicin, linezolid, vancomycin, trimethoprim-sulfamethoxazole, daptomycin, or rifampicin. Only two isolates (Patient D and his television remote control) showed intermediate to ciprofloxacin, levofloxacin, moxifloxacin. Additionally, one isolate (Patient B) was resistant to tetracycline. As for fosfomycin, 94.1% (16/17) of strains were susceptible. Moreover, 70.6% (12/17) of the MRSA isolates were resistant to erythromycin, while 29.4% (5/17) were intermediate. According to the D-test method, 17.6% (3/17) of these strains showed inducible clindamycin resistance, whereas constitutive clindamycin resistance was detected in 47.1% (8/17) of all MRSA strains.
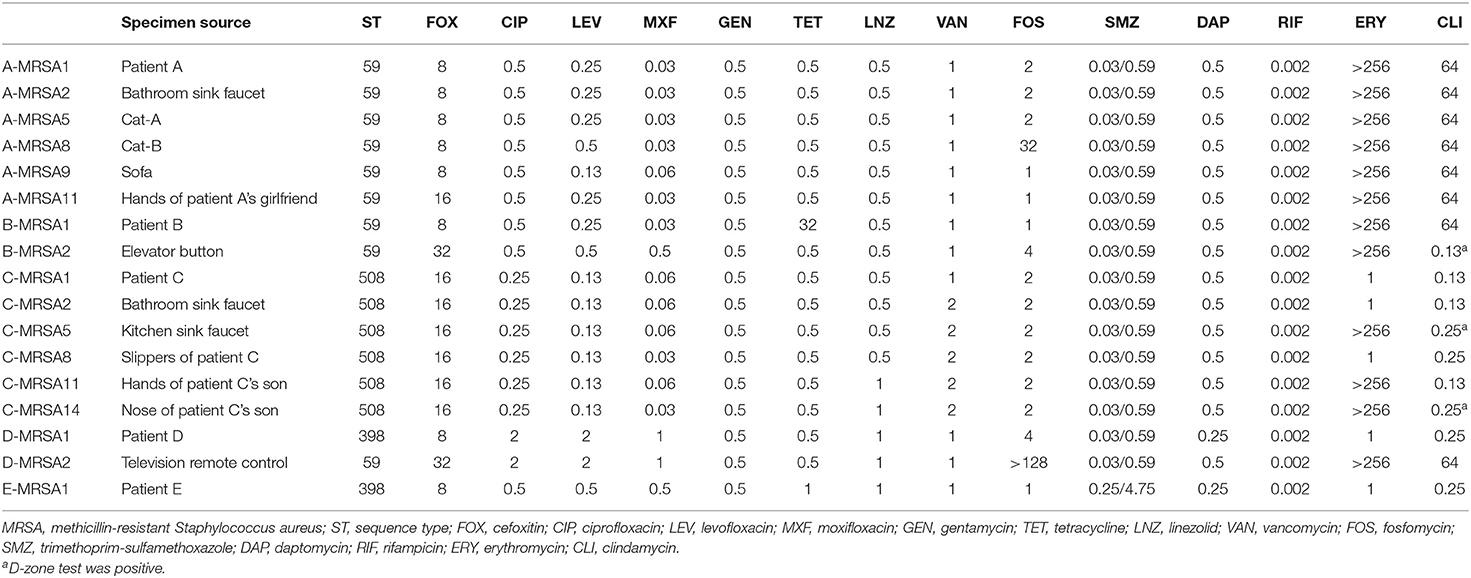
Table 2. Minimum inhibitory antibiotic concentrations of MRSA isolates from patients and their households.
To determine the MRSA transmission pattern, we performed whole-genome sequencing and molecular typing of the S. aureus isolates recovered from the patients and their households. We found that half of the MRSA isolates were predominantly CA-MRSA clones, including ST59 and ST72 (14). The ST of the MRSA clones from patients A and C matched that of the positive household samples. Patient A was infected with MRSA ST59, and the MRSA isolates from the handles of his sink handles of bathroom, sofa, and cats had the identical ST. According to the cgMLST analysis, the MRSA isolates from patient A and his household were closely related with no allelic difference or only one allelic difference from each other. Three allelic differences between two MSSA isolates from patient A's cat were identified (Figure 2). According to the molecular typing results, the two MSSA strains were not related to the isolated MRSA strain. Similarly, the MRSA isolate ST from patient C was ST508 which was identical to the ST of the MRSA isolates from the corresponding environmental surfaces and contacts. The cgMLST results showed that the number of allele differences among these isolates varied from 1 to 19, indicating that they were related (Figure 2). The results of SCCmec typing are shown in Supplementary Table 2.
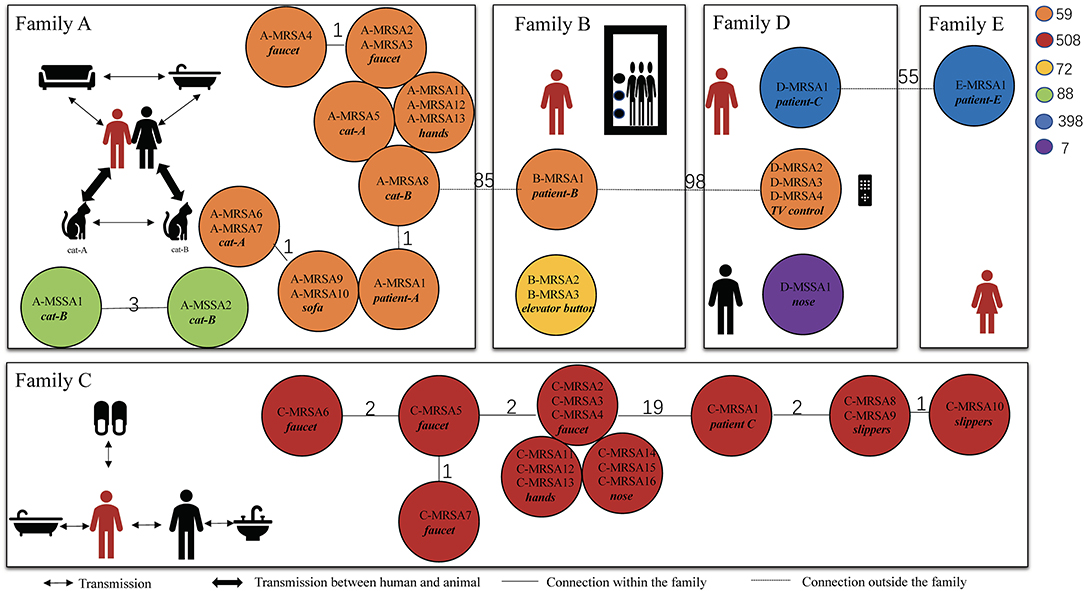
Figure 2. Transmission pattern suggested by core genome multilocus sequence typing of methicillin-resistant Staphylococcus aureus. These isolates were collected from the five patients and their households. The persons in red are patients with CA-MRSA infection, and the ones in black are family members. The different-colored circles represent different ST-type CA-MRSA strains. The number on the line between the circles of different colors represents the number of core allele differences between the isolates. The source of specific specimens is shown in Table 3. CA, community acquired; MRSA, methicillin-resistant Staphylococcus aureus; ST, sequence type; MSSA, methicillin-susceptible Staphylococcus aureus.
In patients B and D, the STs of the MRSA isolates recovered from the household environmental samples did not match those of the MRSA isolates from the patients. The MRSA isolates from patient B were ST59; however, the MRSA isolates recovered from the elevator button of his apartment were ST72. ST398 MRSA isolates were identified in patients D and E; however, ST59 MRSA on the television remote control was the only ST identified in the environmental samples from patient D. The cgMLST results confirmed that there was no relationship between the MRSA isolates from patients B and D and the isolates from their household samples. In addition, no MRSA isolates were found in the environmental samples from patient E's household.
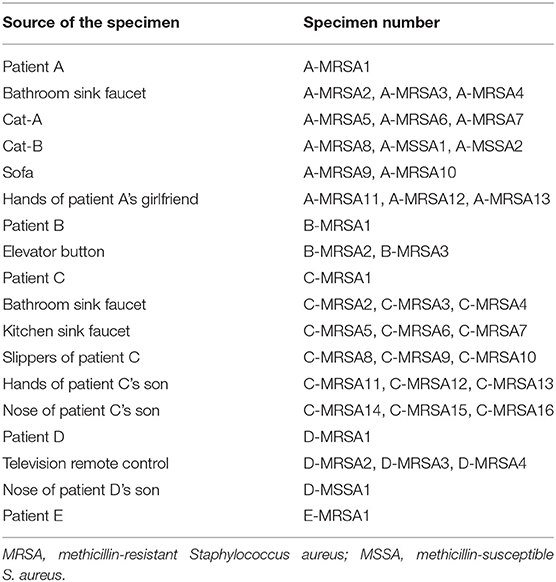
Table 3. Specific sources of specimens in Figure 2.
Discussion
The household environment is considered an important reservoir of MRSA (15). In our study, environmental MRSA contamination was found in four of the five households of patients with CA-MRSA infections. In contrast, in healthy subjects, no MRSA was identified in their households. Our data demonstrated that the household environment plays an important role in CA-MRSA infection. It has been reported that epidemic clones tend to “ping-pong” among family members, leading to a high incidence of repeated infections (16). In a cross-sectional study by Fritz et al., MRSA was detected on environmental surfaces in 46% of households. The bed linens, television remote controls, and refrigerator door handles were most commonly colonized (17). In another study by Shahbazian et al., which included 95 homes of patients diagnosed with CA-MRSA infection, MRSA was recovered from 68% (65/95) of the homes at baseline (18). MRSA prevalence in the household environment in the present study was relatively higher than that reported in previous studies, which might be explained by the limited number of cases included in our study.
A previous study found animals to be an important reservoir of MRSA (19). Although no MRSA isolates were identified from livestock in our study, Bi et al. reported that CA-MRSA isolates with a common genotype could be isolated from humans and pigs, suggesting that human-to-pig transmission of CA-MRSA could occur (20). In our study, two cats belonging to patient A and one cat belonging to healthy subject J were colonized by S. aureus. MRSA isolates belonging to the epidemic CA-MRSA clone were identified from two cats recognized as the infection source according to the cgMLST findings. Our results indicated that pets can be reservoirs and play a role in S. aureus and MRSA transmission. In the HOME study conducted by Mork et al. in the United States, 44% (68/154) of pets sampled were colonized with S. aureus and 29% with MRSA; the molecular typing results showed that the pets were often transmission recipients (9). In our study, MRSA was found in two of the three pets tested, which is a higher prevalence than in other studies.
In addition to the traditional molecular type, we applied cgMLST based on whole-genome sequencing, which provides improved resolution, to trace the MRSA strains' transmission. The MRSA isolates from households were predominantly CA-MRSA clones, such as ST59, ST72, and ST508 (single-locus variants of CA-MRSA clone ST45). Specifically, two patients were infected with MRSA ST59, the predominant clone in China and other Asian countries (21). The cgMLST results also confirmed the relationships among the patients' strains, contacts, environmental surfaces, and pets. In recent years, MRSA ST59 has been recognized as an epidemic lineage in Asia which accounts for 56% of pediatric CA-MRSA infections in Taiwan (22). Moreover, it has been associated with an increasing proportion of CA-MRSA infections in mainland China (23). Pang et al. provided evidence for cross-country transmission potential of MRSA ST59 through the food chain (24). Our data demonstrated that environmental contamination and pets also contribute to the success of MRSA ST59 in China. MRSA ST398, another important lineage confirmed as a cause of livestock-associated MRSA in Asia, Australia, and the Americas, was identified in two patients in this study. Although no MRSA ST398 was recovered from the corresponding households, we could not rule out the possibility of MRSA ST398 contamination in the household because of the limited number of samples collected during a single visit. Previous studies have shown that this new CA-MRSA ST has the potential for serious and fatal infections and should be monitored for its potential spread. Environmental decontamination should be considered as a strategy to prevent the future spread of MRSA ST398.
In our study, MRSA colonization was detected in pets and household environments; thus, decolonization is important. Research about the decolonization of MRSA has been published. Ho et al. revealed that decolonization may be more likely to succeed because most CA-MRSA carriers are otherwise healthy and do not have any indwelling medical equipment that could be used as permanent settlements (25). As for methods of decolonization, Hogan et al. demonstrated a 5-day intervention that consists of intranasal mupirocin application and dilute bleach water baths could reduce MRSA colonization in the months following the one-time visit; however, this effect waned over time (26).
Our study has several limitations. Because it is a pilot study, only five patients were enrolled. A relatively small number of samples were collected from each patient's household, limiting our statistical analysis of the epidemiology of MRSA transmission and risk factors of MRSA infection. However, because epidemiological surveys of MRSA in households are lacking in China, this study provides important preliminary evidence to explain the transmission mechanism of MRSA in Chinese communities. Second, a one-time visit is insufficient to obtain a comprehensive survey of the household environment of patients with MRSA infection. However, the isolates and genome data obtained in this pilot study will help establish a household MRSA database as a basis for further research.
Conclusions
In conclusion, this study confirmed that CA-MRSA is transmitted in the home environments of patients with CA-MRSA. Household environmental decontamination should be considered as a strategy to prevent CA-MRSA from spreading, particularly among households where an infection has occurred.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://bigd.big.ac.cn/bioproject/browse/PRJCA004387.
Ethics Statement
The studies involving human participants were reviewed and approved by local ethics committees of Sir Run Run Shaw Hospital. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YC, YY, and FZ designed the study. FZ, HZ, and ShuJ drafted the article. FZ and HZ obtained the data. EX, LD, ZW, SheJ, PS, and HW did the experiments. LS, YY, and YC revised the article. All authors have approved the final article.
Funding
This work was supported by the National Natural Science Foundation of China (grant 81971977), the Natural Science Foundation of Zhejiang Province (grant LQ20H190005), and the National Science and Technology Major Project of the 13th 5-year Plan (grant 2018ZX10714002). The funders were not involved in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Editage for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2021.658638/full#supplementary-material
References
1. Lister JL, Horswill AR. Staphylococcus aureus biofilms: recent developments in biofilm dispersal. Front Cell Infect Microbiol. (2014) 4:178. doi: 10.3389/fcimb.2014.00178
2. David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. (2010) 23:616–87. doi: 10.1128/CMR.00081-09
3. Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. (2018) 31:e00020–18. doi: 10.1128/CMR.00020-18
4. Nerby JM, Gorwitz R, Lesher L, Juni B, Jawahir S, Lynfield R, et al. Risk factors for household transmission of community-associated methicillin-resistant Staphylococcus aureus. Pediatr Infect Dis J. (2011) 30:927–32. doi: 10.1097/INF.0b013e31822256c3
5. Masterton RG, Coia JE, Notman AW, Kempton-Smith L, Cookson BD. Refractory methicillin-resistant Staphylococcus aureus carriage associated with contamination of the home environment. J Hosp Infect. (1995) 29:318–9. doi: 10.1016/0195-6701(95)90284-8
6. Kniehl E, Becker A, Forster DH. Bed, bath and beyond: pitfalls in prompt eradication of methicillin-resistant Staphylococcus aureus carrier status in healthcare workers. J Hosp Infect. (2005) 59:180–7. doi: 10.1016/j.jhin.2004.06.016
7. Scott E, Duty S, Callahan M. A pilot study to isolate Staphylococcus aureus and methicillin-resistant S aureus from environmental surfaces in the home. Am J Infect Control. (2008) 36:458–60. doi: 10.1016/j.ajic.2007.10.012
8. Scott E, Duty S, McCue K. A critical evaluation of methicillin-resistant Staphylococcus aureus and other bacteria of medical interest on commonly touched household surfaces in relation to household demographics. Am J Infect Control. (2009) 37:447–53. doi: 10.1016/j.ajic.2008.12.001
9. Mork RL, Hogan PG, Muenks CE, Boyle MG, Thompson RM, Sullivan ML, et al. Longitudinal, strain-specific Staphylococcus aureus introduction and transmission events in households of children with community-associated meticillin-resistant S aureus skin and soft tissue infection: a prospective cohort study. Lancet Infect Dis. (2020) 20:188–98. doi: 10.1016/S1473-3099(19)30570-5
10. Miller LG, Eells SJ, David MZ, Ortiz N, Taylor AR, Kumar N, et al. Staphylococcus aureus skin infection recurrences among household members: an examination of host, behavioral, and pathogen-level predictors. Clin Infect Dis. (2015) 60:753–63. doi: 10.1093/cid/ciu943
11. Chen Y, Sun L, Wu D, Wang H, Ji S, Yu Y. Using core-genome multilocus sequence typing to monitor the changing epidemiology of methicillin-resistant Staphylococcus aureus in a teaching hospital. Clin Infect Dis. (2018) 67:S241–8. doi: 10.1093/cid/ciy644
12. Ji S, Jiang S, Wei X, Sun L, Wang H, Zhao F, et al. In-host evolution of daptomycin resistance and heteroresistance in methicillin-resistant Staphylococcus aureus strains from three endocarditis patients. J Infect Dis. (2020) 221:S243–52. doi: 10.1093/infdis/jiz571
13. Cunningham SA, Chia N, Jeraldo PR, Quest DJ, Johnson JA, Boxrud DJ, et al. Comparison of whole-genome sequencing methods for analysis of three methicillin-resistant Staphylococcus aureus outbreaks. J Clin Microbiol. (2017) 55:1946–53. doi: 10.1128/JCM.00029-17
14. Xiao M, Wang H, Zhao Y, Mao LL, Brown M, Yu YS, et al. National surveillance of methicillin-resistant Staphylococcus aureus in China highlights a still-evolving epidemiology with 15 novel emerging multilocus sequence types. J Clin Microbiol. (2013) 51:3638–44. doi: 10.1128/JCM.01375-13
15. Davis MF, Iverson SA, Baron P, Vasse A, Silbergeld EK, Lautenbach E, et al. Household transmission of methicillin-resistant Staphylococcus aureus and other staphylococci. Lancet Infect Dis. (2012) 12:703–16. doi: 10.1016/S1473-3099(12)70156-1
16. Knox J, Uhlemann AC, Miller M, Hafer C, Vasquez G, Vavagiakis P, et al. Environmental contamination as a risk factor for intra-household Staphylococcus aureus transmission. PLoS ONE. (2012) 7:e49900. doi: 10.1371/journal.pone.0049900
17. Fritz SA, Hogan PG, Singh LN, Thompson RM, Wallace MA, Whitney K, et al. Contamination of environmental surfaces with Staphylococcus aureus in households with children infected with methicillin-resistant S aureus. JAMA Pediatr. (2014) 168:1030–8. doi: 10.1001/jamapediatrics.2014.1218
18. Shahbazian JH, Hahn PD, Ludwig S, Ferguson J, Baron P, Christ A, et al. Multidrug and mupirocin resistance in environmental methicillin-resistant Staphylococcus aureus (MRSA) isolates from homes of people diagnosed with community-onset MRSA infection. Appl Environ Microbiol. (2017) 83:e01369–17. doi: 10.1128/AEM.01369-17
19. Aires-de-Sousa M. Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect. (2017) 23:373–80. doi: 10.1016/j.cmi.2016.11.002
20. Bi Z, Sun C, Börjesson S, Chen B, Ji X, Berglund B, et al. Identical genotypes of community-associated MRSA (ST59) and livestock-associated MRSA (ST9) in humans and pigs in rural China. Zoonoses Public Health. (2018) 65:367–71. doi: 10.1111/zph.12443
21. Li S, Ning X, Song W, Dong F, Zheng Y, Chen Q, et al. Clinical and molecular characteristics of community-acquired methicillin-resistant Staphylococcus aureus infections in Chinese neonates. APMIS. (2015) 123:28–36. doi: 10.1111/apm.12304
22. Chuang YY, Huang YC. Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in Asia. Lancet Infect Dis. (2013) 13:698–708. doi: 10.1016/S1473-3099(13)70136-1
23. Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. (2019) 17:203–18. doi: 10.1038/s41579-018-0147-4
24. Pang R, Wu S, Zhang F, Huang J, Wu H, Zhang J, et al. The genomic context for the evolution and transmission of community-associated Staphylococcus aureus ST59 through the food chain. Front Microbiol. (2020) 11:422. doi: 10.3389/fmicb.2020.00422
25. Ho PL, Cheung C, Mak GC, Tse CW, Ng TK, Cheung CH, et al. Molecular epidemiology and household transmission of community-associated methicillin-resistant Staphylococcus aureus in Hong Kong. Diagn Microbiol Infect Dis. (2007) 57:145–51. doi: 10.1016/j.diagmicrobio.2006.08.003
26. Hogan PG, Parrish KL, Mork RL, Boyle MG, Muenks CE, Thompson RM, et al. HOME2: household vs. personalized decolonization in households of children with methicillin-resistant Staphylococcus aureus skin and soft tissue infection - a randomized clinical trial. Clin Infect Dis. (2020) 1–10. doi: 10.1093/cid/ciaa752
Keywords: methicillin-resistant Staphylococcus aureus, whole-genome sequencing, households transmission, pets, colonization
Citation: Zhu F, Zhuang H, Ji S, Xu E, Di L, Wang Z, Jiang S, Wang H, Sun L, Shen P, Yu Y and Chen Y (2021) Household Transmission of Community-Associated Methicillin-Resistant Staphylococcus Aureus. Front. Public Health 9:658638. doi: 10.3389/fpubh.2021.658638
Received: 28 January 2021; Accepted: 07 May 2021;
Published: 31 May 2021.
Edited by:
Richard V. Goering, Creighton University, United StatesReviewed by:
Paulo J. M. Bispo, Harvard Medical School, United StatesEdet E. Udo, Kuwait University, Kuwait
Copyright © 2021 Zhu, Zhuang, Ji, Xu, Di, Wang, Jiang, Wang, Sun, Shen, Yu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Chen, Y2hlbnlhbkB6anUuZWR1LmNu; Yunsong Yu, eXZ5czExOUB6anUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Feiteng Zhu1,2†
Feiteng Zhu1,2† Hemu Zhuang
Hemu Zhuang Shengnan Jiang
Shengnan Jiang Yunsong Yu
Yunsong Yu Yan Chen
Yan Chen