
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Public Health, 10 June 2021
Sec. Infectious Diseases – Surveillance, Prevention and Treatment
Volume 9 - 2021 | https://doi.org/10.3389/fpubh.2021.599285
This article is part of the Research TopicApplication of Next Generation Sequencing (NGS) in Infection PreventionView all 12 articles
 Lindsay A. Rogers1
Lindsay A. Rogers1 Kayla Strong1
Kayla Strong1 Susan C. Cork1
Susan C. Cork1 Tim A. McAllister2
Tim A. McAllister2 Karen Liljebjelke1
Karen Liljebjelke1 Rahat Zaheer2
Rahat Zaheer2 Sylvia L. Checkley1*
Sylvia L. Checkley1*Enterococcus spp. have arisen as important nosocomial pathogens and are ubiquitous in the gastrointestinal tracts of animals and the environment. They carry many intrinsic and acquired antimicrobial resistance genes. Because of this, surveillance of Enterococcus spp. has become important with whole genome sequencing emerging as the preferred method for the characterization of enterococci. A scoping review was designed to determine how the use of whole genome sequencing in the surveillance of Enterococcus spp. adds to our knowledge of antimicrobial resistance in Enterococcus spp. Scoping review design was guided by the PRISMA extension and checklist and JBI Reviewer's Guide for scoping reviews. A total of 72 articles were included in the review. Of the 72 articles included, 48.6% did not state an association with a surveillance program and 87.5% of articles identified Enterococcus faecium. The majority of articles included isolates from human clinical or screening samples. Significant findings from the articles included novel sequence types, the increasing prevalence of vancomycin-resistant enterococci in hospitals, and the importance of surveillance or screening for enterococci. The ability of enterococci to adapt and persist within a wide range of environments was also a key finding. These studies emphasize the importance of ongoing surveillance of enterococci from a One Health perspective. More studies are needed to compare the whole genome sequences of human enterococcal isolates to those from food animals, food products, the environment, and companion animals.
A variety of Enterococcus spp. are commensals within the gastrointestinal tract (GIT) of humans and animals, while others exist within the broader environment; some enterococcal species have also emerged as important human pathogens, especially in nosocomial infections (1). Two enterococcal species, Enterococcus faecium and Enterococcus faecalis, are most commonly implicated in human disease (2). These species, in particular, can acquire antimicrobial resistance (AMR) and harbor virulence genes that give them advantages as opportunistic pathogens (3). Their acquisition of antimicrobial resistance genes (ARGs) can be chromosomally- and plasmid- mediated, arising from selection pressure through antimicrobial use and the transfer of ARGs on mobile genetic elements (MGEs) such as plasmids and transposons (4). Of particular concern is the rising emergence of vancomycin-resistant enterococci (VRE) (4). Acquired vancomycin resistance is mediated by various gene clusters termed VanA/B/D/E/G/L/M/N (4, 5). Each gene cluster codes for a different resistance mechanism. VanA and VanB are the most common clusters seen, are often hospital-acquired, and can be plasmid- or chromosomally- mediated (5–7). Two species, Enterococcus gallinarum and Enterococcus casseliflavus, have intrinsic vancomycin resistance that is chromosomally-mediated by the VanC gene cluster (4, 5).
These two species, along with others such as Enterococcus villorum, Enterococcus thailandicus, Enterococcus durans, and Enterococcus hirae, are more typical of animal- and environmentally- adapted species (8, 9). Their genomes reflect adaptations to specific niches; for example, clusters of orthologous groups (COGs) have been found for ethanolamine utilization as a carbon source in environmental species that are not found in E. faecium (8).
Other antimicrobials can co-select for vancomycin-resistance genes if there are multiple ARGs on a single mobile genetic element. This means that the use of other antimicrobials can lead to the acquisition of vancomycin-resistance genes, even if vancomycin is not used (10). Thus, the VRE that arise in this manner are also multi-drug resistant (MDR) or multi-class resistant (7) referring to organisms that have acquired resistance to two or more antimicrobials or those organisms that have acquired resistance to two or more classes of antimicrobials, respectively.
As AMR pathogens have emerged, the use of antimicrobials for prophylaxis and metaphylaxis in food producing animals has come under scrutiny for its potential to apply a selective pressure that contributes to the dissemination of AMR and MDR enterococci (7, 11). Perhaps the most classic example is in poultry and swine where the previous use of a vancomycin-related glycopeptide, avoparcin, as a growth promoter was associated with carriage of vancomycin-resistant E. faecium (VREfm) in treated herds or flocks through cross-resistance (7, 12). The occurrence of VREfm in food animals decreased after the avoparcin use in animals was banned (12); however, prolonged persistence of specific VREfm clusters in agricultural settings (e.g., VanA gene cluster on the Tn1546 transposon) were observed possibly due to co-selection for VRE through continued use of other antimicrobials, such as macrolide use in swine (7, 12, 13). Retrospective molecular and genetic studies have demonstrated that the VREfm isolated from hospital and agricultural setting are usually separate sequence types (7, 12, 13). In contrast, the same sequence types of vancomycin-resistant E. faecalis can be isolated from hospital settings and from farm animals (7, 12, 14). In human medicine, enterococci are often opportunistic pathogens that acquire resistance and arise in immunocompromised individuals in hospital settings. Often these patients have been treated with multiple classes of antimicrobials in an effort to control difficult to treat infections (6). These antimicrobials may include those deemed critically important for use in human medicine by WHO (10), leading to enterococci resistant to these antimicrobials circulating in the human population (10).
Due to the importance of Enterococcus spp. as potential human pathogens, their ability to easily acquire ARGs, and their ubiquitous nature in the GIT and the broader environment, many countries have added enterococcal species to their list of pathogens under surveillance. Their role as a GIT commensal also make enterococci useful as fecal indicator bacteria (15). Surveillance for Enterococcus spp. and the other so-called ESKAPE pathogens (ESKAPE is an acronym for the following six nosocomial pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) (16) is done on a national level in several countries. These include programs such as the Danish Integrated Antimicrobial Resistance Monitoring and Research Program (DANMAP) (17), USA's National Antimicrobial Resistance Monitoring System (NARMS) (18), the Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) (19), and the Colombian Integrated Program for Antimicrobial Resistance Surveillance (COIPARS) (15). Many hospitals and laboratories also have surveillance systems in place for specific organisms independent of national surveillance programs. Examples include JMI Laboratories' SENTRY Antimicrobial Surveillance Program and the University of Pittsburgh Medical Center-Presbyterian Hospital's Enhanced Detection System for Hospital-Acquired Transmission (UPMC EDS-HAT) (6, 20, 21). The European Antimicrobial Resistance Surveillance Network (EARS-Net) is a large surveillance program based on clinical antimicrobial resistance data from laboratories across Europe (22). The pharmaceutical industry also runs some important post-marketing surveillance programs to comply with licensing requirements of new antimicrobials, looking at potency and spectrum. Examples of these programs are the Zyvox Annual Appraisal of Potency and Spectrum (ZAAPS) and the Linezolid Experience and Accurate Determination of Resistance (LEADER) (23). Integrated surveillance programs survey and address AMR in humans, animals, and the environment from a One Health perspective, emphasizing the interfaces within the system. These programs collect samples and process isolates from various sources, including animal fecal samples, human screening samples, retail meats, wastewater, surface water, groundwater and soils (15). Human screening samples include those from hospital surveillance programs of in-patients and samples submitted to laboratories performing surveillance (6, 24–26). The data generated from the wide range of samples can be integrated with antimicrobial use data allowing for the monitoring of changes in antimicrobial resistance found in bacteria important to public and animal health (15). Information gleaned from surveillance can then inform policy and risk mitigation strategies to combat increasing AMR and protect antimicrobials important to human health (15). For example, surveillance of VRE through DANMAP allowed for the detection of VRE strains in broiler chickens and human isolates connected to the use of avoparcin for growth promotion in broilers. This detection led to the ban of avoparcin use in food animal production (7, 15).
Early surveillance was primarily based on traditional microbiology to determine phenotypic antimicrobial susceptibility profiles, molecular genomics methods such as polymerase chain reactions (PCR) to assess for the presence of resistance genes, and pulsed-field gel electrophoresis (PFGE) for DNA fingerprinting Multi-locus sequence typing (MLST) arose more recently to better assess genetic relationships among isolates (7, 27). The use of these technologies allowed for the phylogenetic study of sequence types, epidemiologic investigation and determination of the presence of specific ARGs. However, the development of whole genome sequencing (WGS) has provided a more in-depth and detailed analysis of enterococcal ARGs, phylogenetics, and virulence (7, 27). As whole genome sequencing has become more widely available and less expensive, many archived isolate collections are being reanalyzed and their genomes compared with new isolates (13, 28). Following WGS, it has become possible to utilize new sequence typing methods for enterococci, such as core-genome multi-locus sequence typing (cgMLST), allowing for better analysis of isolate relatedness across sample sources (28). WGS also allows for the identification of emerging strains, analysis of outbreaks, and the characterization of resistance and virulence genes and their locations and context in the bacterial genome. Due to these advantages many surveillance research groups have been transitioning to WGS-based approaches of isolate characterization. Sequencing compliments traditional microbiology approaches and offers a reliable method of characterizing ARGs and sequence types (27).
With the increasing popularity of WGS for bacterial pathogen surveillance, it is now imperative to review the progress that has been made toward surveillance methods and identify gaps in our surveillance knowledge. We have undertaken this scoping review to investigate and summarize the extent to which whole genome sequencing in surveillance studies has advanced our understanding of AMR in Enterococcus spp.
To investigate our research question, a scoping review was designed following the PRISMA-ScR extension for scoping reviews (29) and the guidelines laid out by the JBI Reviewer's Manual (30). This protocol was not registered with an online registration platform.
The Population, Concept, Context (PCC) framework (30) was used to develop the research question and search strategy. A search strategy was developed to return a broad range of studies that fit within the following population, concept, and context:
- Population: Enterococcus spp. that underwent whole genome sequencing
- Context: Enterococcus spp. isolates derived from surveillance-type studies (as described below)
- Concept: use of whole genome sequencing to better understand antimicrobial resistance.
After consultation with a librarian, three separate databases were selected for searching: PubMed (NCBI)1, Web of Science2, and CAB Abstracts3 All databases within Web of Science were included in order to include both the Web of Science Core Collection and BIOSIS. Searches were performed by a single reviewer (LR) on November 17, 2019, and email search alerts from each database were implemented to inform the reviewer of any new articles. New articles from alerts up to and including December 31, 2020 were included. The same search terms were used for all three databases, except for differences due to specific database formatting.The search terms and resulting number of articles for one database (Web of Science) are described in Table 1. To capture relevant gray literature, a manual search of the reference lists of included articles was completed during data extraction. Gray literature is information produced and distributed outside of academic publications such as government reports. The gray literature underwent the same screening process; however, the full text was read for screening if no abstract was available. The web application, Rayyan, was used for the organization of articles during the screening process (31).
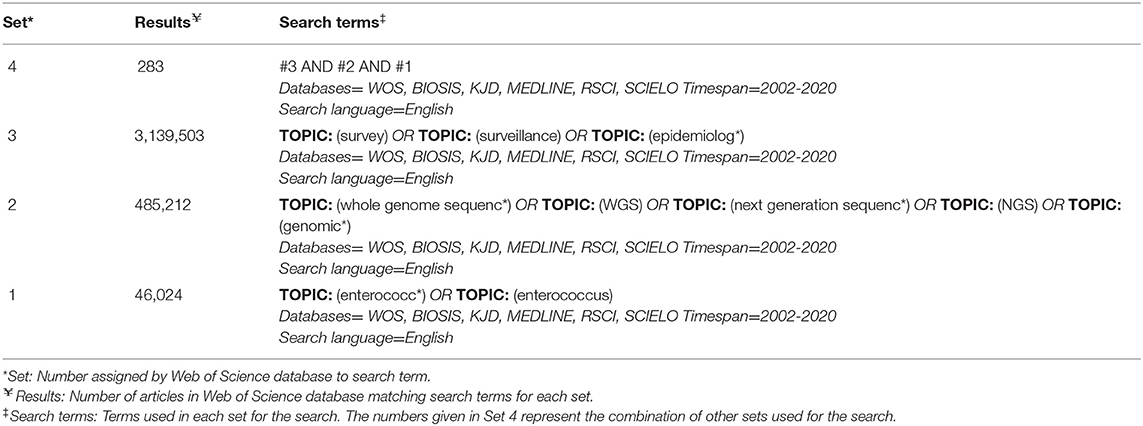
Table 1. Search results table from Web of Science database search with search terms and resulting number of articles.
Only journal articles and abstracts in the English language, published after 2002 were eligible for screening. The entire genome of E. faecium was first sequenced in 2000 (32); however, the assembly was not completed until 2012 (33, 34). The earliest available genome sequence of an Enterococcus sp. from the NCBI database is from 2002 (35). Given this information, 2002 was considered the earliest year that a publication would contain the information relevant to this scoping review. Any publication that was not an article or abstract (e.g., textbook, poster, or conference presentation) was excluded. Relevant gray literature articles were included and searched for, as described above. All screening was done independently by two reviewers (LR and KS). Any discrepancies were resolved in discussion with two other reviewers (SLC and SCC).
The article titles were screened initially and any article that was clearly about bacteria other than Enterococcus spp. was excluded. These articles needed to explicitly include the name of bacteria other than Enterococcus spp. in the title and not include terms relating to the taxonomy of enterococci. All articles that did not meet this exclusion criterion were included for the next screening step. The title screening was intentionally left broad to maximize the number of articles included.
Two screening steps were applied to the included abstracts. The first abstract screening step was performed to exclude any articles that did not include whole genome sequencing and antimicrobial resistance of Enterococcus spp. The abstract had to include all three pieces of information (i.e., WGS, AMR, and Enterococcus) in the abstract text. This step was also intentionally left broad and any articles that mentioned sequencing without providing information about whether or not the whole genome was sequenced were included to be screened based on methodology (described below). Following this, the abstracts were screened a second time using the following question, “did all or a portion of the Enterococcus spp. isolates in this study result from surveillance or screening for enterococci?” The following criteria for surveillance or screening were used:
• Isolates were from a collection maintained by a surveillance group (a surveillance group is defined as an organization collecting and analyzing bacterial isolates for surveillance of those particular bacteria such as CIPARS, SENTRY, or DANMAP). OR,
• A statement was included that the isolates were collected for screening or surveillance purposes. OR,
• Isolates were collected for the sole purpose of genomic comparison. OR,
• The article was published in a journal which included “Surveillance” in the journal name.
Articles needed to meet one or more criteria. Articles that did not meet these criteria, such as those with only clinical isolates, were excluded. Articles that were unclear if they met the surveillance inclusion criteria through their abstract were screened based on methodology as described below.
As stated previously, some abstracts did not contain enough detail to determine if they met the inclusion/exclusion criteria. These articles were further screened through the reading of their methods sections, following the same criteria as for abstract screening. Articles that did not meet the inclusion criteria during the methods review were excluded.
The number of articles excluded at each screening step is displayed in Figure 1.
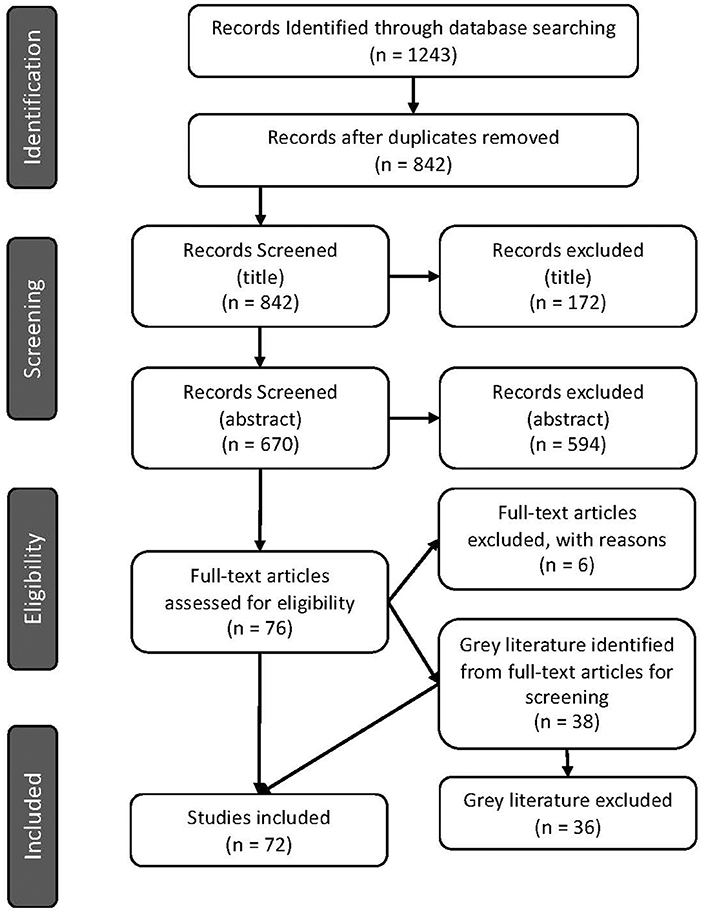
Figure 1. PRISMA flow diagram indicating number of articles screened and identified for inclusion in the scoping review (29).
Data was extracted independently by two reviewers (LR and KS) to answer the research question. The chart was trialed with 15 articles to ensure the reviewers were extracting comparable information. The completed tables from data extraction were compared by one reviewer (LR), and any discrepancies in information were resolved in discussion between the two original reviewers (LR and KS). No critical appraisal of articles was performed and all articles were included regardless of study quality. The methodology of data extraction is outlined in Table 2.
Seventy-two articles were included after the full-text review (Figure 1). Of these, 70 were primary research articles and two were gray literature (government reports) (25, 36). All articles were published in 2015 or later. The corresponding authors were from seventeen (17) different countries with most from Australia (16.7%), Denmark (13.9%), and Germany (13.9%), followed by the USA (12.5%), then the UK (6.9%) and 5.6% from each of Canada, China, the Netherlands and Portugal. Corresponding authors were also from Brazil (4.2%), Italy (1.4%), Japan (1.4%), Colombia (1.4%), Spain (1.4%), South Africa (1.4%), South Korea (1.4%), and Saudi Arabia (1.4%) (Figure 2). Additional information is available in Supplementary Table 1.
Just under half of the studies were not associated with a specified surveillance group (48.6%). The remaining articles were associated with government funded programs (e.g., DANMAP, NARMS), within hospital screening or surveillance programs, or private/industry funded surveillance programs (Table 3 and Supplementary Table 1).
The study objectives could be broadly divided into five categories, as shown in Table 4. These include (i) epidemiology or prevalence studies, (ii) genetic and/or molecular characterization, (iii) comparative genomics, (iv) the description of a novel finding, and (v) the comparison of techniques or development of a new technique. The categories were formed by assessing keywords within the stated objectives. For example, objectives to “determine the prevalence of…” or “perform comparative genomics of…” would fit into the categories of epidemiology or prevalence studies and comparative genomics, respectively. The categories were further broken down into the sample source location, and whether or not the article was studying a specific resistance gene (e.g., vanA), AMR profile (e.g., VRE), or another specific category (e.g., another specific genetic element such as a transposon). Articles could fall into more than one type of objective but generally stayed within the same category as the source location. The majority of articles were an epidemiological or prevalence study (54.2%), most commonly conducted within a defined region. Often these studies included human clinical data and hospital isolates but were not specific to just one hospital. Only three studies compared techniques or developed new ones, and this was in conjunction with other objectives of the study. Details of the objectives for each study are presented in Table 4 and Supplementary Table 2.
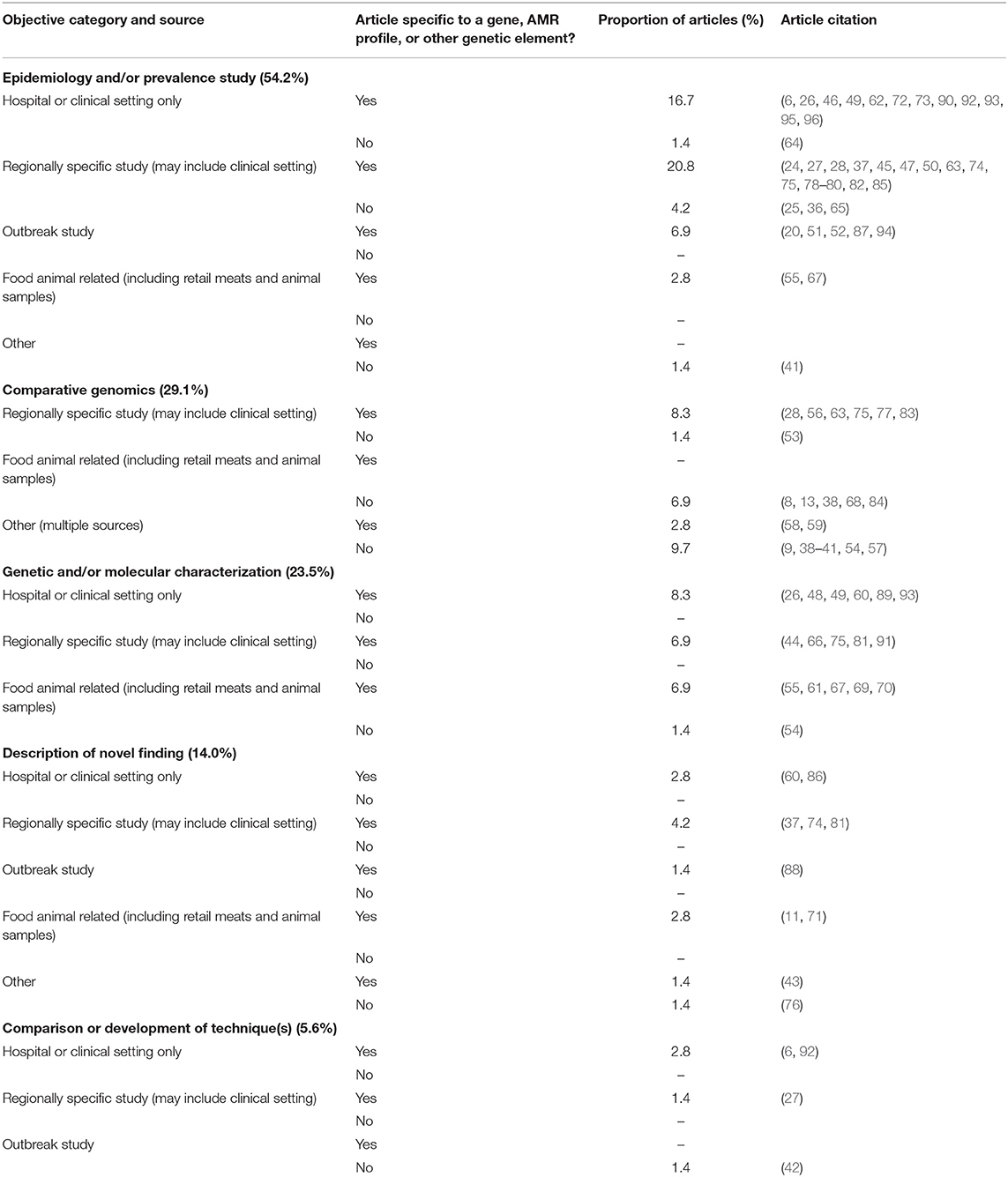
Table 4. Categorization of stated objectives for articles included in scoping review and proportion of included articles in each category.
Nine different source types were sampled for surveillance of Enterococcus spp. isolates, with the majority of articles using isolates from human clinical infections (47 articles, 65.3%). Just over half of the articles (38 articles, 52.8%) used isolates from human screening samples. For animal samples, 12 articles (16.7%) pulled isolates directly from food animals (fecal, cecal, or other samples), 12 articles (16.7%) had retail meat samples, and three articles (4.2%) included isolates from milk products. Isolates were also from environmental samples with eight articles (11.1%) including samples from the hospital environment and eight articles (11.1%) using wastewater samples. Five articles (6.9%) included sequences from GenBank to supplement their isolates from sample sources (Table 5). No studies sampled from companion animals or equids.
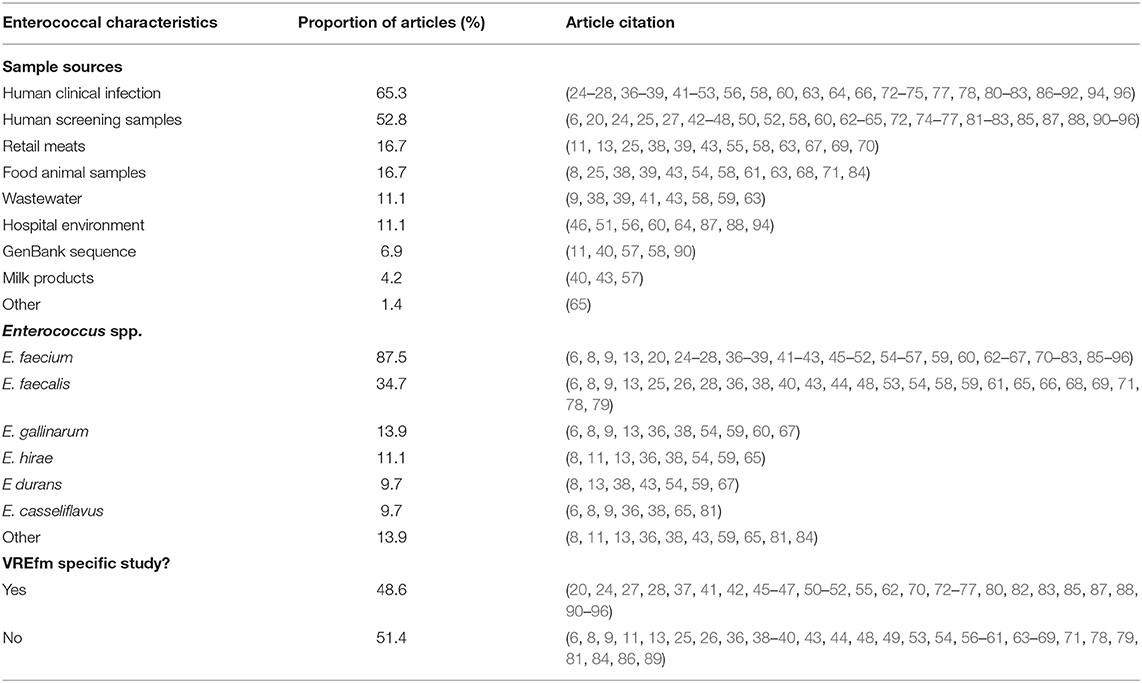
Table 5. Summary of enterococcal source and species from articles included in scoping review and proportion of included articles identifying each source or species.
At least 14 different species of Enterococcus were described in the 72 articles (one article did not identify all species). The majority of articles (87.5%) described Enterococcus faecium with 35 (48.6%) of these articles specifically studying VREfm. Twenty-five articles (34.7%) identified E. faecalis, 10 articles (13.9%) E. gallinarum, eight articles (11.1%) E. hirae, and seven studies (9.7%) each described E. casseliflavus and E. durans. Ten articles described other species, including E. villorum, E. thailandicus, E. cecorum, E. mundtii, E. pseudoavium, E. ratti, E. avium, and E. raffinosus.
All 72 articles provided some information on the AMR phenotypes of their isolates. The majority of articles (80.6%) describe isolates with resistance to glycopeptide antibiotics (vancomycin or teicoplanin). Twenty-one articles (29.2%) described isolates with resistance to oxazolidinones (linezolid or tedizolid). Other antimicrobial classes with identified phenotypic resistance included fluoroquinolones, macrolides, aminoglycosides, penicillins, and tetracyclines (Figure 3 and Supplementary Table 3). Methods used to assess phenotypic resistance in each article are outlined in Supplementary Table 4. Nineteen articles (26.4%) provided no information on the methodology used to define phenotypic resistance.
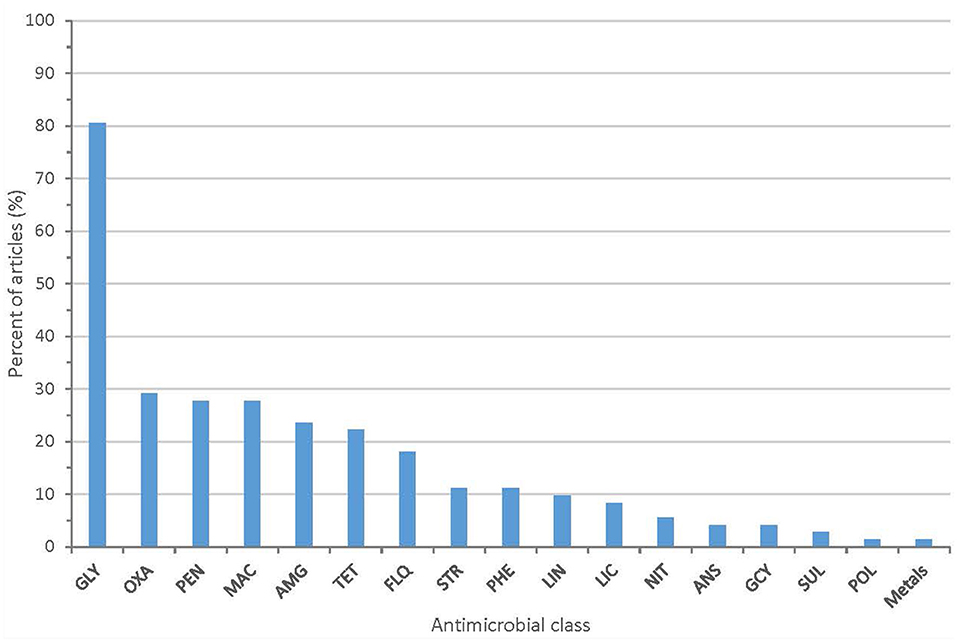
Figure 3. Proportion of articles included in scoping review identifying resistance to each antimicrobial class (GLY, glycopeptides; PEN, penicillins; MAC, macrolides; AMG, aminoglycosides; OXA, oxazolidinones; TET, tetracyclines; FLQ, fluoroquinolones; PHE, phenicols; LIN, lincopeptides; STR, streptogramins; LIC, lincosamides; ANS, ansamycins; GCY, glycylcyclines; NIT, nitrofurans; SUL, sulfonamides; POL, polypeptides).
All articles selected performed whole genome sequencing of the isolates (as a part of the inclusion criteria) and Illumina was the most commonly used platform for sequencing. Sixty-four articles (88.9%) used a version of Illumina for sequencing, whereas four articles did not provide sequencing methodology. Illumina MiSeq was the most commonly used version of Illumina employed. Other platforms included PacBio, Ion Torrent PGM, or Illumina in combination with PacBio, Ion Torrent PGM, or MinIon platforms (Table 6). Sixty-two articles (86.1%) provided archive accession numbers for access to their resultant sequences.
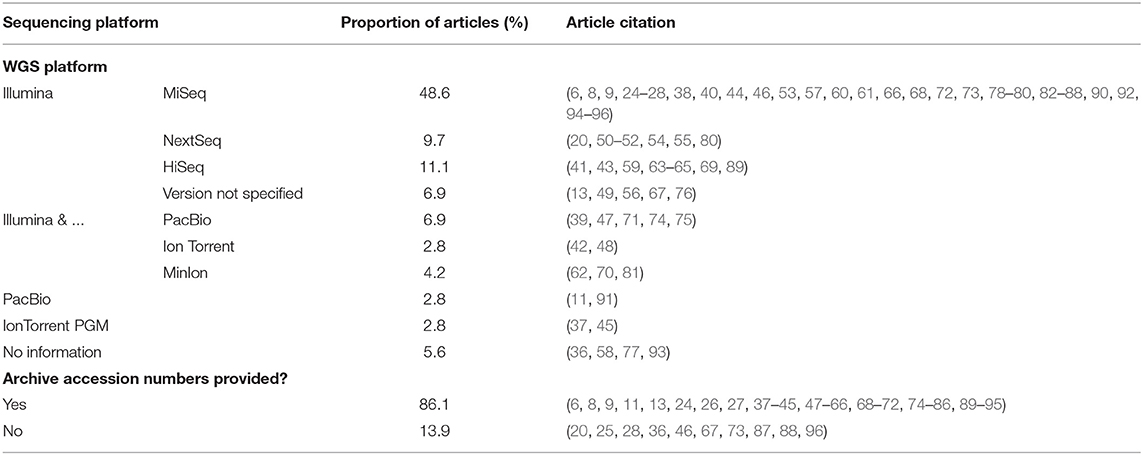
Table 6. Summary of WGS platforms used in each article included in scoping review and whether archive accession numbers were provided in the article.
Whole genome sequencing generated information about AMR genes that was described in almost all articles, with two articles (2.8%) failing to report AMR genes (Figure 4 and Supplementary Table 3). Forty-one articles (56.9%) reported the vanA gene cluster, 26 articles (36.1%) reported vanB, and 15 articles (20.8%) reported optrA.
From the sequencing, 61 articles (84.7%) described the sequence type (ST) and/or clonal complex (CC) of isolates. Over 60 sequence types were reported, including ST203 (25 articles), ST80 (27 articles), ST78 (19 articles), and ST18 (15 articles). Seven articles reported a pstS-null isolate. The summary of clonal complexes is in Figure 5 and Supplementary Table 3.
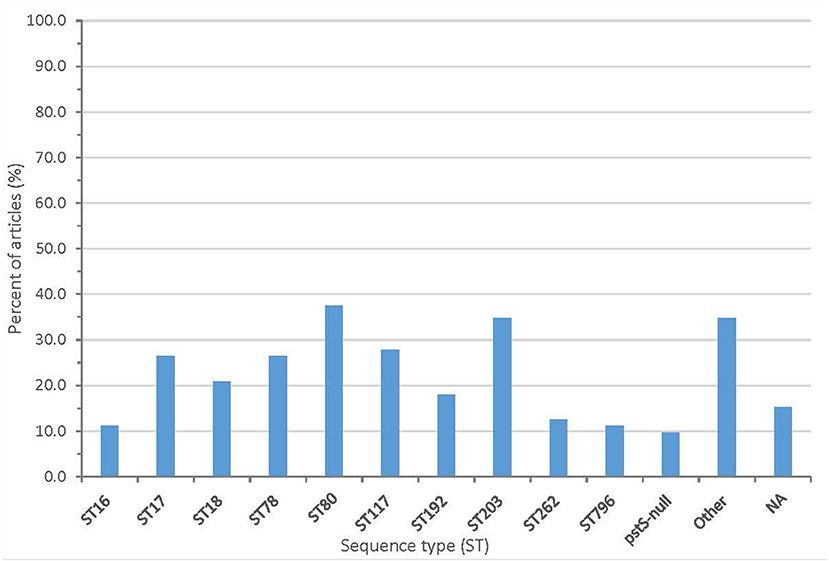
Figure 5. Proportion of articles included in scoping review finding different sequence types (NA, no sequence type described).
Many articles (75.0%) reported other molecular techniques, including PCR, PFGE, and MALDI-TOF MS. These techniques were primarily used for speciation or screening for AMR genes (Supplementary Table 4).
The articles' findings or conclusions related to antimicrobial resistance were summarized into categories, and articles could fit into more than one category. Of all articles, 31.9% reported a new or uncommon finding, which could include a novel strain or gene, or a previously described finding in a novel location. A fifth (20.8%) of articles reported how Enterococcus spp. were optimized for adaptation and survival in their environment. Twelve articles (16.7%) specifically stated that WGS was a better means of detecting or differentiating Enterococcus spp. than other genomic methods (such as PCR or PFGE). The other summarized findings are described in Table 7 with more details are available in Supplementary Table 2.
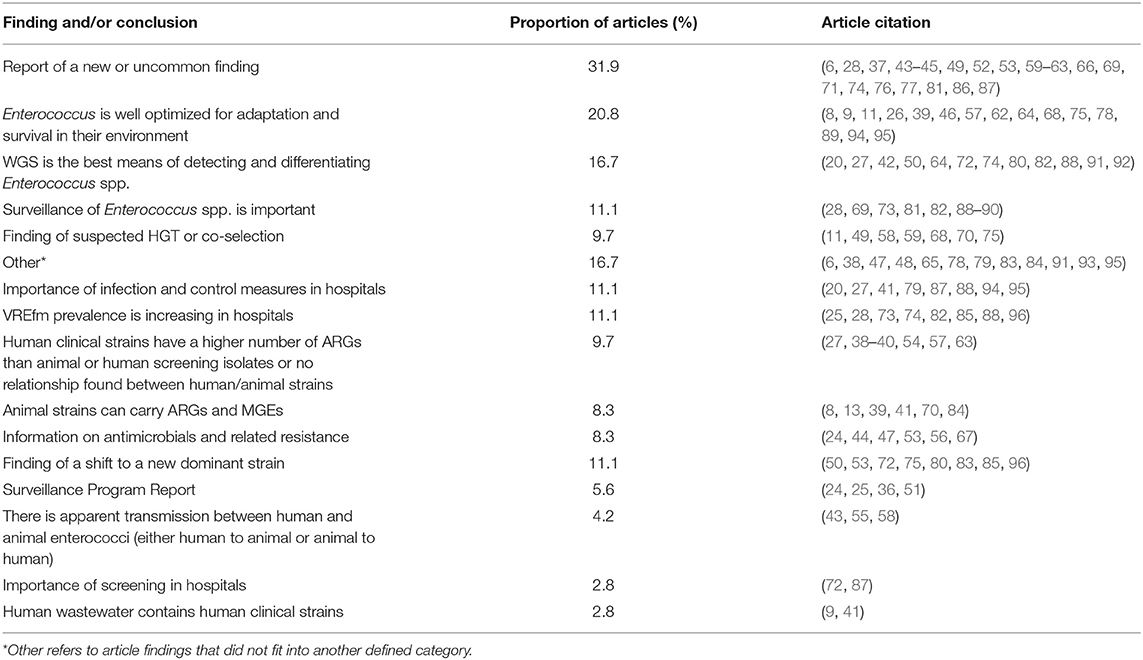
Table 7. Main categories of findings and conclusions and proportion of articles included in scoping review within each category.
In this scoping review, we aimed to assess the value added by the use of WGS in the surveillance of enterococcal AMR. The use of WGS for the surveillance of Enterococcus spp. has added to our knowledge about Enterococcus spp. through the detection of previously unidentified strains and finding that WGS was better at detecting and differentiating Enterococcus spp. than other genomic methods. European countries provided the most surveillance studies, with many of these coming from the Danish DANMAP program. This is perhaps unsurprising as DANMAP is an extensive and well-established program implemented in 1995 (25).
While the majority of Enterococcus spp. are adapted to the natural environment and animal GITs, rarely causing disease in humans, E. faecium and E. faecalis are the species most likely to cause human disease (2). Thus, it was not surprising that they were the most studied species in the included articles and the majority of articles were conducted in hospital settings. Nearly half of the included articles were studies specific to VREfm, showing the importance of vancomycin resistance in enterococci. WGS was important in studies to better understand VRE, especially in the assessment for vancomycin-variable enterococci (VVE). These are enterococcal isolates that are phenotypically sensitive to vancomycin but carry vancomycin-resistance genes. These isolates become phenotypically resistant to vancomycin when exposed to the antibiotic in vivo (25). Even though the resistance genes could be identified via PCR, the importance of WGS to further characterize VVE isolates was shown in the 2018 DANMAP report. The use of WGS and cgMLST allowed for the identification of new complexes and sequence types. DANMAP can now perform surveillance specific to these VVE strains. This should allow for earlier detection of VVE in patients and more appropriate treatment (25). WGS also allowed for a better understanding of E. faecium as it determined new sequence types, including pstS-null types, through cgMLST (37). The pstS gene locus is a housekeeping gene used for MLST but is missing in some VREfm strains. The use of WGS and cgMLST allowed for more robust sequence typing and identification of these isolates (37). Surveillance of enterococci within a hospital using WGS also allowed for the identification of a VREfm outbreak. A combination of sequencing data and an epidemiological investigation allowed for the identification of transmission route and the implementation of measures to prevent further outbreaks (20).
The ubiquitous nature of Enterococcus spp. naturally requires a One-Health approach to the surveillance of AMR in enterococci (38). This means a transdisciplinary approach across the human-animal-environment continuum in order to better understand the problem of AMR in enterococci (97). While enterococcal species other than E. faecium and E. faecalis were discussed in a few studies, in general, there were relatively few studies using a One-Health approach to compare animal, environmental (e.g., water and soil), and human samples (9, 11, 13, 39–41, 68–71). No studies sampled companion animals or equids, even though these animals live in close proximity to humans. This could be due to the complexity and cost of coordinating a study with so many sample sources or that human health studies are more easily funded. A collaborative transdisciplinary approach would bring a broad perspective to study design and allow for a better interpretation of the results. This would ease the complexity of designing and coordinating a One-Health surveillance study and create a more robust understanding of the issue of AMR in enterococci. Two studies included in this review did produce very informative results across multiple sample sources to address the One-Health continuum (38, 39). These studies pulled samples from livestock, retail meat, wastewater, and human bloodstream infections and showed limited sharing of genes between isolates from humans and animals (38, 39). Research using this One-Health approach will provide a means to assess the risk of AMR enterococci moving from food animals, through the food chain, into human populations as well as through the environment.
Many articles included in this study stressed the importance of the surveillance of Enterococcus spp., especially in hospital settings. This is because enterococci are optimized for adaptation and survival in their environment, whether the hospital environment or natural environment (42, 72). Both targeted surveillance of at-risk patients (e.g., immunocompromised) and passive surveillance of incoming hospital patients allowed for early recognition of outbreaks. Outbreaks could be controlled before becoming a significant problem and new hospital protocols surrounding cleaning and isolation could be developed (20, 72, 87). WGS allowed for more accurate sequence typing and identification of AMR genes (27, 73, 88).
From this review, Illumina sequencing platforms are currently the most popular for whole genome sequencing studies. They are historically reliable, with low error rates and have become accessible, abundant, and cost-effective (98). The combination of Illumina short-read with long-read sequencing (usually PacBio) was occasionally used to close a chromosome or a plasmid to accomplish genomic integrity as well as complete understanding of MGEs and their context. Unfortunately, the cost of running large numbers of isolates on a PacBio system is prohibitive (99). The use of long-read sequencing is likely to increase as inexpensive and portable bench-top platforms such as the Nanopore MinION become more reliable with lower error rates (100, 101). This will allow for the rapid identification of an isolate and its genetic composition, including MGEs (99). The majority of papers (86.1%) also provided archive accession numbers for their sequences, highlighting the importance of sharing raw genomic data with the scientific community and the requirements for publication in many journals.
This scoping review held limitations similar to other review papers in the possible omission of relevant literature, such as gray literature or articles written in a language other than English. Findings from government surveillance programs may be published online or as peer-reviewed articles, but in order to maintain an efficient and reproducible search method, gray literature was only searched from the reference lists in the primary research articles included in the review. No separate gray literature search was performed, which may have resulted in the omission of relevant information. Two non-English articles were excluded in our search which otherwise might have been included.
In order to minimize the omission of articles, several databases were searched and inclusion criteria were intentionally left broad until the abstract screening steps. The authors did maintain a rigid definition of surveillance, which could have excluded epidemiological articles that did not fit the selection criteria. This largely eliminated studies on human clinical isolates of Enterococcus spp. as the isolates would have been selected for a study based on certain characteristics.
Another limitation of the study is that a critical appraisal of the included articles was not conducted. This was intentional as one of the objectives of the present study was to identify gaps in the literature, but it means that studies of lower and higher quality would carry equal weight. The findings of some studies may not share the same validity based on their study design, but this was not determined in this review.
Whole genome sequencing has added value to the surveillance efforts of Enterococcus spp. by identifying new genes and strains, adding to the knowledge about its prevalence in various settings, and finding that WGS is a better means of detecting and differentiating Enterococcus spp. than other molecular methods. The ability of Enterococcus spp. to adapt and survive in its environment was frequently stated as a reason for the importance of using WGS for the surveillance of this bacterium. Future studies should focus on the state of Enterococcus spp. in companion animal veterinary medicine and determining the link between humans, animals, food products, and the environment for a better One-Health approach to Enterococcus spp. surveillance.
LR, KS, SLC, and SCC developed the research question and scoping review protocol. LR and KS performed the literature search, article screening, and data extraction of included articles. LR drafted the complete manuscript. SCC and SLC were secondary reviewers of articles and primary reviewers of the manuscript. All authors assisted with editing and content review of the manuscript.
LR and KS stipend funding was through the University of Calgary, Faculty of Veterinary Medicine One Health Award, and the University of Calgary Master's Award. This research is part of the AMR – One Health Consortium, partially funded by the Major Innovation Fund program of the Ministry of Jobs, Economy and Innovation, Government of Alberta. The funders had no role in the study design, data interpretation, or writing of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to acknowledge Lorraine Toews for her advice on search databases and Dr. Sheryl Gow and Dr. Barb Wilhelm for their advice on review papers.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2021.599285/full#supplementary-material
1. ^https://www.ncbi.nlm.nih.gov/pubmed/
2. ^https://apps.webofknowledge.com/WOS_GeneralSearch_input.do?product=WOS&search_mode=GeneralSearch&SID=5D5o7Naqz9xk9WXkIZ8&preferencesSaved=
3. ^http://web.a.ebscohost.com.ezproxy.lib.ucalgary.ca/ehost/search/advanced?vid=0&sid=59def56a-2134-4464-bd34-368a168a49ab%40sessionmgr4008
1. Klein G. Taxonomy, ecology and antibiotic resistance of enterococci from food and the gastro-intestinal tract. Int J Food Microbiol. (2003) 88:123–31. doi: 10.1016/S0168-1605(03)00175-2
2. Strateva T, Atanasova D, Savov E, Petrova G, Mitov I. Incidence of virulence determinants in clinical Enterococcus faecalis and Enterococcus faecium isolates collected in Bulgaria. Braz J Infect Dis. (2016) 20:127–33. doi: 10.1016/j.bjid.2015.11.011
3. Arias CA, Murray BE. Emergence and management of drug-resistant enterococcal infections. Expert Rev Anti Infect Ther. (2008) 6:637–55. doi: 10.1586/14787210.6.5.637
4. Miller WR, Munita JM, Arias CA. Mechanisms of antibiotic resistance in enterococci. Expert Rev Anti Infect Ther. (2014) 12:1221–36. doi: 10.1586/14787210.2014.956092
5. Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Eurosurveillance. (2008) 13:19046–. doi: 10.2807/ese.13.47.19046-en
6. Flipse J, von Wintersdorf CJH, van Niekerk JM, Jamin C, van Tiel FH, Hasman H, et al. Appearance of vanD-positive Enterococcus faecium in a tertiary hospital in the Netherlands: prevalence of vanC and vanD in hospitalized patients. Sci Rep. (2019) 9:6949. doi: 10.1038/s41598-019-42824-4
7. Hammerum AM. Enterococci of animal origin and their significance for public health. Clin Microbiol Infect. (2012) 18:619–25. doi: 10.1111/j.1469-0691.2012.03829.x
8. Beukers AG, Zaheer R, Goji N, Amoako KK, Chaves AV, Ward MP, et al. Comparative genomics of Enterococcus spp. isolated from bovine feces. BMC Microbiol. (2017) 17:52. doi: 10.1186/s12866-017-0962-1
9. Sanderson H, Ortega-Polo R, Zaheer R, Goji N, Amoako KK, Brown RS, et al. Comparative genomics of multidrug-resistant Enterococcus spp. isolated from wastewater treatment plants. BMC Microbiol. (2020) 20:20. doi: 10.1186/s12866-019-1683-4
10. WHO. Critically Important Antimicrobials for Human Medicine, 6th Revision. Geneva: World Health Organization (2019).
11. Peng Z, Li M, Wang W, Liu H, Fanning S, Hu Y, et al. Genomic insights into the pathogenicity and environmental adaptability of Enterococcus hirae R17 isolated from pork offered for retail sale. Microbiol Open. (2017) 6:e514. doi: 10.1002/mbo3.514
12. Nilsson O. Vancomycin resistant enterococci in farm animals - occurrence and importance. Infect Ecol Epidemiol. (2012) 2:1. doi: 10.3402/iee.v2i0.16959
13. Manson AL, Van Tyne D, Straub TJ, Clock S, Crupain M, Rangan U, et al. Chicken meat-associated enterococci: influence of agricultural antibiotic use and connection to the clinic. Appl Environ Microbiol. (2019) 85:e01559–19. doi: 10.1128/AEM.01559-19
14. Rushton-Green R, Darnell RL, Taiaroa G, Carter GP, Cook GM, Morgan XC. Agricultural origins of a highly persistent lineage of vancomycin-resistant Enterococcus faecalis in New Zealand. Appl Environ Microbiol. (2019) 85:e00137–19. doi: 10.1128/AEM.00137-19
15. Donado-Godoy P, Castellanos R, Leon M, Arevalo A, Clavijo V, Bernal J, et al. The establishment of the colombian integrated program for antimicrobial resistance surveillance (COIPARS): a pilot project on poultry farms, slaughterhouses and retail market. Zoonoses Public Health. (2015) 62(Suppl. 1):58–69. doi: 10.1111/zph.12192
16. Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front Microbiol. (2019) 10:539. doi: 10.3389/fmicb.2019.00539
17. About DANMAP. (2018). Available online at: https://www.danmap.org/about-danmap (accessed February 17, 2020).
18. About NARMS: US Centers for Disease Control. (2019). Available online at: https://www.cdc.gov/narms/about/index.html (accessed February 17, 2020).
19. About CIPARS Canada: Government of Canada. (2007). Available online at: https://www.canada.ca/en/public-health/services/surveillance/canadian-integrated-program-antimicrobial-resistance-surveillance-cipars/background.html (accessed February 17, 2020).
20. Sundermann AJ, Babiker A, Marsh JW, Shutt KA, Mustapha MM, Pasculle AW, et al. Outbreak of vancomycin-resistant Enterococcus faecium in interventional radiology: detection through whole genome sequencing-based surveillance. Clin Infect Dis. (2019) doi: 10.1093/cid/ciz666
21. SENTRY Antimicrobial Surveillance Program USA. (2019). Available online at: https://www.jmilabs.com/sentry-surveillance-program/ (accessed February 17, 2020).
22. European Antimicrobial Resistance Surveillance Network (EARS-Net): European Centre for Disease Prevention Control. (2021). Available online at: https://www.ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/ears-net (accessed April 15, 2020).
23. Sader HS, Rhomberg PR, Fuhrmeister AS, Mendes RE, Flamm RK, Jones RN. Antimicrobial resistance surveillance and new drug development. Open Forum Infect Dis. (2019) 6(Suppl. 1):S5–s13. doi: 10.1093/ofid/ofy345
24. Zhou X, Garcia-Cobos S, Ruijs G, Kampinga GA, Arends JP, Borst DM, et al. Epidemiology of extended-spectrum beta-lactamase-producing E. coli and vancomycin-resistant enterococci in the Northern Dutch-German cross-border region. Front Microbiol. (2017) 8:1914. doi: 10.3389/fmicb.2017.01914
25. DANMAP. DANMAP 2018 - Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Denmark: Statens Serum Institut (2018).
26. Deshpande LM, Castanheira M, Flamm RK, Mendes RE. Evolving oxazolidinone resistance mechanisms in a worldwide collection of enterococcal clinical isolates: results from the SENTRY Antimicrobial Surveillance Program. J Antimicrob Chemother. (2018) 73:2314–22. doi: 10.1093/jac/dky188
27. Pinholt M, Larner-Svensson H, Littauer P, Moser CE, Pedersen M, Lemming LE, et al. Multiple hospital outbreaks of vanA Enterococcus faecium in Denmark, 2012-13, investigated by WGS, MLST and PFGE. J Antimicrob Chemother. (2015) 70:2474–82. doi: 10.1093/jac/dkv142
28. Hammerum AM, Baig S, Kamel Y, Roer L, Pinholt M, Gumpert H, et al. Emergence of vanA Enterococcus faecium in Denmark, 2005-15. J Antimicrob Chemother. (2017) 72:2184–90. doi: 10.1093/jac/dkx138
29. Tricco AC, Lillie E, Zarin W, O'Brien KK, Colquhoun H, Levac D, et al. PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann Intern Med. (2018) 169:467–73. doi: 10.7326/M18-0850
30. Aromataris E, Munn Z. Joanna Briggs Institute Reviewer's Manual. Adelaide, SA: Joanna Briggs Institute (2017).
31. Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan - a web and mobile app for systematic reviews. Syst Rev. (2016) 5:210. doi: 10.1186/s13643-016-0384-4
33. Qin X, Galloway-Pena JR, Sillanpaa J, Roh JH, Nallapareddy SR, Chowdhury S, et al. Complete genome sequence of Enterococcus faecium strain TX16 and comparative genomic analysis of Enterococcus faecium genomes. BMC Microbiol. (2012) 12: 135. doi: 10.1186/1471-2180-12-135
34. Lam MMC, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, et al. Comparative analysis of the first complete Enterococcus faecium genome. J Bacteriol. (2012) 194:2334–41. doi: 10.1128/JB.00259-12
35. Genome. Bethesda, MD: National Library of Medicine (US); National Center for Biotechnology Information. (2004).
36. Coombs G, Daley D. Australian Enterococcal Sepsis Outcome Program (AESOP) 2016: Final Report. Australian Group on Antimicrobial Resistance (2016).
37. Lemonidis K Salih TS Dancer SJ Hunter IS Tucker NP. Emergence of an Australian-like pstS-null vancomycin resistant Enterococcus faecium clone in Scotland. PLoS ONE. (2019) 14:e0218185. doi: 10.1371/journal.pone.0218185
38. Zaheer R, Cook SR, Barbieri R, Goji N, Cameron A, Petkau A, et al. Surveillance of Enterococcus spp. reveals distinct species and antimicrobial resistance diversity across a One-Health continuum. Sci Rep. (2020) 10:3937. doi: 10.1038/s41598-020-69044-5
39. Gouliouris T, Raven KE, Ludden C, Blane B, Corander J, Horner CS, et al. Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. Mbio. (2018) 9:e01780–18. doi: 10.1128/mBio.01780-18
40. He Q, Hou Q, Wang Y, Li J, Li W, Kwok L-Y, et al. Comparative genomic analysis of Enterococcus faecalis: insights into their environmental adaptations. BMC Genomics. (2018) 19:527. doi: 10.1186/s12864-018-4887-3
41. Gouliouris T, Raven KE, Moradigaravand D, Ludden C, Coll F, Blane B, et al. Detection of vancomycin-resistant Enterococcus faecium hospital-adapted lineages in municipal wastewater treatment plants indicates widespread distribution and release into the environment. Genome Res. (2019) 29:626–34. doi: 10.1101/gr.232629.117
42. Janes VA, Notermans DW, Spijkerman IJB, Visser CE, Jakobs ME, van Houdt R, et al. Amplified fragment length polymorphism and whole genome sequencing: a comparison of methods in the investigation of a nosocomial outbreak with vancomycin resistant enterococci. Antimicrob Resist Infect Control. (2019) 8:153. doi: 10.1186/s13756-019-0604-5
43. Freitas AR, Elghaieb H, Leon-Sampedro R, Abbassi MS, Novais C, Coque TM, et al. Detection of optrA in the African continent (Tunisia) within a mosaic Enterococcus faecalis plasmid from urban wastewaters. J Antimicrob Chemother. (2017) 72:3245–51. doi: 10.1093/jac/dkx321
44. Holzknecht BJ, Pinholt M, Gumpert H, Hammerum AM, Westh H. Genetic characterisation confirms sporadic occurrence of vancomycin-resistant Enterococcus faecalis in Copenhagen, Denmark. Int J Antimicrob Agents. (2017) 50:501–2. doi: 10.1016/j.ijantimicag.2017.07.001
45. van Hal SJ, Espedido BA, Coombs GW, Howden BP, Korman TM, Nimmo GR, et al. Polyclonal emergence of vanA vancomycin-resistant Enterococcus faecium in Australia. J Antimicrob Chemother. (2017) 72:998–1001. doi: 10.1093/jac/dkw539
46. Lee AS, White E, Monahan LG, Jensen SO, Chan R, van Hal SJ. Defining the role of the environment in the emergence and persistence of vana vancomycin-resistant Enterococcus (VRE) in an intensive care unit: a molecular epidemiological study. Infect Control Hosp Epidemiol. (2018) 39:668–75. doi: 10.1017/ice.2018.29
47. Lee RS, da Silva AG, Baines SL, Strachan J, Ballard S, Carter GP, et al. The changing landscape of vancomycin-resistant Enterococcus faecium in Australia: a population-level genomic study. J Antimicrob Chemother. (2018) 73:3268–78. doi: 10.1093/jac/dky331
48. Marchi AP, Perdigao Neto LV, Martins RCR, Rizek CF, Camargo CH, Moreno LZ, et al. Vancomycin-resistant enterococci isolates colonizing and infecting haematology patients: clonality, and virulence and resistance profile. J Hosp Infect. (2018) 99:346–55. doi: 10.1016/j.jhin.2017.10.010
49. Santona A, Taviani E, Hoang HM, Fiamma M, Deligios M, Ngo TVQ, et al. Emergence of unusual vanA/vanB2 genotype in a highly mutated vanB2-vancomycin-resistant hospital-associated E. faecium background in Vietnam. Int J Antimicrob Agents. (2018) 52:586–92. doi: 10.1016/j.ijantimicag.2018.07.006
50. van Hal SJ, Beukers AG, Timms VJ, Ellem JA, Taylor P, Maley MW, et al. Relentless spread and adaptation of non-typeable vanA vancomycin-resistant Enterococcus faecium: a genome-wide investigation. J Antimicrob Chemother. (2018) 73:1487–91. doi: 10.1093/jac/dky074
51. Andersson P, Beckingham W, Gorrie CL, Kennedy K, Daveson K, Ballard SA, et al. Vancomycin-resistant Enterococcus (VRE) outbreak in a neonatal intensive care unit and special care nursery at a tertiary-care hospital in Australia—A retrospective case-control study. Infect Control Hosp Epidemiol. (2019) 40:551–8. doi: 10.1017/ice.2019.41
52. Douglas AP, Marshall C, Baines SL, Ritchie D, Szer J, Madigan V, et al. Utilizing genomic analyses to investigate the first outbreak of vanA vancomycin-resistant Enterococcus in Australia with emergence of daptomycin non-susceptibility. J Med Microbiol. (2019) 68:303–8. doi: 10.1099/jmm.0.000916
53. Farman M, Yasir M, Al-Hindi RR, Farraj SA, Jiman-Fatani AA, Alawi M, et al. Genomic analysis of multidrug-resistant clinical Enterococcus faecalis isolates for antimicrobial resistance genes and virulence factors from the western region of Saudi Arabia. Antimicrob Resist Infect Control. (2019) 8:55. doi: 10.1186/s13756-019-0508-4
54. O'Dea M, Sahibzada S, Jordan D, Laird T, Lee T, Hewson K, et al. Genomic, antimicrobial resistance, and public health insights into Enterococcus spp. from Australian chickens. J Clin Microbiol. (2019) 57 e00319–19. doi: 10.1128/JCM.00319-19
55. Sacramento AG, Fernandes MR, Sellera FP, Dolabella SS, Zanella RC, Cerdeira L, et al. VanA-type vancomycin-resistant Enterococcus faecium ST1336 isolated from mussels in an anthropogenically impacted ecosystem. Mar Pollut Bull. (2019) 142:533–6. doi: 10.1016/j.marpolbul.2019.04.014
56. Wardenburg KE, Potter RF, D'Souza AW, Hussain T, Wallace MA, Andleeb S, et al. Phenotypic and genotypic characterization of linezolid-resistant Enterococcus faecium from the USA and Pakistan. J Antimicrob Chemother. (2019) 74:3445–52. doi: 10.1093/jac/dkz367
57. Zhong Z, Kwok L-Y, Hou Q, Sun Y, Li W, Zhang H, et al. Comparative genomic analysis revealed great plasticity and environmental adaptation of the genomes of Enterococcus faecium. BMC Genomics. (2019) 20:602. doi: 10.1186/s12864-019-5975-8
58. Freitas AR, Tedim AP, Novais C, Lanza VF, Peixe L. Comparative genomics of global optrA-carrying Enterococcus faecalis uncovers a common chromosomal hotspot for optrA acquisition within a diversity of core and accessory genomes. Microb Genom. (2020) 6:e000350. doi: 10.1099/mgen.0.000350
59. Ekwanzala MD, Dewar JB, Kamika I, Momba MNB. Comparative genomics of vancomycin-resistant Enterococcus spp. revealed common resistome determinants from hospital wastewater to aquatic environments. Sci Total Environ. (2020) 719:137275. doi: 10.1016/j.scitotenv.2020.137275
60. Ahmed MO, Elramalli AK, Baptiste KE, Daw MA, Zorgani A, Brouwer E, et al. Whole genome sequence analysis of the first vancomycin-resistant Enterococcus faecium isolates from a Libyan Hospital in Tripoli. Microb Drug Resist. (2020) 26:1390–8. doi: 10.1089/mdr.2019.0095
61. Almeida LM, Gaca A, Bispo PM, Lebreton F, Saavedra JT, Silva RA, et al. Coexistence of the oxazolidinone resistance-associated genes cfr and optrA in Enterococcus faecalis from a healthy piglet in Brazil. Front Public Health. (2020) 8:518. doi: 10.3389/fpubh.2020.00518
62. Chilambi GS, Nordstrom HR, Evans DR, Ferrolino JA, Hayden RT, Maron GM, et al. Evolution of vancomycin-resistant Enterococcus faecium during colonization and infection in immunocompromised pediatric patients. Proc Natl Acad Sci. USA. (2020) 117:11703–14. doi: 10.1073/pnas.1917130117
63. Freitas AR, Tedim AP, Duarte B, Elghaieb H, Abbassi MS, Hassen A, et al. Linezolid-resistant (Tn6246::fexB-poxtA) Enterococcus faecium strains colonizing humans and bovines on different continents: similarity without epidemiological link. J Antimicrob Chemother. (2020) 75:2416–23. doi: 10.1093/jac/dkaa227
64. Gouliouris T, Coll F, Ludden C, Blane B, Raven KE, Naydenova P, et al. Quantifying acquisition and transmission of Enterococcus faecium using genomic surveillance. Nat Microbiol. (2020) 6:103–111. doi: 10.1038/s41564-020-00806-7
65. Gu J, Xie X-J, Liu J-X, Shui J-R, Zhang H-Y, Feng G-Y, et al. Prevalence and transmission of antimicrobial-resistant Staphylococci and Enterococci from shared bicycles in Chengdu, China. Sci Total Environ. (2020) 738:139735. doi: 10.1016/j.scitotenv.2020.139735
66. Ruiz-Ripa L, Fessler AT, Hanke D, Eichhorn I, Azcona-Gutierrez JM, Perez-Moreno MO, et al. Mechanisms of linezolid resistance among enterococci of clinical origin in spain-detection of optrA- and cfr(D)-carrying E. faecalis. Microorganisms. (2020) 8:1155. doi: 10.3390/microorganisms8081155
67. Sabenca C, de Sousa T, Oliveira S, Viala D, Theron L, Chambon C, et al. Next-generation sequencing and MALDI mass spectrometry in the study of multiresistant processed meat vancomycin-resistant enterococci (VRE). Biol. Basel. (2020) 9:89. doi: 10.3390/biology9050089
68. Zhang S, Wang D, Wang Y, Hasman H, Aarestrup FM, Alwathnani HA, et al. Genome sequences of copper resistant and sensitive Enterococcus faecalis strains isolated from copper-fed pigs in Denmark. Stand Genomic Sci. (2015) 10:35. doi: 10.1186/s40793-015-0021-1
69. Cavaco LM, Bernal JF, Zankari E, Leon M, Hendriksen RS, Perez-Gutierrez E, et al. Detection of linezolid resistance due to the optrA gene in Enterococcus faecalis from poultry meat from the American continent (Colombia). J Antimicrob Chemother. (2016) 72:678–83. doi: 10.1093/jac/dkw490
70. Leinweber H, Alotaibi SMI, Overballe-Petersen S, Hansen F, Hasman H, Bortolaia V, et al. Vancomycin resistance in Enterococcus faecium isolated from Danish chicken meat is located on a pVEF4-like plasmid persisting in poultry for 18 years. Int J Antimicrob Agents. (2018) 52:283–6. doi: 10.1016/j.ijantimicag.2018.03.019
71. Tyson GH, Sabo JL, Hoffmann M, Hsu C-H, Mukherjee S, Hernandez J, et al. Novel linezolid resistance plasmids in Enterococcus from food animals in the USA. J Antimicrob Chemother. (2018) 73:3254–8. doi: 10.1093/jac/dky369
72. Leong KWC, Cooley LA, Anderson TL, Gautam SS, McEwan B, Wells A, et al. Emergence of vancomycin-resistant Enterococcus faecium at an Australian hospital: a whole genome sequencing analysis. Sci Rep. (2018) 8:6274. doi: 10.1038/s41598-018-24614-6
73. Landerslev KG, Jakobsen L, Olsen SS, Pedersen MB, Kristensen B, Lemming LE, et al. Polyclonal spread of vanA Enterococcus faecium in Central Denmark Region, 2009-2013, investigated using PFGE, MLST and WGS. Int J Antimicrob Agents. (2016) 48:767–8. doi: 10.1016/j.ijantimicag.2016.09.001
74. Pinholt M, Gumpert H, Bayliss S, Nielsen JB, Vorobieva V, Pedersen M, et al. Genomic analysis of 495 vancomycin-resistant Enterococcus faecium reveals broad dissemination of a vanA plasmid in more than 19 clones from Copenhagen, Denmark. J Antimicrob Chemother. (2017) 72:40–7. doi: 10.1093/jac/dkw360
75. Pinholt M, Bayliss SC, Gumpert H, Worning P, Jensen VVS, Pedersen M, et al. WGS of 1058 Enterococcus faecium from Copenhagen, Denmark, reveals rapid clonal expansion of vancomycin-resistant clone ST80 combined with widespread dissemination of a vanA-containing plasmid and acquisition of a heterogeneous accessory genome. J Antimicrob Chemother. (2019) 74:1776–85. doi: 10.1093/jac/dkz118
76. Boyd DA, Lalancette C, Levesque S, Golding GR. Characterization of a genomic island harbouring a new vanD allele from Enterococcus faecium N15-508 isolated in Canada. J Antimicrob Chemother. (2016) 71:2052–4. doi: 10.1093/jac/dkw063
77. Falgenhauer L, Fritzenwanker M, Imirzalioglu C, Steul K, Scherer M, Rhine-Main VSG, et al. Near-ubiquitous presence of a vancomycin-resistant Enterococcus faecium ST117/CT71/vanB -clone in the Rhine-Main metropolitan area of Germany. Antimicrob Resist Infect Control. (2019) 8:128. doi: 10.1186/s13756-019-0573-8
78. Bender JK, Fleige C, Lange D, Klare I, Werner G. Rapid emergence of highly variable and transferable oxazolidinone and phenicol resistance gene optrA in German Enterococcus spp. clinical isolates. Int J Antimicrob Agents. (2018) 52:819–27. doi: 10.1016/j.ijantimicag.2018.09.009
79. Wang J, Foxman B, Pirani A, Lapp Z, Mody L, Snitkin E. Application of combined genomic and transfer analyses to identify factors mediating regional spread of antibiotic resistant bacterial lineages. Clin Infect Dis. (2020) 71: e642–9. doi: 10.1093/cid/ciaa364
80. Lee T, Pang S, Stegger M, Sahibzada S, Abraham S, Daley D, et al. A three-year whole genome sequencing perspective of Enterococcus faecium sepsis in Australia. PLoS ONE. (2020) 15:e0228781. doi: 10.1371/journal.pone.0228781
81. Hashimoto Y, Kita I, Suzuki M, Hirakawa H, Ohtaki H, Tomita H. First report of the local spread of vancomycin-resistant enterococci ascribed to the interspecies transmission of a vanA gene cluster-carrying linear plasmid. mSphere. (2020) 5:e00102–20. doi: 10.1128/mSphere.00102-20
82. Leong KWC, Kalukottege R, Cooley LA, Anderson TL, Wells A, Langford E, et al. State-wide genomic and epidemiological analyses of vancomycin-resistant Enterococcus faecium in Tasmania's Public Hospitals. Front Microbiol. (2019) 10:2940. doi: 10.3389/fmicb.2019.02940
83. Eisenberger D, Tuschak C, Werner M, Bogdan C, Bollinger T, Hossain H, et al. Whole-genome analysis of vancomycin-resistant Enterococcus faecium causing nosocomial outbreaks suggests the occurrence of few endemic clonal lineages in Bavaria, Germany. J Antimicrob Chemother. (2020) 75:1398–404. doi: 10.1093/jac/dkaa041
84. Sharma P, Gupta SK, Barrett JB, Hiott LM, Woodley TA, Kariyawasam S, et al. Comparison of antimicrobial resistance and pan-genome of clinical and non-clinical Enterococcus cecorum from poultry using whole-genome sequencing. Foods. (2020) 9:686. doi: 10.3390/foods9060686
85. Xanthopoulou K, Peter S, Tobys D, Behnke M, Dinkelacker AG, Eisenbeis S, et al. Vancomycin-resistant Enterococcus faecium colonizing patients on hospital admission in Germany: prevalence and molecular epidemiology. J Antimicrob Chemother. (2020) 75:2743–51. doi: 10.1093/jac/dkaa271
86. Deshpande LM, Ashcraft DS, Kahn HP, Pankey G, Jones RN, Farrell DJ, et al. Detection of a new cfr-Like Gene, cfr(B), in Enterococcus faecium isolates recovered from human specimens in the United States as Part of the SENTRY Antimicrobial Surveillance Program. Antimicrob Agents Chemother. (2015) 59:6256–61. doi: 10.1128/AAC.01473-15
87. Kampmeier S, Knaack D, Kossow A, Willems S, Schliemann C, Berdel WE, et al. Weekly screening supports terminating nosocomial transmissions of vancomycin-resistant enterococci on an oncologic ward - a retrospective analysis. Antimicrob Resist Infect Control. (2017) 6:48. doi: 10.1186/s13756-017-0206-z
88. Lister DM, Kotsanas D, Ballard SA, Howden BP, Carse E, Tan K, et al. Outbreak of vanB vancomycin-resistant Enterococcus faecium colonization in a neonatal service. Am J Infect Control. (2015) 43:1061–5. doi: 10.1016/j.ajic.2015.05.047
89. Moradigaravand D, Gouliouris T, Blane B, Naydenova P, Ludden C, Crawley C, et al. Within-host evolution of Enterococcus faecium during longitudinal carriage and transition to bloodstream infection in immunocompromised patients. Genome Medicine. (2017) 9:119. doi: 10.1186/s13073-017-0507-0
90. Rios R, Reyes J, Carvajal LP, Rincon S, Panesso D, Echeverri AM, et al. Genomic epidemiology of vancomycin-resistant Enterococcus faecium (VREfm) in latin america: revisiting the global VRE population structure. Sci Rep. (2020) 10:5636. doi: 10.1038/s41598-020-62371-7
91. Kim HM, Chung DR, Cho SY, Huh K, Kang CI, Peck KR. Emergence of vancomycin-resistant Enterococcus faecium ST1421 lacking the pstS gene in Korea. Eur J Clin Microbiol Infect Dis. (2020) 39:1349–56. doi: 10.1007/s10096-020-03853-4
92. Eichel V, Klein S, Bootsveld C, Frank U, Heeg K, Boutin S, et al. Challenges in interpretation of WGS and epidemiological data to investigate nosocomial transmission of vancomycin-resistant Enterococcus faecium in an endemic region: incorporation of patient movement network and admission screening. J. Antimicrob Chemother. (2020) 75:1716–21. doi: 10.1093/jac/dkaa122
93. Heininger A, Zimmermann S, Bootsveld C, Boutin S, Nurjadi D. Low prevalence of combined linezolid- and vancomycin-resistant Enterococcus faecium from hospital admission screening in an endemic region in Germany. J Glob Antimicrob Resist. (2020) 22:646–50. doi: 10.1016/j.jgar.2020.05.003
94. Kampmeier S, Toennies H, Correa-Martinez CL, Mellmann A, Schwierzeck V. A nosocomial cluster of vancomycin resistant enterococci among COVID-19 patients in an intensive care unit. Antimicrob Resist Infect Control. (2020) 9:154. doi: 10.1186/s13756-020-00820-8
95. Neumann B, Bender JK, Maier BF, Wittig A, Fuchs S, Brockmann D, et al. Comprehensive integrated NGS-based surveillance and contact-network modeling unravels transmission dynamics of vancomycin-resistant enterococci in a high-risk population within a tertiary care hospital. PLoS ONE. (2020) 15:e0235160. doi: 10.1371/journal.pone.0235160
96. Weber A, Maechler F, Schwab F, Gastmeier P, Kola A. Increase of vancomycin-resistantEnterococcus faeciumstrain type ST117 CT71 at Charite-Universitatsmedizin Berlin, 2008 to 2018. Antimicrob Resist Infect Control. (2020) 9:109. doi: 10.1186/s13756-020-00754-1
97. Hall DC, Cork SC. Chapter 1: introduction to one health concepts. In: Cork S, Hall D, Liljebjelke K, editors. One Health Case Studies, Addressing Complex Problems in a Changing World. Sheffield: 5M Publishing, Ltd. (2016).
98. Quainoo SC, Jordy PM, van Hijum SAFT, Huynen MA, Melchers WJG, van Schaik W, et al. Whole-genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. (2017) 30:1015–63. doi: 10.1128/CMR.00016-17
99. George S, Pankhurst L, Hubbard A, Votintseva A, Stoesser N, Sheppard AE, et al. Resolving plasmid structures in Enterobacteriaceae using the MinION nanopore sequencer: assessment of MinION and MinION/Illumina hybrid data assembly approaches. Microb Genom. (2017) 3:e000118. doi: 10.1099/mgen.0.000118
100. Tyler AD, Mataseje L, Urfano CJ, Schmidt L, Antonation KS, Mulvey MR, et al. Evaluation of Oxford Nanopore's MinION sequencing device for microbial whole genome sequencing applications. Sci Rep. (2018) 8:10931. doi: 10.1038/s41598-018-29334-5
Keywords: Enterococcus, whole genome sequencing, scoping review, antimicrobial resistance, surveillance, One Health, nosocomial infection
Citation: Rogers LA, Strong K, Cork SC, McAllister TA, Liljebjelke K, Zaheer R and Checkley SL (2021) The Role of Whole Genome Sequencing in the Surveillance of Antimicrobial Resistant Enterococcus spp.: A Scoping Review. Front. Public Health 9:599285. doi: 10.3389/fpubh.2021.599285
Received: 26 August 2020; Accepted: 04 May 2021;
Published: 10 June 2021.
Edited by:
Michael Kemp, University of Southern Denmark, DenmarkReviewed by:
Claire Gorrie, The University of Melbourne, AustraliaCopyright © 2021 Rogers, Strong, Cork, McAllister, Liljebjelke, Zaheer and Checkley. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sylvia L. Checkley, c2xjaGVja2xAdWNhbGdhcnkuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.