
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health , 05 February 2021
Sec. Infectious Diseases: Epidemiology and Prevention
Volume 9 - 2021 | https://doi.org/10.3389/fpubh.2021.570650
 Xiaoyu Ma1,2
Xiaoyu Ma1,2 Fan Zhang1,3
Fan Zhang1,3 Bing Bai1,2
Bing Bai1,2 Zhiwei Lin1,2
Zhiwei Lin1,2 Guangjian Xu1,2
Guangjian Xu1,2 Zhong Chen1
Zhong Chen1 Xiang Sun1,2
Xiang Sun1,2 Jinxin Zheng1,2
Jinxin Zheng1,2 Qiwen Deng1,2
Qiwen Deng1,2 Zhijian Yu1,2*
Zhijian Yu1,2*Background: Enterococcus faecalis has been commonly considered as one of the major pathogens of the urinary tract infection (UTI) in human host worldwide, whereas the molecular characteristics of E. faecalis clinical isolates from the patients with UTI in China remains seldomly reported. This study aimed to investigate the resistance mechanism, molecular characteristics and risk factors of E. faecalis clinical isolates from patients with UTI in China.
Methods: A total of 115 non-duplicated E. faecalis clinical isolates from patients with UTI were retrospectively collected in a tertiary hospital in China and their clinical data was further analyzed. The linezolid and tedizolid susceptibility were determined by agar dilution. The resistance genes, including erm(A), erm(B), erm(C), tet(M), optrA, cfr, cfr(B), poxtA, and MLST-based housekeeping genes were investigated by PCR.
Results: In 115 non-duplicated E. faecalis clinical isolates from the patients with UTI in this hospital setting, the frequency of linezolid or tedizolid-resistant/intermediate isolates were 22.61 and 13.04%, respectively, and the frequency of linezolid-resistant/intermediate E. faecalis clinical isolates carrying with erm(A) were 86%. Among the five linezolid-resistant E. faecalis strains found in this study, three optrA-positive isolates and the other two linezolid-resistant strains were G2576U genetic mutations in the V domain of the 23S rRNA genes. The ST clonality analysis indicated that 31.42% (11/35) of ST16 E. faecalis UTI isolates were not susceptible to linezolid. Moreover, the univariable analysis indicated that the high risk factors of linezolid-resistant/intermediate E. faecalis infections involved the indwelling catheter, trachea cannula catheter and the carriage of erm(A) or optrA. Furthermore, the indwelling catheter and trachea cannula catheter were demonstrated as the independent predictors of linezolid-resistant/intermediate E. faecalis strains in patients with UTI by multivariable analysis.
Conclusion: Linezolid-resistant/intermediate E. faecalis associated with urinary tract infections of patients in this hospital setting from China might be explained by the high carriage frequency of optrA genes and moreover, indwelling catheter and trachea cannula should be considered as the independent predictors of linezolid-resistant/intermediate E. faecalis infections. The transmission mechanism of linezolid-resistant/intermediate E. faecalis in this hospital setting should be further studied.
Enterococcus faecalis has been widely considered as the commensal inhabitants of the intestinal tract of both humans and animals (1). E. faecalis is the most prevalent species of Enterococcus genus that is isolated from the clinical specimens among human hosts with a series of infectious diseases, such as sepsis, abdominal infections, endocarditis, cholecystitis, peritonitis, and neonatal meningitis (2). Moreover, E. faecalis has been regarded as one of the major pathogens from patients with the urinary tract infection (UTI) in clinics worldwide (3, 4). Because of the inherent resistance of E. faecalis to several antibiotic agents and their natural competence for acquired resistance, the treatment difficulty of E. faecalis infections has gradually increased in recent years (5). Linezolid, the first synthetic antimicrobial agent of oxazolidinone class, inhibits the initial ribosome assembly and protein synthesis of multiple gram-positive bacteria species by targeting the 50S ribosome subunits and impacting its binding affinity with formylmethionyl-tRNA (6). Due to its broad antimicrobial spectrum, linezolid has been widely used as one of the most important options for the treatment of infectious diseases caused by multi-drug resistant gram-positive pathogens, especially including methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant enterococci (VRE), penicillin-resistant streptococci and mycobacteria (7). In recent years, with the widespread application of linezolid in clinics, the gradual increasing reports of linezolid resistant gram-positive pathogens highlights the enhanced risk of linezolid resistance transmission (8, 9). Our previous data indicated the possible presence of high frequency of linezolid resistance in E. faecalis clinical isolates. However, the frequency and clinical significance of linezolid-resistant/intermediate E. faecalis in patients with UTI remains elusive (10).
In this study, the E. faecalis clinical isolates from the patients with UTI were collected from a tertiary hospital in China. Subsequently, the clinical data of the patients with UTI was further analyzed. The antimicrobial susceptibility of linezolid and tedizolid was determined by agar dilution. The resistance genes, including erm(A), erm(B), erm(C), tet(M), the domain V region of the 23S rRNA gene, cfr, cfr(B), poxtA, as well as optrA and several commonly detected virulence factors were investigated by PCR. The ST genotype was determined by detecting MLST-based housekeeping genes and their relationship with linezolid-resistant/intermediate E. faecalis infections was further analyzed.
A total of 115 non-duplicate clinical E. faecalis UTI isolates were collected from January 1, 2010 to September 30, 2015 in Nanshan People's Hospital (A teaching hospital) of Shenzhen, China(It is a grade A class three general hospital located in Nanshan District, Shenzhen, with more than 1,300 open beds). E. faecalis clinical strains were isolated from the urine samples and identified by the VITEK 2 system (BioMérieux, Marcyl'Etoile, France). Species-appropriate quality control strains were used to ensure laboratory standards, as directed by the Clinical and Laboratory Standards Institute (CLSI 2020) (11). E. faecalis ATCC29212 and OG1RF (ATCC47077), obtained from the American Type Culture Collection (ATCC), were used as quality control strains. Patient clinical data including age, gender, admission to intensive care unit (ICU), venous catheter, indwelling catheter, D-J tube catheter, trachea cannula catheter and antibiotics therapy, were collected from hospital information system. E. faecalis clinical isolates of inpatients in Nanshan People's Hospital were analyzed retrospectively and approved by the institutional ethical committee of Shenzhen Nanshan people's hospital. This trial followed the ethical principles of the Declaration of the Chinese Ethical Guidelines. All procedures involving human participants were performed in accordance with the ethical standards of Shenzhen University and the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
The antimicrobial susceptibility of several commonly-used antibiotics, such as tetracycline, erythromycin, vancomycin, minocycline, tigecycline, vancomycin, tedizolid, linezolid and doxycycline in E. faecalis were automatically tested through VITEK 2 Compact system (BioMérieux, France). The susceptibility breakpoints of these antibiotics in E. faecalis were recommended by CLSI 2020 (11). The MIC values of linezolid, tedizolid and tigecycline were further determined by agar dilution according to related reports (10, 12, 13). The linezolid susceptible breakpoint recommended in E. faecalis by CLSI was adopted: ≤ 2 μg/mL for susceptibility, 4 μg/mL for intermediate status, and ≥8 μg/mL for resistance. The susceptible breakpoint of tedizolid to E. faecalis was defined as MIC ≤ 0.5 μg/mL (11). The MIC breakpoints for tigecycline recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST), the strains with MIC > 0.25 μg/mL was classified as resistant to tigecycline (14).
The genomic DNA of the bacteria was extracted by DNeasy Blood & Tissue Kit DNA extraction kit (MGI Tech Co, Ltd, Shenzhen, China) according to the performance procedure of gram-positive bacteria, and the extracted DNA was stored at −20°C. The primers in this study listed in Supplementary Table 1 were synthesized by BGI company (13, 15, 16). PCR was carried out for the detection of the following resistance genes: erm(A), erm(B), erm(C), tet(M), the domain V region of the 23S rRNA gene, cfr, cfr(B), poxtA as well as the ABC transporter optrA (13, 15, 16). Several commonly found virulence factors in the E. faecalis, including asal, esp, gelE, cyl, hyl, efaA, and ace, were amplified by PCR based on published documents (17, 18).
On the basis of established Multilocus sequence typing (MLST) schemes (http://www.mlst.net), seven housekeeping genes of E. faecalis (gdh, gyd, pstS, gki, aroE, xpt, and yiqL) were amplified and sequenced as described previously and the primers of the housekeeping genes were listed in Supplementary Table 3 (13). Sequence types (STs) were determined by comparison with published locus types in the E. faecalis MLST.net database (http://efaecalis.mlst.net/). A. Allelic profile or STs were assigned seven integers, corresponding to the allele numbers at the seven loci. STs were assigned to isolates in such a way that the same ST names were kept as far as possible for the same analyzed strains.
The prevalence of antibiotic susceptibility among the isolates is presented as the number (percentage). This prevalence was compared between groups using the chi-square test or Fisher's exact test. Univariable and multivariable conditional logistic regression were performed to determine patient characteristics associated with the development of infection. The tests were performed using SPSS software (version 19.0, Chicago, IL, USA). P < 0.05 were regarded as statistically significant.
The 115 non-duplicated E. faecalis clinical isolates were obtained from urine samples in the patients with UTI and the distribution characteristics of E. facecalis clinical isolates from the hospital wards was shown in Supplementary Figure 1. Our data indicated that linezolid-resistant/intermediate E. faecalis was found in 23% (26/115) of the patients with E. faecalis UTI. Only five linezolid-resistant E. faecalis were detected with linezolid MIC ≥ 8 μg/mL. The relationship of antibiotic susceptibility between linezolid and several commonly used antibiotics was analyzed in Table 1, indicating the frequency of E. faecalis UTI isolates with resistance toward erythromycin, doxycycline, tetracycline and minocycline were 99.13% (114/115), 93.91% (108/115), 93.04% (107/115), and 92.17% (106/115), respectively. Worthy of our concern, although the high frequency of the linezolid-resistant/intermediate E. faecalis strains were found, these strains exhibited the susceptibility phenotype toward ampicillin, tigecycline and vancomycin (Supplementary Table 4). Moreover, 13.04%(15/115) of E. faecalis clinical isolates in this study were non-susceptible to tedizolid.
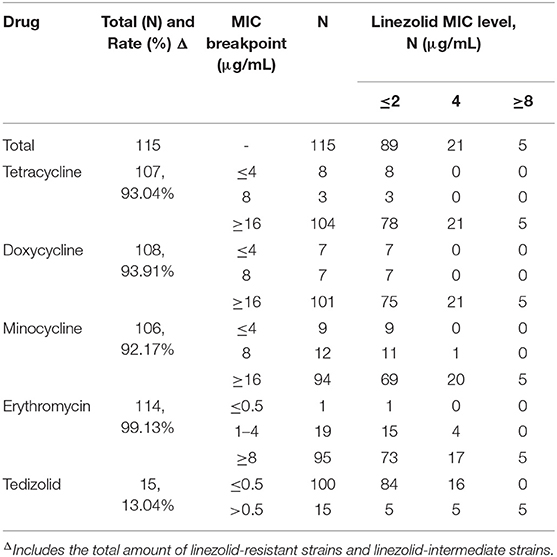
Table 1. The relationship of linezolid susceptibility and antimicrobial susceptibility of commonly used antibiotics in E. faecalis causing UTI.
The detection of several resistance genes and virulence factors in this study was shown in Tables 2, 3, Figure 1, and Supplementary Figure 2, indicating that the carriage frequency of the virulence genes, including esp, hyl, asal, cyl, ace, gelE, and efaA, were 68.70% (79/115), 20.87% (24/115), 83.48% (96/115), 75.65% (87/115), 100% (115 /115), 63.48% (73/115), and 98.26%(113/115), respectively. Moreover, 8.70% of E. feacalis UTI isolates were shown with the carriage of all seven virulence factors (esp/efaA/asa1/ace/cyl/gelE/hyl).
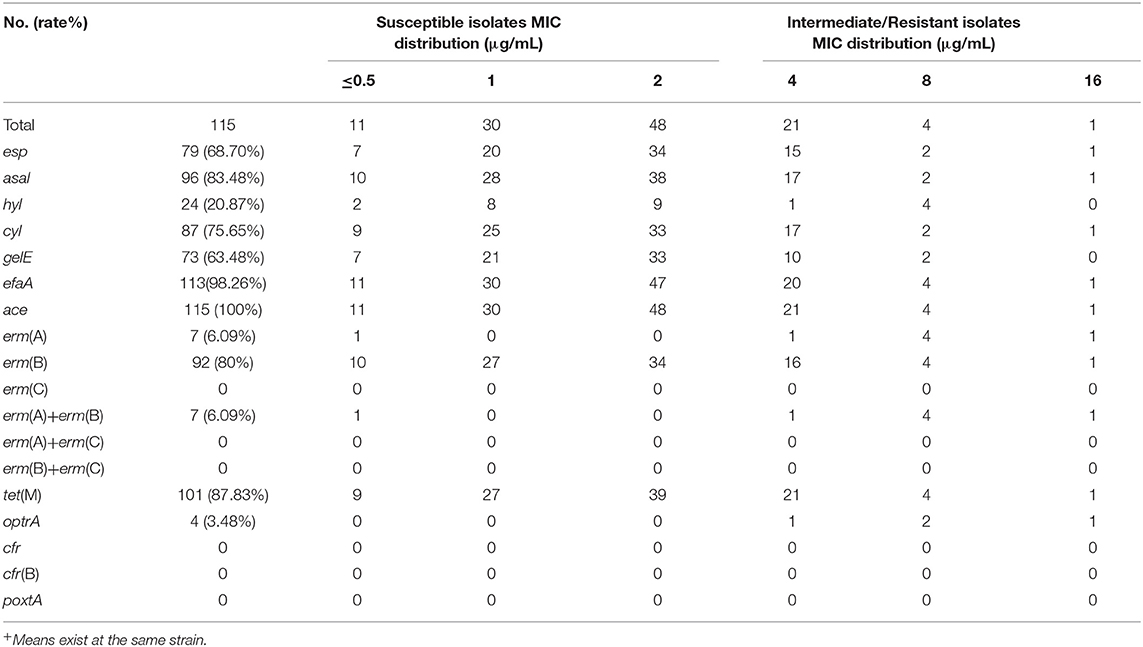
Table 2. The distribution of the antimicrobial resistance genes and virulence genes in linezolid-Intermediate/Resistant E. faecalis.
Eighty percent of E. faecalis UTI isolates in this study were found positively with erm(B), but only 7 strains (6.09%) were carried positively with erm(A), and no erm(C) gene was found. Moreover, our data indicated 68.53% (61/89) of E. faecalis UTI isolates with gelE was susceptible to linezolid, which was significantly high in comparison to that in the gelE-negative isolates. Furthermore, the frequency of the linezolid-resistant/intermediate E. faecalis with erm(A) was 86%, indicating the high frequency of this resistance genes in linezolid-resistant/intermediate E. faecalis UTI isolates (Figure 1 and Supplementary Figure 2).
The plasmid-borne resistance genes, including optrA, poxtA, cfr, or cfr(B) genes were detected in all E. faecalis UTI isolates, suggesting four E. faecalis UTI isolates, including one linezolid-intermediate isolate and three linezolid-resistant isolates, were found with positive carriage of the optrA gene and none was found with poxtA, cfr, or cfr(B) gene (shown in Table 3). Their features of four optrA-positive E. faecalis were shown in Table 3, indicating all three E. faecalis with linezolid resistance were detected with the carriage of optrA and just one linezolid-intermediate strain was positive. The genetic mutation of linezolid target sites, including the V domain of the 23S rRNA genes and 50S ribosome protein L3 and L4, were detected in linezolid-resistant/intermediate E. faecalis, indicating just G2576U genetic mutations in the V domain of the 23S rRNA genes were found in two linezolid-resistant E. faecalis isolates. That is to say, among the five linezolid-resistant E. faecalis found in this study, three optrA-positive isolates and the other two linezolid-resistant strains were explained by G2576U genetic mutations in the V domain of the 23S rRNA genes. The relationship of ST genotype with linezolid susceptibility, the carriage of several virulence factors and resistance genes in E. faecalis UTI isolates was shown in Table 4 and Supplementary Table 5. Overall, these E. faecalis isolates from UTI were classified into 21 ST types, in which the dominant clones were ST16 (35 strains, 30.43%) and ST179 (34 strains, 29.57%). Moreover, 31.42% (11/35) of ST16-E. faecalis and 2/34 (5.88%) of ST179-E. faecalis were not susceptible to linezolid.
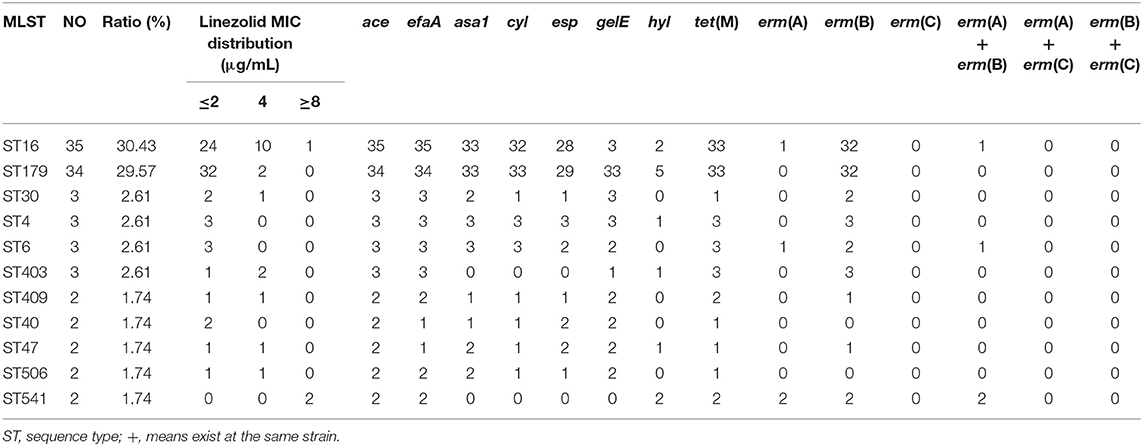
Table 4. Relationship of MLST phenotype with linezolid susceptibility, virulence factors, and resistance genes.
The basic clinical data of the patients with E. faecalis UTI was shown in Table 5. Univariate analysis showed that indwelling catheter, trachea cannula catheter, erm(A) and optrA genes were the risk factors for patients with UTI caused by linezolid-resistant/intermediate E. faecalis isolates (P < 0.05; Table 5). Moreover, multivariable conditional logistic regression model indicated that indwelling catheter and trachea cannula should be considered as the independent predictors of linezolid-resistant/intermediate E. faecalis infections (Table 6).
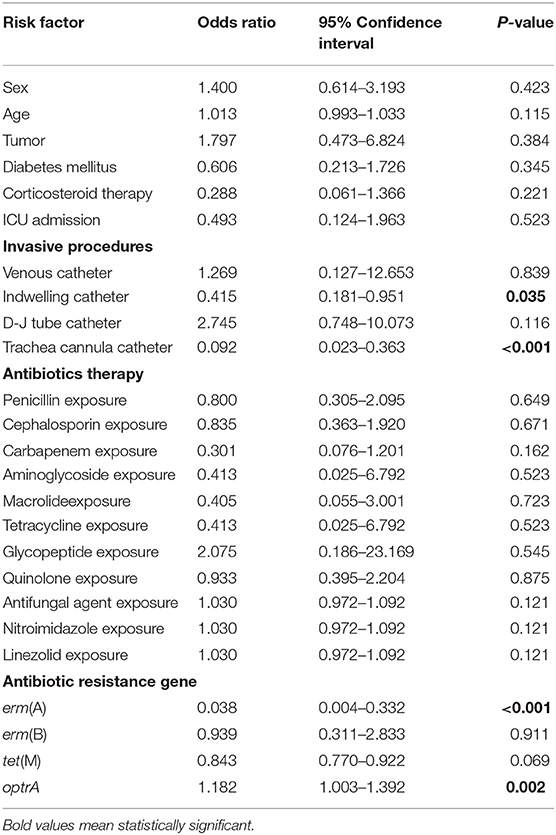
Table 5. Univariate analysis of potential risk factors of linezolid-resistant/intermediate E. faecalis.
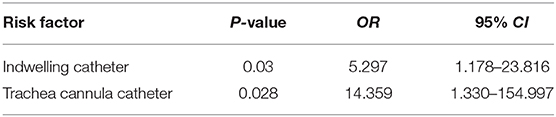
Table 6. Multivariable conditional logistic regression model for potential risk factors of linezolid-resistant/intermediate E. faecalis UTI infection.
E. faecalis is one of the major causative pathogens of UTI among gram-positive bacteria (19). Due to acquired and intrinsic resistance, E. faecalis exhibits a high level of resistance to many commonly used antibiotics, including cephalosporin and macrolides. In this study, our data also indicated a high frequency of E. faecalis clinical isolates from UTI with antibiotics resistance toward tetracyclines, minocycline, and erythromycin. Worthy of attention, the frequency of E. faecalis UTI isolates with linezolid or tedizolid-resistant/intermediate E. faecalis isolates in this hospital setting was significantly high compared with other articles reported (20). Moreover, all E. faecalis UTI isolates in this study, including linezolid-resistant/intermediate isolates, remained still susceptible to ampicillin, vancomycin and tigecycline, indicating rarely cross resistance between linezolid and other antibiotics in E. faecalis clinical isolates isolated from this hospital setting. Our data also indicated that linezolid or tedizolid might not be the first-line choices of the antibiotics suitable for the antimicrobial treatment of E. faecalis infections in this hospital settings. Therefore, the resistance mechanism and risk factors of linezolid-resistant/intermediate E. faecalis infections deserve our attention and need to be further studied.
Several previous reports have indicated the clonality characteristics of linezolid resistance in Staphylococci and E. faecalis (21, 22). ST16 is the predominant STs of linezolid-resistant/intermediate E. faecalis clinical isolates in this study. Multiple reports have demonstrated that ST16 might become more adaptable to the hospital environment and acquire the multi-drug resistance (10, 13). Whereas, whether ST16 E. faecalis with linezolid resistance has been widely transmitted in this hospital setting or this district needs to be further studied.
The reports have indicated the outbreak of the high detection frequency of some linezolid-resistant gram-positive bacteria in the hospital settings and this may be explained by different causes, such as antibiotics exposure, environmental contamination factors, person-to-person contact transmission (23–25). The complex mechanism of linezolid resistance in E. faecalis can be explained by three mechanisms: (1) genetic mutations in linezolid target sites, including the domain V region of 23S rRNA genes; (2) mutations in rpl(D) or rpl(C) genes encoding 50S ribosomal proteins L3 and L4; and (3) acquisition and dissemination of the plasmid-borne genes cfr, cfr(B), poxtA, and ATP-binding cassette (ABC) transporter gene optrA (13, 16, 26, 27). The plasmid-borne genes cfr and cfr(B) haven't been found in this study and moreover, two linezolid-resistant strains have the G2576U genetic mutations in the V domain of the 23S rRNA, which is consistent with previous reports in our laboratory (28). Our data indicated the high carriage of erm(A) in linezolid-resistant/intermediate E. faecalis UTI isolates and three linezolid-resistant E. faecalis isolates were positive with optrA. The mechanism of erm(A) that participate in macrolide or clindamycin resistance is mainly mediated by methylating the V domain of 23S rRNA gene. No report supported linezolid resistance could be caused by erm(A) and we hypothesized linezolid-resistant/intermediate isolates might facilitate the carriage or transmission of this resistance gene in E. faecalis. Therefore, the correlation of erm(A) with linezolid susceptibility needs to be further studied. The plasmid-borne optrA can result in cross resistance to multiple antibiotics in gram-positive bacteria, including oxazolidinones (linezolid and tedizolid) and phenicols (13). The optrA gene was firstly demonstrated for the explanation of linezolid-resistant/intermediate E. faecalis and subsequently the rapid and transmission of this gene among Enterococcus spp. and other gram-positive bacteria was further reported worldwide (13, 15, 16, 23, 25). Recently, the carriage frequency of optrA in Enterococcus spp. from the animals of human food chain in China was reported to be higher than that from human host (15.9% vs. 2–2.9%, respectively) and then, this phenomenon was mainly explained by the continuous and wide application of florfenicol in the food-producing animals or the environment in China from 1999 (15, 29). Considering the high frequency of linezolid resistance in this study, we presume that the transmission of optrA may exist in the environment, food products, medical device surface and so on (30). Overall, the transmission routine and mechanism of optrA in linezolid-resistant/intermediate E. faecalis in this hospital setting should be further elucidated. It is worthy of note that one linezolid-intermediate strains was found optrA gene and it's still unknown for the mechanism explanation of linezolid-intermediate/resistance in majority of E. faecalis UTI isolates. We hypothesized that some unknown proteins or other resistance mechanisms, including the efflux pumps or some membrane proteins, might participate in the MIC enhancement of linezolid. Therefore, the mechanisms of linezolid-intermediate/resistance E. faecalis in this hospital setting need to be further studied. Some reports have shown that linezolid exposure is an independent risk factor for linezolid-resistant/intermediate E. faecalis infections in UTI (Case-control Studies) (31, 32). The univariate and multivariable conditional logistic regression of the risk factors of E. faecalis infection with linezolid resistance in this study suggested indwelling catheter and trachea cannula catheter as the independent predictors of linezolid-resistant/intermediate E. faecalis infections. It's well-known that indwelling catheter and trachea cannula are invasive operations in clinics and they are considered as one of the important causes of nosocomial infection, indicating the hospital environment and invasive operation might prompt the occurrence or dissemination of linezolid resistance in this hospital setting. However, linezolid exposure was not considered as a risk factor in this study, which could be explained by the narrow application of this drug in this hospital. Our data indicated that linezolid resistance, even in some medical environments without the wide application of linezolid, should be alert and might exhibit the high level due to the environmental transmission of linezolid-resistant/intermediate bacteria that possibly caused by invasive operations.
Conclusively, this study demonstrated the high frequency of linezolid-resistant/intermediate E. faecalis in patients with UTI in this hospital setting. These isolates showed the characteristics of clonality to ST16 and ST179. Moreover, E. faecalis with linezolid resistance in this study might be explained by the high carriage frequency of optrA genes and the genetic mutation of linezolid target site. The invasive operations, especially indwelling catheter and trachea cannula catheter might facilitate the development of linezolid-resistant/intermediate E. faecalis UTI infection in hospital setting. The transmission routine of optrA in linezolid-resistant E. faecalis and the mechanisms of linezolid-intermediate/resistant E. faecalis in this hospital setting should be further elucidated.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
XM: participated in the design of the study, carried out RNA silencing test, analyzed, and interpreted the data, and drafted the manuscript. FZ and BB: performed antibiotic susceptibility testing, detected virulence genes by PCR, carried out the RNA silencing test, and participated in the data analysis. ZL and GX: conducted the MLST and CC analysis, and provided a critical revision of the manuscript. ZC, XS, and JZ: participated in the acquisition of the samples, isolated DNA, conducted MLST. QD and ZY: designed the study, participated in the data analysis, and provided critical revisions of the manuscript for important intellectual content. All authors contributed to the article and approved the submitted version.
This work was supported by the grants from National Natural Science Foundation of China (Nos. 81170370 and 81601797); Shenzhen Scientific Research Program (Nos. JCYJ20170412143551332, JCYJ20170307153714512, JCYJ201703 07153919735, and JCYJ20170307153425389); Shenzhen Health and Family Planning Commission (Nos. SZFZ2017063, SZXJ2017032, SZFZ2017036, and 201601058); Sanming Project of Medicine in Shenzhen; the Shenzhen Nanshan District Scientific Research Program of the People's Republic of China (Nos. 2016010, 2017013, 2017015, 2017026, and 2017027), and provincial medical funds of Guangdong (Nos. B2017019, 2014A031313718, and A2018163).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2021.570650/full#supplementary-material
1. Tien BYQ, Goh HMS, Chong KKL, Bhaduri-Tagore S, Holec S, Dress R, et al. Enterococcus faecalis promotes innate immune suppression and polymicrobial catheter-associated urinary tract infection. Infect Immun. (2017) 85:e00378–17. doi: 10.1128/IAI.00378-17
2. Seno Y, Kariyama R, Mitsuhata R, Monden K, Kumon H. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med Okayama. (2005) 59:79–87. doi: 10.1128/microbiolspec.GPP3-0053-2018
3. Armbruster CE, Prenovost K, Mobley HL, Mody L. How often do clinically diagnosed catheter-associated urinary tract infections in nursing homes meet standardized criteria? Am Geriatr Soc. (2017) 65:395–401. doi: 10.1111/jgs.14533
4. Abat C, Huart M, Garcia V, Dubourg G, Raoult D. Enterococcus faecalis urinary-tract infections: do they have a zoonotic origin? J Infect. (2016) 73:305–13. doi: 10.1016/j.jinf.2016.07.012
5. Coombs GW, Pearson JC, Daley DA, Le T, Robinson OJ, Gottlieb T, et al. Molecular epidemiology of enterococcal bacteremia in Australia. J Clin Microbiol. (2014) 52:897–905. doi: 10.1128/JCM.03286-13
6. Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci USA. (2008) 105:13339–44. doi: 10.1073/pnas.0804276105
7. Paterson DL, Pasculle AW, McCurry K. Linezolid: the first oxazolidinone antimicrobial. Ann Intern Med. (2003) 139:863–4. doi: 10.7326/0003-4819-139-10-200311180-00017
8. Torres C, Alonso CA, Ruiz-Ripa L, León-Sampedro R, DelCampo R, Coque TM. Antimicrobial resistance in Enterococcus spp.of animal origin. Microbiol Spectr. (2018) 6:1–41. doi: 10.1128/microbiolspec.ARBA-0032-2018
9. Bi R, Qin T, Fan W, Ma P, Gu B. The emerging problem of linezolid-resistant enterococci. J Glob Antimicrob Resist. (2017) 13:11–9. doi: 10.1016/j.jgar.2017.10.018
10. Zheng JX, Bai B, Lin ZW, Pu ZY, Yao WM, Chen Z, et al. Characterization of biofilm formation by Enterococcus faecalis isolates derived from urinary tract infections in China. J Med Microbiol. (2018) 67:60–7. doi: 10.1099/jmm.0.000647
11. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2020).
12. Zheng JX, Wu Y, Lin ZW, Pu ZY, Yao WM, Chen Z, et al. Characteristics of and virulence factors associated with biofilm formation in clinical Enterococcus faecalis isolates in China. Front Microbiol. (2017) 8:2338. doi: 10.3389/fmicb.2017.02338
13. Bai B, Hu KT, Li H, Yao WM, Li DY, Chen Z, et al. Effect of tedizolid on clinical Enterococcus isolates: in vitro activity, distribution of virulence factor, resistance genes and multilocus sequence typing. FEMS Microbio Lett. (2018) 365:fnx284. doi: 10.1093/femsle/fnx284
14. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0 (2021). Available online at: http://www.eucast.org
15. Wang Y, Lv Y, Cai J, Schwarz S, Cui L, Hu Z, et al. A novel gene optrA that confers transferable resistance to oxazolidinones and phenicols and its presence in Enterococcus faecalis and Enterococcus faecium of human and animal origin. J Antimicrob Chemother. (2015) 70:2182–90. doi: 10.1093/jac/dkv116
16. Egan SA, Shore AC, Brian OC, Brennan GI, Coleman DC. Linezolid resistance in Enterococcus faecium and Enterococcus faecalis from hospitalized patients in Ireland: high prevalence of the MDR genes optrA and poxtA in isolates with diverse genetic backgrounds. J Antimicrob Chemother. (2020) 75:1704–11. doi: 10.1093/jac/dkaa075
17. Vankerckhoven V, Van Autgaerden T, Vael C, Lammens C, Chapelle S, Rossi R, et al. Development of a multiplex PCR for the detection of asa1, gelE, cylA, esp, and hyl genes in enterococci and survey for virulence determinants among European hospital isolates of Enterococcus faecium. J Clin Microbiol. (2004) 42:4473–9. doi: 10.1128/JCM.42.10.4473-4479.2004
18. Barbosa-Ribeiro M, De-Jesus-Soares A, Zaia AA, Ferraz CC, Almeida JF, Gomes BP, et al. Antimicrobial susceptibility and characterization of virulence genes of Enterococcus faecalis isolates from teeth with failure of the endodontic treatment. J Endod. (2016) 42:1022–8. doi: 10.1016/j.joen.2016.03.015
19. Floresmireles A L, Walker J N, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. (2015) 13:269–84. doi: 10.1038/nrmicro3432
20. Klupp EM, Both A, Belmar Campos C, Büttner H, König C, Christopeit M, et al. Tedizolid susceptibility in linezolid- and vancomycin-resistant Enterococcus faecium isolates. Eur J Clin Microbiol Infect Dis. (2016) 35:1957–61. doi: 10.1007/s10096-016-2747-0
21. Azhar A, Rasool S, Haque A, Shan S, Saeed M, Ehsan B, et al. Detection of high levels of resistance to linezolid and vancomycin in Staphylococcus aureus. J Med Microbiol. (2017) 66:1328–31. doi: 10.1099/jmm.0.000566
22. Silva-DelToro SL, Greenwood-Quaintance KE, Patel R. In vitro activity of tedizolid against linezolid-resistant staphylococci and enterococci. Diagn Microbiol Infect Dis. (2016) 85:102–4. doi: 10.1016/j.diagmicrobio.2016.02.008
23. Pfaller MA, Mendes RE, Streit JM, Hogan PA, Flamm RK. Five-year summary of in vitro activity and resistance mechanisms of linezolid against clinically important gram-positive cocci in the United States from the LEADER Surveillance Program (2011 to 2015). Antimicrob Agents Chemother. (2017) 61:e00609–17. doi: 10.1128/AAC.00609-17
24. Pourakbari B, Mahmoudi S, Moradzadeh M, Mahzari M, Ashtiani MTH, Tanzifi P, et al. Antimicrobial resistance patterns of the gram-positive bacteria isolated from children with bloodstream infection in an Iranian referral Hospital a 6-year study. Infect Disord Drug Targets. (2018) 18:136–44. doi: 10.2174/1871526517666170821164343
25. Cai J, Wang Y, Schwarz S, Lv H, Li Y, Liao K, et al. Enterococcal isolates carrying the novel oxazolidinone resistance gene optrA from hospitals in Zhejiang, Guangdong, and Henan, China, 2010-2014. Clin Microbiol Infect. (2015) 21:e1–4. doi: 10.1016/j.cmi.2015.08.007
26. Asadollahi P, Razavi S, Asadollahi K, Pourshafie MR, Talebi M. Rise of antibiotic resistance in clinical enterococcal isolates during 2001-2016 in Iran: a review. New Microbes New Infect. (2018) 26:92–9. doi: 10.1016/j.nmni.2018.08.018
27. Long KS, Vester B. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob Agents Chemother. (2012) 56:603–12. doi: 10.1128/AAC.05702-11
28. Li DY, Yu ZJ, Xu J, Liu XJ, Deng MG, Pan WG, et al. Genetic mutations in the V domain of the 23S ribosomal RNA gene of linezolid-resistant Enterococcus faecalis. J Pathogen Biol. (2014) 9:316–7.
29. He T, Shen Y, Schwarz S, Cai J, Lv Y, Li J, et al. Genetic environment of the transferable oxazolidinone/phenicol resistance gene optrA in Enterococcus faecalis isolates of human and animal origin. J Antimicrob Chemother. (2016) 71:1466–73. doi: 10.1093/jac/dkw016
30. Matthew HG, Bryan DH, Whitney JN, Marley LW, Patty WW, Thomas RT, et al. Risk factors and outcomes associated with acquisition of daptomycin and linezolid-nonsusceptible vancomycin-resistant Enterococcus. Open Forum Infect Dis. (2018) 5:ofy185. doi: 10.1093/ofid/ofy185
31. Pai MP, Rodvold KA, Schreckenberger PC, Gonzales RD, Petrolatti JM, Quinn JP. Risk factors associated with the development of infection with linezolid-and vancomycin-resistant Enterococcus faecium. Clin Infect Dis. (2002) 35:1269–72. doi: 10.1086/344177
Keywords: linezolid resistance, urinary tract infection, virulence factor, resistance genes, Enterococcus faecalis
Citation: Ma X, Zhang F, Bai B, Lin Z, Xu G, Chen Z, Sun X, Zheng J, Deng Q and Yu Z (2021) Linezolid Resistance in Enterococcus faecalis Associated With Urinary Tract Infections of Patients in a Tertiary Hospitals in China: Resistance Mechanisms, Virulence, and Risk Factors. Front. Public Health 9:570650. doi: 10.3389/fpubh.2021.570650
Received: 08 June 2020; Accepted: 13 January 2021;
Published: 05 February 2021.
Edited by:
Richard V. Goering, Creighton University, United StatesReviewed by:
Stefan Paul Schwarz, Freie Universität Berlin, GermanyCopyright © 2021 Ma, Zhang, Bai, Lin, Xu, Chen, Sun, Zheng, Deng and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijian Yu, eXV6aGlqaWFuc211QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.