- 1International Centre for Diarrhoeal Disease Research, Dhaka, Bangladesh
- 2WASH Division, UNICEF Bangladesh, Dhaka, Bangladesh
Introduction: Community-acquired infections due to extended-spectrum beta-lactamase (ESBL) producing Escherichia coli are rising worldwide, resulting in increased morbidity, mortality, and healthcare costs, especially where poor sanitation and inadequate hygienic practices are very common.
Objective: This study was conducted to investigate the prevalence and characterization of multidrug-resistant (MDR) and ESBL-producing E. coli in drinking water samples collected from Rohingya camps, Bangladesh.
Methods: A total of 384 E. coli isolates were analyzed in this study, of which 203 were from household or point-of-use (POU) water samples, and 181 were from source water samples. The isolates were tested for virulence genes, ESBL-producing genes, antimicrobial susceptibility by VITEK 2 assay, plasmid profiling, and conjugal transfer of AMR genes.
Results: Of the 384 E. coli isolates tested, 17% (66/384) were found to be ESBL producers. The abundance of ESBL-producers in source water contaminated with E. coli was observed to be 14% (27/181), whereas, 19% (39/203) ESBL producers was found in household POU water samples contaminated with E. coli. We detected 71% (47/66) ESBL-E. coli to be MDR. Among these 47 MDR isolates, 20 were resistant to three classes, and 27 were resistant to four different classes of antibiotics. Sixty-four percent (42/66) of the ESBL producing E. coli carried 1 to 7 plasmids ranging from 1 to 103 MDa. Only large plasmids with antibiotic resistance properties were found transferrable via conjugation. Moreover, around 7% (29/384) of E. coli isolates harbored at least one of 10 virulence factors belonging to different E. coli pathotypes.
Conclusions: The findings of this study suggest that the drinking water samples analyzed herein could serve as an important source for exposure and dissemination of MDR, ESBL-producing and pathogenic E. coli lineages, which therewith pose a health risk to the displaced Rohingya people residing in the densely populated camps of Bangladesh.
Introduction
Extended-spectrum beta-lactamase (ESBL)-producing E. coli have been recognized as a major multidrug-resistant bacteria implicated in serious hospital and community-acquired infections worldwide, especially in places where poor sanitation, and inadequate hygienic practices are very common (1–4). Infections caused by MDR-E. coli incur huge medical costs and limit treatment options (5–7).
Multidrug-resistant E. coli has been detected in different ecological niches in the community and environment (8, 9). For instance, ESBLs and New Delhi Metallo beta-lactamase 1 (NDM-1) producing E. coli were detected in drinking water and retail meat, respectively (10, 11). In Bangladesh, ESBL producing-E. coli were reported from drinking water as well as from river water samples (12, 13). Though E. coli has had a significant role in water microbiology as an indicator of fecal pollution, it is of greater public health concern when these E. coli isolates turn out to be multidrug-resistant pathogens (14). Detection of ESBL-producing E. coli in drinking water samples is important to recognize the risk of transmission of antimicrobial resistance (AMR) and gastrointestinal diseases. Transmission of ESBL-encoding genes among bacteria is often plasmid-mediated (15), and aquatic environments provide ideal settings for horizontal transfer of AMR genes encoded on various forms of mobile genetic elements (16).
Though the majority of E. coli are typically innocuous, some E. coli variants are virulent and may inflict varying severity of enteric infections. Currently, there are six different E. coli pathotypes that have been documented to cause intestinal infections, they include, enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteropathogenic E. coli (EPEC), shiga toxin-producing E. coli (STEC), enteroaggregative E. coli (EAEC), and diffusely adhering E. coli (DAEC) (17). In Bangladesh, following rotavirus, the second most leading cause of diarrheal infections are caused by pathogenic E. coli (18). Several virulence genes such as st, lt (ETEC); bfp, eae (EPEC); aat, aai (EAEC) are associated with diarrheagenic E. coli pathotypes (19), which can be used to detect these pathotypes using PCR based gene amplification. Watery diarrhea is caused by the secretion of heat-labile (LT) and/or heat-stable (ST) enterotoxins from ETEC. Shiga toxin (Stx) expression is the unique feature of EHEC where systemic absorption of this toxin leads to possibly life-threatening complications. Multiple putative virulence factors expression for typical EAEC strains, containing the aggregative adherence fimbriae (AAF), dispersin, the dispersin translocator Aat, and the Aai type VI secretion system directs to adherence and triggering diarrhea. EPEC adhesion is associated with attaching and effacing adhesion and intestinal colonization, which also include bundle-forming pili (BFP), EspA filaments and intimin (19, 20).
The contaminated drinking water was found to be responsible for a number of waterborne gastroenteritis outbreaks due to diarrheagenic E. coli (21–23). Therefore, it is pertinent to analyze the prevalence and properties of ESBL-E. coli from drinking waters in community settings, particularly, from human habitations that are projected to pose exceptionally high risks of waterborne diseases due to overcrowding, scarcity of safe drinking water, and unhygienic living conditions. In Bangladesh, the displaced Rohingya people are one such community with a population of ~1.16 million who are living in 32 congested camps in a challenging hilly landscape of Cox's Bazar district (290,000 persons per square kilometer). This displacement, of a large population are facing compounding problems, particularly related to water, sanitation, hygiene and health care (24–27). Water from hand-pumped tube wells is the primary water supply for the people in Rohingya camps. Around 6057 water points and 50087 emergency latrines have been built (during the study in 2018). Moreover, in the absence of efficient community sanitation, insufficient sewage disposal, and treatment facilities, the risks of transmission of enteric pathogens become extremely high, and the community as a whole face serious public health concerns (28–30).
In our previous study, we analyzed source water (tubewell) samples, as well as POU drinking water samples, from Rohingya camps and found 10.5% source water and 34.7% POU water samples were contaminated with E. coli, which could cause waterborne diseases in the camps (26). An outbreak of ESBL-producing E. coli might create a medical emergency in a large congested habitation like Rohingya camps because of limited treatment options. The AMR surveillance, especially with regard to ESBL-producing E. coli has never been carried out in these camps. Therefore, this study aims to determine the prevalence of ESBL-producing, MDR, and virulent E. coli in drinking water samples. Furthermore, plasmid profiling and horizontal transfer of resistant gene analyses of isolated ESBL-producing E. coli will provide important insights in understanding the dissemination of resistance determinants.
Materials and Methods
Bacterial Isolates
We employed drinking water samples from Rohingya camps collected in our previous study to culture E. coli isolates (26). From a total of 2512 E. coli contaminated water samples, 421 water samples were randomly selected for the present study. One random E. coli isolate was taken as a representative from each sample, which was further tested using the API-20E test kit (Biomerieux SA, Marcy-I'Etoile, France), and 384 API-20E confirmed E. coli were stored at −70°C in 30% LB glycerol-broth for downstream analysis. Out of 384 E. coli isolates, 203 isolates were from the household water samples and 181 from source water samples. In brief, for each sample, 100 ml water was filtered through a 0.22 μm membrane filter (Sartorius Stedim, Goettingen, Germany), the membrane filter paper was then firmly laid on the mTEC agar plate. Later, the culture plate was incubated for 2 h at 35 ± 0.5°C, followed by further incubation for 22 ± 2 h at 44.5 ± 0.2°C. After incubation, red to magenta-colored colonies, typical of E. coli colony was picked and subcultured on MacConkey agar plate and incubated at 35 ± 2°C for 18 h to 24 h. After incubation, the characteristics of dark pink colonies typical of E. coli were obtained and confirmed using API-20E kit.
Isolation and Confirmation of ESBL-Producing and Carbapenem-Resistant E. coli Using Chromagar
All 384 confirmed E. coli isolates were cultured on CHROMagar ESBL and CHROMagar KPC media at 37°C for 18–24 h. The production of extended-spectrum beta-lactamases and carbapenemase was confirmed by observing the growth and characteristic colony morphology on respective culture media. Dark pink to reddish colonies on CHROMagar ESBL plate indicate ESBL producing E. coli whereas pink to reddish colonies on CHROMagar KPC media suggest carbapenem-resistant E. coli.
Confirmation of E. coli by VITEK 2
ESBL positive E. coli isolated from the ESBL CHROMagar plate were further confirmed by the VITEK 2 system (bioMérieux, Marcy I'Etoile, France) using VITEK 2 GN ID card. Enterobacter hormacchi (ATCC-700323) was used as a positive control for the identification in this system. For VITEK 2 assays, pure isolates were streaked on MacConkey agar plates and incubated at 35°C overnight. One to three isolated colonies were selected from each MacConkey agar plate and suspended in saline for preparation of inoculum to obtain an absorbency of ~0.5 McFarland Units before being subjected to VITEK 2 analysis.
Detection of Diarrheagenic and ExPEC Associated Virulence Genes
Several virulence genes such as st, lt (ETEC); bfp, eae (EPEC); aat, aai (EAEC) are associated with diarrheagenic E. coli pathotypes (19) are used to detect the pathotypes using PCR based gene amplification. In the present study, PCR based screening of diarrheagenic virulence genes was carried out for all 384 E. coli isolates. Gene-specific primers entailing heat-labile (lt), heat-stable (st), attaching and effacing (eae), anti-aggregation protein transporter (aat), bundle forming pilus (bfp) and aggR-activated island (aaiC) were used to detect the respective genes employing a multiplex PCR setup (31–34). The boiling lysis method was used to obtain the DNA template (35). A 3 μl template DNA was taken for a 25 μl PCR reaction containing 12.5 μl of 2X GoTaq G2 Green Master Mix (Promega, USA) with 0.44 μl of eae primer, the primers lt, st, aaiC, aat, bfp were taken in 0.4 μl volume each. The PCR was carried out at standard cycling conditions with an annealing temperature of 57°C for 20 s. A separate PCR was performed for Shiga toxin genes (stx1 and stx2), which was described previously (36, 37). PCR for invasion plasmid antigen H (ipaH) and the invasion associated locus (ial) was performed according to the procedure described elsewhere (38–41). Primer details are tabulated in Supplementary Table 1.
To examine the presence of seven ExPEC associated virulence factors, we performed two multiplex PCRs on 55 non diarrheagenic ESBL-E. coli isolates (42). Among these, the first one was done to screen the presence of kpsMII (group II capsule), papA (pilus-associated protein A), sfaS (S-fmbrial adhesine), and focG (F1C fmbriae protein) genes; whereas the second multiplex was performed to detect hlyD (haemolysin D), afa (afmbrial adhesine), and iutA genes (aerobactin siderophore ferric receptor protein).
Antibiotic Susceptibility by VITEK 2
Antibiotic susceptibility testing (AST) was performed using VITEK 2 system with VITEK 2 cards (AST-N280) for 19 antimicrobial agents according to the CLSI guidelines and manufacturer's recommendations; two additional antimicrobial agents (cefixime and ceftazidime) were also incorporated. E. coli ATCC 25922, susceptible to all drugs, was used for AST in each VITEK testing step as quality control. The 21 antibiotics tested included amikacin, amoxicillin/clavulanic acid, ampicillin, cefepime, cefixime, cefoperazone/sulbactam, ceftazidime, ceftriaxone, cefuroxime, cefuroxime axetil, ciprofloxacin, colistin, ertapenem, gentamicin, imipenem, meropenem, nalidixic acid, nitrofurantoin, piperacillin, tigecycline, and trimethoprim/sulfamethoxazole. Minimum inhibitory concentrations (MIC) were determined, and the isolates were classified into resistant, intermediate and susceptible as per CLSI criteria. The raw MIC data from the VITEK 2 assay are shown in Supplementary Table 2.
Detection of ESBL, Quinolone, and Carbapenemase Resistance Genes by PCR
All CHROMagar confirmed ESBL-producing E. coli isolates were screened for molecular determinants of ESBL and carbapenem resistance comprising of blaSHV, blaTEM, blaCTX−M−15 and all CTX-M-groups (blaCTX−M−1−group, blaCTX−M−2−group, blaCTX−M−8−group, blaCTX−M−9−group) including, blaOXA−1−group, blaOXA−47, and blaNDM−1 were screened (34, 43–48). The gene blaCMY−2 encoding for AmpC β-lactamase was also screened by PCR as per a published protocol (12, 49). Besides, all 66 isolates were tested for the three qnr (quinolone) resistance genes; qnrA, qnrB, and qnrS according to the methods described by others (50–52).
Plasmid Profiling and Conjugal Transfer
Plasmids from ESBL-E. coli isolates were extracted employing the Kado alkaline lysis method (53) and visualized after separation in low percent agarose gel (0.7%) electrophoresis. The size of extracted plasmids was determined comparing with size standard plasmids ran alongside. The following plasmids were used as size standards; Sa (23 MDa), RP4 (34 MDa), R1 (62 MDa), pDK9 (140 MDa), and E. coli V517 plasmids (1.4, 1.8, 2.0, 2.6, 3.4, 3.7, 4.8, and 35.8 MDa) (54). ESBL-producing E. coli were used as donors, and the sodium azide resistant strain E. coli-J53 was employed as a recipient for conjugation using broth mating assays at 30°C for 19 ± 1 h. Transconjugants obtained were plated on MacConkey plates prepared with cefotaxime (20 μg/L) and sodium azide (100 mg/L), transconjugants were selected observing their growth and colony morphology. Transconjugants were analyzed for antibiotic susceptibility tests using VITEK 2 assay, plasmid profiling and presence of ESBL genes (53).
Phylogenetic Analysis
As per the method described by Clermont and colleagues, the distribution of phylogenetic groups among ESBL- E. coli isolates was determined by performing multiplex PCRs after DNA extraction by boiling lysis (55).
Results
ESBL-Positive but Carbapenem Sensitive E. coli Recovered From Drinking Water
To screen for ESBL-producing E. coli, 384 isolates were cultured on ESBL CHROMagar. The typical growth of pink colonies on the CHROMagar plate was considered as ESBL positive E. coli. Of the 384 isolates tested, 17.2% (n = 66) were found to be ESBL-producing E. coli (Table 1). About 15% (27/181), and 19% (39/203) ESBL producing E. coli originated from E. coli contaminated source water and POU water samples, respectively. Further, the ESBL producing E. coli was investigated for carbapenem resistance on CHROMagar KPC media, and none of the isolates was able to grow on CHROMagar KPC media. Therefore, we did not detect any carbapenem-resistance in our collection of ESBL-E. coli isolates.
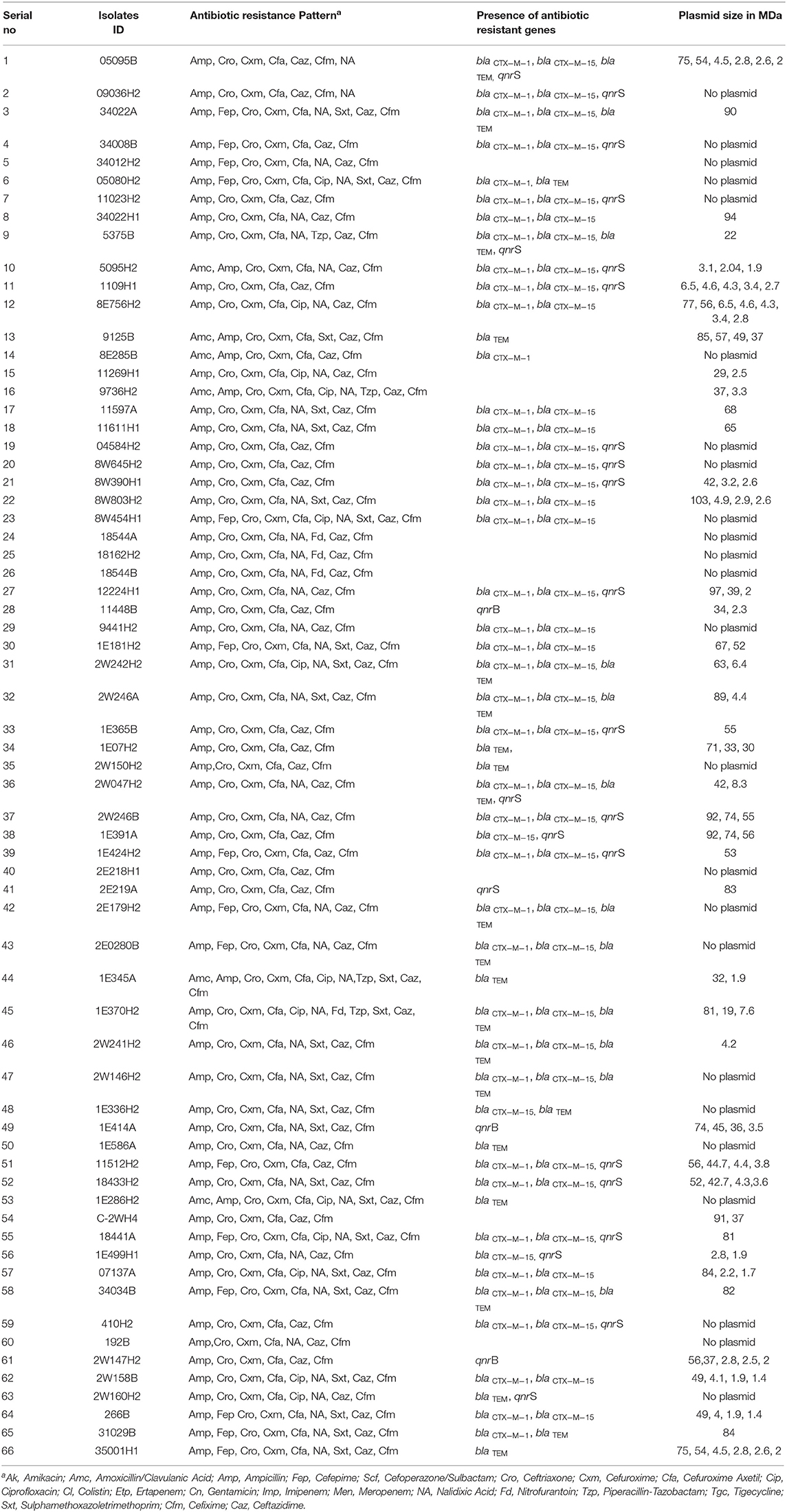
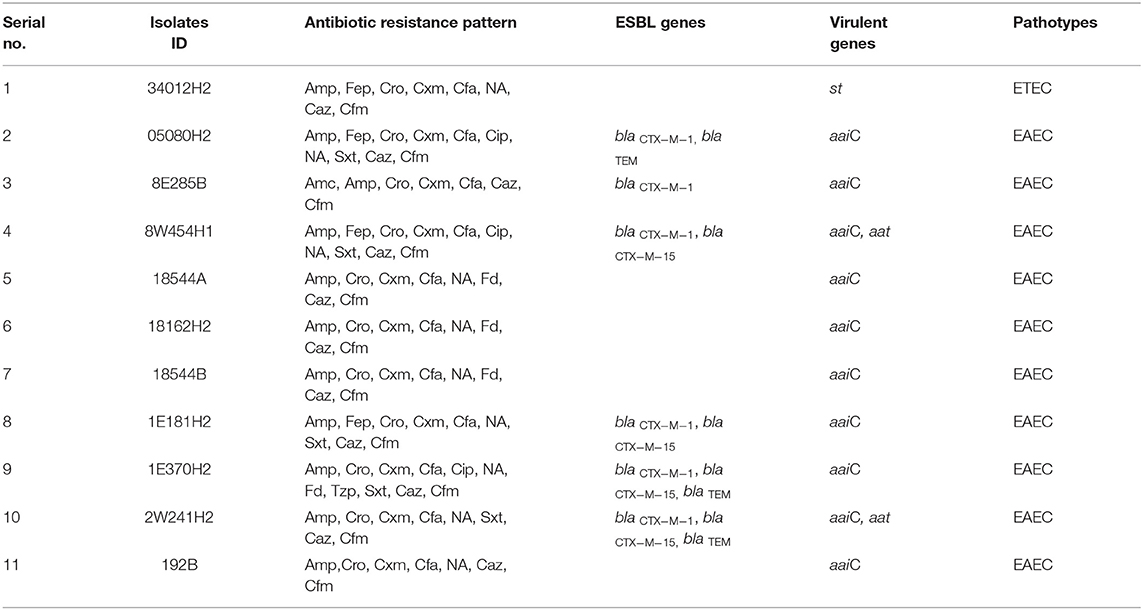
Table 1. Antibiotic resistance pattern, presence of antibiotic resistance genes and plasmid patterns of ESBL-producing Escherichia coli isolated from water sample.
High Prevalence of MDR in ESBL-Producing E. coli
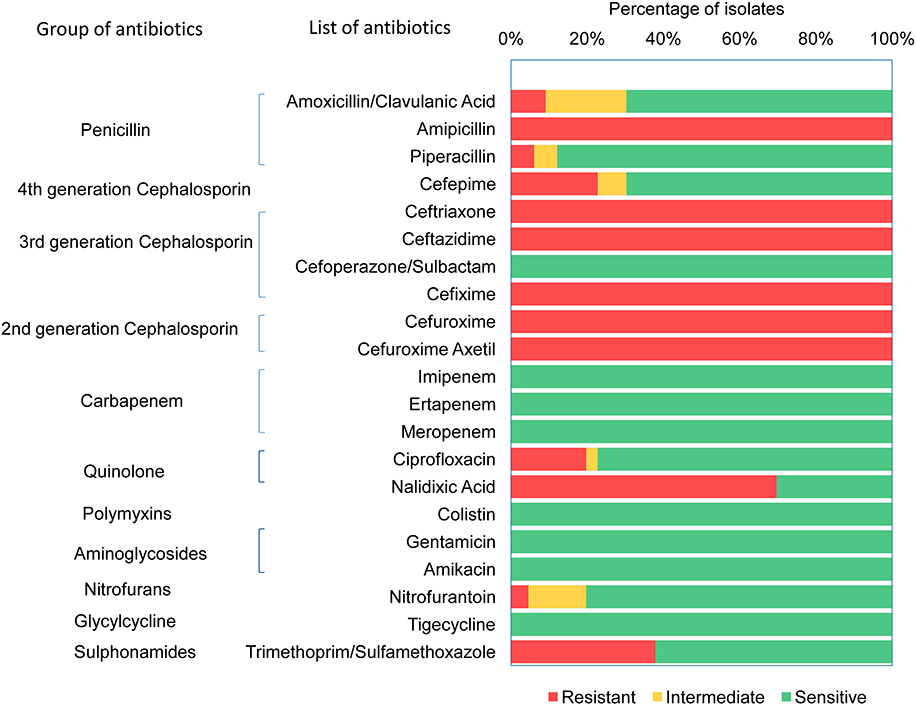
Antimicrobial susceptibility of 66 ESBL-E. coli isolates (Supplementary Table 3) were tested against 21 different antibiotics (Supplementary Table 4) using VITEK 2 assay. All 66 CHROMagar confirmed ESBL positive E. coli isolates demonstrated resistance against ampicillin, ceftriaxone, ceftazidime, cefixime, cefuroxime, and cefuroxime axetil (Figure 1). About 70% (46/66) of the isolates were found to be resistant to nalidixic acid, 37.9% (25/66) isolates were resistant to trimethoprim/sulfamethoxazole, whereas 22.7% (15/66) and 19.7% (13/66) isolates were resistant for cefepime and ciprofloxacin, respectively. However, no resistance was detected in any of the tested strains to the antibiotics used of aminoglycosides (amikacin and gentamicin), cefoperazone/sulbactam, glycylcycline, carbapenem, and polymyxins groups. It was found that 71% (47/66) of E. coli isolates were MDR that were resistant to at least three classes of antibiotics. Among the 47 MDR isolates, 20 were resistant to three different classes, and 27 were resistant to four different classes of antibiotics.
blaCTX-M-1 Group Is the Predominant ESBL Gene Detected
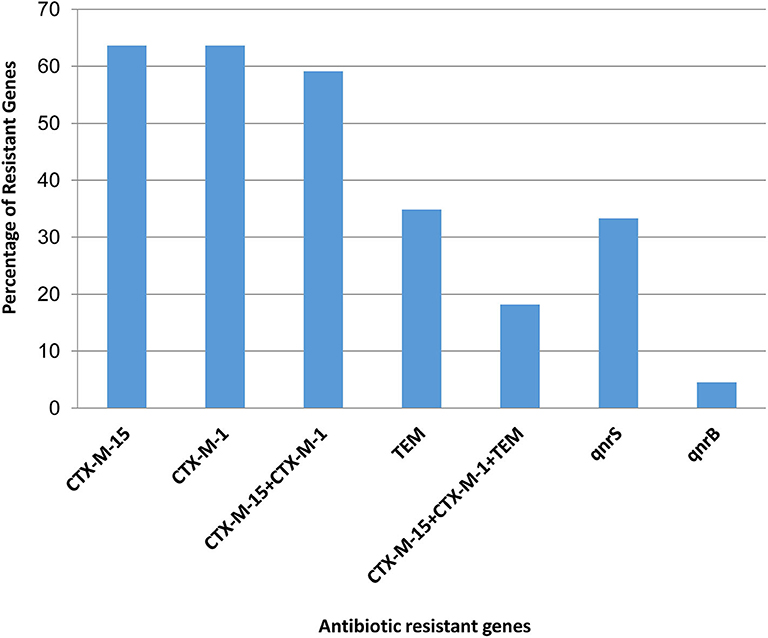
The presence of molecular determinants of ESBLs was tested on all ESBL-producing E. coli isolates. Out of the 66 ESBL-E. coli isolates, 59% (39/66) isolates harbored both blaCTX−M−1 group and blaCTX−M−15 gene. However, 4.5% (3/66) isolates harbored either blaCTX−M−1group or blaCTX−M−15 gene. The blaTEM β-lactamase gene was present in 35% (23/66) of isolates, and none of the isolates harbored other β-lactamase genes such as blaSHV, blaCTX−M−2−group, blaCTX−M−8−group, and blaCTX−M−9−group. The two genes, blaOXA−1−group and blaOXA−47 were screened among 66 ESBL- E. coli isolates, but none of the isolates was found to be positive. In addition, the New Delhi metallo-β-lactamase gene, blaNDM−1 as well as plasmid-mediated ampC-type β-lactamase gene the blaCMY−2 was not present in any of the isolates. The quinolone resistance gene; qnrS, and qnrB were found in 34% (22/66) and 5% (3/66) of the ESBL-E. coli isolates, respectively (Figure 2, Supplementary Table 5).
Distribution of E. coli Pathotypes
Screening for the presence of virulence factors demonstrated that 7% (29/384) of E. coli isolates were positive for at least one virulence factor out of the 10 E. coli pathotype-specific virulence genes tested. Ten isolates were positive for only aaiC gene and five isolates were positive for both aaiC and aat; whereas four isolates were positive for both bfp and eae genes. Heat labile (lt) gene was present in 7 isolates, whereas the heat stable (st) gene was found in 2 isolates and a single isolate was found positive for stx1. None of the isolates was positive for ipaH and iaa genes. Among the 29 pathogenic E. coli isolates detected; 52% (15/29) were EAEC, 31% (9/29) were ETEC, 14% (4/29) were EPEC and 4% (1/29) were EHEC (Figure 3).
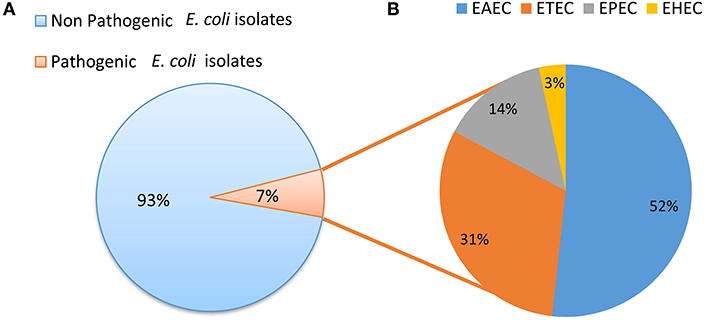
Figure 3. (A) Distribution of pathogenic and non pathogenic E. coli. (B) Distribution of different E. coli pathotypes.
When the ExPEC virulence genes were screened only three out of the seven virulence factors were detected that comprised of KpsMII, sfaS, and iutA genes, their prevalence rates were, 21.8% (12/55), 5.4% (3/55), and 16.4 % (9/55), respectively. Most of isolates (12/18) harboring ExPEC genes were affiliated to phylogenetic group D. However, out of the 55 isolates tested, only 6 isolates qualify as ExPEC as per the inclusion criteria (strains harboring at least two ExPEC associated virulence factors) 5 out of these 6 isolates were from phylogroup D.
The potential pathogenic (diarrheagenic and ExPEC) E. coli isolates showed high resistance rates 83% (39/47) to ampicillin, followed by 74% (35/47) to nalidixic acid, 68% (32/47) to cefuroxime, cefuroxime axetil, 61% (29/47) to cefixime, ceftazidime, and 51% (24/47) to sulfonamides. Out of 29 pathogenic E. coli 11 were found to be ESBL producing in this study (Table 2). Of note, all the pathogenic isolates detected were found to be susceptible to carbapenems, aminoglycosides (amikacin and gentamicin), and polymixin.
Phylogrouping
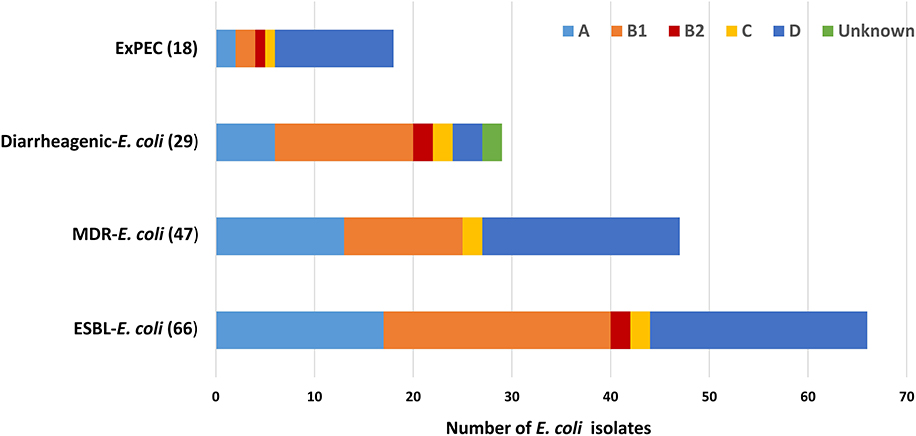
Among the ESBL-E. coli isolates, all phylogenetic groups were represented except for phylogroup F. The predominant phylogenetic group identified was B1 (23/66; 34.8%), followed by D (22/66; 33.3%), E (17/66; 25.7%), B2 and C (2/66; each 3%). Among the 47 multidrug resistant ESBL-E. coli (20/47) were of phylogroup D followed by A (13/47), B1 (12/47), C (2/47). Majority of isolates carrying diarrheagenic virulence genes were from B1(14/29; 48%) followed by A(6/29; 20.6%), B2, C, and unknown groups (2/29; each 7%) (Figure 4).
Plasmid Analysis of ESBL-Producing E. coli
Plasmid profiling and conjugation analysis were performed to see whether the antibiotic-resistance genes of the 66 ESBL producing isolates were plasmid-mediated and can they be horizontally transferred. Plasmid number and size were determined using conventional lysis and agarose gel electrophoresis. About 63% (n = 42) isolates carried 1 to 7 plasmids ranging in size from ~1 to 103 MDa (Figure 5), and the distribution of plasmids was heterogeneous (Table 1). Further, the plasmid containing isolates that showed the ESBL phenotype were tested for their ability to transfer the ESBL determinant by conjugation experiments. Nine isolates were able to transfer the cefotaxime resistance marker to a susceptible E. coli recipient with transfer rates ranging from 4.75 × 10−6 to 1.19 × 10−4 per donor cell (Table 3). Large plasmids (30–103 MDa) were transferred to the sodium azide resistant E. coli-J53 recipient. Whereas, the smaller plasmids (<30 MDa) were not seen to be transferred during conjugation. Among the nine donor isolates, two were able to transfer two plasmids each whereas seven isolates transferred single plasmids (Table 3).
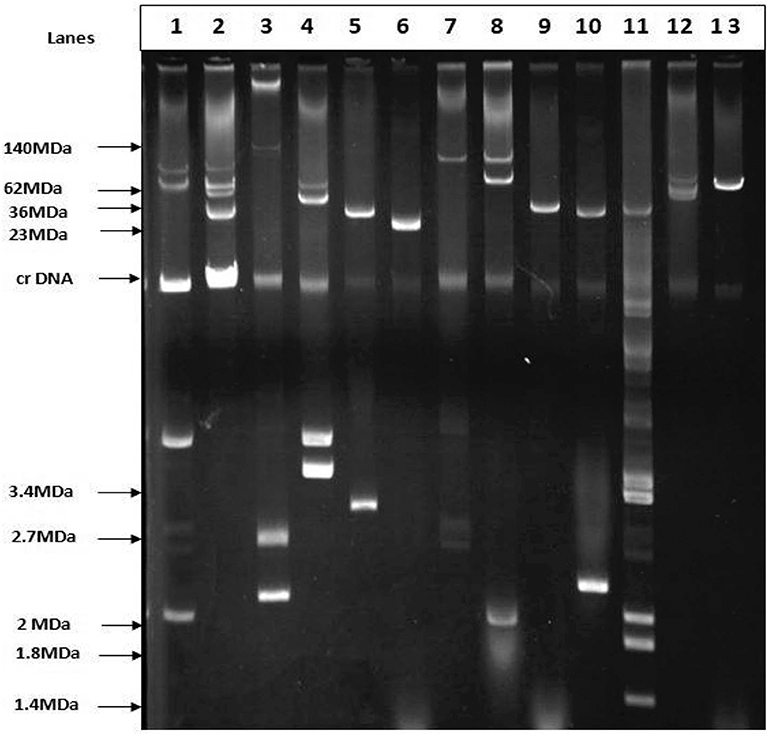
Figure 5. Agarose gel electrophoresis of plasmid DNA showing the patterns among the ESBL positive isolates. Lane-3: E. coli strain pDK9 (140, 105, 2.7, 2.1 MDa), Lane-6: V517 (23MDa), Lane-9:RP4 (36MDa), Lane-11: V517 (35.8, 3.4,3.7, 2,1.8,1.4), Lane-13: R1 (62MDa), Lane-1,2,4,5,7,8,10, and 12 are ESBL positive E. coli. The molecular weight of the markers is shown in the picture.
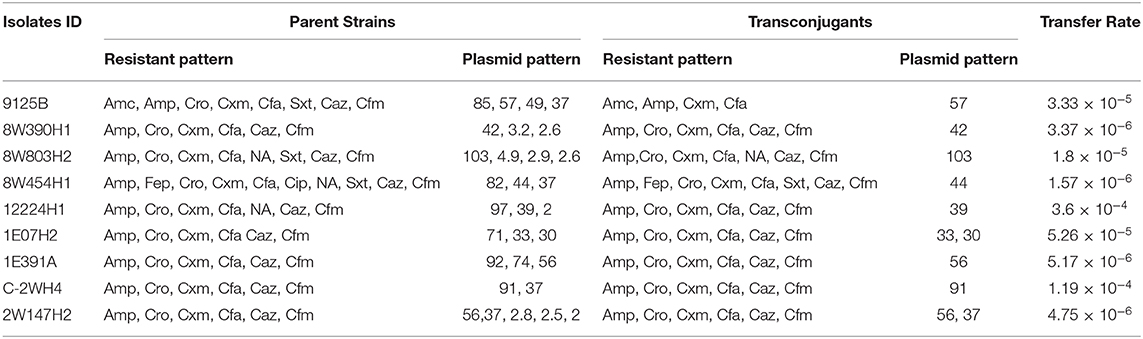
Table 3. Results of conjugation assays between antibiotic resistant E. coli isolates obtained from water samples and the recipient E. coli J53 strain.
Discussion
In our previous study, we investigated the occurrence of E. coli and fecal coliforms in source and household drinking water samples in Rohingya camps, wherein 10.5% tubewell water and 34.7% POU water samples were found to be contaminated with E. coli (26). In the current study, the ESBL-producing E. coli isolates from the contaminated drinking water samples of our previous study were characterized concerning antimicrobial susceptibility, dissemination of drug resistance, and pathogenic potential to comprehend the extent of public health threat due to the exposure of contaminated drinking water in Rohingya camps of Bangladesh.
ESBL-producing E. coli has been increasingly reported globally, it is not only restricted to clinical settings but also recovered from environmental niches like livestock, wildlife and particularly water (56–58). The pandemic spread of ESBL-producing Gram-negative bacteria is a serious health concern. Human habitation and the surrounding environment in Bangladesh are reportedly contaminated by antimicrobial-resistant bacteria (59, 60). Reports emanating from developing countries like Bangladesh indicated a high prevalence of ESBL-producing E. coli in hospital and community drinking water samples (34, 61).
In the current study, we detected 17.2% (66/384) ESBL-producing E. coli, of which 71% (47/66) was multidrug-resistant. The high prevalence of multidrug-resistant E. coli among the ESBL-producing isolates implies that not only β-lactam antibiotics but resistance to other classes of antibiotics is being co-selected. In Bangladesh, cephalosporins and penicillins are the most commonly used antibiotics (62), which explain why all the 66 ESBL-producing isolates are found to be resistant to both the classes of antibiotics. Besides, the majority of ESBL-producing isolates were found to be resistant to the quinolone class of antibiotics (46 to nalidixic acid). This may reflect the overuse and misuse of antibiotics (63) as these drugs are often sold and distributed over the counter (64). The uncontrolled and unregulated use of antibiotics severely limits the therapeutic options as well as aids the rapid dissemination of resistance in such overpopulated Rohingya camps.
We found ESBL-producing E. coli isolates are 100% susceptible to the antibiotics tested of carbapenem, aminoglycoside (amikacin and gentamicin), glycylcycline, and polymyxin groups. This finding was similar to a study in Jordan, where all the E. coli isolates from drinking water were sensitive to carpapenem and glycylcycline (65). There might be several factors responsible for susceptibility, such as these drugs are rarely prescribed in Bangladesh (64) and are not readily available in the hard to reach hilly terrain like Rohingya camps.
In this study, most of the isolates were positive for blaCTX−M−1 group and blaCTX−M−15, genes that concur the previous reports from Bangladesh (59, 60, 66). All blaCTX−M−1 group, blaCTX−M−15 and blaTEM ESBL-positive E. coli isolates showed MDR phenotype ranging from 3 to 4 classes of antibiotics. Similar to our observation, a previous study from Bangladesh showed a high prevalence of CTX-M-15 in ESBL-producing E. coli cultured from drinking water samples from different households (34). The CTX-M group of beta-lactamases are a group of rapidly emerging ESBL genes globally, which has been predominantly detected in E. coli and Klebsiella spp. (56, 67–70). The NDM-1 producing bacteria have been reported in clinical isolates from Bangladesh (71), but in the current study, no NDM-1-producing E. coli was detected in the water samples.
The gene qnrB has been recognized in various enterobacterial species, such as E. coli and Klebsiella spp. (72–74). Plasmid-mediated quinolone resistance is intervened by the genes (qnr) encoding proteins that protect DNA gyrase and topoisomerase IV against quinolone compounds (75). Among the nonclinical sources, qnr gene was reported in E. coli isolated from swine, livestock and poultry (76, 77). In the present study, 25 isolates harbored plasmid-mediated qnr genes comprising of 22 isolates positive for qnrS and 3 isolates for qnrB genes. Isolates harboring qnrS gene also demonstrated co-existence of blaCTX−M−15 and blaCTX−M−1 group gene. Additionally, they were resistant to a range of 7–11 antibiotics, including ciprofloxacin and nalidixic acid. In contrast, all isolates containing qnrS gene also harbored the two ESBL genes, blaCTX−M−1group and blaCTX−M−15; they were resistant to 8 different antibiotics; most interestingly, they were susceptible to ciprofloxacin. Hence, the presence of qnrS gene alone may not be indicative of the isolate being resistant to fluoroquinolones as also been observed in a previous study (78).
Using PCR for virulence genes, 29 (7%) was found to be pathogenic out of 384 isolates from drinking water. The most prevalent pathotype was EAEC, accountable for 52% of the pathogenic isolates; followed by ETEC, EPEC, and EHEC responsible for 31, 14, and 4% of the pathogenic isolates, respectively. In addition to the diarrheagenic E. coli around 22% of E. coli isolates were at least positive for 1 ExPEC associated virulence genes, moreover 11% (6/55) of the isolates were dectected to be potential ExPEC strains. This indicates that the drinking water samples present potential risk of disease epidemic particularly, the diarreaheal disease, this assumes more importants as in this particular setting where the dirinking water is not treated before consumption. Though the reservoir for EAEC is still unclear, it is generally considered to be human (79–81). The transmission of EAEC is often described as waterborne or foodborne; therefore, it is assumed to be transmitted by the fecal-oral route (82). The presence of ETEC in drinking water and environmental water has been reported previously in Bangladesh; viability after long term water incubation and capacity of biofilm formation might imply that the water is an important transmission route of ETEC (83–85). From the 29 pathogenic isolates, 11 were found to be ESBL positive and surprisingly, 10 of them were of EAEC pathotype. In recent studies of Iran and China, a high prevalence of ESBL in EAEC was also reported (86, 87). The alarming rate of ESBL-producing EAEC isolates recommends strict infection control policies to prevent additional spreading of the virulent and resistant EAEC strains. All phylogenetic groups were represented in the E. coli isolates indicating that they were not homogenous in their population structure instead they belonged to diverse phylogenetic backgrounds, mainly the phylogenetic groups that are associated with commensal (group B1) as well as pathogenic and antimicrobial resistant E. coli lineages (group D) were detected. The presence of ESBL producing E. coli among the phylogenetic group D strains represents major public health risks due to the spread of such strains via drinking water.
In this study, 64% of the isolates were observed to harbor plasmids ranging from 1 to 103 MDa and a negligible similarity of plasmids pattern among the isolates inferred their clonal diversity due to heterogenous people of diverse geographical origin. Conjugation experiments are important to understand the transfer potential of plasmids conferring extended-spectrum-β-lactamase resistance. It is reported that only plasmids above 35 MDa contain and transfer antibiotic resistance genes via conjugation (18, 88), and In line with other studies, we have also observed plasmid-mediated transfer of antibiotic resistance genes (18, 88). Conjugative plasmids, carrying cefotaxime resistance phenotype among different isolates, ranged from 42 to 103 MDa in size. These findings imply that horizontal gene transfer might worsen the existing antibiotic resistance scenario by speeding up the spread of antimicrobial resistance (AMR) within heterogeneous bacterial communities in environment (89).
Lack of proper sanitation and hygiene in a densely-populated area like Rohingya camps (26) might play a key role in the development and dissemination of AMR. Open defecation with poor personal hygiene, poor community sanitation and lack of controlled antibiotic usage has been reported to exacerbate the transmission of AMR infections (90). A previous study in four middle-income countries, Brazil, Indonesia, India, and Nigeria showed that improvement in water quality and sanitation alone could lead to reduction in antibiotic usage (90). The contamination of drinking water with ESBL-producing E. coli, as observed in this study could be due to poor sanitation and hygiene, including, open defecation, inappropriate fecal sludge management, etc. This study has shown that the environmental E. coli pose public health threat by being carriers of ESBL-genes. Moreover, these ESBL-producing E. coli were harboring virulence factors corresponding to major E. coli pathotypes. Limitations of this study include lack of genetic fingerprinting analysis of the antibiotic-resistant E. coli from drinking water, lack of exhaustive antimicrobial resistance gene, and extraintestinal pathogenic E. coli (ExPEC) virulence gene screening.
In conclusion, the findings of this study suggest that the drinking water samples analyzed herein could serve as an important source for exposure and dissemination of MDR, ESBL-producing and pathogenic E. coli variants, which also pose a health risk to the displaced Rohingya population residing in the densely populated camps in Cox's Bazar. Based on the results of this work we recommend that the policymakers should make considerable efforts in implementing strong infection control strategies by focusing on providing good quality water and ensuring water quality monitoring programs in the Rohingya camps.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Ethics Statement
The Ethical Review Committee of International Centre for Diarrhoeal Disease Research, Bangladesh has approved the study.
Author Contributions
ZM, MI, SH, MW, KI, DJ, and NA planed and organized the study. ZM, MI, SA, MK, MM, TN, SH, MW, DJ, DA, and NA were involved to implement the study. ZM, SA, MK, KI, DA, and NA carried out laboratory work. ZM, MM, MI, SH, DA, and NA were involved to interpret the data. ZM, MK, SA, MM, MI, and NA had a major contribution in writing the manuscript. All authors contributed in proofreading the manuscript.
Funding
This research study was funded by United Nations International Children's Emergency Fund (unicef), grant number BCO/PCA/2017/035-2018/001. International Centre for Diarrhoeal Disease Research, Bangladesh (icddr,b) acknowledges with gratitude the commitment of unicef to its research efforts. icddr,b is also grateful to the Government of Bangladesh, Canada, Sweden and the UK for providing core/unrestricted support. The authors are grateful to Frederic Geser of unicef Bangaldesh for his constructive input and cooperation during the study. The authors are also grateful to Department of Public Health Engineering (DPHE) Cox's Bazar and unicef Cox's Bazar, Bangladesh for their valuable guidance and cooperation during the study.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpubh.2020.00228/full#supplementary-material
References
1. Aruna K, Mobashshera T. Prevalence of extended spectrum beta-lactamase production among uropathogens in south Mumbai and its antibiogram pattern. EXCLI J. (2012) 11:363–72.
2. Hussain A, Shaik S, Ranjan A, Nandanwar N, Tiwari SK, Majid M, et al. Risk of transmission of antimicrobial resistant Escherichia coli from commercial broiler and free-range retail chicken in India. Front Microbiol. (2017) 8:2120. doi: 10.3389/fmicb.2017.02120
3. Hussain A, Shaik S, Ranjan A, Suresh A, Sarker N, Semmler T, et al. Genomic and functional characterization of poultry Escherichia coli from India revealed diverse Extended-spectrum β-lactamase-producing lineages with shared virulence profiles. Front Microbiol. (2019) 10:2766. doi: 10.3389/fmicb.2019.02766
4. Pitout JDD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J Antimicrob Chemother. (2005) 56:52–9. doi: 10.1093/jac/dki166
5. Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. (2012) 18:413–31. doi: 10.1111/j.1469-0691.2012.03821.x
6. Harris PNA, Tambyah PA, Paterson DL. β-lactam and β-lactamase inhibitor combinations in the treatment of extended-spectrum β-lactamase producing Enterobacteriaceae: time for a reappraisal in the era of few antibiotic options? Lancet Infect Dis. (2015) 15:475–85. doi: 10.1016/S1473-3099(14)70950-8
7. Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. (2005) 18:657–86. doi: 10.1128/CMR.18.4.657-686.2005
8. Muñoz-Miguel J, Roig-Sena J, Saa-Casal A, Salvador-Aguilá M, Bediaga-Collado A. Multidrug-resistant E. coli in the community: assessment and proposal of a feasible indicator. Eur J Public Health. (2018) 28:cky213.709. doi: 10.1093/eurpub/cky212.709
9. Odonkor ST, Addo KK. Prevalence of multidrug-resistant Escherichia coli isolated from drinking water sources. Int J Microbiol. (2018) 2018:7204013. doi: 10.1155/2018/7204013
10. Overdevest I, Willemsen I, Rijnsburger M, Eustace A, Xu L, Hawkey P, et al. Extended-spectrum β-lactamase genes of Escherichia coli in chicken meat and humans, The Netherlands. Emerg Infect Dis. (2011) 17:1216. doi: 10.3201/eid1707.110209
11. Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. (2011) 11:355–62. doi: 10.1016/S1473-3099(11)70059-7
12. Islam MA, Talukdar PK, Hoque A, Huq M, Nabi A, Ahmed D, et al. Emergence of multidrug-resistant NDM-1-producing Gram-negative bacteria in Bangladesh. Eur J Clin Microbiol Infect Dis. (2012) 31:2593–600. doi: 10.1007/s10096-012-1601-2
13. Rashid M, Rakib MM, Hasan B. Antimicrobial-resistant and ESBL-producing Escherichia coli in different ecological niches in Bangladesh. Infect Ecol Epidemiol. (2015) 5:26712. doi: 10.3402/iee.v5.26712
14. Hunter PR. Drinking water and diarrhoeal disease due to Escherichia coli. J Water Health. (2003) 1:65–72. doi: 10.2166/wh.2003.0008
15. Sirot D. Extended-spectrum plasmid-mediated beta-lactamases. J Antimicrob Chemother. (1995) 36:19–34. doi: 10.1093/jac/36.suppl_A.19
16. Taylor NGH, Verner-Jeffreys DW, Baker-Austin C. Aquatic systems: maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol Evol. (2011) 26:278–84. doi: 10.1016/j.tree.2011.03.004
17. Levine MM. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. (1987) 155:377–89. doi: 10.1093/infdis/155.3.377
18. Talukder KA, Islam Z, Dutta DK, Islam MA, Khajanchi BK, Azmi IJ, et al. Antibiotic resistance and genetic diversity of Shigella sonnei isolated from patients with diarrhoea between 1999 and 2003 in Bangladesh. J Med Microbiol. (2006) 55:1257–63. doi: 10.1099/jmm.0.46641-0
19. Nataro JP, Kaper JB. Diarrheagenic escherichia coli. Clin Microbiol Rev. (1998) 11:142–201. doi: 10.1128/CMR.11.1.142
20. Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. (2004) 2:123–40. doi: 10.1038/nrmicro818
21. Coleman BL, Louie M, Salvadori MI, McEwen SA, Neumann N, Sibley K, et al. Contamination of Canadian private drinking water sources with antimicrobial resistant Escherichia coli. Water Res. (2013) 47:3026–36. doi: 10.1016/j.watres.2013.03.008
22. Park J, Kim JS, Kim S, Shin E, Oh K-H, Kim Y, et al. A waterborne outbreak of multiple diarrhoeagenic Escherichia coli infections associated with drinking water at a school camp. Int J Infect Dis. (2018) 66:45–50. doi: 10.1016/j.ijid.2017.09.021
23. Swerdlow DL, Woodruff BA, Brady RC, Griffin PM, Tippen S, Donnell HDJ, et al. A waterborne outbreak in Missouri of Escherichia coli O157:H7 associated with bloody diarrhea and death. Ann Intern Med. (1992) 117:812–9. doi: 10.7326/0003-4819-117-10-812
24. Chan EYY, Chiu CP, Chan GKW. Medical and health risks associated with communicable diseases of Rohingya refugees in Bangladesh 2017. Int J Infect Dis. (2018) 68:39–43. doi: 10.1016/j.ijid.2018.01.001
25. Islam MM, Nuzhath T. Health risks of Rohingya refugee population in Bangladesh: a call for global attention. J Glob Health. (2018) 8:20309. doi: 10.7189/jogh.08.020309
26. Mahmud ZH, Islam MS, Imran KM, Ibna Hakim SA, Worth M, Ahmed A, et al. Occurrence of Escherichia coli and faecal coliforms in drinking water at source and household point-of-use in Rohingya camps, Bangladesh. Gut Pathog. (2019) 11:52. doi: 10.1186/s13099-019-0333-6
27. Rahman MR. Rohingya Crisis–Health issues. Delta Med Coll J. (2018) 6:1–3. doi: 10.3329/dmcj.v6i1.35960
28. Finley R, Glass-Kaastra SK, Hutchinson J, Patrick DM, Weiss K, Conly J. Declines in outpatient antimicrobial use in Canada (1995–2010). PLoS ONE. (2013) 8:e76398. doi: 10.1371/journal.pone.0076398
29. Levy S. The Antibiotic Paradox: How Misuse of Antibiotics Destroys Their Curative Powers (Perseus Cambridge, 2002).
31. Mohamed JA, DuPont HL, Jiang Z-D, Flores J, Carlin LG, Belkind-Gerson J, et al. A single-nucleotide polymorphism in the gene encoding osteoprotegerin, an anti-inflammatory protein produced in response to infection with diarrheagenic Escherichia coli, is associated with an increased risk of nonsecretory bacterial diarrhea in North Ame. J Infect Dis. (2009) 199:477–85. doi: 10.1086/596319
32. Nguyen TV, Le Van P, Le Huy C, Gia KN, Weintraub A. Detection and characterization of diarrheagenic Escherichia coli from young children in Hanoi, Vietnam. J Clin Microbiol. (2005) 43:755–60. doi: 10.1128/JCM.43.2.755-760.2005
33. Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun. (2000) 68:64–71. doi: 10.1128/IAI.68.1.64-71.2000
34. Talukdar PK, Rahman M, Rahman M, Nabi A, Islam Z, Hoque MM, et al. Antimicrobial resistance, virulence factors and genetic diversity of Escherichia coli isolates from household water supply in Dhaka, Bangladesh. PLoS ONE. (2013) 8:e61090. doi: 10.1371/journal.pone.0061090
35. Mahmud ZH, Shirazi FF, Hossainey MRH, Islam MI, Ahmed MA, Nafiz TN, et al. Presence of virulence factors and antibiotic resistance among Escherichia coli strains isolated from human pit sludge. J Infect Dev Ctries. (2019) 13:195–203. doi: 10.3855/jidc.10768
36. Heuvelink AE, Van de Kar N, Meis J, Monnens LAH, Melchers WJG. Characterization of verocytotoxin-producing Escherichia coli O157 isolates from patients with haemolytic uraemic syndrome in Western Europe. Epidemiol Infect. (1995) 115:1–3. doi: 10.1017/S0950268800058064
37. Islam MA, Heuvelink AE, De Boer E, Sturm PD, Beumer RR, Zwietering MH, et al. Shiga toxin-producing Escherichia coli isolated from patients with diarrhoea in Bangladesh. J Med Microbiol. (2007) 56:380–5. doi: 10.1099/jmm.0.46916-0
38. Frankel G, Giron JA, Valmassoi J, Schoolnik GK. Multi-gene amplification: simultaneous detection of three virulence genes in diarrhoeal stool. Mol Microbiol. (1989) 3:1729–34. doi: 10.1111/j.1365-2958.1989.tb00158.x
39. Lüscher D, Altwegg M. Detection of shigellae, enteroinvasive and enterotoxigenic Escherichia coli using the polymerase chain reaction (PCR) in patients returning from tropical countries. Mol Cell Probes. (1994) 8:285–90. doi: 10.1006/mcpr.1994.1040
40. Talukder KA, Islam MA, Khajanchi BK, Dutta DK, Islam Z, Safa A, et al. Temporal shifts in the dominance of serotypes of Shigella dysenteriae from 1999 to 2002 in Dhaka, Bangladesh. J Clin Microbiol. (2003) 41:5053–8. doi: 10.1128/JCM.41.11.5053-5058.2003
41. Thong KL, Hoe SLL, Puthucheary SD, Yasin RM. Detection of virulence genes in Malaysian Shigella species by multiplex PCR assay. BMC Infect Dis. (2005) 5:8. doi: 10.1186/1471-2334-5-8
42. Johnson JR, Kuskowski MA, Smith K, O'Bryan TT, Tatini S. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis. (2005) 191:1040–9. doi: 10.1086/428451
43. Ahmed AM, Motoi Y, Sato M, Maruyama A, Watanabe H, Fukumoto Y, et al. Zoo animals as reservoirs of gram-negative bacteria harboring integrons and antimicrobial resistance genes. Appl Environ Microbiol. (2007) 73:6686–90. doi: 10.1128/AEM.01054-07
44. Guessennd N, Bremont S, Gbonon V, Kacou-Ndouba A, Ekaza E, Lambert T, et al. Qnr-type quinolone resistance in extended-spectrum beta-lactamase producing enterobacteria in Abidjan, Ivory Coast. Pathol Biol. (2008) 56:439–46. doi: 10.1016/j.patbio.2008.07.025
45. Jouini A, Vinué L, Slama K, Ben SY, Klibi N, Hammami S, et al. Characterization of CTX-M and SHV extended-spectrum β-lactamases and associated resistance genes in Escherichia coli strains of food samples in Tunisia. J Antimicrob Chemother. (2007) 60:1137–41. doi: 10.1093/jac/dkm316
46. Leflon-Guibout V, Jurand C, Bonacorsi S, Espinasse F, Guelfi MC, Duportail F, et al. Emergence and spread of three clonally related virulent isolates of CTX-M-15-producing Escherichia coli with variable resistance to aminoglycosides and tetracycline in a French geriatric hospital. Antimicrob Agents Chemother. (2004) 48:3736–42. doi: 10.1128/AAC.48.10.3736-3742.2004
47. Muzaheed YD, Adams-Haduch JM, Endimiani A, Sidjabat HE, Gaddad SM, Paterson DL. High prevalence of CTX-M-15-producing Klebsiella pneumoniae among inpatients and outpatients with urinary tract infection in Southern India. J Antimicrob Chemother. (2008) 61:1393. doi: 10.1093/jac/dkn109
48. Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. (2011) 49:718–21. doi: 10.1128/JCM.01773-10
49. Zhao S, White DG, McDermott PF, Friedman S, English L, Ayers S, et al. Identification and expression of cephamycinasebla CMY Genes in Escherichia coli and Salmonella Isolates from food animals and ground meat. Antimicrob Agents Chemother. (2001) 45:3647–50. doi: 10.1128/AAC.45.12.3647-3650.2001
50. Jacoby GA, Chow N, Waites KB. Prevalence of plasmid-mediated quinolone resistance. Antimicrob Agents Chemother. (2003) 47:559–62. doi: 10.1128/AAC.47.2.559-562.2003
51. Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, et al. qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother. (2006) 50:1178–82. doi: 10.1128/AAC.50.4.1178-1182.2006
52. Pu X-Y, Pan J-C, Wang H-Q, Zhang W, Huang Z-C, Gu Y-M. Characterization of fluoroquinolone-resistant Shigella flexneri in Hangzhou area of China. J Antimicrob Chemother. (2009) 63:917–20. doi: 10.1093/jac/dkp087
53. Kado C, Liu S. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. (1981) 145:1365–73. doi: 10.1128/JB.145.3.1365-1373.1981
54. Macrina FL, Kopecko DJ, Jones KR, Ayers DJ, McCowen SM. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. (1978) 1:417–20. doi: 10.1016/0147-619X(78)90056-2
55. Clermont O, Christenson JK, Denamur E, Gordon DM. The C lermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. (2013) 5:58–65. doi: 10.1111/1758-2229.12019
56. Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. (2001) 14:933–51. doi: 10.1128/CMR.14.4.933-951.2001
57. Ensor VM, Shahid M, Evans JT, Hawkey PM. Occurrence, prevalence and genetic environment of CTX-M β-lactamases in Enterobacteriaceae from Indian hospitals. J Antimicrob Chemother. (2006) 58:1260–3. doi: 10.1093/jac/dkl422
58. Kamruzzaman M, Shoma S, Bari SMN, Ginn AN, Wiklendt AM, Partridge SR, et al. Genetic diversity and antibiotic resistance in Escherichia coli from environmental surface water in Dhaka City, Bangladesh. Diagn Microbiol Infect Dis. (2013) 76:222–6. doi: 10.1016/j.diagmicrobio.2013.02.016
59. Hasan B, Drobni P, Drobni M, Alam M, Olsen B. Dissemination of NDM-1. Lancet Infect Dis. (2012) 12:99–100. doi: 10.1016/S1473-3099(11)70333-4
60. Hasan B, Melhus Å, Sandegren L, Alam M, Olsen B. The gull (Chroicocephalus brunnicephalus) as an environmental bioindicator and reservoir for antibiotic resistance on the coastlines of the Bay of Bengal. Microb Drug Resist. (2014) 20:466–71. doi: 10.1089/mdr.2013.0233
61. Rahman MM, Haq JA, Hossain MA, Sultana R, Islam F, Islam AHMS. Prevalence of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in an urban hospital in Dhaka, Bangladesh. Int J Antimicrob Agents. (2004) 24:508–10. doi: 10.1016/j.ijantimicag.2004.05.007
62. Chouduri AU, Biswas M, Haque MU, Arman MSI, Uddin N, Kona N, et al. Cephalosporin-3G, highly prescribed antibiotic to outpatients in Rajshahi, Bangladesh: prescription errors, carelessness, irrational uses are the triggering causes of antibiotic resistance. J Appl Pharm Sci. (2018) 8:105–12. doi: 10.7324/JAPS.2018.8614
63. Ahmed I, Rabbi MB, Sultana S. Antibiotic resistance in Bangladesh: a systematic review. Int J Infect Dis. (2019) 80:54–61. doi: 10.1016/j.ijid.2018.12.017
64. Biswas M, Roy DN, Tajmim A, Rajib SS, Hossain M, Farzana F, et al. Prescription antibiotics for outpatients in Bangladesh: a cross-sectional health survey conducted in three cities. Ann Clin Microbiol Antimicrob. (2014) 13:15. doi: 10.1186/1476-0711-13-15
65. Swedan S, Abu Alrub H. Antimicrobial resistance, virulence factors, and pathotypes of escherichia coli isolated from drinking water sources in Jordan. Pathogens. (2019) 8:86. doi: 10.3390/pathogens8020086
66. Hasan B, Islam K, Ahsan M, Hossain Z, Rashid M, Talukder B, et al. Fecal carriage of multi-drug resistant and extended spectrum beta-lactamases producing E. coli in household pigeons, Bangladesh. Vet Microbiol. (2014) 168:221–4. doi: 10.1016/j.vetmic.2013.09.033
67. Bonnet R. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. (2004) 48:1–14. doi: 10.1128/AAC.48.1.1-14.2004
68. Doublet B, Granier SA, Robin F, Bonnet R, Fabre L, Brisabois A, et al. Novel plasmid-encoded ceftazidime-hydrolyzing CTX-M-53 extended-spectrum β-lactamase from Salmonella enterica serotypes Westhampton and Senftenberg. Antimicrob Agents Chemother. (2009) 53:1944–51. doi: 10.1128/AAC.01581-08
69. Liu W, Chen L, Li H, Duan H, Zhang Y, Liang X, et al. Novel CTX-M β-lactamase genotype distribution and spread into multiple species of Enterobacteriaceae in Changsha, Southern China. J Antimicrob Chemother. (2009) 63:895–900. doi: 10.1093/jac/dkp068
70. Radice M, Power P, Di Conza J, Gutkind G. Early dissemination of CTX-M-derived enzymes in South America. Antimicrob Agents Chemother. (2002) 46:602–4. doi: 10.1128/AAC.46.2.602-604.2002
71. Islam MA, Huq M, Nabi A, Talukdar PK, Ahmed D, Talukder KA, et al. Occurrence and characterization of multidrug-resistant New Delhi metallo-β-lactamase-1-producing bacteria isolated between 2003 and 2010 in Bangladesh. J Med Microbiol. (2013) 62:62–8. doi: 10.1099/jmm.0.048066-0
72. Kao C-Y, Wu H-M, Lin W-H, Tseng C-C, Yan J-J, Wang M-C, et al. Plasmid-mediated quinolone resistance determinants in quinolone-resistant Escherichia coli isolated from patients with bacteremia in a university hospital in Taiwan, 2001-2015. Sci Rep. (2016) 6:32281. doi: 10.1038/srep32281
73. Poirel L, Cattoir V, Nordmann P. Is plasmid-mediated quinolone resistance a clinically significant problem? Clin Microbiol Infect. (2008) 14:295–7. doi: 10.1111/j.1469-0691.2007.01930.x
74. Takasu H, Suzuki S, Reungsang A, Viet PH. Fluoroquinolone (FQ) contamination does not correlate with occurrence of FQ-resistant bacteria in aquatic environments of Vietnam and Thailand. Microbes Environ. (2011) 26:135–43. doi: 10.1264/jsme2.me10204
75. Strahilevitz J, Jacoby GA, Hooper DC, Robicsek A. Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev. (2009) 22:664–89. doi: 10.1128/CMR.00016-09
76. Ma J, Zeng Z, Chen Z, Xu X, Wang X, Deng Y, et al. High prevalence of plasmid-mediated quinolone resistance determinants qnr, aac (6′)-Ib-cr, and qepA among ceftiofur-resistant Enterobacteriaceae isolates from companion and food-producing animals. Antimicrob Agents Chemother. (2009) 53:519–24. doi: 10.1128/AAC.00886-08
77. Yue L, Jiang H-X, Liao X-P, Liu J-H, Li S-J, Chen X-Y, et al. Prevalence of plasmid-mediated quinolone resistance qnr genes in poultry and swine clinical isolates of Escherichia coli. Vet Microbiol. (2008) 132:414–20. doi: 10.1016/j.vetmic.2008.05.009
78. Cattoir V, Poirel L, Aubert C, Soussy C-J, Nordmann P. Unexpected occurrence of plasmid-mediated quinolone resistance determinants in environmental Aeromonas spp. Emerg Infect Dis. (2008) 14:231. doi: 10.3201/eid1402.070677
79. Beutin L, Martin A. Outbreak of Shiga toxin–producing Escherichia coli (STEC) O104: H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J Food Prot. (2012) 75:408–18. doi: 10.4315/0362-028X.JFP-11-452
80. Huppertz HI, Rutkowski S, Aleksic S, Karch H. Acute and chronic diarrhoea and abdominal colic associated with enteroaggregative Escherichia coli in young children living in western Europe. Lancet. (1997) 349:1660–2. doi: 10.1016/S0140-6736(96)12485-5
81. Oundo JO, Kariuki SM, Boga HI, Muli FW, Iijima Y. High incidence of enteroaggregative Escherichia coli among food handlers in three areas of Kenya: a possible transmission route of travelers' diarrhea. J Travel Med. (2008) 15:31–8. doi: 10.1111/j.1708-8305.2007.00174.x
82. Jiang Z-D, Greenberg D, Nataro JP, Steffen R, DuPont HL. Rate of occurrence and pathogenic effect of enteroaggregative Escherichia coli virulence factors in international travelers. J Clin Microbiol. (2002) 40:4185–90. doi: 10.1128/JCM.40.11.4185-4190.2002
83. Ahmed D, Islam MS, Begum YA, Janzon A, Qadri F, Sjöling Å. Presence of enterotoxigenic Escherichia coli in biofilms formed in water containers in poor households coincides with epidemic seasons in D haka. J Appl Microbiol. (2013) 114:1223–9. doi: 10.1111/jam.12109
84. Begum YA, Talukder KA, Nair GB, Qadri F, Sack RB, Svennerholm A-M. Enterotoxigenic Escherichia coli isolated from surface water in urban and rural areas of Bangladesh. J Clin Microbiol. (2005) 43:3582–3. doi: 10.1128/JCM.43.7.3582-3583.2005
85. Lothigius Å, Janzon A, Begum Y, Sjöling Å, Qadri F, Svennerholm A, et al. Enterotoxigenic Escherichia coli is detectable in water samples from an endemic area by real-time PCR. J Appl Microbiol. (2008) 104:1128–36. doi: 10.1111/j.1365-2672.2007.03628.x
86. Mansour Amin MS, Javaherizadeh H, Motamedifar M, Saki M, Veisi H, Ebrahimi S, et al. Antibiotic resistance pattern and molecular characterization of extended-spectrum β-lactamase producing enteroaggregative Escherichia coli isolates in children from southwest Iran. Infect Drug Resist. (2018) 11:1097. doi: 10.2147/IDR.S167271
87. Zhang R, Gu D, Huang Y, Chan EW-C, Chen G-X, Chen S. Comparative genetic characterization of Enteroaggregative Escherichia coli strains recovered from clinical and non-clinical settings. Sci Rep. (2016) 6:24321. doi: 10.1038/srep24321
88. Talukder KA, Islam MA, Dutta DK, Hassan F, Safa A, Nair GB, et al. Phenotypic and genotypic characterization of serologically atypical strains of Shigella flexneri type 4 isolated in Dhaka, Bangladesh. J Clin Microbiol. (2002) 40:2490–7. doi: 10.1128/JCM.40.7.2490-2497.2002
89. Dionisio F, Matic I, Radman M, Rodrigues OR, Taddei F. Plasmids spread very fast in heterogeneous bacterial communities. Genetics. (2002) 162:1525–32.
Keywords: ESBL-producing E. coli, multidrug-resistant, drinking water, Rohingya camps, Bangladesh
Citation: Mahmud ZH, Kabir MH, Ali S, Moniruzzaman M, Imran KM, Nafiz TN, Islam MS, Hussain A, Hakim SAI, Worth M, Ahmed D, Johnston D and Ahmed N (2020) Extended-Spectrum Beta-Lactamase-Producing Escherichia coli in Drinking Water Samples From a Forcibly Displaced, Densely Populated Community Setting in Bangladesh. Front. Public Health 8:228. doi: 10.3389/fpubh.2020.00228
Received: 31 October 2019; Accepted: 14 May 2020;
Published: 18 June 2020.
Edited by:
Ilana L. B. C. Camargo, University of São Pãulo, BrazilReviewed by:
Rebecca Ashley Gladstone, University of Oslo, NorwaySebastian Guenther, University of Greifswald, Germany
Copyright © 2020 Mahmud, Kabir, Ali, Moniruzzaman, Imran, Nafiz, Islam, Hussain, Hakim, Worth, Ahmed, Johnston and Ahmed. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zahid Hayat Mahmud, emhtYWhtdWRAaWNkZHJiLm9yZw==
 Zahid Hayat Mahmud
Zahid Hayat Mahmud Mir Himayet Kabir
Mir Himayet Kabir Sobur Ali1
Sobur Ali1 M. Moniruzzaman
M. Moniruzzaman Khan Mohammad Imran
Khan Mohammad Imran Arif Hussain
Arif Hussain Dara Johnston
Dara Johnston Niyaz Ahmed
Niyaz Ahmed