- 1Office of Population Health Genomics, Public and Aboriginal Health Division, Western Australian Department of Health, East Perth, WA, Australia
- 2Office of the Chief Health Officer, Public and Aboriginal Health Division, Western Australian Department of Health, East Perth, WA, Australia
- 3School of Biomedical Sciences, Faculty of Health and Medical Sciences, University of Western Australia, Crawley, WA, Australia
- 4Harry Perkins Institute of Medical Research, QEII Medical Centre, Nedlands, WA, Australia
- 5Sir Walter Murdoch School of Policy and International Affairs, Murdoch University, Murdoch, WA, Australia
- 6School of Public Health, Curtin University of Technology, Bentley, WA, Australia
Public health genomics has evolved to responsibly integrate advancements in genomics into the fields of personalized medicine and public health. Appropriate, effective and sustainable integration of genomics into healthcare requires an organized approach. This paper outlines the history that led to the emergence of public health genomics as a distinguishable field. In addition, a range of activities are described that illustrate how genomics can be incorporated into public health practice. Finally, it presents the evolution of public health genomics into the new era of “precision public health.”
Public Health: The Past
The field of public health emerged as a means to “protect” the health of the individual and the community, and thereby minimize morbidity and mortality associated with disease (1, 2). Differentiating itself from the medical field, public health places an emphasis on improving the health of society as a whole through the use of organized, population-wide approaches. Instances of public health efforts have been documented throughout history. For example, by the eighteenth century, it was common practice to isolate and quarantine sick individuals, in order to contain the spread of contagious diseases such as leprosy and the plague (1, 3). Developing and delivering appropriate public health services requires an understanding about health and wellbeing, the presence and absence of disease, how health outcomes are distributed within populations and the factors that determine these outcomes.
Over the last two centuries, the essential activities of public health have evolved significantly. In the nineteenth century, the primary focus of public health was on managing the physical environment, such as the provision of safe drinking water and the development of effective sewerage systems. In the twentieth century, the scope of public health was increased to include social factors (such as housing, employment, income, educational level, and access to transportation and health care) and lifestyle behaviors (such as physical activity, diet, smoking, and alcohol consumption) that are now known to influence health outcomes. This led to the emergence of the “health promotion” era of public health, which stemmed from a movement aimed at providing evidence-based education that would enable people to increase control over and improve their health (4). The health promotion movement drove action in a range of areas of public health including: developing public policy, creating environments that support healthy behaviors, and empowering people to develop personal skills to make choices that lead to healthier lives.
At the heart of public health today is the recognition that health outcomes are influenced by a range of social, cultural, political, economic, environmental, behavioral, and biological (e.g., genetic) factors (5). Otherwise known as the “determinants of health,” these factors may favor health or be harmful to it. Further, while some factors cannot be changed (such as age or ethnic background), others may be modifiable (for example, weight or smoking status). Understanding how these factors influence health outcomes is key to informing public health approaches for promoting health and wellbeing, through the implementation of practices that aim to “prevent” poor health. These prevention strategies can be categorized into three levels, being:
• Primary: where the aim is to prevent disease and injury from occurring, which will reduce their incidence in the population. This is done largely through interventions to eliminate risk factors. For example, seatbelts, sunscreen, tobacco-use cessation.
• Secondary: where the aim is to reduce the more immediate impact of disease and injury if it does occur. The focus of interventions is on early detection and treatment to alter or slow progress of the disease or injury, and thereby prevent the onset of long-term or permanent adverse consequences such as complications and disabilities. For example, population screening programs.
• Tertiary: where the aim is to help people manage the longer term impact of ongoing disease or injury. The focus of interventions is to improve, as much as possible, factors such as physical and mental functioning, quality of life, and life expectancy. For example, chemotherapy, rehabilitation.
A fourth category of prevention, known as quaternary prevention, has also been proposed (6, 7). This is defined as: “action taken to identify a patient or a population at risk of over-medicalization, to protect them from invasive medical interventions and provide for them care procedures which are ethically acceptable” [(6), p. 3]. In other words, the aim of quaternary prevention is to identify and protect those individuals for whom medical interventions are likely to cause more harm than good (7, 8).
Core Functions of Public Health
Following from the health promotion movement was a growing recognition that public health had changed significantly over the years, and that the governmental role in providing public health services needed to be clearly defined, adequately supported, and fully understood. This led the Institute of Medicine (IOM; now known as the Health and Medicine Division) of the United States of America's National Academies of Sciences, Engineering and Medicine in 1988 to identify and define the three core functions to be provided by all public health agencies (3), being:
• Assessment: to assess and monitor the health of communities and populations at risk, and to identify health problems and priorities. This requires the regular and systematic collection, assembly, analysis, and dissemination of information on the health of populations.
• Policy development: to formulate public policies, plans, standards, guidelines, and resources in collaboration and partnership with stakeholders, and to solve identified local and national health problems and priorities.
• Assurance: to assure that all populations have access to appropriate and cost effective care (including health promotion and disease prevention services), and to evaluate the effectiveness of healthcare and public health interventions.
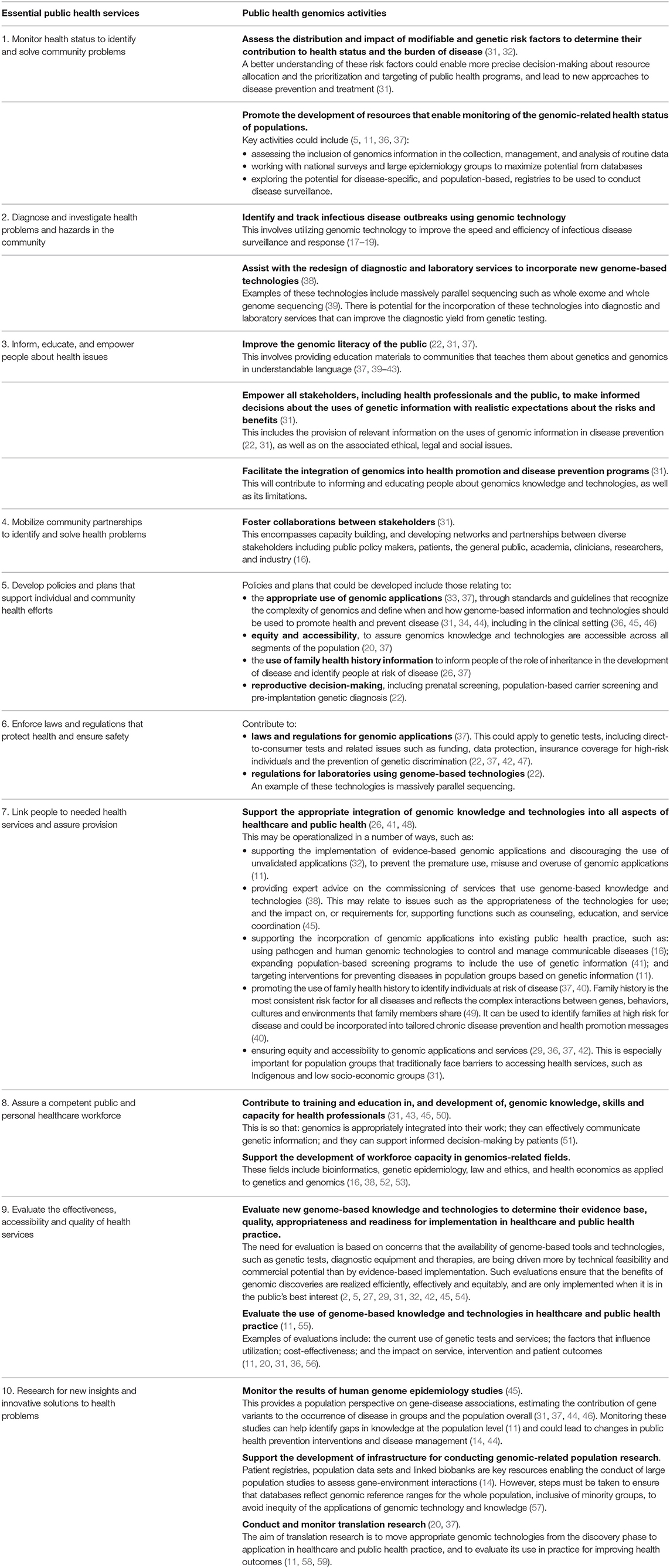
Building on this work, in 1994 the three core functions of public health were further elaborated into 10 essential public health services, to support the application of the core functions in practice (9, 10). These 10 essential public health services are presented in Box 1.
Box 1. The 10 essential public health services (9).
1. Monitor and evaluate health status to identify community health problems.
2. Diagnose and investigate health problems and health hazards in the community.
3. Inform, educate, and empower people about health issues.
4. Mobilize community partnerships to identify and solve health problems.
5. Develop policies and plans that support individual and community health efforts.
6. Enforce laws and regulations that protect health and ensure safety.
7. Link people to needed personal health services and assure the provision of health care when otherwise unavailable.
8. Assure a competent public and personal health care workforce.
9. Evaluate effectiveness, accessibility, and quality of personal and population-based health services.
10. Research for new insights and innovative solutions to health problems.
Public Health Genomics: The Present
In the two decades since the core functions and essential services of public health were defined, rapid developments have occurred in the field of genomics (see Box 2 to understand the distinction between the terms “genetics” and “genomics”). These developments have enhanced our knowledge of how human genes interact with each other and the environment to influence health. A notable example is the completion of the Human Genome Project in 2003, which led to the proliferation, in volume and complexity, of knowledge about the human genome. It also resulted in a significant reduction in the estimated number of genes expected to be found in the human genome, down from previous estimates of as many as 140,000 genes to a probable 20,500 (13). The impact that advancements in genomics have had on our understanding of disease is discussed in Box 3.
Box 2. The difference between genetics and genomics.
Genetics is the science of inheritance and tends to look at the functioning and composition of a single gene at a time. Thus genetic studies into diseases tend to focus on those that are associated with variants in one gene only (11). These “single gene disorders” (or Mendelian disorders) are relatively rare in the population and examples include Fragile X syndrome, cystic fibrosis, muscular dystrophy, and Huntington's disease.
Most common diseases are multi-factorial, caused by variants in numerous genes that interact with each other and with a range of environmental factors (12). To gain more knowledge about these diseases, researchers work in the field of genomics. This field involves the study of the genome, that is, all the genes in the cells of an organism and how these genes interact with each other and with environmental factors to affect an organism's growth and development (11). Hence genomics researchers are able to explore the causes of diseases such as cancer, diabetes, and heart disease, which have multi-factorial determinants including genes, lifestyle behaviors, and other environmental influences.
Box 3. How genomics improved the understanding of diseases.
“Genomic knowledge” refers to the information that is obtained from studying the complete genetic makeup of a cell or organism. In recent years, scientific research in this area has contributed significantly to our knowledge about the human genome, improving our ability to understand disease etiology, risk, prevention, diagnosis, and treatment. The ways in which these areas can be enhanced by genomic knowledge are outlined below. Based on these improved understandings, genomic tools, and technologies are being developed to enable better health not just for the individual, but for populations as well.
Etiology
Increased genomic knowledge about a disease can provide insights into how the disease may develop. This can occur through a better understanding of the function of genes that make up the genome, how different genetic variants contribute to the phenotype of diseases, the role of gene expression, and the role of the interaction between genes (10, 14).
Risk
Genomic knowledge is expected to improve our understanding of why some individuals remain healthy while others are more susceptible to disease. For example, information on the genetic variants associated with an increased risk of common diseases, such as cardiovascular disease and diabetes, might at some point be used to make predictions about the likelihood a person will get these diseases (15). This knowledge could then be applied to develop new tools for risk prediction or predictive testing (5) in relation to the onset or recurrence of disease (16).
Prevention
Understanding how the genome influences the etiology and risk of diseases may lead to improved understanding of how diseases, or the symptoms of disease, can be prevented (5, 16). Genomic tools and technologies can also identify infectious diseases with greater speed and precision to enable rapid responses to disease outbreaks and more efficient surveillance (17–19).
Diagnosis
Historically, clinicians generally used a set of observable or measurable characteristics as the basis for diagnosing disease. Genomic knowledge takes this one step further, by enabling clinicians to look at a person's genes to provide a molecular diagnosis. In line with this, diagnostic technologies have been developed that include a plethora of clinical genetic tests (5, 14, 16).
Treatment
To date, genomic knowledge has mostly been used to inform disease treatments. Pharmacogenetics and pharmacogenomics are two fields where new and improved therapies and treatments have been developed, including hundreds of new drugs which are advancing disease management (16). The expectation is that genomic knowledge will further improve the ability to assess treatment responses, such as how different people metabolize drugs and which people are more likely to experience adverse drug reactions (11, 16, 20). Based on genetic profiles, tailored therapies may be developed for an individual and across individuals within specific patient populations to deliver the right drug in the right dose at the right time (12, 16, 21, 22).
From these advancements comes the increasing recognition of the potential applications of genomic knowledge and related technologies to improve population health. For example, genomic knowledge can offer new ways of differentiating individuals and sub-groups within populations, taking public health beyond the traditional correlates of disease risk such as gender, age, and socio-economic status (20). Specifically, it can enable the stratification and subsequent screening of individuals and sub-groups of populations based on their level of genetic risk for developing a disease. This can then lead to the development of more targeted prevention approaches to reduce the burden of disease (11).
It should be noted that advances in genomics have been dependent on, and facilitated by, progress in related fields such as informatics, and the development of novel technologies capable of evolving to meet the increasing demands of genomic medicine. Specifically, the huge volume of data generated by next generation sequencing has created significant challenges relating to data storage and analysis. These challenges are explored further in Box 4.
Box 4. The data informatics puzzle.
The concept of Moore's Law is useful to consider, when exploring the limitations of current computation and storage in genomics medicine. Moore's Law was proposed in 1965 to describe the long term trend whereby for every doubling of time of ~18 months, there is an exponential increase in the capacity for disk storage and computation (23). Historically, this growth meant that data storage and computation were able to stay ahead of the demands of genomics. However, the advent of next generation sequencing in 2005 and the rapid decline in associated costs have resulted in demands on the informatics capacity outpacing developments in the informatics ecosystem (24). The 100,000 Genomes Project in the United Kingdom (UK) has highlighted the limitations of digital infrastructure in the progression of genomic medicine, and the UK government has committed to fund sufficient digital infrastructure in order achieve successful rollout of their Genomic Medicine Service (25). Future integration of genomics in population medicine must emphasize the development of sustainable computational analytics and storage infrastructure.
Many tools and technologies based on emerging genomic knowledge have been developed. However, for a range of complex reasons, only a small proportion of these tools and technologies have so far been fully translated into healthcare and public health practice from the discovery research phase, beyond the introduction of newborn screening for genetic conditions (20, 26). The literature refers to two key reasons why this may be so. Firstly, in genomic studies, most genetic variants that have been identified as contributing to common diseases are only associated with small increases in relative risk and explain only a little about the relationship between disease and genetic inheritance (10, 21, 27, 28). This is because most common diseases are the result of complex interactions between multiple genes and environmental factors. Furthermore, the genetic variants that contribute to a given disease, and how they are expressed, may vary among different people and sub-populations, as might the relative significance of genetic and non-genetic factors (12). Consequently, attention has shifted toward rare and monogenic diseases where the gene and phenotype(s) may result in more clear causal pathologies. Nevertheless, obtaining a definitive association between a single-gene variant and a distinct disease phenotype remains a complex process.
Secondly, while tools and techniques based on genomic knowledge have been developed, there has often been limited evidence regarding their validity and utility (26). In part, this is due to a lack of investment in the infrastructure required to collect and evaluate tools and technologies in a systematic manner (10, 29) and also to the complexity of conducting evaluations (27). The recognition of this lack of evidence gave rise to the discipline of “public health genomics,” defined in 2005 as “the responsible and effective translation of genome-based knowledge and technologies for the benefit of the population” (30).
While there are expectations that genomic knowledge, tools, and technologies benefit population health, it is essential that they are applied only when the benefits outweigh the potential harms. New tools and technologies that are prematurely introduced without the evidence demonstrating that they are valid and useful run the risk of posing harm to individuals, families, and the broader health system. Such issues might include the potential for over-, under-, or mis-diagnoses, or psychosocial harms. It is also critical to consider the ethical, legal, and social issues inherent in the field of genomics. These issues are particularly relevant in the context of genomic information relative to other medical information due to the fact that variants in genes, by nature, are shared within families. Uncovering genomic determinants of health therefore has implications not only for the individual but for genetic relatives as well. Moreover, genomic information can be obtained in the absence of clinical symptoms and therefore in isolation it may have a weaker predictive association with health outcomes compared to most other health information. In addition, determining who, what, and when to test is fraught with ramifications for service capacity and financial responsibility, and can also have implications for patient autonomy and privacy as evident in the case study presented in Box 5.
Box 5. case study—ethical, legal, and social implications to consider for applications of public health genomics.
Consider the hypothetical scenario in which a newborn screening program performs whole genome sequencing on every newborn within a population. The ethical, legal, and social implications to consider include:
• Which variants should be reported? Should they be limited to known pathogenic variants, or further limited to only those that have an available treatment? Should variants of unknown significance be reported?
• Who decides what information, such as variants of unknown significance or secondary findings, should be reported? Should this be a decision for parents, or for an independent governance body?
• Which conditions should be screened for? Would parents want to know their baby's risk of developing a late-onset disorder such as dementia, or an untreatable condition? Would a child want to know of their risk?
• How should genetic counseling be offered to all parents of newborns such that they can give informed consent for the tests?
• Does the population have sufficient genetic literacy to be able to fully understand the consent process, and implications of the results, for benign, pathogenic, and uncertain variants?
• Should the genomic data be kept, and if so, for how long?
• Should the data be re-interrogated, particularly with inevitable advances in technology? If so, at what time intervals?
• Should these genomic data be available to all healthcare providers?
• What are the implications on health and life insurance if disease risk can be stratified at birth?
• If a baby is shown to be stratified at higher risk for certain lifestyle diseases, what is their responsibility for mitigating that risk? What is the government's responsibility for mitigating that risk?
It is therefore essential to consider existing and emerging knowledge, tools, and technologies in order to determine which are actually beneficial to population health and how they could be appropriately implemented. This requires an objective evaluation of the potential benefits against the potential harms, and the resources required for implementing them (12). Public health genomics bridges this gap between new scientific discoveries and technologies, and the application of genomic knowledge to benefit population health (31, 32).
With genomics being increasingly integrated into population-level health initiatives, it has been internationally recognized that maintaining efficiency, effectiveness, ethics, and equity into the future requires a strategic approach. In line with this, there has been a call for the cooperative development and harmonization of policy on genomics in healthcare between 28 of the European Union member states and Norway (33). Of these nations, Italy has been a leader in the development of public health genomics policy, developing a National Plan for Public Health Genomics that includes consideration of translation of genomics into public health practice (34). An international working group on “Beyond Health Genomics” also recommended the improved facilitation of translation research through greater engagement between public health professionals, geneticists, and scientists (20).
Similarly, the Australian Government's Department of Health has released the National Health Genomics Policy Framework 2018–2021 to harness the health benefits of genomic knowledge and technology into the Australian health system. This framework provides a shared direction and commitment between all governments in Australia to consistently and strategically integrate genomics into the Australian health system through five strategic priority areas: person-centered approach, workforce, financing, services, and data (35). The cohesive strategy is expected to ensure that the integration of genomics in healthcare is not only appropriate for the health of populations, but is also sustainable for the health system.
Public Health Genomics in Practice
The ways in which genomics can contribute in public health practice are clear. However, capitalizing on genomic advances requires a coordinated approach in order to integrate the benefits of associated knowledge and technologies into each aspect of public health service delivery. Beskow et al. (31) were the first to link the 10 essential public health services—provided in Box 1—with genomics, in 2001. Integral to each essential service is the role of “system management” in ensuring the responsible, equitable, and sustainable integration of genomics into healthcare and public health practice.
Almost 20 years after the link between public health and genomics was established, Table 1 furthers Beskow et al.'s. (31) work to provide examples from the literature of how genomics can be incorporated into public health practice. These examples reflect the rapid developments made in genomics and the significant impact the field has had to improve population health.
It should be noted that the ability of individuals to directly access health-related genetic and genomic tests, otherwise referred to as direct-to-consumer (DTC) or “personal genomic” tests, is one particular issue for public health that requires consideration. DTC tests may detect individuals at increased risk of certain diseases. However, the clinical utility and validity of DTC tests is largely uncertain (60–63). Furthermore, there are a range of ethical, legal, and social issues associated with such tests, such as challenges relating to the provision of information about the test and associated results, and obtaining informed consent (61, 64). Given that consumers are able to access some tests without clinical oversight, appropriate regulatory mechanisms need to be implemented to ensure public access to such tests is appropriate and that where possible, results are interpreted and communicated with caution (65). For those individuals with results of clinical significance, quaternary prevention principles should be applied to avoid their over-medicalization, particularly where results are uncertain or not based on evidence (63, 66, 67). Also for consideration is the possibility of under-medicalization of individuals if their genomic results are inappropriately interpreted or actioned.
Precision Public Health: The Future
The integration of genomic knowledge and technologies into healthcare is revolutionizing the way we approach clinical and public health practice. In clinical practice, advances in genomics are allowing information about an individual's genetic and biochemical composition, as determined by the interactions between their genes, environment, and lifestyle, to be used in the delivery of targeted interventions; a field known as “precision medicine” (68). This then enables clinicians to tailor medical treatments better suited to the genetic composition of their patient.
An example of a current initiative that is anticipated to have significant implications for advancing precision medicine is the 100,000 Genomes Project in the United Kingdom, which is briefly discussed in Box 4. This project is sequencing genomes from people with a rare disease and their families, and patients with cancer, in order to improve diagnosis, treatment and care (69). Additionally, in the United States of America, the National Institutes of Health's “All of Us” research program aims to sequence at least 1 million Americans and analyse their health data (70). Launched as part of the US Precision Medicine Initiative, the program will gather environmental and biological information from participants to facilitate and advance research, technology, policies, and individualized medical care (71). The program presents a number of ethical, legal, and social challenges (72) and will serve as a guide for future precision medicine initiatives.
Parallel to the developments in precision medicine has been the advancement of technologies that enable the production, aggregation, analysis, and dissemination of extremely large volumes of individual- and population-level data on genes, environment, behavior, and other social and economic determinants of health. These data have proven useful in finding new correlations, patterns and trends, particularly those involving complex interactions, in relation to diseases, pathogens, exposures, behaviors, susceptibility (risk), and health outcomes in populations (73–75). These technologies and data, such as massively parallel sequencing and genomic reference databases, are now being further utilized to complement and extend the vision of precision medicine, to consider how they can be used to improve health outcomes at the population level (74, 76). This emerging field, of utilizing big data to guide the right intervention to the right people at the right time, has been termed “precision public health” (77). Another way precision public health has been defined is as “the application and combination of new and existing technologies, which more precisely describe and analyse individuals and their environment over the life course, in order to tailor preventive interventions for at-risk groups and improve the overall health of a population” (78).
Building upon the work of public health genomics from the last 20 years, precision public health enables the integration of genomics into public health strategies within the wider context of other determinants of health, such as socioeconomic, behavioral, and environmental factors. This can then lead to more precise individual and population-based interventions (74, 77, 79), and ultimately, improve population health outcomes (78).
The ways in which public health interventions and activities may become more “precise” as a result of technological innovations and the data they produce are evident in a number of areas including: epidemiology; knowledge of the determinants of health; targeting of healthcare disparities; screening and prevention; diagnosis; surveillance; and response to communicable diseases (10, 29, 73, 74, 76, 79–81). For example, genomic technologies could be applied using a precision public health approach to identify the impact of genomic variants in different population subgroups. Each subgroup could then be targeted with tailored interventions that are more relevant to their level of risk, resulting in more efficient and effective disease prevention, screening, and surveillance strategies. Such work is critical given current recognition that a lack of appropriate reference data for ancestral population subgroups could be contributing to disparities in access to effective health interventions (82). This is more likely to occur in minority or disadvantaged populations because they are commonly underrepresented in genomic research (82, 83). Consideration of genetic diversity helps to prevent the misclassification of benign genetic variants as pathogenic for these subgroups, and vice versa, which may otherwise lead to inappropriate care and management (84).
Conclusion
Public health genomics has been successfully integrated into existing paradigms for the provision of traditional public health services. The continued alignment of genomics with public health promises to deliver more precise, personalized health care to benefit the population. Governments and policy makers in this arena have a unique role to play in guiding this activity in such a way that ensures effective and equitable implementation of genomic knowledge and technologies into health systems. A national, coordinated approach to provide centralized governance of decision-making is required to ensure responsible delivery, universality, and equity of access. In addition, investment in important enabling infrastructure such as data informatics and a genomics-literate workforce will be critical to the sustainability of public health genomics and will prepare health systems to reap the valuable benefits of precision public health.
Author Contributions
All authors contributed substantially to the conception of the work and have given final approval for the manuscript to be published. CM and FB drafted the manuscript. GB, AC, BB, KN, and HD revised the manuscript and provided critical feedback.
Funding
In-kind support was provided through the Western Australian Department of Health.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Emily Coles for providing valuable feedback and constructive criticism in the development of this paper.
References
1. Awofeso N. What's new about the “new public health?” Am J Public Health (2004) 94:705–9. doi: 10.2105/AJPH.94.5.705
2. Burton H, Jackson C, Abubakar I. The impact of genomics on public health practice. Br Med Bull. (2014) 112:37–46. doi: 10.1093/bmb/ldu032
3. Institute of Medicine. The Future of Public Health. Washington, DC: The National Academies Press (1988).
4. World Health Organization. Milestones in Health Promotion: Statements from Global Conferences. Geneva: WHO (2009).
5. Wilkinson J, Ells L, Pencheon D, Flowers J, Burton H. Public health genomics: the interface with public health intelligence and the role of public health observatories. Public Health Genomics (2011) 14:35–42. doi: 10.1159/000294170
6. Jamoulle M, Roland M. Quaternary prevention. In: Hong-Kong Wonca Classification Committee. Brussels (1995).
7. Martins C, Godycki-Cwirko M, Heleno B, Brodersen J. Quaternary prevention: reviewing the concept. Eur J Gen Pract. (2018) 24:106–11. doi: 10.1080/13814788.2017.1422177
8. Brodersen J, Schwartz LM, Woloshin S. Overdiagnosis: how cancer screening can turn indolent pathology into illness. APMIS (2014) 122:683–9. doi: 10.1111/apm.12278
9. Public Health Functions Steering Committee. The Vision of Public Health in America: “Healthy People in Healthy Communities”. (1994)
10. Burke W, Burton H, Hall A, Karmali M, Khoury M, Knoppers B, et al. Extending the reach of public health genomics: what should be the agenda for public health in an era of genome-based and “personalized” medicine? Genet Med. (2010) 12:785–91. doi: 10.1097/GIM.0b013e3182011222
11. Cleeran E, Van der Heyden J, Brand A, Van Oyen H. Public health in the genomic era: will Public Health Genomics contribute to major changes in the prevention of common diseases? Arch Public Health (2011) 69:8. doi: 10.1186/0778-7367-69-8
12. Burke W, Khoury M, Stewart A, Zimmern R, Bellagio Group. The path from genome-based research to population health: development of an international public health genomics network. Genet Med. (2006) 8:451–8. doi: 10.1097/01.gim.0000228213.72256.8c
14. Knoppers B, Leroux T, Doucet H, Godard B, Laberge C, Stanton-Jean M, et al. Framing genomics, public health research and policy: points to consider. Public Health Genom. (2010) 163:224–34. doi: 10.1159/000279624
15. Rogowski W, Grosse S, Khoury M. Challenges of translating genetic tests into clinical and public health practice. Nat Rev Genet. (2009) 10:489–94. doi: 10.1038/nrg2606
16. Manamperi A. Current developments in genomics and personlized health care: impact on public health. Asia-Pacific J Public Health (2008) 20:242–50. doi: 10.1177/1010539508316783
17. Gilmour M, Graham M, Reimer A, Van Domselaar G. Public health genomics and the new molecular epidemiology of bacterial pathogens. Public Health Genom. (2013) 16:25–30. doi: 10.1159/000342709
18. Lecuit M, Eloit M. The potential of whole genome NGS for infectious disease diagnosis. Expert Rev Mol Diagn. (2015) 15:1517–9. doi: 10.1586/14737159.2015.1111140
19. Gire SK, Goba A, Andersen KG, Sealfon RS, Park DJ, Kanneh L, et al. Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak. Science (2014) 345:1369–72. doi: 10.1126/science.1259657
20. Boccia S, McKee M, Adany R, Boffetta P, Burton H, Cambon-Thomsen A, et al. Beyond public health genomics: proposals from an international working group. Eur J Public Health (2014) 24:877–9. doi: 10.1093/eurpub/cku142
21. El-Sayed A, Koenen K, Galea S. Rethinking our public health genetics research paradigm. Am J Public Health (2013) 103(Suppl. 1): S14–8. doi: 10.2105/AJPH.2012.301127
22. Burton H, Adams M, Bunton R, Schroder-Back P. Developing stakeholder involvement for introducing public health genomics into public policy. Public Health Genom. (2009) 12:11–9. doi: 10.1159/000153426
23. Stein LD. The case for cloud computing in genome informatics. Genome Biol. (2010) 11:207. doi: 10.1186/gb-2010-11-5-207
24. Sboner A, Mu XJ, Greenbaum D, Auerbach RK, Gerstein MB. The real cost of sequencing: higher than you think! Genome Biol. (2011) 12:125. doi: 10.1186/gb-2011-12-8-125
25. House of Commons Science and Technology Committee. Genomics and Genome Editing in the NHS: Third Report of Session 2017-19. In: House of Commons, London (2018).
26. Bowen M, Kolor K, Dotson W, Ned R, Khoury M. Public health action in genomics is now needed beyond newborn screening. Public Health Genomics (2012) 15:327–34. doi: 10.1159/000341889
27. Boccia S, Brand A, Brand H, Ricciardi G. The integration of genome-based information for common diseases into health policy and healthcare as a major challenge for Public Health Genomics: the example of the methylenetetrahydrofolate reductase gene in non-cancer diseases. Mutat Res. (2009) 667:27–34. doi: 10.1016/j.mrfmmm.2008.10.003
28. Aarden E, Van Hoyweghen I, Horstman K. The paradox of public health genomics: definition and diagnosis of familial hypercholesterolaemia in three European countries. Scand J Public Health (2011) 39:634–9. doi: 10.1177/1403494811414241
29. McGrath B. Advancing the post-genomic era agenda: contributions from public health. Public Health Genom. (2012) 15:125–31. doi: 10.1159/000335551
30. Genome-based research and population health. Report of an Expert Workshop Held at the Rockefeller Foundation Study and Conference Center. Bellagio (2005). Available online at: http://www.phgfoundation.org/documents/74_1138619841.pdf
31. Beskow L, Khoury M, Baker T, Thrasher J. The integration of genomics into public health research, policy and practice in the United States. Commun Genet. (2001) 4:2–11. doi: 10.1159/000051150
32. Khoury M, Bowen M, Burke W, Coates R, Dowling N, Evan J, et al. Current priorities for public health practice in addressing the role of human genomics in improving population health. Am J Prevent Med. (2011) 40:486–93. doi: 10.1016/j.amepre.2010.12.009
33. Mazzucco W, Pastorino R, Lagerberg T, Colotto M, d'Andrea E, Marotta C, et al. Current state of genomic policies in healthcare among EU member states: results of a survey of chief medical officers. Eur J Public Health (2017) 27:931–7. doi: 10.1093/eurpub/ckw155
34. Simone B, Mazzucco W, Gualano MR, Agodi A, Coviello D, Dagna Bricarelli F, et al. The policy of public health genomics in Italy. Health Policy (2013) 110:214–9. doi: 10.1016/j.healthpol.2013.01.015
35. Department of Health. National Health Genomics Policy Framework 2018-2021. Canberra, ACT: Australian Health Ministers' Advisory Council (2017).
36. Khoury M, Valdez R. Rare diseases, genomics and public health: an expanding intersection. In: Genomics and Health Impact Blog. (2016)
37. McWalter K, Gaviglio A. Introduction to special issue: public health genetics and genomics. J Genet Counsel. (2015) 24:375–80. doi: 10.1007/s10897-015-9825-9
38. Zimmern R. Genomics and individuals in public health practice: are we luddites or can we meet the challenge? J Public Health (2011) 33:477–82. doi: 10.1093/pubmed/fdr080
39. Roberts J, Dolinoy D, Tarini B. Emerging issues in public health genomics. Annu Rev Genom Human Genet. (2014) 15:461–80. doi: 10.1146/annurev-genom-090413-025514
40. St Pierre J, Bach J, Duquette D, Oehlke K, Nystrom R, Silvey K, et al. Strategies, actions and outcomes of pilot state programs in public health genomics, 2003-2008. Prevent Chronic Dis. (2014) 11:130267. doi: 10.5888/pcd11.130267
41. Horn E, Baxter K, O'Leary J, Terry S. Exploring priorities for public health genomics. Genet Test Mol Biomark. (2011) 15:741–2. doi: 10.1089/gtmb.2011.1525
42. Noonan A. Key roles of government in genomics and proteomics: a public health perspective. Genet Med. (2002) 4(Suppl. 6):72S−6S. doi: 10.1097/00125817-200211001-00016
43. Marzuillo C, De Vito C, D'Andrea E, Rosso A, Villari P. Predictive genetic testing for complex disease: a public health perspective. Quar J Med. (2014) 107:93–7. doi: 10.1093/qjmed/hct190
44. Gwinn M, Khoury M. Genomics and public health in the United States: signposts on the translation highway. Commun Genet. (2006) 9:21–6 doi: 10.1159/000090689
45. Noonan A. Integrating genomics into US public health. Genet Med. (2002) 4(Suppl. 6):68S−71S. doi: 10.1097/00125817-200211001-00015
46. Khoury M, Gwinn M, Burke W, Bowen S, Zimmern R. Will genomics widen or help heal the schism between medicine and public health? Am J Prevent Med. (2007) 33:310–6. doi: 10.1016/j.amepre.2007.05.010
47. Metcalfe S, Bittles A, O'Leary P, Emery J. Australia: public health genomics. Public Health Genom. (2009) 12:121–8. doi: 10.1159/000160666
48. Khoury M, Bowen S, Bradley L, Coates R, Dowling N, Gwinn M, et al. A decade of public health genomics in the United States: centers for disease control and prevention 1997-2007. Public Health Genom. (2009) 12:20–9. doi: 10.1159/000153427
49. Khoury M, Mensah G. Genomics and the prevention and control of common chronic diseases: emerging priorities for public health action. Prevent Chronic Dis. (2005) 2:A05. Available online at: http://www.cdc.gov/pcd/issues/2005/apr/05_0011.htm
50. Ianuale C, Leoncini E, Mazzucco W, Marzuillo C, Villari P, Ricciardi W, et al. Public Health Genomics education in post-graduate schools of hygiene and preventive medicine: a cross-sectional survey. BMC Med Educ. (2014) 14:213. doi: 10.1186/1472-6920-14-213
51. Guttmacher A, Porteous M, McInerney J. Educating health-care professionals about genetics and genomics. Nat Rev Genet. (2007) 8:151–7. doi: 10.1038/nrg2007
52. Mazzucco W, Ricciardi W, Boccia S. Addressing the gap between genetics knowledge and clinical practice: a pilot study to implement genetics education among physicians in Italy. Italian J Public Health (2012) 9:e8673. doi: 10.2427/8673
53. Michelazzo MB, Pastorino R, Mazzucco W, Boccia S. Distance learning training in genetics and genomics testing for Italian health professionals: results of a pre and post-test evaluation. Epidemiol Biostat Public Health (2015) 12:e11516. doi: 10.2427/11516
54. D'Andrea E, Lagerberg T, De Vito C, Pitini E, Marzuillo C, Massimi A, et al. Patient experience and utility of genetic information: a cross-sectional study among patients tested for cancer susceptibility and thrombophilia. Eur J Hum Genet. (2018) 26:518–26. doi: 10.1038/s41431-017-0083-1
55. Silvia B, Boccia A, Boccia S, Casella C, Ciminello A, Cocchella A, et al. HTA of genetic testing for susceptibility to venous thromboembolism in Italiy. Italian J Public Health (2012) 9:(2, suppl. 1) doi: 10.2427/6348
56. Khoury M, Clauser S, Freedman A, Gillanders E, Glasgow R, Klein W, et al. Population sciences, translational research and the opportunities and challenges for genomics to reduce the burden of cancer in the 21st Century. Cancer Epidemiol Biomark Prevent. (2011) 20:2105–14. doi: 10.1158/1055-9965.EPI-11-0481
57. Nowak KJ, Bauskis A, Dawkins HJ, Baynam G. Incidental inequity. Eur J Human Genet. (2018) 26:616–7. doi: 10.1038/s41431-018-0101-y
58. Khoury M, Gwinn M, Ioannidis J. The emergence of translational epidemiology: from scientific discovery to population health impact. Am J Epidemiol. (2010) 172:517–24. doi: 10.1093/aje/kwq211
59. Khoury M, Gwinn M, Yoon P, Dowling N, Moore C, Bradley L. The continuum of translation research in genomic medicine: how can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention. Genet Med. (2007) 9:665–74. doi: 10.1097/GIM.0b013e31815699d0
60. Camp KM, Trujillo E. Position of the Academy of Nutrition and Dietetics: nutritional genomics. J Acad Nutr Diet (2014) 114:299–312. doi: 10.1016/j.jand.2013.12.001
61. Caulfield T, McGuire AL. Direct-to-consumer genetic testing: perceptions, problems, and policy responses. Annu Rev Med. (2012) 63:23–33. doi: 10.1146/annurev-med-062110-123753
62. Covolo L, Rubinelli S, Ceretti E, Gelatti U. Internet-based direct-to-consumer genetic testing: a systematic review. J Med Internet Res. (2015) 17:e279. doi: 10.2196/jmir.4378
63. McGuire AL, Burke W. Health system implications of direct-to-consumer personal genome testing. Public Health Genom. (2011) 14:53–8. doi: 10.1159/000321962
64. Bunnik EM, Schermer MH, Janssens AC. Personal genome testing: test characteristics to clarify the discourse on ethical, legal and societal issues. BMC Med Ethics (2011) 12:11. doi: 10.1186/1472-6939-12-11
65. Fears R, ter Meulen V, Group E-FW. The perspective from EASAC and FEAM on direct-to-consumer genetic testing for health-related purposes. Eur J Hum Genet. (2013) 21:703–7. doi: 10.1038/ejhg.2012.238
66. Alber K, Kuehlein T, Schedlbauer A, Schaffer S. Medical overuse and quaternary prevention in primary care - a qualitative study with general practitioners. BMC Fam Pract. (2017) 18:99. doi: 10.1186/s12875-017-0667-4
67. Tesser CD. Why is quaternary prevention important in prevention? Rev Saude Public (2017) 51:116. doi: 10.11606/S1518-8787.2017051000041
68. Williamson R, Anderson W, Duckett S, Frazer I, Hillyard C, Kowal E, et al. The Future of Precision Medicine in Australia. Victoria: Australian Council of Learned Academies (2018).
70. Dyer O. “All of Us” study begins to sequence and follow a million Americans. BMJ (2018) 361:k2001. doi: 10.1136/bmj.k2001
72. Sankar PL, Parker LS. The Precision Medicine Initiative's All of Us Research Program: an agenda for research on its ethical, legal, and social issues. Genet Med. (2017) 19:743–50. doi: 10.1038/gim.2016.183
73. Galea S. Precision Medicine and Population Health: Forging a Consensus. School of Public Health; Boston University (2016).
74. Khoury M, Galea S. Will precision medicine improve population health? J Am Med Assoc. (2016) 316:1357–8. doi: 10.1001/jama.2016.12260
75. Khoury M. Planning for the future of epidemiology in the era of big data and precision medicine. Am J Epidemiol. (2015) 182:977–9. doi: 10.1093/aje/kwv228
76. Precision Public Health Summit. Summit Report. San Francisco, CA: University of California (2016).
77. Khoury MJ, Bowen MS, Clyne M, Dotson WD, Gwinn ML, Green RF, et al. From public health genomics to precision public health: a 20-year journey. Genet Med. (2017) 20:574–82. doi: 10.1038/gim.2017.211
78. Weeramanthri TS, Dawkins HJS, Baynam G, Bellgard M, Gudes O, Semmens JB. Editorial: precision public health. Front Public Health (2018) 6:121. doi: 10.3389/fpubh.2018.00121
79. Khoury M. Precision public health: more precision ahead for individual and population interventions. In: Genomics and Health Impact Blog. Centers for Disease Control and Prevention (2016).
80. Riley W, Nilsen W, Manolio T, Masys D, Lauer M. News from the NIH: potential contributions of the behavioral and social sciences to the precision medicine initiative. Trans Behav Med. (2015) 5:243–6. doi: 10.1007/s13142-015-0320-5
81. Fisk Green R, Dotson W, Bowen S, Kolor K, Khoury M. Genomics in public health: perspectives from the Office of Public Health Genomics at the Centers for Disease Control and Prevention (CDC). Healthcare (2015) 3:830–7. doi: 10.3390/healthcare3030830
82. Baynam G, Molster C, Bauskis A, Kowal E, Savarirayan R, Kelaher M, et al. Indigenous genetics and rare diseases: harmony, diversity and equity. In: Posada M, Taruscio D, Groft S, editors. Rare Diseases Epidemiology: Update and Overview. Cham: Springer International Publishing (2017). p. 511–520.
83. Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature (2011) 475:163–5. doi: 10.1038/475163a
84. Manrai AK, Funke BH, Rehm HLOlesen MS, Maron BA, Szolovits P, Margulies DM, et al. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. (2016) 375:655–65. doi: 10.1056/NEJMsa1507092
Glossary
Keywords: public health genomics, precision public health, genomics, population genetics, population health
Citation: Molster CM, Bowman FL, Bilkey GA, Cho AS, Burns BL, Nowak KJ and Dawkins HJS (2018) The Evolution of Public Health Genomics: Exploring Its Past, Present, and Future. Front. Public Health 6:247. doi: 10.3389/fpubh.2018.00247
Received: 16 May 2018; Accepted: 15 August 2018;
Published: 04 September 2018.
Edited by:
Paul Lacaze, Monash University, AustraliaReviewed by:
Sobia Raza, PHG Foundation, United KingdomWalter Mazzucco, Università degli Studi di Palermo, Italy
Copyright © 2018 Molster, Bowman, Bilkey, Cho, Burns, Nowak and Dawkins. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Faye L. Bowman, ZmF5ZS5ib3dtYW5AaGVhbHRoLndhLmdvdi5hdQ==
†These authors have contributed equally to this work
 Caron M. Molster
Caron M. Molster Faye L. Bowman
Faye L. Bowman Gemma A. Bilkey
Gemma A. Bilkey Angela S. Cho
Angela S. Cho Belinda L. Burns
Belinda L. Burns Kristen J. Nowak1,3,4
Kristen J. Nowak1,3,4 Hugh J. S. Dawkins
Hugh J. S. Dawkins