
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Public Health , 01 May 2017
Sec. Digital Public Health
Volume 5 - 2017 | https://doi.org/10.3389/fpubh.2017.00084
This article is part of the Research Topic Real-time Point-of-Care Quantitative Biomarkers: Trekkie Hype or the Future of Personalized Medicine? View all 6 articles
 Swinburne A. J. Augustine1*
Swinburne A. J. Augustine1* Kaneatra J. Simmons2
Kaneatra J. Simmons2 Tarsha N. Eason3
Tarsha N. Eason3 Clarissa L. Curioso4
Clarissa L. Curioso4 Shannon M. Griffin1
Shannon M. Griffin1 Timothy J. Wade5
Timothy J. Wade5 Alfred Dufour1
Alfred Dufour1 G. Shay Fout1
G. Shay Fout1 Ann C. Grimm1
Ann C. Grimm1 Kevin H. Oshima1
Kevin H. Oshima1 Elizabeth A. Sams5
Elizabeth A. Sams5 Mary Jean See4
Mary Jean See4 Larry J. Wymer1
Larry J. Wymer1
Waterborne infectious diseases are a major public health concern worldwide. Few methods have been established that are capable of measuring human exposure to multiple waterborne pathogens simultaneously using non-invasive samples such as saliva. Most current methods measure exposure to only one pathogen at a time, require large volumes of individual samples collected using invasive procedures, and are very labor intensive. In this article, we applied a multiplex bead-based immunoassay capable of measuring IgG antibody responses to six waterborne pathogens simultaneously in human saliva to estimate immunoprevalence in beachgoers at Boquerón Beach, Puerto Rico. Further, we present approaches for determining cutoff points to assess immunoprevalence to the pathogens in the assay. For the six pathogens studied, our results show that IgG antibodies against antigens from noroviruses GI.I and GII.4 were more prevalent (60 and 51.6%, respectively) than Helicobacter pylori (21.4%), hepatitis A virus (20.2%), Campylobacter jejuni (8.7%), and Toxoplasma gondii (8%) in the saliva of the study participants. The salivary antibody multiplex immunoassay can be used to examine immunoprevalence of specific pathogens in human populations.
Acute gastrointestinal illness (AGI) has long been associated with swimming in fecally contaminated waters (1–12). Epidemiological surveys and enzyme-linked immunosorbent assays are often used to ascertain the cause of these illnesses. These approaches are time consuming, costly, and suffer from challenges such as selection bias and patient recollection of symptoms (13, 14). Further, AGIs may be caused by many different pathogens, including a number of viruses, bacteria, and protozoa.
Multiplexed immunoassays have been primarily used to assess antibodies in serum; however, oral fluid represents a promising alternative to serum in studies to determine the presence and incidence of certain infections (15–26). Saliva is simple and safe to collect, well tolerated by children, non-invasive, inexpensive compared to serum, and importantly can usually be self-collected. Antibodies in saliva have been detected against bacteria (27–32), protozoa (33–36), and viruses (20, 37–44). The combination of saliva samples and a multiplex assay provides a powerful tool that could enhance health outcome assessment and measurement for certain types of epidemiological studies (17, 19, 35, 45, 46).
Several potentially waterborne pathogens, including Campylobacter jejuni, Helicobacter pylori, Toxoplasma gondii, hepatitis A virus (HAV), and noroviruses, were selected for this study based on their relative importance for public health and availability of immunogenic proteins. These organisms may have multiple routes of transmission, but all have been implicated as possible sources of waterborne disease. Specifically, C. jejuni is a common cause of acute bacterial gastrointestinal illness, often associated with foodborne disease, primarily from poultry and raw milk (47). However, outbreaks of campylobacteriosis due to contamination of drinking (47–49) and recreational (50, 51) waters have been reported. H. pylori is a bacterium uniquely adapted to chronically colonize the human stomach, which causes transient acute dyspeptic symptoms following initial colonization (52), but chronic infection can cause chronic gastritis, gastric or duodenal ulcers, and gastric cancer (53). Although most H. pylori infections are believed to be acquired via person-to-person transmission, waterborne transmission is also possible (54) due to the ability of Helicobacter to contaminate water supplies (55) and survive in distribution system biofilms (56). T. gondii is a parasite of felines, which also infects a wide variety of intermediate hosts, including livestock and humans (57). Infections are benign in most immunocompetent individuals (57), but Toxoplasma infection in a previously uninfected pregnant woman can cause miscarriage or neurological damage to the fetus (58). The predominant route of toxoplasmosis infection in humans is ingestion of undercooked meat; however, disinfection-resistant oocysts excreted by cats can also cause waterborne outbreaks (59–61). Furthermore, epidemiological associations have been reported between well water use and T. gondii antibody prevalence (62–64). HAV is an RNA virus that causes a highly contagious liver infection. It is typically transmitted by the fecal–oral route, either through consumption of contaminated food or water or via person-to-person contact (65). It was previously demonstrated that areas with inadequate water supply and poor wastewater facilities and hygienic conditions generally have high HAV prevalence (65, 66). HAV outbreaks have also been associated with drinking (67) and recreational water exposures (68). Finally, noroviruses are a diverse group of RNA viruses, which are a major cause of acute gastroenteritis worldwide. Transmission of noroviruses may occur via ingestion of contaminated food or water, exposure to contaminated fomites, and person-to-person contact. Noroviruses can contaminate surface waters (69) and cause outbreaks associated with chlorinated water supplies (70, 71) and untreated ground water (67, 72). Outbreaks have also been associated with recreational swimming exposures in lakes (50, 73, 74).
To study human exposure to these potentially waterborne pathogens, we previously developed a Luminex xMAP™ bead-based, salivary IgG antibody multiplex immunoassay to measure antibodies to C. jejuni, H. pylori, T. gondii, HAV, and noroviruses GI.1 and GII.4 (14, 19). Assay parameters such as antigen concentration and coupling, cross-reactivity, sensitivity, and specificity, as well as anti-human detection antibody and reporter concentrations, were optimized and tested using both characterized human plasma and saliva samples (14, 19).
Here, we used our optimized multiplex immunoassay (14) consisting of recombinant proteins, whole cells, and cell lysates to MicroPlex™ beads to indirectly capture human antibodies in saliva samples collected from visitors to Boquerón Beach, Puerto Rico. Methods were presented to examine potential patterns and estimate exposure in the population. The approaches described here allowed us to determine the prevalence of waterborne infections by using IgG antibodies as biomarkers of exposure in non-invasively collected human saliva samples. The goal of this study was to describe the method and overall prevalence of exposure. Subsequent manuscripts will address exposure routes and rates of new infections (immunoconversions).
Bead sets were obtained from Luminex Corp. (Austin, TX, USA) at a concentration of 12.5 × 106 beads/ml each. Biotin-labeled affinity purified goat anti-human IgG (λ) secondary detection antibody was obtained from KPL (Gaithersburg, MD, USA). Antigens were purchased (as shown in Table 1) and coupled to the beads in accordance with the optimized multiplex immunoassay presented in the study by Augustine et al. (14). The optimized conditions were applied to the antigens multiplexed in this study.
The Luminex beads were activated and coupled, as previously described (14, 16, 19). Coupling confirmation was performed using serial dilutions of commercially available, animal-derived primary capture antibodies specific to each antigen to ensure that the beads were sufficiently coupled and that the dynamic range of the assay could be defined (14). Briefly, a working bead mixture was prepared by diluting the coupled bead stocks to a final concentration of 100 beads/μl of each unique bead set in phosphate-buffered saline containing 1% bovine serum albumin (PBS-1% BSA). Twofold serial dilutions of anti-species IgG primary antibody were prepared per the manufacturer’s recommendations. Then 5 × 103 beads from each bead set were added to each well of a prewet filter plate. An equal volume of the serially diluted species-specific antibody was added to the beads, mixed gently, covered, and allowed to incubate in the dark at room temperature for 30 min at 500 rpm on a VWR™ microplate shaker (Radnor, PA, USA).
The supernatant was removed by vacuum, the wells were washed twice with 100 µl of PBS pH 7.4 containing 0.05% Tween 20 (PBS-T) (Sigma, St. Louis, MO, USA), and excess buffer was removed by vacuum. The beads were gently resuspended in PBS-1% BSA buffer, and 0.8 µg of biotinylated anti-species IgG secondary detection antibody was added to each well. The filter plates were covered and allowed to incubate in the dark at room temperature for 30 min on a plate shaker. After incubation, the wells were washed twice with 100 µl of PBS-T, and excess buffer was removed as mentioned earlier. Finally, 1.2 µg of streptavidin-R-phycoerythrin was added to each well, mixed gently, incubated for 30 min, and washed twice as mentioned earlier. Excess buffer was removed by vacuum, the beads were resuspended in 100 µl of PBS-1% BSA, and the plate was analyzed on a Luminex 100 analyzer (Luminex Corporation, Austin, TX, USA).
Approval was obtained from the institutional review board (# 08-1844, University of North Carolina, Chapel Hill, NC, USA) for the collection of saliva samples from beachgoers at Boquerón Beach, Puerto Rico, as a part of the USEPA National Epidemiological and Environmental Assessment of Recreational Water Study (75). Households were offered enrollment on a first-come, first-served basis each day until a goal of 100 was reached. Study subjects (n = 2,091) provided informed consent and were instructed on the use of the Oracol™ saliva collection device (Malvern Medical Developments, Worcester, UK). Briefly, the Oracol™ sampler was rubbed against the gingival crevices of the oral mucosa (between the gums and teeth) to absorb saliva. Infants younger than 1 year were not included due to the possible presence of maternal antibodies and high rates of non-waterborne infections. Individuals who reported dental or other illnesses at the time of the initial collection were also excluded.
The baseline samples were collected on the beach by trained study staff members. Upon receipt, samples were cataloged using FreezerWorks™ software (Dataworks Development, Inc., Mountlake Terrace, WA, USA) and stored at −80°C until tested. Oracol™ saliva collection devices were thawed at room temperature and centrifuged at 491 × g, 10°C, for 5 min to recover the saliva off the collection sponge, followed by another centrifugation at 1,363 × g, 10°C, for an additional 5 min to pellet debris from the saliva. After centrifugation, the saliva was aspirated from the tubes and transferred to 1.5-ml microcentrifuge tubes. The saliva containing tubes were centrifuged at 1,500 × g for 3 min, and the supernatant was transferred to a clean 1.5-ml microcentrifuge tube and either used immediately for analysis or stored at −80°C.
Before analysis, samples were diluted 1:4 in PBS-1% BSA in a 96-well round bottom plate (Corning™, Corning, NY, USA) and gently mixed. MultiScreen BV 96-well filter plates (Millipore, Billerica, MA, USA) were prewetted with 100 µl of PBS-1% BSA buffer, and excess buffer was removed by vacuum. Then 5 × 103 beads from each bead set and an equal volume of diluted saliva were loaded onto each well of the filter plates resulting in a final dilution of 1:8 for a total volume of 100 µl per well. The loaded filter plates were processed, as previously described (16, 19), and reporter fluorescence was measured using a Luminex 100 analyzer and expressed as median fluorescence intensity (MFI) of at least 100 beads per bead set.
One set of Luminex beads was used as a no-antigen (uncoupled) control to assess the extent of non-specific binding and sample-to-sample variability. These beads were treated identically to antigen-conjugated beads with the exception that control beads were not incubated with any antigen during the coupling step. As with the antigen-coupled beads, the control beads were blocked with BSA, a protein that is a key reagent in the buffers used to perform the assay.
Of the 2,091 saliva samples obtained from study participants, 13 samples were observed to react with the control beads. These samples, likely outliers in the 99th percentile of the control beads, were subsequently removed from analyses due to the potential for contamination of the saliva by serum from bleeding gums. The remaining samples (n = 2,078) were used in the assessment. Cutoff points for immunopositivity were derived from the MFI values of these uncoupled beads (control). In addition, background fluorescence and cross-reactivity of antigen-coupled beads to secondary detection antibodies and/or reporter were evaluated by adding PBS-1% BSA buffer to control wells instead of saliva. The MFI values from the background controls (antigen-coupled beads with PBS-1% BSA) were subtracted from the MFI obtained from every antigen-coupled bead set for each saliva sample.
Additional tests of cross-reactivity were performed with animal-derived primary detection antibodies, including monoplex (one antigen-coupled bead set with its specific primary antibody and controls), duplex (two antigen-coupled bead sets and one primary antibody and controls), and multiplex (six antigen-coupled bead sets and uncoupled control beads with one primary antibody). Bead coupling and confirmation, cross-reactivity, and method validation using characterized plasma samples were performed, as previously described (14).
As a surrogate measure of assay sensitivity, a SNR was calculated by dividing the MFI of the specific antigen signals by the MFI of the uncoupled control beads for each sample (14, 76). We computed a SNR for each reading as a ratio of the MFI value for sample i when tested for antibodies to antigen j(Rij) and the MFI value of the uncoupled control for the sample (Ci):
We explored two approaches to establish a means of estimating immunoprevalence to all targets in the Luminex multiplex assay in the absence of information regarding true infection status of the participants. To help determine immunoprevalence, researchers have used a variety of methods including three times the mean (77) and mean plus 3 times the standard deviation (SD) of the control beads (16, 78). Cutoff point criteria 1 (CC1) is defined as the mean plus 3 SDs (μ + 3σ) of the control beads. As a basic test for normality, histograms were generated, which reflected positive skewness in the salivary antibody responses observed in this study population. Cutoff point criteria 2 (CC2) was established to account for this non-normality and is defined as the antilog of the mean plus 3 SDs of the log10-transformed data [10mean(h)+3SD(h), where h = log10(control MFI)]. All data analysis was performed using Microsoft Excel 2013 and MATLAB Release 2016a.
Since we were interested in only assessing the immunoprevalence for the antigens in the multiplex for this study, IgG levels against the six antigens in the Luminex multiplex immunoassay were measured in baseline saliva samples collected from 2,078 consenting individuals at Boquerón Beach, Puerto Rico.
Figures 1 and 2 summarize the study participants’ antibody profiles to the six waterborne pathogens. In Figure 1A, each row of the heat map represents the MFI values of the target pathogens for an individual saliva sample and provides insight on potential exposure patterns. The MFI values for each target is right skewed, reflecting a non-normal distribution (Shapiro–Wilk test p-values <0.05) (Figure 1B). Figure 2A shows that each antigen is characterized by a broad range of MFI values showing noroviruses with the largest median MFIs. The maximum SNRs for each individual antigen in the multiplex assay ranged from 145:1 (C. jejuni) to 9,004:1 for norovirus GI.1 (Figure 2B). Here, we define immunoprevalence as a baseline sample (n = 2,078) with a MFI reading greater than or equal to the cutoff point established.
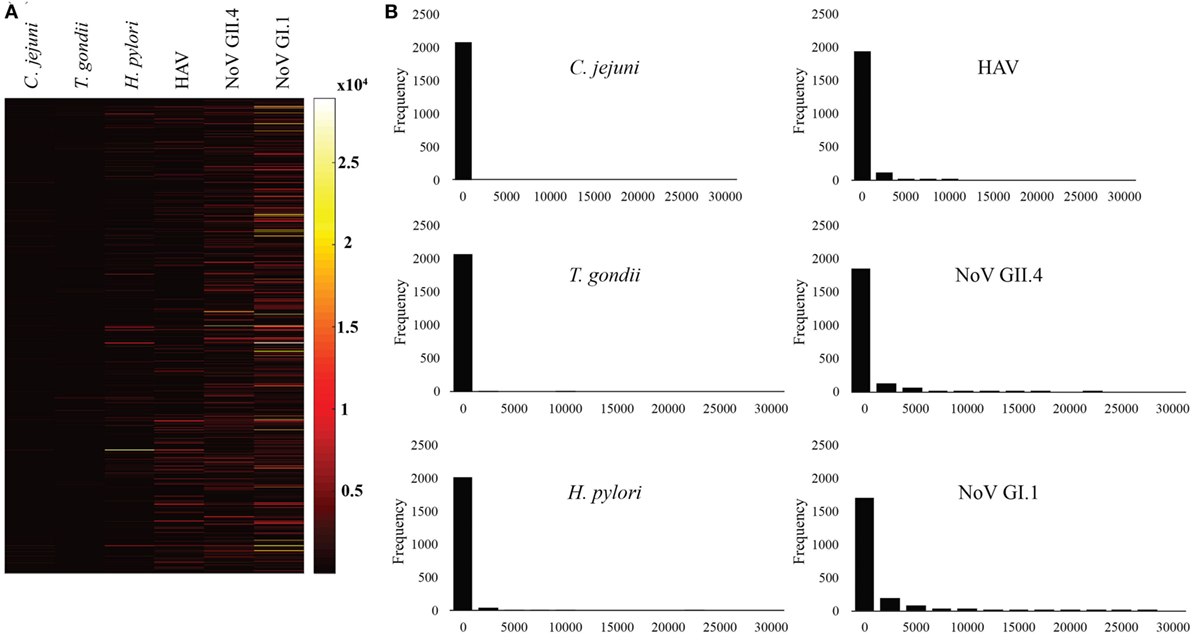
Figure 1. Median fluorescence intensity (MFI) profile of study participants. (A) Heat map visualization of study participants’ antibody profiles to six waterborne pathogens provides insight on possible exposure patterns. Each row presents the MFI values of the target pathogens for the individual saliva sample collected from beachgoers. (B) Histograms showing the non-normal distribution of target pathogen MFIs for the study participants.
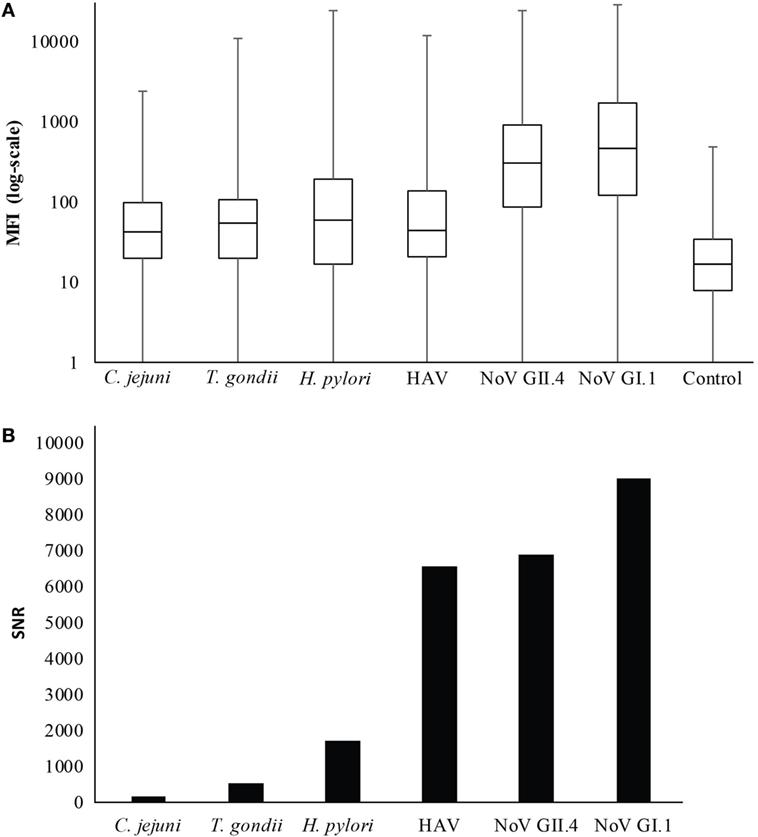
Figure 2. Distribution of median fluorescence intensity (MFI) results. (A) Boxplot of MFI results in log scale. The whiskers represent the minimum and maximum values. (B) Maximum signal-to-noise ratio (SNR) for each antigen in the multiplex immunoassay.
Table 2 provides the cutoff points and immunoprevalence results for CC1 (87.6%) and CC2 (67.7%), and Figure 3 shows the corresponding visualization. Table 3 (upper panel) provides a breakdown of the immunoprevalence to specific pathogens with evidence of the greatest amount of exposure to noroviruses GI.1 (48.6–71.2%) and GII.4 (37.6–65.7%) followed by H. pylori (14–28.7%), HAV (16.2–24.3%), T. gondii (2–14%), and C. jejuni (2.3–15.2%).
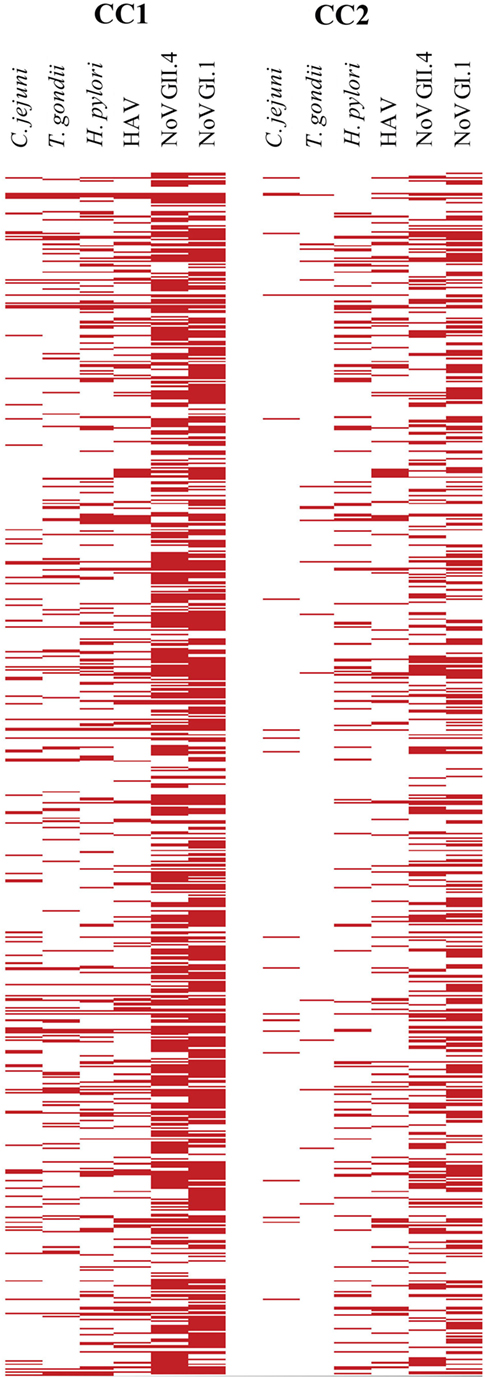
Figure 3. Visualization of cutoff point ratios. The left panel shows the positive samples based on cutoff point criteria 1 (CC1) [150 median fluorescence intensity (MFI)], and the right panel indicates positive samples based on cutoff point criteria 2 (CC2) (505 MFI). Ratios over 1 (MFI ≥ cutoff point) are red in the plots. MFIs below cutoff point are white.
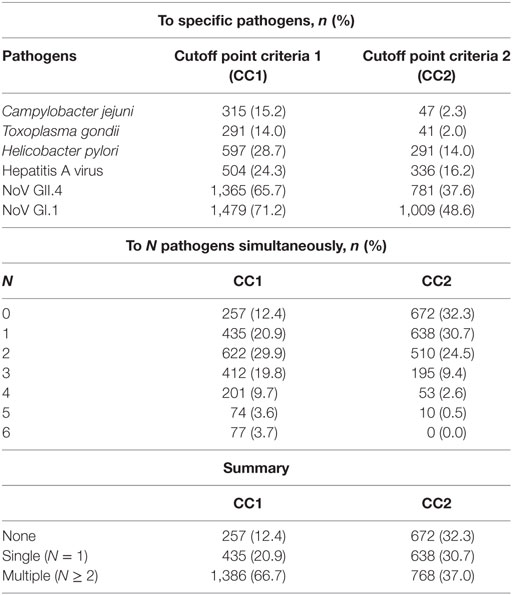
Table 3. Immunoprevalence rates to specific (upper panel) and multiple pathogens simultaneously (lower panel).
Based on our cutoff points, 1,386 (CC1) and 768 (CC2) samples showed salivary antibody responses to multiple (N) antigens (Table 3, lower panel). This prompted us to investigate whether this was a result of cross-reactivity or true coimmunopositivity. The MFI values of the complete baseline data set (n = 2,078) were used to calculate coimmunopositivity and assess correlation for all the multiplexed antigens in the immunoassay. Table 4 shows the numbers and percentages of pairwise analyses of the multiplexed antigens using both the less stringent cutoff point criteria (CC1, upper panel) and a more conservative cutoff point criteria (CC2, lower panel). The Spearman rank-order correlation coefficient (rho) assessing the linear relationship between the MFI of the antigens in the multiplex ranged from 0.20 for H. pylori/norovirus GII.4 to 0.55 for C. jejuni/H. pylori (p-value < 0.05 for each pairing), indicating a statistically significant, weak to moderate positive correlation.
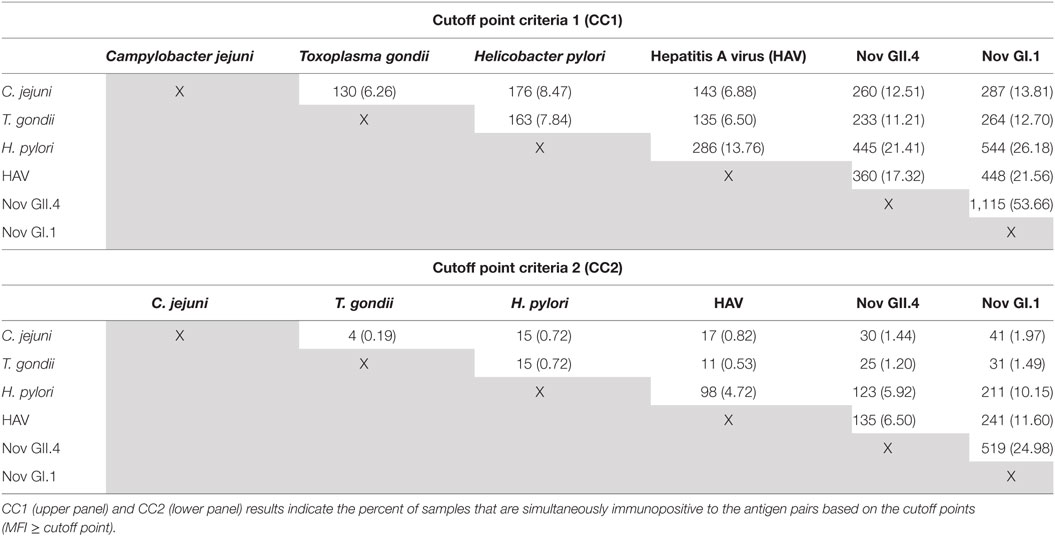
Table 4. Numbers and percentages of coimmunopositive samples observed in baseline samples based on cutoff point.
This is the first multiplex immunoassay study aimed at describing the immunoprevalence of circulating antibodies to six waterborne pathogens in beachgoers in Puerto Rico. The identification of waterborne pathogen immunoprevalence is a necessary and highly important first step in assessing and managing recreational water-associated AGI risks. Analysis of the initial saliva samples for antibodies to these six pathogens revealed that more than two-thirds of the beachgoers were previously infected by at least one of the pathogens studied. On average, the results indicated that more than three-fifths of the beachgoers had evidence of prior exposure to noroviruses, while over a fifth were previously exposed to H. pylori and HAV. Antibodies against T. gondii and C. jejuni were less common. These results are relatively consistent with previously published data that show the prevalence of infections from noroviruses GII.4 and GI.1 at >80% worldwide (79). H. pylori prevalence in developed countries ranged from 30 to 40% and 80 to 90% in the developing world (80). The seroprevalence of HAV in the United States was reportedly 34.9% overall (81), while C. jejuni and T. gondii were 15–30% and 7–22.5% prevalent, respectively (63, 82, 83). Taken together, the initial sample results suggest that these individuals had a historical exposure to the pathogens being studied. Future studies will investigate immunoconversions related to swimming-associated exposures.
Of the beachgoers who were previously exposed, many (37–67%) were found to have evidence of detectable antibodies to two or more antigens from our samples. This finding is consistent with previous studies of AGI performed in multiplex (84–87). These observations demonstrate the potential for multiplex assays to identify multiple infections and may be applied either crosssectionally or longitudinally to provide insight into the epidemiology of these conditions, as well as understanding the role of potential risk factors.
Selection of the definition to determine the cutoff points and immunoprevalence may depend on the study question being answered. For example, a less stringent criterion may be acceptable for a simple screening of samples requiring further clinical diagnostic testing. However, for studies where the immunological status of the subjects is not known or the data are not normally distributed, as is the case here, the more stringent definition is strongly preferred to reduce false positives. Although the more conservative cutoff point may reduce false positives, it may have the opposite effect of increasing false negatives because we do not know the immunological status of the study participants.
In a previous study (14), we described the performance of the multiplex immunoassay to measure IgG antibodies using characterized human plasma samples against the antigens that we have used here. In that study, we described, in detail, the process used to address cross-reactivity using characterized (diagnostically positive and negative) plasma samples. Briefly, multiplexed antigens that showed cross-reactivity >10% to antigen-specific, animal-derived antibodies were removed from the immunoassay. A clear case of cross-reactivity was observed with the rabbit anti-Giardia duodenalis antibody. That particular antibody bound to every antigen in the multiplex immunoassay at MFI levels that ranged from 59 to 180% (14) of the target antigen. Furthermore, we found the sensitivity of the assay to be ~92% based on the number of characterized samples that were correctly identified as being either positive or negative.
We have also reported on the development of multiplex immunoassays using saliva samples (16, 19). We observed that a 1:8 dilution of the saliva, together with the addition of 0.05% Tween 20 to the wash buffer, resulted in decreased non-specific binding. The optimal concentrations for each antigen used in the multiplex are shown in Table 1. Two approaches to determine cutoff points to define immunopositivity were considered for interpreting results of the multiplexed immunoassay to measure salivary IgG antibodies. The purpose was to examine the effects of the two options on the interpretation of the results and to make recommendations on which cutoff point might be the most appropriate for measuring immunoprevalence in a population. The cutoff point will also help inform which approach is most useful for the assessment of incident infections and health effects studies for waterborne infections.
We recognize that there are several limitations to our study. First, the possibility of promiscuous antibody binding with other antigens cannot be totally eliminated due to cross-reactivity (binding to similar or overlapping ligands) or multispecificity (binding of distinctly different ligands or different conformations of the same antibody) (88–91). The data presented in Tables 3 and 4 provide some evidence that for each pair of antigens, there is a statistically significant weak to moderate positive correlation (p-value < 0.05), suggesting either the possibility that these infections were occurring concomitantly as a feature of the study population or some level of cross-reactivity. The percentages of pairwise binding obtained using CC1 appear to show more coimmunopositivity, but those numbers are drastically reduced when the more stringent CC2 is employed. We explored using PVX (polyvinylalcohol + polyvinylpyrrolidone + 0.05% Tween™ 20) buffer to reduce non-specific binding (92–94) and found that PVX buffer had a dramatic effect on reducing non-specific binding in plasma, which was not observed in saliva (data not shown). These results agree with observations made in a previous study (20).
Another limitation of the study is that antibody levels in saliva samples are typically lower than levels observed in sera or plasma. In this study, we found that the maximum SNR ranged from 145:1 to 9004:1, indicating a wide dynamic range for antigens in the assay. Some individuals with low specific serum IgG will have even lower saliva IgG concentrations, and therefore, these individuals may not be considered when reporting immunoprevalence. For example, in studies where vaccination coverage rates were examined, it was noted that a number of participants fell within a “gray zone” (above the negative cutoff point but below the positive cutoff point) using saliva (39). This was attributed to differences in antibody levels between individuals with wild-type virus or vaccine-induced immunity. It was found that sensitivity of the oral fluid method increased as antibody titers in serum increased. The implication of this finding is that immunoprevalence may be underestimated by oral fluid IgG detection because the assay may not have sufficient sensitivity when the serum antibody titer range is low.
The largest concentration of IgG found in the oral cavity gains access to this region by passive diffusion from the blood stream using the crevicular epithelium of the gingiva, while the remaining concentration of IgG is due to the presence of plasma cells in the crevicular epithelium (95). To account for the lower concentrations of IgG in saliva, especially compared to IgA, we employed the Oracol oral fluid sampler, which is designed to collect crevicular fluid enriched with serum antibodies (21). Our study focused on measuring systemic rather than mucosal immune responses in saliva, exclusively of the IgG isotype. This is, in part, because salivary IgA levels are especially influenced by a donor’s age, secretory flow rate, acute and chronic stresses, and other methodological issues (18). In addition, our previous research indicated that IgA from saliva samples produced weaker responses compared to IgG in saliva (19, 20).
In spite of these limitations, the utility of the multiplex saliva assay is that it can be used for population-based immunoprevalence studies, which can be further exploited to understand the contemporary epidemiology of common environmental pathogens. When used in conjunction with large epidemiological and survey type studies of exposure to microbes in water, soil, and food, the assay described here may provide valuable information to improve our understanding of the transmission of environmental pathogens. Furthermore, salivary antibody data obtained from our assay can be used to improve previously described dynamic and static risk assessment models (1, 5, 96, 97).
In conclusion, the multiplexed immunoassay presented here, together with the cutoff points established, allowed us to measure immunoprevalence rates and coinfections to six waterborne pathogens among beachgoers in Puerto Rico. A follow-up study will explore incident infections and attempt to elucidate the sources and health effects of exposure. Future work will exploit these techniques to examine exposure patterns in other communities. This assay, along with the approaches presented here, may enhance our knowledge and understanding of environmental microbial pathogenesis and assist risk assessment modelers.
The IRB # 08-1844, University of North Carolina, Chapel Hill, NC, USA. Approval was obtained from the IRB # 08-1844, University of North Carolina, Chapel Hill, NC, USA, for the collection of saliva samples from beachgoers at Boquerón Beach, Puerto Rico, as part of the USEPA NEEAR Water Study. Study subjects provided informed consent and were instructed on the use of the Oracol™ saliva collection device. Infants younger than 1 year were not included. Informed consent was obtained from parents of minors.
SA designed the study with TW, AD, SG, GF, KO, and AG. TW, KO, AD, and ES provided the (NEEAR) saliva samples. SA, KS, SG, CC, and MS conducted the experiments. SA, TE, KS, SG, and LW were responsible for data analyses.
Mention of trade names or commercial products does not constitute endorsement or recommendation by the United States Environmental Protection Agency for use.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Thanks to Dr. Alec Sutherland, Arizona State University (AZ, USA) for kindly providing rabbit anti-GII.4 antibodies and to Mr. Jatin Mistry for his help in organizing the project’s initial steering committee.
The United States Environmental Protection Agency through its Office of Research and Development funded and managed the research described here. It has been subjected to Agency’s administrative review and approved for publication. CC and MS were supported through an appointment to the Research Participation Program at the US Environmental Protection Agency administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and US EPA.
1. Ashbolt NJ, Bruno M. Application and refinement of the WHO risk framework for recreational waters in Sydney, Australia. J Water Health (2003) 1(3):125–31.
2. Ashbolt NJ. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology (2004) 198(1–3):229–38. doi: 10.1016/j.tox.2004.01.030
3. Cabelli VJ, Dufour AP, McCabe LJ, Levin MA. Swimming-associated gastroenteritis and water quality. Am J Epidemiol (1982) 115(4):606–16. doi:10.1093/oxfordjournals.aje.a113342
4. Corbett SJ, Rubin GL, Curry GK, Kleinbaum DG. The health effects of swimming at Sydney beaches. The Sydney Beach Users Study Advisory Group. Am J Public Health (1993) 83(12):1701–6. doi:10.2105/AJPH.83.12.1701
5. Eisenberg JN, Soller JA, Scott J, Eisenberg DM, Colford JM Jr. A dynamic model to assess microbial health risks associated with beneficial uses of biosolids. Risk Anal (2004) 24(1):221–36. doi:10.1111/j.0272-4332.2004.00425.x
6. Fleisher JM, Jones F, Kay D, Stanwell-Smith R, Wyer M, Morano R. Water and non-water-related risk factors for gastroenteritis among bathers exposed to sewage-contaminated marine waters. Int J Epidemiol (1993) 22(4):698–708. doi:10.1093/ije/22.4.698
7. Fleisher JM, Fleming LE, Solo-Gabriele HM, Kish JK, Sinigalliano CD, Plano L, et al. The BEACHES Study: health effects and exposures from non-point source microbial contaminants in subtropical recreational marine waters. Int J Epidemiol (2010) 39(5):1291–8. doi:10.1093/ije/dyq084
8. Gammie A, Morris R, Wyn-Jones AP. Antibodies in crevicular fluid: an epidemiological tool for investigation of waterborne disease. Epidemiol Infect (2002) 128(2):245–9. doi:10.1017/S095026880100663X
9. Haile RW, Witte JS, Gold M, Cressey R, McGee C, Millikan RC, et al. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology (1999) 10(4):355–63. doi:10.1097/00001648-199907000-00004
10. Marion JW, Lee J, Lemeshow S, Buckley TJ. Association of gastrointestinal illness and recreational water exposure at an inland U.S. beach. Water Res (2010) 44(16):4796–804. doi:10.1016/j.watres.2010.07.065
11. Wade TJ, Calderon RL, Sams E, Beach M, Brenner KP, Williams AH, et al. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ Health Perspect (2006) 114(1):24–8. doi:10.1289/ehp.8273
12. Wade TJ, Sams E, Brenner KP, Haugland R, Chern E, Beach M, et al. Rapidly measured indicators of recreational water quality and swimming-associated illness at marine beaches: a prospective cohort study. Environ Health (2010) 9:66. doi:10.1186/1476-069X-9-66
13. Lilienfeld AM. Practical limitations of epidemiologic methods. Environ Health Perspect (1983) 52:3. doi:10.1289/ehp.83523
14. Augustine SA, Simmons KJ, Eason TN, Griffin SM, Curioso CL, Wymer LJ, et al. Statistical approaches to developing a multiplex immunoassay for determining human exposure to environmental pathogens. J Immunol Methods (2015) 425(2015):1–9. doi:10.1016/j.jim.2015.06.002
15. Augustine SA, Simmons KJ. Saliva-based immunoassay of waterborne pathogen exposure. In: Rennie J, editor. Yearbook of Science and Technology 2013. New York, NY: McGraw-Hill (2013). p. 330–3.
16. Augustine SA, Eason TN, Simmons KJ, Curioso CL, Griffin SM, Ramudit MK, et al. Developing a salivary antibody multiplex immunoassay to measure human exposure to environmental pathogens. J Vis Exp (2016) 115:e54415. doi:10.3791/54415
17. Augustine SA. Towards universal screening for toxoplasmosis: rapid, cost-effective, and simultaneous detection of anti-Toxoplasma IgG, IgM, and IgA antibodies by use of very small serum volumes. J Clin Microbiol (2016) 54(7):1684–5. doi:10.1128/JCM.00913-16
18. Brandtzaeg P. Do salivary antibodies reliably reflect both mucosal and systemic immunity? Ann N Y Acad Sci (2007) 1098:288–311. doi:10.1196/annals.1384.012
19. Griffin SM, Chen IM, Fout GS, Wade TJ, Egorov AI. Development of a multiplex microsphere immunoassay for the quantitation of salivary antibody responses to selected waterborne pathogens. J Immunol Methods (2011) 364(1–2):83–93. doi:10.1016/j.jim.2010.11.005
20. Griffin SM, Converse RR, Leon JS, Wade TJ, Jiang X, Moe CL, et al. Application of salivary antibody immunoassays for the detection of incident infections with Norwalk virus in a group of volunteers. J Immunol Methods (2015) 424:53–63. doi:10.1016/j.jim.2015.05.001
21. McKie A, Vyse A, Maple C. Novel methods for the detection of microbial antibodies in oral fluid. Lancet Infect Dis (2002) 2(1):18–24. doi:10.1016/S1473-3099(01)00169-4
22. Moorthy M, Daniel HD, Kurian G, Abraham P. An evaluation of saliva as an alternative to plasma for the detection of hepatitis C virus antibodies. Indian J Med Microbiol (2008) 26(4):327–32. doi:10.4103/0255-0857.42116
23. Nokes DJ, Enquselassie F, Nigatu W, Vyse AJ, Cohen BJ, Brown DWG, et al. Has oral fluid the potential to replace serum for the evaluation of population immunity levels? A study of measles, rubella and hepatitis B in rural Ethiopia. Bull World Health Organ (2001) 79(7):588–95. doi:10.1590/S0042-96862001000700003
24. O’Connell T, Thornton L, O’Flanagan D, Staines A, Connell J, Dooley S, et al. Oral fluid collection by post for viral antibody testing. Int J Epidemiol (2001) 30(2):298–301. doi:10.1093/ije/30.2.298
25. Parry JV, Perry KR, Mortimer PP. Sensitive assays for viral antibodies in saliva – an alternative to tests on serum. Lancet (1987) 2(8550):72.–75. doi:10.1016/S0140-6736(87)92737-1
26. Shillitoe EJ, Lehner T. Immunoglobulins and complement in crevicular fluid, serum and saliva in man. Arch Oral Biol (1972) 17(2):241–7. doi:10.1016/0003-9969(72)90206-3
27. Ballam LD, Mendall MA, Asante M, Morris J, Strachan DP, Whincup PH, et al. Western blotting is useful in the salivary diagnosis of Helicobacter pylori infection. J Clin Pathol (2000) 53(4):314–7. doi:10.1136/jcp.53.4.314
28. Chart H, Perry NT, Willshaw GA, Cheasty T. Analysis of saliva for antibodies to the LPS of Escherichia coli O157 in patients with serum antibodies to E. coli O157 LPS. J Med Microbiol (2003) 52(Pt 7):569–72. doi:10.1099/jmm.0.05126-0
29. Choo S, Zhang Q, Seymour L, Akhtar S, Finn A. Primary and booster salivary antibody responses to a 7-valent pneumococcal conjugate vaccine in infants. J Infect Dis (2000) 182(4):1260–3. doi:10.1086/315834
30. El-Mekki A, Kumar A, Alknawy B, Al-Ammari O, Moosa R, Quli S, et al. Comparison of enzyme immunoassays detecting Helicobacter pylori specific IgG in serum and saliva with endoscopic and biopsy findings in patients with dyspepsia. Indian J Med Microbiol (2011) 29(2):136–40. doi:10.4103/0255-0857.81793
31. Herath HM. Early diagnosis of typhoid fever by the detection of salivary IgA. J Clin Pathol (2003) 56(9):694–8. doi:10.1136/jcp.56.9.694
32. Marshall B, Howat AJ, Wright PA. Oral fluid antibody detection in the diagnosis of Helicobacter pylori infection. J Med Microbiol (1999) 48(11):1043–6. doi:10.1099/00222615-48-11-1043
33. Abd-Alla MD, Jackson TF, Reddy S, Ravdin JI. Diagnosis of invasive amebiasis by enzyme-linked immunosorbent assay of saliva to detect amebic lectin antigen and anti-lectin immunoglobulin G antibodies. J Clin Microbiol (2000) 38(6):2344–7.
34. Estevez PT, Satoguina J, Nwakanma DC, West S, Conway DJ, Drakeley CJ. Human saliva as a source of anti-malarial antibodies to examine population exposure to Plasmodium falciparum. Malar J (2011) 10:104. doi:10.1186/1475-2875-10-104
35. Moss DM, Montgomery JM, Newland SV, Priest JW, Lammie PJ. Detection of cryptosporidium antibodies in sera and oral fluids using multiplex bead assay. J Parasitol (2004) 90(2):397–404. doi:10.1645/GE-3267
36. Toyoguchi A, Omata Y, Koyama T, Kamiyoshi T, Takeda K, Furuoka H, et al. Antibody reactivity to Cryptosporidium parvum in saliva of calves after experimental infection. J Vet Med Sci (2000) 62(11):1231–4. doi:10.1292/jvms.62.1231
37. Corstjens PL, Abrams WR, Malamud D. Detecting viruses by using salivary diagnostics. J Am Dent Assoc (2012) 143(10 Suppl):12S–8S. doi:10.14219/jada.archive.2012.0338
38. Hodinka RL, Nagashunmugam T, Malamud D. Detection of human immunodeficiency virus antibodies in oral fluids. Clin Diagn Lab Immunol (1998) 5(4):419–26.
39. Kremer JR, Muller CP. Evaluation of commercial assay detecting specific immunoglobulin g in oral fluid for determining measles immunity in vaccinees. Clin Diagn Lab Immunol (2005) 12(5):668–70. doi:10.1128/CDLI.12.5.668-670.2005
40. Moe CL, Sair A, Lindesmith L, Estes MK, Jaykus LA. Diagnosis of Norwalk virus infection by indirect enzyme immunoassay detection of salivary antibodies to recombinant Norwalk virus antigen. Clin Diagn Lab Immunol (2004) 11(6):1028–34. doi:10.1128/CDLI.11.6.1028-1034.2004
41. Morris-Cunnington MC, Edmunds WJ, Miller E, Brown DW. A population-based seroprevalence study of hepatitis A virus using oral fluid in England and Wales. Am J Epidemiol (2004) 159(8):786–94. doi:10.1093/aje/kwh107
42. Schramm W, Angulo GB, Torres PC, Burgess-Cassler A. A simple saliva-based test for detecting antibodies to human immunodeficiency virus. Clin Diagn Lab Immunol (1999) 6(4):577–80.
43. Stuart JM, Majeed FA, Cartwright KA, Room R, Parry JV, Perry KR, et al. Salivary antibody testing in a school outbreak of hepatitis A. Epidemiol Infect (1992) 109(1):161–6.
44. Yap G, Sil BK, Ng LC. Use of saliva for early dengue diagnosis. PLoS Negl Trop Dis (2011) 5(5):e1046. doi:10.1371/journal.pntd.0001046
45. Moss DM, Priest JW, Boyd A, Weinkopff T, Kucerova Z, Beach MJ, et al. Multiplex bead assay for serum samples from children in Haiti enrolled in a drug study for the treatment of lymphatic filariasis. Am J Trop Med Hyg (2011) 85(2):229–37. doi:10.4269/ajtmh.2011.11-0029
46. Nie S, Henley WH, Miller SE, Zhang H, Mayer KM, Dennis PJ, et al. An automated integrated platform for rapid and sensitive multiplexed protein profiling using human saliva samples. Lab Chip (2014) 14(6):1087–98. doi:10.1039/c3lc51303c
47. Kuhn KG, Falkenhorst G, Emborg HD, Ceper T, Torpdahl M, Krogfelt KA, et al. Epidemiological and serological investigation of a waterborne Campylobacter jejuni outbreak in a Danish town. Epidemiol Infect (2017) 145(4):701–9. doi:10.1017/S0950268816002788
48. Clark CG, Price L, Ahmed R, Woodward DL, Melito PL, Rodgers FG, et al. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg Infect Dis (2003) 9(10):1232–41. doi:10.3201/eid0910.020584
49. Hanninen ML, Haajanen H, Pummi T, Wermundsen K, Katila ML, Sarkkinen H, et al. Detection and typing of Campylobacter jejuni and Campylobacter coli and analysis of indicator organisms in three waterborne outbreaks in Finland. Appl Environ Microbiol (2003) 69(3):1391–6. doi:10.1128/AEM.69.3.1391-1396.2003
50. Yoder J, Roberts V, Craun GF, Hill V, Hicks LA, Alexander NT, et al. Surveillance for waterborne disease and outbreaks associated with drinking water and water not intended for drinking – United States, 2005-2006. MMWR Surveill Summ (2008) 57(9):39–62.
51. Schonberg-Norio D, Takkinen J, Hanninen ML, Katila ML, Kaukoranta SS, Mattila L, et al. Swimming and Campylobacter infections. Emerg Infect Dis (2004) 10(8):1474–7. doi:10.3201/eid1008.030924
52. Graham DY, Opekun AR, Osato MS, El-Zimaity HM, Lee CK, Yamaoka Y, et al. Challenge model for Helicobacter pylori infection in human volunteers. Gut (2004) 53(9):1235–43. doi:10.1136/gut.2003.037499
53. Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med (2002) 347:1175–86. doi:10.1056/NEJMra020542
54. Bellack NR, Koehoorn MW, MacNab YC, Morshed MG. A conceptual model of water’s role as a reservoir in Helicobacter pylori transmission: a review of the evidence. Epidemiol Infect (2006) 134(3):439–49. doi:10.1017/S0950268806006005
55. Horiuchi T, Ohkusa T, Watanabe M, Kobayashi D, Miwa H, Eishi Y. Helicobacter pylori DNA in drinking water in Japan. Microbiol Immunol (2001) 45(7):515–9. doi:10.1111/j.1348-0421.2001.tb02652.x
56. Watson CL, Owen RJ, Said B, Lai S, Lee JV, Surman-Lee S, et al. Detection of Helicobacter pylori by PCR but not culture in water and biofilm samples from drinking water distribution systems in England. J Appl Microbiol (2004) 97(4):690–680. doi:10.1111/j.1365-2672.2004.02360.x
57. Dodds EM. Toxoplasmosis. Curr Opin Ophthalmol (2006) 17(6):557–61. doi:10.1097/ICU.0b013e32801094ca
59. Bowie WR, King AS, Werker DH, Isaac-Renton JL, Bell A, Eng SB, et al. Outbreak of toxoplasmosis associated with municipal drinking water: the BC Toxoplasma Investigation Team. Lancet (1997) 350(9072):173–7. doi:10.1016/S0140-6736(96)11105-3
60. de Moura L, Bahia-Oliveira LM, Wada MY, Jones JL, Tuboi SH, Carmo EH, et al. Waterborne toxoplasmosis, Brazil, from field to gene. Emerg Infect Dis (2006) 12(2):326–9. doi:10.3201/eid1202.041115
61. Jones JL, Dubey JP. Waterborne toxoplasmosis – recent developments. Exp Parasitol (2010) 124(1):10–25. doi:10.1016/j.exppara.2009.03.0132010
62. Jones JL, Lopez B, Mury MA, Wilson M, Klein R, Luby S, et al. Toxoplasma gondii infection in rural Guatemalan children. Am J Trop Med Hyg (2005) 72(3):295–300. doi:10.4269/ajtmh.2005.72.295
63. Krueger WS, Hilborn ED, Converse RR, Wade TJ. Drinking water source and human Toxoplasma gondii infection in the United States: a cross-sectional analysis of NHANES data. BMC Public Health (2014) 14:711. doi:10.1186/1471-2458-14-711
64. Sroka J, Wojcik-Fatla A, Dutkiewicz J. Occurrence of Toxoplasma gondii in water from wells located on farms. Ann Agric Environ Med (2006) 13(1):169–75.
65. Jacobsen KH, Koopman JS. The effects of socioeconomic development on worldwide hepatitis A virus seroprevalence patterns. Int J Epidemiol (2005) 34(3):600–9. doi:10.1093/ije/dyi062
66. Ouardani I, Manso CF, Aouni M, Romalde JL. Efficiency of hepatitis A virus removal in six sewage plants from central Tunisia. Appl Microbiol Biotechnol (2015) 99(24):10759–69. doi:10.1007/s00253-015-6902-9
67. Wallender EK, Ailes EC, Yoder JS, Roberts VA, Brunkard JM. Contributing factors to disease outbreaks associated with untreated groundwater. Ground Water (2014) 52(6):886–97. doi:10.1111/gwat.12121
68. Sinclair RG, Jones EL, Gerba CP. Viruses in recreational water-borne disease outbreaks: a review. J Appl Microbiol (2009) 107(6):1769–80. doi:10.1111/j.1365-2672.2009.04367.x
69. Lodder WJ, de Roda Husman AM. Presence of noroviruses and other enteric viruses in sewage and surface waters in The Netherlands. Appl Environ Microbiol (2005) 71(3):1453–61. doi:10.1128/AEM.71.3.1453-1461.2005
70. Kukkula M, Arstila P, Klossner ML, Maunula L, Bonsdorff CH, Jaatinen P. Waterborne outbreak of viral gastroenteritis. Scand J Infect Dis (1997) 29(4):415–8. doi:10.3109/00365549709011840
71. Kukkula M, Maunula L, Silvennoinen E, von Bonsdorff CH. Outbreak of viral gastroenteritis due to drinking water contaminated by Norwalk-like viruses. J Infect Dis (1999) 180(6):1771–6. doi:10.1086/315145
72. Parshionikar SU, Willian-True S, Fout GS, Robbins DE, Seys SA, Cassady JD, et al. Waterborne outbreak of gastroenteritis associated with a norovirus. Appl Environ Microbiol (2003) 69(9):5263–8. doi:10.1128/AEM.69.9.5263-5268.2003
73. Baron RC, Murphy FD, Greenberg HB, Davis CE, Bregman DJ, Gary GW, et al. Norwalk gastrointestinal illness: an outbreak associated with swimming in a recreational lake and secondary person-to-person transmission. Am J Epidemiol (1982) 115:163–72. doi:10.1093/oxfordjournals.aje.a113287
74. Zlot A, Simckes M, Vines J, Reynolds L, Sullivan A, Scott AK, et al. Norovirus outbreak associated with a natural lake used for recreation—Oregon, 2014. MMWR Morb Mortal Wkly Rep (2015) 64(18):485–90. doi:10.1111/ajt.13404
75. Wade TJ, Sams EA, Haugland R, Brenner KP, Li Q, Wymer W, et al. Report on 2009 National Epidemiologic and Environmental Assessment of Recreational Water Epidemiology Studies. Chapel Hill, NC: USEPA (2009). p. 1–449.
76. Cretich M, Daaboul GG, Sola L, Unlu MS, Chiari M. Digital detection of biomarkers assisted by nanoparticles: application to diagnostics. Trends Biotechnol (2015) 33(6):343–51. doi:10.1016/j.tibtech.2015.03.002
77. Clotilde LM, Bernard CT, Hartman GL, Lau DK, Carter JM. Microbead-based immunoassay for simultaneous detection of Shiga toxins and isolation of Escherichia coli O157 in foods. J Food Prot (2011) 74(3):373–9. doi:10.4315/0362-028X.JFP-10-344
78. Baughman AL, Bisgard KM, Edwards KM, Guris D, Decker MD, Holland K, et al. Establishment of diagnostic cutoff points for levels of serum antibodies to pertussis toxin, filamentous hemagglutinin, and fimbriae in adolescents and adults in the United States. Clin Diagn Lab Immunol (2004) 11(6):1045–53. doi:10.1128/CDLI.11.6.1045-1053.2004
79. Ajami NJ, Kavanagh OV, Ramani S, Crawford SE, Atmar RL, Jiang ZD, et al. Seroepidemiology of norovirus-associated travelers’ diarrhea. J Travel Med (2014) 21(1):6–11. doi:10.1111/jtm.12092
80. Nikakhlagh S, Samarbafzadeh AR, Jahani M, Poostchi H, Kayedani GA, Naghashpoor M, et al. Determining the role of Helicobacter pylori in chronic sinus infections using the polymerase chain reaction. Jundishapur J Microbiol (2015) 8(3):e20783. doi:10.5812/jjm.20783
81. Klevens RM, Kruszon-Moran D, Wasley A, Gallagher K, McQuillan GM, Kuhnert W, et al. Seroprevalence of hepatitis A virus antibodies in the U.S.: results from the National Health and Nutrition Examination Survey. Public Health Rep (2011) 126(4):522–32. doi:10.1177/003335491112600408
82. Sivadon-Tardy V, Orlikowski D, Porcher R, Ronco E, Caudie C, Roussi J, et al. Detection of Campylobacter jejuni by culture and real-time PCR in a French cohort of patients with Guillain-Barre syndrome. J Clin Microbiol (2010) 48(6):2278–81. doi:10.1128/JCM.00381-10
83. Jones JL, Kruszon-Moran D, Wilson M, McQuillan G, Navin T, McAuley JB. Toxoplasma gondii infection in the United States: seroprevalence and risk factors. Am J Epidemiol (2001) 154(4):357–65. doi:10.1093/aje/154.4.357
84. Spina A, Kerr KG, Cormican M, Barbut F, Eigentler A, Zerva L, et al. Spectrum of enteropathogens detected by FilmArray(R) GI Panel in a multi-centre study of community-acquired gastroenteritis. Clin Microbiol Infect (2015) 21(8):719–28. doi:10.1016/j.cmi.2015.04.007
85. Thongprachum A, Takanashi S, Kalesaran AF, Okitsu S, Mizuguchi M, Hayakawa S, et al. Four-year study of viruses that cause diarrhea in Japanese pediatric outpatients. J Med Virol (2015) 87(7):1141–8. doi:10.1002/jmv.24155
86. Osborne CM, Montano AC, Robinson CC, Schultz-Cherry S, Dominguez SR. Viral gastroenteritis in children in Colorado 2006-2009. J Med Virol (2015) 87(6):931–939. doi:10.1002/jmv.24022
87. Xue Y, Pan H, Hu J, Wu H, Li J, Xiao W, et al. Epidemiology of norovirus infections among diarrhea outpatients in a diarrhea surveillance system in Shanghai, China: a cross-sectional study. BMC Infect Dis (2015) 15(1):183. doi:10.1186/s12879-015-0922-z
88. Arevalo JH, Hassig CA, Stura EA, Sims MJ, Taussig MJ, Wilson IA. Structural analysis of antibody specificity. Detailed comparison of five Fab’-steroid complexes. J Mol Biol (1994) 241(5):663–90. doi:10.1006/jmbi.1994.1543
89. James LC, Tawfik DS. The specificity of cross-reactivity: promiscuous antibody binding involves specific hydrogen bonds rather than nonspecific hydrophobic stickiness. Protein Sci (2003) 12(10):2183–93. doi:10.1110/ps.03172703
90. Kramer A, Keitel T, Winkler K, Stocklein W, Hohne W, Schneider-Mergener J. Molecular basis for the binding promiscuity of an anti-p24 (HIV-1) monoclonal antibody. Cell (1997) 91(6):799–809. doi:10.1016/S0092-8674(00)80468-7
91. Trinh CH, Hemmington SD, Verhoeyen ME, Phillips SE. Antibody fragment Fv4155 bound to two closely related steroid hormones: the structural basis of fine specificity. Structure (1997) 5(7):937–48. doi:10.1016/S0969-2126(97)00247-5
92. Waterboer T, Sehr P, Pawlita M. Suppression of non-specific binding in serological Luminex assays. J Immunol Methods (2006) 309(1–2):200–4. doi:10.1016/j.jim.2005.11.008
93. Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, et al. Multiplex human papillomavirus serology based on in situ-purified glutathione S-transferase fusion proteins. Clin Chem (2005) 51(10):1845–53. doi:10.1373/clinchem.2005.052381
94. Studentsov YY, Schiffman M, Strickler HD, Ho GY, Pang YY, Schiller J, et al. Enhanced enzyme-linked immunosorbent assay for detection of antibodies to virus-like particles of human papillomavirus. J Clin Microbiol (2002) 40(2002):1755. doi:10.1128/JCM.40.5.1755-1760.2002
95. Brandtzaeg P. Secretory immunity with special reference to the oral cavity. J Oral Microbiol (2013) 5:1. doi:10.3402/jom.v5i0.20401
96. Ashbolt NJ. Risk analysis of drinking water microbial contamination versus disinfection by-products (DBPs). Toxicology (2004) 198(1–3):255–62. doi:10.1016/j.tox.2004.01.034
Keywords: multiplex, immunoassay, salivary antibody, saliva, exposure, bead-based, immunoprevalence, coimmunopositivity
Citation: Augustine SAJ, Simmons KJ, Eason TN, Curioso CL, Griffin SM, Wade TJ, Dufour A, Fout GS, Grimm AC, Oshima KH, Sams EA, See MJ and Wymer LJ (2017) Immunoprevalence to Six Waterborne Pathogens in Beachgoers at Boquerón Beach, Puerto Rico: Application of a Microsphere-Based Salivary Antibody Multiplex Immunoassay. Front. Public Health 5:84. doi: 10.3389/fpubh.2017.00084
Received: 07 September 2016; Accepted: 03 April 2017;
Published: 01 May 2017
Edited by:
Paul Desmond Slowey, Oasis Diagnostics, USACopyright: © 2017 Augustine, Simmons, Eason, Curioso, Griffin, Wade, Dufour, Fout, Grimm, Oshima, Sams, See and Wymer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Swinburne A. J. Augustine, YXVndXN0aW5lLnN3aW5idXJuZUBlcGEuZ292
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.