
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Psychiatry , 09 January 2024
Sec. Autism
Volume 14 - 2023 | https://doi.org/10.3389/fpsyt.2023.1318637
 Ping Lin1,2†
Ping Lin1,2† Qianwen Zhang3,4†
Qianwen Zhang3,4† Junyu Sun5†
Junyu Sun5† Qingtian Li6
Qingtian Li6 Dan Li2
Dan Li2 Mengyuan Zhu2
Mengyuan Zhu2 Xiaomei Fu2
Xiaomei Fu2 Ling Zhao2
Ling Zhao2 Mengxia Wang2
Mengxia Wang2 Xiaoyan Lou2
Xiaoyan Lou2 Qing Chen2
Qing Chen2 Kangyi Liang2
Kangyi Liang2 Yuxin Zhu2
Yuxin Zhu2 Caiwei Qu2
Caiwei Qu2 Zhenhua Li2
Zhenhua Li2 Peijun Ma2
Peijun Ma2 Renyu Wang2
Renyu Wang2 Huafen Liu3,4
Huafen Liu3,4 Ke Dong7
Ke Dong7 Xiaokui Guo7
Xiaokui Guo7 Xunjia Cheng1*
Xunjia Cheng1* Yang Sun8*
Yang Sun8* Jing Sun9,10*
Jing Sun9,10*Introduction: Autism spectrum disorder (ASD) is a multifaceted developmental condition that commonly appears during early childhood. The etiology of ASD remains multifactorial and not yet fully understood. The identification of biomarkers may provide insights into the underlying mechanisms and pathophysiology of the disorder. The present study aimed to explore the causes of ASD by investigating the key biomedical markers, trace elements, and microbiota factors between children with autism spectrum disorder (ASD) and control subjects.
Methods: Medline, PubMed, ProQuest, EMBASE, Cochrane Library, PsycINFO, Web of Science, and EMBSCO databases have been searched for publications from 2012 to 2023 with no language restrictions using the population, intervention, control, and outcome (PICO) approach. Keywords including “autism spectrum disorder,” “oxytocin,” “GABA,” “Serotonin,” “CRP,” “IL-6,” “Fe,” “Zn,” “Cu,” and “gut microbiota” were used for the search. The Joanna Briggs Institute (JBI) critical appraisal checklist was used to assess the article quality, and a random model was used to assess the mean difference and standardized difference between ASD and the control group in all biomedical markers, trace elements, and microbiota factors.
Results: From 76,217 records, 43 studies met the inclusion and exclusion criteria and were included in this meta-analysis. The pooled analyses showed that children with ASD had significantly lower levels of oxytocin (mean differences, MD = −45.691, 95% confidence interval, CI: −61.667, −29.717), iron (MD = −3.203, 95% CI: −4.891, −1.514), and zinc (MD = −6.707, 95% CI: −12.691, −0.722), lower relative abundance of Bifidobacterium (MD = −1.321, 95% CI: −2.403, −0.238) and Parabacteroides (MD = −0.081, 95% CI: −0.148, −0.013), higher levels of c-reactive protein, CRP (MD = 0.401, 95% CI: 0.036, 0.772), and GABA (MD = 0.115, 95% CI: 0.045, 0.186), and higher relative abundance of Bacteroides (MD = 1.386, 95% CI: 0.717, 2.055) and Clostridium (MD = 0.281, 95% CI: 0.035, 0.526) when compared with controls. The results of the overall analyses were stable after performing the sensitivity analyses. Additionally, no substantial publication bias was observed among the studies.
Interpretation: Children with ASD have significantly higher levels of CRP and GABA, lower levels of oxytocin, iron, and zinc, lower relative abundance of Bifidobacterium and Parabacteroides, and higher relative abundance of Faecalibacterium, Bacteroides, and Clostridium when compared with controls. These results suggest that these indicators may be a potential biomarker panel for the diagnosis or determining therapeutic targets of ASD. Furthermore, large, sample-based, and randomized controlled trials are needed to confirm these results.
Autism spectrum disorder (ASD) is a multifaceted developmental condition that commonly appears during early childhood. It is marked by difficulties in social interaction, challenges in communication, and the presence of repetitive behaviors or fixations (1). According to the estimation by the American Centers for Disease Control and Prevention Center (CDC), one in 44 children aged 8 years was diagnosed with autism in 2018 (2). The prevalence of autism was approximately 1% globally (3). The risk in boys is 3–4 times higher than that in girls, and male autistic patients tend to exhibit more obvious signs than female counterparts (4). The current clinical diagnosis of autism is largely dependent on the fifth version of Diagnostic and Statistical Manual of Mental Disorder (DSM-5) by the American Psychiatric Association (1). ASD is usually first diagnosed in toddlerhood, with many of the most obvious signs (e.g., stop acquiring or losing previously gained skills) presenting around 2–3 years old; however, clear predictions of later cognitive impairment are sometimes difficult in children at 2–3 years of age (1).
Despite extensive research efforts, the etiology of ASD remains multifactorial and not yet fully understood (5–9). In recent years, studies in biomarkers have found that patients with ASD have a higher level of inflammatory factors, such as C-reactive protein (CRP) (10, 11), a lower tracer elements level including iron (Fe) (12, 13) and zinc (Zn) (14, 15), and a lower abundance of beneficial microbiota bacteria (16–19) than healthy patients. The identification of biomarkers may provide insights into the underlying mechanisms and pathophysiology of the disorder (20–22). However, there is no study that has systematically assessed the characteristics of ASD patients in comparison with healthy people. This study aimed to fill in this research gap by comprehensively evaluating a wide range of potential biomarkers associated with ASD, including biomedical markers, trace elements, and microbiota factors, using the meta-analysis method. This method included potential biomedical markers (oxytocin, γ-aminobutyric acid, i.e., GABA, serotonin, CRP, and IL-6), trace elements (Fe, Zn, and Cu), and microbiota factors (Bifidobacterium, Faecalibacterium, Parabacteroides, Bacteroides, and Clostridium) in ASD. The inclusion of specific biomarkers in this study was predicated upon their established roles within biological pathways pertinent to ASD. Oxytocin, GABA, serotonin, CRP, and IL-6 were selected due to their documented involvement in neurodevelopmental processes, neurotransmission modulation, and immune system regulation—all of which are fundamental elements intertwined with ASD pathophysiology (10, 23–25). These biomarkers are integral to understanding the neurobiological substrates governing social cognition, emotional regulation, and behavioral responses, all of which are significantly impaired in individuals with ASD. Furthermore, the incorporation of trace elements, i.e., Fe, Zn, and Cu, stems from their crucial roles as cofactors in essential enzymatic reactions pivotal for brain development and synaptic functioning (26–29). Perturbations in their levels have exhibited correlations with ASD-related traits, necessitating their examination within this context. Additionally, the exploration of microbiota factors, including Bifidobacterium, Faecalibacterium, Parabacteroides, Bacteroides, and Clostridium, aligns with burgeoning research elucidating the bidirectional communication between the gut microbiome and the central nervous system (30–34). These factors potentially influence neurodevelopment and behavior, prompting their investigation in relation to ASD. By examining and synthesizing the existing literature on these biomarkers, this study aimed to enhance the understanding of the biological underpinnings of ASD and potentially contribute to the development of improved diagnostic and therapeutic approaches.
This meta-analysis was prepared in accordance with the preferred reporting items for systematic reviews and meta-analyses (PRISMA) standards (35) (Supplementary eMethods 1). Medline, PubMed, ProQuest, EMBASE, Cochrane Library, PsycINFO, Web of Science, and EMBSCO databases have been searched for publications from 2012 to 2023 with the following keywords: “autism spectrum disorder,” “oxytocin,” “GABA,” “Serotonin,” “CRP,” “IL-6,” “Fe,” “Zn,” “Cu,” and “gut microbiota” (Supplementary eMethods 2).
Using population, intervention, control, and outcome (PICO) approach, the study included the following criteria: (i) P: studies conducted with ASD participants aged 18 years or younger, (ii) C: assessment of participants with ASD and controls, and (iii) O: reporting oxytocin, GABA, serotonin, CRP, IL-6, Fe, Zn, Cu, and gut microbiota levels with available data for the meta-analysis. Studies with full-text available were included regardless of language. As intervention studies are not available, the present study focused on observational studies, including cross-sectional, case-control, or prospective cohort studies. Any studies that did not meet the aforementioned criteria, e.g., P: patients older than 18 years old; C: control groups who are not healthy or have other mental disorders; and O: outcomes focusing on other types of biomarkers, were excluded.
The characteristics of the studies, including the following data, were extracted from each study: first author’s surname, year of publication, country, study design, total population, number of male participants, mean age, age range, biomarker type, contributed biomarker, study outcome, and statistics for meta-analysis (mean, standard deviation, and participant number in ASD and control groups).
Finally, for outcomes including biomedical markers and trace elements, the mean and standard deviation of concentrations were extracted and for microbiota factors, the genera level of relative abundance (%) was extracted.
The Joanna Briggs Institute (JBI) critical appraisal checklist was used to assess the article quality (36). The total JBI scores possible for a cohort study is 12 (37), for case-control study is 10 (38), and for cross-sectional study is 8 (39).
For biomedical markers and trace elements, the mean and standard deviation of concentrations were collected; for microbiota factors, the genera level of relative abundance (%) was recorded. Mean differences (MDs) were used as the primary index of comparative results between ASD and the control group, and standard mean differences (SMDs) were used as the effect size. Heterogeneity across the studies was examined using the I2 statistics. Low heterogeneity was indicated by I2 less than 25%, moderate heterogeneity by I2 of approximately 50%, and substantial heterogeneity by I2 of 75% (40). The significance was quantified with Cochrane’s Q statistics. Random-effects models were chosen if heterogeneity by I2 is approximately 50% across studies resulting from differences in subjects and measurements. Sensitivity analysis was performed to avoid any single study influencing the results of the meta-analysis. To access the publication bias, Egger’s test was performed (41), and forest plots were performed to visualize the results. A two-tailed p-value of <0.05 was considered significant. The analyses were performed in a comprehensive meta-analysis (version 3) (42).
The selection process for the included studies is shown in Figure 1. From the 76,217 potentially eligible articles published between January 1967 and May 2023, 3,386 publications remained after removing duplicates and restricting the publication year from 2012 to 2023. Following the screening of the title and abstracts, the full-text of 227 publications were assessed, and 184 publications were excluded for the following reasons: review or meta-analysis, unavailability of the full-text, inappropriate sample format, wrong study design, wrong study population, insufficient data, low quality, incomparability of the comparison and control groups, and other diseases. Finally, 43 publications satisfied the eligibility criteria and were included in the meta-analysis.
The study characteristics are summarized in Table 1. A total of 17 articles had the results of biomedical markers (10, 11, 24, 43–56); 8 articles had the results of trace elements (12–15, 57–60); 16 articles had the results of microbiota factors (16–19, 63–74); and 2 articles had the results for both biomedical markers and microbiota factors (61, 62). These studies were performed in 16 countries, including 16 studies in China (10, 11, 14, 17, 18, 47, 55, 58, 61, 63, 64, 66, 67, 69, 70), 5 studies in the USA (52, 59, 71, 72, 74), 4 studies in Iraq (43, 44, 48, 49), 3 studies in Egypt (13, 45, 51), 3 studies in Italy (16, 19, 68), 2 studies in Jordan (15, 53), 2 studies in Turkey (54, 56), and 1 study each in Ecuador (62), India (73), Japan (46), Malaysia (60), Qatar (12), Russia (57), Saudi Arabia (24), Spain (65), and Tunisia (50). In this pooled review, 35 articles were case-control studies and their JBI scores ranged from 6 to 10; 7 articles were cross-sectional studies and their JBI scores ranged from 5 to 6; and 1 article was a cohort study and its JBI score was 10.
Three studies (10, 11, 54), with a total of 616 participants, provided sufficient information for a meta-analysis of CRP (Table 2). There was a statistically significant difference in the mean CRP concentration between ASD and control, with an overall mean difference of 0.401 (95% CI: 0.036, 0.772, p = 0.034), indicating that the ASD group had a higher CRP concentration than the healthy control group. The effect size was moderate, and the SMD was 0.497 (95% CI: 0.219, 0.775, p < 0.001). The heterogeneity was significant (I2 = 91.968%, p < 0.0001). The forest plot of the effect is shown in Figure 2A.
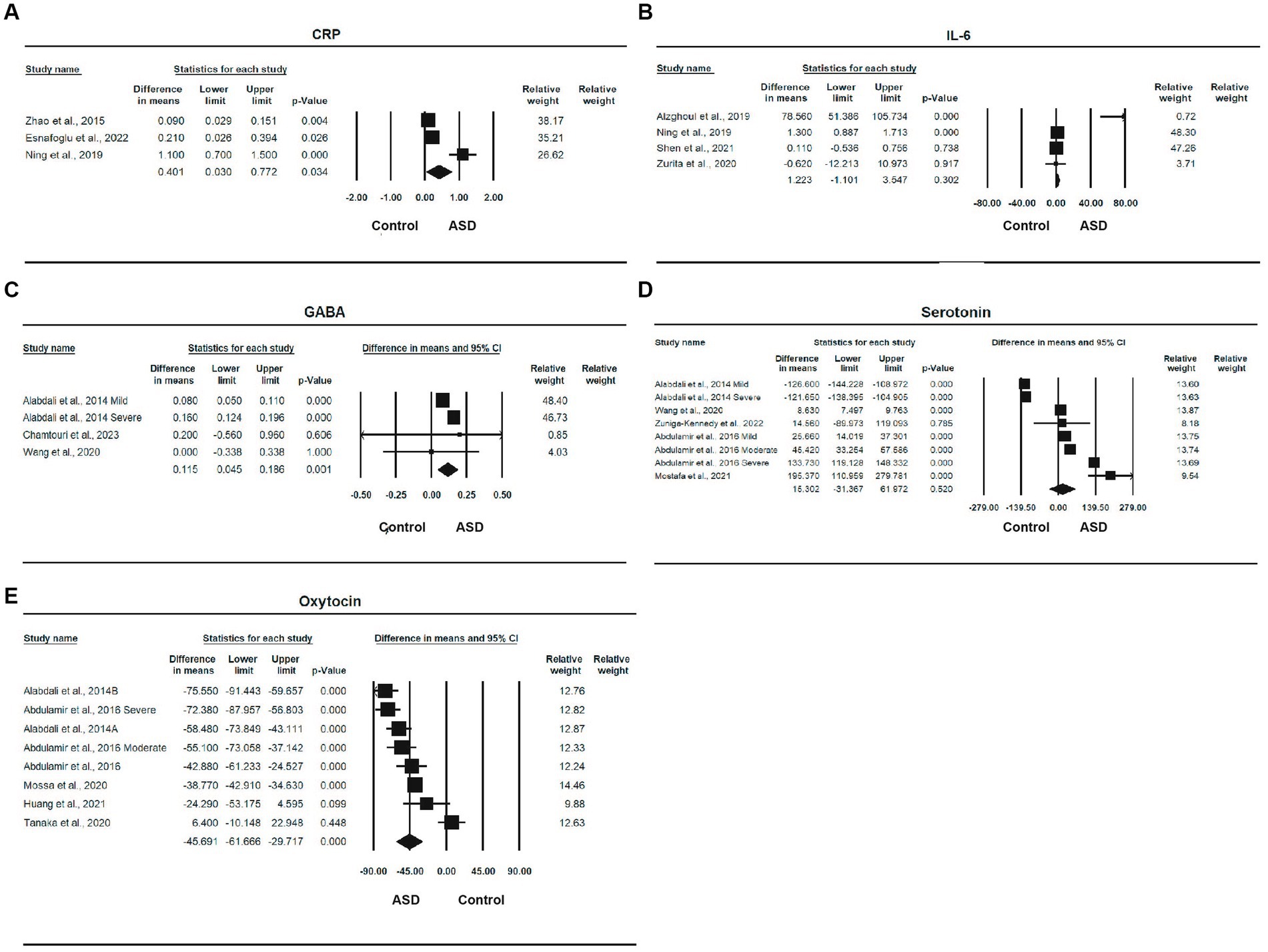
Figure 2. Forest plots of biomedical markers between ASD and control; (A) CRP; (B) IL-6; (C) GABA; (D) Serotonin; (E) Oxytocin.
Four studies (10, 53, 55, 62), with a total of 510 participants, provided sufficient information for a meta-analysis of IL-6. The MD between ASD and control was 1.223 (95% CI, −1.101, 3.547, p = 0.302), and the ASD group had a higher level of IL-6 than the control group. The effect size was moderate, with an SMD of 0.485 (95% CI, 0.034, 0.936, p < 0.05). The heterogeneity was significant (I2 = 92.622%, p < 0.0001). The forest plot of the MD is shown in Figure 2B.
Three studies (24, 50, 61) involving a total of 166 participants provided sufficient information for a meta-analysis of GABA. There was a statistically significant difference in the mean GABA concentration between ASD and control, with an overall MD of 0.115 (95% CI: 0.045, 0.186, p = 0.001), and the ASD group had a higher GABA concentration than the control group. There effect size was large, the SMD was 1.163 (95% CI, −0.022, 2.347, p = 0.054), and the heterogeneity was also statistically significant (I2 = 74.260%, p < 0.0001). One study was removed due to sensitivity analysis (45). The forest plot of the effect is shown in Figure 2C.
Five studies (24, 44, 51, 52, 61) involving a total of 283 participants provided sufficient information for a meta-analysis of serotonin (Table 2). The MD between ASD and control was 15.302 (95% CI: −31.367, 61.972), indicating that the ASD group had a higher serotonin level than the control group, but the difference was not statistically significant (p = 0.520). The effect size was large, and the SMD was 0.825 (95% CI: −1.127, 2.776, p = 0.407); although the difference was also not statistically significant, heterogeneity was significant (I2 = 99.128%, p < 0.0001). The forest plot of serotonin is shown in Figure 2D.
Five studies (24, 43, 46, 47, 49) involving a total of 419 participants provided sufficient information for a meta-analysis of oxytocin. There was a statistically significant difference in the mean oxytocin concentration between ASD and control, with a mean difference of −45.691 (95% CI: −61.667, −29.717, p < 0.0001), and the ASD group had a lower oxytocin level than the control group and a very large effect size, with an SMD of −1.849 (95% CI: 2.796, −0.903, p < 0.0001). One study was removed due to sensitivity analysis (48). The heterogeneity was also significant (I2 = 90.646%, p < 0.0001). The forest plot of the effect is shown in Figure 2E.
Four studies (14, 15, 57, 58) involving a total of 577 participants provided sufficient information for a meta-analysis of Cu (Table 2). The MD between ASD and control was 0.293 (95% CI: −1.349, 1.935, p = 0.726), indicating that the ASD group had a high Cu level than the control and a small effect size, with an SMD of 0.059 (95% CI: −0.368, 0.485, p = 0.788). The heterogeneity was significant (I2 = 84.559%, p < 0.0001). The forest plot of the effect is shown in Figure 3A.
Six studies (12, 13, 15, 57, 58, 60) involving a total of 1,313 participants provided sufficient information for a meta-analysis of Fe (Table 2). There was a statistically significant difference in the mean iron concentration between ASD and control, with an MD of −3.203 (95% CI: −4.891, −1.514, p < 0.0001), and the ASD group had a lower iron level than the control and a smaller effect size, with a standardized mean difference of −0.375 (95% CI: −0.743, −0.007, p = 0.046). The heterogeneity was also significant (I2 = 95.032%, p < 0.0001). The forest plot of the effect is shown in Figure 3B.
Five studies (14, 15, 57–59), with a total of 706 participants, provided sufficient information for a meta-analysis of Zn (Table 2). There was a statistically significant difference in the mean zinc concentration between ASD and control, with a mean difference of −6.707 (95% CI: −12.691, −0.722, p = 0.028), and the ASD group had a lower zinc level than the control and a moderate effect size, with a standardized mean difference of −0.498 (95% CI: −0.958, −0.037, p = 0.034). The heterogeneity was also significant (I2 = 95.055%, p < 0.0001). The forest plot of the effect is shown in Figure 3C.
Eleven studies (16–19, 61, 63–66, 72, 74) involving a total of 1,067 participants provided sufficient information for a meta-analysis of Bifidobacterium (Table 2). There was a statistically significant difference in the relative abundance of Bifidobacterium between ASD and control, with a mean difference of −1.321 (95% CI: −2.403, −0.238, p = 0.019), and the ASD group had a smaller abundance than the control group. The effect size was small, with an SMD of −0.311 [95% CI: −1.005, 0.383, which was not statistically significant (p = 0.380)]. The heterogeneity was also significant (I2 = 99.184%, p < 0.0001). The forest plot of the effect is shown in Figure 4A.
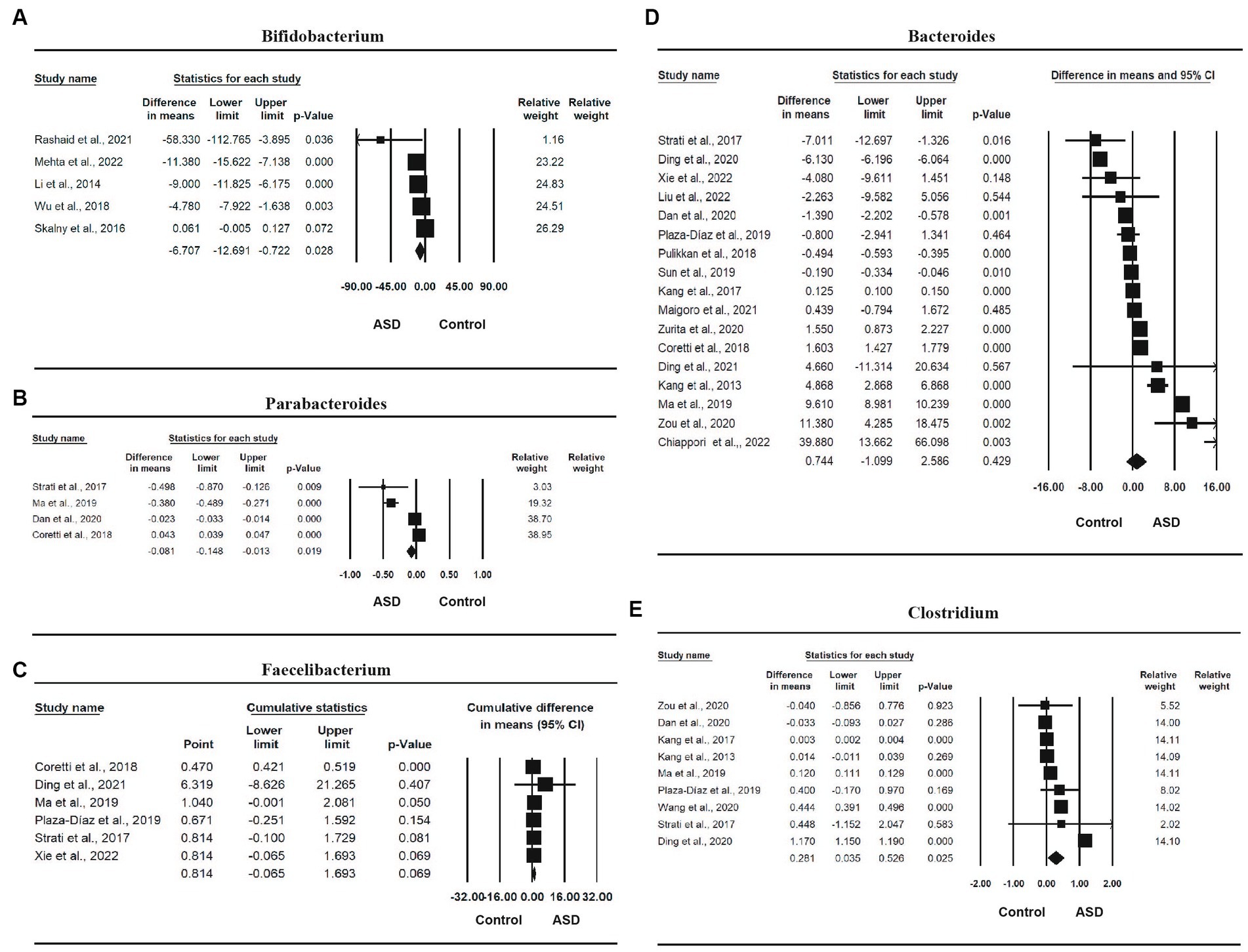
Figure 4. Forest plots of the relative abundance of microbiota factors between ASD and control; (A) Bifidobacterium; (B) Faecalibacterium; (C) Parabacteroides; (D) Bacteroides; (E) Clostridium.
Four studies (16, 18, 19, 66) involving a total of 481 participants were included in the meta-analysis of Parabacteroides. There was a statistically significant difference in the relative abundance of Parabacteroides between ASD and control, with a mean difference of −0.081 (95% CI: −0.148, −0.013, p = 0.019), and the ASD group had a smaller abundance than the control group in Parabacteroides. The effect size was small, with an SMD of 0.287 (95% CI: −0.837, 1.410), which was not statistically significant (p = 0.617). One study was removed due to sensitivity analysis (65). The heterogeneity was also significant (I2 = 98.625%, p < 0.0001). The forest plot of the effect is presented in Figure 4B.
Six studies (16, 17, 19, 64–66) involving a total of 501 participants were included in the meta-analysis of Faecalibacterium. The mean difference of relative abundance between ASD and control was 0.814 (95% CI: −0.065, 1.693, p = 0.069), the ASD group had a higher abundance than the control group, and the effect size was large, with an SMD of 1.144 (95% CI: 0.296, 1.992), which was statistically significant (p = 0.008). One study (63) was removed due to sensitivity analysis, and another study (18) was removed due to publication bias and Egger’s test being less than 0.05. The heterogeneity was significant (I2 = 90.051%, p < 0.0001). The forest plot of the effect is shown in Figure 4C.
Sixteen studies (16–19, 62, 64–74) involving a total of 1,257 participants were included in the meta-analysis of Bacteroides. There was a statistically significant difference in the relative abundance of Bacteroides between ASD and control, with a mean difference of 1.386 (95% CI: 0.717, 2.055, p < 0.0001), and the ASD group had a higher abundance than the control group, and the effect size is large, with a standardized mean difference of 0.287 (95% CI: −0.837, 1.410, p = 0.01). One study was removed due to sensitivity analysis (63). The heterogeneity was also significant (I2 = 98.923%, p < 0.0001). The forest plot of the effect is shown in Figure 4D.
Finally, nine studies (18, 19, 61, 63, 65, 66, 69, 72, 74) involving a total of 912 participants were included in the meta-analysis of Clostridium (Table 2). There was a statistically significant difference in the relative abundance of Clostridium between ASD and control, with a mean difference of 0.281 (95% CI: 0.035, 0.526, p = 0.025), and the ASD group had a higher abundance than the control group, and a large effect size, with a standardized mean difference of 2.994 (95% CI: 1.724, 4.265, p < 0.0001). The heterogeneity was also significant (I2 = 99.942%, p < 0.0001). The forest plot of the effect is shown in Figure 4E.
Findings from the Egger’s test (Table 3), it can be suggested that there was no publication bias for all markers (CRP: p = 0.70; GABA: p = 1.00; serotonin: p = 0.99; oxytocin: p = 0.14; Cu: p = 0.56, Fe: p = 0.86; Zn: p = 0.92; Bifidobacterium: p = 0.71; Parabacteroides: p = 0.25; Faecalibacterium: p = 0.16; Bacteroides: p = 0.31; and Clostridium: p = 0.23), except IL-6 (p = 0.05).
The pooled effects based on our meta-analyses found that ASD youth have significantly higher levels of CRP and GABA, lower levels of oxytocin, iron, and zinc, lower relative abundance of Bifidobacterium and Parabacteroides, and higher relative abundance of Faecalibacterium, Bacteroides, and Clostridium when compared with control youth. To our knowledge, this is the first meta-analysis providing a wide range of potential biomarkers for children with ASD.
Inflammatory processes have gained increasing attention as potential contributors to the pathophysiology of ASD. Inflammation can impact brain development and function, and alterations in immune system markers have been observed in individuals with ASD (75, 76). CRP and IL-6 are two of the most commonly studied inflammatory markers and have shown associations with ASD in previous research (25, 77). CRP is an acute-phase protein that reflects systemic inflammation (78, 79). IL-6, a pro-inflammatory cytokine, is involved in immune reactions and synaptic plasticity (80, 81). In this meta-analysis, we have found that the CRP levels were significantly higher in ASD compared with controls, which is consistent with previous studies (77, 82). High levels of CRP could increase the blood-brain barrier (BBB) paracellular permeability, activating the microglia to impair the central nervous system (CNS) (83). Previous studies have pointed out that microglia play key roles in the pathogenesis of ASD (84–86). Thus, there might be a causal relationship between CRP levels and ASD, and CRP could be a potential biomarker for autism diagnosis. Elevated IL-6 levels have been documented not solely in autoimmune disorders but also in certain neurodegenerative conditions, such as Alzheimer’s disease, as well as diverse mental disorders (42, 87). Additionally, studies also reported that children who were later diagnosed with ASD have elevated levels of IL-6 during gestation, as IL-6 may have transferred across the placental barriers and accumulated in the fetuses (88, 89).
Neurotransmitters have also been implicated in the regulation of various neurodevelopmental processes, including social cognition, emotional regulation, and sensory integration (90, 91). Studies have suggested alterations in the levels and functioning of these neurotransmitters in individuals with ASD (92). Oxytocin, known for its role in social bonding and affiliation, has shown promise as a potential biomarker for ASD (93–95). Similarly, dysregulation of gamma-aminobutyric acid (GABA), the primary inhibitory neurotransmitter in the brain, has been implicated in the pathogenesis of ASD (96–98). Serotonin, involved in mood regulation and sensory processing, has also been the subject of investigation, with studies reporting variations in serotonin levels in individuals with ASD (52, 99, 100). In this meta-analysis, the ASD groups have significantly higher levels of GABA and lower levels of oxytocin compared with control groups. A preliminary meta-analysis has provided support for a potential link between autism spectrum disorder and variations in the oxytocin receptor gene, OXTR (94). However, this association does not reach the level of significance when considering the entire genome. Additionally, numerous studies have been conducted to explore the potential of oxytocin therapy in individuals with autism spectrum disorders (93, 101–105). Despite the fact that the majority of these investigations have yielded inconclusive results, oxytocin could be developed as a potential biomarker for autism diagnosis.
Trace elements play crucial roles in various biological processes, including neurotransmitter synthesis (106), antioxidant defense (107), and immune function (108). Imbalances in these trace elements have been reported in individuals with ASD, suggesting a potential link between their dysregulation and the pathogenesis of the disorder (109, 110). Iron deficiency, for instance, has been associated with impaired cognitive function and socioemotional difficulties observed in ASD (111, 112). Similarly, alterations in Zn and Cu levels have been reported, potentially affecting neurotransmitter systems and oxidative stress (113, 114). In this meta-analysis, we have found lower levels of Fe and Zn in the ASD groups. While it is acknowledged that there is a mineral imbalance in ASD, we anticipate the implementation of pertinent examinations and the establishment of standard levels for trace elements as potential biomarkers that can be valuable for diagnosing, preventing, and treating ASD.
Emerging research has also highlighted the potential involvement of the gut microbiome in ASD (115, 116). The gut microbiota, a diverse community of microorganisms residing in the gastrointestinal tract, is known to influence brain development and function through bidirectional communication with the central nervous system (117, 118). Altered microbial compositions and imbalances in microbial metabolites have been reported in individuals with ASD, suggesting a potential role for the gut microbiome as a biomarker and therapeutic target for the disorder (119–121). Specific bacterial genera, such as Bifidobacterium, Faecalibacterium, Parabacteroides, Bacteroides, and Clostridium, have been investigated in this meta-analysis. As a result, Bifidobacterium and Parabacteroides as protective bacteria showed a significantly lower abundance in the ASD group, while Faecalibacterium, Bacteroides, and Clostridium showed a significantly higher abundance in the ASD group, suggesting that there is indeed dysbiosis in ASD patients. Several studies have reported a lesser abundance of beneficial bacteria, such as Bifidobacterium in autism (16, 61, 65), leading to clinical trials with probiotic treatment (122–125). With the intervention of probiotics, children with ASD have shown improved autistic behavioral scores as well as gastrointestinal symptoms. In addition to the low abundance of beneficial bacteria, an increased abundance of bacteria such as Faecalibacterium, Bacteroides, and Clostridium in autism was also reported (126). Butyrate-producing bacteria, Faecalibacterium, were reported to affect physiological functions and homeostasis in the gut and were closely associated with autism core symptoms (127). Bacteroides are the main producers of propionic acid (PPA), which is one of the neurotoxic short-chain fatty acids, and have been reported to cause autistic characteristics in animal models (128). Additional studies suggested that Bacteroides are the distinguishing differences between autism and control children (129). Notwithstanding their non-invasive nature, Clostridium possess the ability to elicit deleterious effects on distant organs or tissues, such as the brain, by means of their secreted molecules traversing the intestinal barrier and spreading via the systemic circulation to remote locations, where they exert their biological activity (126). It has been hypothesized that there is a potential association between Clostridium and symptoms related to ASD (130), with a particular emphasis on the significance of Clostridium’s ability to form spores in relation to the recurrence of ASD symptoms (131).
This study has several limitations. First, this meta-analysis was based on observational studies; therefore, we cannot establish a causal relationship between the proposed markers and ASD. These changes in the levels of the abovementioned markers could be either a cause or a consequence of the pathogenesis of ASD. Second, some of the markers have limited numbers of studies, i.e., CRP only had three studies. While the results of the meta-analysis markers were consistent with previous studies (77, 82), further research is recommended. Third, heterogeneity was high in this meta-analysis. Due to the paucity of information from each of the included studies, we were unable to investigate the underlying confounding factors for the reason of high heterogeneity by doing subgroup analysis. For example, this meta-analysis encountered complexities in categorizing participants into children and adolescents due to heterogeneous age reporting across the included studies. Ambiguities in age ranges and missing mean age data hindered robust subgroup analyses, highlighting the need for standardized and comprehensive age reporting in future ASD biomarker studies to elucidate age-specific trends more effectively. Future studies will be conducted to systematically assess the randomized controlled trial on these markers, and subgroup analysis will be conducted to identify the sources of high heterogeneity.
Despite the limitations, this meta-analysis also has several major strengths. First, we conducted a comprehensive literature search through a list of electronic databases. Second, our literature selection workflow was based on rigorous inclusion and exclusion criteria. Third, the publication bias for each type of biomarker in this meta-analysis was minimal. Finally, we have included a wide range of popular but controversial biomarkers, ranging from biomedical trace elements to gut microbiota.
In conclusion, the present meta-analysis found an association between the levels of CRP, GABA, oxytocin, Fe, Zn and the relative abundance of Bifidobacterium, Parabacteroides, Bacteroides, Clostridium, and ASD, suggesting that these indications may be promising biomarkers for ASD. Future investigation is necessary to determine whether these biomarker profiles are specific to ASD. Additionally, our study also indicated that enhancing the levels of these biomarkers could serve as an effective intervention and treatment strategies for ASD.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
PL: Conceptualization, Data curation, Writing – original draft, Supervision, Formal analysis, Investigation. QZ: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. JuS: Formal analysis, Writing – original draft, Data curation, Investigation. QL: Data curation, Formal analysis, Writing – original draft, Investigation. DL: Data curation, Writing – review & editing. MZ: Data curation, Writing – review & editing. XF: Data curation, Writing – review & editing. LZ: Data curation, Writing – review & editing. MW: Data curation, Writing – review & editing. XL: Data curation, Writing – review & editing. QC: Data curation, Writing – review & editing. KL: Data curation, Writing – review & editing. YZ: Data curation, Writing – review & editing. CQ: Data curation, Writing – review & editing. ZL: Data curation, Writing – review & editing. PM: Data curation, Writing – review & editing. RW: Data curation, Writing – review & editing. HL: Supervision, Writing – review & editing. KD: Data curation, Writing – review & editing. XG: Supervision, Writing – original draft, Data curation, Investigation. XC: Conceptualization, Formal analysis, Supervision, Writing – original draft, Data curation, Investigation. YS: Conceptualization, Investigation, Methodology, Supervision, Writing – review & editing, Data curation. JiS: Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study is supported by the China Public Health Alliance Batch 1 Research Project (GWLM202014); the Shanghai Mental Health Center Project (2017-YJ-14 and 2021-YJ-10); and the Shanghai Natural Science Fund Project under grant 20ZR1447700.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2023.1318637/full#supplementary-material
1. Lord, C, Elsabbagh, M, Baird, G, and Veenstra-Vanderweele, J. Autism spectrum disorder. Lancet. (2018) 392:508–20. doi: 10.1016/S0140-6736(18)31129-2
2. Maenner, MJ, Shaw, KA, Bakian, AV, Bilder, DA, Durkin, MS, Esler, A, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2018. MMWR Surveill Summ. (2021) 70:1–16. doi: 10.15585/mmwr.ss7011a1
3. Elsabbagh, M, Divan, G, Koh, YJ, Kim, YS, Kauchali, S, Marcin, C, et al. Global prevalence of autism and other pervasive developmental disorders. Autism Res. (2012) 5:160–79. doi: 10.1002/aur.239
4. Messinger, DS, Young, GS, Webb, SJ, Ozonoff, S, Bryson, SE, Carter, A, et al. Early sex differences are not autism-specific: a Baby Siblings Research Consortium (BSRC) study. Mol Autism. (2015) 6:32. doi: 10.1186/s13229-015-0027-y
5. Wiśniowiecka-Kowalnik, B, and Nowakowska, BA. Genetics and epigenetics of autism spectrum disorder-current evidence in the field. J Appl Genet. (2019) 60:37–47. doi: 10.1007/s13353-018-00480-w
6. Tye, C, Runicles, AK, Whitehouse, AJO, and Alvares, GA. Characterizing the interplay between autism spectrum disorder and comorbid medical conditions: an integrative review. Front Psychiatry. (2018) 9:751. doi: 10.3389/fpsyt.2018.00751
7. Nazeen, S, Palmer, NP, Berger, B, and Kohane, IS. Integrative analysis of genetic data sets reveals a shared innate immune component in autism spectrum disorder and its co-morbidities. Genome Biol. (2016) 17:228. doi: 10.1186/s13059-016-1084-z
8. Kim, JY, Son, MJ, Son, CY, Radua, J, Eisenhut, M, Gressier, F, et al. Environmental risk factors and biomarkers for autism spectrum disorder: an umbrella review of the evidence. Lancet Psychiatry. (2019) 6:590–600. doi: 10.1016/S2215-0366(19)30181-6
9. Fatemi, SH, Aldinger, KA, Ashwood, P, Bauman, ML, Blaha, CD, Blatt, GJ, et al. Consensus paper: pathological role of the cerebellum in autism. Cerebellum. (2012) 11:777–807. doi: 10.1007/s12311-012-0355-9
10. Ning, J, Xu, L, Shen, CQ, Zhang, YY, and Zhao, Q. Increased serum levels of macrophage migration inhibitory factor in autism spectrum disorders. Neurotoxicology. (2019) 71:1–5. doi: 10.1016/j.neuro.2018.11.015
11. Zhao, HX, Yin, SS, and Fan, JG. High plasma neopterin levels in Chinese children with autism spectrum disorders. Int J Dev Neurosci. (2015) 41:92–7. doi: 10.1016/j.ijdevneu.2015.02.002
12. Bener, A, Khattab, AO, Bhugra, D, and Hoffmann, GF. Iron and vitamin D levels among autism spectrum disorders children. Ann Afr Med. (2017) 16:186–91. doi: 10.4103/aam.aam_17_17
13. Higazi, AM, Kamel, HM, Abdel-Naeem, EA, Abdullah, NM, Mahrous, DM, and Osman, AM. Expression analysis of selected genes involved in tryptophan metabolic pathways in Egyptian children with autism spectrum disorder and learning disabilities. Sci Rep. (2021) 11:6931. doi: 10.1038/s41598-021-86162-w
14. Li, SO, Wang, JL, Bjørklund, G, Zhao, WN, and Yin, CH. Serum copper and zinc levels in individuals with autism spectrum disorders. Neuroreport. (2014) 25:1216–20. doi: 10.1097/WNR.0000000000000251
15. Rashaid, AHB, Nusair, SD, Alqhazo, MT, Adams, JB, Abu-Dalo, MA, and Bashtawi, MA. Heavy metals and trace elements in scalp hair samples of children with severe autism spectrum disorder: a case-control study on Jordanian children. J Trace Elem Med Biol. (2021) 67:126790. doi: 10.1016/j.jtemb.2021.126790
16. Coretti, L, Paparo, L, Riccio, MP, Amato, F, Cuomo, M, Natale, A, et al. Gut microbiota features in young children with autism spectrum disorders. Front Microbiol. (2018) 9:3146. doi: 10.3389/fmicb.2018.03146
17. Ding, H, Yi, X, Zhang, X, Wang, H, Liu, H, and Mou, WW. Imbalance in the gut microbiota of children with autism spectrum disorders. Front Cell Infect Microbiol. (2021) 11:572752. doi: 10.3389/fcimb.2021.572752
18. Dan, Z, Mao, X, Liu, Q, Guo, M, Zhuang, Y, Liu, Z, et al. Altered gut microbial profile is associated with abnormal metabolism activity of autism spectrum disorder. Gut Microbes. (2020) 11:1246–67. doi: 10.1080/19490976.2020.1747329
19. Strati, F, Cavalieri, D, Albanese, D, De Felice, C, Donati, C, Hayek, J, et al. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. (2017) 5:24. doi: 10.1186/s40168-017-0242-1
20. Shen, L, Liu, X, Zhang, H, Lin, J, Feng, C, and Iqbal, J. Biomarkers in autism spectrum disorders: current progress. Clin Chim Acta. (2020) 502:41–54. doi: 10.1016/j.cca.2019.12.009
21. Ristori, MV, Mortera, SL, Marzano, V, Guerrera, S, Vernocchi, P, Ianiro, G, et al. Proteomics and metabolomics approaches towards a functional insight onto autism spectrum disorders: phenotype stratification and biomarker discovery. Int J Mol Sci. (2020) 21:6274. doi: 10.3390/ijms21176274
22. Shen, L, Zhao, Y, Zhang, H, Feng, C, Gao, Y, Zhao, D, et al. Advances in biomarker studies in autism spectrum disorders. Adv Exp Med Biol. (2019) 1118:207–33. doi: 10.1007/978-3-030-05542-4_11
23. Marotta, R, Risoleo, MC, Messina, G, Parisi, L, Carotenuto, M, Vetri, L, et al. The neurochemistry of autism. Brain Sci. (2020) 10:3. doi: 10.3390/brainsci10030163
24. Alabdali, A, Al-Ayadhi, L, and El-Ansary, A. Association of social and cognitive impairment and biomarkers in autism spectrum disorders. J Neuroinflammation. (2014) 11:4. doi: 10.1186/1742-2094-11-4
25. Williams, JA, Burgess, S, Suckling, J, Lalousis, PA, Batool, F, Griffiths, SL, et al. Inflammation and brain structure in schizophrenia and other neuropsychiatric disorders: a Mendelian randomization study. JAMA Psychiatry. (2022) 79:498–507. doi: 10.1001/jamapsychiatry.2022.0407
26. Ma, J, Wu, J, Li, H, Wang, J, Han, J, and Zhang, R. Association between essential metal elements and the risk of autism in Chinese Han population. Biol Trace Elem Res. (2022) 200:505–15. doi: 10.1007/s12011-021-02690-6
27. Santos, G, Borges, JMP, Avila-Rodriguez, M, Gaíno, SB, Barreto, GE, Rúbio, ÉP, et al. Copper and neurotoxicity in autism spectrum disorder. Curr Pharm Des. (2019) 25:4747–54. doi: 10.2174/1381612825666191217091939
28. Skalny, AV, Aschner, M, and Tinkov, AA. Zinc. Adv Food Nutr Res. (2021) 96:251–310. doi: 10.1016/bs.afnr.2021.01.003
29. Georgieff, MK . Iron deficiency in pregnancy. Am J Obstet Gynecol. (2020) 223:516–24. doi: 10.1016/j.ajog.2020.03.006
30. Taniya, MA, Chung, HJ, Al Mamun, A, Alam, S, Aziz, MA, Emon, NU, et al. Role of gut microbiome in autism spectrum disorder and its therapeutic regulation. Front Cell Infect Microbiol. (2022) 12:915701. doi: 10.3389/fcimb.2022.915701
31. Rivière, A, Selak, M, Lantin, D, Leroy, F, and De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. (2016) 7:979. doi: 10.3389/fmicb.2016.00979
32. Eicher, TP, and Mohajeri, MH. Overlapping mechanisms of action of brain-active bacteria and bacterial metabolites in the pathogenesis of common brain diseases. Nutrients. (2022) 14:2661. doi: 10.3390/nu14132661
33. Levkova, M, Chervenkov, T, and Pancheva, R. Genus-level analysis of gut microbiota in children with autism spectrum disorder: a mini review. Children. (2023) 10:1103. doi: 10.3390/children10071103
34. Soltysova, M, Tomova, A, and Ostatnikova, D. Gut microbiota profiles in children and adolescents with psychiatric disorders. Microorganisms. (2022) 10:2009. doi: 10.3390/microorganisms10102009
35. Moher, D, Liberati, A, Tetzlaff, J, and Altman, DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6:e1000097. doi: 10.1371/journal.pmed.1000097
36. Munn, Z, Tufanaru, C, and Aromataris, E. JBI’s systematic reviews: data extraction and synthesis. Am J Nurs. (2014) 114:49–54. doi: 10.1097/01.NAJ.0000451683.66447.89
37. Checklist for cohort studies. Available at: https://jbi.global/sites/default/files/2020-08/Checklist_for_Cohort_Studies.pdf
38. Checklist for case control studies. Available at: https://jbi.global/sites/default/files/2020-08/Checklist_for_Case_Control_Studies.pdf
39. Checklist for analytical cross sectional studies. Available at: https://jbi.global/sites/default/files/2020-07/Checklist_for_Analytical_Cross_Sectional_Studies.pdf
40. Higgins, JP, Thompson, SG, Deeks, JJ, and Altman, DG. Measuring inconsistency in meta-analyses. BMJ. (2003) 327:557–60. doi: 10.1136/bmj.327.7414.557
41. Egger, M, Davey Smith, G, Schneider, M, and Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ. (1997) 315:629–34. doi: 10.1136/bmj.315.7109.629
42. Borenstein, M . Comprehensive meta-analysis software In: JPTH Matthias Egger and GD Smith, editors. Systematic reviews in health research: meta-analysis in context. 3rd ed. Hoboken, NJ, USA: John Wiley & Sons Ltd. (2022) Available at: https://onlinelibrary.wiley.com/doi/book/10.1002/9781119099369
43. Abdulamir, HA, Abdul-Rasheed OF, and Abdulghani, EA . Low oxytocin and melatonin levels and their possible role in the diagnosis and prognosis in Iraqi autistic children. Saudi Med J. (2016) 37:29–36. doi: 10.15537/smj.2016.1.13183
44. Abdulamir, HA, Abdul-Rasheed OF, and Abdulghani, EA . Serotonin and serotonin transporter levels in autistic children. Saudi Med J. (2018) 39:487–94. doi: 10.15537/smj.2018.5.21751
45. El-Ansary, A, Zayed, N, Al-Ayadhi, L, Qasem, H, Anwar, M, Meguid, NA, et al. GABA synaptopathy promotes the elevation of caspases 3 and 9 as pro-apoptotic markers in Egyptian patients with autism spectrum disorder. Acta Neurol Belg. (2021) 121:489–501. doi: 10.1007/s13760-019-01226-z
46. Tanaka, S, Komagome, A, Iguchi-Sherry, A, Nagasaka, A, Yuhi, T, Higashida, H, et al. Participatory art activities increase salivary oxytocin secretion of ASD children. Brain Sci. (2020) 10:680. doi: 10.3390/brainsci10100680
47. Huang, M, Liu, K, Wei, Z, Feng, Z, Chen, J, Yang, J, et al. Serum oxytocin level correlates with gut microbiome dysbiosis in children with autism spectrum disorder. Front Neurosci. (2021) 15:721884. doi: 10.3389/fnins.2021.721884
48. Al-Ali, Z, Yasseen, AA, Al-Dujailli, A, Al-Karaqully, AJ, McAllister, KA, and Jumaah, AS. The oxytocin receptor gene polymorphism rs2268491 and serum oxytocin alterations are indicative of autism spectrum disorder: a case-control paediatric study in Iraq with personalized medicine implications. PLoS One. (2022) 17:e0265217. doi: 10.1371/journal.pone.0265217
49. Mossa, ME, Ahmed, AS, and Khedhair, KA. Evaluation of serum oxytocin hormone level in children with autism. Indian J Public Health Res Dev. (2020) 11:305–9. doi: 10.37506/ijphrd.v11i10.11161
50. Chamtouri, M, Merghni, A, Salazar, N, Redruello, B, Gaddour, N, Mastouri, M, et al. An overview on fecal profiles of amino acids and related amino-derived compounds in children with autism spectrum disorder in Tunisia. Molecules. (2023) 28:3269. doi: 10.3390/molecules28073269
51. Mostafa, GA, Meguid, NA, Shehab, AAS, Elsaeid, A, and Maher, M. Plasma levels of nerve growth factor in Egyptian autistic children: relation to hyperserotonemia and autoimmunity. J Neuroimmunol. (2021) 358:577638. doi: 10.1016/j.jneuroim.2021.577638
52. Zuniga-Kennedy, M, Davoren, M, Shuffrey, LC, Luna, RA, Savidge, T, Prasad, V, et al. Intestinal predictors of whole blood serotonin levels in children with or without autism. J Autism Dev Disord. (2022) 52:3780–9. doi: 10.1007/s10803-022-05597-w
53. Alzghoul, L, Abdelhamid, SS, Yanis, AH, Qwaider, YZ, Aldahabi, M, and Albdour, SA. The association between levels of inflammatory markers in autistic children compared to their unaffected siblings and unrelated healthy controls. Turk J Med Sci. (2019) 49:1047–53. doi: 10.3906/sag-1812-167
54. Esnafoglu, E, and Subaşı, B. Association of low 25-OH-vitamin D levels and peripheral inflammatory markers in patients with autism spectrum disorder: vitamin D and inflammation in autism. Psychiatry Res. (2022) 316:114735. doi: 10.1016/j.psychres.2022.114735
55. Shen, Y, Li, Y, Shi, L, Liu, M, Wu, R, Xia, K, et al. Autism spectrum disorder and severe social impairment associated with elevated plasma interleukin-8. Pediatr Res. (2021) 89:591–7. doi: 10.1038/s41390-020-0910-x
56. Kartalcı, G, Çalışkan Demir, A, Kartalcı, Ş, Üremiş, N, and Türköz, Y. Evaluation of blood Zonulin levels, inflammatory processes and neuronal changes in children with autism spectrum disorder. Psychiatr Danub. (2022) 34:279–87. doi: 10.24869/psyd.2022.279
57. Skalny, AV, Simashkova, NV, Klyushnik, TP, Grabeklis, AR, Radysh, IV, Skalnaya, MG, et al. Assessment of serum trace elements and electrolytes in children with childhood and atypical autism. J Trace Elem Med Biol. (2017) 43:9–14. doi: 10.1016/j.jtemb.2016.09.009
58. Wu, LL, Mao, SS, Lin, X, Yang, RW, and Zhu, ZW. Evaluation of whole blood trace element levels in Chinese children with autism spectrum disorder. Biol Trace Elem Res. (2019) 191:269–75. doi: 10.1007/s12011-018-1615-4
59. Mehta, SQ, Behl, S, Day, PL, Delgado, AM, Larson, NB, Stromback, LR, et al. Evaluation of Zn, Cu, and Se levels in the north American autism spectrum disorder population. Front Mol Neurosci. (2021) 14:665686. doi: 10.3389/fnmol.2021.665686
60. Abd Wahil, MS, Ja’afar, MH, and Md, IZ. Assessment of urinary Lead (Pb) and essential trace elements in autism spectrum disorder: a case-control study among preschool children in Malaysia. Biol Trace Elem Res. (2022) 200:97–121. doi: 10.1007/s12011-021-02654-w
61. Wang, Y, Li, N, Yang, JJ, Zhao, DM, Chen, B, Zhang, GQ, et al. Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharmacol Res. (2020) 157:104784. doi: 10.1016/j.phrs.2020.104784
62. Zurita, MF, Cárdenas, PA, Sandoval, ME, Peña, MC, Fornasini, M, Flores, N, et al. Analysis of gut microbiome, nutrition and immune status in autism spectrum disorder: a case-control study in Ecuador. Gut Microbes. (2020) 11:453–64. doi: 10.1080/19490976.2019.1662260
63. Ding, X, Xu, Y, Zhang, X, Zhang, L, Duan, G, Song, C, et al. Gut microbiota changes in patients with autism spectrum disorders. J Psychiatr Res. (2020) 129:149–59. doi: 10.1016/j.jpsychires.2020.06.032
64. Xie, X, Li, L, Wu, X, Hou, F, Chen, Y, Shi, L, et al. Alteration of the fecal microbiota in Chinese children with autism spectrum disorder. Autism Res. (2022) 15:996–1007. doi: 10.1002/aur.2718
65. Plaza-Díaz, J, Gómez-Fernández, A, Chueca, N, Torre-Aguilar, MJ, Gil, Á, Perez-Navero, JL, et al. Autism spectrum disorder (ASD) with and without mental regression is associated with changes in the fecal microbiota. Nutrients. (2019) 11:337. doi: 10.3390/nu11020337
66. Ma, B, Liang, J, Dai, M, Wang, J, Luo, J, Zhang, Z, et al. Altered gut microbiota in Chinese children with autism spectrum disorders. Front Cell Infect Microbiol. (2019) 9:40. doi: 10.3389/fcimb.2019.00040
67. Liu, ZC, Wu, D, Qu, AN, and Wang, LL. Diversity and functional prediction of gut microbiota in children with autism spectrum disorder. Chin J Contemp Pediat. (2022) 24:1356–64. doi: 10.7499/j.issn.1008-8830.2207130
68. Chiappori, F, Cupaioli, FA, Consiglio, A, Di Nanni, N, Mosca, E, Licciulli, VF, et al. Analysis of faecal microbiota and small ncRNAs in autism: detection of miRNAs and piRNAs with possible implications in host-gut microbiota cross-talk. Nutrients. (2022) 14:1340. doi: 10.3390/nu14071340
69. Zou, R, Xu, F, Wang, Y, Duan, M, Guo, M, Zhang, Q, et al. Changes in the gut microbiota of children with autism spectrum disorder. Autism Res. (2020) 13:1614–25. doi: 10.1002/aur.2358
70. Sun, H, You, Z, Jia, L, and Wang, F. Autism spectrum disorder is associated with gut microbiota disorder in children. BMC Pediatr. (2019) 19:516. doi: 10.1186/s12887-019-1896-6
71. Maigoro, AY, and Lee, S. Gut microbiome-based analysis of lipid a biosynthesis in individuals with autism spectrum disorder: an in silico evaluation. Nutrients. (2021) 13:688. doi: 10.3390/nu13020688
72. Kang, DW, Adams, JB, Gregory, AC, Borody, T, Chittick, L, Fasano, A, et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. (2017) 5:10. doi: 10.1186/s40168-016-0225-7
73. Pulikkan, J, Maji, A, Dhakan, DB, Saxena, R, Mohan, B, Anto, MM, et al. Gut microbial dysbiosis in Indian children with autism spectrum disorders. Microb Ecol. (2018) 76:1102–14. doi: 10.1007/s00248-018-1176-2
74. Kang, DW, Park, JG, Ilhan, ZE, Wallstrom, G, Labaer, J, Adams, JB, et al. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. (2013) 8:e68322. doi: 10.1371/journal.pone.0068322
75. Liu, X, Lin, J, Zhang, H, Khan, NU, Zhang, J, Tang, X, et al. Oxidative stress in autism spectrum disorder-current progress of mechanisms and biomarkers. Front Psychiatry. (2022) 13:813304. doi: 10.3389/fpsyt.2022.813304
76. Matta, SM, Hill-Yardin, EL, and Crack, PJ. The influence of neuroinflammation in autism spectrum disorder. Brain Behav Immun. (2019) 79:75–90. doi: 10.1016/j.bbi.2019.04.037
77. Nadeem, R, Hussain, T, and Sajid, H. C reactive protein elevation among children or among mothers’ of children with autism during pregnancy, a review and meta-analysis. BMC Psychiatry. (2020) 20:251. doi: 10.1186/s12888-020-02619-8
78. Lambertsen, KL, Soares, CB, Gaist, D, and Nielsen, HH. Neurofilaments: the C-reactive protein of neurology. Brain Sci. (2020) 10:56. doi: 10.3390/brainsci10010056
79. Li, D, Zhou, X, Shi, Y, Li, P, Chen, S, Jin, W, et al. Immunofluorescence lateral chromatography in combination with detection of hypersensitive C-reactive protein and procalcitonin levels in patients with mental disease associated with coronavirus 2019 disease infection. Mater Express. (2021) 11:248, 10.1166/mex.2021.1886–54.
80. Gruol, DL . IL-6 regulation of synaptic function in the CNS. Neuropharmacology. (2015) 96:42–54. doi: 10.1016/j.neuropharm.2014.10.023
81. Huang, J, Song, Z, Wei, B, Li, Q, Lin, P, Li, H, et al. Immunological evaluation of patients with Alzheimer’s disease based on mitogen-stimulated cytokine productions and mitochondrial DNA indicators. BMC Psychiatry. (2023) 23:145. doi: 10.1186/s12888-023-04634-x
82. Yin, F, Wang, H, Liu, Z, and Gao, J. Association between peripheral blood levels of C-reactive protein and autism spectrum disorder in children: a systematic review and meta-analysis. Brain Behav Immun. (2020) 88:432–41. doi: 10.1016/j.bbi.2020.04.008
83. Hsuchou, H, Kastin, AJ, Mishra, PK, and Pan, W. C-reactive protein increases BBB permeability: implications for obesity and neuroinflammation. Cell Physiol Biochem. (2012) 30:1109–19. doi: 10.1159/000343302
84. Andoh, M, Ikegaya, Y, and Koyama, R. Microglia as possible therapeutic targets for autism spectrum disorders. Prog Mol Biol Transl Sci. (2019) 167:223–45. doi: 10.1016/bs.pmbts.2019.06.012
85. Xu, ZX, Kim, GH, Tan, JW, Riso, AE, Sun, Y, Xu, EY, et al. Elevated protein synthesis in microglia causes autism-like synaptic and behavioral aberrations. Nat Commun. (2020) 11:1797. doi: 10.1038/s41467-020-15530-3
86. Liao, X, Yang, J, Wang, H, and Li, Y. Microglia mediated neuroinflammation in autism spectrum disorder. J Psychiatr Res. (2020) 130:167–76. doi: 10.1016/j.jpsychires.2020.07.013
87. Gadient, RA, and Otten, UH. Interleukin-6 (IL-6)—a molecule with both beneficial and destructive potentials. Prog Neurobiol. (1997) 52:379–90. doi: 10.1016/S0301-0082(97)00021-X
88. Majerczyk, D, Ayad, EG, Brewton, KL, Saing, P, and Hart, PC. Systemic maternal inflammation promotes ASD via IL-6 and IFN-γ. Biosci Rep. (2022) 42:BSR20220713. doi: 10.1042/BSR20220713
89. Ozaki, K, Kato, D, Ikegami, A, Hashimoto, A, Sugio, S, Guo, Z, et al. Maternal immune activation induces sustained changes in fetal microglia motility. Sci Rep. (2020) 10:21378. doi: 10.1038/s41598-020-78294-2
90. Teleanu, RI, Niculescu, AG, Roza, E, Vladâcenco, O, Grumezescu, AM, and Teleanu, DM. Neurotransmitters-key factors in neurological and neurodegenerative disorders of the central nervous system. Int J Mol Sci. (2022) 23:5954. doi: 10.3390/ijms23115954
91. Lin, P, Sun, J, Lou, X, Li, D, Shi, Y, Li, Z, et al. Consensus on potential biomarkers developed for use in clinical tests for schizophrenia. Gen Psychiatr. (2022) 35:e100685. doi: 10.1136/gpsych-2021-100685
92. Walker, MA . Treatment of autism spectrum disorders: neurotransmitter signaling pathways involved in motivation and reward as therapeutic targets. Expert Opin Ther Targets. (2008) 12:949–67. doi: 10.1517/14728222.12.8.949
93. Sikich, L, Kolevzon, A, King, BH, McDougle, CJ, Sanders, KB, Kim, SJ, et al. Intranasal oxytocin in children and adolescents with autism spectrum disorder. N Engl J Med. (2021) 385:1462–73. doi: 10.1056/NEJMoa2103583
94. LoParo, D, and Waldman, ID. The oxytocin receptor gene (OXTR) is associated with autism spectrum disorder: a meta-analysis. Mol Psychiatry. (2015) 20:640–6. doi: 10.1038/mp.2014.77
95. Insel, TR, O’Brien, DJ, and Leckman, JF. Oxytocin, vasopressin, and autism: is there a connection? Biol Psychiatry. (1999) 45:145–57. doi: 10.1016/S0006-3223(98)00142-5
96. Sears, SM, and Hewett, SJ. Influence of glutamate and GABA transport on brain excitatory/inhibitory balance. Exp Biol Med (Maywood). (2021) 246:1069–83. doi: 10.1177/1535370221989263
97. Howes, OD, Rogdaki, M, Findon, JL, Wichers, RH, Charman, T, King, BH, et al. Autism spectrum disorder: consensus guidelines on assessment, treatment and research from the British Association for Psychopharmacology. J Psychopharmacol. (2018) 32:3–29. doi: 10.1177/0269881117741766
98. Cochran, DM, Sikoglu, EM, Hodge, SM, Edden, RA, Foley, A, Kennedy, DN, et al. Relationship among glutamine, γ-aminobutyric acid, and social cognition in autism spectrum disorders. J Child Adolesc Psychopharmacol. (2015) 25:314–22. doi: 10.1089/cap.2014.0112
99. Kheirouri, S, Kalejahi, P, and Noorazar, SG. Plasma levels of serotonin, gastrointestinal symptoms, and sleep problems in children with autism. Turk J Med Sci. (2016) 46:1765–72. doi: 10.3906/sag-1507-68
100. Dölen, G . Autism: oxytocin, serotonin, and social reward. Soc Neurosci. (2015) 10:450–65. doi: 10.1080/17470919.2015.1087875
101. Kong, XJ, Liu, J, Liu, K, Koh, M, Sherman, H, Liu, S, et al. Probiotic and oxytocin combination therapy in patients with autism spectrum disorder: a randomized, double-blinded, placebo-controlled pilot trial. Nutrients. (2021) 13:1552. doi: 10.3390/nu13051552
102. Kanat, M, Spenthof, I, Riedel, A, van Elst, LT, Heinrichs, M, and Domes, G. Restoring effects of oxytocin on the attentional preference for faces in autism. Transl Psychiatry. (2017) 7:e1097. doi: 10.1038/tp.2017.67
103. Althaus, M, Groen, Y, Wijers, AA, Noltes, H, Tucha, O, and Hoekstra, PJ. Oxytocin enhances orienting to social information in a selective group of high-functioning male adults with autism spectrum disorder. Neuropsychologia. (2015) 79:53–69. doi: 10.1016/j.neuropsychologia.2015.10.025
104. Guastella, AJ, Einfeld, SL, Gray, KM, Rinehart, NJ, Tonge, BJ, Lambert, TJ, et al. Intranasal oxytocin improves emotion recognition for youth with autism spectrum disorders. Biol Psychiatry. (2010) 67:692–4. doi: 10.1016/j.biopsych.2009.09.020
105. Watanabe, T, Abe, O, Kuwabara, H, Yahata, N, Takano, Y, Iwashiro, N, et al. Mitigation of socio communicational deficits of autism through oxytocin-induced recovery of medial prefrontal activity: a randomized trial. JAMA Psychiatry. (2014) 71:166–75. doi: 10.1001/jamapsychiatry.2013.3181
106. Kawahara, M, Kato-Negishi, M, and Tanaka, KI. Dietary trace elements and the pathogenesis of neurodegenerative diseases. Nutrients. (2023) 15:2067. doi: 10.3390/nu15092067
107. Wołonciej, M, Milewska, E, and Roszkowska-Jakimiec, W. Trace elements as an activator of antioxidant enzymes. Postepy Hig Med Dosw. (2016) 70:1483–98. doi: 10.5604/17322693.1229074
108. Maggini, S, Wintergerst, ES, Beveridge, S, and Hornig, DH. Selected vitamins and trace elements support immune function by strengthening epithelial barriers and cellular and humoral immune responses. Br J Nutr. (2007) 98:S29–35. doi: 10.1017/S0007114507832971
109. Baj, J, Flieger, W, Flieger, M, Forma, A, Sitarz, E, Skórzyńska-Dziduszko, K, et al. Autism spectrum disorder: trace elements imbalances and the pathogenesis and severity of autistic symptoms. Neurosci Biobehav Rev. (2021) 129:117–32. doi: 10.1016/j.neubiorev.2021.07.029
110. Zhang, J, Li, X, Shen, L, Khan, NU, Zhang, X, Chen, L, et al. Trace elements in children with autism spectrum disorder: a meta-analysis based on case-control studies. J Trace Elem Med Biol. (2021) 67:126782. doi: 10.1016/j.jtemb.2021.126782
111. Pivina, L, Semenova, Y, Doşa, MD, Dauletyarova, M, and Bjørklund, G. Iron deficiency, cognitive functions, and neurobehavioral disorders in children. J Mol Neurosci. (2019) 68:1–10. doi: 10.1007/s12031-019-01276-1
112. Gunes, S, Ekinci, O, and Celik, T. Iron deficiency parameters in autism spectrum disorder: clinical correlates and associated factors. Ital J Pediatr. (2017) 43:86. doi: 10.1186/s13052-017-0407-3
113. Bjorklund, G . The role of zinc and copper in autism spectrum disorders. Acta Neurobiol Exp. (2013) 73:225–36. doi: 10.55782/ane-2013-1932
114. Fluegge Ba, K . Zinc and copper metabolism and risk of autism: a reply to Sayehmiri et al. J Child Neurol. (2017) 11:66–9.
115. Ristori, MV, Quagliariello, A, Reddel, S, Ianiro, G, Vicari, S, Gasbarrini, A, et al. Autism, gastrointestinal symptoms and modulation of gut microbiota by nutritional interventions. Nutrients. (2019) 11:2812. doi: 10.3390/nu11112812
116. Cao, X, Lin, P, Jiang, P, and Li, C. Characteristics of the gastrointestinal microbiome in children with autism spectrum disorder: a systematic review. Shanghai Arch Psychiatry. (2013) 25:342, 10.3969/j.issn.1002-0829.2013.06.003–53.
117. Rutsch, A, Kantsjö, JB, and Ronchi, F. The gut-brain axis: how microbiota and host inflammasome influence brain physiology and pathology. Front Immunol. (2020) 11:604179. doi: 10.3389/fimmu.2020.604179
118. Lin, P, Li, D, Shi, Y, Li, Q, Guo, X, Dong, K, et al. Dysbiosis of the gut microbiota and kynurenine (Kyn) pathway activity as potential biomarkers in patients with major depressive disorder. Nutrients. (2023) 15:1752. doi: 10.3390/nu15071752
119. Iglesias-Vázquez, L, Van Ginkel, RG, Arija, V, and Canals, J. Composition of gut microbiota in children with autism spectrum disorder: a systematic review and meta-analysis. Nutrients. (2020) 12:792. doi: 10.3390/nu12030792
120. Fattorusso, A, Di Genova, L, Dell’Isola, GB, Mencaroni, E, and Esposito, S. Autism spectrum disorders and the gut microbiota. Nutrients. (2019) 11:521. doi: 10.3390/nu11030521
121. Srikantha, P, and Mohajeri, MH. The possible role of the microbiota-gut-brain-axis in autism spectrum disorder. Int J Mol Sci. (2019) 20:2115. doi: 10.3390/ijms20092115
122. Arnold, LE, Luna, RA, Williams, K, Chan, J, Parker, RA, Wu, Q, et al. Probiotics for gastrointestinal symptoms and quality of life in autism: a placebo-controlled pilot trial. J Child Adolesc Psychopharmacol. (2019) 29:659–69. doi: 10.1089/cap.2018.0156
123. Santocchi, E, Guiducci, L, Prosperi, M, Calderoni, S, Gaggini, M, Apicella, F, et al. Effects of probiotic supplementation on gastrointestinal, sensory and core symptoms in autism spectrum disorders: a randomized controlled trial. Front Psychiatry. (2020) 11:550593. doi: 10.3389/fpsyt.2020.550593
124. Sanctuary, MR, Kain, JN, Chen, SY, Kalanetra, K, Lemay, DG, Rose, DR, et al. Pilot study of probiotic/colostrum supplementation on gut function in children with autism and gastrointestinal symptoms. PLoS One. (2019) 14:e0210064. doi: 10.1371/journal.pone.0210064
125. Billeci, L, Callara, AL, Guiducci, L, Prosperi, M, Morales, MA, Calderoni, S, et al. A randomized controlled trial into the effects of probiotics on electroencephalography in preschoolers with autism. Autism. (2022) 27:117–32. doi: 10.1177/13623613221082710
126. Argou-Cardozo, I, and Zeidán-Chuliá, F. Clostridium bacteria and autism spectrum conditions: a systematic review and hypothetical contribution of environmental glyphosate levels. Med Sci. (2018) 6:29. doi: 10.3390/medsci6020029
127. Hua, X, Zhu, J, Yang, T, Guo, M, Li, Q, Chen, J, et al. The gut microbiota and associated metabolites are altered in sleep disorder of children with autism spectrum disorders. Front Psychiatry. (2020) 11:855. doi: 10.3389/fpsyt.2020.00855
128. MacFabe, DF, Cain, DP, Rodriguez-Capote, K, Franklin, AE, Hoffman, JE, Boon, F, et al. Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behav Brain Res. (2007) 176:149–69. doi: 10.1016/j.bbr.2006.07.025
129. Finegold, SM, Dowd, SE, Gontcharova, V, Liu, C, Henley, KE, Wolcott, RD, et al. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. (2010) 16:444–53. doi: 10.1016/j.anaerobe.2010.06.008
130. Finegold, SM, Summanen, PH, Downes, J, Corbett, K, and Komoriya, T. Detection of Clostridium perfringens toxin genes in the gut microbiota of autistic children. Anaerobe. (2017) 45:133–7. doi: 10.1016/j.anaerobe.2017.02.008
Keywords: autism spectrum disorder, biomarkers, biomedical, trace elements, microbiota
Citation: Lin P, Zhang Q, Sun J, Li Q, Li D, Zhu M, Fu X, Zhao L, Wang M, Lou X, Chen Q, Liang K, Zhu Y, Qu C, Li Z, Ma P, Wang R, Liu H, Dong K, Guo X, Cheng X, Sun Y and Sun J (2024) A comparison between children and adolescents with autism spectrum disorders and healthy controls in biomedical factors, trace elements, and microbiota biomarkers: a meta-analysis. Front. Psychiatry. 14:1318637. doi: 10.3389/fpsyt.2023.1318637
Received: 11 October 2023; Accepted: 13 December 2023;
Published: 09 January 2024.
Edited by:
Antonio M. Persico, University of Modena and Reggio Emilia, ItalyReviewed by:
Angela Caruso, National Institute of Health (ISS), ItalyCopyright © 2024 Lin, Zhang, Sun, Li, Li, Zhu, Fu, Zhao, Wang, Lou, Chen, Liang, Zhu, Qu, Li, Ma, Wang, Liu, Dong, Guo, Cheng, Sun and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xunjia Cheng, eGpjaGVuZ0BzaG11LmVkdS5jbg==; Yang Sun, c2htenhfc3lAYWxpeXVuLmNvbQ==; Jing Sun, ai5zdW5AZ3JpZmZpdGguZWR1LmF1
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.