
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Psychiatry , 02 May 2023
Sec. Autism
Volume 14 - 2023 | https://doi.org/10.3389/fpsyt.2023.1079683
 Qi Ji1,2†
Qi Ji1,2† Si-Jia Li1,2†
Si-Jia Li1,2† Jun-Bo Zhao1,2†
Jun-Bo Zhao1,2† Yun Xiong1,2†
Yun Xiong1,2† Xiao-Hui Du1
Xiao-Hui Du1 Chun-Xiang Wang1
Chun-Xiang Wang1 Li-Ming Lu3
Li-Ming Lu3 Jing-Yao Tan3
Jing-Yao Tan3 Zhi-Ru Zhu1*
Zhi-Ru Zhu1*Background: The incidence of sleep disorders in children with autism spectrum disorder (ASD) is very high. Sleep disorders can exacerbate the development of ASD and impose a heavy burden on families and society. The pathological mechanism of sleep disorders in autism is complex, but gene mutations and neural abnormalities may be involved.
Methods: In this review, we examined literature addressing the genetic and neural mechanisms of sleep disorders in children with ASD. The databases PubMed and Scopus were searched for eligible studies published between 2013 and 2023.
Results: Prolonged awakenings of children with ASD may be caused by the following processes. Mutations in the MECP2, VGAT and SLC6A1 genes can decrease GABA inhibition on neurons in the locus coeruleus, leading to hyperactivity of noradrenergic neurons and prolonged awakenings in children with ASD. Mutations in the HRH1, HRH2, and HRH3 genes heighten the expression of histamine receptors in the posterior hypothalamus, potentially intensifying histamine’s ability to promote arousal. Mutations in the KCNQ3 and PCDH10 genes cause atypical modulation of amygdala impact on orexinergic neurons, potentially causing hyperexcitability of the hypothalamic orexin system. Mutations in the AHI1, ARHGEF10, UBE3A, and SLC6A3 genes affect dopamine synthesis, catabolism, and reuptake processes, which can elevate dopamine concentrations in the midbrain. Secondly, non-rapid eye movement sleep disorder is closely related to the lack of butyric acid, iron deficiency and dysfunction of the thalamic reticular nucleus induced by PTCHD1 gene alterations. Thirdly, mutations in the HTR2A, SLC6A4, MAOA, MAOB, TPH2, VMATs, SHANK3, and CADPS2 genes induce structural and functional abnormalities of the dorsal raphe nucleus (DRN) and amygdala, which may disturb REM sleep. In addition, the decrease in melatonin levels caused by ASMT, MTNR1A, and MTNR1B gene mutations, along with functional abnormalities of basal forebrain cholinergic neurons, may lead to abnormal sleep–wake rhythm transitions.
Conclusion: Our review revealed that the functional and structural abnormalities of sleep–wake related neural circuits induced by gene mutations are strongly correlated with sleep disorders in children with ASD. Exploring the neural mechanisms of sleep disorders and the underlying genetic pathology in children with ASD is significant for further studies of therapy.
According to the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5), autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by social communication deficits, restricted interests, and repetitive behaviors. Approximately 1/100 children are diagnosed with ASD around the world, and the prevalence has been increasing over time (1).
Sleep disorders are common clinical symptoms in children with ASD, with a rate of nearly 50% (2). Clinical evidence has shown that sleep disorders can exacerbate ASD symptomatology (3–6). Firstly, sleep disorders are closely related to the impairment of social ability in children with ASD. Children with ASD with sleep disorders have further diminished social functioning and lower quality of life than those without sleep disorders (7). Sleep problems in autistic adolescents may lead to difficulties regulating social interactions and cause disharmonious relationships with peers (8). Secondly, sleep disorders can aggravate repetitive behaviors in children with ASD. The parental questionnaire revealed a significant correlation between Children’s Sleep Habits Questionnaire scores and Repetitive Behavioral Questionnaire-2 scores (9). Children with ASD with poor sleep quality have more repetitive behaviors (10, 11). Thirdly, sleep disorders may be a mediator of cognitive function deficits in children with ASD. Sleep is important for many complex physiological processes, such as cognitive development, learning and memory processes (12, 13). Sleep disorders can exacerbate the impairment of memory consolidation in children with ASD (5). Children with ASD who slept longer performed better on working memory tests and had higher rates of correct hits in the attention task. There was a linear relationship between poor working memory and sleep disorders in children with ASD (14). Children with ASD showed worse narrative abilities than their healthy peers (15). In addition, sleep disorders can also affect the language function of children with ASD (4). At the same time, sleep disorders of children with ASD increase the difficulty and cost of care, placing a heavy burden on family and society (16).
The main types of sleep disorders in children with ASD are difficulties in falling asleep and maintaining sleep, which manifest as long sleep latency, nighttime waking, and reduced sleep efficiency (17). Compared to healthy controls, children with ASD experience a 30–45 min prolongation of sleep latency on weekdays, generally lower sleep efficiency, and an average nighttime waking time of 2–3 h (17). Healthy individuals’ sleep–wake cycle is divided into three parts based on the electroencephalogram (EEG) and behavioral characteristics: the awakening period, non-rapid eye movement (NREM) sleep, and rapid eye movement (REM) sleep (18). Characteristics of NREM sleep include sharp-wave ripples, cortical slow oscillations, delta waves, and spindles. REM sleep is associated with theta oscillations (19, 20). Excess synapses are removed during NREM sleep and hippocampal neural activity during REM sleep is critically involved in memory consolidation (19, 20). Polysomnography analysis showed high rates of EEG abnormalities during sleep in children with ASD. Paroxysmal slowing and epileptiform abnormalities in EEG recordings were found in children with ASD with or without a history of seizures (21). Abnormalities of NREM sleep include a reduction in NREM sleep duration, K-complex density, and density of spindle activity. In addition, children with ASD also exhibit shorter REM sleep duration, prolonged REM latency, increased theta activity during REM sleep, and circadian rhythm sleep–wake disorders (22). The pathological mechanisms of sleep disorders in children with autism are still unclear. The lack of discussion on genetic and neural mechanisms has hindered the exploration of clinical treatment.
The purpose of this review is to summarize the genetic and neural mechanisms of sleep disorders in children with ASD from 2013 until now. We searched the databases of PubMed and Scopus for eligible studies and limited our search to publications between 2013 and 2023. We included articles that were peer-reviewed, written in English, purposefully addressed genetic and neural mechanisms, focused on sleep disorders, and included autism. Articles were excluded if they were not written in English, were non-peer reviewed, did not primarily focus on individuals with both autism and sleep disorders, did not clearly address genetic and neural mechanisms, or were abstracts, dissertations, methodological papers, or conference papers. The search strategy identified 397 documents, which were reduced to 65 included articles after applying exclusion criteria.
The nucleus locus coeruleus (LC) is a brainstem nucleus located on the dorsal side of the pons (Figure 1), which maintains the desynchronization of brain electrical activity. LC is the main nucleus that releases norepinephrine (NE), which can improve arousal level. Locus coeruleus-norepinephrine (LC-NE) neurons project widely to the cortex, hippocampus, thalamus, cerebellum, pons and medulla. The firing frequency of LC is highest during wakefulness, decreases during slow-wave sleep, and almost stops during REM sleep (23). The excitation of LC neurons promotes wakefulness, while inhibition of LC neurons reduces wakening and promotes REM sleep (24, 25).
Clinical data showed an abnormally enhanced activity of the LC-NE system in children with ASD. Resting eye-tracking task confirmed that the resting pupil diameter of children with ASD was significantly increased, suggesting increased tonic activity of LC-NE (26). Moreover, the NE levels are increased in the cortex, cerebellum and pons of the valproic acid rat model of autism, suggesting abnormal activation of NE system in the brain (27). The hyperactivity of the LC-NE system in the brain may lead to increased arousal and reduced sleep time in children with ASD. However, conflicting data showed the blockage of norepinephrine synthetase and reduced urine norepinephrine metabolites in children with ASD, which may suggest the reduced NE level in the blood (28). This implies that the NE level changes are complicated.
LC-NE hyperactivity in ASD patients may be related to decreased inhibition of γ-aminobutyric acid (GABA). LC neuronal activity is regulated by GABAergic interneurons. Animal experiments showed that presynaptic GABA release of LC neurons in ASD model mice was reduced, leading to hyperactivity of LC neurons (29). Changes in several genes participating in the synthesis, storage, and release of GABA may lead to a decrease in GABAergic inhibition. Mutations in the MECP2 gene, which encodes the transcriptional regulator methyl-CpG-binding protein 2 (MECP2), cause autism-like stereotypies and Rett syndrome (30). MECP2-deficient GABAergic neurons show a reduction of presynaptic glutamic acid decarboxylase (GAD) levels, indicating a decrease in GABA synthesis (31). Experiments on Epac2−/− mice, a model of ASD, showed an alteration of vesicular GABA transporter (VGAT) expression, which may affect the release of GABA (32). Additionally, a recent study showed that mutations in solute carrier family 6 member 1 (SLC6A1) are associated with autism. The GABA transporter1 (GAT-1) encoded by SLC6A1 is responsible for GABA reuptake into presynaptic neurons and glial cells to regulate neurotransmission (33). Thus, mutations in the SLC6A1 gene in certain types of ASD patients may affect the reuptake of GABA.
The posterior hypothalamus is one of the brain regions involved in arousal (Figure 1), in which the excitability of histaminergic neurons can promote and maintain the state of arousal. Histaminergic neurons are active during wakefulness and inactive during NREM and REM sleep. Histamine receptor agonists induce arousal, while antagonists promote sleep (34, 35). Wright et al. compared histamine receptor-related genes in the brains of ASD patients with healthy controls, including histidine decarboxylase gene (HDC), histamine N-methyltransferase (HNMT), histamine receptor H1 (HRH1), histamine receptor H2 (HRH2), histamine receptor H3 (HRH3), histamine receptor H4 (HRH4). They found that the expression of HRH1, HRH2, and HRH3 genes in ASD patients was higher than in healthy controls (36). The changes of gene encoding histamine receptor lead to the increase of histamine receptors in sleep-related nuclei in the brain, which amplifies the effect of histamine on promoting arousal, and this may be the reason for the prolongation of the arousal period in children with ASD.
The posterolateral hypothalamus is a brain region that promotes and maintains arousal, which is rich in orexinergic neurons (Figure 1). Orexinergic neurons are necessary for maintaining waking and behavioral arousal and widely project to sleep-related brain regions, including the LC basal forebrain (BF), tuberomammillary nucleus (TMN), and dorsal raphe nuclei (DRN) (18). These projections can significantly improve the excitability of arousal-related neurons, shorten sleep duration, and maintain the arousal state. Clinical data have shown that children with ASD have higher plasma orexin levels than healthy controls (control = 6, ASD = 18) (37). The increased activity of the orexinergic system is thought to be involved in insomnia in ASD (38).
The activity of orexinergic neurons is regulated by the amygdala, and abnormal amygdala function can cause hyper excitation of orexinergic neurons. Some ASD patients have structural and functional abnormalities in the bilateral amygdala, including an increase in volume and cell density in the amygdala (39), as well as inhibition of amygdala neuron synchronization activity (38). The functional magnetic resonance imaging (fMRI) results showed that the functional connectivity between the amygdala and other brain regions was reduced (40), leading to a weakening of its regulatory role in various physiological functions (41). Some gene mutations may lead to structural and functional abnormalities of the amygdala in the brain of patients with ASD. Missense variants at R230 and R227 in the potassium voltage-gated channel subfamily Q Member 3 (KCNQ3) have been reported in some autistic patients. Further patch clamp analysis in amygdala neurons showed that variants of the KCNQ3 gene resulted in abnormal function of voltage-gated potassium channels (42), which may disturb the modulating effects of amygdala neurons on orexin neurons and prolong waking time (43). In addition, the deletion of the protocadherin 10 (PCDH10) gene has also been reported in ASD patients (44). PCDH10 is an activity-regulated gene that is expressed at high levels in olfactory and limbic regions, including the basolateral amygdala. It is implicated in social and emotional behavior phenotypes in ASD (45). Mice lacking one copy of PCDH10 (PCDH10+/−) show reduced levels of N-methyl-D-aspartate receptor subunits in the amygdala (45), which wakens the regulatory effects of the amygdala on orexinergic neurons. Thus, orexinergic neurons show hyperexcitation, leading to prolonged awakenings in ASD patients.
The substantia nigra pars compacta and ventral tegmental areas are arousal-related nuclei, where most human dopaminergic neurons are located, and the dopamine secreted by them can effectively promote and maintain arousal (Figure 1). It was found that the increase of dopamine concentrations in substantia nigra pars compacta in midbrain significantly prolonged wakening time and shortened REM sleep. The treatment of dopamine receptor agonists significantly increased wakefulness, while antagonists augmented REM sleep (46).
Mutations of several genes participating in dopamine synthesis, catabolism and reuptake were found in ASD patients. The Abelson-helper integration site 1 (AHI1) gene is a candidate gene of ASD (47). Down-regulation of the expression of the rate-limiting enzyme in dopamine biosynthesis, tyrosine hydroxylase (TH), in AHI1-knockout (KO) mice is responsible for AHI1-deficiency-mediated autism symptoms. The AHI1-knockout autism mouse model showed low expression of TH and a decrease in dopamine synthesis (48). In addition, the deficiency of the Rho Guanine Nucleotide Exchange Factor 10 (ARHGEF10) gene decreased the expression of Monoamine oxidase A (MAOA), one of the enzymes in the catabolism of dopamine, and increased the level of dopamine in ASD model mice (49). The Ubiquitin protein ligase E3A (UBE3A) gene is another candidate gene of ASD (47). The deletion of the UBE3A gene is the cause of the disease in certain children with ASD, which reduces dopamine transporter (DAT) function and thus affects dopamine reuptake, increasing dopamine levels and wakefulness (50). Both clinical evidence and animal model experiments have shown that certain types of syndromic autism are associated with mutations in genes encoding DAT (50–53). Apart from that, mutations in the solute carrier family 6 member 3 (SLC6A3) gene also occur in clinical ASD patients (52). Mutation in the SLC6A3 gene causes abnormal DAT function and continuous outflow of dopamine from cells, leading to an abnormal increase in extracellular dopamine concentrations (53). Therefore, in ASD patients with mutations in the AHI1, UBE3A and SLC6A3 genes, abnormal dopamine synthesis, catabolism and reuptake result in increased dopamine concentrations in the brain, which may lead to a prolonged wakening period.
Studies showed that the functional connection between the cortex and thalamus is important for sleep regulation, which is regulated by nerves and exhibits dynamic changes (54, 55). The data from fMRI showed that the thalamocortical functional connectivity of healthy adults decreased during NREM sleep, and the information from the thalamic afferent cortex was reduced. This ensures that people enter NREM sleep from an arousal state (54). However, fMRI data showed a lack of regulation of thalamo-cortical functional connectivity in the brain of children with ASD, resulting in an abnormal increase in thalamo-cortical functional connectivity, which led to an inability to transition smoothly from wakefulness to NREM sleep (55). The thalamic reticular nucleus (TRN) is specifically excited by the thalamus cortex system and then inhibits the activity of the whole dorsal thalamus through its GABAergic neuron fiber projection, which makes TRN become the inhibitory gate to control the thalamo-cortical circuit. TRN can effectively control the thalamo-cortical functional connection and plays a sedative and sleep-promoting effect (56).
TRN is mainly composed of GABAergic neurons. The slow synchronous oscillation generated by TRN neurons can block the processing of sensory information in the cerebral cortex and limbic system, thus promoting sleep. After photogenetic activation of TRN neurons, the NREM sleep period of mice was prolonged, and the frequency and amplitude of δ waves were also increased, while muscular tension and activity in the mice decreased (57). Moreover, TRN neuronal activity is important in regulating the formation of spindle oscillations during NREM sleep. Spindle oscillation is a characteristic brainwave during the NREM sleep period, which can reduce the influence of external stimulation on the brain during sleep and stabilize sleep. Spindle oscillation is caused by the low-threshold Ca2+ current induced by the periodic firing of TRN cells. Therefore, spindle oscillation disappears after cutting off the connection between cortical-thalamic circuit cells and TRN neurons (58).
Patched domain containing protein 1 (PTCHD1) gene maps to chromosome Xp22.11 and encodes PTCHD1, which is closely related to TRN neuronal activity (59). During early development in mice, PTCHD1 is selectively expressed in the TRN and continues to be highly expressed into adulthood (60). The firing activity of TRN neurons in PTCHD1 knockout mice was reduced during sleep, leading to a reduction in NREM sleep spindle waves (60). In clinical practice, about 1% of ASD patients have PTCHD1 gene mutations, which cause a significant reduction in gene transcription activity (61). Furthermore, the volume of TRN neurons in ASD model mice is larger than that in wild-type mice (62). These results suggest that the abnormal structure and function of TRN neurons may be responsible for the shortened sleep duration and reduced spindle wave in NREM sleep in some children with ASD.
Short-chain fatty acids are important metabolites produced by human colonic flora, including acetate, propionate, butyrate and so on (63). Butyrate can prolong the time of NREM sleep through hepatoportal butyrate-sensitive mechanisms (64). A gut flora metabolism study found that many patients with ASD have intestinal microbiota disorders, and the level of short-chain fatty acids such as butyric acid in the brain is generally lower than that of healthy controls (65). The level of butyric acid may be associated with complex changes of butyric acid-producing bacteria. A study on butyric acid-producing bacteria showed that the abundance of butyric acid-producing bacteria Faecalibacterium and Agathobacter were significantly reduced in children with ASD who have sleep disorders (65). However, Roseburia intestinalis, another butyrate-producing bacterium, has a significantly higher abundance in children with ASD compared to control groups (66). Furthermore, it was reported that the abundance of genes related to butyric acid production decreased in the metagenome of ASD patients (67). Therefore, the complex changes of butyric acid-producing bacteria and the decreased abundance of genes associated with butyrate production may result in the decrease of butyric acid, which subsequently shortens NREM sleep.
Iron is important for sleep maintenance and the synthesis of neurotransmitters, such as serotonin, noradrenaline, dopamine, glutamate and γ-aminobutyric acid (GABA) (68). Iron deficiency can lead to abnormal metabolism of sleep-related neurotransmitters and worse sleep quality (69). Children with ASD had significantly lower serum ferritin levels compared to healthy controls (70). The analysis results of a retrospective chart review also showed that the serum iron level of ASD patients was lower than that of the control group (70). The above evidence suggests that iron deficiency in ASD patients can lead to abnormal metabolism of sleep-related transmitters and abnormal formation of sleep spindles, which may contribute to NREM sleep abnormalities in ASD patients.
The 5-hydroxytryptamine-ergic (5-HT-ergic) neurons in the dorsal raphe nucleus (DRN) have the function of promoting wakefulness and reducing REM sleep, which are known as REM-off neurons (Figure 1). The firing frequency of DRN 5-HTergic neurons during REM sleep was significantly lower than that during wakefulness. What’s more, SEP-363856, an agonist of 5-HT, could suppress REM sleep with very large effect sizes (71).
Elevated blood 5-HT levels are a biomarker of ASD. Thirty percent of ASD patients had significantly higher blood 5-HT levels compared to healthy controls (72). However, it is reported that levels of whole blood 5-HT were lower in some ASD patients compared to healthy controls (73). Mutations of multiple 5-HT related genes, such as 5-Hydroxytryptamine Receptor 2A (HTR2A), Solute Carrier Family 6 Member 4 (SLC6A4), Monoamine Oxidase A (MAOA), Monoamine Oxidase B (MAOB), Tryptophan Hydroxylase 2 (TPH2), and Vesicular Monoamine Transporters (VMATs), were found in certain types of ASD patients. These mutations result in an abnormal synthesis, transport, or inactivation of 5-HT and sleep disorders in children with ASD (74–78).
In addition to 5-HT-ergic neurons, there are also a large number of GABAergic neurons in the DRN that can regulate their activity. GABAergic cells projecting to the DRN are mainly located in the hypothalamus, ventral tegmental area (VTA), and locally within the DRN (79). Studies have shown that SH3 and multiple Ankyrin repeat domains 3 (SHANK3) -deficient mice, a model of autism, had significantly lower levels of GABA synthesis in the hypothalamus and VTA compared to controls (80). This may lead to reduced inhibition of DRN 5-HT-ergic neurons and increased activity in the DRN of ASD patients. Therefore, it is speculated that the enhanced REM-off function resulting from increased 5-HT-ergic neuron activity in the DRN may be why some children with ASD experience shorter REM sleep duration.
An article recently published in Science reported that the initiation of REM sleep is modulated by the amygdala (Figure 1) (81). When the dopamine level in the amygdala was instantaneously increased through photogenetic activation, NREM sleep in mice was terminated immediately, and REM sleep began simultaneously. The study confirmed that dopamine induces a transition from NREM sleep to REM sleep by binding to dopamine type II receptors on amygdala neurons (81), while inhibiting amygdala activity results in a decrease in REM sleep duration (82).
The structure and function of the amygdala in ASD patients can undergo complex changes. Abnormal changes in dopamine concentrations in the brain of ASD patients were described in section 2.4. Calcium-dependent secretion activator 2 (CADPS2) gene encodes a calcium binding protein that regulates the exocytosis of synaptic secretory granules, including monoamines and neuropeptides (83). Some ASD patients have a missense variant of the CADPS2 gene in the amygdala, which can block the release of dopamine (84, 85). Additionally, as described in section 2.3, amygdala neurons in ASD have abnormal structure and function. Therefore, the complex changes in the amygdala and the dopamine system in ASD may result in abnormal binding of dopamine to type II dopamine receptors in amygdala neurons, which could be responsible for abnormal REM sleep in some children with ASD.
Melatonin is an important hormone involved in the regulation of the sleep–wake cycle and the circadian rhythm, which is synthesized by the pineal gland at night (Figure 1). After binding to melatonin receptors in the suprachiasmatic nucleus, melatonin can promote sleep initiation and attenuate the arousal of the circadian clock. Studies have shown a sharp increase in nighttime sleep tendency in the hours after endogenous melatonin production (86).
Melatonin has been shown to be released at lower levels in individuals with ASD compared to healthy individuals (87). However, other research shows no significant difference in melatonin levels between 40% of ASD individuals and healthy controls (88). Melatonin treatment can increase sleep duration in ASD patients (89), suggesting that their sleep disorders are related to pineal dysfunction and lack of melatonin. Mutations in the Acetylserotonin O-Methyltransferas (ASMT), Melatonin Receptor 1A (MTNR1A), and Melatonin Receptor 1B (MTNR1B) genes can induce sleep disorders in ASD patients by affecting melatonin production and utilization (90–92). ASMT is an enzyme gene involved in melatonin synthesis, while MTNR1A and MTNR1B are melatonin receptor genes. Therefore, decreased melatonin levels due to pineal gland dysfunction may contribute to circadian rhythm sleep–wake disorders in children with ASD.
Acetylcholine is an important neurotransmitter associated with arousal. There are two groups of acetylcholinergic neurons in the human body: one is in the brainstem, while the other is in the basal forebrain (BF) (Figure 1). Cholinergic neurons in the basal forebrain play a role in cortical activation during the sleep–wake cycle, promoting the transition from NREM sleep to the waking state or to REM sleep. The level of acetylcholine in the basal forebrain is high during the arousal period and REM sleep, and low during slow wave-sleep (93).
Neuroanatomical analysis revealed that autistic patients show smaller neurons and increased cell density in the BF as compared to age-matched healthy individuals (94). Moreover, diminished activity of cholinergic neurons in the basal forebrain was reported in mice (95). Therefore, BF lesions in children with ASD may cause abnormal function of BF cholinergic neurons, resulting in their abnormal regulation of phase conversion during sleep. This may be one of the neural mechanisms causing circadian rhythm sleep–wake disorders in children with ASD.
In the present review, we limited our search to articles published in English. Thus, we may have missed related articles published in other languages. In addition, terminology related to genetic and neurological abnormalities varies, which may result in missing related articles due to variability in keyword use. Moreover, we did not restrict our review to studies with human subjects. Therefore, further investigation in ASD patients is required to replicate some of the reported findings in animal models of ASD. Finally, we did not consider sex differences in sleep disorders in children with ASD.
There is an established relationship between genetic mutations/neurological abnormalities and sleep disorders in children with ASD. As shown in Figures 1, 2, mutations in the MECP2, VGAT, SLC6A1, SLC6A3, HRH1-3, KCNQ3, PCDH10, AHI1, UBE3A and ARHGEF10 genes can cause atypical activity of wake-related neural circuits, which may lead to sleep problems such as prolonged sleep latency, short total sleep duration, and waking up at night. Additionally, butyric acid, iron deficiency, and TRN dysfunction are also linked to NREM sleep disorder in children with ASD. Mutations in the HTR2A, SLC6A4, MAOA, MAOB, TPH2, VMATs, SHANK3, and CADPS2 genes induce abnormal reactions of REM-off and REM-on neurons, which may lead to NREM disorder. Mutations in the ASMT, MTNR1A and MTNR1B genes induce the decreased synthesis and secretion of melatonin, which may cause abnormal sleep–wake rhythm transition. However, more research is needed on the clinical intervention of sleep disorders in children with ASD, as there are few intervention studies available. Overall, the genetic and neural mechanisms of sleep disorders in children with ASD warrant further investigation.
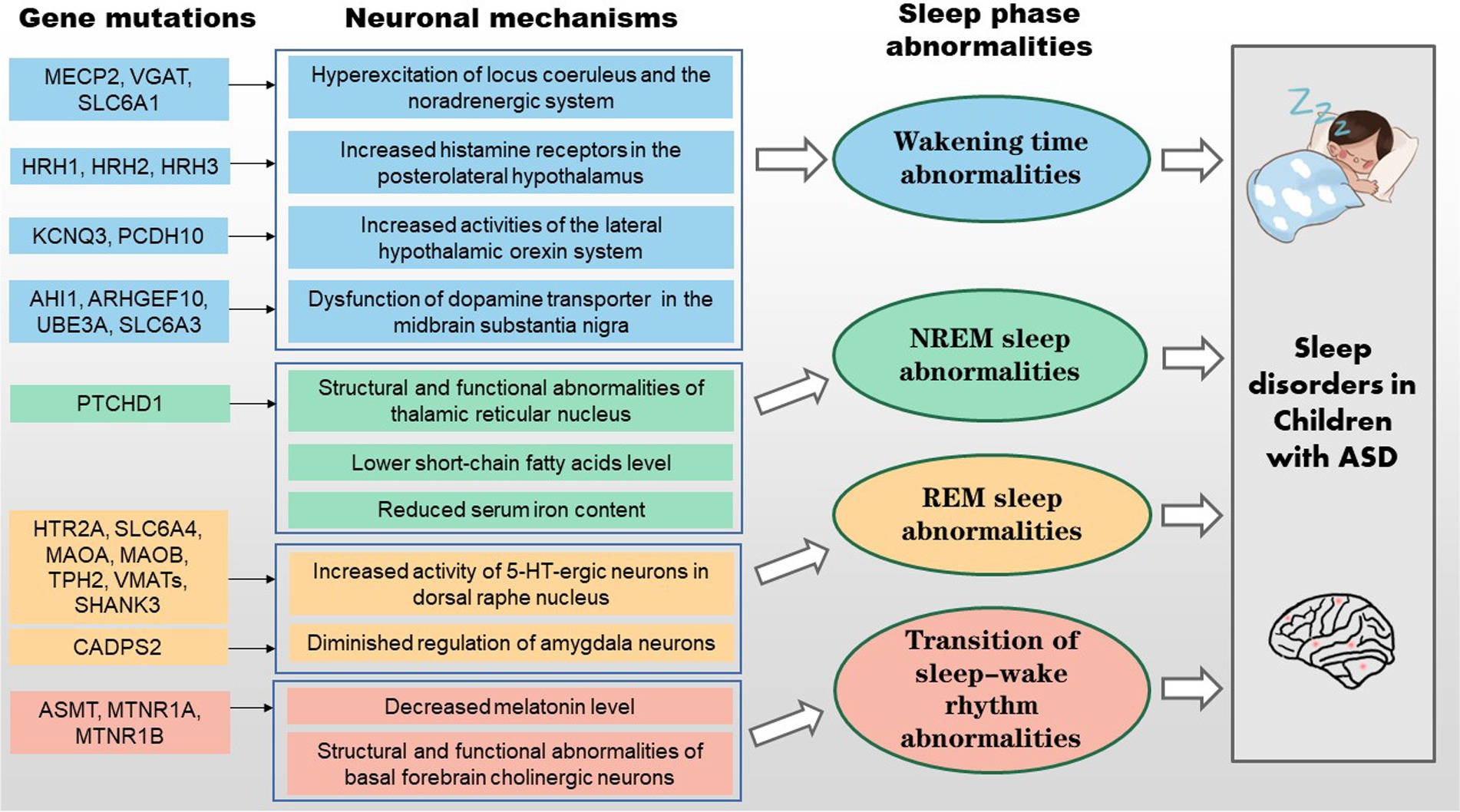
Figure 2. Schematic representation of the genetic and neural mechanisms of sleep disorders in children with ASD.
QJ, S-JL, J-BZ, YX, X-HD, C-XW, L-ML, J-YT and Z-RZ contributed to the writing and editing of this review. X-HD and Z-RZ conceived the scope of the review, coordinated efforts among authors, and wrote the bulk of the wake sections. QJ, S-JL, J-BZ and YX wrote the bulk of the sleep section. QJ and LS-J wrote the bulk of the sleep–wake rhythm section. All authors contributed to the article and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (31671106), the Scientific Foundation of Chongqing (Cstc2019jcyj-msxmX0019), and the Science Foundation of Army Medical University (2019XYY08).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Zeidan, J, Fombonne, E, Scorah, J, Ibrahim, A, Durkin, MS, Saxena, S, et al. Global prevalence of autism: a systematic review update. Autism Res. (2022) 15:778–90. doi: 10.1002/aur.2696
2. Parvataneni, T, Srinivas, S, Shah, K, and Patel, RS. Perspective on melatonin use for sleep problems in autism and attention-deficit hyperactivity disorder: a systematic review of randomized clinical trials. Cureus. (2020) 12:e8335. doi: 10.7759/cureus.8335
3. Wang, G, Chen, J, Zhang, K, Tang, S, and Wang, G. The mediating role of gaze patterns in the association of child sleep disturbances and core symptoms of autism spectrum disorder. Autism Res. (2022) 15:1719–31. doi: 10.1002/aur.2737
4. Souders, MC, Zavodny, S, Eriksen, W, Sinko, R, Connell, J, Kerns, C, et al. Sleep in children with autism Spectrum disorder. Curr Psychiatry Rep. (2017) 19:34. doi: 10.1007/s11920-017-0782-x
5. Tse, CYA, Lee, HP, Chan, KSK, Edgar, VB, Wilkinson-Smith, A, and Lai, WHE. Examining the impact of physical activity on sleep quality and executive functions in children with autism spectrum disorder: a randomized controlled trial. Autism. (2019) 23:1699–710. doi: 10.1177/1362361318823910
6. Hunter, JE, McLay, LK, France, KG, and Blampied, NM. Sleep and stereotypy in children with autism: effectiveness of function-based behavioral treatment. Sleep Med. (2021) 80:301–4. doi: 10.1016/j.sleep.2021.01.062
7. Malhi, P, Kaur, A, Singhi, P, and Sankhyan, N. Sleep dysfunction and behavioral daytime problems in children with autism Spectrum disorders: a comparative study. Indian J Pediatr. (2019) 86:12–7. doi: 10.1007/s12098-018-2731-z
8. Whelan, S, Mannion, A, Madden, A, Berger, F, Costello, R, Ghadiri, S, et al. Examining the relationship between sleep quality, social functioning, and behavior problems in children with autism Spectrum disorder: a systematic review. Nat Sci Sleep. (2022) 14:675–95. doi: 10.2147/NSS.S239622
9. Kang, YQ, Song, XR, Wang, GF, Su, YY, Li, PY, and Zhang, X. Sleep problems influence emotional/behavioral symptoms and repetitive behavior in preschool-aged children with autism Spectrum disorder in the unique social context of China. Front Psych. (2020) 11:273. doi: 10.3389/fpsyt.2020.00273
10. Deliens, G, and Peigneux, P. Sleep-behaviour relationship in children with autism spectrum disorder: methodological pitfalls and insights from cognition and sensory processing. Dev Med Child Neurol. (2019) 61:1368–76. doi: 10.1111/dmcn.14235
11. Mason, GM, Lokhandwala, S, Riggins, T, and Spencer, RMC. Sleep and human cognitive development. Sleep Med Rev. (2021) 57:101472. doi: 10.1016/j.smrv.2021.101472
12. Mazurek, MO, and Sohl, K. Sleep and behavioral problems in children with autism Spectrum disorder. J Autism Dev Disord. (2016) 46:1906–15. doi: 10.1007/s10803-016-2723-7
13. Karthikeyan, R, Cardinali, DP, Shakunthala, V, Spence, DW, Brown, GM, and Pandi-Perumal, SR. Understanding the role of sleep and its disturbances in autism spectrum disorder. Int J Neurosci. (2020) 130:1033–46. doi: 10.1080/00207454.2019.1711377
14. Mughal, R, Hill, CM, Joyce, A, and Dimitriou, D. Sleep and cognition in children with fetal alcohol Spectrum disorders (FASD) and children with autism Spectrum disorders (ASD). Brain Sci. (2020) 10:863. doi: 10.3390/brainsci10110863
15. Westby, C. Nature and effects of autobiographical memory issues in persons with autism Spectrum disorders. Neuropsychiatr Dis Treat. (2022) 18:2279–93. doi: 10.2147/NDT.S332521
16. Baraskewich, J, von Ranson, KM, McCrimmon, A, and McMorris, CA. Feeding and eating problems in children and adolescents with autism: a scoping review. Autism. (2021) 25:1505–19. doi: 10.1177/1362361321995631
17. Van der Heijden, KB, Stoffelsen, RJ, Popma, A, and Swaab, H. Sleep, chronotype, and sleep hygiene in children with attention-deficit/hyperactivity disorder, autism spectrum disorder, and controls. Eur Child Adolesc Psychiatry. (2018) 27:99–111. doi: 10.1007/s00787-017-1025-8
18. Schwartz, MD, and Kilduff, TS. The neurobiology of sleep and wakefulness. Psychiatr Clin North Am. (2015) 38:615–44. doi: 10.1016/j.psc.2015.07.002
19. Girardeau, G, and Lopes-Dos-Santos, V. Brain neural patterns and the memory function of sleep. Science (New York, NY). (2021) 374:560–4. doi: 10.1126/science.abi8370
20. Siegel, JM. Sleep function: an evolutionary perspective. Lancet Neurol. (2022) 21:937–46. doi: 10.1016/S1474-4422(22)00210-1
21. Santarone, ME, Zambrano, S, Zanotta, N, Mani, E, Minghetti, S, Pozzi, M, et al. EEG features in autism Spectrum disorder: a retrospective analysis in a cohort of preschool children. Brain Sci. (2023) 13:345. doi: 10.3390/brainsci13020345
22. Devnani, PA, and Hegde, AU. Autism and sleep disorders. J Pediatr Neurosci. (2015) 10:304–7. doi: 10.4103/1817-1745.174438
23. Van Egroo, M, Koshmanova, E, Vandewalle, G, and Jacobs, HIL. Importance of the locus coeruleus-norepinephrine system in sleep-wake regulation: implications for aging and Alzheimer's disease. Sleep Med Rev. (2022) 62:101592. doi: 10.1016/j.smrv.2022.101592
24. Szabo, ST, Thorpy, MJ, Mayer, G, Peever, JH, and Kilduff, TS. Neurobiological and immunogenetic aspects of narcolepsy: implications for pharmacotherapy. Sleep Med Rev. (2019) 43:23–36. doi: 10.1016/j.smrv.2018.09.006
25. Khanday, MA, Somarajan, BI, Mehta, R, and Mallick, BN. Noradrenaline from locus Coeruleus neurons acts on Pedunculo-Pontine neurons to prevent REM sleep and induces its loss-associated effects in rats. eNeuro. (2016) 3:ENEURO.0108–16.2016. doi: 10.1523/ENEURO.0108-16.2016
26. Keehn, B, Kadlaskar, G, Bergmann, S, McNally Keehn, R, and Francis, A. Attentional disengagement and the locus Coeruleus-norepinephrine system in children with autism Spectrum disorder. Front Integr Neurosci. (2021) 15:716447. doi: 10.3389/fnint.2021.716447
27. Ali, EH, and Elgoly, AH. Combined prenatal and postnatal butyl paraben exposure produces autism-like symptoms in offspring: comparison with valproic acid autistic model. Pharmacol Biochem Behav. (2013) 111:102–10. doi: 10.1016/j.pbb.2013.08.016
28. Gevi, F, Belardo, A, and Zolla, L. A metabolomics approach to investigate urine levels of neurotransmitters and related metabolites in autistic children. Biochim Biophys Acta Mol basis Dis. (2020) 1866:165859. doi: 10.1016/j.bbadis.2020.165859
29. Jin, X, Cui, N, Zhong, W, Jin, XT, and Jiang, C. GABAergic synaptic inputs of locus coeruleus neurons in wild-type and Mecp 2-null mice. Am J Physiol Cell Physiol. (2013) 304:C844–57. doi: 10.1152/ajpcell.00399.2012
30. Liu, Z, Li, X, Zhang, JT, Cai, YJ, Cheng, TL, Cheng, C, et al. Autism-like behaviours and germline transmission in transgenic monkeys overexpressing MeCP2. Nature. (2016) 530:98–102. doi: 10.1038/nature16533
31. El-Khoury, R, Panayotis, N, Matagne, V, Ghata, A, Villard, L, and Roux, JC. GABA and glutamate pathways are spatially and developmentally affected in the brain of Mecp 2-deficient mice. PLoS One. (2014) 9:e92169. doi: 10.1371/journal.pone.0092169
32. Jones, KA, Sumiya, M, Woolfrey, KM, Srivastava, DP, and Penzes, P. Loss of EPAC2 alters dendritic spine morphology and inhibitory synapse density. Mol Cell Neurosci. (2019) 98:19–31. doi: 10.1016/j.mcn.2019.05.001
33. Mermer, F, Poliquin, S, Rigsby, K, Rastogi, A, Shen, W, Romero-Morales, A, et al. Common molecular mechanisms of SLC6A1 variant-mediated neurodevelopmental disorders in astrocytes and neurons. Brain J Neurol. (2021) 144:2499–512. doi: 10.1093/brain/awab207
34. Kamei, C. Effects of histamine and related compounds on the central nervous system. Yakugaku Zasshi. (2021) 141:93–110. doi: 10.1248/yakushi.20-00197
35. Dauvilliers, Y, Bassetti, C, Lammers, GJ, Arnulf, I, Mayer, G, Rodenbeck, A, et al. Pitolisant versus placebo or modafinil in patients with narcolepsy: a double-blind, randomised trial. Lancet Neurol. (2013) 12:1068–75. doi: 10.1016/S1474-4422(13)70225-4
36. Wright, C, Shin, JH, Rajpurohit, A, Deep-Soboslay, A, Collado-Torres, L, Brandon, NJ, et al. Altered expression of histamine signaling genes in autism spectrum disorder. Transl Psychiatry. (2017) 7:e1126. doi: 10.1038/tp.2017.87
37. Kobylinska, L, Gabreanu, G, Simion, A, Mihailescu, I, and Zagrean, LJEN. Measuring orexin B values in a Romanian population of children with Autism Spectrum Disorders. Eur Neuropsychopharmacol. (2016) 26:206. doi: 10.1016/S0924-977X(16)31052-5
38. Kohyama, J. Possible neuronal mechanisms of sleep disturbances in patients with autism spectrum disorders and attention-deficit/hyperactivity disorder. Med Hypotheses. (2016) 97:131–3. doi: 10.1016/j.mehy.2016.11.001
39. Howard, MA, Cowell, PE, Boucher, J, Broks, P, Mayes, A, Farrant, A, et al. Convergent neuroanatomical and behavioural evidence of an amygdala hypothesis of autism. Neuroreport. (2000) 11:2931–5. doi: 10.1097/00001756-200009110-00020
40. Ibrahim, K, Eilbott, JA, Ventola, P, He, G, Pelphrey, KA, McCarthy, G, et al. Reduced amygdala-prefrontal functional connectivity in children with autism Spectrum disorder and co-occurring disruptive behavior. Biol Psychiatry Cogn Neurosci Neuroimaging. (2019) 4:1031–41. doi: 10.1016/j.bpsc.2019.01.009
41. Seo, M, and Anderson, G. Gut-amygdala interactions in autism Spectrum disorders: developmental roles via regulating mitochondria, Exosomes, immunity and micro RNAs. Curr Pharm Des. (2019) 25:4344–56. doi: 10.2174/1381612825666191105102545
42. Sands, TT, Miceli, F, Lesca, G, Beck, AE, Sadleir, LG, Arrington, DK, et al. Autism and developmental disability caused by KCNQ3 gain-of-function variants. Ann Neurol. (2019) 86:181–92. doi: 10.1002/ana.25522
43. Cheng, P, Qiu, Z, and Du, Y. Potassium channels and autism spectrum disorder: an overview. Int J Dev Neurosci. (2021) 81:479–91. doi: 10.1002/jdn.10123
44. Ferri, SL, Dow, HC, Schoch, H, Lee, JY, Brodkin, ES, and Abel, T. Age-and sex-specific fear conditioning deficits in mice lacking Pcdh 10, an autism associated gene. Neurobiol Learn Mem. (2021) 178:107364. doi: 10.1016/j.nlm.2020.107364
45. Schoch, H, Kreibich, AS, Ferri, SL, White, RS, Bohorquez, D, Banerjee, A, et al. Sociability deficits and altered amygdala circuits in mice lacking Pcdh 10, an autism associated gene. Biol Psychiatry. (2017) 81:193–202. doi: 10.1016/j.biopsych.2016.06.008
46. Monti, JM, and Jantos, H. The effects of local microinjection of selective dopamine D1 and D2 receptor agonists and antagonists into the dorsal raphe nucleus on sleep and wakefulness in the rat. Behav Brain Res. (2018) 339:11–8. doi: 10.1016/j.bbr.2017.11.006
47. Benvenuto, A, Moavero, R, Alessandrelli, R, Manzi, B, and Curatolo, P. Syndromic autism: causes and pathogenetic pathways. World J Pediatr. (2009) 5:169–76. doi: 10.1007/s12519-009-0033-2
48. Guo, D, Zhang, S, Sun, H, Xu, X, Hao, Z, Mu, C, et al. Tyrosine hydroxylase down-regulation after loss of Abelson helper integration site 1 (AHI1) promotes depression via the circadian clock pathway in mice. J Biol Chem. (2018) 293:5090–101. doi: 10.1074/jbc.RA117.000618
49. Lu, DH, Liao, HM, Chen, CH, Tu, HJ, Liou, HC, Gau, SS, et al. Impairment of social behaviors in Arhgef 10 knockout mice. Mol Autism. (2018) 9:11. doi: 10.1186/s13229-018-0197-5
50. Kosillo, P, and Bateup, HS. Dopaminergic Dysregulation in Syndromic autism Spectrum disorders: insights from genetic mouse models. Front Neural Circuits. (2021) 15:700968. doi: 10.3389/fncir.2021.700968
51. Mariggiò, MA, Palumbi, R, Vinella, A, Laterza, R, Petruzzelli, MG, Peschechera, A, et al. DRD1 and DRD2 receptor polymorphisms: genetic Neuromodulation of the dopaminergic system as a risk factor for ASD, ADHD and ASD/ADHD overlap. Front Neurosci. (2021) 15:705890. doi: 10.3389/fnins.2021.705890
52. Saha, S, Chatterjee, M, Shom, S, Sinha, S, and Mukhopadhyay, K. Functional SLC6A3 polymorphisms differentially affect autism spectrum disorder severity: a study on Indian subjects. Metab Brain Dis. (2022) 37:397–410. doi: 10.1007/s11011-021-00876-4
53. DiCarlo, GE, Aguilar, JI, Matthies, HJ, Harrison, FE, Bundschuh, KE, West, A, et al. Autism-linked dopamine transporter mutation alters striatal dopamine neurotransmission and dopamine-dependent behaviors. J Clin Invest. (2019) 129:3407–19. doi: 10.1172/JCI127411
54. Picchioni, D, Pixa, ML, Fukunaga, M, Carr, WS, Horovitz, SG, Braun, AR, et al. Decreased connectivity between the thalamus and the neocortex during human nonrapid eye movement sleep. Sleep. (2014) 37:387–97. doi: 10.5665/sleep.3422
55. Linke, AC, Chen, B, Olson, L, Ibarra, C, Fong, C, Reynolds, S, et al. Sleep problems in preschoolers with autism Spectrum disorder are associated with sensory sensitivities and Thalamocortical Overconnectivity. Biol Psychiatry Cogn Neurosci Neuroimaging. (2021)
56. Takata, N. Thalamic reticular nucleus in the thalamocortical loop. Neurosci Res. (2020) 156:32–40. doi: 10.1016/j.neures.2019.12.004
57. Lewis, LD, Voigts, J, Flores, FJ, Schmitt, LI, Wilson, MA, Halassa, MM, et al. Thalamic reticular nucleus induces fast and local modulation of arousal state. elife. (2015) 4:e08760. doi: 10.7554/eLife.08760
58. Golomb, D, Wang, XJ, and Rinzel, J. Synchronization properties of spindle oscillations in a thalamic reticular nucleus model. J Neurophysiol. (1994) 72:1109–26. doi: 10.1152/jn.1994.72.3.1109
59. Pastore, SF, Ko, SY, Frankland, PW, Hamel, PA, and Vincent, JB. PTCHD1: identification and neurodevelopmental contributions of an autism Spectrum disorder and intellectual disability susceptibility gene. Genes. (2022) 13:527. doi: 10.3390/genes13030527
60. Wells, MF, Wimmer, RD, Schmitt, LI, Feng, G, and Halassa, MM. Thalamic reticular impairment underlies attention deficit in Ptchd 1 (Y/−) mice. Nature. (2016) 532:58–63. doi: 10.1038/nature17427
61. Torrico, B, Fernàndez-Castillo, N, Hervás, A, Milà, M, Salgado, M, Rueda, I, et al. Contribution of common and rare variants of the PTCHD1 gene to autism spectrum disorders and intellectual disability. Eur J Hum Genet. (2015) 23:1694–701. doi: 10.1038/ejhg.2015.37
62. Pirone, A, Viaggi, C, Cantile, C, Giannessi, E, Pardini, C, Vaglini, F, et al. Morphological alterations of the reticular thalamic nucleus in Engrailed-2 knockout mice. J Anat. (2020) 236:883–90. doi: 10.1111/joa.13150
63. Tan, J, McKenzie, C, Potamitis, M, Thorburn, AN, Mackay, CR, and Macia, L. The role of short-chain fatty acids in health and disease. Adv Immunol. (2014) 121:91–119. doi: 10.1016/B978-0-12-800100-4.00003-9
64. Szentirmai, É, Millican, NS, Massie, AR, and Kapás, L. Butyrate, a metabolite of intestinal bacteria, enhances sleep. Sci Rep. (2019) 9:7035. doi: 10.1038/s41598-019-43502-1
65. Hua, X, Zhu, J, Yang, T, Guo, M, Li, Q, Chen, J, et al. The gut microbiota and associated metabolites are altered in sleep disorder of children with autism Spectrum disorders. Front Psych. (2020) 11:855. doi: 10.3389/fpsyt.2020.00855
66. Andreo-Martínez, P, García-Martínez, N, Sánchez-Samper, EP, and Martínez-González, AE. An approach to gut microbiota profile in children with autism spectrum disorder. Environ Microbiol Rep. (2020) 12:115–35. doi: 10.1111/1758-2229.12810
67. Averina, OV, Kovtun, AS, Polyakova, SI, Savilova, AM, Rebrikov, DV, and Danilenko, VN. The bacterial neurometabolic signature of the gut microbiota of young children with autism spectrum disorders. J Med Microbiol. (2020) 69:558–71. doi: 10.1099/jmm.0.001178
68. Shah, HE, Bhawnani, N, Ethirajulu, A, Alkasabera, A, Onyali, CB, Anim-Koranteng, C, et al. Iron deficiency-induced changes in the hippocampus, corpus striatum, and monoamines levels that Lead to anxiety, depression, sleep disorders, and psychotic disorders. Cureus. (2021) 13:e18138. doi: 10.7759/cureus.18138
69. Murat, S, Ali, U, Serdal, K, Süleyman, D, İlknur, P, Mehmet, S, et al. Assessment of subjective sleep quality in iron deficiency anaemia. Afr Health Sci. (2015) 15:621–7. doi: 10.4314/ahs.v15i2.40
70. Youssef, J, Singh, K, Huntington, N, Becker, R, and Kothare, SV. Relationship of serum ferritin levels to sleep fragmentation and periodic limb movements of sleep on polysomnography in autism spectrum disorders. Pediatr Neurol. (2013) 49:274–8. doi: 10.1016/j.pediatrneurol.2013.06.012
71. Hopkins, SC, Dedic, N, and Koblan, KS. Effect of TAAR1/5-HT (1A) agonist SEP-363856 on REM sleep in humans. Transl Psychiatry. (2021) 11:228. doi: 10.1038/s41398-021-01331-9
72. Gabriele, S, Sacco, R, and Persico, AM. Blood serotonin levels in autism spectrum disorder: a systematic review and meta-analysis. Eur Neuropsychopharmacol. (2014) 24:919–29. doi: 10.1016/j.euroneuro.2014.02.004
73. Carpita, B, Stagnari, R, Palego, L, Baroni, D, Massimetti, G, Nardi, B, et al. Circulating levels of 5-HT and BDNF in adults with autism Spectrum conditions: an investigation in a sample of subjects with autism Spectrum disorder, their first-degree relatives and controls. Curr Med Chem. (2023) 30:15031. doi: 10.2174/0929867330666230131115031
74. Singh, AS, Chandra, R, Guhathakurta, S, Sinha, S, Chatterjee, A, Ahmed, S, et al. Genetic association and gene-gene interaction analyses suggest likely involvement of ITGB3 and TPH2 with autism spectrum disorder (ASD) in the Indian population. Prog Neuro-Psychopharmacol Biol Psychiatry. (2013) 45:131–43. doi: 10.1016/j.pnpbp.2013.04.015
75. Noroozi, R, Ghafouri-Fard, S, Omrani, MD, Habibi, M, Sayad, A, and Taheri, M. Association study of the vesicular monoamine transporter 1 (VMAT1) gene with autism in an Iranian population. Gene. (2017) 625:10–4. doi: 10.1016/j.gene.2017.05.003
76. Jaiswal, P, Guhathakurta, S, Singh, AS, Verma, D, Pandey, M, Varghese, M, et al. SLC6A4 markers modulate platelet 5-HT level and specific behaviors of autism: a study from an Indian population. Prog Neuro-Psychopharmacol Biol Psychiatry. (2015) 56:196–206. doi: 10.1016/j.pnpbp.2014.09.004
77. Chakraborti, B, Verma, D, Karmakar, A, Jaiswal, P, Sanyal, A, Paul, D, et al. Genetic variants of MAOB affect serotonin level and specific behavioral attributes to increase autism spectrum disorder (ASD) susceptibility in males. Prog Neuro-Psychopharmacol Biol Psychiatry. (2016) 71:123–36. doi: 10.1016/j.pnpbp.2016.07.001
78. Chen, C, and Van Horn, JD. Developmental neurogenetics and multimodal neuroimaging of sex differences in autism. Brain Imaging Behav. (2017) 11:38–61. doi: 10.1007/s11682-015-9504-3
79. Soiza-Reilly, M, and Commons, KG. Unraveling the architecture of the dorsal raphe synaptic neuropil using high-resolution neuroanatomy. Front neural circuits. (2014) 8:105. doi: 10.3389/fncir.2014.00105
80. Bukatova, S, Renczes, E, Reichova, A, Filo, J, Sadlonova, A, Mravec, B, et al. Shank 3 deficiency is associated with altered profile of neurotransmission markers in pups and adult mice. Neurochem Res. (2021) 46:3342–55. doi: 10.1007/s11064-021-03435-6
81. Hasegawa, E, Miyasaka, A, Sakurai, K, Cherasse, Y, Li, Y, and Sakurai, T. Rapid eye movement sleep is initiated by basolateral amygdala dopamine signaling in mice. Science (New York, NY). (2022) 375:994–1000. doi: 10.1126/science.abl6618
82. Wellman, LL, Lonart, G, Adkins, AM, and Sanford, LD. Regulation of dark period sleep by the amygdala: a microinjection and optogenetics study. Brain Res. (2022) 1781:147816. doi: 10.1016/j.brainres.2022.147816
83. Sadakata, T, Shinoda, Y, Oka, M, Sekine, Y, and Furuichi, T. Autistic-like behavioral phenotypes in a mouse model with copy number variation of the CAPS2/CADPS2 gene. FEBS Lett. (2013) 587:54–9. doi: 10.1016/j.febslet.2012.10.047
84. Bonora, E, Graziano, C, Minopoli, F, Bacchelli, E, Magini, P, Diquigiovanni, C, et al. Maternally inherited genetic variants of CADPS2 are present in autism spectrum disorders and intellectual disability patients. EMBO Mol Med. (2014) 6:795–809. doi: 10.1002/emmm.201303235
85. Vardarajan, BN, Eran, A, Jung, JY, Kunkel, LM, and Wall, DP. Haplotype structure enables prioritization of common markers and candidate genes in autism spectrum disorder. Transl Psychiatry. (2013) 3:e262. doi: 10.1038/tp.2013.38
86. Zisapel, N. New perspectives on the role of melatonin in human sleep, circadian rhythms and their regulation. Br J Pharmacol. (2018) 175:3190–9. doi: 10.1111/bph.14116
87. Rossignol, DA, and Frye, RE. Melatonin in autism spectrum disorders: a systematic review and meta-analysis. Dev Med Child Neurol. (2011) 53:783–92. doi: 10.1111/j.1469-8749.2011.03980.x
88. da Silveira, C-MS, Guissoni Campos, LM, Fadini, CC, Anderson, G, Markus, RP, and Pinato, L. Disrupted nocturnal melatonin in autism: association with tumor necrosis factor and sleep disturbances. J Pineal Res. (2021) 70:e12715
89. Rzepka-Migut, B, and Paprocka, J. Efficacy and safety of melatonin treatment in children with autism Spectrum disorder and attention-deficit/hyperactivity disorder-a review of the literature. Brain Sci. (2020) 10:219. doi: 10.3390/brainsci10040219
90. Abel, EA, Schwichtenberg, AJ, Mannin, OR, and Marceau, K. Brief report: a gene enrichment approach applied to sleep and autism. J Autism Dev Disord. (2020) 50:1834–40. doi: 10.1007/s10803-019-03921-5
91. Veatch, OJ, Pendergast, JS, Allen, MJ, Leu, RM, Johnson, CH, Elsea, SH, et al. Genetic variation in melatonin pathway enzymes in children with autism spectrum disorder and comorbid sleep onset delay. J Autism Dev Disord. (2015) 45:100–10. doi: 10.1007/s10803-014-2197-4
92. Wu, ZY, Huang, SD, Zou, JJ, Wang, QX, Naveed, M, Bao, HN, et al. Autism spectrum disorder (ASD): disturbance of the melatonin system and its implications. Biomed Pharmacother. (2020) 130:110496. doi: 10.1016/j.biopha.2020.110496
93. Irmak, SO, and de Lecea, L. Basal forebrain cholinergic modulation of sleep transitions. Sleep. (2014) 37:1941–51. doi: 10.5665/sleep.4246
94. Griguoli, M, and Pimpinella, D. Medial septum: relevance for social memory. Front Neural Circuits. (2022) 16:965172. doi: 10.3389/fncir.2022.965172
95. Rapanelli, M, Wang, W, Hurley, E, Feltri, ML, Pittenger, C, Frick, LR, et al. Cholinergic neurons in the basal forebrain are involved in behavioral abnormalities associated with Cul3 deficiency: role of prefrontal cortex projections in cognitive deficits. Transl Psychiatry. (2023) 13:22. doi: 10.1038/s41398-023-02306-8
Keywords: sleep disorders, autism spectrum disorder, neural mechanism, wakening time, NREM sleep, REM sleep, sleep–wake rhythm, genetic mechanism
Citation: Ji Q, Li S-J, Zhao J-B, Xiong Y, Du X-H, Wang C-X, Lu L-M, Tan J-Y and Zhu Z-R (2023) Genetic and neural mechanisms of sleep disorders in children with autism spectrum disorder: a review. Front. Psychiatry. 14:1079683. doi: 10.3389/fpsyt.2023.1079683
Received: 25 October 2022; Accepted: 13 April 2023;
Published: 02 May 2023.
Edited by:
Anthony Russo, Hartwick College, United StatesReviewed by:
Lifei Xiao, Ningxia Medical University, ChinaCopyright © 2023 Ji, Li, Zhao, Xiong, Du, Wang, Lu, Tan and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Ru Zhu, emh1enIyMDEzQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.