
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Psychiatry, 24 November 2022
Sec. Mood Disorders
Volume 13 - 2022 | https://doi.org/10.3389/fpsyt.2022.1020530
This article is part of the Research TopicCommunity Series in Bio-Psycho-Social Indicators of Suicide Risk - Volume IIView all 5 articles
 Britta Stapel1*†
Britta Stapel1*† Ke Xiao2,3†
Ke Xiao2,3† Nataliya Gorinski4
Nataliya Gorinski4 Kevin Schmidt5
Kevin Schmidt5 Angelika Pfanne5
Angelika Pfanne5 Jan Fiedler2,3
Jan Fiedler2,3 Imke Richter4
Imke Richter4 Anna-Lena Vollbrecht4
Anna-Lena Vollbrecht4 Thomas Thum2,3,5
Thomas Thum2,3,5 Kai G. Kahl1‡
Kai G. Kahl1‡ Evgeni Ponimaskin4*‡
Evgeni Ponimaskin4*‡Objective: Major depressive disorder (MDD) constitutes a main risk factor for suicide. Suicide risk in psychiatric patients is primarily determined by often unreliable, self-reported information. We assessed serum levels of three microRNAs (miRNAs), previously demonstrated to be dysregulated in post-mortem brain samples of suicide victims, as potential peripheral biomarkers for suicidality.
Methods: All study participants were diagnosed with MDD according to Diagnostic and Statistical Manual of Mental Disorders, 5th edition criteria. Suicidality, defined as acute suicide risk or suicide attempt within one week prior to study entry, was assessed by clinical interview. Relative serum levels of miR-30a, miR-30e, and miR-200a, normalized to U6, were measured by quantitative real-time PCR in MDD inpatients with (MDD/SI, N = 19) and without (MDD, N = 31) acute suicide risk. Median age and gender distribution were comparable in both groups.
Results: Levels of miR-30a, miR-30e, and miR-200a were significantly elevated in MDD/SI compared to MDD. Subgroup analysis of the MDD/SI group showed that levels of miR-30e and miR-200a were significantly higher and miR-30a was increased by trend in patients admitted following a suicide attempt (N = 7) compared to patients with acute suicide risk but without recent suicide attempt (N = 12). Additionally, use of two databases for in silico transcription factor–miRNA interaction prediction indicated early growth response protein (EGR) 1 as potential transcriptional regulator for all three miRNAs.
Conclusion: This study demonstrates suicide risk in MDD patients to be associated with increased levels of miR-30a, miR-30e, and miR-200a. Thus, these miRNAs might constitute potential biomarkers to predict suicidal behavior in MDD patients.
Major depressive disorder (MDD), defined as at least one discrete depressive episode that lasts for more than two weeks, is a debilitating mental disorder that is estimated to affect about 6% of the adult population worldwide every year (1). When measured as “years lived with disability,” MDD constitutes the second leading burden of disease (2). Importantly, MDD is one of the main risk factors for death by suicide. According to the World Health Organization, every year over 800,000 people die due to self-inflicted injuries (3). This accounts for 1.5% of total death and makes suicide one of the leading causes of death in people 15–34 years of age (4). In this regard, it is also noteworthy that the number of non-fatal suicide attempts significantly exceeds the number of suicides, i.e., for each death by suicide, there are an estimated 25 suicide attempts (5, 6). In the context of MDD, results from the ESEMED study, a European cross-sectional household survey, indicate that attributable risk for lifetime suicide attempt was 28% for MDD and additionally, major depressive episode together with alcohol dependency determined most strongly the progression from suicidal ideation to suicide attempt (7).
Currently, clinicians determine suicide risk in psychiatric patients primarily based on information regarding prior suicidal behavior and a family history of suicide as provided by patients. However, reliability of self-reported information concerning suicidal ideation is limited (8) and information regarding the family history of suicidal behavior might be inaccurate due to the associated stigma.
Given the significant personal as well as economic impact that suicide imposes especially in patients with MDD, reliable markers to objectively identify suicide risk are required to improve diagnosis, classifications and outcomes.
The etiology of MDD and suicidality is currently considered to involve a complex interplay of genetic and environmental factors (9). Accordingly, research targeting the identification of potential biomarkers for suicide risk in patients with MDD has focused on structural and functional brain alterations assessed by brain imaging as well as on biochemical and genomic markers associated with systems implicated in depression and suicidal behavior (10). However, based on the well-established association of early as well as of acute adverse life events with MDD and suicide risk (11–16), epigenetic markers have recently come into focus (17). While epigenetic mechanisms are classically defined as modulations of DNA organization in form of DNA methylation and histone modifications (18–20), more recently, non-coding RNAs have been identified as post-transcriptional regulators that mediate stress-induced changes in the context of psychiatric disorders (20). Within the group of non-coding RNAs, microRNAs (miRNAs) were first described as single-stranded RNA molecules with a length of 19–25 base-pairs that bind to 3’-untranslated regions (3’UTRs) of their respective target mRNAs resulting in repression of translation or alterations in mRNA stability (21–23). Since then, various studies have investigated the involvement of miRNAs in the etiopathology of psychiatric disorders including MDD and suicidal ideation (24). Importantly, alterations in miRNA levels are not limited to the brain but distinct changes were also detected in blood samples of depressed patients highlighting their potential as putative biomarkers for MDD and suicidal behavior (24, 25).
We previously described alterations in miR-30a and miR-30e as well as in miR-200a levels in post-mortem brain samples of individuals with MDD that died by suicide compared to mentally healthy individuals that died of unrelated causes (26). Given the high prevalence of suicidality in MDD patients and the need for reliable biomarkers to determine suicide risk, we aimed to assess whether these observed dysregulations in miRNA expression could be replicated in a pilot study with depressed patients with and without suicidality and suicide attempt.
The present study was performed in accordance with principles expressed in the Declaration of Helsinki and was approved by the local ethics committee at Hannover Medical School. Written, informed consent was given by all participants before study entry. Fifty patients diagnosed with MDD in accordance to the Diagnostic and Statistical Manual of Mental Disorders 5th edition (DSM-V) criteria were included in the study. All patients were treated in the Department of Psychiatry, Social Psychiatry and Psychotherapy at Hannover Medical School at the time of study entry.
Suicidality, defined as acute suicide risk or suicide attempt within 1 week prior to study entry, was determined as follows: (1) patients self-report of acute suicidality, (2) clinician’s impression of acute suicidality, (3) necessity of psychiatric intensive care unit (PICU) treatment (either patients were transferred to PICU internally after admission following a suicide attempt, or were directly admitted to PICU as emergencies from outside). Nineteen depressed patients fulfilled the criterion suicidality (MDD/SI). Of these, N = 12 patients were at acute suicide risk, and N = 7 patients were assessed following a suicide attempt. Blood of these patients was drawn within 1 week of admission to the clinics. Thirty-one depressed inpatients without the criterion suicidality according to clinical impression and chart analysis were included as a control group (MDD).
Exclusion criteria for both the both groups included current or lifetime substance use disorder (excluding nicotine), schizophrenia, bipolar disorder, eating disorders, intellectual disability, any acute physical disorder, in particular current or lifetime (auto-) immune disease, type-2 diabetes mellitus, cardiac diseases, systemic diseases, and current infectious disease and an age < 18 years.
Blood was drawn under fasting conditions. Samples were collected in serum S-Monovettes (Sarstedt, Nümbrecht, Germany), centrifuged at 2,000 rpm for 15 min. Finally, 200 μl of blood serum was transferred into fresh tube and stored at –80°C. Isolation of miRNAs from samples was performed using the miRNeasy Serum/Plasma Advanced Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol and miRNAs were eluted in water. Reverse transcriptions were performed with the TaqMan™ MicroRNA Reverse Transcription Kit. (Applied Biosystems, Waltham, MA, USA). Subsequently, quantitative real time polymerase chain reaction (qPCR) of miR-30a, miR-30e and miR-200a was conducted with respective TaqMan™ Assays (hsa-miR-30a-5p, ID 000417; hsa-miR-30e, ID 002223; hsa-miR-200a, ID 000502; all Thermo Scientific, Waltham, MA, USA) using the ABsolute Blue QPCR Mix (Thermo Scientific, Waltham, MA, USA). Additionally, U6 snRNA levels were measured (ID: 001973, Thermo Scientific) for expression level normalization. All qPCRs were run on a QuantStudio™ 7 Flex System (Applied Biosystems, Waltham, MA, USA) amplification and detection machine. Results were analyzed with the QuantStudio™ Real-Time PCR Software v1.3 (Applied Biosystems, Waltham, MA, USA). In corresponding graphs, 2–ΔCt values are displayed.
TransmiR v2.0 database (27) and the RegNetwork database (28) were used individually to predict transcription factors (TFs) that can putatively regulate the three miRNAs investigated in this study. Additionally, Targetscan was used to identify potential target genes of the assessed miRNAs (29).
All statistical analyses were performed using SPSS version 26 (IBM, Armonk, NY, USA). Extreme outliers defined as values three times the interquartile range (IQR) were excluded from the respective analyses. Kolmogorov-Smirnoff test was used to test for normal distribution of data and subsequently, Mann-Whitney U test was applied to assess group differences. To analyze group differences of nominal data, Chi Square test followed by Fisher’s Exact test was utilized. Two-tailed P-values are depicted and P-values < 0.05 were considered statistically significant.
No significant differences with regard to age (U = 233.00, Z = –1.230, P = 0.223) and gender distribution [χ2(1) = 0.002, P = 1.00] were observed between the MDD/SI and the MDD group (Table 1).
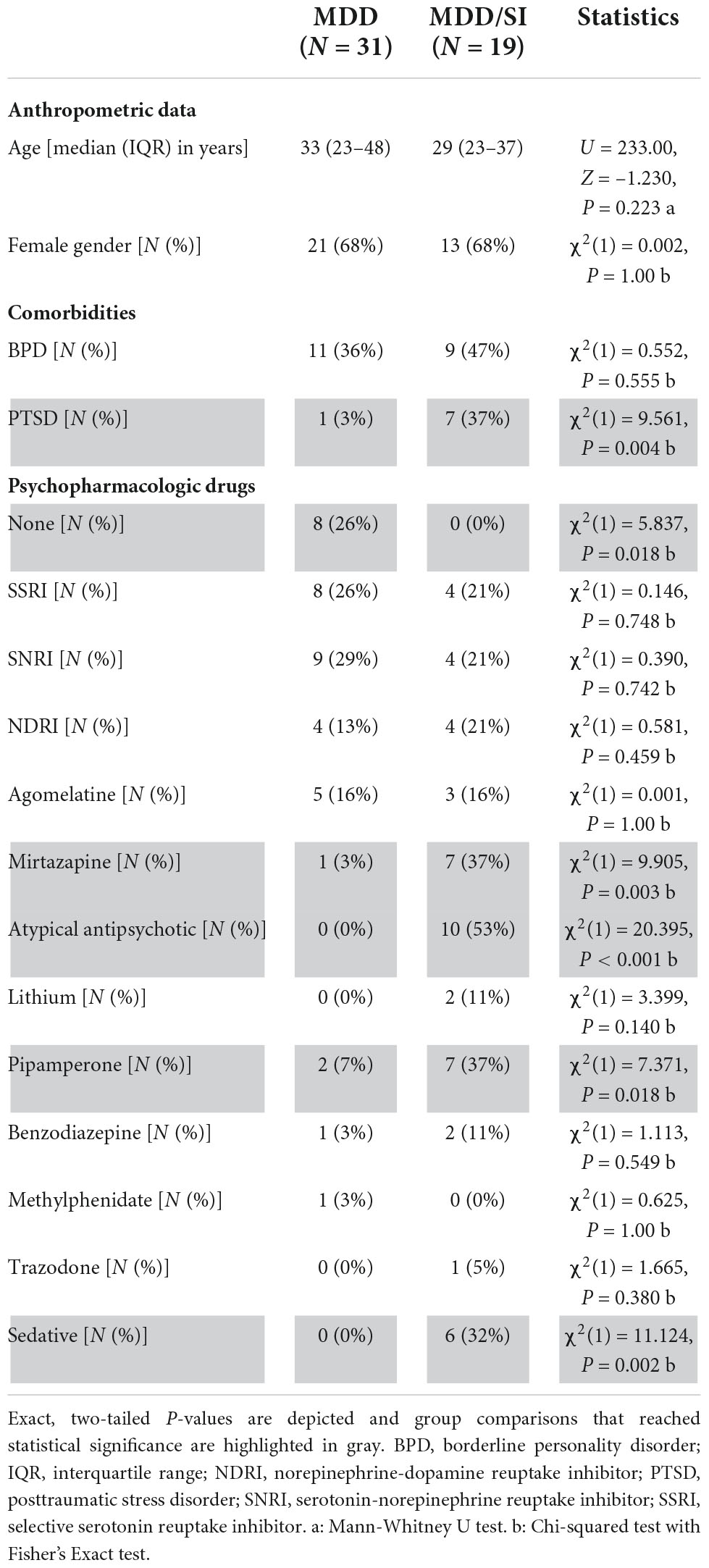
Table 1. Anthropometric data and psychopharmacologic drug treatment in the MDD and the MDD/SI group.
Relevant psychiatric comorbidities included borderline personality disorder (BPD) that was similar in frequency in the MDD/SI and in the MDD group [χ2(1) = 0.552, P = 0.555] and posttraumatic stress disorder (PTSD) that was more frequent in the MDD/SI group than in the MDD group [χ2(1) = 9.561, P = 0.004] (Table 1).
Concerning psychopharmacologic drug treatment, all patients in the MDD/SI group [N = 19 (100%)], and 23 out of 31 patients in the MDD group (74.2%) received at least one psychopharmacologic drug [χ2(1) = 5.837, P = 0.018] (Table 1). In this regard, use of mirtazapine [χ2(1) = 9.905, P = 0.003], of pipamperone [χ2(1) = 7.371, P = 0.018], and of atypical antipsychotics [χ2(1) = 20.395, P < 0.001] was more frequent in the MDD/SI group when compared to the MDD group (Table 1). Finally, N = 6 patients in the MDD/SI group received a sedative, compared to none of the patients in the MDD group [χ2(1) = 11.124, P = 0.002] (Table 1).
Relative serum levels of miR-30a (U = 123.00, Z = –3.235, P = 0.001), miR-30e (U = 114.00, Z = –3.223, P = 0.001) and of miR-200a (U = 150.00, Z = –2.888, P = 0.003) normalized to U6 were significantly higher in the MDD/SI group compared to the MDD group (Figures 1A–C and Supplementary Table 1).
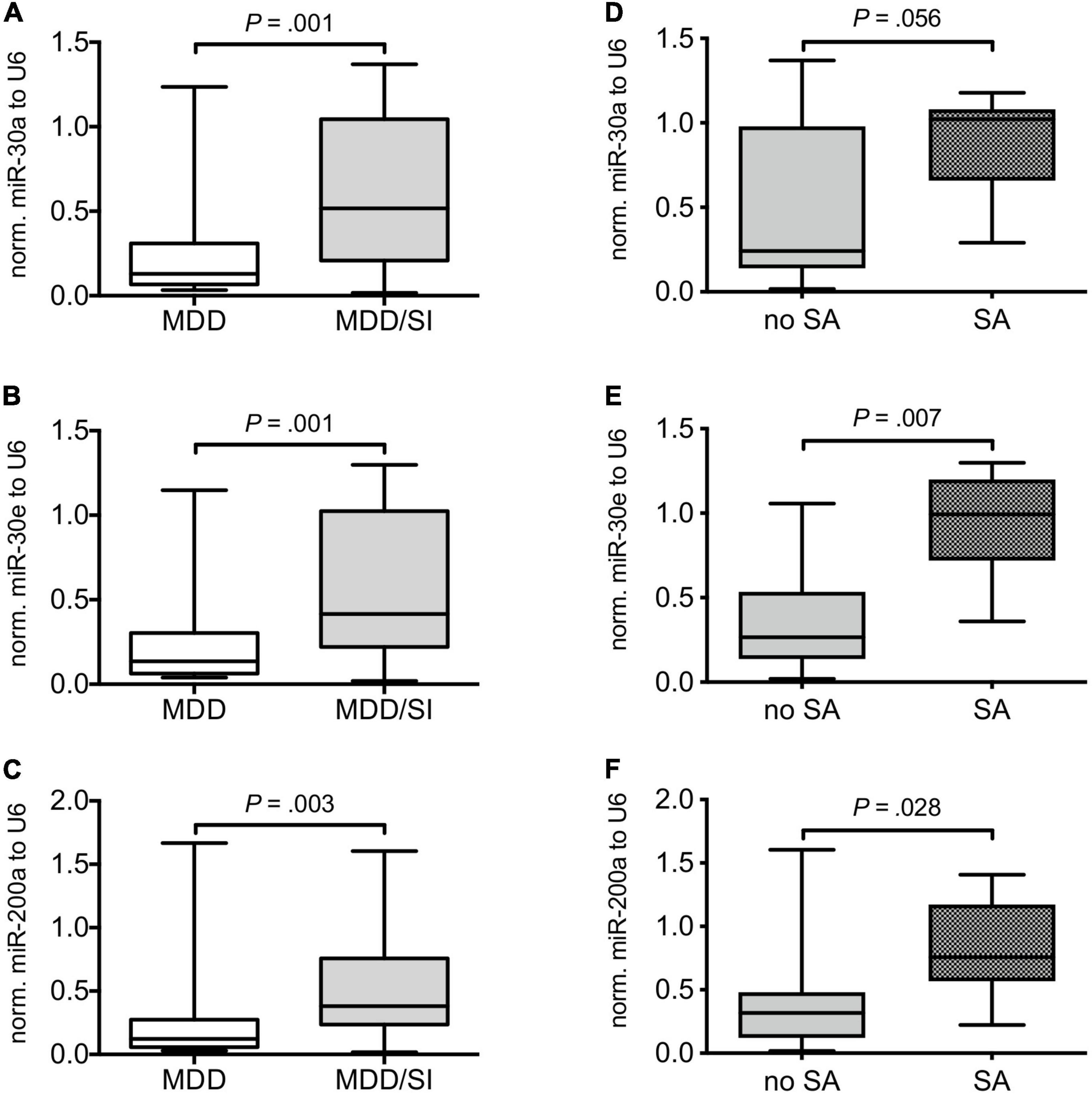
Figure 1. Comparison of relative miRNA expression in MDD patients with and without acute suicide risk and in suicidal MDD patients with and without suicide attempt. Boxplots depict median with interquartile range and whiskers depict minimum and maximum values. The left panel shows relative miR-30a (A), miR-30e (B), and miR-200a (C) expression normalized to U6 in MDD patients without (MDD) and in MDD patients with (MDD/SI) acute suicide risk. The right panel depicts relative levels of miR-30a (D), miR-30e (E), and miR-200a (F) in suicidal patients with (SA) and without (no SA) suicide attempt. Exact, two-tailed P-values were computed by use of Whitney-Mann U test and P < 0.05 was considered statistically significant.
As the MDD/SI group displayed heterogeneous values with regard to miRNA expression, sub-group analyses of the MDD/SI group were performed to compare suicide attempters (SA) to individuals with an acute suicide risk, but without suicide attempt (no SA). Both subgroups were comparable regarding mean age (U = 31.00, Z = –0.931, P = 0.372) and gender distribution [χ2(1) = 0.046, P = 0.622] (Table 2). Additionally, no significant differences in psychopharmacological medication were present (Table 2). However, comorbid BPD tended to be more frequent in the no SA subgroup [χ2(1) = 4.866, P = 0.057], while frequency of PTSD diagnosis was comparable between the SA and the no SA subgroups [χ2(1) = 0.326, P = 0.656] (Table 2).
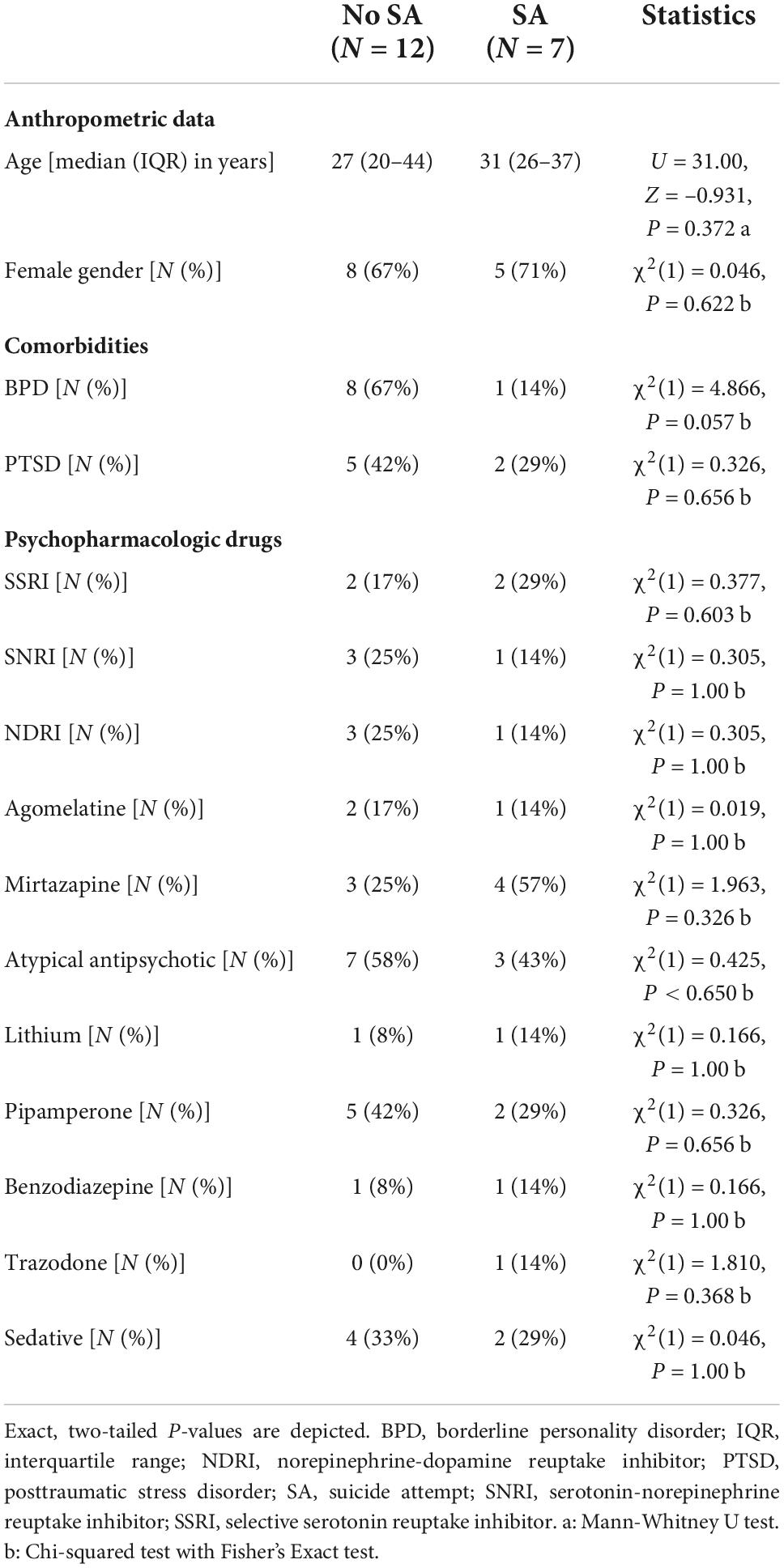
Table 2. Anthropometric data and psychopharmacologic drug treatment in MDD/SI subgroups with and without suicide attempt (SA).
Subsequent subgroup analyses of the MDD/SI group showed significantly higher values of miR-30e (U = 8.00, Z = –2.635, P = 0.007) and of miR-200a (U = 16.00, Z = –2.197, P = 0.028) in SA patients when compared to the no SA group. Additionally, a trend toward higher values of miR-30a was observed in the SA group compared to the no SA group (U = 17.00, Z = –1.947, P = 0.056) (Figures 1D–F and Supplementary Table 2).
To identify potential transcriptional regulators of the differentially expressed miRNAs, miR-30a, miR-30e, and miR-200a were submitted to the TransmiR v2.0 database (27) to predict putative transcription factor (TF) binding sites within their respective promoter regions. The approach indicated 77 TFs that were predicted to putatively regulate at least two of the three miRNAs, and 18 TFs, including early growth response 1 (EGR1), that were implicated in the regulation of all three miRNAs (Figures 2A,B). To confirm the obtained results, RegNetwork as a second TF-miRNA database was utilized in parallel (28). Using this approach, 14 TFs were identified to be able to regulate one of the three miRNAs with high confidence (Figure 2C and Supplementary Table 3). Among them, EGR1 was found to putatively interact with miR-30a and miR-30e. Overall, comparing results from both approaches, EGR1 was predicted as a potential transcriptional regulator of the relevant miRNAs with high confidence.
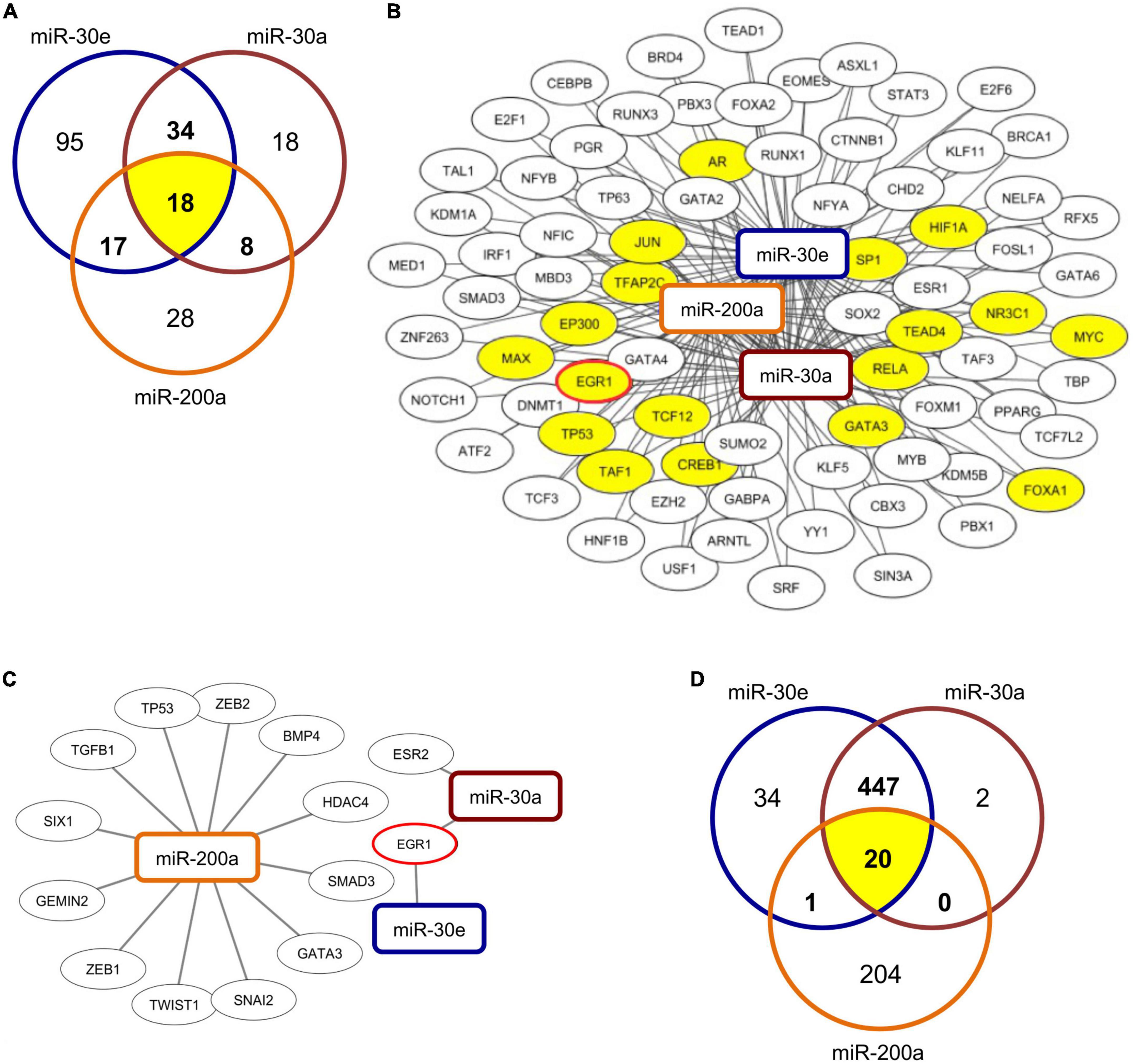
Figure 2. Prediction of TF-miRNA binding and of potential miRNA targets. (A) Venn diagram depicts number of predicted transcription factors (TF) that are able to interact with the respective miRNAs and their overlap and (B) shows network of TF-miRNA interactions of 77 TFs predicted to interact with at least two of the three assessed miRNAs. Visualization was carried out using the Cytoscape software with the respective miRNAs predicted by TransmiR database. The 18 common TFs predicted to interact with all three miRNAs are highlighted in yellow. (C) Network of predicted TF-miRNA interactions from RegNetwork database analysis. 14 TFs were predicted to interact with at least one of the assessed miRNAs. Visualization was carried out by use of the Cytoscape software. (D) Venn diagram depicts number of predicted targets of respective miRNAs and their overlap.
Targetscan (29) was used to identify potential target genes of the assessed miRNAs. In silico analyses identified 469 genes that can potentially be targeted by miR-30a, 502 genes that are potential targets of miR-30e, and 225 genes that constitutes potential targets of miR-200a (Figure 2D) at the 3′UTR region. Among them 20 genes were predicted to be targeted by all three miRNAs (Supplementary Table 4).
In the present study, we report significantly higher relative expression levels of three miRNAs (i.e., miR-30a, miR-30e, and miR-200a) in blood serum samples from suicidal MDD patients compared to an MDD group that did not display current suicide risk. It is noteworthy that our previous study demonstrated that expression of these miRNAs was dysregulated in post-mortem brain samples of MDD patients that died by suicide (26). Additionally, we propose transcription factor EGR1 as potential regulator of miRNA expression.
Studies dedicated to the identification of suitable biomarkers for MDD and therapeutic responses to psychopharmacologic treatment are numerous and subsequently potential molecular markers have been identified on the genomic, epigenomic, transcriptomic, and proteomic level (30, 31).
Contrarily, studies that aim to identify markers for suicidality are less frequent. In this regard, several studies assessed alterations in miRNA expression in different brain regions of suicide victims (32, 33). However, while these findings offer important insights in potential pathomechanisms underlying suicidal behavior, interpretation of results is complicated by an often unclear psychiatric diagnose and/or by the lack of an appropriate control group (32). Consequently, post-mortem studies often do not allow for a differentiation between miRNA regulation linked to suicidality and changes brought forward by the underlying psychiatric illness. Therefore, a replication of post-mortem results in clinical samples is of significant importance.
In the context of central nervous system (CNS)-related disorders, including psychiatric disorders and MDD, numerous studies have described alterations in miRNA levels in peripheral blood samples (25), and it has been shown that alterations in brain miRNA expression appear to be reflected in the periphery (34, 35). Albeit the need for reliable biomarkers that objectively predict suicide risk in MDD patients and the demonstrated potential of peripheral miRNAs as suitable biomarkers in the context of CNS-related diseases (34), a recent systemic review identified only two studies that assessed miRNAs as potential biomarkers for suicidality in peripheral blood samples (33) and neither study included patients with recent suicide attempt (36, 37).
This is of importance, as the observed higher levels of miR-30a, miR-30e, and miR-200a in suicidal MDD patients compared to those without current suicide risk in this study, could be mostly attributed to a subgroup of patients that had recently attempted suicide compared to those without a recent suicide attempt.
Our observation is in line with the understanding that suicidal behavior, defined as suicide attempt or suicide completion, is characterized by overlapping, psychobiological abnormalities that are discernable from suicidal ideation (38). Accordingly, it is generally thought that vulnerability to suicidal behavior is mediated partly by an underlying genetic predisposition, which interacts with environmental and probable epigenetic factors (39), and might be reflected by changes in miRNA expression. Indeed, several studies demonstrated changed miRNA expression profiles in post-mortem as well as peripheral blood samples from MDD patients (40, 41). In particular, our previous study identified altered expression levels of miR-30a, miR-30e, and miR-200a in prefrontal cortex (PFC) samples of suicide victims diagnosed with depression compared to mentally healthy controls that died of unrelated causes (26). Interestingly, an association analyses in MDD patients and control subjects identified polymorphisms within the miR-30e precursor to be positively correlated with depression and its symptomatic onset (42). Moreover, results from a social defeat stress mouse model indicated a downregulation of miR-30a and miR-30e in the context of stress and depression-like behavior and in hippocampal neurogenesis (43). Intriguingly, we have recently found that the higher prevalence for MDD and panic disorders in women with peripartum cardiomyopathy was associated with elevated levels of miR-30e (44). Finally, alterations in miR-200a expression have been reported in a rodent model of unpredictable chronic mild stress as well as in plasma samples of patients with Alzheimer’s disease (45–47).
Regarding potential upstream regulators of miR-30a, miR-30e, and miR-200a transcription, predictions based on two independent database searches identified EGR1 as a potential transcriptional regulator of the assessed miRNAs. EGR1, also known as NGFI-A, constitutes a nuclear phosphoprotein attributed with an activation potential as well as with a negative regulatory function (48–51). Among others, EGR1 was found to be critically involved in neuroplasticity (52). Decreased EGR1 expression was found in post-mortem PFC samples of MDD patients in comparison to healthy individuals (53) and its downregulation was also detected in animal models of depression and anxiety in specific brain areas including the hippocampus (53, 54). Importantly, a study based on transcriptome sequencing data derived from post-mortem PFC samples found a gradual downregulation of a gene cluster that included EGR1 in non-suicidal MDD patients compared to control with a further decrease being detected in suicidal MDD patients (55). Given the opposite regulation of EGR1 (i.e., decreased in the context of MDD and suicidality) and of the miRNAs assessed in this study (i.e., increased in the context of SI and SA), one might speculate that EGR1 could function as a negative regulator of these miRNAs. In this regard, repressor function of EGR1 has been previously shown in neuronal cells (56–58) and positive as well as negative regulatory effects of EGR1 on miRNA expression have been described in various tissues and disease states (59–63) and EGR1 binding sites within the predicted promoter region of miR-30a and miR-30e were identified (64). A potential transcriptional regulation of the relevant miRNAs by EGR1 and its direction should be concern of future studies.
Concerning potential targets of miRNA-30a, miR-30e, and miR-200a, in silico analysis identified 20 mRNAs that were predicted to be regulated by all three miRNAs via their respective 3’ UTRs. To our knowledge, out of these predicted targets, only three [i.e., muscleblind like splicing regulator 1 (MBNL1), receptor accessory protein 3 (REEP3), retinoic acid receptor beta (RARB)] have previously been reported in the context of depression or depression-like phenotypes in the literature and none were described to be associated with suicidality (65–68). In this regard, only RARB has been reported in the context of MDD in humans (68) and rodents (66), while MBNL1 and REEP3 were reported only in rodent models (65, 67). The respective studies utilized screening approaches and we are unaware of studies that have investigated the function of the respective genes in the context of depression and/or suicidality. While our in silico analysis gives a first indication of potential targets of the assessed miRNAs in the context of depression and suicidality, further studies are needed to confirm these targets in vitro and to assess their respective relevancy in the context of suicidality in patients with MDD.
In conclusion, we found that miRNAs previously described to be dysregulated in post-mortem brain samples of MDD patients that died by suicide, were elevated in serum samples of MDD patients that displayed current suicide risk in comparison to MDD patients that did not show acute suicidal ideation. Our results suggest that the observed miRNAs might function as a trait marker of suicidal behavior (i.e., suicide attempt and suicide completion) as the overall higher levels could be mainly attributed to a patient subgroup that attempted suicide in the week prior to study entry. Therefore, further studies might focus not only on the potential application of these miRNAs as biomarkers for suicidal behavior, but also on their respective involvement in the pathomechanisms that underlie suicidal behavior in general and in the context of MDD in particular.
This pilot study has several limitations. Firstly, we do not provide data from a second validation cohort. Additionally, the respective MDD groups in this study are heterogeneous regarding psychopharmacological treatment and psychiatric comorbidities. This is of importance as pharmacological drugs might differentially impact miRNA expression at the subcellular level which we also cannot specify herein (69). Next to intracellular miRNA biogenesis, we can only speculate on miRNA secretion to patient’s plasma, which is often a directed mechanism via extracellular vesicles (70). Additionally, it remains unclear whether the neurobiology of suicidality is consistent across different diagnoses. For instant, an imaging study found mGluR5 availability to be associated with suicidality in patients with PTSD but not in those with MDD (71). On the other hand, it has also been demonstrated that suicidal behavior (suicide attempt and completion) are characterized by psychobiological abnormalities that are independent of co-occurring psychiatric disorders (72). As PTSD was more frequent in the MDD/SI group compared to the MDD group, we cannot exclude that PTSD comorbidity contributed to the observed effects. However, given the fact that frequency of PTSD comorbidity as well as medication did not significantly differ in the respected MDD/SI subgroups it appears likely that the observed differential miRNA levels are indeed brought forward by suicidal behavior. Finally, identification of suicidal ideation might lack accuracy as its diagnosis in our study solely relied on self-report in the MDD/SI group, and on chart analysis in the MDD group without suicidality. This might be of importance as it has been shown that prior suicide attempts partly predict future suicidality (73).
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by Ethics Committee, Hanover Medical School. The patients/participants provided their written informed consent to participate in this study.
BS: formal analysis and writing—original draft. KX and NG: investigation, formal analysis, methodology, and visualization. KS: investigation and formal analysis. AP: methodology and writing—review and editing. JF: supervision, methodology, and writing—review and editing. IR and A-LV: resources. TT, KK, and EP: supervision, funding acquisition, project administration, conceptualization, formal analysis, and writing—review and editing. All authors contributed to the article and approved the submitted version.
This study was supported by Deutsche Forschungsgemein-schaft (DFG) grant PO732 to EP and GO3411/1-1 to NG. Additionally, this work was supported by the Fraunhofer Cluster of Excellence Immune-Mediated Diseases (CIMD). The funding sources were not involved in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
KK received speaker honoraria by Janssen, Otsuka, Neuraxpharm, EliLilly, Schwabe, Servier, and Trommsdorff/Ferrer; he received an unrestricted grant by Ferrer, ADDISCA. TT was founder and shareholder of Cardior Pharmaceuticals GmbH. TT filed and licensed patents regarding noncoding RNAs. TT received speaker honoraria and/or was in advisory boards by/of Takeda, Boehringer Ingelheim, Novo Nordisk, Sanofi-Genzyme, Amicus Therapeutics, Ksilink.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2022.1020530/full#supplementary-material
1. Bromet E, Andrade LH, Hwang I, Sampson NA, Alonso J, de Girolamo G, et al. Cross-national epidemiology of DSM-IV major depressive episode. BMC Med. (2011) 9:90. doi: 10.1186/1741-7015-9-90
2. Global Burden of Disease Study 2013 Collaborators. Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990-2013: a systematic analysis for the global burden of disease study 2013. Lancet. (2015) 386:743–800.
3. World Health Organization [WHO]. Preventing Suicide: A Global Imperative. Geneva: World Health Organization (2014).
4. Murray CJL, Lopez AD, World Health Organization, World Bank, Harvard School of Public Health. The Global Burden of Disease : A Comprehensive Assessment of Mortality and Disability from Diseases, Injuries, and Risk Factors in 1990 and Projected to 2020 : Summary / edited by Christopher J. L. Murray, Alan D. Lopez. Geneva: World Health Organization (1996).
5. Crosby AE, Han B, Ortega LA, Parks SE, Gfroerer J, Centers for Disease Control and Prevention [CDC], et al. Suicidal thoughts and behaviors among adults aged >/=18 years–United States, 2008-2009. MMWR Surveill Summ. (2011) 60:1–22.
6. Nock MK, Borges G, Bromet EJ, Cha CB, Kessler RC, Lee S. Suicide and suicidal behavior. Epidemiol Rev. (2008) 30:133–54. doi: 10.1093/epirev/mxn002
7. Bernal M, Haro JM, Bernert S, Brugha T, de Graaf R, Bruffaerts R, et al. Risk factors for suicidality in Europe: results from the ESEMED study. J Affect Disord. (2007) 101:27–34. doi: 10.1016/j.jad.2006.09.018
8. Busch KA, Fawcett J, Jacobs DG. Clinical correlates of inpatient suicide. J Clin Psychiatry. (2003) 64:14–9. doi: 10.4088/JCP.v64n0105
9. Klengel T, Binder EB. Gene-environment interactions in major depressive disorder. Can J Psychiatry. (2013) 58:76–83. doi: 10.1177/070674371305800203
10. Sudol K, Mann JJ. Biomarkers of suicide attempt behavior: toward a biological model of risk. Curr Psychiatry Rep. (2017) 19:31. doi: 10.1007/s11920-017-0781-y
11. Osvath P, Voros V, Fekete S. Life events and psychopathology in a group of suicide attempters. Psychopathology. (2004) 37:36–40. doi: 10.1159/000077018
12. Heikkinen M, Aro H, Lonnqvist J. Recent life events, social support and suicide. Acta Psychiatr Scand Suppl. (1994) 377:65–72. doi: 10.1111/j.1600-0447.1994.tb05805.x
13. Paykel ES, Prusoff BA, Myers JK. Suicide attempts and recent life events. A controlled comparison. Arch Gen Psychiatry. (1975) 32:327–33. doi: 10.1001/archpsyc.1975.01760210061003
14. Sarchiapone M, Carli V, Cuomo C, Roy A. Childhood trauma and suicide attempts in patients with unipolar depression. Depress Anxiety. (2007) 24:268–72. doi: 10.1002/da.20243
15. Molnar BE, Berkman LF, Buka SL. Psychopathology, childhood sexual abuse and other childhood adversities: relative links to subsequent suicidal behaviour in the US. Psychol Med. (2001) 31:965–77. doi: 10.1017/S0033291701004329
16. Davidson JR, Hughes DC, George LK, Blazer DG. The association of sexual assault and attempted suicide within the community. Arch Gen Psychiatry. (1996) 53:550–5. doi: 10.1001/archpsyc.1996.01830060096013
17. Allen L, Dwivedi Y. MicroRNA mediators of early life stress vulnerability to depression and suicidal behavior. Mol Psychiatry. (2020) 25:308–20. doi: 10.1038/s41380-019-0597-8
18. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. (2008) 9:465–76. doi: 10.1038/nrg2341
19. Kouzarides T. Chromatin modifications and their function. Cell. (2007) 128:693–705. doi: 10.1016/j.cell.2007.02.005
20. Pena CJ, Bagot RC, Labonte B, Nestler EJ. Epigenetic signaling in psychiatric disorders. J Mol Biol. (2014) 426:3389–412. doi: 10.1016/j.jmb.2014.03.016
22. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. (2008) 9:102–14. doi: 10.1038/nrg2290
23. Crabtree JS. Chapter 3 - Fundamentals of epigenetics. In: Crabtree JS editor. Clinical Precision Medicine. Cambridge: Academic Press (2020). p. 27–37. doi: 10.1016/B978-0-12-819834-6.00003-3
24. Serafini G, Trabucco A, Corsini G, Escelsior A, Amerio A, Aguglia A, et al. The potential of microRNAs as putative biomarkers in major depressive disorder and suicidal behavior. Biomark Neuropsychiatr. (2021) 5:100035. doi: 10.1016/j.bionps.2021.100035
25. Yuan H, Mischoulon D, Fava M, Otto MW. Circulating microRNAs as biomarkers for depression: many candidates, few finalists. J Affect Disord. (2018) 233:68–78. doi: 10.1016/j.jad.2017.06.058
26. Gorinski N, Bijata M, Prasad S, Wirth A, Abdel Galil D, Zeug A, et al. Attenuated palmitoylation of serotonin receptor 5-HT1A affects receptor function and contributes to depression-like behaviors. Nat Commun. (2019) 10:3924. doi: 10.1038/s41467-019-11876-5
27. Tong Z, Cui Q, Wang J, Zhou Y. TransmiR v2.0: an updated transcription factor-microRNA regulation database. Nucleic Acids Res. (2019) 47:D253–8. doi: 10.1093/nar/gky1023
28. Liu ZP, Wu C, Miao H, Wu H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database. (2015) 2015:bav095. doi: 10.1093/database/bav095
29. McGeary SE, Lin KS, Shi CY, Pham TM, Bisaria N, Kelley GM, et al. The biochemical basis of microRNA targeting efficacy. Science. (2019) 366:eaav1741. doi: 10.1126/science.aav1741
30. Strawbridge R, Young AH, Cleare AJ. Biomarkers for depression: recent insights, current challenges and future prospects. Neuropsychiatr Dis Treat. (2017) 13:1245–62. doi: 10.2147/NDT.S114542
31. Zhou L, Zhu Y, Chen W, Tang Y. Emerging role of microRNAs in major depressive disorder and its implication on diagnosis and therapeutic response. J Affect Disord. (2021) 286:80–6. doi: 10.1016/j.jad.2021.02.063
32. Dwivedi Y. MicroRNAs in depression and suicide: recent insights and future perspectives. J Affect Disord. (2018) 240:146–54. doi: 10.1016/j.jad.2018.07.075
33. Kouter K, Paska AV. Biomarkers for suicidal behavior: miRNAs and their potential for diagnostics through liquid biopsy - a systematic review. Epigenomics. (2020) 12:2219–35. doi: 10.2217/epi-2020-0196
34. Rao P, Benito E, Fischer A. MicroRNAs as biomarkers for CNS disease. Front Mol Neurosci. (2013) 6:39. doi: 10.3389/fnmol.2013.00039
35. Sheinerman KS, Umansky SR. Circulating cell-free microRNA as biomarkers for screening, diagnosis and monitoring of neurodegenerative diseases and other neurologic pathologies. Front Cell Neurosci. (2013) 7:150. doi: 10.3389/fncel.2013.00150
36. Wang Q, Roy B, Turecki G, Shelton RC, Dwivedi Y. Role of complex epigenetic switching in tumor necrosis factor-alpha upregulation in the prefrontal cortex of suicide subjects. Am J Psychiatry. (2018) 175:262–74. doi: 10.1176/appi.ajp.2017.16070759
37. Belzeaux R, Fiori LM, Lopez JP, Boucekine M, Boyer L, Blier P, et al. Predicting worsening suicidal ideation with clinical features and peripheral expression of messenger RNA and MicroRNA during antidepressant treatment. J Clin Psychiatry. (2019) 80:18m12556. doi: 10.4088/JCP.18m12556
38. Courtet P, Gottesman II, Jollant F, Gould TD. The neuroscience of suicidal behaviors: what can we expect from endophenotype strategies? Transl Psychiatry. (2011) 1:e7. doi: 10.1038/tp.2011.6
39. Mann JJ, Currier DM. Stress, genetics and epigenetic effects on the neurobiology of suicidal behavior and depression. Eur Psychiatry. (2010) 25:268–71. doi: 10.1016/j.eurpsy.2010.01.009
40. Lopez JP, Lim R, Cruceanu C, Crapper L, Fasano C, Labonte B, et al. miR-1202 is a primate-specific and brain-enriched microRNA involved in major depression and antidepressant treatment. Nat Med. (2014) 20:764–8. doi: 10.1038/nm.3582
41. Roy B, Dunbar M, Shelton RC, Dwivedi Y. Identification of MicroRNA-124-3p as a putative epigenetic signature of major depressive disorder. Neuropsychopharmacology. (2017) 42:864–75. doi: 10.1038/npp.2016.175
42. Xu Y, Liu H, Li F, Sun N, Ren Y, Liu Z, et al. A polymorphism in the microRNA-30e precursor associated with major depressive disorder risk and P300 waveform. J Affect Disord. (2010) 127:332–6. doi: 10.1016/j.jad.2010.05.019
43. Khandelwal N, Dey SK, Chakravarty S, Kumar A. miR-30 family mirnas mediate the effect of chronic social defeat stress on hippocampal neurogenesis in mouse depression model. Front Mol Neurosci. (2019) 12:188. doi: 10.3389/fnmol.2019.00188
44. Pfeffer TJ, Herrmann J, Berliner D, Konig T, Winter L, Ricke-Hoch M, et al. Assessment of major mental disorders in a German peripartum cardiomyopathy cohort. ESC Heart Fail. (2020) 7:4394–8. doi: 10.1002/ehf2.12967
45. Satyanarayanan SK, Shih YH, Wen YR, Palani M, Lin YW, Su H, et al. miR-200a-3p modulates gene expression in comorbid pain and depression: molecular implication for central sensitization. Brain Behav Immun. (2019) 82:230–8. doi: 10.1016/j.bbi.2019.08.190
46. Wang L, Liu J, Wang Q, Jiang H, Zeng L, Li Z, et al. MicroRNA-200a-3p mediates neuroprotection in alzheimer-related deficits and attenuates amyloid-beta overproduction and tau hyperphosphorylation via coregulating BACE1 and PRKACB. Front Pharmacol. (2019) 10:806. doi: 10.3389/fphar.2019.00806
47. Nagaraj S, Laskowska-Kaszub K, Debski KJ, Wojsiat J, Dabrowski M, Gabryelewicz T, et al. Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget. (2017) 8:16122–43. doi: 10.18632/oncotarget.15109
48. Svaren J, Sevetson BR, Apel ED, Zimonjic DB, Popescu NC, Milbrandt J. NAB2, a corepressor of NGFI-A (Egr-1) and Krox20, is induced by proliferative and differentiative stimuli. Mol Cell Biol. (1996) 16:3545–53. doi: 10.1128/MCB.16.7.3545
49. Gashler AL, Swaminathan S, Sukhatme VP. A novel repression module, an extensive activation domain, and a bipartite nuclear localization signal defined in the immediate-early transcription factor Egr-1. Mol Cell Biol. (1993) 13:4556–71. doi: 10.1128/mcb.13.8.4556-4571.1993
50. Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci U S A. (1995) 92:6873–7. doi: 10.1073/pnas.92.15.6873
51. Russo MW, Matheny C, Milbrandt J. Transcriptional activity of the zinc finger protein NGFI-A is influenced by its interaction with a cellular factor. Mol Cell Biol. (1993) 13:6858–65. doi: 10.1128/MCB.13.11.6858
52. Duclot F, Kabbaj M. The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front Behav Neurosci. (2017) 11:35. doi: 10.3389/fnbeh.2017.00035
53. Covington HE III, Lobo MK, Maze I, Vialou V, Hyman JM, Zaman S, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci. (2010) 30:16082–90. doi: 10.1523/JNEUROSCI.1731-10.2010
54. Xu Y, Pan J, Sun J, Ding L, Ruan L, Reed M, et al. Inhibition of phosphodiesterase 2 reverses impaired cognition and neuronal remodeling caused by chronic stress. Neurobiol Aging. (2015) 36:955–70. doi: 10.1016/j.neurobiolaging.2014.08.028
55. Zeng D, He S, Ma C, Wen Y, Song W, Xu Q, et al. Network-based approach to identify molecular signatures in the brains of depressed suicides. Psychiatry Res. (2020) 294:113513. doi: 10.1016/j.psychres.2020.113513
56. James AB, Conway AM, Morris BJ. Genomic profiling of the neuronal target genes of the plasticity-related transcription factor – Zif268. J Neurochem. (2005) 95:796–810. doi: 10.1111/j.1471-4159.2005.03400.x
57. Duclot F, Kabbaj M. The estrous cycle surpasses sex differences in regulating the transcriptome in the rat medial prefrontal cortex and reveals an underlying role of early growth response 1. Genome Biol. (2015) 16:256. doi: 10.1186/s13059-015-0815-x
58. James AB, Conway AM, Morris BJ. Regulation of the neuronal proteasome by Zif268 (Egr1). J Neurosci. (2006) 26:1624–34. doi: 10.1523/JNEUROSCI.4199-05.2006
59. Ramasamy S, Velmurugan G, Rekha B, Anusha S, Shanmugha Rajan K, Shanmugarajan S, et al. Egr-1 mediated cardiac miR-99 family expression diverges physiological hypertrophy from pathological hypertrophy. Exp Cell Res. (2018) 365:46–56. doi: 10.1016/j.yexcr.2018.02.016
60. Chen J, Zhan Y, Xu J, Wang Y, Gao Q. EGR1 overexpression inhibits the occurrence of preeclampsia by binding to MicroRNA-574 promoter and upregulating GAB1. Reprod Sci. (2021) 28:1112–21. doi: 10.1007/s43032-020-00392-3
61. Fan K, Huang W, Qi H, Song C, He C, Liu Y, et al. The Egr-1/miR-15a-5p/GPX4 axis regulates ferroptosis in acute myocardial infarction. Eur J Pharmacol. (2021) 909:174403. doi: 10.1016/j.ejphar.2021.174403
62. Zhang L, Ren R, Yang X, Ge Y, Zhang X, Yuan H. Oncogenic role of early growth response-1 in liver cancer through the regulation of the microRNA-675/sestrin 3 and the Wnt/beta-catenin signaling pathway. Bioengineered. (2021) 12:5305–22. doi: 10.1080/21655979.2021.1964889
63. Yang Y, Wu F, Zhang J, Sun R, Li F, Li Y, et al. EGR1 interacts with DNMT3L to inhibit the transcription of miR-195 and plays an anti-apoptotic role in the development of gastric cancer. J Cell Mol Med. (2019) 23:7372–81. doi: 10.1111/jcmm.14597
64. Wang W, Zhou D, Shi X, Tang C, Xie X, Tu J, et al. Global Egr1-miRNAs binding analysis in PMA-induced K562 cells using ChIP-Seq. J Biomed Biotechnol. (2010) 2010:867517. doi: 10.1155/2010/867517
65. Czibere L, Baur LA, Wittmann A, Gemmeke K, Steiner A, Weber P, et al. Profiling trait anxiety: transcriptome analysis reveals cathepsin B (Ctsb) as a novel candidate gene for emotionality in mice. PLoS One. (2011) 6:e23604. doi: 10.1371/journal.pone.0023604
66. Lisowski P, Juszczak GR, Goscik J, Stankiewicz AM, Wieczorek M, Zwierzchowski L, et al. Stress susceptibility-specific phenotype associated with different hippocampal transcriptomic responses to chronic tricyclic antidepressant treatment in mice. BMC Neurosci. (2013) 14:144. doi: 10.1186/1471-2202-14-144
67. de Jong TV, Kim P, Guryev V, Mulligan MK, Williams RW, Redei EE, et al. Whole genome sequencing of nearly isogenic WMI and WLI inbred rats identifies genes potentially involved in depression and stress reactivity. Sci Rep. (2021) 11:14774. doi: 10.1038/s41598-021-92993-4
68. Huang TL, Sung ML, Chen TY. 2D-DIGE proteome analysis on the platelet proteins of patients with major depression. Proteome Sci. (2014) 12:1. doi: 10.1186/1477-5956-12-1
69. Lin C-C, Tsai M-C, Lee C-T, Sun M-H, Huang T-L. Antidepressant treatment increased serum miR-183 and miR-212 levels in patients with major depressive disorder. Psychiatry Res. (2018) 270:232–7. doi: 10.1016/j.psychres.2018.09.025
70. Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, et al. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. (2014) 124:2136–46. doi: 10.1172/JCI70577
71. Davis MT, Hillmer A, Holmes SE, Pietrzak RH, DellaGioia N, Nabulsi N, et al. In vivo evidence for dysregulation of mGluR5 as a biomarker of suicidal ideation. Proc Natl Acad Sci U S A. (2019) 116:11490–5. doi: 10.1073/pnas.1818871116
72. Mann JJ. Neurobiology of suicidal behaviour. Nat Rev Neurosci. (2003) 4:819–28. doi: 10.1038/nrn1220
Keywords: major depressive disorder, suicidal ideation, suicide, microRNA, biomarker, early growth response protein 1 (EGR1)
Citation: Stapel B, Xiao K, Gorinski N, Schmidt K, Pfanne A, Fiedler J, Richter I, Vollbrecht A-L, Thum T, Kahl KG and Ponimaskin E (2022) MicroRNAs as novel peripheral markers for suicidality in patients with major depressive disorder. Front. Psychiatry 13:1020530. doi: 10.3389/fpsyt.2022.1020530
Received: 16 August 2022; Accepted: 02 November 2022;
Published: 24 November 2022.
Edited by:
Gianluca Serafini, Department of Neuroscience, San Martino Hospital (IRCCS), ItalyReviewed by:
Chuanjun Zhuo, Tianjin Anding Hospital, ChinaCopyright © 2022 Stapel, Xiao, Gorinski, Schmidt, Pfanne, Fiedler, Richter, Vollbrecht, Thum, Kahl and Ponimaskin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Britta Stapel, c3RhcGVsLmJyaXR0YUBtaC1oYW5ub3Zlci5kZQ==; Evgeni Ponimaskin, cG9uaW1hc2tpbi5ldmdlbmlAbWgtaGFubm92ZXIuZGU=
†These authors share first authorship
‡These authors share senior authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.