
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Psychiatry , 08 March 2021
Sec. Behavioral and Psychiatric Genetics
Volume 12 - 2021 | https://doi.org/10.3389/fpsyt.2021.631475
This article is part of the Research Topic Genetic and Epigenetic Mechanisms Underpinning Vulnerability to Developing Psychiatric Disorders View all 17 articles
 Hiroshi Komatsu1,2*
Hiroshi Komatsu1,2* Hikaru Takeuchi3
Hikaru Takeuchi3 Chiaki Ono4
Chiaki Ono4 Zhiqian Yu4
Zhiqian Yu4 Yoshie Kikuchi4
Yoshie Kikuchi4 Yoshihisa Kakuto2
Yoshihisa Kakuto2 Shunichi Funakoshi2,5
Shunichi Funakoshi2,5 Takashi Ono2
Takashi Ono2 Ryuta Kawashima3,6
Ryuta Kawashima3,6 Yasuyuki Taki3,7,8
Yasuyuki Taki3,7,8 Hiroaki Tomita1,4,9,10,11
Hiroaki Tomita1,4,9,10,11Recent evidence has indicated that the disruption of oligodendrocytes may be involved in the pathogenesis of depression. Genetic factors are likely to affect trait factors, such as characteristics, rather than state factors, such as depressive symptoms. Previously, a negative self-schema had been proposed as the major characteristic of constructing trait factors underlying susceptibility to depression. Thus, the association between a negative self-schema and the functional single nucleotide polymorphism (SNP) rs1059004 in the OLIG2 gene, which influences OLIG2 gene expression, white matter integrity, and cerebral blood flow, was evaluated. A total of 546 healthy subjects were subjected to genotype and psychological evaluation using the Beck Depression Inventory-II (BDI-II) and the Brief Core Schema Scale (BCSS). The rs1059004 SNP was found to be associated with the self-schema subscales of the BCSS and scores on the BDI-II in an allele dose-dependent manner, and to have a predictive impact on depressive symptoms via a negative-self schema. The results suggest the involvement of a genetic factor regulating oligodendrocyte function in generating a negative-self schema as a trait factor underlying susceptibility to depression.
Major depressive disorder (MDD) is a common disease with a 12-month and lifetime prevalence of 6.6 and 16.2%, respectively (1). Depressive disorders, including MDD and dysthymia, are also a leading cause of the global disease burden (2). Although effective treatments (e.g., antidepressants and cognitive behavioral therapy) are available for MDD, remission rates for antidepressants are not currently optimally achieved (3). Therefore, the development of new and more effective drugs is required in the future and elucidating the pathology of MDD could help us develop these novel medicines. Depressive disorders are multifactorial diseases that are thought to develop through interactions between genetic and psychosocial factors. Although the neurobiological mechanism underlying MDD has been extensively investigated, the definitive biological mechanism remains unclear.
Prior studies based on post-mortem brain, neuroimaging, and animal studies have indicated the involvement of disrupted oligodendrocytes in the pathology of MDD. Histological examination of the post-mortem brain revealed that patients with MDD have a substantially reduced density and number of oligodendrocytes in their prefrontal cortex (4, 5). Oligodendrocyte-related genes were also found to be reduced in the temporal frontal cortex and in the white matter of the ventral prefrontal cortex in patients with MDD (6, 7). More recent studies have reported that psychological stress results in the downregulation of oligodendrocyte-related genes and reduced myelination fiber length and density in the brains of mice (8, 9). Cathomas et al. (8) revealed that after psychological stress, emotion, and microglial activity were altered in mice that were heterozygous for the cyclic nucleotide phosphodiesterase (Cnp1) oligodendrocyte gene compared to wild-type mice; this suggests that oligodendrocyte gene expression affects behavioral changes after psychosocial stress. Several diffusion tensor imaging studies have documented decreased white matter integrity in the corpus callosum and several frontal, temporal, and parietal regions in patients with MDD (10–13). In addition, other studies have found microstructural abnormalities in some fiber tracts, such as anterior callosal fibers, in patients with MDD (14, 15). Patients with MDD also show a reduced magnetic transfer ratio, which reflects demyelination in brain regions, such as the frontal and striatal regions, limbic areas, occipital white matter, and the genu and splenium of the corpus callosum (16–18). Evidence found in post-mortem brains, neuroimaging, and animal studies suggests that disrupted oligodendrocyte function may be involved in impaired mood regulation in MDD.
The OLIG2 gene is a basic helix-loop-helix transcription factor that is expressed exclusively in oligodendrocytes and oligodendrocyte precursors and is involved in oligodendrocyte differentiation (19, 20). One post-mortem brain study found that OLIG2 gene expression was downregulated in the temporal cortex of patients with MDD (6). The OLIG2 gene single nucleotide polymorphism (SNP) rs1059004 is a functional SNP that influences OLIG2 gene expression and influences white matter integrity in Caucasian populations (21–23). A more recent study revealed that SNP rs1059004 affects resting-state cerebral blood flow in a broad region of the brain as well as white matter integrity in the Japanese population (23). Genetic association studies have shown that the OLIG2 SNP rs1059004 is associated with several psychiatric disorders such as schizophrenia and obsessive-compulsive disorder in a certain population (24–28). However, no previous studies have investigated the genetic association between this SNP and MDD.
Beck proposed the cognitive model of depression, in which negative self-schema individuals are vulnerable to developing depression in the future (29). Previous studies have shown a marked association between the schemata concerning the self and others, and the severity of depressive symptoms (30, 31). Evans et al. revealed that a negative self-schema was a risk factor for the development of depression in women (32). The results of these studies support the cognitive model of depression hypothesized by Beck (29).
Based on prior evidence indicating the involvement of oligodendrocyte abnormalities in the pathology of MDD, the present study hypothesized the association between OLIG2 SNP rs1059007 and the negative-self core schema, as it plays a major role in constructing trait factors underlying susceptibility to MDD by influencing the function of oligodendrocytes. To verify this hypothesis, we investigated whether the OLIG2 SNP rs1059007 was associated with self-negative core schema in healthy subjects.
A total of 777 healthy, right-handed individuals were recruited, and their genotyping data, mood states, core beliefs, brain imaging data, cognitive function, aging, genetics, and daily habits were examined as detailed below and elsewhere (33–36). Of the 777 participants, all gene polymorphism data, scores on the Beck Depression Inventory-II (BDI-II), and scores on the Brief Core Schema Scale (BCSS) were successfully obtained from 546 subjects (313 men and 233 women; 20.5 ± 1.8 years of age). All subjects had normal vision and were university, college or postgraduate students, or subjects who had graduated from these institutions within 1 year prior to the experiment. None of the participants had a history of neurological or psychiatric illness. Handedness was evaluated using the Edinburgh Handedness Inventory (37). High-molecular-weight DNA was isolated from saliva specimens using Oragene containers (DNA Genotek Inc., Ottawa, Canada), according to the manufacturer's instructions. After the study procedures were fully explained, written informed consent was obtained from all participants in accordance with the Declaration of Helsinki (1991). This study was approved by the Ethics Committee of Tohoku University.
Participants were administered the Japanese version of the Brief Core Schema Scale (BCSS) to assess core schemas about the self and others. The BCSS is a 24-item self-report scale developed by Fowler et al. to measure core schemata with regard to the self and others (30). The BCSS includes four dimensions of self- and other-assessment: negative self, positive self, negative others, and positive others. Each dimension comprised six items that were evaluated on a five-point rating scale (0–4), and respondents were asked to indicate “yes” or “no” for whether they held each belief. If they held the belief, they were then asked to indicate the degree of their conviction on a 4-point scale ranging from 1 to 4 (1: believe slightly, 2: believe it moderately, 3: believe it very much, 4: believe it totally).
Participants were also tested using the Japanese version of the BDI-II to evaluate the degree of depressive symptoms. BDI-II, developed by Beck et al. (38), is a 21-item self-report questionnaire used to evaluate the severity of depression in normal and psychiatric subjects (39). Each item consisted of a 4-point scale from 0 (symptoms absent) to 3 (severe symptoms). The minimum and maximum BDI-II scores were 0 and 63, respectively with higher scores indicating greater severity of depressive symptoms. In normal subjects, scores above 20 have been reported to indicate depression (40).
Genotyping of OLIG2 SNP rs1059004 was carried out using TaqMan assays (Applied Biosystems, Foster City, CA, USA). Polymerase chain reactions (PCRs) were performed using 20 ng of genomic DNA, 40 × TaqMan Probe Assay Mix (Probe ID: C_2442961_10) (Applied Biosystems, Waltham, MA, USA), 2 × Universal PCR Master Mix (Applied Biosystems), and nuclease-free water in a 10 μL total reaction volume; allele-specific fluorescence was measured using the CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA). Information about the TaqMan Probe sequence can be obtained from https://www.thermofisher.com/order/genome-database/details/genotyping/C___2442961_20?CID=&ICID=&subtype=. The PCR cycle conditions consisted of an initial denaturation at 95°C for 10 min, followed by 50 cycles at 92°C for 15 s and at 57°C for 1 min. Then, PCR products spanning the SNP were amplified with primers (forward: gagcgctgtctggctttaac, reverse: gaggaacggccacagttcta) from two representative subjects for each of the three genotypes of this SNP and were subjected to direct sequencing to validate the TaqMan assay-based genotyping.
Statistical evaluations were performed using SPSS statistics 24 and Amos 23 (Japan IBM, Tokyo, Japan) software packages. Demographic variables among groups were compared using χ2-tests, analysis of variance, t-tests, or Kruskal-Wallis test, where appropriate. Deviations in genotype distribution from the Hardy-Weinberg equilibrium (HWE) were assessed using the χ2-test for goodness of fit. Spearman's bivariate correlation analysis was performed to examine the correlations between BDI-II scores and the four BCSS subscales. The Jonckheere-Terpstra test was performed to investigate whether the OLIG2 SNP rs1059004 was associated with the scores on the four BCSS subscales or those of BDI-II in an allele dose-dependent manner. After the Jonckheere-Terpstra test, the post-hoc test was used to compare the differences between each genotype using the Bonferroni corrections.
The associations between the OLIG2 SNP rs1059004, self-schema, and depressive symptoms were assessed using path analysis. Structural equation modeling was performed to evaluate the association between the above-mentioned three variables using SPSS and Amos 23, and model fits were estimated using the maximum likelihood method. The two initial hypothetical models shown in Figure 1 were constructed based on the results of the trend test, bivariate correlation analysis, and a previous study (32). In the two initial models shown in Figure 1, the self-schema affects depressive symptoms, and OLIG2 SNP rs1059004 impacts schema and depressive symptoms in one direction. In contrast, a negative schema influences or is affected by a positive schema about the self. We removed paths scoring p > 0.05 from the initial models and examined whether the model fit improved. The best-fitting path model was ultimately adopted in the path analysis. Chi-square statistics were used to test the goodness-of-fit model, and the following fit indices were calculated: goodness-of-fit index (GFI), adjusted GFI (AGFI), comparative fit index (CFI), Akaike information criterion (AIC), and root mean square error of approximation (RMSEA).
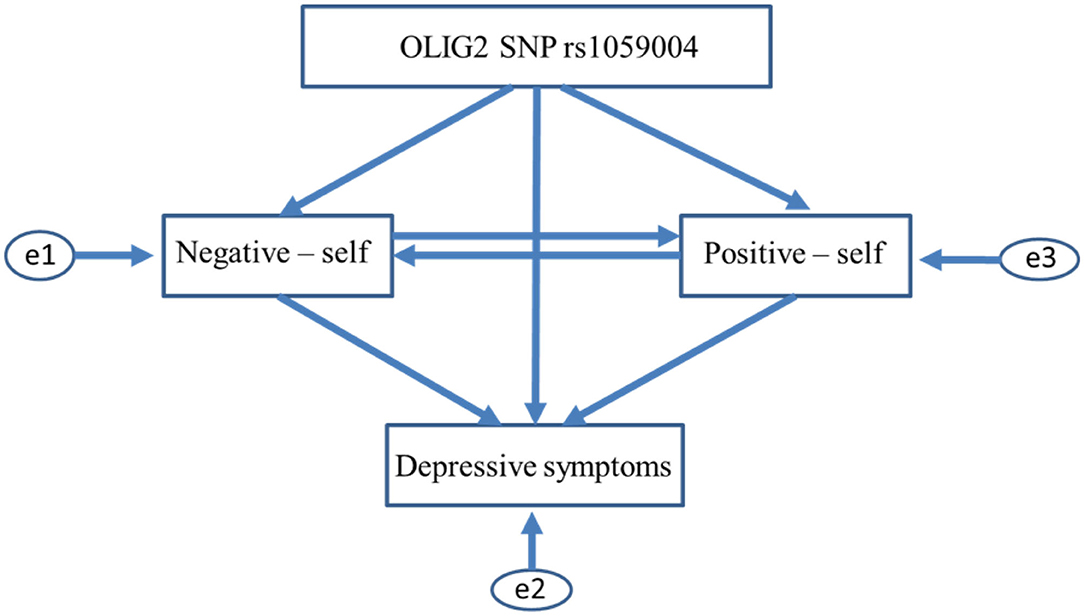
Figure 1. Initial path models between OLIG2 gene SNP rs1059004, self-core schema, and depressive symptoms. The two initial path models (AIC = 20, CFI = 1, RMSEA = 0.363 in both models) shown in Figure 1 assume that OLIG2 SNP rs1059004 affects negative self-schema (negative self), positive self-schema (positive self), and depressive symptoms in one direction. In contrast, hypothetical paths were not created among each self-schema. SNP, single nucleotide polymorphism; AIC, Akaike information criterion; CFI, comparative fit index; RMSEA, root mean square error of approximation.
To investigate whether the OLIG2 SNP rs1059004 influenced the severity of depressive symptoms and core schema about the self and others in a gender-dependent manner, two-way analysis of covariance (ANCOVA) controlling for age, with genotype and gender as independent variables, was used to assess differences in scores on the BDI-II and on the four BCSS subscales between A-allele carriers (AA genotype + AC genotype) and non-A-allele carriers (CC genotype). As few individuals carried the AA genotype, subjects carrying the AA genotype were combined with those carrying the AC genotype, and the differences in scores on the BDI-II and BCSS subscales were compared between carriers and non-carriers of the A allele. Post-hoc analyses were performed using Bonferroni corrections. Statistical significance was defined as a two-tailed p < 0.05.
There were significant differences in age and in positive, negative, and other subscale scores between men and women (Table 1).
The genotype distribution of the subjects was as follows: homozygous A allele (n = 14, 2.5%), heterozygous A/C (n = 155, 28.3%), and homozygous C allele (n = 377, 69%), which did not deviate from the HWE (χ2 = 0.33, p > 0.05). There were no significant differences in age or sex among the three OLIG2 genotypic groups (Table 2).
Meanwhile, the Kruskal-Wallis test showed significant differences in BDI-II and the negative and positive self subscales of the BCSS among OLIG2 genotypes (p = 0.03, p = 0.042, p = 0.015, respectively, Table 2). The Cronbach's α values for the BDI-II total score, the negative and positive self subscale scores, and the negative and positive other subscale scores were 0.82, 0.80, 0.82, 0.78, and 0.86, respectively.
Spearman's bivariate correlation analysis revealed a positive association between BDI-II score and the negative self and negative other subscales of the BCSS (r = 0.636 and r = 0.296, respectively, p < 0.001, Table 3). There was a significant negative association between BDI-II score and the positive self and positive other subscales of the BCSS (r = −0.459 and r = −0.306, respectively, p < 0.001, Table 3).
The present study found a significant association between the OLIG2 SNP rs1059004 and BDI-II score in an allele dose-dependent manner (Jonckheere-Terpstra test, p = 0.022, Figure 2A). Post-hoc analysis showed that AA genotype carriers had a significantly higher BDI-II score than the CC genotype carriers (Bonferroni-corrected p = 0.041, Figure 2A). Two-way ANCOVA adjusted for age, with genotype and gender as the fixed factors showed that there was a significant main effect of genotype on BDI-II score (Table 4). However, the genotype-gender interaction was not significant for the BDI-II score (Table 4).
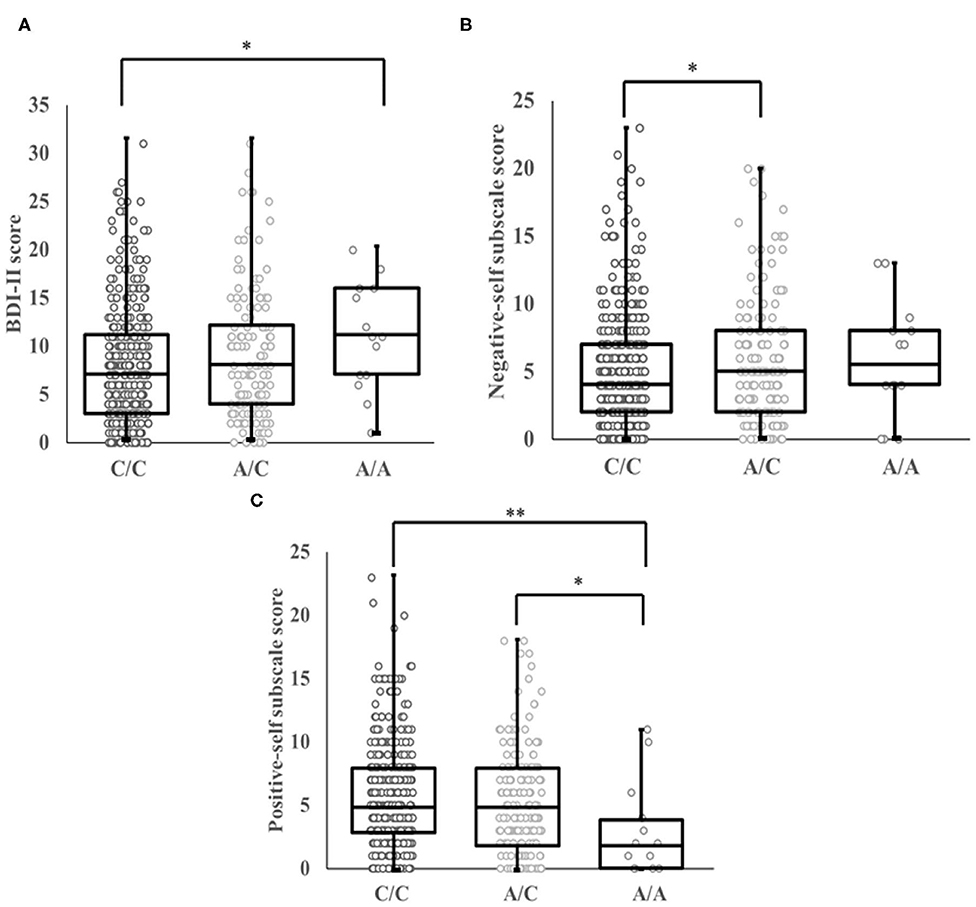
Figure 2. (A) Association between genotype at SNP rs1059004 in the OLIG2 gene and BDI-II scores. There were significant correlations between A allele dosage and BDI-II scores among the three genotype groups. BDI-II scores decreased in a C-allele dose-dependent manner (p = 0.022, Jonckheere-Terpstra test). AA genotype carriers had a significantly higher BDI-II score than the CC genotype carriers (Bonferroni-corrected p = 0.041). (B) Association between genotype at SNP rs1059004 in the OLIG2 gene and negative-self subscale scores. Significant gene-dose associations between the SNP rs1089004 and negative-self subscale scores were observed among the three genotype groups. Negative-self subscale scores decreased in a C-allele dose-dependent manner (p = 0.012, Jonckheere-Terpstra test). Subjects with AC genotype had a significantly higher negative-self subscale score compared to those with CC genotype (p = 0.026). (C) Association between genotype at SNP rs1059004 and positive-self subscale scores. The Jonckheere-Terpstra test also showed significant associations between A allele dosage and positive-self subscale scores among the three genotype groups. Positive-self subscale scores increased in a C-allele dose-dependent manner (p = 0.037, Jonckheere-Terpstra test). People carrying the AA genotype had a significantly lower positive-self subscale score than those with the CC genotype and AC genotype, respectively (Bonferroni-corrected p = 0.009, p = 0.049). The bars of the box plot show the range from minimum scores to maximum scores. SNP, single nucleotide polymorphism. *p < 0.05, **p < 0.01 compared to the CC genotype.
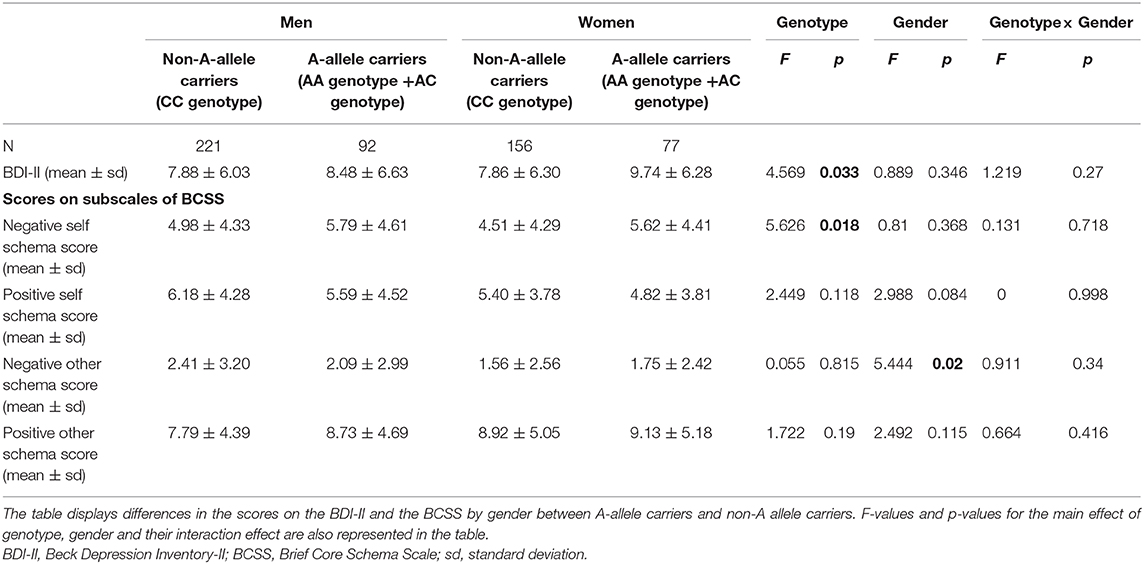
Table 4. The results of two-way ANCOVA controlling for age, with genotype and gender as independent variables.
The Jonckheere-Terpstra test showed that the OLIG2 SNP rs1059004 was associated with both negative and positive self-schema in an allele dose-dependent manner (Jonckheere-Terpstra test, p = 0.012, p = 0.037; Figures 2B,C). Subjects with the AC genotype had a significantly higher negative-self subscale score than those with the CC genotype (Bonferroni-corrected p = 0.026, Figure 2B). Individuals carrying the AA genotype had a significantly lower positive-self subscale score than those with the CC genotype and AC genotype, respectively (Bonferroni-corrected p = 0.009, p = 0.049, Figure 2C).
Two-way ANCOVA controlling for age and with genotype and gender as the fixed factors showed that there was a significant main effect of genotype on negative-self subscale score (Table 4). However, no significant genotype-gender interactions were observed for scores on the four BCSS subscales (Table 4).
We removed paths scoring p > 0.05 from the initial models (AIC = 20, CFI = 1, RMSEA = 0.363 in both models) and examined whether the model fit improved. Ultimately, the path model shown in Figure 3 produced the best results in the model fit evaluations (Chi-square = 2.669, df = 2, P = 0.263; GFI = 0.998; AGFI = 0.988; CFI = 0.998; AIC = 18.669; RMSEA = 0.025). The number of C alleles of the OLIG2 gene SNP rs1059004 was negatively associated with the negative self-schema, and the schema was assumed to affect the positive self-schema and lead to a higher severity of depressive symptoms (Figure 3).
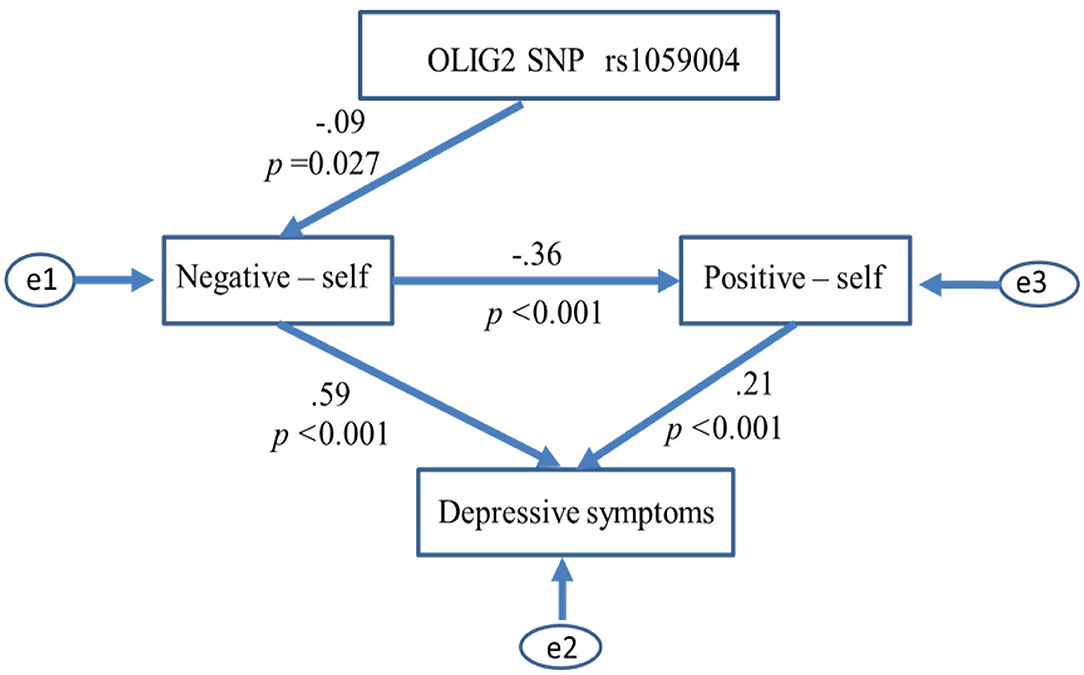
Figure 3. Relationships between OLIG2 SNP rs1059004, self-core schema and depressive symptoms. Paths scoring p > 0.05 were removed from the initial models, and the best-fitting path model was ultimately adopted in the path analysis. The path model produced the best model fit results, as shown below: Chi-square = 2.669, df = 2, P = 0.263; GFI = 0.998; AGFI = 0.988; CFI = 0.998; AIC = 18.669; and RMSEA = 0.025. This path model indicates the predictive effect of the OLIG2 SNP rs1059004 on depressive symptoms via a negative self-schema (negative self) that influences a positive self-schema (positive self). SNP, single nucleotide polymorphism; GFI, goodness-of-fit index; AGFI, adjusted GFI; CFI, comparative fit index; AIC, Akaike information criterion; RMSEA, root mean square error of approximation.
Based on prior evidence that the disruption of oligodendrocytes may be implicated in the pathology of MDD, the present study hypothesized an association between the OLIG2 SNP rs1059004 and the negative self-schema, constructing trait factors underlying susceptibility to MDD. To verify this hypothesis, the associations between the OLIG2 SNP rs1059004, BCSS and BDI-II scores were examined in 546 healthy subjects. Consistent with the above hypothesis, the number of C alleles in the OLIG2 gene SNP rs1059004 was associated with decreased BDI-II scores and the negative BCSS self-schema subscale in an allele dose-dependent manner. Path analysis revealed that a negative self-schema mediated the association between the OLIG2 SNP rs1059004 and the severity of depressive symptoms. The results of the present study indicate that the OLIG2 SNP rs1059004 has a predictive impact on depressive symptoms via a negative schema of the self. In addition, the results suggest the involvement of a genetic factor regulating oligodendrocyte function in generating a negative-self schema that plays a major role in constructing factors underlying susceptibility to depression.
Although previous studies have not found significant differences in scores for any BCSS subscale between men and women (30, 31), the current study found that gender created significant differences in positive, negative, and “other” BCSS subscale scores. This discrepancy in the results may be due to differences in the ethnicities, number, and/or ages of the participants. The findings of the present study indicate that the positive self-schema may be affected by a variety of experiences during young adulthood in the Japanese population. In contrast, four of the BCSS subscales were significantly correlated with the severity of depressive symptoms, consistent with previous reports (30, 31).
Previous genetic association studies have exclusively focused on genes related to monoamines, the hypothalamic-pituitary-adrenal axis, and glutamatergic neurotransmitters. For example, polymorphisms of the glucocorticoid receptor gene, monoamine oxidase A gene, and group-2 metabotropic glutamate receptor gene have been previously reported to be associated with MDD (41–44). Although polymorphisms of oligodendrocyte-related genes have been shown to be associated with schizophrenia (24), no previous studies have investigated the genetic association between the polymorphisms of oligodendrocyte-related genes and MDD. This is the first study to reveal that the OLIG2 SNP rs1059004 was associated with the severity of depressive symptoms and with negative self-schema. Whether oligodendrocyte disruption is a causative factor in mood dysregulation remains unclear, but several studies have suggested that changes in oligodendrocyte function and structure can influence neural circuits that mediate mood regulation in mice (8, 45–47). For example, mice exposed to cuprizone, a mouse model of demyelination, showed deceased anxiety-like behavior. Moreover, mice lacking the Cnp1 oligodendrocyte-related gene were found to show depressive-like behaviors (45). Therefore, the OLIG2 SNP rs1059004 may be associated with the severity of depressive symptoms and negative self-schema by influencing the function of oligodendrocytes. One functional magnetic resonance imaging-based study indicated that the activities of the anterior cingulate cortex and the inferior frontal cortex are involved in negative self-reflection (48), and prior investigations have found that the OLIG2 polymorphism rs1059004 affects not only white matter integrity but also resting-state cerebral blood flow in widespread brain regions, including the anterior cingulate and inferior frontal regions (23). Therefore, the effect of SNP rs1059004 on brain perfusion may be a biological mechanism underlying the association between the variant and negative self-schema. Further studies are needed to reveal the biological basis of the association between the OLIG2 SNP rs1059004 and negative self-schema.
Beck proposed the cognitive model of depression, in which individuals who hold negative self-schemas are vulnerable to developing depression in the future (29). Evans et al. revealed that a negative self-schema was a risk factor for the onset of depression in women in a longitudinal study (32). This finding may support the cognitive model of depression proposed by Beck. Therefore, negative self-schema could play a major role in constructing trait factors underlying susceptibility to MDD. The results of the present study indicate a significant association between the OLIG2 SNP rs1059004 and a negative self-schema, constructing trait factors concerning susceptibility to MDD. Considering the significant association between the OLIG2 SNP rs1059004 and a negative self-schema as the trait factor underlying the development of depression, it would be worthwhile to conduct a longitudinal study investigating whether SNP rs1059004 affects the risk of the onset of MDD by influencing negative self-schema.
The present study has several limitations. First, the participants were limited in terms of age and education history; that is, only young adults and university, college, or postgraduate students were examined; therefore, the results of the present study may not apply to the general population. The second limitation is the cross-sectional nature, which cannot disentangle the direction of effects between variables. The third limitation is that some problems have recently arisen regarding single-gene studies of multifactorial phenotypes, such as depression, and many studies have failed to be replicable. For example, Culverhouse et al. found no proof that the thousands of studies on the relationship between 5-HTTLPR and depression provided evidence of a genuine genetic difference (49). More recently, Border et al. found no evidence for “any candidate gene polymorphism associations with depression phenotypes or any polymorphism-by-environment moderator effects (50).” Although a negative self-schema can relatively reflect trait factors compared with depression, the phenotype is also highly polygenic, and the genetic effect expected from a single gene can be small. However, in the present study, the number of participants was small, the AA genotype was relatively low compared to other genotypes, the power was insufficient, and cross-validation was not performed. Therefore, it will be necessary to replicate these results with a much larger sample size in the future. The fourth limitation is that it is not clear whether the genetic association in the present study is applied to ethnicities other than Japanese. The fifth limitation is that although prior studies indicated that SNP rs1059004 predicted OLIG2 gene expression in Caucasian subjects, it is unclear whether a significant association exists between the SNP rs1059004 and OLIG2 gene expression in Japanese subjects. This study is the first to indicate a significant genetic association between the OLIG2 SNP rs1059004 and negative self-schema as a trait factor in susceptibility to MDD in Japanese subjects. The results of the present study suggest an association between the OLIG2 SNP rs1059004 and negative self-schema by influencing the function of oligodendrocytes. These results may support the involvement of a genetic factor regulating oligodendrocyte function in generating a negative self-schema as a trait factor underlying susceptibility to depression in the pathology of MDD.
The SNP data presented in the study are publicly available. This data can be found in dbSNP (https://www.ncbi.nlm.nih.gov/SNP/snp_viewTable.cgi?handle=TOHOKUPSY).
The studies involving human participants were reviewed and approved by The Ethics Committee of Tohoku University. The patients/participants provided their written informed consent to participate in this study.
HK, HTa, YKi, CO, ZY, and HTo contributed to the acquisition of data or the analysis and interpretation of data. HK and HTa were involved in drafting the manuscript. HTa, YKa, SF, TO, and HTo critically revised the manuscript for important scientific content. HTa, RK, YT, and HTo made substantial contributions to the conception and design of the study. All authors read and approved the final version of the manuscript and agreed on the order in which their names would be listed in the manuscript.
This work was partially supported by a grant-in-aid for scientific research on innovative areas (No. 24116007) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Strategic Research Program for Brain Sciences from the Japan Agency for Medical Research and Development, AMED (JP20dm0107099). This study was also partially supported by JST/RISTEX, JST/CREST, a grant-in-aid for young scientists (B) (KAKENHI 23700306), a grant-in-aid for young scientists (A) (KAKENHI 25700012), and Tohoku University Advanced Research Center for Innovations in Next-Generation Medicine, Japan. None of the funding sources mentioned above were involved in the study design; collection, analysis, or interpretation of data; writing of the report, or decision to submit the article for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Haruka Nouchi for conducting the psychological tests, all other assistants for helping with the experiments in the study, and the study participants and all our other colleagues at Tohoku University for their support.
1. Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, et al. The epidemiology of major depressive disorder: results from the National Comorbidity Survey Replication (NCS-R). JAMA. (2003) 289:3095–105. doi: 10.1001/jama.289.23.3095
2. Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, et al. Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLOS Med. (2013) 10:e1001547. doi: 10.1371/journal.pmed.1001547
3. Rush AJ, Trivedi MH, Wisniewski SR, Nierenberg AA, Stewart JW, Warden D, et al. Acute and longer-term outcomes in depressed outpatients requiring one or several treatment steps: a STAR*D report. Am J Psychiatry. (2006) 163:1905–17. doi: 10.1176/ajp.2006.163.11.1905
4. Hayashi Y, Tatebayashi Y. A flow cytometric postmortem brain study for major depressive disorders: implication for oligodendroglial differentiation and functions. Nihon Shinkei Seishin Yakurigaku Zasshi. (2012) 32:211–8.
5. Vostrikov VM, Uranova NA, Orlovskaya DD. Deficit of perineuronal oligodendrocytes in the prefrontal cortex in schizophrenia and mood disorders. Schizophr Res. (2007) 94:273–80. doi: 10.1016/j.schres.2007.04.014
6. Aston C, Jiang L, Sokolov BP. Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry. (2005) 10:309–22. doi: 10.1038/sj.mp.4001565
7. Rajkowska G, Mahajan G, Maciag D, Sathyanesan M, Iyo AH, Moulana M, et al. Oligodendrocyte morphometry and expression of myelin - related mRNA in ventral prefrontal white matter in major depressive disorder. J Psychiatr Res. (2015) 65:53–62. doi: 10.1016/j.jpsychires.2015.04.010
8. Cathomas F, Azzinnari D, Bergamini G, Sigrist H, Buerge M, Hoop V, et al. Oligodendrocyte gene expression is reduced by and influences effects of chronic social stress in mice. Genes Brain Behav. (2019) 18:e12475. doi: 10.1111/gbb.12475
9. Lehmann ML, Weigel TK, Elkahloun AG, Herkenham M. Chronic social defeat reduces myelination in the mouse medial prefrontal cortex. Sci Rep. (2017) 7:46548. doi: 10.1038/srep46548
10. de Diego-Adeliño J, Pires P, Gómez-Ansón B, Serra-Blasco M, Vives-Gilabert Y, Puigdemont D, et al. Microstructural white-matter abnormalities associated with treatment resistance, severity and duration of illness in major depression. Psychol Med. (2014) 44:1171–82. doi: 10.1017/S003329171300158X
11. Jiang J, Zhao YJ, Hu XY, Du MY, Chen ZQ, Wu M, et al. Microstructural brain abnormalities in medication-free patients with major depressive disorder: a systematic review and meta-analysis of diffusion tensor imaging. J Psychiatry Neurosci. (2017) 42:150–63. doi: 10.1503/jpn.150341
12. Ma N, Li L, Shu N, Liu J, Gong G, He Z, et al. White matter abnormalities in first-episode, treatment-naive young adults with major depressive disorder. Am J Psychiatry. (2007) 164:823–6. doi: 10.1176/ajp.2007.164.5.823
13. Matsuoka K, Yasuno F, Kishimoto T, Yamamoto A, Kiuchi K, Kosaka J, et al. Microstructural differences in the corpus callosum in patients with bipolar disorder and major depressive disorder. J Clin Psychiatry. (2017) 78:99–104. doi: 10.4088/JCP.15m09851
14. Osoba A, Hänggi J, Li M, Horn DI, Metzger C, Eckert U, et al. Disease severity is correlated to tract specific changes of fractional anisotropy in MD and CM thalamus–a DTI study in major depressive disorder. J Affect Disord. (2013) 149:116–28. doi: 10.1016/j.jad.2012.12.026
15. Yamada S, Takahashi S, Ukai S, Tsuji T, Iwatani J, Tsuda K, et al. Microstructural abnormalities in anterior callosal fibers and their relationship with cognitive function in major depressive disorder and bipolar disorder: a tract-specific analysis study. J Affect Disord. (2015) 174:542–8. doi: 10.1016/j.jad.2014.12.022
16. Chen Z, Zhang H, Jia Z, Zhong J, Huang X, Du M, et al. Magnetization transfer imaging of suicidal patients with major depressive disorder. Sci Rep. (2015) 5:9670. doi: 10.1038/srep09670
17. Gunning-Dixon FM, Hoptman MJ, Lim KO, Murphy CF, Klimstra S, Latoussakis V, et al. Macromolecular white matter abnormalities in geriatric depression: a magnetization transfer imaging study. Am J Geriatr Psychiatry. (2008) 16:255–62. doi: 10.1097/JGP.0000300628.33669.03
18. Kumar A, Gupta RC, Albert Thomas M, Alger J, Wyckoff N, Hwang S. Biophysical changes in normal-appearing white matter and subcortical nuclei in late-life major depression detected using magnetization transfer. Psychiatry Res. (2004) 130:131–40. doi: 10.1016/j.pscychresns.2003.12.002
19. Ligon KL, Fancy SP, Franklin RJ, Rowitch DH. Olig gene function in CNS development and disease. Glia. (2006) 54:1–10. doi: 10.1002/glia.20273
20. Meijer DH, Kane MF, Mehta S, Liu H, Harrington E, Taylor CM, et al. Separated at birth? The functional and molecular divergence of OLIG1 and OLIG2. Nat Rev Neurosci. (2012) 13:819–31. doi: 10.1038/nrn3386
21. Mitkus SN, Hyde TM, Vakkalanka R, Kolachana B, Weinberger DR, Kleinman JE, et al. Expression of oligodendrocyte-associated genes in dorsolateral prefrontal cortex of patients with schizophrenia. Schizophr Res. (2008) 98:129–38. doi: 10.1016/j.schres.2007.09.032
22. Prata DP, Kanaan RA, Barker GJ, Shergill S, Woolley J, Georgieva L, et al. Risk variant of oligodendrocyte lineage transcription factor 2 is associated with reduced white matter integrity. Hum Brain Mapp. (2013) 34:2025–31. doi: 10.1002/hbm.22045
23. Komatsu H, Takeuchi H, Kikuchi Y, Ono C, Yu Z, Iizuka K, et al. Ethnicity-dependent effects of schizophrenia risk variants of the OLIG2 gene on OLIG2 transcription and white matter integrity. Schizophr Bull. (2020) 46:1619–28. doi: 10.1093/schbul/sbaa049
24. Georgieva L, Moskvina V, Peirce T, Norton N, Bray NJ, Jones L, et al. Convergent evidence that oligodendrocyte lineage transcription factor 2 (OLIG2) and interacting genes influence susceptibility to schizophrenia. Proc Natl Acad Sci USA. (2006) 103:12469–74. doi: 10.1073/pnas.0603029103
25. Huang K, Tang W, Tang R, Xu Z, He Z, Li Z, et al. Positive association between OLIG2 and schizophrenia in the Chinese Han population. Hum Genet. (2008) 122:659–60. doi: 10.1007/s00439-007-0434-z
26. Sims R, Hollingworth P, Moskvina V, Dowzell K, O'Donovan MC, Powell J, et al. Evidence that variation in the oligodendrocyte lineage transcription factor 2 (OLIG2) gene is associated with psychosis in Alzheimer's disease. Neurosci Lett. (2009) 461:54–9. doi: 10.1016/j.neulet.2009.05.051
27. Stewart SE, Platko J, Fagerness J, Birns J, Jenike E, Smoller JW, et al. A genetic family-based association study of OLIG2 in obsessive-compulsive disorder. Arch Gen Psychiatry. (2007) 64:209–14. doi: 10.1001/archpsyc.64.2.209
28. Zhang X, Liu J, Guo Y, Jiang W, Yu J. populationAssociation. Depress Anxiety. (2015) 32:720–7. doi: 10.1002/da.22394
29. Beck AT. Thinking and depression. ii. theory and therapy. Arch Gen Psychiatry. (1964) 10:561–71. doi: 10.1001/archpsyc.1964.01720240015003
30. Fowler D, Freeman D, Smith B, Kuipers E, Bebbington P, Bashforth H, et al. The Brief Core Schema Scales (BCSS): psychometric properties and associations with paranoia and grandiosity in non-clinical and psychosis samples. Psychol Med. (2006) 36:749–59. doi: 10.1017/S0033291706007355
31. Uchida T, Kawamura C, Mifune N, Hamaie Y, Matsumoto K, Ambo H, et al. The Japnanese version of the brief core schema scale for schemata concerning the self and others: indentification of Shema patterns and relationschip with depression. Jpn J Pers. (2012) 20:143–54. doi: 10.2132/personality.20.143
32. Evans J, Heron J, Lewis G, Araya R, Wolke D, ALSPAC Study Team. Negative self-schemas and the onset of depression in women: longitudinal study. Br J Psychiatry. (2005) 186:302–7. doi: 10.1192/bjp.186.4.302
33. Takeuchi H, Taki Y, Nouchi R, Hashizume H, Sekiguchi A, Kotozaki Y, et al. Anatomical correlates of self-handicapping tendency. Cortex. (2013) 49:1148–54. doi: 10.1016/j.cortex.2013.01.014
34. Takeuchi H, Taki Y, Nouchi R, Hashizume H, Sekiguchi A, Kotozaki Y, et al. Effects of working memory training on functional connectivity and cerebral blood flow during rest. Cortex. (2013) 49:2106–25. doi: 10.1016/j.cortex.2012.09.007
35. Takeuchi H, Taki Y, Nouchi R, Sekiguchi A, Kotozaki Y, Miyauchi CM, et al. Regional gray matter density is associated with achievement motivation: evidence from voxel-based morphometry. Brain Struct Funct. (2014) 219:71–83. doi: 10.1007/s00429-012-0485-3
36. Takeuchi H, Taki Y, Sassa Y, Hashizume H, Sekiguchi A, Fukushima A, et al. Brain structures associated with executive functions during everyday events in a non-clinical sample. Brain Struct Funct. (2013) 218:1017–32. doi: 10.1007/s00429-012-0444-z
37. Oldfield RC. The assessment and analysis of handedness: the Edinburgh inventory. Neuropsychologia. (1971) 9:97–113. doi: 10.1016/0028-3932(71)90067-4
38. Beck AT, Ward CH, Mendelson M, Mock J, Erbaugh J. An inventory for measuring depression. Arch Gen Psychiatry. (1961) 4:561–71. doi: 10.1001/archpsyc.1961.01710120031004
39. Beck AT, Steer RA, Brown GK. Manual for the Beck Depression Inventory - II. San Antonio, TX: The Psychological Corporation (1996) doi: 10.1037/t00742-000
40. Kendall PC, Hollon SD, Beck AT, Hammen CL, Ingram RE. Issues and recommendations regarding use of the beck depression inventory. Cognit Ther Res. (1987) 11:289–99. doi: 10.1007/BF01186280
41. Fan M, Liu B, Jiang T, Jiang X, Zhao H, Zhang J. Meta-analysis of the association between the monoamine oxidase-A gene and mood disorders. Psychiatr Genet. (2010) 20:1–7. doi: 10.1097/YPG.0b013e3283351112
42. Tsunoka T, Kishi T, Ikeda M, Kitajima T, Yamanouchi Y, Kinoshita Y, et al. Association analysis of group II metabotropic glutamate receptor genes (GRM2 and GRM3) with mood disorders and fluvoxamine response in a Japanese population. Prog Neuropsychopharmacol Biol Psychiatry. (2009) 33:875–9. doi: 10.1016/j.pnpbp.2009.04.007
43. van Rossum EF, Binder EB, Majer M, Koper JW, Ising M, Modell S, et al. Polymorphisms of the glucocorticoid receptor gene and major depression. Biol Psychiatry. (2006) 59:681–8. doi: 10.1016/j.biopsych.2006.02.007
44. Yoon HK, Kim YK. Association between glycogen synthase kinase-3beta gene polymorphisms and major depression and suicidal behavior in a Korean population. Prog Neuropsychopharmacol Biol Psychiatry. (2010) 34:331–4. doi: 10.1016/j.pnpbp.2009.12.009
45. Edgar NM, Touma C, Palme R, Sibille E. Resilient emotionality and molecular compensation in mice lacking the oligodendrocyte-specific gene Cnp1. Transl Psychiatry. (2011) 1:e42. doi: 10.1038/tp.2011.40
46. Franco-Pons N, Torrente M, Colomina MT, Vilella E. Behavioral deficits in the cuprizone-induced murine model of demyelination/remyelination. Toxicol Lett. (2007) 169:205–13. doi: 10.1016/j.toxlet.2007.01.010
47. Xu H, Yang HJ, Zhang Y, Clough R, Browning R, Li XM. Behavioral and neurobiological changes in C57BL/6 mice exposed to cuprizone. Behav Neurosci. (2009) 123:418–29. doi: 10.1037/a0014477
48. Ma Y, Li B, Wang C, Shi Z, Sun Y, Sheng F, et al. 5-HTTLPR polymorphism modulates neural mechanisms of negative self-reflection. Cereb Cortex. (2014) 24:2421–9. doi: 10.1093/cercor/bht099
49. Culverhouse RC, Saccone NL, Horton AC, Ma Y, Anstey KJ, Banaschewski T, et al. Collaborative meta-analysis finds no evidence of a strong interaction between stress and 5-HTTLPR genotype contributing to the development of depression. Mol Psychiatry. (2018) 23:133–42. doi: 10.1038/mp.2017.44
Keywords: depressive symptoms, negative self-schema, single nucleotide polymorphism, OLIG2, oligodendrocyte
Citation: Komatsu H, Takeuchi H, Ono C, Yu Z, Kikuchi Y, Kakuto Y, Funakoshi S, Ono T, Kawashima R, Taki Y and Tomita H (2021) Association Between OLIG2 Gene SNP rs1059004 and Negative Self-Schema Constructing Trait Factors Underlying Susceptibility to Depression. Front. Psychiatry 12:631475. doi: 10.3389/fpsyt.2021.631475
Received: 20 November 2020; Accepted: 05 February 2021;
Published: 08 March 2021.
Edited by:
Elena Martín-García, Pompeu Fabra University, SpainReviewed by:
Shwetal Mehta, Barrow Neurological Institute (BNI), United StatesCopyright © 2021 Komatsu, Takeuchi, Ono, Yu, Kikuchi, Kakuto, Funakoshi, Ono, Kawashima, Taki and Tomita. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroshi Komatsu, aGtvbWF0c3UxMDE5QGdtYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.