
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Psychiatry , 25 February 2021
Sec. Child and Adolescent Psychiatry
Volume 12 - 2021 | https://doi.org/10.3389/fpsyt.2021.560948
Objective: To explore the association of the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism with birth body mass and risk of autism in Chinese Han population.
Methods: A total 1,505 Chinese Han autism patients were recruited, using the Diagnostic and Statistical Manual of Mental Disorders, 4th revised version (DSM-IV-R) diagnostic criteria for autism, and 1,308 sex-matched healthy controls were also enrolled for the study. All the participants' birth body masses were counted according to the medical records. The MTHFR C677T genotypes were detected using the polymerase chain reaction-restrict fragment length polymorphism (PCR-RFLP) method. The association between C677T polymorphism, birth body mass, and risk of autism were analyzed using the chi-square tests.
Results: The present study found that the MTHFR 677T was significantly associated with risk of autism [P = 0.004, odds ratio (OR) = 1.18, 95% CI = 1.02–1.29). The autism children more frequently showed low birth body mass (<2.5 kg) than healthy control subjects (8.6 vs. 5.3%, P = 0.001, OR = 1.67, 95% CI = 1.24–2.26). The interactive effects between MTHFR 677T and low birth body mass (P = 0.0001, OR = 2.18, 95% CI = 1.44–3.32) were also significantly associated with risk of autism.
Conclusions: The MTHFR C677T polymorphism and low birth body mass may be associated with risk of autism in Chinese Han population.
Autism is a common neurodevelopmental disorder, which is clinically characterized by social communication and language communication disorders and repetitive and stereotyped interests and hobbies. At present, exact molecular and biochemical mechanism of autism has not been fully understood. Nevertheless, multiple genetic factors, environmental factors, and their interactions are believed to be involved in autism (1–4).
A large number of family and twin studies show that autism is highly inheritable, with heritability ranging from 60 to 90% (5–8). Genetic risk factors are considered to explain most risks of autism. In a large-scale cross-country cohort study of more than two million people carried out by Ban et al. (9), 22,156 of them were diagnosed with autism, with heritability of ~80% but without support for maternal effect and environmental impact, indicating that the changes in the occurrence rate of autism in this population were mainly attributed to genetic impact. However, new evidence show that environmental factors also increase the risk of autism, such as prenatal estrogen, maternal body mass index (BMI), perinatal complications, birth season, birth body mass, and other environmental risk factors, as well as the interaction between heredity and environment, which are also related to the risk of autism (10–16).
A literature reports that the risk of autism may be related to a variety of key genes involved in neurodevelopmental regulation, among which methylenetetrahydrofolate reductase (MTHFR) gene may be one of the candidate genes for neurodevelopmental disorders such as autism (17). The MTHFR is a key enzyme of folate metabolism in the process of one-carbon metabolism, converting 5,10-methylenetetrahydrofolate into 5-methyltetrahydrofolate, and participating in folate and homocysteine conversion correlated to DNA methylation (18). The active form of MTHFR could impact on the generation of 5-methyltetrahydrofolate, which is the active form of folate in vivo. Folate metabolism plays an extremely important role in the risk of neurodevelopmental disorders such as autism (19, 20). In a case–control cohort study of 45,300 offspring by Levine et al. (21), folate supplementation before and/or during pregnancy was observed to reduce the risk of autism in offspring. Methylation is a common regulation process of gene expression that influences cellular development and function (22), which is dependent on S-adenosylmethionine as a methyl donor. S-Adenosylmethionine originated from methionine cycle in which 5-methyltetrahydrofolate transfers methyl groups to homocysteine in a reaction catalyzed by methionine synthase to produce methionine (23).
The MTHFR gene has been identified as having 14 common or rare single nucleotide polymorphisms related to enzyme deficiency (24), of which C677T (rs1801133) located in exon 5 of the gene is one of the most reported (25), and its polymorphism TT genotype leads to a decrease in enzyme activity, which is only 40–50% of the CC-type enzyme activity at 37°C (17, 26). As consequences of polymorphism of MTHFR, reduction in MTHFR enzymatic activity would cause impaired methylation, deficiency of folate, as well as increase in homocysteine. Individuals with the 677T MTHFR alleles are predisposed to homocysteinemia and low plasma folate (27) and DNA hypomethylation (28). Individuals with the 677TT genotype manifest the highest homocysteine and lowest plasma folate levels (29) and lowest DNA methylation (27, 30). Children with autism have been found to have high plasma levels of homocysteine (31) and a biochemical profile of reduced methylation capacity (32). High levels of plasma homocysteine and increased oxidative stress have generally been associated in the pathophysiology of autism (33).
However, whether the above environmental factors and genetic susceptibility increase the risk of autism synergistically still needs further verification. This study intends to explore the association between MTHFR gene C677T polymorphism, birth body mass, and risk of autism in Chinese Han population.
This study included a total of 1,505 autistic children of Han nationality in China, including 998 boys and 507 girls. Admission criteria were the following: meet the diagnostic criteria for autism in the 4th edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-IV-TR), Han nationality, regardless of gender, age 3–18 years old, outpatient children, and with written informed consent form signed by the children themselves or their guardians. Exclusion criteria were the following: patients with mental retardation, schizophrenia, affective disorders, and other mental disorders that meet the DSM-IV Axis I diagnostic criteria; patients with severe physical diseases; and those unwilling to sign the informed consent form.
The control group consisted of 1,318 Chinese Han healthy controls, including 792 male and 526 female. Admission criteria were the following: Han nationality, regardless of gender, age 3–18 years old, database of normal subjects recruited from the community or physical examination center, with written informed consent form signed by the subjects themselves or their guardians. Exclusion criteria were the following: other mental disorders conforming to axis I diagnostic criteria such as autism, mental retardation, schizophrenia, and affective disorders in DSM-IV; patients with severe physical diseases; and those unwilling to sign the informed consent form.
All the participants' birth body masses were counted according to the medical records. According to the World Health Organization (WHO) standards, the birth body mass of full-term normal delivery children is divided into three groups: low birth body mass infants (<2.5 kg), normal birth body mass infants (2.5–4.0 kg), and fetal macrosomias (≥4.0 kg) (34).
Five milliliters of peripheral venous blood was extracted from all subjects, which was placed in ethylenediaminetetraacetic acid (EDTA) anticoagulation tube and stored at 4°C. A small amount of blood genomic DNA extraction kit from Qiagen Company was used to extract genomic DNA from samples within 1 week. After quality control, the samples were stored at −70°C for later use.
The PCR amplification primer sequences were as follows: upstream, 5′ AGC CCA GCC ACT CAC TGT TTT 3′; downstream, 5′ CAG CGA ACT CAG CAC TCC A 3′. Twenty-five microliters PCR amplification reaction system consist of 0.4 μM primer, 10 mM Tris–HCl (pH 8.3), 1.5 mM magnesium chloride, 50 mM potassium chloride, 200 μM deoxyribonucleotide triphosphates (dNTPs), 30–50 ng genomic DNA, and 1 U Taq DNA polymerase. PCR amplification conditions were predenaturation at 94°C for 5 min, denaturation at 94°C for 30 s, annealing at 64°C for 30 s, extension at 72°C for 1 min, a total of 35 cycles, and finally extension at 72°C for 10 min. The PCR products were digested with restriction enzyme Hinf I at 37°C overnight and observed by 4% agarose gel electrophoresis.
All data were managed and analyzed by Statistical Packages for Social Science 16.0 (SPSS 16.0) software. The Hardy–Weinberg equilibrium test was carried out on the genotype distribution of patients with fitness test. Chi-square analysis was used to test the association between the MTHFR C677T polymorphism site and birth body mass and risk of autism, and logistic regression analysis was used to comprehensively evaluate the contribution of genetic and environmental factors to autism risk. The statistical significance level of bilateral tests was set at P < 0.05.
The genotype frequency of MTHFR C677T (rs1801133) polymorphism in the case group and the control group both passed the Hardy–Weinberg equilibrium test, and there was no significant difference between the observed value and the expected value (P > 0.05) (Table 1). This showed that the research subjects were collected from a large population, and the individuals were randomly matched. There were no significant natural selection, migration, and other factors affecting the genetic balance, and the data were reliable.
The genotype and allele distributions are all shown in Table 2. There were statistically significant differences both in genotype frequencies (x2 = 8.088, df = 2, P = 0.018) and in allele frequencies (x2 = 8.251, df = 1, P = 0.004), respectively (Table 2). The results suggested that the risk allele 677T of MTHFR gene was significantly associated with risk of autism [odds ratio (OR) = 1.18, 95% CI = 1.02–1.29].
The average birth body mass in autism group was (3.15 ± 0.74) kg, male (3.24 ± 0.33) kg, and female (3.06 ± 0.62) kg. The average birth body mass of the healthy control group was (3.24 ± 0.52) kg, male (3.35 ± 0.49) kg, female (3.22 ± 0.46) kg, respectively. According to the WHO standards, newborn body mass is divided into low birth weight infants <2.5 kg, normal birth weight infants 2.5–4.0 kg, and fetal macrosomia ≥4.0 kg, respectively. The frequency of autism children with low birth body mass in the autism group (8.6%) was higher than that in the healthy control group (5.3%) (P = 0.003) (Table 3 and Figure 1). Compared with non-low birth body mass infants, low birth body mass infants increased the effect value of autism risk (OR = 1.67, 95% CI = 1.24–2.26), suggesting that low birth body mass infants may increase risk of autism.
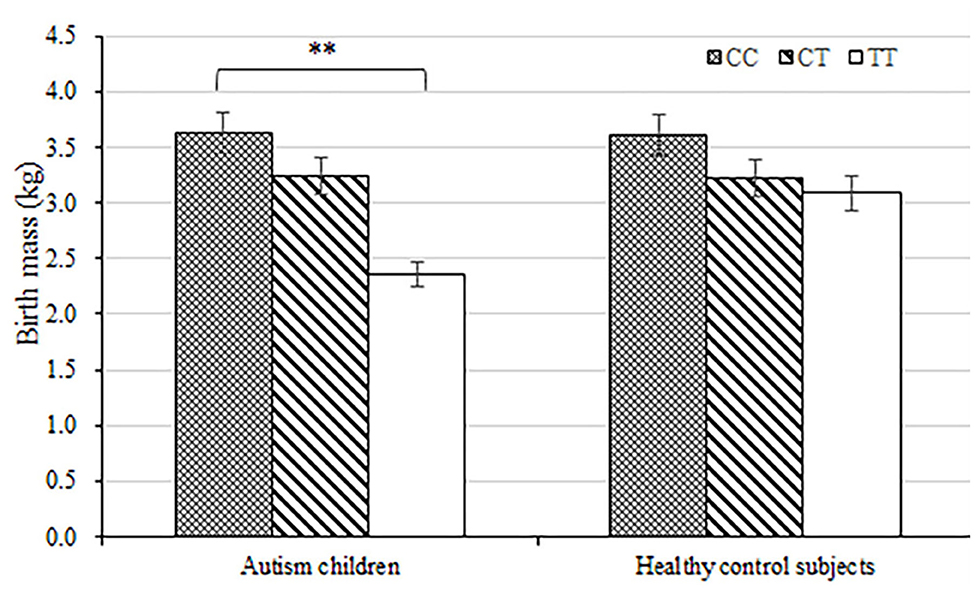
Figure 1. Load-dependent effects of MTHFR C677 genotypes on the birth body mass (kg) in autism children but not in healthy control subjects. Error bars ± 1.00 standard error. **P < 0.01.
To explore the potential interaction effects of the risk allele 677T of MTHFR gene and low birth body mass on the risk of autism, the number of children with autism who both carried MTHFR 677T and low birth body mass, were counted then were compared to the other children. The results indicated that the frequency of autism children both carrying MTHFR 677T and with low birth body mass (n = 75, 5.0%) was statistically higher than that of other subjects (n = 30, 2.3%). Although the number of patients carrying risk factors of 677T allele and low birth body mass infants in the two groups was relatively small (n = 46 in total), there was also a strong statistical correlation effect (χ2 = 14.38, P = 0.0001; OR = 2.18, 95% CI = 1.44–3.32). The logistic multifactor regression analyses were conducted to explore the effects of potential risk factors for autism. The diagnosis of autism was set as dependent variable, and the independent variables includes age, gender, MTHFR C677T polymorphism, birth body mass, and interaction between MTHFR C677T polymorphism and birth body mass, respectively. The results showed that sex, MTHFR C677T polymorphism, birth body mass, and interaction between MTHFR C677T polymorphism and birth body mass all significantly increased the risk of autism (Table 4).
The causes of autism, as a common neurodevelopmental disorder, are complex, containing both genetic and environmental factors. It has been reported in the literatures that multiple neurodevelopmental-related susceptibility genes might be associated with the risk of autism, such as neuregulin genes NRX1/3, NLGN3/4, etc. (35–38). The folic acid metabolic-rate-limiting enzyme, methylenetetrahydrofolate reductase (MTHFR), also may play an important regulatory role in neurodevelopmental disorders such as autism (19, 20). At the same time, there were also many reports that perinatal environmental factors such as low birth body mass may increase the risk of autism (10–16). The results of the present study also further confirmed some above-mentioned findings.
As one of the key enzymes in metabolic regulation, MTHFR is also one of the important candidate genes for common neurodevelopmental disorders. More than 10 mutations have been found contributing to effects of the MTHFR enzyme, of which C677T is one of the most common mutations found up to the present (24, 25). Decreased enzyme activity and heat tolerance caused by mutations can lead to folic acid metabolic abnormalities, methylation abnormalities, and neurodevelopmental disorders (17). This study suggested that carriers of MTHFR 677T allele were more likely to show an increased risk of autism than carriers of CC homozygotes (P = 0.004, OR = 1.18, 95% CI = 1.02–1.29), to some extent, which verifies that MTHFR may be one of the predisposing genes for autism. Paşca et al. (39) first reported the association between MTHFR C677T and risk of autism; however, it was a small sample study that just was carried out in 15 cases of autism and 25 cases of autism spectrum disorders. However, dos Santos (40) did not find any association between MTHFR C677T and risk of autism in Brazilian population. Sener et al. (41) also failed to find a significant association between MTHFR C677T and autism in a small sample population in Turkey (98 autism children and 70 healthy control subjects). Guo et al. (42) found that MTHFRC677T may be associated with risk of autism in a small Han Chinese sample of 186 autistic patients and 186 controls. It suggests that the association between MTHFR C677T and autism may be influenced by genetic heterogeneity among different populations. Shaik et al. (43) attempted to use six polymorphic loci of folate metabolism-related genes including MTHFR C677T in 138 pairs of autism spectrum disorders and control samples for artificial neural network calculation, and combined with meta-analysis of 1,361 autism spectrum disorders and 6,591 normal subjects, they finally found that C677T might be one of the important risk factors for autism.
In 2019, Sadeghiyeh et al. (44) in a meta-analysis of 18 studies evaluated the association between MTHFR 677C>T polymorphism and autism risk. Of the 18 studies, eight were conducted in Caucasian countries, three studies were in African population, and six studies were in Asian population. Their results suggested that MTHFR 677C>T was associated with increased autism risk in overall and by ethnicity among Caucasians, Asians, and Africans. In another recent meta-analysis by Razi et al. (45), 17 studies were included, and their results also suggested a significant association between C667T polymorphism and ASD risk in overall. However, in subgroup analysis, the significant association was only found among Caucasians but not in East Asians and Africans. The results of their subgroup analysis were inconsistent with the above meta-analysis (44), probably because only three studies were included in East-Asian countries and the sample size was insufficient. However, Sadeghiyeh et al. (44) also searched some Chinese databases, so there were more studies conducted in Asia. Therefore, the finding that there was a significant association between C677T and autism risk among Asians was more convincing, which is in line with our results as well.
Shaik Mohammad et al. (43) developed an artificial neural network (ANN) model as the predictors of autism risk using GCPII C1561T, SHMT1 C1420T, MTHFR C677T, MTR A2756G, and MTRR A66G. The ANN model showed 63.8% accuracy in predicting the risk of autism. In addition to our research, many other studies on Chinese Han children also showed MTHFR C677T to be a genetic risk factor for autism (46–48). Thus, MTHFR C677T may be considered as a potential biomarker of children with autism. Future research can also use MTHFR C677T to develop a prediction model to evaluate its prediction accuracy.
In addition, this study also found that low birth body mass (<2.5 kg) might increase risk of autism. There have been many reports that low birth body mass may be one of the risk factors for autism (16, 49), and the meta-analysis by Hisle-Gorman et al. (16) also suggests that epilepsy, maternal mental history during pregnancy, low birth body mass, etc. may increase the risk of autism. Willfors et al. (50) found that the correlation between birth body mass and autism was not significant but showed a similar trend (β = −0.01, P = 0.05) in the analysis of 54 pairs of identical twins with inconsistent autistic characteristics, indicating that low birth body mass was related to the risk of autism. A meta-analysis reported by Wang et al. (51) investigated the risk factors of childhood autism before, during, and after birth, which also confirmed that the increased risk of autism was related to low birth body mass [relative risk (RR) = 1.26; 95% CI: 1.20–1.34; P < 0.001].
In our study, we further explore the interactive effects of MTHFR 677T and low birth body mass on the risk of autism. The results indicated that the frequency of autism children both carrying MTHFR 677T and with low birth body mass (n = 75, 5.0%) was much higher than that of other subjects (n = 30, 2.3%) (χ2 = 14.38, P = 0.0001; OR = 2.18, 95% CI = 1.44–3.32). Moreover, the logistic regression analysis showed that gender, MTHFR C677T polymorphism, birth body mass, interaction between MTHFR C677T polymorphism, and birth body mass significantly increased risk of autism in children. El-Baz et al. (52) found that low birth body mass increased the risk of autism in 31 cases of autism spectrum disorders and 39 healthy controls in Egypt, and the frequency of MTHFR 677T in case group was higher than that in healthy controls (31.1 vs. 5.13%) (P < 0.001), which was basically consistent with the conclusion of this study. A meta-analysis showed association between maternal MTHFR C677T polymorphism with low birth body mass under most of the genetic models, respectively. Specifically, the carriers of the TT genotype increased the risk of low birth body mass (53). One explanation is that when MTHFR C677T causes the decrease in enzyme activity, the plasma homocysteine concentration increases, which leads to oxidative stress, arteriolar constriction, endothelial damage, and placental thrombosis (54, 55). All these conditions might be associated with impaired flow and prothrombotic changes in the vessel wall, inadequate trophoblast invasion into the uterine vasculature, and placental hypoperfusion that subsequently triggers poor pregnancy outcomes including low birth body mass (56). Another explanation is that MTHFR enzyme activity is reduced due to MTHFR C677T, which may cause the deficiency of folate. Folate deficiency is a modifiable nutritional status that has been linked with adverse pregnancy outcomes including low birth body mass (57, 58). Thus, the gene–environmental interaction between MTHFR 677T risk allele and low birth body mass may further increase the susceptibility of autism. It further supports the pathogenesis reported in the literatures that autism may be a complex disease caused by the interaction of genetic and environmental risk factors (13–16).
In the present study, the mean age of healthy control subjects was much older than that of autistic patients. Although there was age mismatch, it could also ensure that there was no risk of autism in the control group, and the birth body mass statistics of normal subjects were relatively accurate. In the future, it is planned to further expand the sample size and include age-matched healthy controls. In addition, the specific neurobiological mechanism of genetic polymorphism affecting risk of autism needs to be further elaborated, and further verification of sample size expansion and long-term follow-up in different populations is needed. In conclusion, this study suggests that MTHFR 677T and low birth body mass may be one of the important influencing factors of autism risk.
The original contributions presented in the study are included in the article/supplementary materials; further inquiries can be directed to the corresponding author/s.
The studies involving human participants were reviewed and approved by the medical ethics committee of Peking University Sixth Hospital and Beijing Children's Hospital. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
JZ, SS, and WY designed the study. JZ, SS, XM, and WY contributed to analysis, interpretation of data, and wrote the first draft of the manuscript. YS and LW contribute to the clinical assessment and experimental examination, as well as the statistical analyses. All authors contributed to drafting the work, revising it critically for important intellectual content, made substantial contributions to the concept, design of the study, acquisition, analysis, and interpretation of the data.
This work was supported by grants from the National Key R&D Program of China (2016YFC1307000), the National Natural Science Foundation of China (81825009, 81571313, 81901358, and 91432304), Natural Science Foundation of Shandong Province (ZR2019BH001), Chinese Academy of Medical Sciences Research Unit (No. 2018RU006), and Peking University Clinical Scientist Program (BMU2019LCKXJ012).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We also thank all subjects who participated in our study.
1. Hertz-Picciotto I, Schmidt RJ, Krakowiak P. Understanding environmental contributions to autism: causal concepts and the state of science. Autism Res. (2018) 11:554–86. doi: 10.1002/aur.1938
2. Schieve LA, Tian LH, Drews-Botsch C, Windham GC, Newschaffer C, Daniels JL, et al. Autism spectrum disorder and birth spacing: Findings from the study to explore early development (SEED). Autism Res. (2018) 11:81–94. doi: 10.1002/aur.1887
3. Kim YS, Leventhal BL. Genetic epidemiology and insights into interactive genetic and environmental effects in autism spectrum disorders. Biol Psychiatry. (2015) 77:66–74. doi: 10.1016/j.biopsych.2014.11.001
4. Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA. (2014) 311:1770–7. doi: 10.1001/jama.2014.4144
5. Colvert E, Tick B, McEwen F, Stewart C, Curran SR, Woodhouse E, et al. Heritability of autism spectrum disorder in a UK population-based twin sample. JAMA Psychiatry. (2015) 72:415–23. doi: 10.1001/jamapsychiatry.2014.3028
6. Tick B, Bolton PF, Happé F, Rutter M, Rijsdijk F. Heritability of autism spectrum disorders: a meta-analysis of twin studies. J Child Psychol Psychiatry Allied Discip. (2016) 57:585–95. doi: 10.1111/jcpp.12499
7. Sandin S, Lichtenstein P, Kuja-Halkola R, Hultman C, Larsson H, Reichenberg A. The heritability of autism spectrum disorder. JAMA. (2017) 318:1182. doi: 10.1001/jama.2017.12141
8. Wang K, Gaitsch H, Poon H, Cox NJ, Rzhetsky A. Classification of common human diseases derived from shared genetic and environmental determinants. Nat Genet. (2017) 49:1319–25. doi: 10.1038/ng.3931
9. Bai D, Yip BHK, Windham GC, Sourander A, Francis R, Yoffe R, et al. Association of genetic and environmental factors with autism in a 5-country cohort. JAMA Psychiatry. (2019) 76:1035–43. doi: 10.1001/jamapsychiatry.2019.1411
10. Baron-Cohen S, Tsompanidis A, Auyeung B, Nørgaard-Pedersen B, Hougaard DM, Abdallah M, et al. Foetal oestrogens and autism. Mol Psychiatry. (2019) 25:2970–8. doi: 10.1038/s41380-019-0454-9
11. Wang Y, Tang S, Xu S, Wen S, Liu Z. Maternal body mass index and risk of autism spectrum disorders in offspring: a meta-analysis. Sci Rep. (2016) 6:34248. doi: 10.1038/srep34248
12. Ismail S, Senna AA, Behiry EG, Ashaat EA, Zaki MS, Ashaat NA, et al. Study of C677T variant of methylene tetrahydrofolate reductase gene in autistic spectrum disorder Egyptian children. Am J Med Genet B Neuropsychiatr Genet. (2019) 180:305–9. doi: 10.1002/ajmg.b.32729
13. Wolford E, Pesonen AK, Heinonen K, Lahti M, Pyhälä R, Lahti J, et al. Autism spectrum traits and visual processing in young adults with very low birth weight: the Helsinki Study of Very Low Birth Weight adults. J Dev Orig Health Dis. (2017) 8:161–7. doi: 10.1017/S2040174416000738
14. Vanya M, Szucs S, Vetro A, Bartfail G. The potential role of oxytocin and perinatal factors in the pathogenesis of autism spectrum disorders - review of the literature. Psychiatry Res. (2017) 247:288–90. doi: 10.1016/j.psychres.2016.12.007
15. Modabbernia A, Velthorst E, Reichenberg A. Environmental risk factors for autism: an evidence-based review of systematic reviews and meta-analyses. Mol Autism. (2017) 8:13. doi: 10.1186/s13229-017-0121-4
16. Hisle-Gorman E, Susi A, Stokes T, Gorman G, Erdie-Lalena C, Nylund CM. Prenatal, perinatal, and neonatal risk factors of autism spectrum disorder. Pediatr Res. (2018) 84:190–8. doi: 10.1038/pr.2018.23
17. Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. (1995) 10:111–3. doi: 10.1038/ng0595-111
18. Froese DS, Huemer M, Suormala T, Burda P, Coelho D, Guéant J, et al. Mutation update and review of severe methylenetetrahydrofolate reductase deficiency. Hum Mutat. (2016) 37:427–38. doi: 10.1002/humu.22970
19. De Vilbiss EA, Gardner RM, Newschaffer CJ, Lee BK. Maternal folate status as a risk factor for autism spectrum disorders: a review of existing evidence. Br J Nutr. (2015) 114:663–72. doi: 10.1017/S0007114515002470
20. Jiang H, Liu L, Sun D, Yin X, Chen Z, Wu C, et al. Interaction between passive smoking and folic acid supplement during pregnancy on autism spectrum disorder behaviors in children aged 3 years. Chinese J Epidemiol. (2016) 37:940–4. doi: 10.3760/cma.j.issn.0254-6450.2016.07.007
21. Levine SZ, Kodesh A, Viktorin A, Smith L, Uher R, Reichenberg A, et al. Association of maternal use of folic acid and multivitamin supplements in the periods before and during pregnancy with the risk of autism spectrum disorder in offspring. JAMA Psychiatry. (2018) 75:176–84. doi: 10.1001/jamapsychiatry.2017.4050
22. Krebs MO, Bellon A, Mainguy G, Jay TM, Frieling H. One-carbon metabolism and schizophrenia: current challenges and future directions. Trends Mol Med. (2009) 15:562–70. doi: 10.1016/j.molmed.2009.10.001
23. de Arruda IT, Persuhn DC, de Oliveira NF. The MTHFR C677T polymorphism and global DNA methylation in oral epithelial cells. Genet Mol Biol. (2013) 36:490–3. doi: 10.1590/S1415-47572013005000035
24. Liew SC, Gupta ED. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: epidemiology, metabolism and the associated diseases. Eur J Med Genet. (2015) 58:1–10. doi: 10.1016/j.ejmg.2014.10.004
25. Yoshimi A, Aleksic B, Kawamura Y, Takahashi N, Yamada S, Usui H, et al. Gene-wide association study between the methylenetetrahydrofolate reductase gene (MTHFR) and schizophrenia in the Japanese population, with an updated meta-analysis on currently available data. Schizophr Res. (2010) 124:216–22. doi: 10.1016/j.schres.2010.07.011
26. Goyette P, Christensen B, Rosenblatt DS, Rozen R. Severe and mild mutations in cis for the Methylenetetrahydrofolate Reductase (MTHFR) gene, and description of five novel mutations in MTHFR. Am J Human Genet. (1996) 59:1268–75.
27. van der Put NM, Gabreels F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Human Genet. (1998) 62:1044–51. doi: 10.1086/301825
28. Castro R, Rivera I, Ravasco P, Camilo ME, Jakobs C, Blom HJ, et al. 5,10-methylenetetrahydrofolate reductase (MTHFR) 677C → T and 1298A → C mutations are associated with DNA hypomethylation. J Med Genet. (2004) 41:454–8. doi: 10.1136/jmg.2003.017244
29. Ulvik A, Ueland PM, Fredriksen A, Meyer K, Vollset SE, Hoff G, et al. Functional inference of the methylenetetrahydrofolate reductase 677C > T and 1298A > C polymorphisms from a large-scale epidemiological study. Human Genet. (2007) 121:57–64. doi: 10.1007/s00439-006-0290-2
30. Friso S, Girelli D, Trabetti E, Olivieri O, Guarini P, Pignatti PF, et al. The MTHFR 1298A > C polymorphism and genomic DNA methylation in human lymphocytes. Cancer Epidemiol Biomark Prevent. (2005) 14:938–43. doi: 10.1158/1055-9965.EPI-04-0601
31. Pasca SP, Nemes B, Vlase L, Gagyi CE, Dronca E, Miu AC, et al. High levels of homocysteine and low serum paraoxonase 1 arylesterase activity in children with autism. Life Sci. (2006) 78:2244–8. doi: 10.1016/j.lfs.2005.09.040
32. James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, et al. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am J Med Genet B Neuropsychiatr Genet. (2006) 141B:947–56. doi: 10.1002/ajmg.b.30366
33. Suh JH, Walsh WJ, McGinnis WR, Lewis A, Ames BN. Altered sulfur amino acid metabolism in immune cells of children diagnosed with autism. Am J Biochem Biotech. (2008) 4:105–13 doi: 10.3844/ajbbsp.2008.105.113
34. Lian Q, Deng H, You C, Chen K, Wang S, Cen C, et al. Study on correlation between gestational age, birth weight and autism spectrum disorder. Chinese J Behav Med Brain Sci. (2017) 26:215–9. doi: 10.3760/cma.j.issn.1674-6554.2017.03.005
35. Tsakos E, Tolikas A, Daniilidis A, Asimakopoulos B. Predictive value of anti-müllerian hormone, follicle-stimulating hormone and antral follicle count on the outcome of ovarian stimulation in women following GnRH-antagonist protocol for IVF/ET. Arch Gynecol Obstet. (2014) 290:1249–53. doi: 10.1007/s00404-014-3332-3
36. Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. (2003) 34:27–9. doi: 10.1038/ng1136
37. Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, et al. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. (2006) 409:10–3. doi: 10.1016/j.neulet.2006.08.017
38. Szatmari P, Paterson AD, Scherer SW, Vieland VJ, Pericak-Vance MA, Betancur C, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. (2007) 39:319–28. doi: 10.1038/ng1985
39. Paşca SP, Dronca E, Kaucsár T, Crǎciun EC, Endreffy E, Ferencz BK, et al. One carbon metabolism disturbances and the C677T MTHFR gene polymorphism in children with autism spectrum disorders. J Cell Mol Med. (2009) 13:4229–38. doi: 10.1111/j.1582-4934.2008.00463.x
40. dos Santos PA, Longo D, Brandalize AP, Schuler-Faccinil L. MTHFR C677T is not a risk factor for autism spectrum disorders in South Brazil. Psychiatr Genet. (2010) 20:187–9. doi: 10.1097/YPG.0b013e32833a2220
41. Sener EF, Oztop DB, Ozkul Y. MTHFR gene C677T polymorphism in autism spectrum disorders. Genet Res Int. (2014):698574. doi: 10.1155/2014/698574
42. Guo T, Chen H, Liu B, Ji W, Yang C. Methylenetetrahydrofolate reductase polymorphisms C677T and risk of autism in the Chinese Han population. Genet Test Mol Biomarkers. (2012) 16:968–73. doi: 10.1089/gtmb.2012.0091
43. Shaik Mohammad N, Sai Shruti P, Bharathi V, Krishna Prasad C, Hussain T, Alrokayan S, et al. Clinical utility of folate pathway genetic polymorphisms in the diagnosis of autism spectrum disorders. Psychiatr Genet. (2016) 26:281–6. doi: 10.1097/YPG.0000000000000152
44. Sadeghiyeh T, Dastgheib SA, Mirzaee-Khoramabadi K, Morovati-Sharifabad M, Akbarian-Bafghi MJ, Poursharif Z, et al. Association of MTHFR 677C>T and 1298A>C polymorphisms with susceptibility to autism: a systematic review and meta-analysis. Asian J Psychiatr. (2019) 46:54–61. doi: 10.1016/j.ajp.2019.09.016
45. Razi B, Imani D, Makoui MH, Rezaei R, Aslani S. Association between MTHFR gene polymorphism and susceptibility to autism spectrum disorders: systematic review and meta-analysis. Res Autism Spectr Disord. (2020) 70:101473. doi: 10.1016/j.asd.2019.101473
46. Xia W, Sun C, Li N, Wu K. Association methylenetetrahydrofolate reductase gene C677T polymorphism with autism children. Chinese J Child Health Care. (2012) 20:585–90. Available online at: https://kns.cnki.net/KXReader/Detail?TIMESTAMP=637487267554525234&DBCODE=CJFD&TABLEName=CJFD2012&FileName=ERTO201207008&RESULT=1&SIGN=buenMRWL9w98P6PRQQqI%2baJUM9w%3d#
47. Zhao D, Sun C, Yang X, Zhang Z, Li N, Hou L, et al. Association of methylenetetrahydrofolate reductase in gene C677T and A1298C polymorphism among children with autism. Chin J Sch Health. (2013) 34 (1):52–8.
48. Zhang H, Shang Q, Gao C, Geng X. Correlation between gene polymorphism of MTHFR gene C677T locus, 5-HTTLPR and autism in Han nationality children. J Clin Pathol Res. (2020) 41:80–5. doi: 10.3978/j.issn.2095-6959.2020.01.014
49. Talmi Z, Mankuta D, Raz R. Birth weight and autism spectrum disorder: a population-based nested case-control study. Autism Res. (2020) 13:655–65. doi: 10.1002/aur.2260
50. Willfors C, Carlsson T, Anderlid B, Nordgren A, Kostrzewa E, Berggren S, et al. Medical history of discordant twins and environmental etiologies of autism. Transl Psychiatry. (2017) 7:e1014. doi: 10.1038/tp.2016.269
51. Wang C, Geng H, Liu W, Zhang G. Prenatal, perinatal, and postnatal factors associated with autism. Medicine. (2017) 96:e6696. doi: 10.1097/MD.0000000000006696
52. El-Baz F, El-Aal MA, Kamal TM, Sadek AA, Othman AA. Study of the C677T and 1298AC polymorphic genotypes of MTHFR Gene in autism spectrum disorder. Electron Physician. (2017) 9:5287–93. doi: 10.19082/5287
53. Wu H, Zhu P, Geng X, Liu Z, Cui L, Gao Z, et al. Genetic polymorphism of MTHFR C677T with preterm birth and low birth weight susceptibility: a meta-analysis. Arch Gynecol Obstet. (2017) 295:1105–18. doi: 10.1007/s00404-017-4322-z
54. Chen H, Yang X, Lu M. Methylenetetrahydrofolate reductase gene polymorphisms and recurrent pregnancy loss in China: a systematic review and meta-analysis. Arch Gynecol Obstet. (2016) 293:283–90. doi: 10.1007/s00404-015-3894-8
55. Kupferminc MJ, Eldor A, Steinman N, Many A, Bar-Am A, Jaffa A, et al. Increased frequency of genetic thrombophilia in women with complications of pregnancy. N Engl J Med. (1999) 340:9–13. doi: 10.1056/NEJM199901073400102
56. Ulukuş M, Eroglu Z, Yeniel AÖ, Toprak E, Kosova B, Turan ÖD, et al. Frequency of factor V Leiden (G1691A), prothrombin (G20210A) and methylenetetrahydrofolate reductase (C677T) genes mutations in woman with adverse pregnancy outcome. J Turk Ger Gynecol Assoc. (2006) 7:195–201. Available online at: https://hdl.handle.net/11454/21662
57. Bhowmik B, Siddique T, Majumder A, Mdala I, Hossain IA, Hassan Z, et al. Maternal BMI and nutritional status in early pregnancy and its impact on neonatal outcomes at birth in Bangladesh. BMC Pregnancy Childb. (2019) 19:413. doi: 10.1186/s12884-019-2571-5
Keywords: autism, MTHFR gene, polymorphism, birth body mass, risk of disease
Citation: Zhang J, Ma X, Su Y, Wang L, Shang S and Yue W (2021) Association Study of MTHFR C677T Polymorphism and Birth Body Mass With Risk of Autism in Chinese Han Population. Front. Psychiatry 12:560948. doi: 10.3389/fpsyt.2021.560948
Received: 11 May 2020; Accepted: 22 January 2021;
Published: 25 February 2021.
Edited by:
David Cohen, Université Pierre et Marie Curie, FranceReviewed by:
Munis Dundar, Erciyes University, TurkeyCopyright © 2021 Zhang, Ma, Su, Wang, Shang and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weihua Yue, ZHJ5dWVAYmptdS5lZHUuY24=; Shaomei Shang, bWVpOTE2QDI2My5uZXQ=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.