
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Psychiatry, 15 December 2020
Sec. Schizophrenia
Volume 11 - 2020 | https://doi.org/10.3389/fpsyt.2020.564277
This article is part of the Research TopicGenetic Mechanisms of Biomarkers in Schizophrenia, Bipolar Disorder and DepressionView all 18 articles
Background: Schizophrenia (SZ) is a severe chronic mental disorder with complex genetic mechanisms. Brain-derived neurotrophic factor (BDNF) is one of promising candidate genes for SZ, and rs6265 is a non-synonymous single nucleotide polymorphism (SNP) in BDNF.
Methods: In this study, we performed a case-control association study of rs6265 in a cohort of Han Chinese population from eastern China, including 1,407 SZ patients and 1,136 healthy controls; and carried out a cis-mQTL (Methylation Quantitative Trait Loci) analysis for BDNF rs6265.
Results: We found a positive association of rs6265 with SZ (P = 0.037), with the minor allele (A) of rs6265 conferring a protecting effect for SZ (OR = 0.89). Furthermore, cis-mQTL analysis indicates that rs6265 is associated with several methylation loci surrounding BDNF.
Conclusions: Together, our findings provide further evidence to support the involvement of BDNF gene in the genesis of SZ.
Schizophrenia (SZ) is a serious mental disorder featured with profound disruption in emotion and cognition, affecting the most basic human properties such as language, thought, perception, and so on. The neurodevelopmental hypothesis is one of dominant hypotheses for SZ study in the last two decades, which supposes that SZ originates from developmental disorders of the nervous system early in brain development, long before the onset of the illness (1).
Brain-derived neurotrophic factor (BDNF) is believed to be involved in the pathophysiology of SZ and has been widely studied as a marker of neuropsychiatric diseases (2). The expression or functional changes of BDNF are proven associates of the pathophysiological process of several brain diseases, including mental diseases and neuro degenerative diseases (3). Animal studies have demonstrated the importance of BDNF in neurodevelopment and survival. Most of the homozygous BDNF mutant mice die within 2 days of birth, with some surviving 2 to 4 weeks. They exhibit distinct behavioral phenotypes as well as lack motor coordination and balance (4). BDNF affects cell level maturation, survival, diffusion, and synaptic function by activating intracellular signaling cascades, including mitogen-activated protein kinase/extracellular signal regulated protein kinase (MAPK/ERK), phosphatidylinositol 3-kinase, and phospholipase Cc pathways (5–7).
The single nucleotide polymorphism (SNP) rs6265 in BDNF, also known as Val66Met or G189A replacement at codon 66 in the pro-region of BDNF, alters the classification of BDNF protein and its availability in the synaptic cleft (2). The rs6265 variant interferes with the activity-dependent secretion of BDNF by inhibiting the sorting of BDNF into secretory granules, thereby affecting its function (8). An association of rs6265 polymorphism with the changes in hippocampal structure and function has been replicated in both human and mice (8, 9). The BDNF rs6265 knock-in mice show decreased BDNF expression, reduced hippocampal neurogenesis (10), decreased hippocampal volume, and abnormal morphology of hippocampal neurons (9). It has been suggested that rs6265 genotypes in the hippocampus and infra limbic medial prefrontal cortex may affect NMDA receptor-mediated neurotransmission and plasticity, which are associated with the production of positive or negative symptoms of schizophrenia (11, 12). Moreover, there is increasing evidence that rs6265 modifies both the clinical presentation and genetic risk architecture of schizophrenia, possibly by influencing cognitive function, brain morphology, age of onset, and treatment response (13, 14).
Prenatal stress is deemed to be a risk factor for SZ as a neurodevelopmental disorder (15). Dong et al. (15) reported both the SZ-like behavioral abnormalities in adult offsprings of mice exposed to prenatal stress mice and the molecular changes in the postmortem brains of SZ patients, with expression levels of DNA-methyltransferase 1 (DNMT1) and 10-11-translocation hydroxylase being significantly increased in the frontal cortex and hippocampus. Moreover, the corresponding reduction of BDNF transcription levels, along with enrichment of 5-methylcytosine and 5-hydroxymethylcytosine in the regulatory regions of BDNF gene, were observed, pointing at important role of epigenetic modification of BDNF in the phenotype and pathogenesis of SZ. It's worth noting that epigenetic modifications, including DNA methylation of the BDNF promoter, are significantly related to the pathophysiology of psychiatric disorders (4).
To further elucidate the role of the BDNF gene as a risk allele or regulator in SZ, we performed a case-control association study of rs6265 in a cohort of Han Chinese population from eastern China, including 1,407 SZ patients and 1,136 healthy controls, and carried out a cis-mQTL (Methylation Quantitative Trait Loci) analysis for BDNF rs6265.
All subjects were unrelated Han Chinese recruited from China. In the patient group, the diagnosis of SZ was in line with criteria in the Diagnostic and Statistical Manual of Mental Disorders, Fourth edition (DSM-IV) and confirmed by two or more experienced psychiatrists using the Structured Clinical Interview for DSM-IV (SCID-I). Exclusion criteria included the presence of other mood or neurodevelopmental disorders, epilepsy, or intellectual disability. For the choice of healthy controls, the Structured Clinical Interview for DSM-IV, Non-patients edition (SCID-NP) was used to interview members of an unrelated general population and exclude those with mental illness by professional psychiatrists.
For genetic association analysis, our study sample includes 1,407 SZ patients (874 men and 533 women, aged 45.8 ± 11.5 years) and 1,136 healthy controls (633 men and 503 women, aged 44.9 ± 10.3 years). Healthy subjects were recruited through advertisement. This study was approved by the Ethics Committees of the Wuxi Health Mental Center, and either patients or their guardians signed informed consents.
Peripheral blood samples were collected from all subjects. Blood samples were collected from all participants using K2EDTA tubes and a Blood Genotyping DNA Extraction Kit. The genotype of the SNP was analyzed by the Shanghai Biowing Applied Biotechnology Co. Ltd. (www.biowing.com.cn) using the Ligase Detection Reaction-Polymerase Chain Reaction method. Genomic DNA extracted from blood samples was first subjected to multiplex RCR to obtain a PCR product including SNPs. The PCR product and LDR probes were then subjected to multiplex LDR reaction, with a DNA sequencer to detect the products (1).
Genetic association tests were analyzed using PLINK v1.07 (16). The data obtained from SZ patients and healthy controls was compared. SNP association analyses were performed to test for possible associations between SNP rs6265 in BDNF and SZ using Plink v1.07. The two-tailed Fisher's exact test was used to compare the polymorphisms' distributions and testing their significance at p < 0.05 (17). The p-values were adjusted by false discovery rate correction for multiple test analysis (17). The allele frequencies and genotype distribution of rs6265 were calculated for the SZ cases and healthy controls and were analyzed for association by Logistic regression with the assumption of an additive genetic model, and Odds ratios (OR) with 95% confidence intervals were calculated (18). We performed the cis-mQTL in the methylation dataset (19) using Genevar 3.3 (20), with which we analyzed the association of rs6265 genotypes with neighboring methylations within 100 Kb distance. Associations between DNA methylation levels and probabilities of imputed genotypes were tested in samples of related individuals by a two-step analysis (19). That is, estimating a linear mixed model of methylation levels, covariates, and a kinship matrix, and then a score test. Age, beadchip, BS conversion efficiency, and BS-treated DNA input were cofactors. Cis analysis was limited to SNPs located within 100 kb of either side of the probe location and false discovery rate for the cis analysis was calculated with the q value package (19).
Genetic association analysis was performed in a study sample comprised of 1,407 patients (874 men and 533 women, aged 45.8 ± 11.5 years) and 1,136 unrelated healthy controls (633 men and 503 women, aged 44.9 ± 10.3 years).
In both the patient and the control groups, genotypic distributions of rs6265 had not deviated from Hardy-Weinberg equilibrium (HWE) (P > 0.05). Allelic distribution of rs6265 was associated with SZ (P = 0.037), with the minor allele (A) of rs6265 conferring a protecting effect for SZ (OR = 0.89). Specifically, OR = 0.89 indicates that minor allele (A) of rs6265 is negatively correlated with SZ, and reduces the risk of SZ in its carriers (Table 1).
When the associations of rs6265 genotypes with neighboring methylations within 100 Kb distance were studied using the cis-mQTL analysis, an association of rs6265 with 4 methylation loci near or within BDNF was detected (Figure 1).
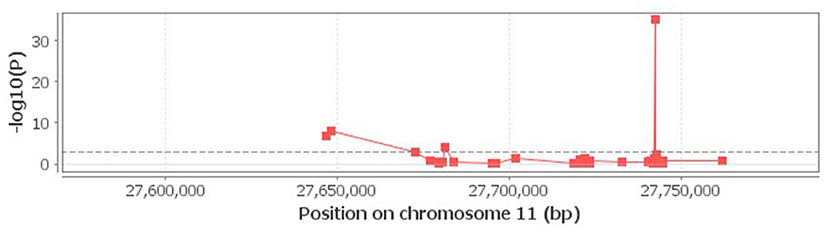
Figure 1. Correlation of rs6265 with neighboring methylations. The 4 loci above the dash lime are deemed to be significant (P < 0.001).
SZ is one of most disabling mental disorders; it affects the whole brain functions. Although the origin of this disorder remains unclear, a bulk of evidence supports that the abnormalities of early brain development play an important role in the pathogenesis of SZ.
In recent years, researches have focused on the effects of BDNF on brain development in the early stages of psychosis. For example, a significant correlation between the rs6265 polymorphism of BDNF and psychosis risk was found (21). An obvious association between the BDNF rs6265 genotype and the onset age of psychosis has also been demonstrated (22). Serum BDNF levels in first-episode psychosis patients were found to be much lower than those in the control group (23). Cumulatively, these findings indicate that BDNF may play an important role in the early stages of SZ.
For genetic association analysis, we found a positive association between rs6265 and SZ, with the minor A-allele of rs6265 conferring a protecting effect for SZ. A meta-analysis found that individuals with the Met/Met genotype had a 19% increased risk of developing SZ compared to individuals with the Val/Met genotype (24), a conclusion consistent with our results. However, it has been suggested that the non-mutated 66Val allele may be a risk locus for SZ, while the 66Met allele may actually be protective (14). For example, it was found that the Val gene conveyed risk in 321 Scottish SZ-spectrum probands (25). Moreover, a significant association was found between Val/Val genotype frequency and male patients with chronic SZ (26). The reasons for the inconsistencies in results may include differences in sample size, ethnic heterogeneity, etc., which still need further investigation.
A meta-analysis of imaging studies in neuropsychiatric patients with SZ, bipolar disorder, major depression or anxiety, showed that psychiatric patients with different BDNF rs6265 genotypes, such as Val/Val homozygous and Met-carrier both had smaller hippocampal volume as compared to that in healthy controls (2). Considering that rs6265 SNP was not associated with decreased hippocampal volume in neuropsychiatric patients, it followed that the Met allele might not be a risk allele (A/Met) for SZ (2). This hypothesis supports our findings pointing that allele A of rs6265 confers a protective effect for SZ. Due to a wide range of psychiatric disorders included in the meta-analysis cited above, whether this conclusion holds for the association between SZ and rs6265 genotype required further investigation.
At present, the body of literature describing results of the risk association studies of rs6265 and SZ remain contradictory due to lack of strong evidence and small sample sizes (14, 27). In view of larger sample size (1,406 SZ patients and 1,136 healthy controls), our findings may be important for bringing more clarity to the association between rs6265 and SZ.
cis-mQTL analysis indicated an association of rs6265 with several methylation loci within BDNF. As our study strongly suggests that rs6265 is associated with SZ, mQTL findings may be used to support the role of BDNF related methylation in the etiology of SZ. The peripheral blood level of DNA methylation of BDNF in SZ were accessed previously, with more methylated alleles and lower expression levels of BDNF found in the patient group than in the control group (28). Our findings are consistent with those observation, suggesting that BDNF related methylation may play an important role in SZ. Ikegame et al. (4) put forth that the down-regulation of BDNF levels is commonly associated with the increase in DNA methylation of the BDNF promoter. Cell and animal models have shown that the expression of BDNF in the neurons may be regulated by DNA methylation of specific promoters, but the mechanism of elevated DNA methylation at those specific sites is still elusive (4).
One previous study investigated peripheral blood lymphocytes of SZ patients and found the changes in DNA methylation of BDNF promoter I, thus, connecting the epigenetic alteration of BDNF locus in peripheral blood cells to the pathophysiology of SZ (29). Dong et al. (30) have shown that mice born from dams stressed during pregnancy develop behavioral deficits similar to those detected in adult SZ patients. Moreover, they showed that clozapine treatment reverses both the behavioral deficits and 5-methylcytosine and 5-hydroxy methylcytosine changes at BDNF promoters, as well as reduces mRNA and protein expression of this gene (30). This provides further evidence that BDNF-related methylation may play an important role in the pathophysiology of SZ.
On the other hand, in a study of the prefrontal cortex samples from 25 SZ patients and 25 healthy controls, the risk of SZ was independent of BDNF mRNA expression levels and the differences in DNA methylation of its promoter (31). Another study examined the DNA methylation levels of BDNF-encoded exons and three different promoter regions in the prefrontal cortex of SZ patients and found no significant differences in DNA methylation in patients with schizophrenia when their brain regions were compared to that of the controls (32). In sum, the above studies did not find an association between BDNF and DNA methylation in SZ. Since these studies were made using samples from the prefrontal cortex of the postmortem brains, these conclusions are vulnerable to possible bias due to their postmortem character, precluding direct comparisons with the conclusions made using living samples from the cells peripheral blood.
Kundakovic et al. (33) have explored a model of environmental exposure to bisphenol A in pregnant mice. Persistent changes of DNA methylation were detected in regions related to the transcription of BDNF gene in the hippocampus and blood of exposed pups. These changes were consistent with those in human umbilical cord blood of the newborns of exposed mothers, suggesting that the DNA methylation of BDNF in blood may serve as indicator for the DNA methylation of BDNF in the brain and the biomarker of behavioral vulnerability. Here we studied first-episode SZ patients and found an association between BDNF polymorphism rs6265 and methylation within BDNF locus. Our findings suggest that abnormal methylation of BDNF-related regions occurs early in SZ, pointing at the utility of these minimally-invasive for the detection of early SZ.
Many studies have found that epigenetic abnormalities in the SZ postmortem brain and the peripheral blood lymphocytes of SZ patients parallel each other. For example, in both types of samples, activities of cytosine modify DNMT1 and ten-eleven methylcytosine dioxygenase 1 increase, while amounts of BDNF-encoding mRNA, which is highly sensitive to the levels of its DNA methylation, decrease (34–41). In addition, studies have shown that, in SZ patients, increased DNMT expression may be seen as a probable cause of the concomitant decrease in BDNF expression, mediated by the DNMT-dependent elevation of cytosine methylation levels in BDNF promoter (36, 42). Davies et al. (43) showed that the DNA methylation patterns in the brain and the blood are highly correlated, including that in genes related to neural differentiation and neurodevelopment, such as BDNF. Furthermore, a similarity between brain- and lymphocyte-specifics changes of DNMT and ten-eleven methylcytosine dioxygenase 1 activities was shown (34, 44). In the case of SZ, the mechanisms of epigenetic regulation in the peripheral blood lymphocytes and the brain may be similar (34, 41, 43, 45). Therefore, the methylation levels in certain genes expressed in cells of the peripheral blood may be used as proxy biomarkers for early identification of SZ, and early interventions.
There are several limitations to consider in our study. The methylation data were derived from peripheral tissues rather than the brain itself, thus caution is needed when extrapolating the conclusions. Said that, detected abnormalities in peripheral tissue may serve as potential indicators of disease pathology even though they distinct from that in the brain. The changes in peripheral biomarkers may parallel pathological processes in the brain only in part, while in other part they would reflect the molecular reaction of the peripheral cells, which is secondary to the disease (46). Because both of these regulatory arms are reflective of the disease, “cause and consequences” arguments have lower applicability to biomarkers, which typically serve as indicators of association rather that causality. This study highlighted methylation biomarkers in the BDNF promoter as possible contributors for SZ detection in first-episode patients.
Our study supports the association between BDNF polymorphism rs6265 and SZ, as well as the relevance between rs6265 and BDNF methylation, providing further evidence to support the involvement of BDNF gene in the genesis of SZ.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: European Variation Archive, accession no: PRJEB41532 and ERZ1685357.
The studies involving human participants were reviewed and approved by Ethics Committees of the Wuxi Health Mental Center. The patients/participants provided their written informed consent to participate in this study.
FZ designed the study and performed data analyses. XF, JW, JD, JS, and AB were responsible for manuscript writing and modification. All authors reviewed and approved the final manuscript.
This work was supported by National Key R&D Program of China (2016YFC1307000), National Natural Science Foundation of China (81471364) and Suzhou Municipal Bureau of Science and Technology Program (SYSD2017136).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We want to take this opportunity to thank all the subjects for their support and participation and all the medical staff involved in collecting blood samples.
1. Wang Z, Yang B, Liu Y, Shugart YY, Cheng Z, Jin C, et al. Further evidence supporting the association of NKAPL with schizophrenia. Neurosci Lett. (2015) 605:49–52. doi: 10.1016/j.neulet.2015.08.023
2. Harrisberger F, Smieskova R, Schmidt A, Lenz C, Walter A, Wittfeld K, et al. BDNF Val66Metpolymorphism and hippocampal volume in neuropsychiatric disorders: a systematic review and meta-analysis. Neurosci Biobehav Rev. (2015) 55:107–18. doi: 10.1016/j.neubiorev.2015.04.017
3. Numakawa T, Richards M, Adachi N, Kishi S, Kunugi H, Hashido K. MicroRNA function and neurotrophin BDNF. Neurochem Int. (2011) 59:551–8. doi: 10.1016/j.neuint.2011.06.009
4. Ikegame T, Bundo M, Murata Y, Kasai K, Kato T, Iwamoto K. DNA methylation of the BDNF gene and its relevance to psychiatric disorders. J Hum Genet. (2013) 58:434–8. doi: 10.1038/jhg.2013.65
5. Minichiello L. TrkB signalling pathways in LTP and learning. Nat Rev Neurosci. (2009) 10:850–60. doi: 10.1038/nrn2738
6. Russo SJ, Mazei-Robison MS, Ables JL, Nestler EJ. Neurotrophic factors and structural plasticity in addiction. Neuropharmacology. (2009) 56:73–82. doi: 10.1016/j.neuropharm.2008.06.059
7. Numakawa T, Suzuki S, Kumamaru E, Adachi N, Richards M, Kunugi H. BDNF function and intracellular signaling in neurons. Histol Histopathol. (2010) 25:237–58. doi: 10.14670/HH-25.237
8. Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, et al. The BDNF Val66Met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. (2003) 112:257–69. doi: 10.1016/S0092-8674(03)00035-7
9. Chen Z, Jing D, Bath KG, Ieraci A, Khan T, Siao C, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. (2006) 314:140–3. doi: 10.1126/science.1129663
10. Bath KG, Jing DQ, Dincheva I, Neeb CC, Pattwell SS, Chao MV, et al. BDNF Val66Met impairs fluoxetine-induced enhancement of adult hippocampus plasticity. Neuropsychopharmacology. (2012) 37:1297–304. doi: 10.1038/npp.2011.318
11. Ninan I, Bath KG, Dagar K, Perez-Castro R, Plummer MR, Lee FS, et al. The BDNF Val66Met polymorphism impairs NMDA receptor-dependent synaptic plasticity in the hippocampus. J Neurosci. (2010) 30:8866–70. doi: 10.1523/JNEUROSCI.1405-10.2010
12. Pattwell SS, Bath KG, Perez-Castro R, Lee FS, Chao MV, Ninan I. The BDNF Val66Met polymorphism impairs synaptic transmission and plasticity in the infralimbic medial prefrontal cortex. J Neurosci. (2012) 32:2410–21. doi: 10.1523/JNEUROSCI.5205-11.2012
13. Schweiger JI, Bilek E, Schafer A, Braun U, Moessnang C, Harneit A, et al. Effects of BDNF Val(66)Met genotype and schizophrenia familial risk on a neural functional network for cognitive control in humans. Neuropsychopharmacology. (2019) 44:590–7. doi: 10.1038/s41386-018-0248-9
14. Notaras M, Hill R, van den Buuse M. A role for the BDNF gene Val66Met polymorphism in schizophrenia? A comprehensive review. Neurosci Biobehav Rev. (2015) 51:15–30. doi: 10.1016/j.neubiorev.2014.12.016
15. Dong E, Dzitoyeva SG, Matrisciano F, Tueting P, Grayson DR, Guidotti A. Brain-derived neurotrophic factor epigenetic modifications associated with schizophrenia-like phenotype induced by prenatal stress in mice. Biol Psychiatry. (2015) 77:589–96. doi: 10.1016/j.biopsych.2014.08.012
16. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. (2007) 81:559–75. doi: 10.1086/519795
17. Landini M, Merelli I, Raggi ME, Galluccio N, Ciceri F, Bonfanti A, et al. Association analysis of noncoding variants in neuroligins 3 and 4X genes with autism spectrum disorder in an Italian Cohort. Int J Mol Sci. (2016) 17:1765. doi: 10.3390/ijms17101765
18. Karroum EG, Saini PS, Trotti LM, Rye DB. TOX3 gene variant could be associated with painful restless legs. Sleep Med. (2020) 65:4–7. doi: 10.1016/j.sleep.2019.07.003
19. Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. (2013) 93:876–90. doi: 10.1016/j.ajhg.2013.10.004
20. Yang TP, Beazley C, Montgomery SB, Dimas AS, Gutierrez-Arcelus M, Stranger BE, et al. Genevar: a database and Java application for the analysis and visualization of SNP-gene associations in eQTL studies. Bioinformatics. (2010) 26:2474–6. doi: 10.1093/bioinformatics/btq452
21. Rosa A, Cuesta MJ, Fatjó-Vilas M, Peralta V, Zarzuela A, Fañanás L. The Val66Met polymorphism of the brain-derived neurotrophic factor gene is associated with risk for psychosis: evidence from a family-based association study. Am J Med Genet B Neuropsychiatr Genet. (2006) 141B:135–8. doi: 10.1002/ajmg.b.30266
22. Decoster J, van Os J, Kenis G, Henquet C, Peuskens J, De Hert M, et al. Age at onset of psychotic disorder: Cannabis, BDNF Val66Met, and sex-specific models of gene-environment interaction. Am J Med Genet B Neuropsychiatr Genet. (2011) 156:363–9. doi: 10.1002/ajmg.b.31174
23. Rizos EN, Siafakas N, Stefanis N, Douzenis A, Lykouras L. Association of serum BDNF and val66met polymorphism of the brain-derived neurotrophic factor in a sample of first psychotic episode patients. Psychiatriki. (2009) 20:297–304. doi: 10.2352/ISSN.2169-4672.2009.2.0.44
24. Gratacòs M, González JR, Mercader JM, Cid RD, Urretavizcaya M, Estivill X. Brain-derived neurotrophic factor Val66Met and psychiatric disorders: meta-analysis of case-control studies confirm association to substance-related disorders, eating disorders, and schizophrenia. Biol Psychiatry. (2007) 61:911–22. doi: 10.1016/j.biopsych.2006.08.025
25. Neves-Pereira M, Cheung JK, Pasdar A, Zhang F, Breen G, Yates P, et al. BDNF gene is a risk factor for schizophrenia in a Scottish population. Mol Psychiatry. (2005) 10:208–12. doi: 10.1038/sj.mp.4001575
26. Golimbet VE, Korovaitseva GI, Abramova LI, Kasparov SV, Uvarova LG. Association of the Val66Met polymorphism of the brain-derived neurotrophic factor gene with schizophrenia in Russians. Mol Biol. (2008)42:531–5. doi: 10.1134/S0026893308040079
27. Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. (2010) 68:182–6.doi: 10.1016/j.neuron.2010.10.003
28. Kordi-Tamandani DM, Sahranavard R, Torkamanzehi A. DNA methylation and expression profiles of the brain-derived neurotrophic factor (BDNF) and dopamine transporter (DAT1) genes in patients with schizophrenia. Mol Biol Rep. (2012) 39:10889–93.doi: 10.1007/s11033-012-1986-0
29. Ikegame T, Bundo M, Sunaga F, Asai T, Nishimura F, Yoshikawa A, et al. DNA methylation analysis of BDNF gene promoters in peripheral blood cells of schizophrenia patients. Neurosci Res. (2013) 77:208–14. doi: 10.1016/j.neures.2013.08.004
30. Dong E, Tueting P, Matrisciano F, Grayson DR, Guidotti A. Behavioral and molecular neuroepigenetic alterations in prenatally stressed mice: relevance for the study of chromatin remodeling properties of antipsychotic drugs. Transl Psychiatry. (2016) 6:e711. doi: 10.1038/tp.2015.191
31. Cheah SY, McLeay R, Wockner LF, Lawford BR, Young RM, Morris CP, et al. Expression and methylation of BDNF in the human brain in schizophrenia. World J Biol Psychiatry. (2017) 18:392–400. doi: 10.1080/15622975.2016.1245443
32. Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. (2008) 82:696–711. doi: 10.1016/j.ajhg.2008.01.008
33. Kundakovic M, Gudsnuk K, Herbstman JB, Tang D, Perera FP, Champagne FA. DNA methylation of BDNF as a biomarker of early-life adversity. Proc Natl Acad Sci USA. (2015) 112:6807–13. doi: 10.1073/pnas.1408355111
34. Auta J, Smith RC, Dong E, Tueting P, Sershen H, Boules S, et al. DNA-methylation gene network dysregulation in peripheral blood lymphocytes of schizophrenia patients. Schizophr Res. (2013) 150:312–8. doi: 10.1016/j.schres.2013.07.030
35. Ruzicka WB, Zhubi A, Veldic M, Grayson DR, Costa E, Guidotti A. Selective epigenetic alteration of layer I GABAergic neurons isolated from prefrontal cortex of schizophrenia patients using laser-assisted microdissection. Mol Psychiatr. (2007) 12:385–97. doi: 10.1038/sj.mp.4001954
36. Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-anpongkul N, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. (2009) 323:1074–7. doi: 10.1126/science.1166859
37. McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat. Neurosci. (2009) 12:342–8. doi: 10.1038/nn.2270
38. Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. (2012) 2:e159. doi: 10.1038/tp.2012.86
39. Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, Beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. (2012) 37:531–42. doi: 10.1038/npp.2011.221
40. Labonté B, Suderman M, Maussion G, Navaro L, Yerko V, Mahar I, et al. Genome-wide epigenetic regulation by early-life trauma. Arch Gen Psychiatry. (2012) 69:722–31. doi: 10.1001/archgenpsychiatry.2011.2287
41. Provencal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci. (2012) 32:15626–42. doi: 10.1523/JNEUROSCI.1470-12.2012
42. Zhang TY, Labonte B, Wen XL, Turecki G, Meaney MJ. Epigenetic mechanisms for the early environmental regulation of hippocampal glucocorticoid receptor gene expression in rodents and humans. Neuropsychopharmacology. (2013) 38:111–23. doi: 10.1038/npp.2012.149
43. Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol. (2012) 13:R43. doi: 10.1186/gb-2012-13-6-r43
44. Zhubi A, Veldic M, Puri NV, Kadriu B, Caruncho H, Loza I, et al. An upregulation of DNA-methyltransferase 1 and 3a expressed in telencephalic GABAergic neurons of schizophrenia patients is also detected in peripheral blood lymphocytes. Schizophr Res. (2009) 111:115–22. doi: 10.1016/j.schres.2009.03.020
45. Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, et al. Allele-specific FKBP5 DNA demethylation mediates gene–childhood trauma interactions. Nat Neurosci. (2013) 16:33–41. doi: 10.1038/nn.3275
Keywords: schizophrenia, BDNF, single nucleotide polymorphism, rs6265, methylation
Citation: Fu X, Wang J, Du J, Sun J, Baranova A and Zhang F (2020) BDNF Gene's Role in Schizophrenia: From Risk Allele to Methylation Implications. Front. Psychiatry 11:564277. doi: 10.3389/fpsyt.2020.564277
Received: 05 June 2020; Accepted: 25 November 2020;
Published: 15 December 2020.
Edited by:
Ingrid Melle, University of Oslo, NorwayReviewed by:
Chunxia Yang, First Hospital of Shanxi Medical University, ChinaCopyright © 2020 Fu, Wang, Du, Sun, Baranova and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuquan Zhang, emhhbmdmcUBuam11LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.