
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Psychiatry , 15 July 2020
Sec. Mood Disorders
Volume 11 - 2020 | https://doi.org/10.3389/fpsyt.2020.00667
This article is part of the Research Topic The Search for Biomarkers in Psychiatry View all 17 articles
Depression is one of the most prevalent and serious mental disorders with a worldwide significant health burden. Metabolic abnormalities and disorders in patients with depression have attracted great research attention. Thirty-six metabolic biomarkers of clinical plasma metabolomics were detected by platform technologies, including gas chromatography–mass spectrometry (GC–MS), liquid chromatography–mass spectrometry (LC–MS) and proton nuclear magnetic resonance (1H-NMR), combined with multivariate data analysis techniques in previous work. The principal objective of this study was to provide valuable information for the pathogenesis of depression by comprehensive analysis of 36 metabolic biomarkers in the plasma of depressed patients. The relationship between biomarkers and enzymes were collected from the HMDB database. Then the metabolic biomarkers-enzymes interactions (MEI) network was performed and analyzed to identify hub metabolic biomarkers and enzymes. In addition, the docking score-weighted multiple pharmacology index (DSWMP) was used to assess the important pathways of hub metabolic biomarkers involved. Finally, validated these pathways by published literature. The results show that stearic acid, phytosphingosine, glycine, glutamine and phospholipids were important metabolic biomarkers. Hydrolase, transferase and acyltransferase involve the largest number of metabolic biomarkers. Nine metabolite targets (TP53, IL1B, TNF, PTEN, HLA-DRB1, MTOR, HRAS, INS and PIK3CA) of potential drug proteins for treating depression are widely involved in the nervous system, immune system and endocrine system. Seven important pathways, such as PI3K-Akt signaling pathway and mTOR signaling pathway, are closely related to the pathology mechanisms of depression. The application of important biomarkers and pathways in clinical practice may help to improve the diagnosis of depression and the evaluation of antidepressant effect, which provides important clues for the study of metabolic characteristics of depression.
Depression is one of the most prevalent and serious mental disorders. In recent years, the number of patients with depression has increased dramatically. In the world’s population, the lifetime prevalence of depressed patients is about 17% with a significant burden of disease (1). Previous research reports have found that in the United States, more than 19 million adults suffer from depression in the USA and spend more than 30 billion annually, directly or indirectly (2). In addition, the incidence of depression is about 3 to 5% and currently 26 million Chinese people suffer from depression (3). Depression is a major cause of neuropsychiatric disability worldwide and the accurate diagnosis of depression before treatment is a hub change (4). Increased studies have reported the serotonin and norepinephrine dysfunction in the central nervous system of patients with depression (5). In addition, studies have also reported that hypothalamic pituitary adrenal (HPA) axis is one of the largest neuroendocrine findings of depression (6). Abnormal changes of inflammatory cytokines and endogenous metabolites are also involved in the molecular mechanism of pathology of depression (7, 8). The occurrence and development of depression is a complicated process, the etiology and pathogenesis mechanism of depression represent challenging issues in scientific and medical research.
A new psychological immune neuroendocrine (PINE) network model on depression has been proposed to provide an in-depth understanding of the pathogenesis of depression and the treatment of the disease with antidepressant drugs (1). The PINE network model is composed of three parts of the central nervous system, immune, and endocrine molecular networks, and the three networks are interconnected (9). The three molecular networks consist of many nodes and edges. The nodes in the network can be small molecules with different properties, such as genes, proteins or metabolites. If different nodes are related in biological function, it can be connected by edges. How to determine the key underlying molecular mechanism from PINE network that plays leading roles in the depression is a difficult problem due to the high complexity of the network composition and the incompletely understanding the complex multi-targets mechanism of depression.
Network pharmacology is constructed by integrating pharmacological data and network analysis methods to provide a comprehensive approach to explain the disease pathogenesis and drug treatment mechanism (10). The network pharmacology technology is considered to be one of the next frontiers of new drug research (11). Recently, network pharmacology has been widely used in the pathogenesis of complex diseases and the mechanism of drug action. For example, Huang decoded the mechanism of traditional Chinese medicine in treating depression by analyzing drug target networks and disease target networks (12).
Increased studies have reported metabolic disorders or abnormal metabolic pathways in the plasma of depressed patients (13, 14). Thirty-six metabolic biomarkers of clinical plasma metabolomics were detected by platform technologies, including gas chromatography–mass spectrometry (GC–MS), liquid chromatography–mass spectrometry (LC–MS) and proton nuclear magnetic resonance (1H-NMR), combined with multivariate data analysis techniques in previous our work (13, 14). These metabolic biomarkers are distributed in disordered metabolic pathways and maybe potential diagnostic biomarkers or therapeutic markers. Therefore, how to use the network pharmacology method to mine the existing markers is of great significance to the study of the metabolic mechanism of depression.
The main purposes of this study were to comprehensively analyze plasma metabolites by network pharmacology method and provide valuable information for the pathogenesis of depression. During this process, the relationship between biomarkers and enzymes were collected from the HMDB database and the metabolic biomarkers–enzymes interactions (MEI) network was performed and analyzed to identify hub metabolic biomarkers and enzymes. In addition, the interactions between each MB and each target of the nervous system, immune system and endocrine system was calculated by systemsDock, and then the docking score-weighted multiple pharmacology index (DSWMP) was used to assess the importance pathways of hub metabolic biomarkers involved. Finally, the important pathways were verified through published literature. The application of important biomarkers and pathways in clinical practice may help to improve the diagnosis of depression and the evaluation of antidepressant effect, which provides important clues for the study of metabolic characteristics of depression.
For the research object, only the metabolic biomarkers of clinical plasma metabolomics in our consideration, because the excavation of metabolites is more meaningful in the same clinical sample. Candidate metabolites refer to the statistical significance found in the original study. Unique metabolites were obtained by removing duplicates. We obtained biological function information and structural data of identified metabolic biomarkers from the Human Metabolome Database (HMDB) (15). The structures of these compounds were downloaded from PubChem (16). Enzymes related to the metabolic biomarkers were summarized from HMDB, while the functional categories of the relevant proteins were found from the UniProt database (17). The target proteins related to the nervous system (Table S1), immune system (Table S2) and endocrine system (Table S3) were retrieved from several Published online databases. The FDA-approved drugs for the nervous system, immune system and endocrine system with their targets were collected from DrugBank (18). The depression-related proteins were retrieved from Genecards (19), OMIM (20) and TTD (21). Finally, we carefully checked the target proteins in the literature. Integrate all target proteins and divide them into three categories, including the nervous system, immunity and endocrine.
Molecular docking was used to investigate the affinity between metabolic biomarkers and targets. The protein 3D structures were achieved from the RCSB protein data bank (http://www.rcsb.org) (22). We chose structures with more complete peptide chains, higher resolution, and better ligands as the selection criteria for the most appropriate protein structure.
SystemsDock (http://systemsdock.unit.oist.jp/) (23) was used for network pharmacology prediction and analysis, including four specific steps, selecting proteins by different parameters, defining binding sites by interactive molecular visualizer, preparing metabolic biomarkers for the test, and performing docking simulation and evaluating the result. The docking score calculated by systemsDock was used to assess the interactions between the metabolic biomarkers and the target proteins.
Metabolic biomarker–enzyme relationships were collected from HMDB and visualized by using Cytoscape 3.4.0 (24). The network topological properties of nodes (metabolic biomarkers and enzymes) were calculated by the NetworkAnalyzer Cytoscape plugin (25). In an undirected network, the degree centrality of a node denotes the number of edges connected to this node is commonly used to measure the importance of a node in a single-layer network (26, 27). Degree centrality is a key indicator in analyzing the network, thereby reflecting the importance and influence of a metabolome biomarker or an enzyme in the metabolic biomarkers–enzyme interactions (MEI) network.
The threshold of the docking score was set to 5.52 (pKd), which was equal to the dissociation constant (Kd) of 3 μM. A docking score greater than the threshold was considered to be a very good correlation between the metabolic biomarker and the target protein (23). Based on previously reported works and experimental findings, a high accuracy level (80–83%) was found to evaluate the binding of a metabolomic biomarker and a protein, when the threshold score was in the range of 4.82 to 6.11 (pKd). The metabolic biomarkers-target interactions (MTI) network was visualized by Cytoscape software.
The target-related pathways were collected from Kyoto Encyclopedia of Genes and Genomes (KEGG) (28). One metabolic biomarker can be combined with many targets, and one target can be enriched to function in many different biological pathways. Therefore, a metabolic biomarker involves multiple different biological pathways. At present, there is no method to directly confirm that the metabolic biomarkers are directly enriched in the pathway through the targets, so we have defined a docking score-weighted multiple pharmacology index (DSWMP):
where DSBiTj is the affinity between metabolic biomarker Bi and target Tj, and TPk is a group of targets involved in pathway Pk. N and M are the numbers of metabolic biomarkers and targets, respectively.
To find experimental evidence for the predicted important pathways of depression by DSWMP calculation, the published literature will be identified by searching Pubmed, Embase, Cochrane Central Register of Controlled Trials and Web of Science. In the end, we chose the literature to verify the important pathways we found and provide references for the next step of research.
In this report, a novel network pharmacology strategy was designed to detect the important pathway and elucidate the molecular mechanisms of depression (Figure 1). Firstly, the relationship between biomarkers and enzymes were collected from the HMDB database and the MEI network was performed and analyzed to identify hub metabolic biomarkers and enzymes. Secondly, the interactions between each MB and each target were calculated by systemsDock, and then the DSWMP was used to assess the important pathways of hub metabolic biomarkers involved. Thirdly, the important pathways verified through published literature.
The information of 36 metabonomic biomarkers and 350 enzymes are shown in Table S4. Most enzymes related to metabolic biomarkers belong to hydrolases, transferases and acyltransferases, which is consistent with our previous results. These enzymes mainly regulate metabolites such as energy metabolism, amino acid metabolism, gut microbe metabolism, glycerophospholipid metabolism and sphingolipid metabolism (13, 14). These enzymes play an important role in the regulation of metabolic pathways, which affect the molecular mechanism of depression. Overall, the results show that functional studies of enzymes related to metabolic biomarkers can reflect the molecular mechanism of depression, which also provides a reliable reference for the next network analysis.
The MEI network was used to characterize the relationship between enzymes and metabolic biomarkers, as shown in Figure 2. In MEI network, the node represents a metabolic biomarker or an enzyme, and the edges represent the interactions between them originated from HMDB. It turns out that the concentrations of some biomarkers in depression patients are up-regulated, while others are down-regulated. Among these regulated biomarkers, their related enzymes form a particularly large cluster around them, suggesting that multiple enzymes regulate the same metabolite. Tables 1 and 2 show the important enzymes and biomarkers identified by the network pharmacology approach, respectively. Among these data, previous literatures reported that cytosolic phospholipase A2 (cPLA2) is an important enzyme for PUFA metabolism and PGE 2 synthesis, which is the pivotal to the mode action of mood stabilizers in animal studies (29, 30). In addition, in peripheral blood cells of patients with depression, the mRNA expression of the gene encoding COX-2 increases sharply, which plays an important role in the pathogenesis of depression (31).
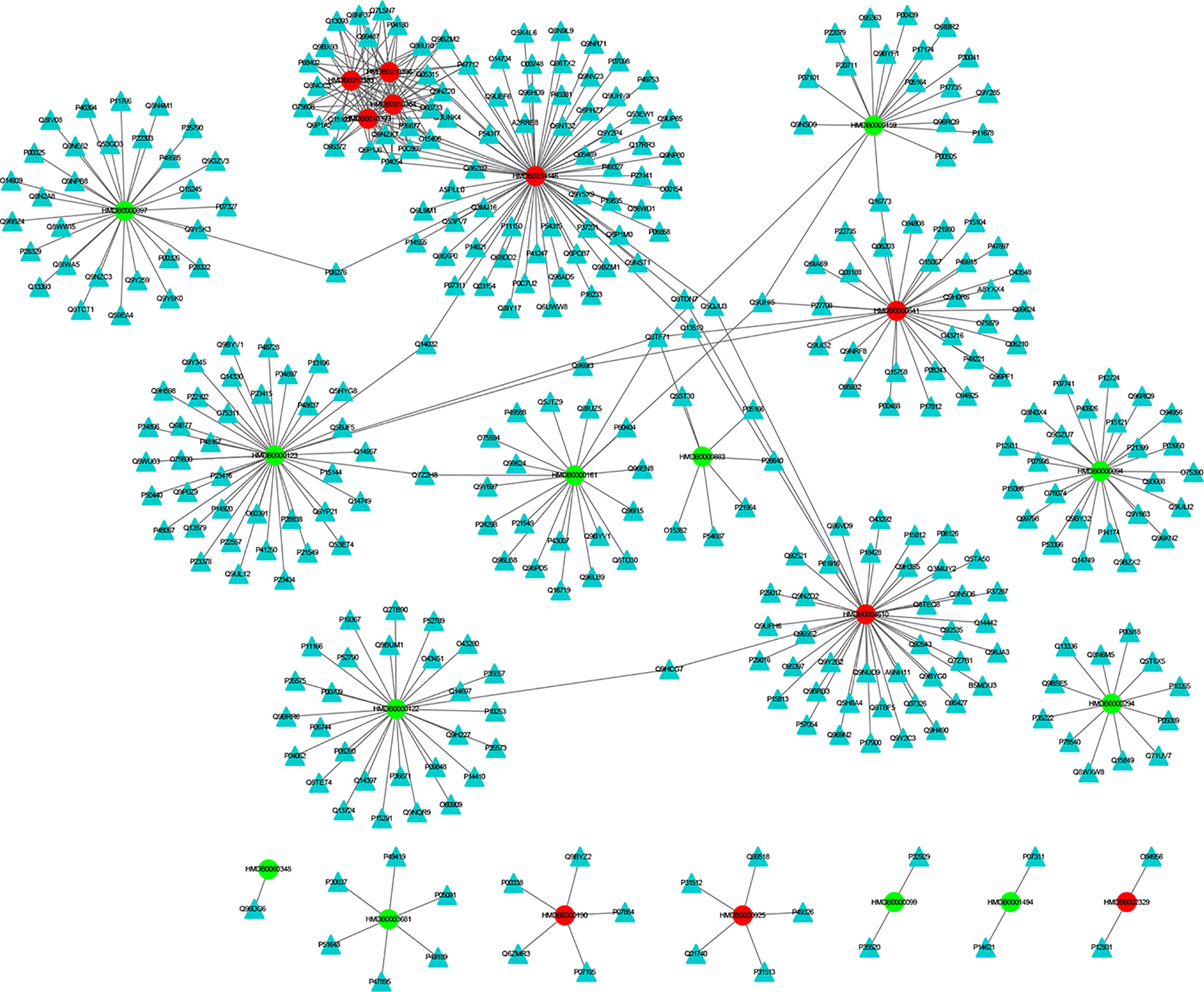
Figure 2 The MEI network. The cycle and triangle represent metabolic biomarker and enzyme, respectively. The color of cycle is green or red if the concentration of the metabonomic biomarker is down-regulated or up-regulated, respectively.
Among these metabolite-related enzymes, the phospholipase A2 family is related to many metabolites, so it is also important to study the function of depression. Previous research reports suggest that different phospholipase A2 types are associated with somatic symptoms of depression (32). It is believed that genetic variation of the phospholipase A2 gene increases the risk factor of depression induced by IFN-α (32). In addition, phospholipase A2 inhibitors have potential therapeutic effects in treating inflammation-related diseases, such as depression (33). Moreover, eicosapentaenoic acid (EPA) could up-regulate the expression of cytosolic phospholipase A2 gene and play an antidepressant effect in clinic, which shows the superiority of EPA antidepressant effect (34). Another research group also found that the same G allele of the PLA2 BanI polymorphism is one of the risk factors for depression in Korean populations (35). The expression levels of acyl protein thioesterase 2 (APT-2) and oleamide are increased in chronic mild stress (CMS). This provides a reference for the treatment of depression by targeting these proteins (36). At present, there is no literature on the relationship between acetylhydrolase and depression, which may be a novel research point.
Fourteen metabolic biomarkers are associated with more than ten enzymes in MEI network (Table 2). The concentrations of most MB were down-regulated, while stearic acid, phytosphingosine, glutamine and phospholipids were up-regulated. It is well known that fatty acids are an important source of human body production and storage. Acetyl-coenzyme A (CoA) produced by oxidation of β-fatty acids can produce adenosine triphosphate (ATP) in the TCA cycle and could be converted to ketone bodies for storage in the kidney and liver (37). There are reports showing that the concentration of stearic acid in plasma of patients with depression is significantly increased, which may lead to blockage of fatty acid transport and inhibition of TCA cycle, which is consistent with the previous research results (38, 39). Moreover, the AUCs for oxalic acid and stearic acid were >0.7, indicating a great clinical diagnostic value. Therefore, stearic acid may be a diagnostic indicator of depression. These results indicate that the metabolic biomarkers in MEI network are involved in the molecular mechanism of depression.
In addition, most metabolic biomarkers belong to long-chain fatty acids and phytosphingosine, which indicates that the lipid metabolism disorder plays a crucial role in the mechanism of depression. These metabolic biomarkers are involved in glycerophospholipid metabolism and sphingolipid metabolism. Among them, sphingosine involves various biological processes, including cell–cell interactions, cell proliferation, differentiation and apoptosis. Studies have reported that elevated levels of hemolytic phosphatidylcholine also increase oxidative stress, which is a key factor in the onset of depression (40). The results indicate that the long-chain fatty acids and phytosphingosine in MEI network is involved in the pathological mechanism of depression.
Glutamine and its related enzymes form an independent part of the MEI network, suggesting that glutamine may play an unusual role in the pathogenesis mechanism of depression. Glutamate is associated with the neurobiology of depression and can cause neurotoxicity if over-released (40). In addition, glutamine and glutamate can be converted between neurons and astrocytes, which is necessary for the steady state of the glutamine-glutamic acid cycle (41). Therefore, the increase in glutamine in the plasma of depression patients may be a compensatory adaptation to glutamate-induced neurotoxicity. The results show that the glutamine in the MEI network is involved in the pathological mechanism of depression.
Tables S1–S3 show the nervous system, immune system and endocrine system target proteins, and the nervous system, immune system and endocrine system target numbers are 155, 60 and 125, respectively. The interactions between each metabolic biomarker and the target protein were performed by molecular docking. The MTI network of nervous system, immune system and endocrine system were visualized by Cytoscape, respectively (Figures 3–5). In MTI network, the node represents a metabolic biomarker or a target, and the edges represent the interactions between them from the docking score meeting the set threshold. The metabolic biomarker links the target to a highly interconnected cluster. In order to analyze the correlation between metabolic biomarkers and targets and to mine important molecules, we calculated the topological parameter of the network, namely degree centrality.
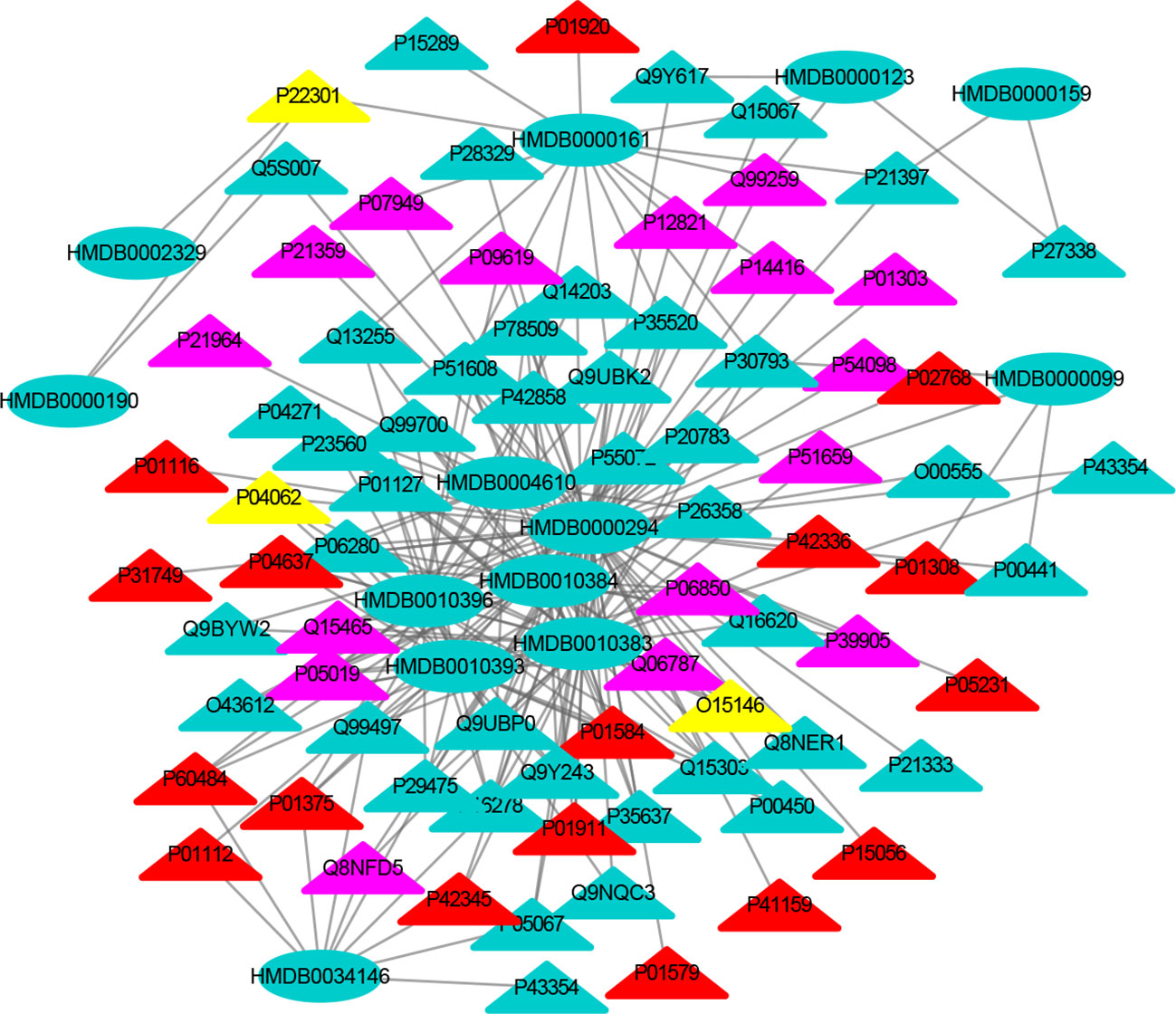
Figure 3 The MTI network of the nervous system. The ellipses refer to metabolic biomarkers, and triangles refer to targets, respectively. The color of the nodes indicates that the three systems connected to depression are different, including nervous system, immune system and endocrine system. The colors of the nodes are blue, yellow, and red, indicating that the target is connected to one system, two systems, and three systems, respectively.
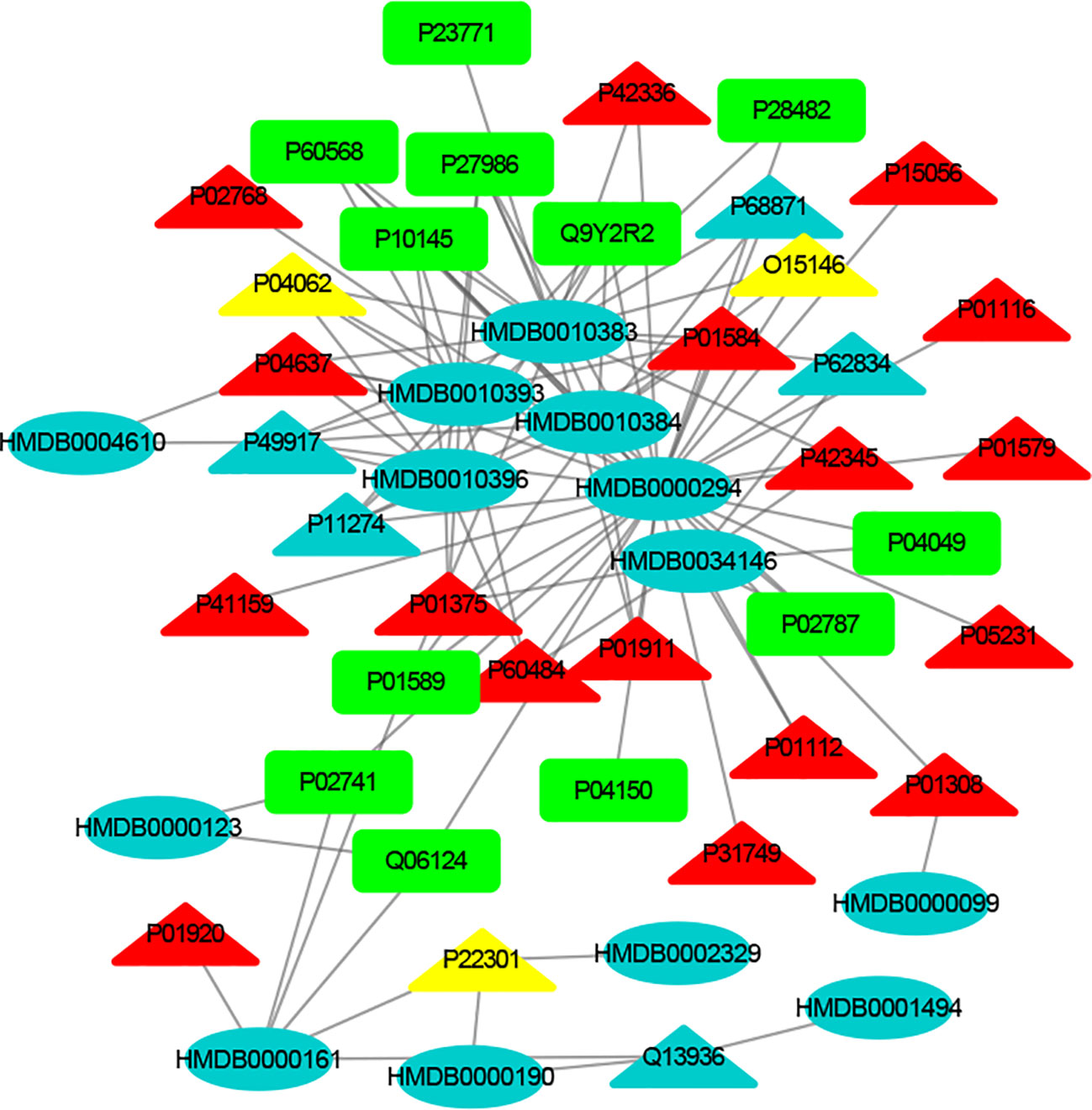
Figure 4 The MTI network of the immune system. The ellipses refer to metabolic biomarkers, and triangles refer to targets, respectively. The color of the nodes indicates that the three systems connected to depression are different, including nervous system, immune system and endocrine system. The colors of the nodes are blue, yellow, and red, indicating that the target is connected to one system, two systems, and three systems, respectively.
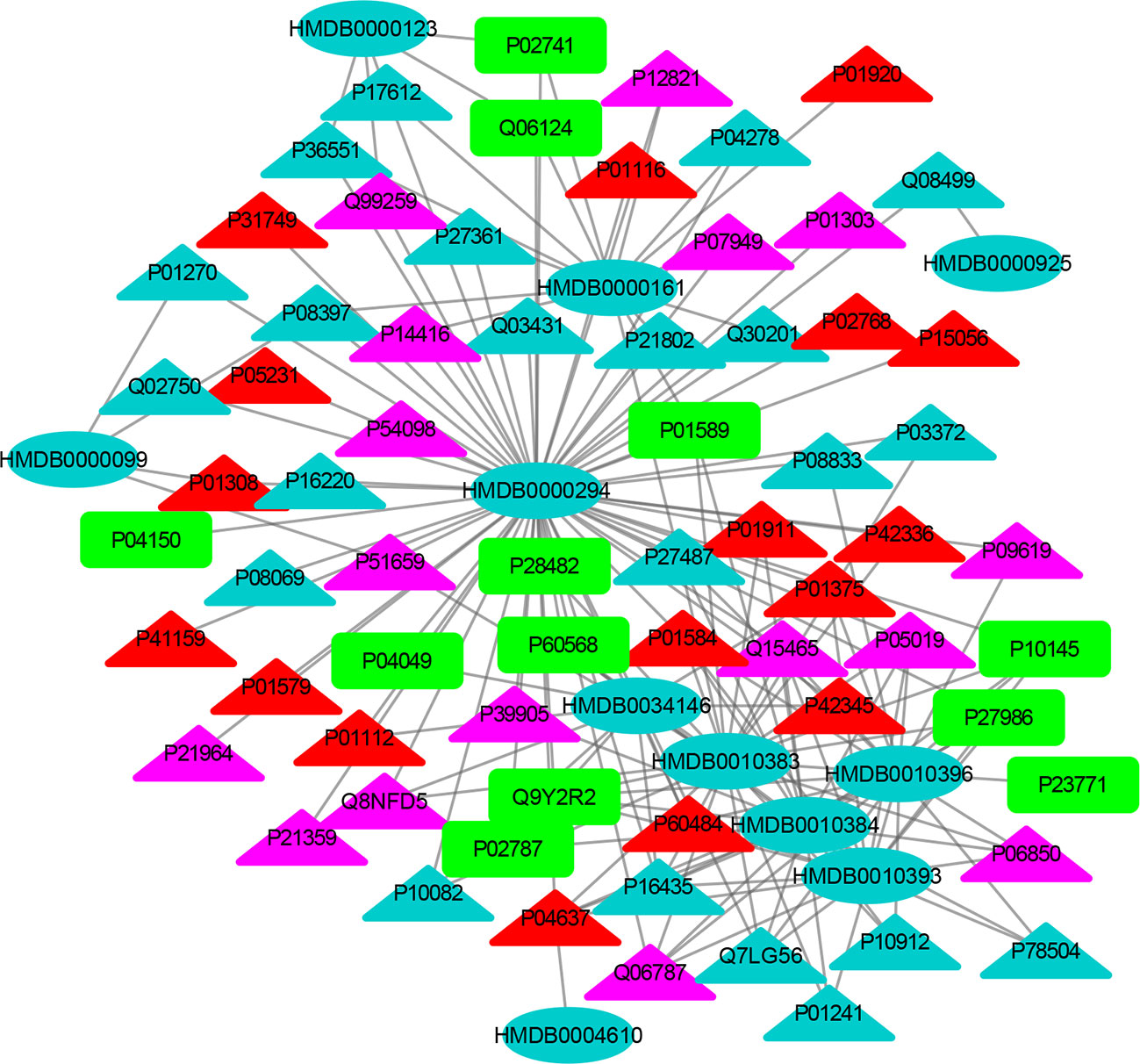
Figure 5 The MTI network of the endocrine system. The ellipses refer to metabolic biomarkers, and triangles refer to targets, respectively. The color of the nodes indicates that the three systems connected to depression are different, including nervous system, immune system and endocrine system. The colors of the nodes are blue, yellow, and red, indicating that the target is connected to one system, two systems, and three systems, respectively.
The MTI network of the nervous system (Figure 3) contains 13 metabolic biomarkers and 78 targets, resulting in 232 edges associations between them. The average number of targets for each metabolic biomarker is 17.85 and the average number of metabolic biomarkers for each target is 2.97. Similarly, the MTI network of the immune system (Figure 4) contains 13 metabolic biomarkers and 37 targets, resulting in 107 edges associations between them. The average number of targets for each metabolic biomarker is 8.23 and the average number of metabolic biomarkers for each target is 2.89. Moreover, the MTI network of the endocrine system (Figure 5) contains 11 metabolic biomarkers and 67 targets, resulting in 78 edges associations between them. The average number of targets for each metabolic biomarker is 15.91 and the average number of metabolic biomarkers for each target is 2.61. This indicates that the molecular mechanism of depression in the nervous system, immune system and endocrine system has the characteristics of multiple metabolic biomarkers and multiple targets.
In these three MTI networks, the three metabolic biomarkers of urea, LPC 16:1 and LPC 18:0 have a higher degree centrality in the network. The number of targets that the three metabolic biomarkers could bind to in the depression-related nervous system, immune system and endocrine system are 68, 33, 61; 40, 18, 25; 29, 13, 20, respectively. The role of these metabolic biomarkers in the pathogenesis of depression is worthy of our in-depth study, because these metabolic biomarkers play a crucial role in the three networks. Among these metabolic biomarkers, urea, LPC 16:1 and LPC 18:0 can be classified as organic carbonic acids or glycerophospholipids, and further research is needed on their role in the pathogenesis of depression.
There are nine identical targets in these three systems, which simultaneously play an important role in the three MTI networks involved in the pathogenesis of depression. Among these targets related to metabolic biomarkers, P04637 (Tumor Protein P53), P01584 (Interleukin 1 Beta), P01375 (Tumor Necrosis Factor), P60484 (Phosphatase And Tensin Homolog), P01911 (Major Histocompatibility Complex, Class II, DR Beta 1), P42345 (Mechanistic Target of Rapamycin Kinase), P01112 (HRas Proto-Oncogene, GTPase), P01308 (Insulin), P42336 (Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha), were widely involved in the nervous system, immune system and endocrine system.
The minor allele 72C of the tumor protein P53 (TP53) gene plays a protective role in the occurrence of depression. It participates in the pathological mechanism of depression through the cell survival and death regulation (42). Previous studies have reported that proinflammatory cytokines such as IL-1 beta (IL1B) (43) and tumor necrosis factor (TNF) (44) have been implicated in the pathogenesis molecular mechanism of depression. The evidence provided by Liu suggests that the rs701848, rs2735343 and rs112025902 polymorphisms in the phosphatase and tensin homologous gene (PTEN) genes may be related to the risk of depression in Chinese (45). Major Histocompatibility Complex (HLA-DRB1) plays a significant role in the immune system by presenting peptides derived from extracellular proteins (46). HRas Proto-Oncogene (HRAS) (46) can encode some genes in signal transduction pathways. These proteins can be linked to GTP and GDP, and they have their own GTPase activity. In addition, rapamycin kinase (mTOR) is a serine/threonine kinase that regulates cell proliferation (47). Li research found that the activation of mTOR in the prefrontal cortex of rats was one of the important mechanisms by which ketamine exerts antidepressant effects (48). Some related studies have also found that acute ketamine administration will activate mTOR in the peripheral blood of patients with depression (49). The antioxidant alpha lipoic acid has been shown to increase insulin (INS) sensitivity and has been used to treat diabetic patients. INS also plays an important role in the pathogenesis of depression. Therefore, the nutrient alpha lipoic acid should be clinically tested as an adjunct treatment for depression. Therefore, some scholars suggested that the nutrient alpha lipoic acid should be used as an adjunct treatment for depression (50). Phosphatidylinositol 4, 5-bisphosphate 3-kinase catalytic subunit alpha isoform (PIK3CA) inhibitors used in bipolar disease and depression (51). Consequently, these results demonstrated that the crucial roles of nine identical targets in the treatment of depression and further confirmed that drug works in a multi-targets manner to treat depression.
In order to improve the therapeutic effect while reducing side effects, drug treatment of diseases usually go through a variety of target methods (52). These targets involve multiple different pathways of disease pathology. The target-pathway approach is a disease-specific research module, which has been widely used in disease pathology exploration and drug development. To find the important pathological mechanism of depression, the strategy of DSWMP index was applied and calculated to select crucial pathway, which provides a methodological reference for the research and development of disease. The DSWMP index is an indicator to evaluate the importance of the pathway. The size of the DSWMP index is determined by the number of metabolic biomarkers binding targets and the binding energy between them. In other words, the DSWMP index method can be used to evaluate the importance of metabolic biomarkers in pathways.
There are 105, 113 and 125 metabolic biomarkers-related targets involved in the nervous system pathways immune system and endocrine system of depression, respectively. In each function, the top 10 pathways of DSWMP index are shown in Table 3 and Figure 6. Among these pathways, the pathways of hsa04151 (PI3K-Akt signaling pathway) and hsa04150 (mTOR signaling pathway) are top-ranked in the pathological mechanism of depression among the nervous system, immune system and endocrine system, indicating that these two pathways play an important role in the pathogenesis of depression. The hsa04151 has been reported involved in the inhibition of apoptosis, cell proliferation and expression of inflammatory cytokines (53, 54). The hsa04150 pathway connects intracellular and extracellular signaling communication, and plays an important role in the protein synthesis process of new synaptic connections (55). Recent research supports the hypothesis that major depression may be the result of disruption of mTOR-dependent translational regulation (13, 14). This result indicates that this pathway plays a crucial role in the molecular mechanism of depression. In addition, there are another five pathways, including hsa04010 (MAPK signaling pathway), hsa04012 (ErbB signaling pathway), hsa04722 (Neurotrophin signaling pathway), hsa04015 (Rap1 signaling pathway) and hsa04014 (Ras signaling pathway), were involved in two of the nervous system, immune system and endocrine system related to depression. Finally, the experimental evidence for the seven important signaling pathways with the most significant therapeutic relationships of depression is shown in Table 4. These pathways could have closely related to the pathological process of depression and need to research in depth.
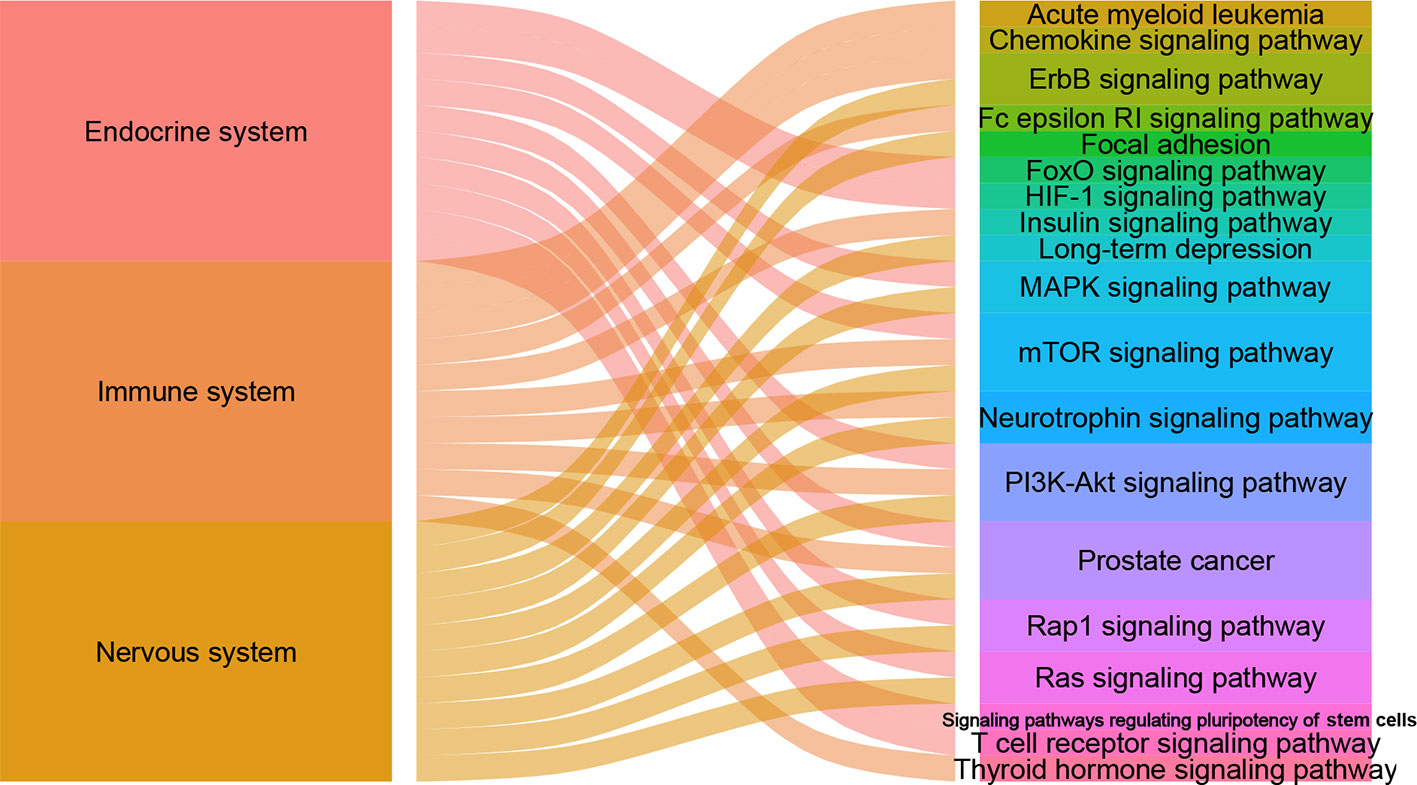
Figure 6 The alluvial plot of systems and pathways. The left column represents nervous system, immune system and endocrine system, the right represents pathways, and the edge represents the relationship between them. A larger edge width indicates the number of pathways-linked systems.

Table 4 Experimental evidence for the seven important signaling pathways with most significant therapeutic relationships of depression.
It is undeniable that there are several limitations in this study. First, it is not sufficient to study based on the data currently available, because the technique of identifying metabolites for depression is still a continuous improvement process. Second, although we have used published literature to verify some results, validation of molecular mechanisms based on clinical samples are necessary to analysis the complex pathogenesis of depression. Our results provide good ideas and methods for studying the pathogenesis of depression.
We identified 36 metabolic biomarkers of clinical plasma metabolomics using NMR and MS. The relationship between biomarkers and enzymes were collected from the HMDB database. The results show that stearic acid, phytosphingosine, glycine, glutamine and phospholipids were important metabolic biomarkers. Hydrolase, transferase and acyltransferase involve the largest number of metabolic biomarkers. The important metabolites and enzymes screened by the topology of the network may play a key role in the underlying molecular mechanism of depression.
The nervous system, immune system and endocrine system are mainly involved in the underlying pathological mechanism of depression. The DSWMP index was used to assess the importance pathways of hub metabolic biomarkers involved. Nine proteins (TP53, IL1B, TNF, PTEN, HLA-DRB1, MTOR, HRAS, INS and PIK3CA) are widely involved in the nervous system, immune system and endocrine system. These targets may be important targets for antidepressants in the treatment of depression. Seven important pathways, such as PI3K-Akt signaling pathway and mTOR signaling pathway, are closely related to the pathogenesis molecular mechanisms of depression and require further investigation. A combination of network pharmacology strategy and metabolomics approach has great potentials in comprehensively and deeply understanding the molecular mechanism of depression. The application of important biomarkers and pathways in clinical practice may help to improve the diagnosis of depression and the evaluation of antidepressant effect, which provides important clues for the study of metabolic characteristics of depression.
All datasets generated for this study are included in the article/Supplementary Material.
X-MQ and J-ST provided the concept and designed the study. YG, TX, Y-XZ, TL-H and S-BL conducted the analyses and wrote the manuscript. YG, TX, Y-XZ, TL-H and S-BL participated in data analysis. X-MQ and J-ST contributed to revise the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National S&T Major Projects for “Major New Drugs Innovation and Development” (2017ZX09301047), the Science and Technology of Shanxi Province (No. 201701D121137 and No. 201903D321210).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2020.00667/full#supplementary-material
1H-NMR, Proton nuclear magnetic resonance; APT-2, Acyl-protein thioesterase 2; ATP, Adenosine triphosphate; CMS, Chronic mild stress; CNS, Central nervous system; CoA, Acetyl-coenzyme A; CPLA2, Cytosolic phospholipase A2; EPA, Eicosapentaenoic acid; DSWP, Docking score-weighted polypharmacological index; HMDB, Human Metabolome Database; HPA, Hypothalamic pituitary adrenal; HRAS, HRas Proto-Oncogene; LC–MS, Liquid chromatography–mass spectrometry; GC–MS, Gas chromatography–mass spectrometer; MEI, Metabolic Biomarker–enzyme interactions; PTEN, Phosphatase and tensin homolog; MTI, Metabolic Biomarker–target interactions; MTOR, Rapamycin Kinase; PIK3CA, Phosphatidylinositol 4, 5-bisphosphate 3-kinase catalytic subunit alpha isoform; PINE, Psycho-immune-neuroendocrine; TP53, Tumor Protein P53; TNF, Tumor necrosis factor.
1. Stapelberg NJC, Pratt R, Neumann DL, Shum DHK, Brandis S, Muthukkumarasamy V, et al. From feedback loop transitions to biomarkers in the psycho-immune-neuroendocrine network: Detecting the critical transition from health to major depression. Neurosci Biobehav Rev (2018) 90:1–15. doi: 10.1016/j.neubiorev.2018.03.005
2. Baglioni C, Battagliese G, Feige B, Spiegelhalder K, Nissen C, Voderholzer U, et al. Insomnia as a predictor of depression: a meta-analytic evaluation of longitudinal epidemiological studies. J Affect Disord (2011) 135(1-3):10–9. doi: 10.1016/j.jad.2011.01.011
3. Peng GJ, Tian JS, Gao XX, Zhou YZ, Qin XM. Research on the Pathological Mechanism and Drug Treatment Mechanism of Depression. Curr Neuropharmacol (2015) 13(4):514–23. doi: 10.2174/1570159x1304150831120428
4. Lopez AD, Mathers CD. Measuring the global burden of disease and epidemiological transitions: 2002-2030. Ann Trop Med Parasitol (2006) 100(5-6):481–99. doi: 10.1179/136485906x97417
5. Nemeroff CB. Recent advances in the neurobiology of depression. Psychopharmacol Bull (2002) 36 Suppl 2:6–23. doi: 10.1016/S0074-7742(06)73005-7
6. Heim C, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. The link between childhood trauma and depression: insights from HPA axis studies in humans. Psychoneuroendocrinology (2008) 33(6):693–710. doi: 10.1016/j.psyneuen.2008.03.008
7. Gao XX, Cui J, Zheng XY, Li ZY, Choi YH, Zhou YZ, et al. An investigation of the antidepressant action of xiaoyaosan in rats using ultra performance liquid chromatography-mass spectrometry combined with metabonomics. Phytother Res : PTR (2013) 27(7):1074–85. doi: 10.1002/ptr.4805
8. Köhler O, Benros ME, Krogh J. Anti-inflammatory Intervention in Depression–Reply. JAMA Psychiatry (2015) 72(5):512–3. doi: 10.1001/jamapsychiatry.2014.3186
9. Stapelberg NJC, Neumann DL, Shum D, Headrick JP. Health, pre-disease and critical transition to disease in the psycho-immune-neuroendocrine network: Are there distinct states in the progression from health to major depressive disorder? Physiol Behav (2019) 198:108–19. doi: 10.1016/j.physbeh.2018.10.014
10. Hopkins AL. Network pharmacology. Nat Biotechnol (2007) 25(10):1110–1. doi: 10.1038/nbt1007-1110
11. Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol (2008) 4(11):682–90. doi: 10.1038/nchembio.118
12. Huang C, Zheng C, Li Y, Wang Y, Lu A, Yang L. Systems pharmacology in drug discovery and therapeutic insight for herbal medicines. Briefings Bioinf (2014) 15(5):710–33. doi: 10.1093/bib/bbt035
13. Liu CC, Wu YF, Feng GM, Gao XX, Zhou YZ, Hou WJ, et al. Plasma-metabolite-biomarkers for the therapeutic response in depressed patients by the traditional Chinese medicine formula Xiaoyaosan: A (1)H NMR-based metabolomics approach. J Affect Disord (2015) 185:156–63. doi: 10.1016/j.jad.2015.05.005
14. Liu X, Liu C, Tian J, Gao X, Li K, Du G, et al. Plasma metabolomics of depressed patients and treatment with Xiaoyaosan based on mass spectrometry technique. J Ethnopharmacol (2020) 246:112219. doi: 10.1016/j.jep.2019.112219
15. Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, et al. HMDB 3.0–The Human Metabolome Database in 2013. Nucleic Acids Res (2013) 41(Database issue):D801–7. doi: 10.1093/nar/gks1065
16. Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem Substance and Compound databases. Nucleic Acids Res (2016) 44(D1):D1202–13. doi: 10.1093/nar/gkv951
17. Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res (2004) 32(Database issue):D115–9. doi: 10.1093/nar/gkh131
18. Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res (2018) 46(D1):D1074–d82. doi: 10.1093/nar/gkx1037
19. Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database : J Biol Database Curation (2010) 2010:baq020. doi: 10.1093/database/baq020
20. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res (2015) 43(Database issue):D789–98. doi: 10.1093/nar/gku1205
21. Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F, et al. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res (2012) 40(Database issue):D1128–36. doi: 10.1093/nar/gkr797
22. Berman HM, Kleywegt GJ, Nakamura H, Markley JL. The Protein Data Bank archive as an open data resource. J computer-aided Mol Design (2014) 28(10):1009–14. doi: 10.1007/s10822-014-9770-y
23. Hsin KY, Matsuoka Y, Asai Y, Kamiyoshi K, Watanabe T, Kawaoka Y, et al. systemsDock: a web server for network pharmacology-based prediction and analysis. Nucleic Acids Res (2016) 44(W1):W507–13. doi: 10.1093/nar/gkw335
24. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res (2003) 13(11):2498–504. doi: 10.1101/gr.1239303
25. Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinf (Oxford England) (2008) 24(2):282–4. doi: 10.1093/bioinformatics/btm554
26. Janga SC, Tzakos A. Structure and organization of drug-target networks: insights from genomic approaches for drug discovery. Mol Biosyst (2009) 5(12):1536–48. doi: 10.1039/B908147j
27. Gu J, Luo F, Chen L, Yuan G, Xu X. A systematic study of chemogenomics of carbohydrates. Mol Biosyst (2014) 10(3):391–7. doi: 10.1039/c3mb70534j
28. Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res (2012) 40(Database issue):D109–14. doi: 10.1093/nar/gkr988
29. Bosetti F, Rintala J, Seemann R, Rosenberger TA, Contreras MA, Rapoport SI, et al. Chronic lithium downregulates cyclooxygenase-2 activity and prostaglandin E(2) concentration in rat brain. Mol Psychiatry (2002) 7(8):845–50. doi: 10.1038/sj.mp.4001111
30. Rao JS, Ertley RN, DeMar JC Jr., Rapoport SI, Bazinet RP, Lee HJ. Dietary n-3 PUFA deprivation alters expression of enzymes of the arachidonic and docosahexaenoic acid cascades in rat frontal cortex. Mol Psychiatry (2007) 12(2):151–7. doi: 10.1038/sj.mp.4001887
31. Gałecki P, Gałecka E, Maes M, Chamielec M, Orzechowska A, Bobińska K, et al. The expression of genes encoding for COX-2, MPO, iNOS, and sPLA2-IIA in patients with recurrent depressive disorder. J Affect Disord (2012) 138(3):360–6. doi: 10.1016/j.jad.2012.01.016
32. Su KP, Huang SY, Peng CY, Lai HC, Huang CL, Chen YC, et al. Phospholipase A2 and cyclooxygenase 2 genes influence the risk of interferon-alpha-induced depression by regulating polyunsaturated fatty acids levels. Biol Psychiatry (2010) 67(6):550–7. doi: 10.1016/j.biopsych.2009.11.005
33. Magrioti V, Kokotos G. Phospholipase A2 inhibitors as potential therapeutic agents for the treatment of inflammatory diseases. Expert Opin Ther patents (2010) 20(1):1–18. doi: 10.1517/13543770903463905
34. Su KP, Yang HT, Chang JP, Shih YH, Guu TW, Kumaran SS, et al. Eicosapentaenoic and docosahexaenoic acids have different effects on peripheral phospholipase A2 gene expressions in acute depressed patients. Prog Neuropsychopharmacol Biol Psychiatry (2018) 80(Pt C):227–33. doi: 10.1016/j.pnpbp.2017.06.020
35. Pae CU, Yu HS, Kim JJ, Lee CU, Lee SJ, Lee KU, et al. BanI polymorphism of the cytosolic phospholipase A2 gene and mood disorders in the Korean population. Neuropsychobiology (2004) 49(4):185–8. doi: 10.1159/000077364
36. Ge L, Zhu MM, Yang JY, Wang F, Zhang R, Zhang JH, et al. Differential proteomic analysis of the anti-depressive effects of oleamide in a rat chronic mild stress model of depression. Pharmacol Biochem Behav (2015) 131:77–86. doi: 10.1016/j.pbb.2015.01.017
37. Lin L, Huang Z, Gao Y, Chen Y, Hang W, Xing J, et al. LC-MS-based serum metabolic profiling for genitourinary cancer classification and cancer type-specific biomarker discovery. Proteomics (2012) 12(14):2238–46. doi: 10.1002/pmic.201200016
38. Gao X, Zheng X, Li Z, Zhou Y, Sun H, Zhang L, et al. Metabonomic study on chronic unpredictable mild stress and intervention effects of Xiaoyaosan in rats using gas chromatography coupled with mass spectrometry. J Ethnopharmacol (2011) 137(1):690–9. doi: 10.1016/j.jep.2011.06.024
39. Tian JS, Peng GJ, Gao XX, Zhou YZ, Xing J, Qin XM, et al. Dynamic analysis of the endogenous metabolites in depressed patients treated with TCM formula Xiaoyaosan using urinary (1)H NMR-based metabolomics. J Ethnopharmacol (2014) 158(Pt A):1–10. doi: 10.1016/j.jep.2014.10.005
40. Nunes SO, Vargas HO, Prado E, Barbosa DS, de Melo LP, Moylan S, et al. The shared role of oxidative stress and inflammation in major depressive disorder and nicotine dependence. Neurosci Biobehav Rev (2013) 37(8):1336–45. doi: 10.1016/j.neubiorev.2013.04.014
41. Schousboe A, Scafidi S, Bak LK, Waagepetersen HS, McKenna MC. Glutamate metabolism in the brain focusing on astrocytes. Adv Neurobiol (2014) 11:13–30. doi: 10.1007/978-3-319-08894-5_2
42. Mahmood S, Evinová A, Škereňová M, Ondrejka I, Lehotský J. Association of EGF, IGFBP-3 and TP53 Gene Polymorphisms with Major Depressive Disorder in Slovak Population. Cent Eur J Public Health (2016) 24(3):223–30. doi: 10.21101/cejph.a4301
43. Ovaskainen Y, Koponen H, Jokelainen J, Keinänen-Kiukaanniemi S, Kumpusalo E, Vanhala M. Depressive symptomatology is associated with decreased interleukin-1 beta and increased interleukin-1 receptor antagonist levels in males. Psychiatry Res (2009) 167(1-2):73–9. doi: 10.1016/j.psychres.2007.12.004
44. Mak A, Tang CS, Ho RC. Serum tumour necrosis factor-alpha is associated with poor health-related quality of life and depressive symptoms in patients with systemic lupus erythematosus. Lupus (2013) 22(3):254–61. doi: 10.1177/0961203312471872
45. Liu LJ, Zhu C, Tian HJ, Zheng TS, Ye MJ, Li H. Correlations of PTEN genetic polymorphisms with the risk of depression and depressive symptoms in a Chinese population. Gene (2016) 595(1):77–82. doi: 10.1016/j.gene.2016.09.034
46. Ortega-Hernandez OD, Cuccia M, Bozzini S, Bassi N, Moscavitch S, Diaz-Gallo LM, et al. Autoantibodies, polymorphisms in the serotonin pathway, and human leukocyte antigen class II alleles in chronic fatigue syndrome: are they associated with age at onset and specific symptoms? Ann New Y Acad Sci (2009) 1173:589–99. doi: 10.1111/j.1749-6632.2009.04802.x
47. Abelaira HM, Réus GZ, Neotti MV, Quevedo J. The role of mTOR in depression and antidepressant responses. Life Sci (2014) 101(1-2):10–4. doi: 10.1016/j.lfs.2014.02.014
48. Li N, Lee B, Liu RJ, Banasr M, Dwyer JM, Iwata M, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Sci (New Y NY) (2010) 329(5994):959–64. doi: 10.1126/science.1190287
49. Yu JJ, Zhang Y, Wang Y, Wen ZY, Liu XH, Qin J, et al. Inhibition of calcineurin in the prefrontal cortex induced depressive-like behavior through mTOR signaling pathway. Psychopharmacology (2013) 225(2):361–72. doi: 10.1007/s00213-012-2823-9
50. Salazar MR. Alpha lipoic acid: a novel treatment for depression. Med Hypotheses (2000) 55(6):510–2. doi: 10.1054/mehy.2000.1103
51. Pilot-Storck F, Chopin E, Rual JF, Baudot A, Dobrokhotov P, Robinson-Rechavi M, et al. Interactome mapping of the phosphatidylinositol 3-kinase-mammalian target of rapamycin pathway identifies deformed epidermal autoregulatory factor-1 as a new glycogen synthase kinase-3 interactor. Mol Cell Proteomics : MCP (2010) 9(7):1578–93. doi: 10.1074/mcp.M900568-MCP200
52. Zhang W, Bai Y, Wang Y, Xiao W. Polypharmacology in Drug Discovery: A Review from Systems Pharmacology Perspective. Curr Pharm Design (2016) 22(21):3171–81. doi: 10.2174/1381612822666160224142812
53. Shi HS, Zhu WL, Liu JF, Luo YX, Si JJ, Wang SJ, et al. PI3K/Akt signaling pathway in the basolateral amygdala mediates the rapid antidepressant-like effects of trefoil factor 3. Neuropsychopharmacology (2012) 37(12):2671–83. doi: 10.1038/npp.2012.131
54. Cunha MP, Budni J, Ludka FK, Pazini FL, Rosa JM, Oliveira Á, et al. Involvement of PI3K/Akt Signaling Pathway and Its Downstream Intracellular Targets in the Antidepressant-Like Effect of Creatine. Mol Neurobiol (2016) 53(5):2954–68. doi: 10.1007/s12035-015-9192-4
Keywords: metabolic biomarkers, depression, network pharmacology, drug-target network, docking score-weighted multiple pharmacology index (DSWMP)
Citation: Gao Y, Xu T, Zhao Y-X, Ling-Hu T, Liu S-B, Tian J-S and Qin X-M (2020) A Novel Network Pharmacology Strategy to Decode Metabolic Biomarkers and Targets Interactions for Depression. Front. Psychiatry 11:667. doi: 10.3389/fpsyt.2020.00667
Received: 03 December 2019; Accepted: 26 June 2020;
Published: 15 July 2020.
Edited by:
Maria S. Garcia-Gutierrez, Miguel Hernández University of Elche, SpainReviewed by:
Miao Qu, Capital Medical University, ChinaCopyright © 2020 Gao, Xu, Zhao, Ling-Hu, Liu, Tian and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun-Sheng Tian, anN0aWFuQHN4dS5lZHUuY24=; Xue-Mei Qin, cWlueG1Ac3h1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.