- 1Department of Psychiatry and Psychotherapy, Medical Center – University of Freiburg, Faculty of Medicine, University of Freiburg, Freiburg, Germany
- 2Center for Basics in NeuroModulation, Faculty of Medicine, University of Freiburg, Freiburg, Germany
Susceptibility and resilience to mental disorders result from a complex choreography of gene–environment interactions with epigenetics at the intersection of external psychological stressors and internal biological systems. Increasing awareness of the growing disease burden influenced by daily life stress (“daily hassles”), work-related stress, and low socioeconomic status (SES) has resulted in a novel interest into their underlying molecular signatures. This review offers a brief outline of psychiatric epigenetics and a comprehensive overview of recent findings exploring the relationship of various occupational stressors and DNA methylation in epigenome-wide association studies (EWAS) and in candidate gene studies including the serotonin transporter (SLC6A4; 5-HTTLPR), melatonin receptor 1A (MTNR1A), brain-derived neurotrophic factor (BDNF), tyrosine hydroxylase (TH), and the protein family of DNA methyltransferases (DNMTs). Conceptual and methodological challenges of epigenetic investigations with a special focus on gene–environment interactions are highlighted and discussed. The findings are integrated into a pathophysiological framework featuring epigenetic plasticity factors and work-related stress as a possible central detrimental component targetable by workplace interventions. Finally, the potential of dynamic epigenetic biomarkers of treatment response to pharmacotherapy or psychotherapy is expanded upon.
Introduction
Chronic exposure to environmental stressors, including but not limited to work-related stress, has been implicated in the patho-etiology of various mental disorders and represents a substantial cause of morbidity, imposing a considerable societal disease burden (1). An individual’s response to detrimental environmental influences is affected by an interplay of its intrinsic genetic configuration and its susceptibility or resilience toward external stimuli, which has led to a growing understanding of gene–environment interactions in the onset, course, and treatment of mental disorders (2). Epigenetics has emerged as a molecular correlate of this intersection, and a converging body of evidence has been collected outlining the effects of early-life stress and traumatic experiences, but less so of chronic psychosocial stressors on the expression of candidate genes involved in psychopathology (3).
Given the known connections between increased work-related stress and poor long-term mental health outcome, this mini review explicitly focuses on summarizing the current state of the literature on the epigenetics of daily life stress, work-related stress, and socioeconomic status (SES) and highlights future challenges and opportunities relevant to the field of psychiatry and psychology. In order to offer a concise overview, PubMed searches combining the terms “work/daily/occupational stress” and “socioeconomic status” with “epigenetics” and “DNA/genome wide methylation” were performed in October 2019, and the resulting titles and abstracts were screened for relevance. Please refer to Table 1 for an overview of primary findings.
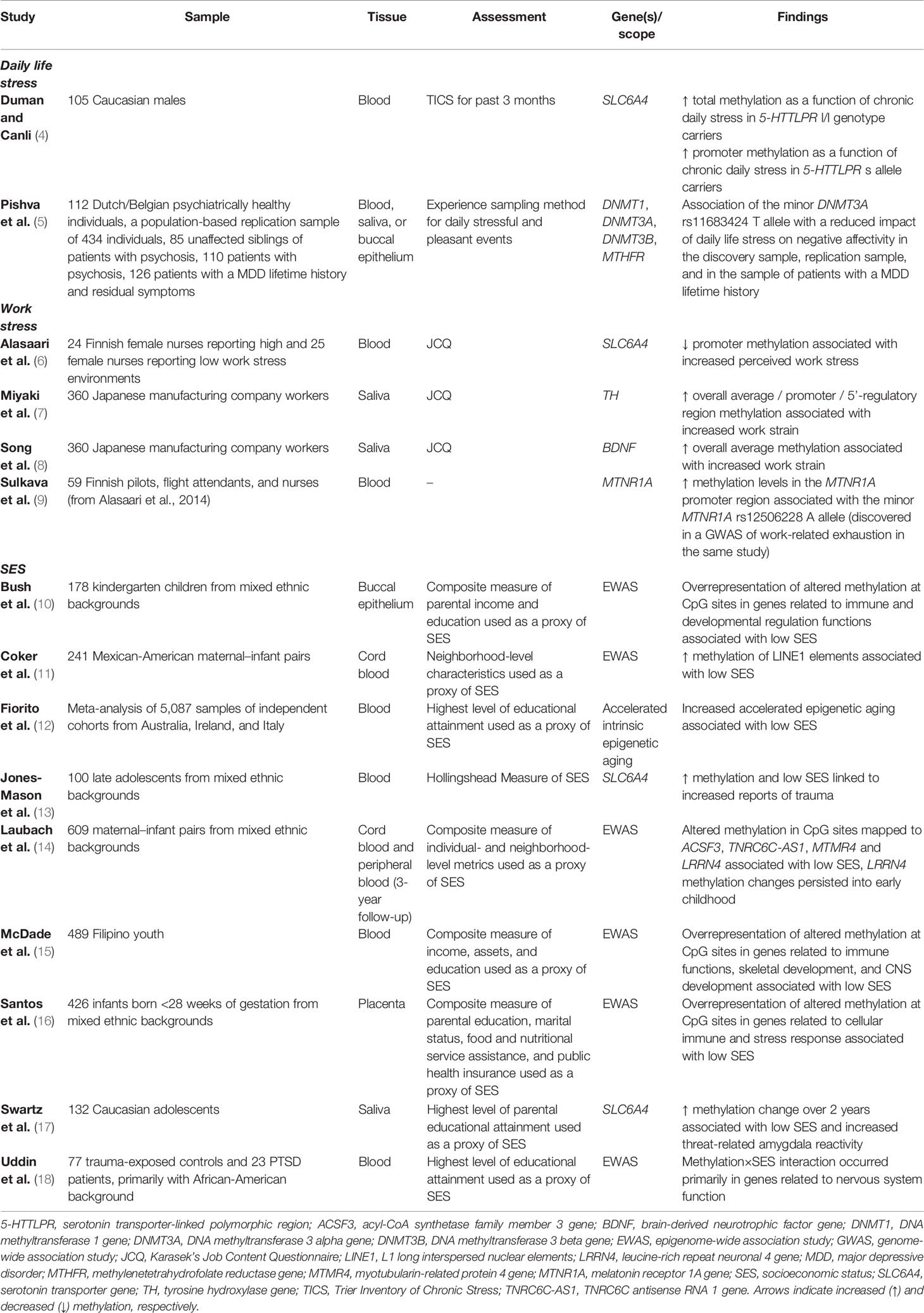
Table 1 Overview of epigenetic studies relating to daily life stress, work stress, and socioeconomic status.
A Brief Background in Epigenetics
Epigenetics constitutes the effects of DNA and histone modifications and of micro RNAs, subsumed as changes to the structure of the DNA strand (but importantly not as changes to the DNA sequence itself), or the interaction of mRNA with noncoding RNAs, affecting transcription and translation. Epigenetic modifications describe dynamic processes with an inherent capacity of intraindividual reversibility. The functionally most well described and major epigenetic modification with relevance to available studies investigating work stress-related phenomena is DNA methylation. Therefore, other epigenetic processes will not be further expanded upon. DNA methylation occurs at cytosine carbon atoms in CpG nucleotide pairs, often accumulating in CpG-rich genomic sites referred to as CpG islands (19). DNA methylations are catalyzed by the enzyme class of DNA-methyltransferases (DNMTs) and become functionally relevant due to other proteins recognizing and binding to methylated DNA sequences and thereby, for example, prohibiting other interaction partners of the transcription start complex to bind or pass, resulting in a de facto silencing of the downstream genomic region. Generally speaking, increased CpG methylation of promoters, enhancer elements, or transcription start sites leads to lower transcription levels of the associated mRNA, whereas decreased methylation of the respective sites would be linked to higher transcript concentrations (20). It should be noted that structural [e.g., single nucleotide polymorphisms (SNPs)] and functional (e.g., CpG methylation) changes of the DNA share an interdependence, insofar as for example, SNPs can interfere with target recognition sites of enzymes and hence affect the local occurrence of epigenetic base pair modifications, while methylation changes can alter the topological susceptibility to regional mutations, both of which may impact higher levels of biological complexity including cognitive processes and behavior (21, 22).
Daily Life Stress and Epigenetics
The serotonin transporter-linked polymorphic region (5-HTTLPR), a promoter region polymorphism of the serotonin transporter gene (SLC6A4)—differentiated into a high transcription (more active) long (l) allele and a less active short (s) allele—as well as SLC6A4 methylation have been commonly investigated in interaction with early life stressors on stress responsivity. Far fewer studies, however, have integrated the role of current stressful events. One such study not only evaluated the interaction between early life stress and 5-HTTLPR genotype on blood-based SLC6A4 methylation, SLC6A4 expression, and cortisol response to the Trier Social Stress Test (TSST) but also analyzed the association of chronic stress experiences, gene expression, and cortisol response as a function of 5-HTTLPR genotypes (4). Current stress experience over the past 3 months as measured by the Trier Inventory of Chronic Stress (TICS) and baseline SLC6A4 expression in 105 psychiatrically healthy Caucasian males did not differ as a function of 5-HTTLPR genotype, whereas l/l genotype carriers displayed significantly higher SLC6A4 expression in response to the TSST, as compared to s allele carriers. Interestingly, l/l genotype carriers further showed increased global promoter methylation levels, both as a function of early-life stress [childhood trauma questionnaire (CTQ)] and in interaction with recent chronic daily stress. Upon closer investigation, s allele carriers, but not l/l genotype carriers, possessed significantly increased methylation of CpG islands flanked upstream by the 5-HTTLPR and downstream by the untranslated first SLC6A4 exon, which showed increased methylation levels as a function of recent chronic daily stress and correlated positively with glucocorticoid receptor (NR3C1) expression [for more evidence of NR3C1 as a key mediator of early adversity in stress-related disorders, please refer to Palma-Gudiel, Cordova-Palomera (23)]. Given that elevated SLC6A4 methylation levels of the very same site have been linked to major depressive disorder (MDD) (24) and increased levels of burnout symptoms (6) (see below), this distinct genomic region might represent a particularly dynamic methylation site, responsive to chronic recent daily stressors.
By way of an indirect epigenetic approach, a multistep gene–environment interaction study tested for main and interaction effects of 31 SNPs in epigenetic regulatory genes and daily life stress or pleasant experiences, respectively, on emotional affectivity in a discovery sample of 112 psychiatrically healthy individuals, a population-based replication sample of 434 individuals and three further samples of unaffected siblings of patients with psychosis (n = 85), patients with psychosis (n = 110), and patients with a MDD lifetime history and residual symptoms (n = 126) (5). All samples showed associations between daily life stress, negative affectivity, pleasant experiences, and positive affectivity, respectively, yet no SNP main effect emerged for either negative or positive affectivity. Following post hoc multiple comparisons correction in the interaction analyses of SNPs and daily life stress or pleasant experiences, respectively, in the discovery sample, only one SNP was validated as significant and directionally similar in the replication sample and furthermore in the sample of patients with a MDD lifetime history: The minor rs11683424 T allele in the gene coding for the DNA methyltransferase 3 alpha (DNMT3A) was associated with a reduced impact of daily life stress on negative affectivity. This result is particularly relevant since DNMT3A is classified as a de novo methyltransferase, dynamically reshaping methylation patterns, rather than statically preserving preexisting methylation patterns during DNA replication, as for example maintained by DNMT1 (25), signifying that genetic variation in regulatory enzymes of DNA methylation might moderate the ad hoc effects of daily life stressors on emotional regulation and thereby psychopathological susceptibility and resilience via downstream epigenetic processes.
Work-Related Stress and Epigenetics
Considering the adverse emotional and physical responses to chronic work-related stress, which are reached when the work demands exceed an individual’s coping ability and capacity (26), it appears surprising how seldom the molecular interplay of perceived occupational stressors and maladaptive outcomes has been examined.
In a female nurses cohort who reported either high (n = 24) or low (n = 25) work stress environments, significantly lower SLC6A4 methylation of the same region described by Duman and Canli (4) above was discerned in the high-stress group (6). Notably, in a multifactorial analysis of covariance including work stress environment, burnout, work control, work demand, age, and 5-HTTLPR genotypes as explanatory variables, only perceived work stress significantly and independently affected overall methylation levels. Higher methylation levels were associated with an increasing intensity of burnout symptom reports when corrected for perceived work stress and vice versa. It could be hypothesized that SLC6A4 promoter hypomethylation reflects an adaptational mechanism independent of the 5-HTTLPR genotype in response to work-related stress, which may initially follow a physiological role to minimize stress exposure and preserve cognitive functioning, but results in dysfunctional long-term consequences manifesting as failed coping mechanisms and increasingly depressed mood.
Another impactful factor of work-related stress is shift work, which has been linked to adverse long-term health effects, including, but not limited to, an increased risk of mental disorders (27), but whose molecular underpinnings have rarely been explored. In a fixed-effects meta-analysis of genome-wide association studies (GWAS) (total n = 753 samples, including subsets of the general population, nurses, and airline employees) of work-related exhaustion, rs12506228 was the only SNP reaching genome-wide significance (p = 4.90 × 10-8) (9). Since rs12506228 is located in a CpG island in the 5’-regulatory region of the gene coding for the melatonin receptor 1A (MTNR1A), a subsequent in silico search of available postmortem brain data suggested reduced MTNR1A central nervous system (CNS) expression levels to be linked to the minor »risk« A allele. Complementing this transcriptomic approach, an analysis of bead chip methylation data of pooled blood samples from the nurse (6) and airline employee sample subsets (total n = 59) further hinted at significantly higher methylation levels in the MTNR1A promoter region to be associated with the rs12506228 MTNR1A A allele. Reduced central melatonin signaling in rs12506228 A allele carriers, especially considering its inhibitory effect via MTNR1A in the suprachiasmatic nucleus, where melatonin acts as a stabilizer of the circadian rhythm against light-induced phase shifts (28), might result in an increased sensitivity to nocturnal lights, for example, because of epigenetically reduced expression of MTNR1A, leading to circadian disruptions and ultimately increased work-related stress due to night shifts.
Additional candidate gene approaches targeting the epigenetics of work-related stress have been conducted in a cohort of 360 Japanese manufacturing company workers, who were subdivided into four quartile-based groups according to their reported job strain. The overall average methylation across CpG sites covering the tyrosine hydroxylase gene (TH) was significantly elevated in workers reporting a higher work strain, as was the methylation of the promoter and the 5’-regulatory region by itself, with methylation and the degree of work-related stress levels displaying a significant positive correlation (7). Furthermore, the overall average methylation of the gene coding for the brain-derived neurotrophic factor (BDNF) was increased in individuals with higher reported work strain (8). These results highlight the need to account for the temporal dynamic of epigenetic modifications associated with work-related stress, for example, BDNF methylation has been demonstrated to be unaltered following acute psychosocial stress (29), while preclinical evidence proposes that chronic stress exposure leads to a glucocorticoid-dependent hypermethylation of TH, but only when combined with additional neuropsychiatric risk mutations (30).
Socioeconomic Status and Epigenetics
Recently, interest has sparked into investigations of epigenetics related to SES, as a combination of low SES and high job demands has been linked to increased morning cortisol and diurnal cortisol secretion (31), supporting the notion that lower SES is associated with various work-related stressors precipitating a higher prevalence of mental disorders and health-impairing behaviors (32).
A meta-analysis of three samples (total n = 5,087) reported a linear increase of accelerated intrinsic epigenetic aging when comparing high to medium and low SES (12). Moreover, individuals who experienced a change in SES over their life span (based on the trajectory of father’s occupational position as a proxy of early-life SES and individual highest occupational position as a proxy for adulthood SES) showed intermediate levels of accelerated epigenetic aging, indicating a potential reversibility of maladaptive epigenetic effects due to low SES. Notably, sex-dependent DNA methylation changes associated with SES have been demonstrated to already exist at the time of birth and to partially persist into early childhood (14, 16), mainly affecting biological processes regulating immunological and developmental functions (10).
Integrating attachment variables into an analysis of SES and SLC6A4 promoter methylation in 100 adolescents, higher methylation and low SES were linked to higher reports of trauma (13). Within the group with unresolved attachment style, yet not in the groups with secure or organized attachment style, low SES was associated with increased methylation, implying that attachment security might act as a protective factor against the effects of low SES on SLC6A4 promoter methylation. This notion appears to be particularly relevant since an explorative epigenome-wide association study (EWAS) including trauma-exposed controls (n = 77) and post-traumatic stress disorder (PTSD) patients (n = 23) has proposed that methylation × SES interaction effects occur primarily in genes related to nervous system function (18), offering a biological pathway by which external stressors might elicit maladaptive central changes. Additionally, biological pathways highlighted by EWAS of early-life SES-related stress have pointed toward altered methylation in genes governing CNS and immune functions (10, 15), some of which appear stable over different developmental stages (14), and with a particular molecular focus on L1 long interspersed nuclear elements (LINE1 elements) hypermethylation (11).
Reasonably, continuous research efforts have been invested into an elucidation of how low SES results in a heightened risk of mental disorders. A longitudinal multilevel approach integrating epigenetic, neuroimaging, and psychometric readouts from 132 adolescents reported that low SES was the only factor among environmental stress variables predicting a greater 2-year increase in SLC6A4 promoter methylation, independent of a MDD family history (17). Besides, increases in proximal SLC6A4 promoter methylation predicted increases in threat-related amygdala reactivity over the same time window, which in turn predicted elevated levels of depressive symptoms another year later, but this time solely in individuals with a MDD family history. A moderated mediation model confirmed that lower SES indirectly affected future increases in depressive symptom intensity in individuals with, but not without, a MDD family history, via SLC6A4 promoter hypermethylation, which in turn predicted heightened amygdala responsivity. These findings add to the notion of elevated peripheral SLC6A4 promoter methylation as an early surrogate stress marker reflecting increased central amygdala reactivity and lower brain expression levels of SLC6A4 (33).
Discussion
Conceptual and Methodological Challenges
As exemplified by the above studies, investigations into stress-related epigenetics not only have to reach adequate sample numbers to allow for the detection of potentially small effect sizes, they also have to specifically monitor the homogeneity of their cohorts, for example, sex (34), ethnicity (35), and smoking status (36) have all been shown to significantly influence the epigenome. Also, it should be kept in mind that, as with most human psychiatric research, the above listed findings constitute correlational evidence in need of further independent replication. Moreover, a notorious challenge is the division into epigenetic responses to recent environmental stressors and methylation changes that date back to conception. The epigenetic modifications resulting from in utero exposure or during critical “windows of vulnerability” in early-life development have been demonstrated to permanently affect methylation patterns for several decades (37, 38). Therefore, longitudinal within-subject designs should be preferred in order to distinguish into hypothetically life-long methylation patterns as a source of susceptibility to work-related stressor and dynamic molecular adaptions to recently experienced occupational adversities.
Another often raised controversy relates to the evaluation of epigenetic patterns in peripheral tissues, mainly leukocytes derived from blood or buccal cells from saliva samples, when the psychiatric main interest lies within the CNS pathophysiology of mental disorders. However, interindividual differences in brain methylome patterns have been proven to be reflected in peripheral samples (39), a positive cross-correlation between blood and saliva methylation has been confirmed (40), and both sampling types have been verified to deliver temporally stable results (41). Nonetheless, direct conclusions from peripheral to central molecular changes should be drawn with caution, and only independent replication and cross-tissue evidence will further support the rise of surrogate epigenetic biomarkers viable to contribute clinical benefits in therapeutic decision making.
Furthermore, accessible measures of daily life stress and work-related stress ultimately represent subjective values and are difficult to accurately quantify. The application of multiple validated retrospective self-report questionnaires assessing comparable stressor constructs, or of repeated recall bias-free real-time evaluations of behaviors and experiences in natural daily environments [ecological momentary assessment; (42)], appears to be the gold standard for future investigations. Also, epigenetic findings should be followed up by in vivo and in vitro evaluation of potential causal mechanisms relating transcriptomic expression changes to higher levels of biological functioning. Eventually, candidate risk gene will be combined into candidate risk haplotypes before hypothesis-driven work will start to complement increasingly accessible hypothesis-generating EWAS approaches (43). As this mini review has demonstrated, currently, there is a reliance on single-locus investigations and comparably small sample sizes (and therefore reduced statistical power), which has resulted in a research focus on mostly established candidate risk genes. Prospective multi-locus study designs will help to disentangle the epigenetic effects of occupational adversities and targeted protective measures in relation to work stress-related susceptibility (44).
From Novel Pathophysiological Frameworks Toward Individualized Treatments
The present state of literature harmonizes with the notion of the “differential susceptibility hypothesis,” stressing that rather to hunt for »risk« genes, one should consider »plasticity« genes, which can render an individual more receptive to environmental stimuli in general, while their interaction can result in added allostatic load or buffer organismal defences (45). As exemplified above, the 5-HTTLPR genotype-dependent SLC6A4 promoter methylation in response to an acute (4) or chronic work-related stressor (6) might boost or diminish an individual’s subsequent reactions to similar or derivative forms of stress. Daily life stress, work-related stress, and SES therefore represent welcome additions to future epigenetic approaches to unravel the ever-elusive point where heredity and nature meet environment and nurture but should always be considered as plasticity variables and be evaluated alongside potential coping factors (46).
The accumulation and integration of multilevel evidence relating to individual epigenetic plasticity markers, for example, from SES-dependent modifications in SLC6A4 promoter methylation to neuronal reactivity and finally to psychometric alterations (17), enhance the accessibility of predictive molecular signatures and finally deepen our insight into interlaced levels of systems biology (47). Biomarkers of disease course development or treatment response hold a particular value due to their inherent epigenetic dynamic. In light of potentially reversible SES-dependent maladaptive DNA methylation patterns, it should be noted that epigenetic markers have already been suggested as predictors and potential mechanistic correlates of pharmacological and psychological therapy responsiveness, with DNA methylation measurements reverting to values comparable to healthy controls over the time course of treatment (48–51). Multiple forms of preventive interventions against stressors like work exhaustion/strain and shift work and associated common mental disorders are currently recognized, including the enhancement of employee control and the promotion of physical activity (52). Future working generations might consequently possibly benefit from indicated preventive interventions or targeted treatment approaches depending on the quantifiable state of epigenetic biomarkers. Particularly, when considering potential long-term effects of transgenerational epigenetic inheritance and in utero exposure influenced by work-related stressors on mental health traits, which creates novel perspectives of social equality and responsibility alike (53).
Conclusion
Notwithstanding the epidemiological and economic impact of workplace stressors, investigations into the relevant epigenetic mechanisms mediating susceptibility or resilience toward mental disorders have been limited. Gathered multilevel evidence specifically favors a role of SLC6A4 promoter methylation patterns as correlates of acute daily stress, chronic work-related stress, and low SES upbringing, although hypothesis-generating studies supporting an involvement of the serotonergic system are still needed. Upcoming epigenetic research should aim to expand beyond an integration of occupational psychometric readouts as additional risk variables and progress to a dimensional understanding of psychopathology accompanied by prognostic molecular plasticity markers regulating individual work stress internalization and allowing for personalized treatment.
Author Contributions
MG conducted the literature search and wrote the first manuscript draft. MS and KD contributed to the literature search and to the revision of the manuscript. All authors contributed to and have approved the final manuscript version.
Funding
This work was partly supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – project number 44541416 – TRR 58 “Fear, Anxiety, Anxiety Disorders” (projects C02 and Z02 to KD), the German Ministry of Research and Education (BMBF, 01EE1402F, PROTECT-AD, project P5 to KD), and the EQUIP Medical Scientist Program of the Medical Faculty, University of Freiburg, Germany (to MAS). The funding sources had no further role in the study design, in the collection, analysis, and interpretation of data, in the writing of the report, and in the decision to submit the paper for publication.
Conflict of Interest
KD is a member of the Janssen Pharmaceuticals, Inc. Steering Committee Neurosciences.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
KD and MS are members of the Anxiety Disorders Research Network (ADRN), European College of Neuropsychopharmacology (ECNP).
References
1. Virtanen M, Honkonen T, Kivimaki M, Ahola K, Vahtera J, Aromaa A, et al. Work stress, mental health and antidepressant medication findings from the Health 2000 Study. J Affect Disord (2007) 98(3):189–97. doi: 10.1016/j.jad.2006.05.034
2. Schiele MA, Domschke K. Epigenetics at the crossroads between genes, environment and resilience in anxiety disorders. Genes Brain Behav (2018) 17(3):e12423. doi: 10.1111/gbb.12423
3. Schuebel K, Gitik M, Domschke K, Goldman D. Making Sense of Epigenetics. Int J Neuropsychopharmacol (2016) 19(11):pyw058. doi: 10.1093/ijnp/pyw058
4. Duman EA, Canli T. Influence of life stress, 5-HTTLPR genotype, and SLC6A4 methylation on gene expression and stress response in healthy Caucasian males. Biol Mood Anxiety Disord (2015) 5:2. doi: 10.1186/s13587-015-0017-x
5. Pishva E, Drukker M, Viechtbauer W, Decoster J, Collip D, van Winkel R, et al. Epigenetic genes and emotional reactivity to daily life events: a multi-step gene-environment interaction study. PloS One (2014) 9(6):e100935. doi: 10.1371/journal.pone.0100935
6. Alasaari JS, Lagus M, Ollila HM, Toivola A, Kivimaki M, Vahtera J, et al. Environmental stress affects DNA methylation of a CpG rich promoter region of serotonin transporter gene in a nurse cohort. PloS One (2012) 7(9):e45813. doi: 10.1371/journal.pone.0045813
7. Miyaki K, Suzuki T, Song Y, Tsutsumi A, Kawakami N, Takahashi M, et al. Epigenetic changes caused by occupational stress in humans revealed through noninvasive assessment of DNA methylation of the tyrosine hydroxylase gene. J Neurol Neurol Disord (2015) 2(2):1–8. doi: 10.15744/2454-4981.2.201
8. Song Y, Miyaki K, Suzuki T, Sasaki Y, Tsutsumi A, Kawakami N, et al. Altered DNA methylation status of human brain derived neurotrophis factor gene could be useful as biomarker of depression. Am J Med Genet B Neuropsychiatr Genet (2014) 165B(4):357–64. doi: 10.1002/ajmg.b.32238
9. Sulkava S, Ollila HM, Alasaari J, Puttonen S, Harma M, Viitasalo K, et al. Common Genetic Variation Near Melatonin Receptor 1A Gene Linked to Job-Related Exhaustion in Shift Workers. Sleep (2017) 40(1):1–10. doi: 10.1093/sleep/zsw011
10. Bush NR, Edgar RD, Park M, MacIsaac JL, McEwen LM, Adler NE, et al. The biological embedding of early-life socioeconomic status and family adversity in children’s genome-wide DNA methylation. Epigenomics (2018) 10(11):1445–61. doi: 10.2217/epi-2018-0042
11. Coker ES, Gunier R, Huen K, Holland N, Eskenazi B. DNA methylation and socioeconomic status in a Mexican-American birth cohort. Clin Epigenet (2018) 10:61. doi: 10.1186/s13148-018-0494-z
12. Fiorito G, Polidoro S, Dugue PA, Kivimaki M, Ponzi E, Matullo G, et al. Social adversity and epigenetic aging: a multi-cohort study on socioeconomic differences in peripheral blood DNA methylation. Sci Rep (2017) 7(1):16266. doi: 10.1038/s41598-017-16391-5
13. Jones-Mason K, Allen IE, Bush N, Hamilton S. Epigenetic marks as the link between environment and development: examination of the associations between attachment, socioeconomic status, and methylation of the SLC6A4 gene. Brain Behav (2016) 6(7):e00480. doi: 10.1002/brb3.480
14. Laubach ZM, Perng W, Cardenas A, Rifas-Shiman SL, Oken E, DeMeo D, et al. Socioeconomic status and DNA methylation from birth through mid-childhood: a prospective study in Project Viva. Epigenomics (2019) 11(12):1413–27. doi: 10.2217/epi-2019-0040
15. McDade TW, Ryan CP, Jones MJ, Hoke MK, Borja J, Miller GE, et al. Genome-wide analysis of DNA methylation in relation to socioeconomic status during development and early adulthood. Am J Phys Anthropol (2019) 169(1):3–11. doi: 10.1002/ajpa.23800
16. Santos HP Jr., Bhattacharya A, Martin EM, Addo K, Psioda M, Smeester L, et al. Epigenome-wide DNA methylation in placentas from preterm infants: association with maternal socioeconomic status. Epigenetics (2019) 14(8):751–65. doi: 10.1080/15592294.2019.1614743
17. Swartz JR, Hariri AR, Williamson DE. An epigenetic mechanism links socioeconomic status to changes in depression-related brain function in high-risk adolescents. Mol Psychiatry (2017) 22(2):209–14. doi: 10.1038/mp.2016.82
18. Uddin M, Galea S, Chang SC, Koenen KC, Goldmann E, Wildman DE, et al. Epigenetic signatures may explain the relationship between socioeconomic position and risk of mental illness: preliminary findings from an urban community-based sample. Biodemography Soc Biol (2013) 59(1):68–84. doi: 10.1080/19485565.2013.774627
19. Fu Y, He C. Nucleic acid modifications with epigenetic significance. Curr Opin Chem Biol (2012) 16(5-6):516–24. doi: 10.1016/j.cbpa.2012.10.002
20. Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet (2008) 9(6):465–76. doi: 10.1038/nrg2341
21. Lester BM, Tronick E, Nestler E, Abel T, Kosofsky B, Kuzawa CW, et al. Behavioral epigenetics. Ann N Y Acad Sci (2011) 1226:14–33. doi: 10.1111/j.1749-6632.2011.06037.x
22. Meaburn EL, Schalkwyk LC, Mill J. Allele-specific methylation in the human genome: implications for genetic studies of complex disease. Epigenetics (2010) 5(7):578–82. doi: 10.4161/epi.5.7.12960
23. Palma-Gudiel H, Cordova-Palomera A, Leza JC, Fananas L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neurosci Biobehav Rev (2015) 55:520–35. doi: 10.1016/j.neubiorev.2015.05.016
24. Zhao J, Goldberg J, Bremner JD, Vaccarino V. Association between promoter methylation of serotonin transporter gene and depressive symptoms: a monozygotic twin study. Psychosom Med (2013) 75(6):523–9. doi: 10.1097/PSY.0b013e3182924cf4
25. Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell (1999) 99(3):247–57. doi: 10.1016/s0092-8674(00)81656-6
26. Folkman S, Lazarus RS, Dunkel-Schetter C, DeLongis A, Gruen RJ. Dynamics of a stressful encounter: cognitive appraisal, coping, and encounter outcomes. J Pers Soc Psychol (1986) 50(5):992–1003. doi: 10.1037//0022-3514.50.5.992
27. Wisetborisut A, Angkurawaranon C, Jiraporncharoen W, Uaphanthasath R, Wiwatanadate P. Shift work and burnout among health care workers. Occup Med (Lond) (2014) 64(4):279–86. doi: 10.1093/occmed/kqu009
28. Arendt J. Melatonin and human rhythms. Chronobiol Int (2006) 23(1-2):21–37. doi: 10.1080/07420520500464361
29. Unternaehrer E, Luers P, Mill J, Dempster E, Meyer AH, Staehli S, et al. Dynamic changes in DNA methylation of stress-associated genes (OXTR, BDNF ) after acute psychosocial stress. Transl Psychiatry (2012) 2:e150. doi: 10.1038/tp.2012.77
30. Niwa M, Jaaro-Peled H, Tankou S, Seshadri S, Hikida T, Matsumoto Y, et al. Adolescent stress-induced epigenetic control of dopaminergic neurons via glucocorticoids. Science (2013) 339(6117):335–9. doi: 10.1126/science.1226931
31. Kunz-Ebrecht SR, Kirschbaum C, Steptoe A. Work stress, socioeconomic status and neuroendocrine activation over the working day. Soc Sci Med (2004) 58(8):1523–30. doi: 10.1016/S0277-9536(03)00347-2
32. Baum A, Garofalo JP, Yali AM. Socioeconomic status and chronic stress. Does stress account for SES effects on health? Ann N Y Acad Sci (1999) 896:131–44. doi: 10.1111/j.1749-6632.1999.tb08111.x
33. Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E, et al. Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nat Neurosci (2014) 17(9):1153–5. doi: 10.1038/nn.3778
34. Bale TL. Sex differences in prenatal epigenetic programming of stress pathways. Stress (2011) 14(4):348–56. doi: 10.3109/10253890.2011.586447
35. Capron LE, Ramchandani PG, Glover V. Maternal prenatal stress and placental gene expression of NR3C1 and HSD11B2: The effects of maternal ethnicity. Psychoneuroendocrinology (2018) 87:166–72. doi: 10.1016/j.psyneuen.2017.10.019
36. Stroud LR, Papandonatos GD, Rodriguez D, McCallum M, Salisbury AL, Phipps MG, et al. Maternal smoking during pregnancy and infant stress response: test of a prenatal programming hypothesis. Psychoneuroendocrinology (2014) 48:29–40. doi: 10.1016/j.psyneuen.2014.05.017
37. Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am J Hum Genet (2016) 98(4):680–96. doi: 10.1016/j.ajhg.2016.02.019
38. Guida F, Sandanger TM, Castagne R, Campanella G, Polidoro S, Palli D, et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet (2015) 24(8):2349–59. doi: 10.1093/hmg/ddu751
39. Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, et al. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biol (2012) 13(6):R43. doi: 10.1186/gb-2012-13-6-r43
40. Thompson TM, Sharfi D, Lee M, Yrigollen CM, Naumova OY, Grigorenko ,EL. Comparison of whole-genome DNA methylation patterns in whole blood, saliva, and lymphoblastoid cell lines. Behav Genet (2013) 43(2):168–76. doi: 10.1007/s10519-012-9579-1
41. Talens RP, Boomsma DI, Tobi EW, Kremer D, Jukema JW, Willemsen G, et al. Variation, patterns, and temporal stability of DNA methylation: considerations for epigenetic epidemiology. FASEB J (2010) 24(9):3135–44. doi: 10.1096/fj.09-150490
42. Shiffman S, Stone AA, Hufford MR. Ecological momentary assessment. Annu Rev Clin Psychol (2008) 4:1–32. doi: 10.1146/annurev.clinpsy.3.022806.091415
43. Goldman D, Domschke K. Making sense of deep sequencing. Int J Neuropsychopharmacol (2014) 17(10):1717–25. doi: 10.1017/S1461145714000789
44. Provenzi L, Brambilla M, Borgatti R, Montirosso R. Methodological Challenges in Developmental Human Behavioral Epigenetics: Insights Into Study Design. Front Behav Neurosci (2018) 12:286. doi: 10.3389/fnbeh.2018.00286
45. Belsky J, Jonassaint C, Pluess M, Stanton M, Brummett B, Williams R. Vulnerability genes or plasticity genes? Mol Psychiatry (2009) 14(8):746–54. doi: 10.1038/mp.2009.44
46. Schiele MA, Ziegler C, Holitschke K, Schartner C, Schmidt B, Weber H, et al. Influence of 5-HTT variation, childhood trauma and self-efficacy on anxiety traits: a gene-environment-coping interaction study. J Neural Transm (Vienna) (2016) 123(8):895–904. doi: 10.1007/s00702-016-1564-z
47. Gottschalk MG, Domschke K. Novel developments in genetic and epigenetic mechanisms of anxiety. Curr Opin Psychiatry (2016) 29(1):32–8. doi: 10.1097/YCO.0000000000000219
48. Ziegler C, Richter J, Mahr M, Gajewska A, Schiele MA, Gehrmann A, et al. MAOA gene hypomethylation in panic disorder-reversibility of an epigenetic risk pattern by psychotherapy. Transl Psychiatry (2016) 6:e773. doi: 10.1038/tp.2016.41
49. Schiele MA, Ziegler C, Kollert L, Katzorke A, Schartner C, Busch Y, et al. Plasticity of Functional MAOA Gene Methylation in Acrophobia. Int J Neuropsychopharmacol (2018) 21(9):822–7. doi: 10.1093/ijnp/pyy050
50. Domschke K, Tidow N, Schwarte K, Deckert J, Lesch KP, Arolt V, et al. Serotonin transporter gene hypomethylation predicts impaired antidepressant treatment response. Int J Neuropsychopharmacol (2014) 17(8):1167–76. doi: 10.1017/S146114571400039X
51. Schiele MA, Gottschalk MG, Domschke K. The applied implications of epigenetics in anxiety, affective and stress-related disorders - A review and synthesis on psychosocial stress, psychotherapy and prevention. Clin Psychol Rev (2020) 77:101830. doi: 10.1016/j.cpr.2020.101830
52. Joyce S, Modini M, Christensen H, Mykletun A, Bryant R, Mitchell PB, et al. Workplace interventions for common mental disorders: a systematic meta-review. Psychol Med (2016) 46(4):683–97. doi: 10.1017/S0033291715002408
Keywords: epigenetics (MeSH), work stress, daily life stress, socioeconomic status (MeSH), susceptibility, resilience (psychological), candidate risk variants, epigenome wide association
Citation: Gottschalk MG, Domschke K and Schiele MA (2020) Epigenetics Underlying Susceptibility and Resilience Relating to Daily Life Stress, Work Stress, and Socioeconomic Status. Front. Psychiatry 11:163. doi: 10.3389/fpsyt.2020.00163
Received: 06 November 2019; Accepted: 20 February 2020;
Published: 20 March 2020.
Edited by:
Christine Allwang, Technical University of Munich, GermanyReviewed by:
Livio Provenzi, Neurological Institute Foundation Casimiro Mondino (IRCCS), ItalyMichael Deuschle, Central Institute for Mental Health, Germany
Copyright © 2020 Gottschalk, Domschke and Schiele. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miriam A. Schiele, bWlyaWFtLnNjaGllbGVAdW5pa2xpbmlrLWZyZWlidXJnLmRl
 Michael G. Gottschalk
Michael G. Gottschalk Katharina Domschke
Katharina Domschke Miriam A. Schiele
Miriam A. Schiele