- 1Department of Pediatrics, Penn State College of Medicine, Hershey, PA, USA
- 2Department of Neuroscience and Physiology, SUNY Upstate Medical University, Syracuse, NY, USA
- 3Department of Psychiatry and Behavioral Sciences, SUNY Upstate Medical University, Syracuse, NY, USA
- 4Department of Biochemistry and Molecular Biology, SUNY Upstate Medical University, Syracuse, NY, USA
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by a wide spectrum of deficits in social interaction, communication, and behavior. There is a significant genetic component to ASD, yet no single gene variant accounts for >1% of incidence. Posttranscriptional mechanisms such as microRNAs (miRNAs) regulate gene expression without altering the genetic code. They are abundant in the developing brain and are dysregulated in children with ASD. Patterns of miRNA expression are altered in the brain, blood, saliva, and olfactory precursor cells of ASD subjects. The ability of miRNAs to regulate broad molecular pathways in response to environmental stimuli makes them an intriguing player in ASD, a disorder characterized by genetic predisposition with ill-defined environmental triggers. In addition, the availability and extracellular stability of miRNAs make them an ideal candidate for biomarker discovery. Here, we discuss 27 miRNAs with overlap across ASD studies, including 3 miRNAs identified in 3 or more studies (miR-23a, miR-146a, and miR-106b). Together, these 27 miRNAs have 1245 high-confidence mRNA targets, a significant number of which are expressed in the brain. Furthermore, these mRNA targets demonstrate over-representation of autism-related genes with enrichment of neurotrophic signaling molecules. Brain-derived neurotrophic factor, a molecule involved in hippocampal neurogenesis and altered in ASD, is targeted by 6 of the 27 miRNAs of interest. This neurotrophic pathway represents one intriguing mechanism by which perturbations in miRNA signaling might influence central nervous system development in children with ASD.
Background
In 1993, the first non-coding antisense RNA sequence was described in Caenorhabditis elegans (1, 2) and termed microRNA (miRNA). Over the next 10 years, the roles of miRNAs in modifying mRNA translation and their potential involvement in human diseases were revealed (3, 4). We now know that miRNAs play an important role in central nervous system (CNS) development and function (5, 6) and that dysregulation of miRNAs is tied to alterations in behavior and cognition seen in a number of neuropsychiatric disorders (7). Here, we focus on miRNAs identified in studies of humans with autism spectrum disorder (ASD).
Brain miRNA expression was first examined in postmortem cerebellum from ASD subjects (8). A succession of ensuing studies reported widespread miRNA dysregulation in the CNS and periphery, including lymphoblasts (9–11), blood (12–14), saliva (15), and olfactory precursor cells (16). These studies have produced vast amounts of information, some unifying and some conflicting. The way in which reports of circulating miRNAs instruct our understanding of miRNAs in the CNS is still being explored (17). To guide that exploration, this review summarizes current knowledge of miRNA in ASD, with the goal of identifying the most consistent findings across studies with potential implications for biomarker discovery.
Clinical Aspects of Autism Spectrum Disorder
Autism spectrum disorder is a heterogeneous disorder typified by deficits in social communication and restricted, repetitive patterns of behavior (18). Children with ASD are often adherent to daily routines, preoccupied with specific topics, and over sensitive to sensory inputs such as sounds or textures (19). In most cases, ASD symptoms are first recognized in early childhood. The average age of ASD diagnosis is 4 years (20). Rates of ASD are increasing. Recent reports from the Centers for Disease Control and Prevention estimate the prevalence of ASD to be 1:68 children (1:45 among males) (21). This increase may be the result of multiple factors including public awareness, changes in diagnostic criteria, and environmental influences. However, genetics also plays an important role in ASD (22).
Genetic Contributions to ASD
A large number of familial studies demonstrate that ASD is a heritable disorder (23, 24), with estimated genetic contributions accounting for >50–60% of ASD risk (25). Indeed, a large number of genetic variants and chromosomal abnormalities are linked to ASD (26). These variants tend to be highly penetrant but rare. However, in some cases, common variants with low penetrance have been reported (27). Neither explains the incidence of ASD in the general population. Genetic epidemiologic studies have also shown that ASD is not a single disease but a constellation of symptoms involving multiple gene networks (28). Posttranscriptional mechanisms, such as miRNA, that broadly influence gene expression without altering the DNA code represent one means of altering entire gene networks (29). Increasingly, investigators have turned toward these mechanisms to explain the dysregulation of neurodevelopmental pathways that occurs in ASD (30, 31).
miRNAs Regulate Posttranscriptional Gene Expression
MicroRNAs are short (18–25 nucleotide), non-coding RNA molecules (32) that influence gene expression and numerous cellular processes, including proliferation, differentiation, and apoptosis (33). Over 2500 mature miRNAs (and 1800 precursors) have been identified in humans (34). Many of these are evolutionarily conserved (35). The genes for miRNAs are found in inter- or intra-genic regions (36). They are transcribed into precursor hairpin structures (pri-miRNAs) by RNA polymerase-II and trimmed into pre-miRNA by the RNase Drosha. After leaving the nucleus, Dicer liberates the pre-miRNA into a single-stranded molecule that can be bound within the miRNA-induced silencing complex (miRISC) (37). The targeting of the mature miRISC to individual mRNAs is based on the “seed sequences” that reside at the 5′ end of the miRNA and are complementary to the 3′ regions of mRNAs. Each seed sequence aligns with hundreds of mRNAs, and there are several miRNAs with shared seed sequences (38). Once bound to a target mRNA, the miRISC complex reduces the efficiency of gene expression by repressing protein translation or promoting degradation of the mRNA transcript (37).
miRNAs in the Brain and Periphery
MicroRNAs regulate approximately two-thirds of human mRNAs (39). Each miRNA has hundreds of potential mRNA targets, giving it the ability to modulate entire gene networks. In addition, miRNAs can circulate within exosomes, microvesicles, or RNA-binding proteins such as high-density lipoproteins, and thereby travel extracellularly to alter gene expression in distant tissues. This renders them exceedingly stable and easily measured in serum, plasma, saliva, urine, and other biofluids (40). However, miRNAs are most ubiquitous in the CNS, which expresses an estimated 70% of all miRNAs (41). Notably, expression of brain miRNAs changes throughout childhood and varies across brain regions (42). Neurons and glia are abundant sources of miRNAs, which can be readily transported across the blood–brain barrier (BBB). Moreover, miRNAs in neurons also help compartmentalize or localize mRNA expression and translation within specific subcellular regions such as axons and dendrites.
The Role of miRNAs in Neurodevelopment
MicroRNAs play important roles in neurogenesis, synaptogenesis, and neuronal migration (43, 44). A major means by which these roles are manifest is the effect that neuronal miRNAs have on spatial localization or compartmentalization of protein translation in different neuronal subregions, such as axons, dendrites, and synapses (45, 46). Recent data support the relevance of these processes for neurodevelopmental disorders. For example, in mice with fragile X-associated tremor/ataxia syndrome (FXTAS), the fragile X mental retardation (FMR1) transcript is targeted by miR-221, miR-101, and miR-129-5p (47). Synaptosomal preparations from FXTAS mice demonstrate dysregulation of transcripts in learning and social interaction along with reduced levels of miR-221. Other examples include studies of disruption of miRNA regulatory genes such as Dicer or DGCR8, which produce alterations in synaptic plasticity within the prefrontal cortex (48). The gene for DGCR8 is located at 22q11.21 and a well-characterized hemi-deletion of this region (referred to as velocardiofacial syndrome) produces a complex phenotype along with ASD traits in approximately one-third of subjects (49). Moreover, mice with DGCR8 haploinsufficiency have reduced dendrite complexity, fewer neuronal spines, and decreased neurogenesis (50). Complementing these data, dysregulation of miRNA expression has been described in a number of developmental, neuropsychiatric, and neurodegenerative disorders including schizophrenia, Alzheimer’s disease, and Parkinson’s disease (36).
The Role of miRNAs in ASD Pathogenesis
Four studies examining postmortem brain tissue of human subjects with ASD have identified 91 miRNAs with differential expression compared with typically developing controls. The exact pathophysiologic role of these miRNAs in ASD remains unclear; however, many are implicated in neurogenesis and synaptogenesis. A study of cerebellar cortex in 13 adults with ASD (8) identified 28 miRNAs with differential expression and 7 of these targeted the SHANK3 transcript. SHANK3 is involved in regulation of synaptic density and has copy number variants (CNVs) and point mutations in nearly 1% of ASD cases (51). A study by Wu et al. (52) examined cerebellar cortex in 28 ASD subjects. The authors found miR-21-3p was upregulated in ASD subjects and demonstrated that its overexpression in human neuroprogenitor cells could repress multiple M16 hub genes, including DLGAP1 (a scaffold protein that interacts with SHANK3). Thus, SHANK3 represents one pathophysiologic target with an established role in ASD that appears to be targeted by multiple miRNAs identified in multiple studies of ASD brain tissue.
Overlapping miRNAs in ASD
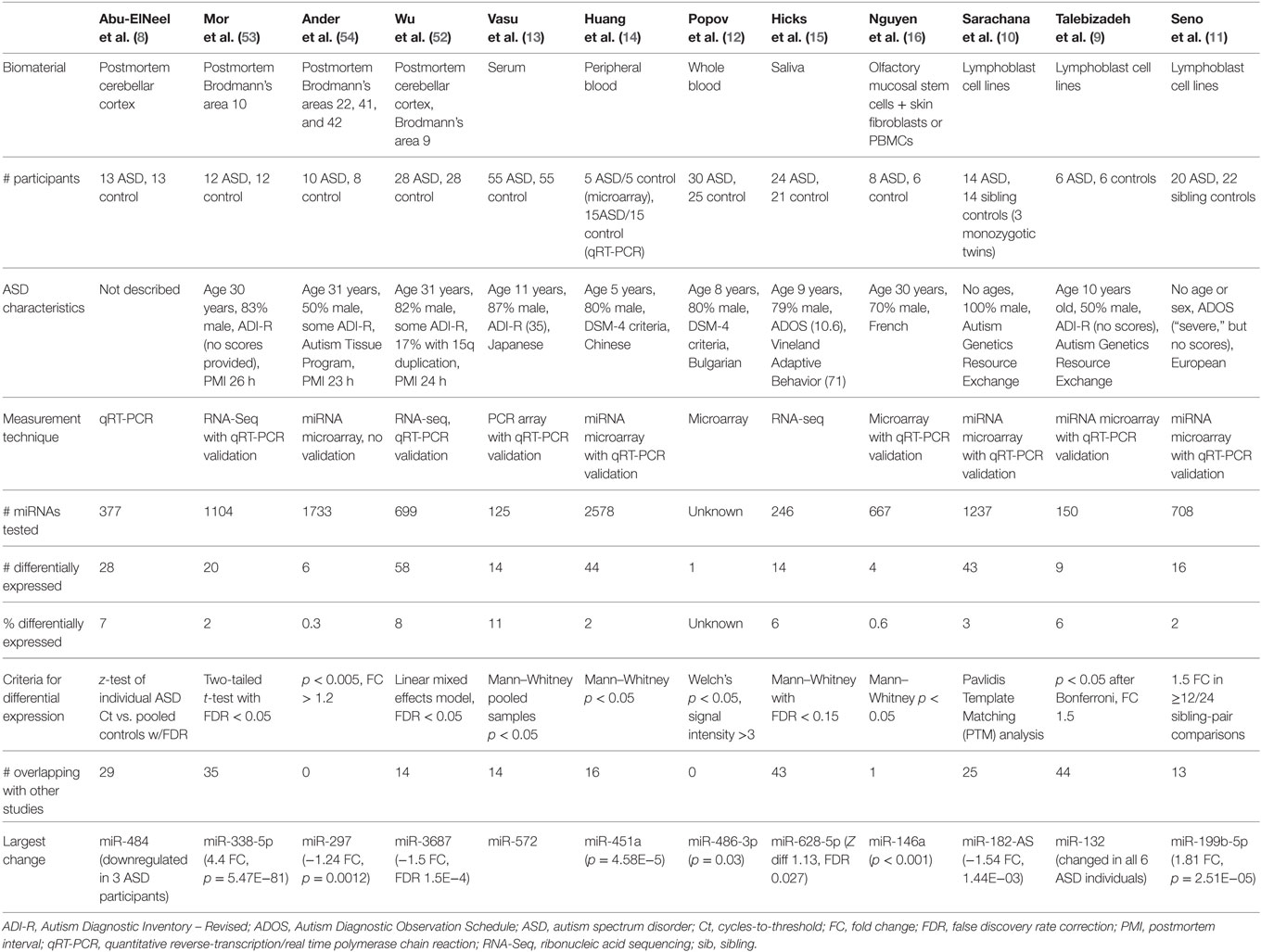
This review synthesizes the findings of 12 studies, which compared miRNA expression in human cases of ASD to miRNA profiles in healthy controls (Table 1). Four of these studies involved brain tissue (8, 52–54), three involved peripheral blood (12–14), one examined saliva (15), one examined olfactory precursor cells (16), and three employed cultured lymphoblasts (9–11).
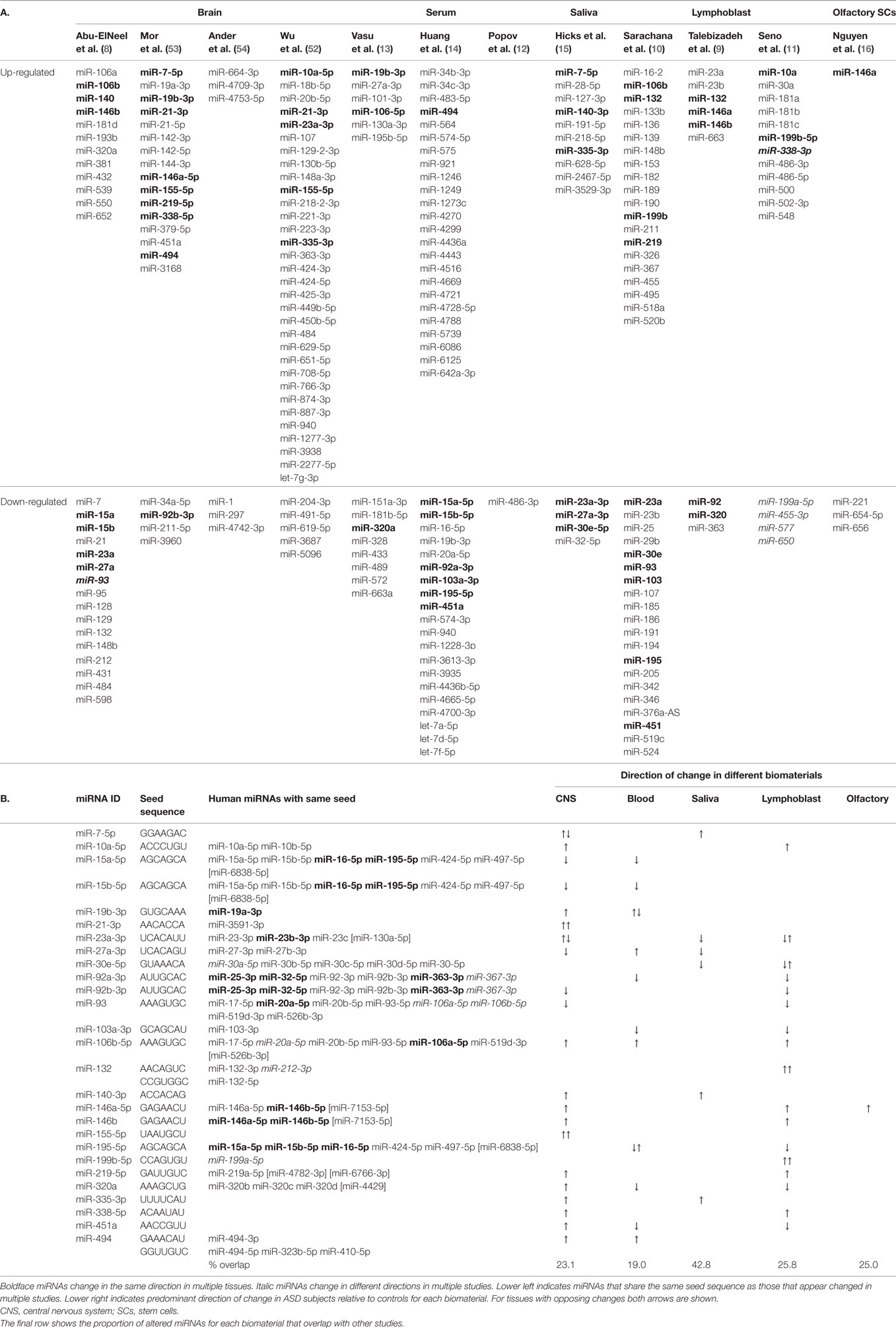
The 12 studies together identified 219 miRNAs with potential implications in ASD (Table 2A), including 185 unique and 34 overlapping miRNAs (15.5%). Twenty-seven overlapping miRNAs changed in the same direction (Table 2B) in ≥2 studies, and 3 changed in the same direction in ≥3 studies (miR-23a, miR-146a, and miR-106b). Notably, 14 of the 27 miRNAs shared seed sequences with other ASD-associated miRNAs, suggesting that the pathophysiology of miRNA dysregulation in ASD might be controlled, at least in part, at the seed level. There is no single miRNA identified in all the 12 studies. This may be a function of ASD heterogeneity, miRNA tissue specificity, or advances in RNA quantification, which have led to varying alignment techniques and identification of new miRNAs.
Functional Significance of Overlapping miRNAs and Their Gene Targets in ASD
Of the 27 overlapping miRNAs, 22 show differential regulation in studies of the CNS. The remaining five are expressed in the developing brain (42, 53). These 27 were predicted to target 1245 mRNAs (Table S1 in Supplementary Material) with target scores ≥93 (55).1 Notably, 86 of these mRNAs overlapped with the 519 human ASD candidate genes with supportive evidence existing in the Simons Foundation Autism Research Initiative (SFARI) database,2 representing a 6.6-fold enrichment for ASD-associated genes compared to chance (odds ratio: 7.7, 95th CI = 6.0–9.9, z = 16.2, p < 0.0001).
We next examined the evidence for functional enrichment of specific biologic processes using the Database for Annotation, Visualization, and Integrated Discovery (DAVID). This revealed 225 total cluster mappings (56), with the top nodes including a KEGG pathway involved in neurotrophin signaling [n = 25 genes, false discovery rate (FDR) = 0.0018]. In fact, this node had the largest fold change (FC) (3.0-fold) among all detected clusters (Table S2 in Supplementary Material). DAVID enrichment analysis also identified 662 (55.9%) of the target mRNAs as expressed in brain tissue, with additional enrichment of genes expressed in epithelium (n = 268), fetal kidney (n = 35), placenta (n = 275), amygdala (n = 61), and T-cells (n = 37) (Table S3 in Supplementary Material).
To further examine functional relevance of the most probable mRNA targets, the Ingenuity Pathway Analysis (IPA) databases for “Canonical Pathways” and “Diseases and Biological Function” were used. These analyses were performed using a restricted set of 293 mRNAs that were predicted to be targets of the 27 overlapping miRNAs with miRDB target scores ≥95. The results indicated enrichment of 20 Canonical Pathways (FDR < 0.05), including two related to neurotrophin signaling (Table S4 in Supplementary Material). Moreover, the “Diseases and Biological Function” analysis yielded a list of 500 networks for the 293 mRNA targets, including 93 (18.6%) involved in brain function, of which 35 survived FDR correction (Table S5 in Supplementary Material). These represented several functions, including brain development, neurogenesis, neural and glial differentiation, synaptic development and function, and cognitive and motor function. The diseases identified among the brain-related networks included schizophrenia, abnormal brain morphology and trigeminal nerve morphology, abnormal posture, cognitive impairment, and seizure disorders.
Upstream Regulatory analysis was performed in IPA considering only experimentally observed and high-confidence miRNA–mRNA interactions, which reduced our list of 27 miRNAs to 20. Notably, 199 mRNAs were targeted by ≥2 miRNAs, and 55 genes were targeted by ≥5 of the 20 miRNAs. The significance of this was determined using a Fisher’s exact test, which confirmed enrichment for 49 mRNAs (Table S6 in Supplementary Material). Protein–protein interaction (PPI) networks were also explored in the 199 target genes using the STRINGv10 database,3 which yielded a significant PPI network containing 137 edges and several nodes involved in brain function (Figure S1 in Supplementary Material). Among this network was an intriguing interaction between neuronal cell adhesion molecule (NRCAM; involved in directional signaling of axonal cone growth and implicated in ASD) (57), semaphorin 3a (SEMA3A; a chemoattractive agent involved in axon guidance) (58), and brain-derived neurotrophic factor (BDNF; essential for synaptogenesis in the developing hippocampus and altered in the periphery of ASD subjects) (59).
Expression of miRNA across Brain Regions
MicroRNA expression is a dynamic process, changing throughout development and across brain regions (43). A study of non-coding RNA in the superior temporal gyrus of ASD subjects demonstrated changes in regional miRNA profiles that could contribute to ASD (60). Specifically, while miRNA patterns in the superior temporal sulcus (a region relevant to social interaction) and primary auditory cortex changed throughout development in the typical brain, many of the changes were blunted in subjects with ASD. Four of the individual miRNAs identified (miR-93-3p, miR-103, miR-132, and miR-320) were among the 219 miRNAs altered in other studies of ASD, with 3 of the 4 changed in multiple studies (Table 2).
Potential Mechanisms for miRNA Dysregulation
The molecular mechanisms underlying miRNA dysregulation in ASD are still being explored. One explanation is that autistic miRNA patterns may represent a permanent response to an early environmental insult, such as perinatal hypoxia (61). In this scenario, miRNA regulation could be disrupted by a layer of epigenetic machinery such as DNA methylation. In a study of 12 subjects with ASD, expression of miR-142 was increased in Brodmann’s area 10 compared with healthy controls – a change accompanied by hypomethylation of the miR-142 promoter region (53).
Another possible mechanism is that individual miRNAs sequences are altered in children with ASD. Pooled analysis of common and rare single nucleotide polymorphisms (SNPs) in 449 cases of ASD using whole exome sequencing has identified 2 miRNA clusters: miR-133b/miR-206 and mir-17/miR/18a/miR-19a/miR20a/miR-19b-1/miR-92a (62). Four of these individual components have disrupted expression levels in children with ASD, and miR-92a is downregulated in multiple studies (9, 14).
Yet another explanation could be that the location of specific miRNAs at CNV loci leads to their dysregulation. Of the 41 CNV loci associated with ASD, >10% contain miRNAs (63). Three of the 10 “hub miRNAs” (i.e., miRNAs at CNV loci with the greatest gene network connectivity) have been previously identified in studies of ASD (miR-34a, miR-548, and miR-195) and target multiple genes involved in neurodevelopment. Alterations in miR-195 have been reported in serum (13, 14) and lymphoblasts (10) of ASD individuals. Furthermore, a key gene involved in miRNA synthesis (Dicer) was identified as a common target for many of the hub miRNAs, suggesting that some CNVs in ASD might cause extensive dysregulation of the entire miRNA machinery.
Systemic miRNA Dysregulation
Although the brain is considered the primary pathological target in ASD, alterations in miRNA expression have been noted in a number of other body sites (13, 15). This may explain (in part) why children with ASD experience medical comorbidities outside the CNS, including gastrointestinal issues such as esophageal reflux and constipation (64). They also have altered oral-motor function (speech apraxia) and sensory processing (65, 66). Given these comorbidities, it should come as no surprise that oral miRNA patterns are altered in children with ASD. In addition, the proximity of the oropharynx to the CNS and its abundant sensory and motor nerve innervation (involving branches of the glossopharyngeal, facial, vagus, and trigeminal nerves) could provide an ample source of miRNA (67, 68).
Alterations in miRNA profiles of peripheral blood sites may be explained by circulating miRNA that has crossed the BBB (especially considering that ASD is not associated with hematologic disorders). In peripheral sources such as blood, brain-related miRNA may be stable and abundant as a result of exosomal transport (13, 14). Thus, while biofluids such as serum and saliva may not represent optimal models for studying CNS pathophysiology, they may represent ideal reservoirs for ASD biomarkers.
Employing miRNAs as Biomarkers
The potential of miRNAs as biomarkers in ASD has been explored in studies of serum and saliva. In a study of serum miRNAs from 55 children with ASD, the authors identified three miRNAs (miR-130-3p, miR-181b-5p, and miR-320a) with an area under the curve (AUC) >0.85 (13). In our own more recent study, involving 24 children with ASD, a set of 14 salivary miRNAs showed more than 95% accuracy (AUC = 0.92) at differentiating control and ASD subjects (15). This study also showed that expression patterns of individual salivary miRNAs were significantly correlated with several measures of adaptive behavior. This highlights how the utility of miRNA extends beyond simple ASD diagnosis and may 1 day be used to predict ASD phenotype and severity.
Conclusion
It is clear that miRNA profiles are dysregulated across multiple tissue types in subjects with ASD. This review encapsulated 219 target miRNAs from 12 human studies of ASD and identified 27 that were dysregulated in ≥2 investigations. Functional pathway analysis supports the idea that these miRNAs target brain-expressed genes related to neurodevelopment and implicated in ASD. Three miRNAs showed consistent dysregulation across ≥3 studies (miR-23a-3p, miR-146a-5p, and miR-106b-5p). The networks of genes targeted by these miRNAs are implicated in ASD and have significant roles in neurotrophin signaling. BDNF, which is important for hippocampal neurogenesis and decreased in the serum of adult ASD patients (59), is targeted by 6 of the 27 miRNAs of interest. Animal and cell models that assess these molecular mechanisms behind miRNAs and their mRNA targets will be critical in advancing the current ASD knowledge-base.
Ethics Approval and Consent
This manuscript involves a review and analysis of current literature collected in preparation for the study “Improving Autism Screening with Brain-Related miRNA,” which has been approved by the Institutional Review Board at the Penn State College of Medicine, Hershey, PA, USA (STUDY00003658). According to the publications on which this review was based, informed consent or assent was obtained from all participants or their parents, unless the study involved exclusively postmortem human brain material. In the case of minor children unable to provide informed assent, parental consent was obtained.
Author Contributions
SH and FM conceived of this study, prepared the figures and tables, and wrote the manuscript.
Conflict of Interest Statement
SH and FM hold a provisional patent for the use of a panel of specific miRNAs in the saliva as biomarkers for autism. The patent was filed by the State University of New York, Upstate Medical University, and has been licensed to Motion Intelligence, Inc. SH also receives consulting fees from Motion Intelligence, Inc. These interests have been disclosed and are under active management by the Penn State Conflict of Interest Committee.
Acknowledgments
We thank R. Uhlig, C. Dowd-Greene, R. Carpenter, C. Ignacio, T. Welch, and K. Gentile for their creative and technical insights and support of this work.
Funding
This work was supported by the Department of Pediatrics at Penn State College of Medicine, the Department of Pediatrics at SUNY Upstate Medical University, and research grants from Motion Intelligence Inc., the National Institute of Mental Health (MH111347), and the Kirson-Kolodner-Fedder Charitable Fund.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fpsyt.2016.00176.
Figure S1. Protein–protein interaction (PPI) network of the genes that are targeted by 2 or more of at least 20 of the 27 conserved miRNAs affected across studies. This analysis involved mapping of 199 mRNA targets in the STRING v10 database (http://string-db.org/). Note the presence of a significant interaction network containing 137 edges for the 197 targets with annotation information available. Notably, only genes with connections are included in the PPI. Also, note that many of these genes were involved in brain-related functions, including some that mapped to a neuronal projection Gene Ontology (highlighted in red).
Abbreviations
ADI-R, autism diagnostic interview – revised; ADOS, Autism Diagnostic Observation Schedule; ASD, autism spectrum disorder; AUC, area under the curve; BBB, blood–brain barrier; CNS, central nervous system; CNV, copy number variant; DAVID, Database for Annotation, Visualization, and Integrated Discovery; HDL, high-density lipoprotein; MeCP2, methyl CpG-binding protein; miRNA, microRNA; PPI, protein–protein interaction; SFARI, Simons Foundation Autism Research Initiative; SNP, single nucleotide polymorphism.
Footnotes
References
1. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell (1993) 75(5):843–54. doi:10.1016/0092-8674(93)90529-Y
2. Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell (1993) 75(5):855–62. doi:10.1016/0092-8674(93)90530-4
3. Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science (2001) 294(5543):858–62. doi:10.1126/science.1065062
4. Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res (2004) 14(10a):1902–10. doi:10.1101/gr.2722704
6. Kos A, Loohuis NO, Meinhardt J, van Bokhoven H, Kaplan BB, Martens GJ, et al. MicroRNA-181 promotes synaptogenesis and attenuates axonal outgrowth in cortical neurons. Cell Mol Life Sci (2016) 73(18):3555–67. doi:10.1007/s00018-016-2179-0
7. Wang W, Kwon EJ, Tsai LH. MicroRNAs in learning, memory, and neurological diseases. Learn Mem (2012) 19(9):359–68. doi:10.1101/lm.026492.112
8. Abu-Elneel K, Liu T, Gazzaniga FS, Nishimura Y, Wall DP, Geschwind DH, et al. Heterogeneous dysregulation of microRNAs across the autism spectrum. Neurogenetics (2008) 9(3):153–61. doi:10.1007/s10048-008-0133-5
9. Talebizadeh Z, Butler MG, Theodoro MF. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism Res (2008) 1(4):240–50. doi:10.1002/aur.33
10. Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome Med (2010) 2(4):23. doi:10.1186/gm144
11. Seno MMG, Hu P, Gwadry FG, Pinto D, Marshall CR, Casallo G, et al. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res (2011) 1380:85–97. doi:10.1016/j.brainres.2010.09.046
12. Popov NT, Madjirova NP, Minkov IN, Vachev TI. MicroRNA HSA-486-3P gene expression profiling in the whole blood of patients with autism. Biotechnol Biotechnol Equip (2012) 26(6):3385–8. doi:10.5504/BBEQ.2012.0093
13. Vasu MM, Anitha A, Thanseem I, Suzuki K, Yamada K, Takahashi T, et al. Serum microRNA profiles in children with autism. Mol Autism (2014) 5:40. doi:10.1186/2040-2392-5-40
14. Huang F, Long Z, Chen Z, Li J, Hu Z, Qiu R, et al. Investigation of gene regulatory networks associated with autism spectrum disorder based on MiRNA expression in China. PLoS One (2015) 10(6):e0129052. doi:10.1371/journal.pone.0129052
15. Hicks SD, Ignacio C, Gentile K, Middleton FA. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr (2016) 16:52. doi:10.1186/s12887-016-0586-x
16. Nguyen LS, Lepleux M, Makhlouf M, Martin C, Fregeac J, Siquier-Pernet K, et al. Profiling olfactory stem cells from living patients identifies miRNAs relevant for autism pathophysiology. Mol Autism (2016) 7(1):1. doi:10.1186/s13229-015-0064-6
17. Jin XF, Wu N, Wang L, Li J. Circulating microRNAs: a novel class of potential biomarkers for diagnosing and prognosing central nervous system diseases. Cell Mol Neurobiol (2013) 33(5):601–13. doi:10.1007/s10571-013-9940-9
18. Volkmar FR, Reichow B, Westphal A, Mandell DS. Autism and the autism spectrum: diagnostic concepts. In: Volkmar FR, editor. Handbook of Autism and Pervasive Developmental Disorders. 4th ed. Hoboken, NJ: John Wiley & Sons Inc. (2014). doi:10.1002/9781118911389.hautc01
19. Tsatsanis KD, Volkmar FR, Paul R, Klin A, Cohen D. Neuropsychological characteristics in autism and related conditions. In: Volkmar FR, Paul R, Klin A, Cohen D, editors. Handbook of Autism and Pervasive Developmental Disorders: Diagnosis, Development, Neurobiology, and Behavior. Vol. 1, 3rd ed. Hoboken, NJ: John Wiley & Sons Inc. (2014). p. 365–81. doi:10.1002/9780470939345.ch13
20. Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators, Centers for Disease Control and Prevention (CDC). Prevalence of autism spectrum disorder among children aged 8 years-autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ (2014) 63(2):1–21.
21. Barad DH, Kushnir VA, Albertini D, Gleicher N. CDC analysis of ICSI/autism: association is not causation. Hum Reprod (2015) 30(7):1745–6. doi:10.1093/humrep/dev104
22. Huguet G, Benabou M, Bourgeron T. The Genetics of Autism Spectrum Disorders. In a Time for Metabolism and Hormones. New York, NY: Springer International Publishing (2016). p. 101–29.
23. Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med (1995) 25(01):63–77. doi:10.1017/S0033291700028099
24. Pickles A, Bolton P, Macdonald H, Bailey A, Le Couteur A, Sim CH, et al. Latent-class analysis of recurrence risks for complex phenotypes with selection and measurement error: a twin and family history study of autism. Am J Hum Genet (1995) 57(3):717.
25. Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet (2014) 46(8):881–5. doi:10.1038/ng.3039
26. Huguet G, Ey E, Bourgeron T. The genetic landscapes of autism spectrum disorders. Annu Rev Genomics Hum Genet (2013) 14:191–213. doi:10.1146/annurev-genom-091212-153431
27. De Rubeis S, Buxbaum JD. Recent advances in the genetics of autism spectrum disorder. Curr Neurol Neurosci Rep (2015) 15(6):1–9. doi:10.1007/s11910-015-0553-1
28. Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res (2009) 37:D832–6. doi:10.1093/nar/gkn835
29. Mazzio EA, Soliman KF. Basic concepts of epigenetics: impact of environmental signals on gene expression. Epigenetics (2012) 7(2):119–30. doi:10.4161/epi.7.2.18764
30. Miyake K, Hirasawa T, Koide T, Kubota T. Epigenetics in autism and other neurodevelopmental diseases. In: Ahmad SI, editor. Neurodegenerative Diseases. USA: Springer (2012). p. 91–8.
31. Rajalakshmi K. Epigenetics as a solution in autism: control above autism genes. Autism Open Access (2015) 5:e130. doi:10.4172/2165-7890.1000e130
32. Ross SA, Davis CD. The emerging role of microRNAs and nutrition in modulating health and disease. Annu Rev Nutr (2014) 34:305–36. doi:10.1146/annurev-nutr-071813-105729
33. Chen F, Hu SJ. Effect of microRNA-34a in cell cycle, differentiation, and apoptosis: a review. J Biochem Mol Toxicol (2012) 26(2):79–86. doi:10.1002/jbt.20412
34. Friedländer MR, Lizano E, Houben AJ, Bezdan D, Báñez-Coronel M, Kudla G, et al. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol (2014) 15(4):R57. doi:10.1186/gb-2014-15-4-r57
35. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife (2015) 4:e05005. doi:10.7554/eLife.05005
36. Sun E, Shi Y. MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp Neurol (2015) 268:46–53. doi:10.1016/j.expneurol.2014.08.005
37. Tritschler F, Huntzinger E, Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of déjà vu. Nat Rev Mol Cell Biol (2010) 11:379–84. doi:10.1038/nrm2885
38. Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA–target recognition. PLoS Biol (2005) 3(3):e85. doi:10.1371/journal.pbio.0030085
39. Friedman RC, Farth KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of miRNAs. Genome Res (2009) 19(1):92–105. doi:10.1101/gr.082701.108
40. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The miRNA spectrum in 12 body fluids. Clin Chem (2010) 56(11):1733–41. doi:10.1373/clinchem.2010.147405
41. Adlakha YK, Saini N. Brain miRNAs and insights into biological functions and therapeutic potential of brain enriched miRNA-128. Mol Cancer (2014) 13:33. doi:10.1186/1476-4598-13-33
42. Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry (2014) 19(7):848–52. doi:10.1038/mp.2013.93
43. Tonelli DDP, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development (2008) 135(23):3911–21. doi:10.1242/dev.025080
44. Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, et al. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J Neurosci (2008) 28(17):4322–30. doi:10.1523/JNEUROSCI.4815-07.2008
45. Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci (2012) 35(5):325–34. doi:10.1016/j.tins.2012.01.004
46. Karam R, Wilkinson M. A conserved microRNA/NMD regulatory circuit controls gene expression. RNA Biol (2012) 9(1):22–6. doi:10.4161/rna.9.1.18010
47. Zongaro S, Hukema R, D’Antoni S, Davidovic L, Barbry P, Catania MV, et al. The 3′ UTR of FMR1 mRNA is a target of miR-101, miR-129-5p and miR-221: implications for the molecular pathology of FXTAS at the synapse. Hum Mol Genet (2013) 22(10):1971–82. doi:10.1093/hmg/ddt044
48. Schofield CM, Hsu R, Barker AJ, Gertz CC, Blelloch R, Ullian EM. Monoallelic deletion of the miRNA biogenesis gene Dgcr8 produces deficits in the development of excitatory synaptic transmission in the prefrontal cortex. Neural Dev (2011) 6(1):11. doi:10.1186/1749-8104-6-11
49. Fénelon K, Mukai J, Xu B, Hsu PK, Drew LJ, Karayiorgou M, et al. Deficiency of Dgcr8, a gene disrupted by the 22q11.2 microdeletion results in altered short-term plasticity in the prefrontal cortex. Proc Natl Acad Sci U S A (2011) 108(1):4447–52. doi:10.1073/pnas.1101219108
50. Issler O, Chen A. Determining the role of microRNAs in psychiatric disorders. Nat Rev Neurosci (2015) 16(4):201–12. doi:10.1038/nrn3879
51. Boccuto L, Lauri M, Sarasua SM, Skinner CD, Buccella D, Dwivedi A, et al. Prevalence of SHANK3 variants in patients with different subtypes of autism spectrum disorders. Eur J Hum Genet (2013) 21(3):310–6. doi:10.1038/ejhg.2012.175
52. Wu YE, Parikshak NN, Belgard TG, Geschwind DH. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. Nat Neurosci (2016). doi:10.1038/nn.4373
53. Mor M, Nardone S, Sams DS, Elliott E. Hypomethylation of miR-142 promoter and upregulation of microRNAs that target the oxytocin receptor gene in the autism prefrontal cortex. Mol Autism (2015) 6(1):1. doi:10.1186/s13229-015-0040-1
54. Ander BP, Barger N, Stamova B, Sharp FR, Schumann CM. Atypical miRNA expression in temporal cortex associated with dysregulation of immune, cell cycle, and other pathways in autism spectrum disorders. Mol Autism (2015) 6:37. doi:10.1186/s13229-015-0029-9
55. Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res (2015) 43(D1):D146–52. doi:10.1093/nar/gku1104
56. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc (2009) 4(1):44–57. doi:10.1038/nprot.2008.211
57. Hutcheson HB, Olson LM, Bradford Y, Folstein SE, Santangelo SL, Sutcliffe JS, et al. Examination of NRCAM, LRRN3, KIAA0716, and LAMB1 as autism candidate genes. BMC Med Genet (2004) 5(1):1. doi:10.1186/1471-2156-5-1
58. Nakamura F, Kalb RG, Strittmatter SM. Molecular basis of semaphorin-mediated axon guidance. J Neurobiol (2000) 44(2):219–29. doi:10.1002/1097-4695(200008)44:2<219::AID-NEU11>3.0.CO;2-W
59. Hashimoto K, Iwata Y, Nakamura K, Tsujii M, Tsuchiya KJ, Sekine Y, et al. Reduced serum levels of brain-derived neurotrophic factor in adult male patients with autism. Prog Neuropsychopharmacol Biol Psychiatry (2006) 30(8):1529–31. doi:10.1016/j.pnpbp.2006.06.018
60. Stamova B, Ander BP, Barger N, Sharp FR, Schumann CM. Specific regional and age-related small Noncoding RNA expression patterns within superior temporal gyrus of typical human brains are less distinct in autism brains. J Child Neurol (2015) 30(14):1930–46. doi:10.1177/0883073815602067
61. Froehlich-Santino W, Tobon AL, Cleveland S, Torres A, Phillips J, Cohen B, et al. Prenatal and perinatal risk factors in a twin study of autism spectrum disorders. J Psychiatr Res (2014) 54:100–8. doi:10.1016/j.jpsychires.2014.03.019
62. Toma C, Torrico B, Hervás A, Salgado M, Rueda I, Valdés-Mas R, et al. Common and rare variants of microRNA genes in autism spectrum disorders. World J Biol Psychiatry (2015) 16(6):376–86. doi:10.3109/15622975.2015.1029518
63. Vaishnavi V, Manikandan M, Tiwary BK, Munirajan AK. Insights on the functional impact of microRNAs present in autism-associated copy number variants. PLoS One (2013) 8(2):e56781. doi:10.1371/journal.pone.0056781
64. Chaidez V, Hansen RL, Hertz-Picciotto I. Gastrointestinal problems in children with autism, developmental delays or typical development. J Autism Devel Disor (2014) 44(5):1117–27. doi:10.1007/s10803-013-1973-x
65. Cermak SA, Curtin C, Bandini L. Sensory sensitivity and food selectivity in children with autism spectrum disorders. In: Patel VB, Preedy VR, Martin CR, editors. Comprehensive Guide to Autism. New York: Springer (2014). p. 2061–76.
66. Tierney C, Mayes S, Lohs SR, Black A, Gisin E, Veglia M. How valid is the checklist for autism spectrum disorder when a child has apraxia of speech? J Dev Behav Pediatr (2015) 36(8):569–74. doi:10.1097/DBP.0000000000000189
67. Miller M, Chukoskie L, Zinni M, Townsend J, Trauner D. Dyspraxia, motor function and visual–motor integration in autism. Behav Brain Res (2014) 269:95–102. doi:10.1016/j.bbr.2014.04.011
Keywords: autism, microRNA, neurodevelopment, biomarker
Citation: Hicks SD and Middleton FA (2016) A Comparative Review of microRNA Expression Patterns in Autism Spectrum Disorder. Front. Psychiatry 7:176. doi: 10.3389/fpsyt.2016.00176
Received: 15 August 2016; Accepted: 11 October 2016;
Published: 04 November 2016
Edited by:
Mark Nicholas Ziats, National Institute of Child Health and Human Development, USAReviewed by:
Antonio Benítez-Burraco, University of Huelva, SpainKimberly Raab-Graham, Wake Forest School of Medicine, USA
Copyright: © 2016 Hicks and Middleton. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven D. Hicks, c2hpY2tzMUBobWMucHN1LmVkdQ==
 Steven D. Hicks
Steven D. Hicks Frank A. Middleton
Frank A. Middleton