
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Psychiatry , 19 April 2012
Sec. Neuroimaging
Volume 3 - 2012 | https://doi.org/10.3389/fpsyt.2012.00032
This article is part of the Research Topic Magnetic resonance imaging of disturbed brain connectivity in psychiatric illness View all 11 articles
Psychiatric disorders are phenotypically heterogeneous entities with a complex genetic basis. To mitigate this complexity, many investigators study so-called intermediate phenotypes (IPs) that putatively provide a more direct index of the physiological effects of candidate genetic risk variants than overt psychiatric syndromes. Magnetic resonance imaging (MRI) is a particularly popular technique for measuring such phenotypes because it allows interrogation of diverse aspects of brain structure and function in vivo. Much of this work however, has focused on relatively simple measures that quantify variations in the physiology or tissue integrity of specific brain regions in isolation, contradicting an emerging consensus that most major psychiatric disorders do not arise from isolated dysfunction in one or a few brain regions, but rather from disturbed interactions within and between distributed neural circuits; i.e., they are disorders of brain connectivity. The recent proliferation of new MRI techniques for comprehensively mapping the entire connectivity architecture of the brain, termed the human connectome, has provided a rich repertoire of tools for understanding how genetic variants implicated in mental disorder impact distinct neural circuits. In this article, we review research using these connectomic techniques to understand how genetic variation influences the connectivity and topology of human brain networks. We highlight recent evidence from twin and imaging genetics studies suggesting that the penetrance of candidate risk variants for mental illness, such as those in SLC6A4, MAOA, ZNF804A, and APOE, may be higher for IPs characterized at the level of distributed neural systems than at the level of spatially localized brain regions. The findings indicate that imaging connectomics provides a powerful framework for understanding how genetic risk for psychiatric disease is expressed through altered structure and function of the human connectome.
Genetic factors play a major role in liability for mental illness. Most psychiatric disorders are familial (Gottesman, 1991; Sullivan et al., 2000; Hettema et al., 2001), and twin studies of disease heritability – the proportion of illness susceptibility attributable to genes – indicate that genetic contributions are as high as 80–90% for schizophrenia and bipolar disorder (Cannon et al., 1998; Cardno et al., 1999; McGuffin et al., 2003); 40–70% for major depression (Kendler et al., 1993, 2006; Sullivan et al., 2000); 37 to >90% for autism and autistic traits (Hallmayer et al., 2011; Robinson et al., 2012); 30–70% for substance addiction (Agrawal and Lynskey, 2008); and ∼30–50% for anxiety disorders and obsessive–compulsive disorder (Hettema et al., 2001; van Grootheest et al., 2005). This liability has a complex genetic basis, arising from the combined effect of multiple (hundreds or thousands) genetic variants of small-effect, rather than one or a few genes of large effect (e.g., Purcell et al., 2009). This complexity is compounded by the substantial phenotypic heterogeneity that characterizes most of these disorders. Some of this heterogeneity may be intrinsic to the disease process itself, but a large proportion likely reflects our current lack of biologically informed diagnostic criteria. In practice, this failure to appropriately “carve nature at its joints” (Kendler, 2006) introduces considerable noise into any attempts to map illness phenotypes to genetic risk mechanisms and positions psychiatric disorders, as currently diagnosed, far downstream of the pathophysiological effects of genetic variants influencing disease susceptibility.
One strategy proposed to mitigate the phenotypic heterogeneity of psychiatric disorders involves studying intermediate phenotypes (IPs; Meyer-Lindenberg and Weinberger, 2006). IPs are quantitative, biological traits that are interposed between gene and clinical phenotype on the causal pathway leading from inherited vulnerability to disease. They putatively provide a more direct index of the physiological effects of genetic risk variants and can be used to parse a phenotypically heterogeneous disorder with complex genetic basis into more homogeneous phenotypes with a presumably simpler genetic architecture (see Gottesman and Gould, 2003; Cannon and Keller, 2006; though, see Flint and Munafo, 2007; Walters and Owen, 2007; Kendler and Neale, 2010 for critiques)1. Various criteria for viable IPs have been proposed (Gottesman and Gould, 2003; Cannon and Keller, 2006; Walters and Owen, 2007), generally stating that such a phenotype should (1) be quantitative and heritable; (2) differentiate patients from controls; (3) be associated with disease causes rather than effects; (4) co-segregate with illness within families; (5) be more frequent in unaffected individuals at increased genetic risk; and (6) be stable over time (although this last criterion has been challenged; see Pantelis et al., 2009; Gogtay et al., 2011).
Psychiatric disorders are disturbances of mental processes mediated by the brain. Neural measures therefore represent a major class of candidate IPs for mental illness, and in vivo neuroimaging has provided a powerful means for identifying and characterizing such IPs for a broad spectrum of psychiatric disease, particularly when related to variation in specific candidate risk genes (Meyer-Lindenberg and Weinberger, 2006). This “imaging genetics” approach has been used to elucidate the mechanisms through which genetically mediated variations in brain structure and function might give risk to psychiatric illness, and provide biological validation of candidate variants (Esslinger et al., 2009; Erk et al., 2010). In some cases, neuroimaging has also been used to augment the search for risk genes (Potkin et al., 2009).
Most imaging genetics work has studied relatively simple IPs, defined using measures of task-related activation or anatomical structure (e.g., gray matter volume or cortical thickness) in specific brain regions. Though the simplicity of these measures is attractive, they likely over-simplify relevant (patho)physiological processes in most cases. Over two decades of neuroimaging research has found that most major psychiatric disorders do not arise from isolated damage to one or a few brain regions, but rather from multiple abnormalities distributed throughout the cerebrum (Phillips et al., 2003b; Belmonte et al., 2004; Menzies et al., 2008; Fornito et al., 2009, 2012; Minzenberg et al., 2009). These abnormalities likely have their origin in disturbed interactions between discrete and distributed neural circuits; i.e., disordered brain connectivity. Moreover, many risk genes for psychiatric disorders are expressed diffusely throughout the brain, acting on physiological pathways involved in synaptic function/regulation, neurotransmitter release, degradation or re-uptake, and the development and maintenance of axonal pathways (Harrison and Weinberger, 2005; Bennett, 2011; Lips et al., 2011; Gai et al., 2012). These considerations indicate that a systems-level approach should prove useful for characterizing the neurophysiological impact of candidate risk variants, and in defining novel IPs.
Attempts to understand genetic influences on distributed brain networks have been greatly facilitated by recent attempts to map the structural and functional properties of the human connectome – the complete set of neural elements and inter-connections comprising the brain (Sporns et al., 2005; Sporns, 2011). The use of neuroimaging to achieve this goal, termed imaging connectomics, has led to the development and proliferation of a rich repertoire of tools for characterizing diverse aspects of human brain connectivity (Bullmore and Sporns, 2009; Bullmore et al., 2009; Fornito and Bullmore, 2010; Margulies et al., 2010). These tools are now increasingly being deployed within genetically informative designs to uncover new connectomic IPs (cIPs) that show promise as sensitive measures of genetic variation in brain structure and physiology (Meyer-Lindenberg, 2009). In this article we overview recent work attempting to understand genetic influences on brain connectivity, focusing principally on studies using magnetic resonance imaging (MRI) that provide illustrative examples of the potential utility of cIPs for characterizing genetic risk mechanisms in mental illness. Our intention is not to identify specific cIPs that fulfill all of the above criteria and which may thus be considered to be comprehensively validated, as this is difficult to achieve in practice. Rather, our aim is to draw attention to emerging evidence suggesting the connectomic measures provide particularly sensitive probes of the functional effects of disease risk genes. As an orientation to relevant concepts, we begin with a brief primer on some basic concepts central to the imaging connectomics approach.
Magnetic resonance imaging studies have used a wide range of techniques to explore different properties of the human connectome. A simplified conceptual overview of how some of the most commonly used imaging techniques relate to these properties is presented in Figure 1.
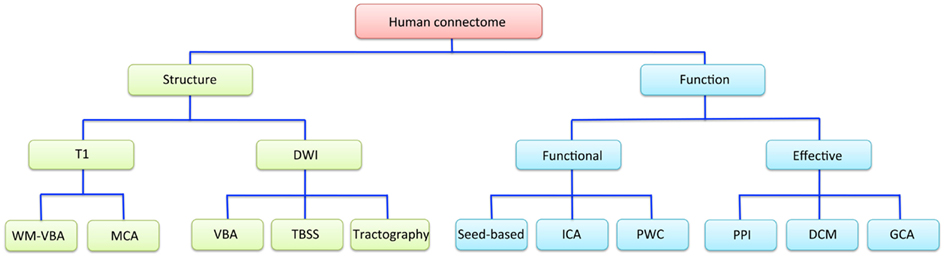
Figure 1. A schematic overview of the different neuroimaging techniques available for measuring structural and functional properties of the human connectome. DCM, dynamic causal modeling; DWI, diffusion-weighted imaging; GCA, Granger causality analysis; ICA, independent component analysis; MCA, morphometric covariance analysis; PPI, psychophysiological interactions; PWC, pair-wise correlations; TBSS, tract-based spatial statistics; T1, T1-weighted imaging; VBA, voxel-based analysis; WM, white matter.
Broadly, the connectome can be studied in terms of structure or function. Connectome structure refers to the anatomical connections between different brain regions, and is measured in MRI studies using either T1- or diffusion-weighted imaging (DWI). The former indirectly infers connectivity between regions either through voxel-wise mapping of variations in white matter density, or covariance in regional morphometric parameters, such as gray matter volume or cortical thickness. These morphometric covariance analyses (MCA) can be performed either between a single seed region and all other brain voxels (Pezawas et al., 2005; Meyer-Lindenberg et al., 2007), between (many) pairs of a priori defined regions (He et al., 2007; Bassett et al., 2008), or following multivariate decomposition of the data using techniques such as independent component analysis (ICA; Xu et al., 2009a,b). DWI provides a more direct measure of the integrity and trajectory of anatomical connections, subject to certain caveats (Mori and Zhang, 2006; Zalesky and Fornito, 2009; Jbabdi and Johansen-Berg, 2011). Voxel-based analysis (VBA) techniques applied to estimates of white matter density/volume or DWI-derived measures (e.g., axial/radial diffusivity and/or fractional anisotropy) allow whole-brain mapping of differences in white matter integrity or morphometry. This includes methods such as tract-based spatial statistics (TBSS; Smith et al., 2006). Alternatively, connectivity may be indexed with DWI by deterministic or probabilistic tractography of fiber trajectories linking different brain regions, either at the level of specific pairs of brain regions, or between every possible pair of regions defined using some comprehensive cerebral parcelation (Hagmann et al., 2008; Zalesky et al., 2010). With this approach, connectivity is estimated either as the number of tracts intersecting each regional pair (Zalesky et al., 2011), or some index of tissue integrity averaged across the extent of the reconstructed trajectory (van den Heuvel et al., 2010). Note that all of these anatomical measures only provide undirected estimates of inter-regional connectivity; i.e., they may imply that two regions are connected, but they cannot resolve whether the connection runs from region A to B or vice-versa.
Magnetic resonance imaging studies of connectome function generally use blood-oxygenation-level dependent (BOLD) imaging. Functional interactions between regions can be categorized into one of two broad classes: functional and effective connectivity (Friston, 1994). Effective connectivity refers to the influence that one neuronal system exerts over another and allows inferences concerning causal (directed) interactions between regions. The most rudimentary model of effective connectivity is provided by psychophysiological interaction (PPI) analysis (Friston et al., 1997), which involves analyzing task-related changes in connectivity between a seed region and all other brain voxels. Dynamic causal modeling (DCM; Friston et al., 2003) and Granger causality analysis (GCA; Bressler and Seth, 2011) provide more elaborate means for inferring causal interactions between brain regions but have seldom been studied in the context of genetically informative designs (though, see Curcic-Blake et al., 2012 for an exception).
Functional connectivity refers to a statistical dependence between regionally distinct neurophysiological recordings and is undirected in nature. Most frequently, this is measured through simple Pearson correlation of regional activity time courses. This covariance is typically studied between a specific seed region and all other brain voxels (termed a seed-based correlation analysis), or in a pair-wise correlation (PWC) analysis where associations between either a single pair of a priori defined regions, or multiple pairs of regions, are computed. Another popular technique for studying functional connectivity involves multivariate spatio-temporal decomposition using techniques such as spatial ICA (Beckmann and Smith, 2004; Calhoun et al., 2004), which offers a data-driven method for identifying spatially independent networks of voxels with temporally coherent activity.
A final important distinction that applies to studies of both connectome function and structure concerns whether inter-regional interactions are studied at the level of connectivity or topology. Studies of connectivity are concerned with measuring variations in the strength and nature (e.g., structural or functional, positive or negative) of connectivity between regions. Studies of topology are concerned with how connections between different brain regions are configured with respect to each other. These analyses require relatively comprehensive mapping of inter-regional connectivity using a PWC approach combined with graph analysis to characterize such topological variations (Bullmore and Sporns, 2009; Bullmore and Bassett, 2011). Briefly, graph analysis involves modeling the brain as a graph of nodes, representing brain regions, connected by edges, representing some measure of inter-regional structural or functional interaction. The method allows a wide range of topological properties representing diverse aspects of connectome organization to be computed (Bullmore and Sporns, 2009; Rubinov and Sporns, 2010). Though topology and connectivity are not necessarily independent (Barrat et al., 2004; Alexander-Bloch et al., 2010; Lynall et al., 2010), the distinction between them provides a useful heuristic for evaluating findings in imaging connectomics. In the following, we first consider studies examining genetic influences on brain network connectivity before discussing studies of network topology.
Most of the connectomic phenotypes studied in imaging genetics studies to date have been relatively specific, focusing on particular neural circuits of theoretical interest. These have typically been investigated by analyzing connectivity between specific pairs of brain regions, or between one seed region and all other brain voxels. Most of this work has used fMRI and therefore focuses on understanding how specific genetic variants influence functional connectivity between regions. Here, we focus on studies of three neural circuits that have been widely studied in imaging genetic research – fronto-limbic, fronto-temporal, and the default mode network. Examples of how genetic variants impact other neural systems can be found elsewhere (e.g., Meyer-Lindenberg et al., 2007; Tan et al., 2007; Kempf et al., 2008; Krugel et al., 2009; Rasetti et al., 2011).
The first MRI study to examine genetic influences on a connectomic phenotype examined how variation in the human serotonin (5-HT) transporter gene SLC6A4 impacted functional connectivity between the amygdala and medial prefrontal cortex (mPFC; Heinz et al., 2005). A variable number of tandem repeats (VNTR) in the 5′ promoter region (5-HTTPLR) of SLC6A4 influences mRNA and protein expression such that a short (s) compared to long (l) allele is associated with reduced transcriptional efficiency (Heils et al., 1996) and reduced 5-HT transporter binding in the brain (Heinz et al., 2000). Accordingly, the s allele has been variably associated with personality traits associated with negative emotionality (Lesch et al., 1996; Munafo et al., 2009), alcohol dependence (McHugh et al., 2010), mood disorders (Clarke et al., 2010), and suicidality (Li and He, 2007). In some cases, these associations may be moderated by environmental factors (Caspi et al., 2010).
Consistent with an association between the s allele and negative emotions, an early fMRI study found that human carriers of this allele showed increased amygdala activation during perception of aversive emotional stimuli (Hariri et al., 2002). In a separate study using a similar task, an association was reported between the s allele and greater functional connectivity between the amygdala and anterior mPFC (Heinz et al., 2005). This finding was noteworthy given the abundant evidence pointing to the amygdala and mPFC as critical nodes within a broader neural network subserving emotional regulation (Phillips et al., 2003a). These findings were subsequently replicated by an independent group, who also reported an association between the s allele and reduced functional connectivity between the amygdala and more posterior medial prefrontal regions located in pre- and sub-genual anterior cingulate cortex (ACC; Pezawas et al., 2005). An anatomical basis for these functional effects was suggested by a corresponding reduction in gray matter volume covariance between the amygdala and pre-genual ACC in s allele carriers compared to l/l homozygotes, a finding replicated in an independent study using DTI-derived measures of anatomical connectivity (Pacheco et al., 2009). The functional connectivity measures predicted approximately 30% of the variance in temperamental measures of anxiety-related traits such as harm avoidance. No such association was found with regional indices of activation or volume, indicating that the connectomic measures provided a more direct physiological marker of clinically relevant behavioral phenotypes. Collectively, these results suggest that variation in SLC6A4 regulates fronto-limbic dynamics in highly specific ways, such that the s allele up-regulates functional connectivity between amygdala and anterior mPFC and down-regulates connectivity between amygdala and perigenual ACC (Figure 2).
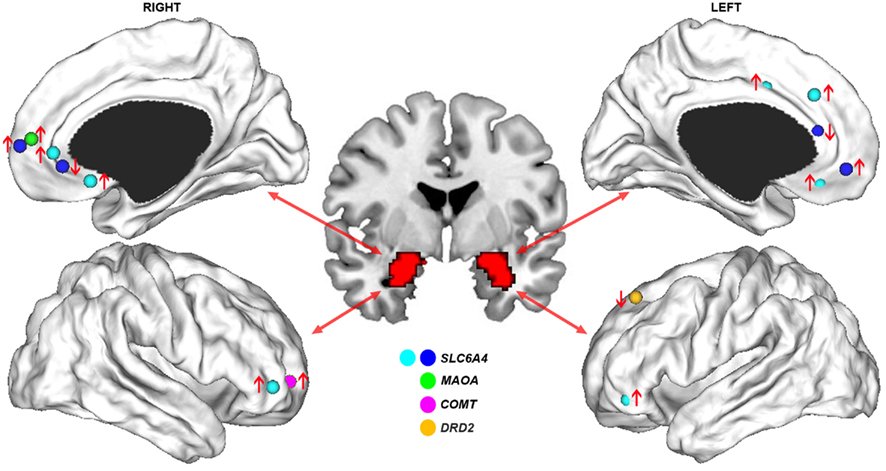
Figure 2. Genetic influences on functional connectivity of fronto-limbic circuits. Cortical surface renderings display the stereotactic peaks of regions in prefrontal cortex where functional connectivity with the amygdala is impacted by common genetic variants. Different colors denote the different genes studied in relation to fronto-limbic functional connectivity; namely, SLC6A4 (Heinz et al., 2005; Pezawas et al., 2005; Schardt et al., 2010), MAOA (Buckholtz et al., 2008), COMT (Rasch et al., 2010), and DRD2 (Blasi et al., 2009). Arrows indicate whether functional connectivity was increased (↑) or decreased (↓) in carriers of the putative risk allele for each gene. The distinction between cyan and blue foci for SLC6A4 studies differentiates a single study examining functional connectivity during cognitive regulation of emotion (Schardt et al., 2010) from others involving passive perception of emotional stimuli. This distinction illustrates how the risk allele of 5-HTTPLR polymorphism of this gene can be associated with either increased or decreased functional connectivity with the amygdala in adjacent regions of right pre-genual ACC, depending on task context.
Another genetic variant shown to impact fronto-limbic connectivity is a VNTR in the upstream promoter region of the X-linked Monoamine Oxidase A (MAOA) gene (Buckholtz et al., 2008). MAOA is the main enzyme responsible for catabolizing synaptic 5-HT and norepinephrine, and MAOA expression is higher when the VNTR comprises 3.5–4 repeats (MAOA-H) than when 2, 3, or 5 repeats are present (MAOA-L; Sabol et al., 1998). The low activity allele has been shown to interact with childhood stressors to increase risk for adverse mental health outcomes such as aggressive behavior and mood disturbances (Caspi et al., 2002; Kim-Cohen et al., 2006; Nikulina et al., 2012). Accordingly, functional connectivity between the amygdala and anterior mPFC is higher in MAOA-L individuals compared to MAOA-H participants performing an emotional perception task (Buckholtz et al., 2008), a finding that parallels the functional connectivity increases between these two regions found in carriers of the 5-HTTPLR-s allele (Heinz et al., 2005; Pezawas et al., 2005; Figure 2).
The common effect of the MAOA-L and 5-HTTPLR-s alleles on amygdala–mPFC circuitry may result from a shared influence on synaptic serotonin levels, as both are thought to be associated with increased brain 5-HT. Greater functional connectivity between the amygdala and anterior mPFC may therefore represent a final common pathway for genetically mediated increases in synaptic 5-HT concentrations. Contrary to this view however, recent imaging evidence suggests that 5-HTPPLR genotype has little effect on basal synaptic serotonin levels (Jedema et al., 2010; Murthy et al., 2010). As such, the precise mechanism through which the polymorphism influences mPFC–amygdala connectivity remains unclear, but may be related to its emerging role in neurodevelopment (Ansorge et al., 2004). Regardless of the precise mechanism, it is noteworthy that the effects seem specific to variants impacting on 5-HT function, as functional polymorphisms in genes that do not directly impact this neurotransmitter show no effect on amygdala–mPFC connectivity. For example, two genetic variants that influence synaptic dopamine levels, the Val108/158 Met polymorphism of the catechol-o-methyl transferase (COMT) gene, and the rs1076560 polymorphism of the D2 receptor gene DRD2, influence functional connectivity between the amygdala and lateral, but not medial, PFC (Buckholtz et al., 2008; Blasi et al., 2009; Rasch et al., 2010; Figure 2). These findings point to highly specific genetic influences on different components of fronto-limbic circuitry.
Some of these genetic effects can be expressed in different ways depending on psychological context. For example, during conditions requiring cognitive regulation of emotion, s allele carriers actually show increased functional connectivity between the amygdala and perigenual ACC (Schardt et al., 2010). This result indicates that the previously reported reduction in functional coupling between these regions does not reflect a hard-wired deficit, despite evidence for anatomical connectivity reductions between these regions (Pezawas et al., 2005). Rather, the connectivity reduction may reflect a bias to process aversive stimuli in maladaptive ways, which may be ameliorated given appropriate environmental circumstances. In this regard, the connectivity increase in s allele carriers during cognitive regulation of emotion may reflect the deployment of greater processing resources to overcome this processing bias. These context-specific effects on connectomic phenotypes are consistent with evidence for environmental factors such as life stress moderating the influence of SLC6A4 genotype on functional connectivity of the amygdala (Canli et al., 2006).
To summarize, studies of the SLC6A4 and MAOA genes provide examples of how genetic influences on brain connectivity can be expressed in circuit specific ways, and in a manner contingent on environmental context. The circuit- and context-specificity of these effects is underscored by work on other genes, such as CA1CNAC (Erk et al., 2010), α5 acetylcholine receptor subunit (CHRNA5; Hong et al., 2010), brain-derived neurotrophic factor (BDNF; Mukherjee et al., 2011), and α2b-adrenergic receptor (ADRA2B; Rasch et al., 2009). Variants in each of these genes have been shown to impact fronto-limbic circuitry in a task-dependent manner, and in regions outside the amygdala–mPFC loop influenced by SLC6A4 and MAOA. Understanding how each of these variants impact different components of fronto-limbic circuits, and under what conditions, will no doubt prove to be a challenging task.
Fronto-temporal connectivity is of particular interest to schizophrenia researchers, as diverse lines of evidence point to altered structural and functional interactions between frontal and temporal brain regions as a core feature of the disease phenotype (Weinberger et al., 1992; Meyer-Lindenberg et al., 2005; Ellison-Wright and Bullmore, 2009; van den Heuvel et al., 2010; Fornito et al., 2011a, 2012; Pettersson-Yeo et al., 2011). Accordingly, variation in several candidate risk genes for schizophrenia has been shown to impact fronto-temporal connectivity, including loci in COMT (Bertolino et al., 2006; Dennis et al., 2010b), RGS4 (Buckholtz et al., 2007), PPP1R1B encoding DAARP-32 (Curcic-Blake et al., 2012), and ErbB4 (Konrad et al., 2009).
One genetic variant receiving increasing attention for its effects on fronto-temporal circuitry is the rs1344706 polymorphism of ZNF804A. The gene encodes a zinc finger domain protein with unknown function, although preliminary evidence suggests a role in oligodendrocyte regulation (Riley et al., 2010) and neurodevelopment (Chung et al., 2010). The rs1344706 polymorphism has now been implicated as a genome-wide significant risk variant for both schizophrenia and bipolar disorder in several independent samples (O’Donovan et al., 2008; Stefansson et al., 2009; Riley et al., 2010; Steinberg et al., 2011), with fine-mapping of nearly all common variants in the gene confirming that rs1344706 is the most strongly associated marker (Williams et al., 2011). The polymorphism is functional, affecting ZNF804A expression (Williams et al., 2011) and DNA–protein binding (Hill and Bray, 2011).
The first study to investigate the effects of the rs1344706 polymorphism on the brain used fMRI to examine activation and functional connectivity during performance of a working memory and emotion perception task (Esslinger et al., 2009). The variant was not found to impact measures of regional activation, but did influence inter-regional functional connectivity, suggesting its effects show higher penetrance at the level of distributed neural systems. Specifically, carriers of the risk associated A allele demonstrated reduced functional connectivity of the dorsolateral PFC with other ipsi- and contra-lateral prefrontal regions, as well as increased functional connectivity between dorsolateral PFC and the hippocampal formation, during working memory performance. Increased fronto-hippocampal functional connectivity had previously been reported in a positron emission tomography study of patients with schizophrenia (Meyer-Lindenberg et al., 2005) and was thus interpreted as a possible cIP mediating the role of ZNF804A in risk for the disorder. The A allele was also associated with enhanced functional connectivity within fronto-limbic circuits during the emotional perception task, which was interpreted as a possible cIP for bipolar disorder. Thus, this study pointed to context-specific and pleiotropic effects of the rs1344706 polymorphism with relevance for understanding its role in risk for both schizophrenia and bipolar disorder.
A subsequent study in an independent sample using the same working memory task replicated the association between the A allele and both increased fronto-hippocampal functional coupling and decreased fronto-frontal connectivity (Rasetti et al., 2011). The lack of a significant effect on regional activation was also replicated. Similar connectivity changes were apparent in schizophrenia patients and their unaffected relatives suggesting that these changes may indeed represent a candidate cIP for the disorder (Figure 3). However, the A allele carriers in this study showed stronger negative fronto-hippocampal functional connectivity, contrasting the original report of higher positive functional connectivity between these two regions (Esslinger et al., 2011). These results suggest distinct interpretations: stronger positive connectivity implies greater functional integration or cooperation between regions; stronger negative connectivity points to antagonistic or competitive dynamics (Clare Kelly et al., 2008). The reasons for this discrepancy are unclear, although the fact that stronger negative functional connectivity has been found in patients and their unaffected relatives (Meyer-Lindenberg et al., 2005; Rasetti et al., 2011) suggests that it may provide a more faithful representation of genetically influenced pathophysiological processes in schizophrenia.
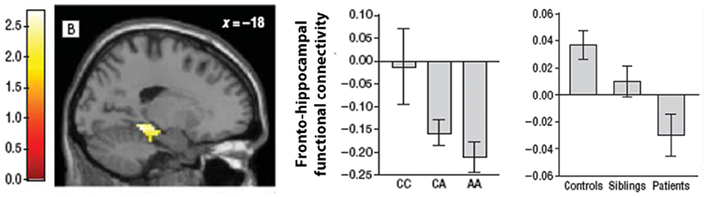
Figure 3. Illustration of the hippocampal region where functional connectivity with a seed in right dorsolateral PFC was influenced by ZNF804A variation and was found to differ between schizophrenia patients, their unaffected relatives and healthy controls. The two bar charts plot parameter estimates for fronto-hippocampal functional connectivity as a function of ZNF804A in healthy controls (middle), and in controls, patients, and siblings (right). Carriers of the A risk allele showed an increasing trend toward greater negative functional connectivity, as did patients with schizophrenia. Image adapted from Rasetti et al. (2011).
The effect of the rs1344706 variant on fronto-hippocampal connectivity may be less robust than its effect on fronto-frontal coupling, and may only be expressed in certain contexts. In one recent study, the variant was found to influence fronto-hippocampal functional connectivity only during working memory performance, whereas it impacted fronto-frontal coupling during working memory, emotion perception, and resting-state paradigms (Esslinger et al., 2011). Thus, the effect on fronto-frontal connectivity may be generalized, possibly resulting from an influence on anatomical connectivity between these regions (Wei et al., 2010), whereas the association with fronto-hippocampal coupling may be context-specific. This result is consistent with research into the effects of SLC6A4 variation on fronto-limbic circuitry (e.g., Schardt et al., 2010) demonstrating the highly context-specific influences of genetic variants of brain functional dynamics. Precisely mapping the circumstances under which these genetic effects are expressed will be an important step in characterizing the functional significance of any putative cIPs related to ZNF804A variation, particularly given recent evidence that schizophrenia may also be characterized by a profile of both generalized and context-specific functional connectivity deficits which affect prefrontal regions in particular (Fornito et al., 2011a).
The DMN comprises regions of posterior cingulate cortex (PCC), precuneus, medial PFC and lateral parietal cortices, and functionally couples with the hippocampus under certain task contexts (Buckner et al., 2008). The DMN characteristically shows elevated activity during passive rest conditions or tasks requiring introspective processing and deactivates during cognitively demanding tasks (Shulman et al., 1997; Buckner et al., 2008; Harrison et al., 2008, 2011). Alterations of DMN connectivity have been found in patients suffering a range of neuropsychiatric disorders including schizophrenia (Bluhm et al., 2007), autism (Kennedy and Courchesne, 2008), attention-deficit hyperactivity disorder (Castellanos et al., 2008), major depression (Greicius et al., 2007), and Alzheimer’s disease (Greicius et al., 2004; see Sonuga-Barke and Castellanos, 2007; Zhang and Raichle, 2010 for reviews). In the case of schizophrenia, similar alterations have been found in patients’ unaffected relatives, suggesting an association between DMN connectivity changes and genetic risk for the disorder (Whitfield-Gabrieli et al., 2009). Accordingly, specific variants in genes linked to risk for psychosis, such as the D-Amino Acid Oxidase (DAAO; Papagni et al., 2011; Prata et al., 2012) and neuregulin-1 (NRG1; Winterer et al., 2008), have been shown to impact structural and functional connectivity of DMN regions.
Confirmation that functional connectivity of the DMN is under genetic control came from a recent resting-state fMRI study of a sample of extended pedigrees comprising 333 individuals selected from 29 families (Glahn et al., 2010). Spontaneous neural dynamics measured during the so-called resting-state, when people lie quietly in the scanner without performing any specific task, are highly organized and correlate in a manner that recapitulates well-known functional networks (Biswal et al., 1995; Smith et al., 2009). The topography of these networks is robust over time and individuals (Damoiseaux et al., 2006; Shehzad et al., 2009) and has been linked to neurophysiological measures (He et al., 2008; Shmuel and Leopold, 2008). In addition, spontaneous BOLD signal fluctuations influence task-evoked activity (Fox et al., 2006; Mennes et al., 2011), perception (Hesselmann et al., 2008), and behavior (Fox et al., 2007), suggesting that they represent an intrinsic and functionally important component of neural dynamics (Fox and Raichle, 2007; though, see Morcom and Fletcher, 2007; Fornito and Bullmore, 2010 for caveats). The extended pedigree design exploited the varying degree of genetic relatedness between participants to estimate genetic and environmental influences on functional connectivity of the DMN, as characterized using ICA. Genetic influences on an averaged estimate of functional connectivity of the entire DMN were significant, with heritability (h2) estimated to be 0.42. The degree to which each constituent region of the DMN was functionally connected with the entire network was also heritable, ranging from 0.33 to 0.42 for key nodes such as posterior cingulate/precuneus, mPFC, and lateral parietal cortex.
Despite the DMN’s involvement in a wide range of diseases, the most frequently studied genetic locus in relation to DMN connectivity has been the Apolipoprotein E (APOE) gene. The gene encodes a lipoprotein that in the central nervous system, plays a role in coordinating mobilization of cholesterol, phospholipids, and fatty acids, and has been implicated in neuronal development, plasticity, and repair (Mahley and Rall, 2000). The gene has three allelic variants – ε2, ε3, and ε4. Homozygosity for the last is the most established genetic risk factor for late-onset Alzheimer’s disease, being associated with a >10-fold increase in risk (Farrer et al., 1997; Bertram et al., 2007). In contrast, average allelic summary odds ratios for non-APOE-related variants are ∼1.25 (Bertram and Tanzi, 2008). APOE has also been implicated in the pathophysiology of schizophrenia and mood disorders (Gibbons et al., 2011).
Several studies have demonstrated an association between the ε4 allele and both increased and decreased functional connectivity of the DMN and medial temporal regions, though reports of increases have been more common in younger samples (Filippini et al., 2009; Dennis et al., 2010a; Sheline et al., 2010a,b; Machulda et al., 2011; Westlye et al., 2011). One hypothesis proposed to explain this age effect is that ε4 carriers inherit a cIP characterized by enhanced DMN connectivity early in life. This enhanced connectivity facilitates the spread of amyloid-β plaques, the primary pathological characteristic of Alzheimer’s disease, throughout the network and exacerbates risk of disease onset and progression (Machulda et al., 2011; Seeley, 2011). With advancing age, these plaques accumulate and result in a deterioration of DMN function and concomitant reduction of network connectivity relative to non-risk allele carriers. Though this postulate requires experimental verification, it is supported by reports that DMN regions are among the first to show aggregation of amyloid-β plaques in Alzheimer’s disease and that these plaques disperse throughout the network with disease progression (Buckner et al., 2005, 2009). It also accords with reports that gray matter volume loss in neurodegenerative disease occurs within functionally connected networks (Seeley et al., 2009).
The effects of APOE genotype on functional connectivity are often apparent in the absence of any differences in gray matter volume, cognitive impairment, or β-amyloid deposition (Dennis et al., 2010a; Sheline et al., 2010a; Machulda et al., 2011). In addition, one recent study found that resting-state connectivity measures of the DMN were able to differentiate between cognitively unimpaired ε4 carriers with a family history of Alzheimer’s disease and individuals without any apparent genetic risk with an effect size approximately three times greater than was possible using measures of task-related activation during a memory encoding paradigm (Fleisher et al., 2009). Such findings provide a powerful demonstration of the enhanced sensitivity provided by connectomic measures for indexing the physiological effects of disease risk variants, and support the contention that these variants often show higher penetrance at the level of distributed neural systems.
The preceding discussion illustrates how genetic variation can influence the nature and strength of connectivity between different brain regions. We now consider studies of genetic influences on how connections are arranged in the brain; i.e., how genes influence the topology of the connectome. Such an investigation is motivated by the sparse character of brain connectivity. The human cerebral cortex comprises an estimated 1011 neurons connected by 1013 fibers (Braitenberg and Schüz, 1991). Though numerous, these connections represent only a small fraction of the total 5 × 1021 connections that are possible. Similarly, neuronal connectivity in the nematode worm Caenorhabditis elegans, which has been comprehensively mapped at the level of each and every synapse, comprises <3% of the total number of connections possible while estimates of the degree of inter-regional connectivity (i.e., connectivity between distinct cytoarchitectonic regions) in the macaque monkey Macaca mulatta and the feline brain range from 18 to 38% (Latora and Marchiori, 2003; Kaiser and Hilgetag, 2006). This sparsity suggests that connections between brain regions develop according to specific constraints and/or design principles that are likely to be under genetic control.
Ramón y Cajal (1995) proposed over a century ago that one critical organizational principle for brain networks involves the minimization of wiring costs. The adaptive benefit of this principle is clear, as less wiring reduces the total energy required to support neuronal communication (Laughlin and Sejnowski, 2003). Accordingly, a substantial body of evidence has accumulated to suggest that pressure to minimize wiring costs can explain numerous aspects of cortical organization, including axonal branching and cortical folding patterns (Van Essen, 1997; Cherniak et al., 1999), neuronal morphology (Buzsaki et al., 2004; Chklovskii, 2004), the spatial location of neurons and cytoarchitectonic fields (Klyachko and Stevens, 2003; Cherniak et al., 2004), and even the fraction of cortical volume occupied by axons and dendrites (Chklovskii et al., 2002). However, wiring cost minimization alone cannot account for all design features of nervous system networks (Chen et al., 2006; Kaiser and Hilgetag, 2006), and recent evidence points to a trade-off between cost minimization and the emergence of topological properties such as communication efficiency, which may be behaviorally advantageous but entail a wiring cost premium (Kaiser and Hilgetag, 2006; Bassett et al., 2010).
The topological efficiency of a network can be readily quantified using graph theoretic techniques. In complex networks, communication is more efficient when fewer connections must be traversed to transfer information between any two nodes; the fewer the connections, the faster the rate of information transmission, and the lower the probability of signal degradation or transmission errors (Latora and Marchiori, 2003). In an economically wired network, long-range projections act as topological short-cuts that reduce the mean path length between regions and dramatically increase communication efficiency (Buzsaki et al., 2004; Kaiser and Hilgetag, 2006). However, these short-cuts come at the cost of increased wiring. In principle it would be possible to maximize network efficiency simply by adding more connections (Latora and Marchiori, 2003; Achard and Bullmore, 2007), though the metabolic costs associated with forming and maintaining each connection limit the total volume of wiring that can be supported (Laughlin and Sejnowski, 2003). This balance, between efficiency maximization and connection cost minimization, may be construed as one concerned with the optimization of connection cost-efficiency. The importance of this balance for the connectome was recently underscored by a study showing that the physical embedding of the brain’s connectivity architecture in the three-dimensional space of the skull is optimally cost-efficient, subject to certain higher-dimensional constraints (Bassett et al., 2010).
We examined the heritability of cost-efficient properties of functional brain network topology using resting-state fMRI in healthy twins (Fornito et al., 2011b). Network efficiency was measured using established graph theoretic methods (Latora and Marchiori, 2001), whereas connection costs were estimated indirectly using a measure of physical distance between regions. We found that the balance between these two measures, taken as an index of network cost-efficiency, was highly heritable at a global level, with genetic factors accounting for approximately 60% of the phenotypic variance. These influences were not uniformly distributed throughout the cortex, with regional estimates of significant genetic influences ranging from 0.30 to 0.80 (Figure 4). Some of the largest effects were seen in regions of lateral PFC and core components of the DMN, such as posterior cingulate and medial prefrontal cortices (Figure 4). Heritability estimates in these regions are comparable to those reported for whole-brain and regional gray matter volume (0.66 < h2 < 0.97; Thompson et al., 2001; Peper et al., 2007), cognitive abilities (0.30 < h2 < 0.80; McClearn et al., 1997; Bouchard, 1998; Boomsma et al., 2002), and various measures of personality and psychopathology (0.50 < h2 < 0.60; Boomsma et al., 2002). They also compare favorably with heritability estimates for simple measures of functional connectivity within the DMN (0.33 < h2 < 0.42; Glahn et al., 2010), as well as regional task-related activation (0.40 < h2 < 0.80; Koten et al., 2009; Blokland et al., 2011) and DTI-derived measures of anatomical connectivity (0.55 < h2 < 0.85; Chiang et al., 2009).
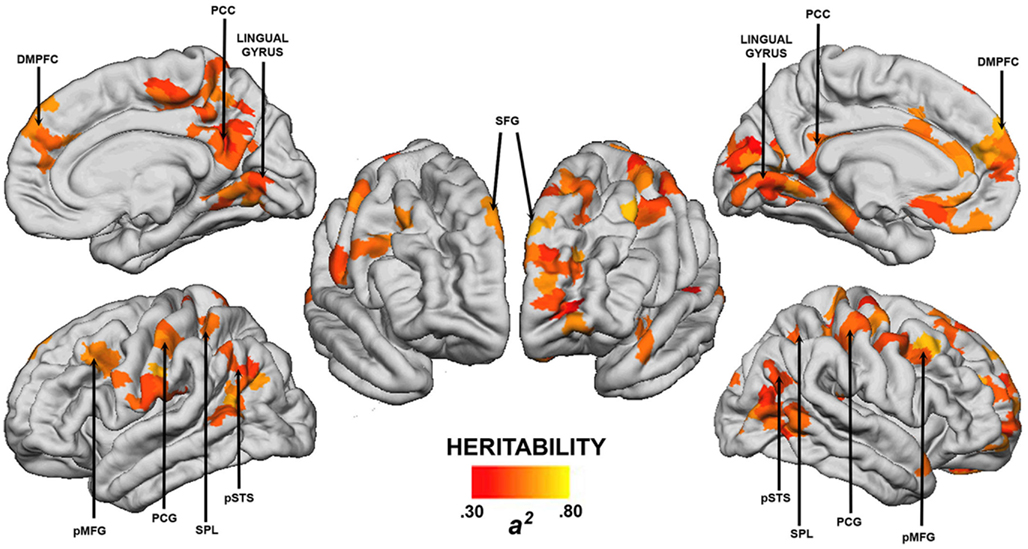
Figure 4. Cortical regions showing statistically significant (p < 0.05, corrected) heritability for regional cost-efficiency of network functional connectivity. Arrows highlight regions showing significant genetic effects in both cerebral hemispheres. DMPFC, dorso-medial prefrontal cortex; PCC, posterior cingulate cortex; SFG, superior frontal gyrus; pMFG, posterior middle frontal gyrus; PCG, post-central gyrus; SPL, superior parietal lobule; pSTS, posterior superior temporal sulcus. Left hemisphere is presented on the right. Image reproduced from Fornito et al. (2011b).
The adaptive benefit of a cost-efficient neural architecture is intuitive, and it is plausible that evolution would favor connections that can provide high communication efficiency for low metabolic cost. Evidence that greater topological efficiency and/or greater cost-efficiency predicts better performance on tests of intelligence and working memory (Bassett et al., 2009; Li et al., 2009; van den Heuvel et al., 2009; Zalesky et al., 2011), and that alterations of network efficiency and cost-efficiency are apparent in a range of patient groups, including schizophrenia (Bassett et al., 2009; Lynall et al., 2010; Zalesky et al., 2011), ADHD (Wang et al., 2009), and Alzheimer’s disease (Lo et al., 2010), indicates that these findings have clear implications for psychiatric disorders. To our knowledge however, no study to date has examined how variations in specific candidate genes influence brain network cost-efficiency, or any other topological properties of the connectome.
Cost-efficiency optimization is but one of many potential organizational principles for the connectome. Indeed, graph analytic studies of neuroimaging data have pointed to several other candidate topological properties that may be under genetic control. For example, both structural and functional human brain networks are characterized by a small-world topology (Achard et al., 2006; Hagmann et al., 2007), which concurrently supports locally segregated and globally integrated connectivity to provide high dynamical complexity, a property that may have been favored by evolutionary processes (Sporns et al., 2000). The human connectome also possesses a hierarchical, modular architecture; i.e., it can be decomposed into subsets of regions, termed modules, that show relatively high connectivity with each other than with other areas at multiple scales of resolution, allowing the formation of modules within modules and so on (Meunier et al., 2009; Bassett et al., 2010). This property has been associated with enhanced functional stability and diversity (Kaiser et al., 2007), the emergence of self-organized, critical dynamics (Rubinov et al., 2011), and may be intimately linked to optimization of connection cost-efficiency (Bassett et al., 2010).
These considerations raise questions as to whether genetic influences are specific to network cost-efficiency, or are related to other topological variations. In our analysis, we found no evidence for genetic influences on a range of other topological properties frequently studied in the graph theoretic literature, including clustering, path length, small-worldness, and global and local efficiency, although we did not examine modularity (Fornito et al., 2011b). Other studies have however reported heritability values as high as 0.80 for some of these measures, computed for functional networks derived from electroencephalographic (EEG) recordings (Smit et al., 2008). The reasons for these inconsistencies are unclear, but may be related to the imaging techniques employed or the enhanced power provided by the much larger sample analyzed in the EEG study. In general, the high degree of inter-correlation between different topological measures of brain network organization (Alexander-Bloch et al., 2010; Lynall et al., 2010), means that multivariate genetic analyses allowing categorization of different topological measures into distinct groupings defined by common genetic influences will be critical for identifying the primary genetic constraints on connectome development and organization.
This brief overview illustrates the potential power of imaging connectomics in the search for IPs for neuropsychiatric disorders. Specifically, the available data indicate that connectomic measures are able to index the physiological effects of disease risk genes when simpler regional measures of activation or volume, or cognition, cannot (Esslinger et al., 2009; Dennis et al., 2010a; Sheline et al., 2010a; Machulda et al., 2011; Rasetti et al., 2011). They also indicate that variations in cIPs correlate with relevant behavioral indices when regional measures do not (Pezawas et al., 2005; Buckholtz et al., 2008), and that certain connectomic phenotypes are highly heritable (Chiang et al., 2009; Glahn et al., 2010; Fornito et al., 2011b). Collectively, these data support the idea that the penetrance of many disease risk genes is higher at the level of distributed brain systems than at the level of isolated brain regions, a conclusion consistent with an emerging consensus that many psychiatric disorders arise from genetically mediated vulnerabilities in discrete neural circuits (Meyer-Lindenberg and Weinberger, 2006; Meyer-Lindenberg, 2009, 2010; Insel, 2011). As such, cIPs may offer greater sensitivity for characterizing genetic effects in imaging genetic designs than more traditional, regionally focused neuroimaging measures.
One potential criticism of cIPs is that, by design, they are more complicated, requiring additional processing steps and assumptions for their derivation. Characterizing activation or volume in a single region is intrinsically univariate, requiring measurement of one property describing the behavior of that region, whereas connectivity measures are intrinsically bi or multivariate as they involve two or more regions by definition. The problem is worse for graph analytic studies, which often incorporate multiple measures and processing steps when computing topological properties. Each additional measure and step can introduce noise into the analysis, making the resulting phenotypic characterizations less reliable (Habeck and Moeller, 2011), and placing an upper limit on power for detecting genetic effects (Kendler and Neale, 2010; Blokland et al., 2011). Moreover, more complex measures presumably have a more complex genetic basis. This emphasis on simplicity and reliability must however be balanced with a need to define valid and pathophysiologically relevant IPs. Most major psychiatric diseases are disorders of brain connectivity, and the physiological effects of putative disease risk variants are likely to be expressed throughout multiple, interconnected neural circuits. Connectomic measures will provide a more sensitive index of these effects. The findings discussed here support these conclusions, and demonstrate that even highly derived topological properties, such as network cost-efficiency, can show high heritability (Fornito et al., 2011b). Moreover, recent studies have shown that connectomic measures, despite their increased complexity, represent more robust biomarkers of schizophrenia (Calhoun et al., 2008; Erhardt et al., 2011; Bassett et al., 2012) and risk for Alzheimer’s disease (Fleisher et al., 2009), than simpler, regional measures. Thus, while parsimony and reliability are always important, candidate IPs should be selected for their reliability and their hypothesized role in relevant pathophysiological processes; not purely for their simplicity.
A more general criticism often leveled at the IP approach concerns whether any candidate IPs studied thus far are truly “intermediate” in the sense that they mediate the effects of candidate genetic variants on disease risk or expression (Walters and Owen, 2007; Kendler and Neale, 2010). Under such a mediation model, the risk gene is assumed to influence the IP, which in turn influences illness susceptibility (i.e., gene > IP > disorder); thus, the IP is truly interposed between gene and disorder. In this case the IP is likely to represent a physiological mechanism through which genetic risk is expressed. An alternative possibility however, is that the gene exerts pleiotropic and possibly independent effects on both the IP and disease risk (i.e., IP < gene > disorder). Under this liability model, the IP is not involved in disease pathophysiology but may represent a biomarker of illness susceptibility. Distinguishing between these two possibilities experimentally is challenging (Walters and Owen, 2007; Kendler and Neale, 2010) and it is thus unclear which category the cIPs discussed here fall into. Our use of the term “IP” in reference to the measures discussed here is therefore subject to this caveat. In practice however, the distinction may be overly simplistic as it assumes near-complete genetic overlap between the IP and psychiatric disorder. A more likely scenario is that IPs and psychiatric disorders posses both common and independent genetic influences which require multivariate analyses in genetically informative samples for complete characterization (Kendler and Neale, 2010). To our knowledge, no such studies of cIPs have yet been conducted.
Though it is as yet unclear whether any of the candidate IPs identified to date should be best conceptualized using mediation, liability, or multivariate models, they can certainly provide important clues as to how genetic risk factors influence variation in key phenotypic properties of psychiatric disease. Mapping differences and commonalities in these properties across disorders may prove useful in developing new, biologically informed diagnostic criteria. In addition, preliminary evidence suggests that IPs can augment the search for novel risk genes (Potkin et al., 2009). The research reviewed here indicates that this kind work will benefit from a greater focus on identifying novel IPs that describe systems-level properties of the brain, and that imaging connectomics provides a powerful methodological and conceptual framework for doing so.
Edward T. Bullmore is employed half-time by GlaxoSmithKline.
Alex Fornito was supported by a National Health and Medical Research Council CJ Martin Fellowship (ID: 454797).
Achard, S., and Bullmore, E. (2007). Efficiency and cost of economical brain functional networks. PLoS Comput. Biol. 3, e17. doi:10.1371/journal.pcbi.0030017
Achard, S., Salvador, R., Whitcher, B., Suckling, J., and Bullmore, E. (2006). A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. J. Neurosci. 26, 63–72.
Agrawal, A., and Lynskey, M. T. (2008). Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction 103, 1069–1081.
Alexander-Bloch, A. F., Gogtay, N., Meunier, D., Birn, R., Clasen, L., Lalonde, F., Lenroot, R., Giedd, J., and Bullmore, E. T. (2010). Disrupted modularity and local connectivity of brain functional networks in childhood-onset schizophrenia. Front. Syst. Neurosci. 4:147. doi:10.3389/fnsys.2010.00147
Ansorge, M. S., Zhou, M., Lira, A., Hen, R., and Gingrich, J. A. (2004). Early-life blockade of the 5-HT transporter alters emotional behavior in adult mice. Science 306, 879–881.
Barrat, A., Barthelemy, M., Pastor-Satorras, R., and Vespignani, A. (2004). The architecture of complex weighted networks. Proc. Natl. Acad. Sci. U.S.A. 101, 3747–3752.
Bassett, D. S., Bullmore, E., Verchinski, B. A., Mattay, V. S., Weinberger, D. R., and Meyer-Lindenberg, A. (2008). Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 28, 9239–9248.
Bassett, D. S., Bullmore, E. T., Meyer-Lindenberg, A., Apud, J. A., Weinberger, D. R., and Coppola, R. (2009). Cognitive fitness of cost-efficient brain functional networks. Proc. Natl. Acad. Sci. U.S.A. 106, 11747–11752.
Bassett, D. S., Greenfield, D. L., Meyer-Lindenberg, A., Weinberger, D. R., Moore, S. W., and Bullmore, E. T. (2010). Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput. Biol. 6, e1000748. doi:10.1371/journal.pcbi.1000748
Bassett, D. S., Nelson, B. G., Mueller, B. A., Camchong, J., and Lim, K. O. (2012). Altered resting state complexity in schizophrenia. Neuroimage 59, 2196–2207.
Beckmann, C. F., and Smith, S. M. (2004). Probabilistic independent component analysis for functional magnetic resonance imaging. IEEE Trans. Med. Imaging 23, 137–152.
Belmonte, M. K., Allen, G., Beckel-Mitchener, A., Boulanger, L. M., Carper, R. A., and Webb, S. J. (2004). Autism and abnormal development of brain connectivity. J. Neurosci. 24, 9228–9231.
Bennett, M. R. (2011). Schizophrenia: susceptibility genes, dendritic-spine pathology and gray matter loss. Prog. Neurobiol. 95, 275–300.
Bertolino, A., Rubino, V., Sambataro, F., Blasi, G., Latorre, V., Fazio, L., Caforio, G., Petruzzella, V., Kolachana, B., Hariri, A., Meyer-Lindenberg, A., Nardini, M., Weinberger, D. R., and Scarabino, T. (2006). Prefrontal-hippocampal coupling during memory processing is modulated by COMT val158met genotype. Biol. Psychiatry 60, 1250–1258.
Bertram, L., McQueen, M. B., Mullin, K., Blacker, D., and Tanzi, R. E. (2007). Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat. Genet. 39, 17–23.
Bertram, L., and Tanzi, R. E. (2008). Thirty years of Alzheimer’s disease genetics: the implications of systematic meta-analyses. Nat. Rev. Neurosci. 9, 768–778.
Biswal, B., Yetkin, F. Z., Haughton, V. M., and Hyde, J. S. (1995). Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541.
Blasi, G., Lo Bianco, L., Taurisano, P., Gelao, B., Romano, R., Fazio, L., Papazacharias, A., Di Giorgio, A., Caforio, G., Rampino, A., Masellis, R., Papp, A., Ursini, G., Sinibaldi, L., Popolizio, T., Sadee, W., and Bertolino, A. (2009). Functional variation of the dopamine D2 receptor gene is associated with emotional control as well as brain activity and connectivity during emotion processing in humans. J. Neurosci. 29, 14812–14819.
Blokland, G. A., McMahon, K. L., Thompson, P. M., Martin, N. G., de Zubicaray, G. I., and Wright, M. J. (2011). Heritability of working memory brain activation. J. Neurosci. 31, 10882–10890.
Bluhm, R. L., Miller, J., Lanius, R. A., Osuch, E. A., Boksman, K., Neufeld, R. W., Theberge, J., Schaefer, B., and Williamson, P. (2007). Spontaneous low-frequency fluctuations in the BOLD signal in schizophrenic patients: anomalies in the default network. Schizophr. Bull. 33, 1004–1012.
Boomsma, D., Busjahn, A., and Peltonen, L. (2002). Classical twin studies and beyond. Nat. Rev. Genet. 3, 872–882.
Bouchard, T. J. Jr. (1998). Genetic and environmental influences on adult intelligence and special mental abilities. Hum. Biol. 70, 257–279.
Braitenberg, V., and Schüz, A. (1991). Anatomy of the Cortex: Statistics and Geometry. Berlin: Springer.
Bressler, S. L., and Seth, A. K. (2011). Wiener-Granger causality: a well established methodology. Neuroimage 58, 323–329.
Buckholtz, J. W., Callicott, J. H., Kolachana, B., Hariri, A. R., Goldberg, T. E., Genderson, M., Egan, M. F., Mattay, V. S., Weinberger, D. R., and Meyer-Lindenberg, A. (2008). Genetic variation in MAOA modulates ventromedial prefrontal circuitry mediating individual differences in human personality. Mol. Psychiatry 13, 313–324.
Buckholtz, J. W., Meyer-Lindenberg, A., Honea, R. A., Straub, R. E., Pezawas, L., Egan, M. F., Vakkalanka, R., Kolachana, B., Verchinski, B. A., Sust, S., Mattay, V. S., Weinberger, D. R., and Callicott, J. H. (2007). Allelic variation in RGS4 impacts functional and structural connectivity in the human brain. J. Neurosci. 27, 1584–1593.
Buckner, R. L., Andrews-Hanna, J. R., and Schacter, D. L. (2008). The brain’s default network: anatomy, function, and relevance to disease. Ann. N. Y. Acad. Sci. 1124, 1–38.
Buckner, R. L., Sepulcre, J., Talukdar, T., Krienen, F. M., Liu, H., Hedden, T., Andrews-Hanna, J. R., Sperling, R. A., and Johnson, K. A. (2009). Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer’s disease. J. Neurosci. 29, 1860–1873.
Buckner, R. L., Snyder, A. Z., Shannon, B. J., LaRossa, G., Sachs, R., Fotenos, A. F., Sheline, Y. I., Klunk, W. E., Mathis, C. A., Morris, J. C., and Mintun, M. A. (2005). Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J. Neurosci. 25, 7709–7717.
Bullmore, E., Barnes, A., Bassett, D. S., Fornito, A., Kitzbichler, M., Meunier, D., and Suckling, J. (2009). Generic aspects of complexity in brain imaging data and other biological systems. Neuroimage 47, 1125–1134.
Bullmore, E., and Sporns, O. (2009). Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198.
Bullmore, E. T., and Bassett, D. S. (2011). Brain graphs: graphical models of the human brain connectome. Annu. Rev. Clin. Psychol. 7, 113–140.
Buzsaki, G., Geisler, C., Henze, D. A., and Wang, X. J. (2004). Interneuron diversity series: circuit complexity and axon wiring economy of cortical interneurons. Trends Neurosci. 27, 186–193.
Calhoun, V. D., Adali, T., and Pekar, J. J. (2004). A method for comparing group fMRI data using independent component analysis: application to visual, motor and visuomotor tasks. Magn. Reson. Imaging 22, 1181–1191.
Calhoun, V. D., Maciejewski, P. K., Pearlson, G. D., and Kiehl, K. A. (2008). Temporal lobe and “default” hemodynamic brain modes discriminate between schizophrenia and bipolar disorder. Hum. Brain Mapp. 29, 1265–1275.
Canli, T., Qiu, M., Omura, K., Congdon, E., Haas, B. W., Amin, Z., Herrmann, M. J., Constable, R. T., and Lesch, K. P. (2006). Neural correlates of epigenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 16033–16038.
Cannon, T. D., Kaprio, J., Lönnqvist, J., Huttunen, M., and Koskenvuo, M. (1998). The genetic epidemiology of schizophrenia in a Finnish twin cohort: a population-based modeling study. Arch. Gen. Psychiatry 55, 67–74.
Cannon, T. D., and Keller, M. C. (2006). Endophenotypes in the genetic analyses of mental disorders. Annu. Rev. Clin. Psychol. 2, 267–290.
Cardno, A. G., Marshall, E. J., Cold, B., MacDonald, A. M., Ribchester, T. R., Davies, N. J., Venturi, P., Jones, L. A., Lewis, S. W., Sham, P. C., Gottesman, I. I., Farmer, A. E., McGuffin, P., Reveley, A. M., and Murray, R. M. (1999). Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch. Gen. Psychiatry 56, 162–168.
Caspi, A., Hariri, A. R., Holmes, A., Uher, R., and Moffitt, T. E. (2010). Genetic sensitivity to the environment: the case of the serotonin transporter gene and its implications for studying complex diseases and traits. Am. J. Psychiatry 167, 509–527.
Caspi, A., McClay, J., Moffitt, T. E., Mill, J., Martin, J., Craig, I. W., Taylor, A., and Poulton, R. (2002). Role of genotype in the cycle of violence in maltreated children. Science 297, 851–854.
Castellanos, F. X., Margulies, D. S., Kelly, C., Uddin, L. Q., Ghaffari, M., Kirsch, A., Shaw, D., Shehzad, Z., Di Martino, A., Biswal, B., Sonuga-Barke, E. J., Rotrosen, J., Adler, L. A., and Milham, M. P. (2008). Cingulate-precuneus interactions: a new locus of dysfunction in adult attention-deficit/hyperactivity disorder. Biol. Psychiatry 63, 332–337.
Chen, B. L., Hall, D. H., and Chklovskii, D. B. (2006). Wiring optimization can relate neuronal structure and function. Proc. Natl. Acad. Sci. U.S.A. 103, 4723–4728.
Cherniak, C., Changizi, M., and Won Kang, D. (1999). Large-scale optimization of neuron arbors. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 59, 6001–6009.
Cherniak, C., Mokhtarzada, Z., Rodriguez-Esteban, R., and Changizi, K. (2004). Global optimization of cerebral cortex layout. Proc. Natl. Acad. Sci. U.S.A. 101, 1081–1086.
Chiang, M. C., Barysheva, M., Shattuck, D. W., Lee, A. D., Madsen, S. K., Avedissian, C., Klunder, A. D., Toga, A. W., McMahon, K. L., de Zubicaray, G. I., Wright, M. J., Srivastava, A., Balov, N., and Thompson, P. M. (2009). Genetics of brain fiber architecture and intellectual performance. J. Neurosci. 29, 2212–2224.
Chklovskii, D. B. (2004). Synaptic connectivity and neuronal morphology: two sides of the same coin. Neuron 43, 609–617.
Chklovskii, D. B., Schikorski, T., and Stevens, C. F. (2002). Wiring optimization in cortical circuits. Neuron 34, 341–347.
Chung, H. J., Lee, J. Y., Deocaris, C. C., Min, H., Kim, S. H., and Kim, M. H. (2010). Mouse homologue of the schizophrenia susceptibility gene ZNF804A as a target of Hoxc8. J. Biomed. Biotechnol. 2010, 231708.
Clare Kelly, A. M., Uddin, L. Q., Biswal, B. B., Castellanos, F. X., and Milham, M. P. (2008). Competition between functional brain networks mediates behavioral variability. Neuroimage 39, 527–537.
Clarke, H., Flint, J., Attwood, A. S., and Munafo, M. R. (2010). Association of the 5-HTTLPR genotype and unipolar depression: a meta-analysis. Psychol. Med. 40, 1767–1778.
Curcic-Blake, B., Swart, M., Ter Horst, G. J., Langers, D. R., Kema, I. P., and Aleman, A. (2012). Variation of the gene coding for DARPP-32 (PPP1R1B) and brain connectivity during associative emotional learning. Neuroimage 59, 1540–1550.
Damoiseaux, J. S., Rombouts, S. A., Barkhof, F., Scheltens, P., Stam, C. J., Smith, S. M., and Beckmann, C. F. (2006). Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. U.S.A. 103, 13848–13853.
Dennis, N. A., Browndyke, J. N., Stokes, J., Need, A., Burke, J. R., Welsh-Bohmer, K. A., and Cabeza, R. (2010a). Temporal lobe functional activity and connectivity in young adult APOE varepsilon4 carriers. Alzheimers Dement. 6, 303–311.
Dennis, N. A., Need, A. C., LaBar, K. S., Waters-Metenier, S., Cirulli, E. T., Kragel, J., Goldstein, D. B., and Cabeza, R. (2010b). COMT val108/158 met genotype affects neural but not cognitive processing in healthy individuals. Cereb. Cortex 20, 672–683.
Ellison-Wright, I., and Bullmore, E. (2009). Meta-analysis of diffusion tensor imaging studies in schizophrenia. Schizophr. Res. 108, 3–10.
Erhardt, E. B., Allen, E. A., Damaraju, E., and Calhoun, V. D. (2011). On network derivation, classification and visualization: a response to habeck and moeller. Brain Connect. 1, 105–110.
Erk, S., Meyer-Lindenberg, A., Schnell, K., Opitz von Boberfeld, C., Esslinger, C., Kirsch, P., Grimm, O., Arnold, C., Haddad, L., Witt, S. H., Cichon, S., Nothen, M. M., Rietschel, M., and Walter, H. (2010). Brain function in carriers of a genome-wide supported bipolar disorder variant. Arch. Gen. Psychiatry 67, 803–811.
Esslinger, C., Kirsch, P., Haddad, L., Mier, D., Sauer, C., Erk, S., Schnell, K., Arnold, C., Witt, S. H., Rietschel, M., Cichon, S., Walter, H., and Meyer-Lindenberg, A. (2011). Cognitive state and connectivity effects of the genome-wide significant psychosis variant in ZNF804A. Neuroimage 54, 2514–2523.
Esslinger, C., Walter, H., Kirsch, P., Erk, S., Schnell, K., Arnold, C., Haddad, L., Mier, D., Opitz von Boberfeld, C., Raab, K., Witt, S. H., Rietschel, M., Cichon, S., and Meyer-Lindenberg, A. (2009). Neural mechanisms of a genome-wide supported psychosis variant. Science 324, 605.
Farrer, L. A., Cupples, L. A., Haines, J. L., Hyman, B., Kukull, W. A., Mayeux, R., Myers, R. H., Pericak-Vance, M. A., Risch, N., and van Duijn, C. M. (1997). Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA 278, 1349–1356.
Filippini, N., MacIntosh, B. J., Hough, M. G., Goodwin, G. M., Frisoni, G. B., Smith, S. M., Matthews, P. M., Beckmann, C. F., and Mackay, C. E. (2009). Distinct patterns of brain activity in young carriers of the APOE-epsilon4 allele. Proc. Natl. Acad. Sci. U.S.A. 106, 7209–7214.
Fleisher, A. S., Sherzai, A., Taylor, C., Langbaum, J. B., Chen, K., and Buxton, R. B. (2009). Resting-state BOLD networks versus task-associated functional MRI for distinguishing Alzheimer’s disease risk groups. Neuroimage 47, 1678–1690.
Flint, J., and Munafo, M. R. (2007). The endophenotype concept in psychiatric genetics. Psychol. Med. 37, 163–180.
Fornito, A., and Bullmore, E. T. (2010). What can spontaneous fluctuations of the blood oxygenation-level-dependent signal tell us about psychiatric disorders? Curr. Opin. Psychiatry 23, 239–249.
Fornito, A., Yoon, J., Zalesky, A., Bullmore, E. T., and Carter, C. S. (2011a). General and specific functional connectivity disturbances in first-episode schizophrenia during cognitive control performance. Biol. Psychiatry 70, 64–72.
Fornito, A., Zalesky, A., Bassett, D. S., Meunier, D., Ellison-Wright, I., Yucel, M., Wood, S. J., Shaw, K., O’Connor, J., Nertney, D., Mowry, B. J., Pantelis, C., and Bullmore, E. T. (2011b). Genetic influences on cost-efficient organization of human cortical functional networks. J. Neurosci. 31, 3261–3270.
Fornito, A., Yucel, M., Patti, J., Wood, S. J., and Pantelis, C. (2009). Mapping grey matter reductions in schizophrenia: an anatomical likelihood estimation analysis of voxel-based morphometry studies. Schizophr. Res. 108, 104–113.
Fornito, A., Zalesky, A., Pantelis, C., and Bullmore, E. T. (2012). Schizophrenia, neuroimaging and connectomics. Neuroimage. doi:10.1016/j.neuroimage.2011.12.090
Fox, M. D., and Raichle, M. E. (2007). Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat. Rev. Neurosci. 8, 700–711.
Fox, M. D., Snyder, A. Z., Vincent, J. L., and Raichle, M. E. (2007). Intrinsic fluctuations within cortical systems account for intertrial variability in human behavior. Neuron 56, 171–184.
Fox, M. D., Snyder, A. Z., Zacks, J. M., and Raichle, M. E. (2006). Coherent spontaneous activity accounts for trial-to-trial variability in human evoked brain responses. Nat. Neurosci. 9, 23–25.
Friston, K. J. (1994). Functional and effective connectivity in neuroimaging: a synthesis. Hum. Brain Mapp. 2, 56–78.
Friston, K. J., Buechel, C., Fink, G. R., Morris, J., Rolls, E., and Dolan, R. J. (1997). Psychophysiological and modulatory interactions in neuroimaging. Neuroimage 6, 218–229.
Friston, K. J., Harrison, L., and Penny, W. (2003). Dynamic causal modelling. Neuroimage 19, 1273–1302.
Gai, X., Xie, H. M., Perin, J. C., Takahashi, N., Murphy, K., Wenocur, A. S., D’Arcy, M., O’Hara, R. J., Goldmuntz, E., Grice, D. E., Shaikh, T. H., Hakonarson, H., Buxbaum, J. D., Elia, J., and White, P. S. (2012). Rare structural variation of synapse and neurotransmission genes in autism. Mol. Psychiatry 17, 402–411.
Gibbons, A. S., Udawela, M., Jeon, W. J., Seo, M. S., Brooks, L., and Dean, B. (2011). The neurobiology of APOE in schizophrenia and mood disorders. Front. Biosci. 16, 962–979.
Glahn, D. C., Winkler, A. M., Kochunov, P., Almasy, L., Duggirala, R., Carless, M. A., Curran, J. C., Olvera, R. L., Laird, A. R., Smith, S. M., Beckmann, C. F., Fox, P. T., and Blangero, J. (2010). Genetic control over the resting brain. Proc. Natl. Acad. Sci. U.S.A. 107, 1223–1228.
Gogtay, N., Vyas, N. S., Testa, R., Wood, S. J., and Pantelis, C. (2011). Age of onset of schizophrenia: perspectives from structural neuroimaging studies. Schizophr. Bull. 37, 504–513.
Gottesman, I. I. (1991). Schizophrenia Genesis: The Origins of Madness. New York: W.H. Freeman and Company.
Gottesman, I. I., and Gould, T. D. (2003). The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry 160, 1–10.
Greicius, M. D., Flores, B. H., Menon, V., Glover, G. H., Solvason, H. B., Kenna, H., Reiss, A. L., and Schatzberg, A. F. (2007). Resting-state functional connectivity in major depression: abnormally increased contributions from subgenual cingulate cortex and thalamus. Biol. Psychiatry 62, 429–437.
Greicius, M. D., Srivastava, G., Reiss, A. L., and Menon, V. (2004). Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc. Natl. Acad. Sci. U.S.A. 101, 4637–4642.
Habeck, C., and Moeller, J. R. (2011). Intinsic functional-connectivity networks for diagnosis: just beautiful pictures? Brain Connect. 1, 99–103.
Hagmann, P., Cammoun, L., Gigandet, X., Meuli, R., Honey, C. J., Wedeen, V. J., and Sporns, O. (2008). Mapping the structural core of human cerebral cortex. PLoS Biol. 6, e159. doi:10.1371/journal.pbio.0060159
Hagmann, P., Kurant, M., Gigandet, X., Thiran, P., Wedeen, V. J., Meuli, R., and Thiran, J. P. (2007). Mapping human whole-brain structural networks with diffusion MRI. PLoS ONE 2, e597. doi:10.1371/journal.pone.0000597
Hallmayer, J., Cleveland, S., Torres, A., Phillips, J., Cohen, B., Torigoe, T., Miller, J., Fedele, A., Collins, J., Smith, K., Lotspeich, L., Croen, L. A., Ozonoff, S., Lajonchere, C., Grether, J. K., and Risch, N. (2011). Genetic heritability and shared environmental factors among twins pairs with autism. Arch. Gen. Psychiatry 68, 1095–1102.
Hariri, A. R., Mattay, V. S., Tessitore, A., Kolachana, B., Fera, F., Goldman, D., Egan, M. F., and Weinberger, D. R. (2002). Serotonin transporter genetic variation and the response of the human amygdala. Science 297, 400–403.
Harrison, B. J., Pujol, J., Contreras-Rodriguez, O., Soriano-Mas, C., Lopez-Sola, M., Deus, J., Ortiz, H., Blanco-Hinojo, L., Alonso, P., Hernandez-Ribas, R., Cardoner, N., and Menchon, J. M. (2011). Task-induced deactivation from rest extends beyond the default mode brain network. PLoS ONE 6, e22964. doi:10.1371/journal.pone.0022964
Harrison, B. J., Pujol, J., Lopez-Sola, M., Hernandez-Ribas, R., Deus, J., Ortiz, H., Soriano-Mas, C., Yucel, M., Pantelis, C., and Cardoner, N. (2008). Consistency and functional specialization in the default mode brain network. Proc. Natl. Acad. Sci. U.S.A. 105, 9781–9786.
Harrison, P. J., and Weinberger, D. R. (2005). Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol. Psychiatry 10, 40–68.
He, B. J., Snyder, A. Z., Zempel, J. M., Smyth, M. D., and Raichle, M. E. (2008). Electrophysiological correlates of the brain’s intrinsic large-scale functional architecture. Proc. Natl. Acad. Sci. U.S.A. 105, 16039–16044.
He, Y., Chen, Z. J., and Evans, A. C. (2007). Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cereb. Cortex 17, 2407–2419.
Heils, A., Teufel, A., Petri, S., Stober, G., Riederer, P., Bengel, D., and Lesch, K. P. (1996). Allelic variation of human serotonin transporter gene expression. J. Neurochem. 66, 2621–2624.
Heinz, A., Braus, D. F., Smolka, M. N., Wrase, J., Puls, I., Hermann, D., Klein, S., Grusser, S. M., Flor, H., Schumann, G., Mann, K., and Buchel, C. (2005). Amygdala-prefrontal coupling depends on a genetic variation of the serotonin transporter. Nat. Neurosci. 8, 20–21.
Heinz, A., Jones, D. W., Mazzanti, C., Goldman, D., Ragan, P., Hommer, D., Linnoila, M., and Weinberger, D. R. (2000). A relationship between serotonin transporter genotype and in vivo protein expression and alcohol neurotoxicity. Biol. Psychiatry 47, 643–649.
Hesselmann, G., Kell, C. A., Eger, E., and Kleinschmidt, A. (2008). Spontaneous local variations in ongoing neural activity bias perceptual decisions. Proc. Natl. Acad. Sci. U.S.A. 105, 10984–10989.
Hettema, J. M., Neale, M. C., and Kendler, K. S. (2001). A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am. J. Psychiatry 158, 1568–1578.
Hill, M. J., and Bray, N. J. (2011). Allelic differences in nuclear protein binding at a genome-wide significant risk variant for schizophrenia in ZNF804A. Mol. Psychiatry 16, 787–789.
Hong, L. E., Hodgkinson, C. A., Yang, Y., Sampath, H., Ross, T. J., Buchholz, B., Salmeron, B. J., Srivastava, V., Thaker, G. K., Goldman, D., and Stein, E. A. (2010). A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc. Natl. Acad. Sci. U.S.A. 107, 13509–13514.
Jbabdi, S., and Johansen-Berg, H. (2011). Tractography: where do we go from here? Brain Connect. 1, 169–183.
Jedema, H. P., Gianaros, P. J., Greer, P. J., Kerr, D. D., Liu, S., Higley, J. D., Suomi, S. J., Olsen, A. S., Porter, J. N., Lopresti, B. J., Hariri, A. R., and Bradberry, C. W. (2010). Cognitive impact of genetic variation of the serotonin transporter in primates is associated with differences in brain morphology rather than serotonin neurotransmission. Mol. Psychiatry 15, 512–522, 446.
Kaiser, M., Gorner, M., and Hilgetag, C. C. (2007). Criticality of spreading dynamics in hierarchical cluster networks without inhibition. New J. Phys. 9, 2–13.
Kaiser, M., and Hilgetag, C. C. (2006). Nonoptimal component placement, but short processing paths, due to long-distance projections in neural systems. PLoS Comput. Biol. 2, e95. doi:10.1371/journal.pcbi.0020095
Kempf, L., Nicodemus, K. K., Kolachana, B., Vakkalanka, R., Verchinski, B. A., Egan, M. F., Straub, R. E., Mattay, V. A., Callicott, J. H., Weinberger, D. R., and Meyer-Lindenberg, A. (2008). Functional polymorphisms in PRODH are associated with risk and protection for schizophrenia and fronto-striatal structure and function. PLoS Genet. 4, e1000252. doi:10.1371/journal.pgen.1000252
Kendler, K. S. (2006). Reflections on the relationship between psychiatric genetics and psychiatric nosology. Am. J. Psychiatry 163, 1138–1146.
Kendler, K. S., Gatz, M., Gardner, C. O., and Pedersen, N. L. (2006). A Swedish national twin study of lifetime major depression. Am. J. Psychiatry 163, 109–114.
Kendler, K. S., and Neale, M. C. (2010). Endophenotype: a conceptual analysis. Mol. Psychiatry 15, 789–797.
Kendler, K. S., Neale, M. C., Kessler, R. C., Heath, A. C., and Eaves, L. J. (1993). The lifetime history of major depression in women. Reliability of diagnosis and heritability. Arch. Gen. Psychiatry 50, 863–870.
Kennedy, D. P., and Courchesne, E. (2008). The intrinsic functional organization of the brain is altered in autism. Neuroimage 39, 1877–1885.
Kim-Cohen, J., Caspi, A., Taylor, A., Williams, B., Newcombe, R., Craig, I. W., and Moffitt, T. E. (2006). MAOA, maltreatment, and gene-environment interaction predicting children’s mental health: new evidence and a meta-analysis. Mol. Psychiatry 11, 903–913.
Klyachko, V. A., and Stevens, C. F. (2003). Connectivity optimization and the positioning of cortical areas. Proc. Natl. Acad. Sci. U.S.A. 100, 7937–7941.
Konrad, A., Vucurevic, G., Musso, F., Stoeter, P., Dahmen, N., and Winterer, G. (2009). ErbB4 genotype predicts left frontotemporal structural connectivity in human brain. Neuropsychopharmacology 34, 641–650.
Koten, J. W. Jr., Wood, G., Hagoort, P., Goebel, R., Propping, P., Willmes, K., and Boomsma, D. I. (2009). Genetic contribution to variation in cognitive function: an FMRI study in twins. Science 323, 1737–1740.
Krugel, L. K., Biele, G., Mohr, P. N., Li, S. C., and Heekeren, H. R. (2009). Genetic variation in dopaminergic neuromodulation influences the ability to rapidly and flexibly adapt decisions. Proc. Natl. Acad. Sci. U.S.A. 106, 17951–17956.
Latora, V., and Marchiori, M. (2001). Efficient behavior of small-world networks. Phys. Rev. Lett. 87, 198701.
Latora, V., and Marchiori, M. (2003). Economic small-world behavior in weighted networks. Eur. Phys. J. B 32, 249–263.
Laughlin, S. B., and Sejnowski, T. J. (2003). Communication in neuronal networks. Science 301, 1870–1874.
Lesch, K. P., Bengel, D., Heils, A., Sabol, S. Z., Greenberg, B. D., Petri, S., Benjamin, J., Muller, C. R., Hamer, D. H., and Murphy, D. L. (1996). Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 274, 1527–1531.
Li, D., and He, L. (2007). Meta-analysis supports association between serotonin transporter (5-HTT) and suicidal behavior. Mol. Psychiatry 12, 47–54.
Li, Y., Liu, Y., Li, J., Qin, W., Li, K., Yu, C., and Jiang, T. (2009). Brain anatomical network and intelligence. PLoS Comput. Biol. 5, e1000395. doi:10.1371/journal.pcbi.1000395
Lips, E. S., Cornelisse, L. N., Toonen, R. F., Min, J. L., Hultman, C. M., Holmans, P. A., O’Donovan, M. C., Purcell, S. M., Smit, A. B., Verhage, M., Sullivan, P. F., Visscher, P. M., and Posthuma, D. (2011). Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol. Psychiatry.
Lo, C. Y., Wang, P. N., Chou, K. H., Wang, J., He, Y., and Lin, C. P. (2010). Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer’s disease. J. Neurosci. 30, 16876–16885.
Lynall, M. E., Bassett, D. S., Kerwin, R., McKenna, P. J., Kitzbichler, M., Muller, U., and Bullmore, E. (2010). Functional connectivity and brain networks in schizophrenia. J. Neurosci. 30, 9477–9487.
Machulda, M. M., Jones, D. T., Vemuri, P., McDade, E., Avula, R., Przybelski, S., Boeve, B. F., Knopman, D. S., Petersen, R. C., and Jack, C. R. Jr. (2011). Effect of APOE epsilon4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch. Neurol. 68, 1131–1136.
Mahley, R. W., and Rall, S. C. Jr. (2000). Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 1, 507–537.
Margulies, D. S., Bottger, J., Long, X., Lv, Y., Kelly, C., Schafer, A., Goldhahn, D., Abbushi, A., Milham, M. P., Lohmann, G., and Villringer, A. (2010). Resting developments: a review of fMRI post-processing methodologies for spontaneous brain activity. MAGMA 23, 289–307.
McClearn, G. E., Johansson, B., Berg, S., Pedersen, N. L., Ahern, F., Petrill, S. A., and Plomin, R. (1997). Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science 276, 1560–1563.
McGuffin, P., Rijsdijk, F., Andrew, M., Sham, P., Katz, R., and Cardno, A. (2003). The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch. Gen. Psychiatry 60, 497–502.
McHugh, R. K., Hofmann, S. G., Asnaani, A., Sawyer, A. T., and Otto, M. W. (2010). The serotonin transporter gene and risk for alcohol dependence: a meta-analytic review. Drug Alcohol Depend. 108, 1–6.
Mennes, M., Zuo, X. N., Kelly, C., Di Martino, A., Zang, Y. F., Biswal, B., Castellanos, F. X., and Milham, M. P. (2011). Linking inter-individual differences in neural activation and behavior to intrinsic brain dynamics. Neuroimage 54, 2950–2959.
Menzies, L., Chamberlain, S. R., Laird, A. R., Thelen, S. M., Sahakian, B. J., and Bullmore, E. T. (2008). Integrating evidence from neuroimaging and neuropsychological studies of obsessive-compulsive disorder: the orb-itofronto-striatal model revisited. Neurosci. Biobehav. Rev. 32, 525–549.
Meunier, D., Lambiotte, R., Fornito, A., Ersche, K. D., and Bullmore, E. T. (2009). Hierarchical modularity in human brain functional networks. Front. Neuroinformatics 3, 37.
Meyer-Lindenberg, A. (2009). Neural connectivity as an intermediate phenotype: brain networks under genetic control. Hum. Brain Mapp. 30, 1938–1946.
Meyer-Lindenberg, A. (2010). From maps to mechanisms through neuroimaging of schizophrenia. Nature 468, 194–202.
Meyer-Lindenberg, A., Straub, R. E., Lipska, B. K., Verchinski, B. A., Goldberg, T., Callicott, J. H., Egan, M. F., Huffaker, S. S., Mattay, V. S., Kolachana, B., Kleinman, J. E., and Weinberger, D. R. (2007). Genetic evidence implicating DARPP-32 in human frontostriatal structure, function, and cognition. J. Clin. Invest. 117, 672–682.
Meyer-Lindenberg, A., and Weinberger, D. R. (2006). Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat. Rev. Neurosci. 7, 818–827.
Meyer-Lindenberg, A. S., Olsen, R. K., Kohn, P. D., Brown, T., Egan, M. F., Weinberger, D. R., and Berman, K. F. (2005). Regionally specific disturbance of dorsolateral prefrontal-hippocampal functional connectivity in schizophrenia. Arch. Gen. Psychiatry 62, 379–386.
Minzenberg, M. J., Laird, A. R., Thelen, S., Carter, C. S., and Glahn, D. C. (2009). Meta-analysis of 41 functional neuroimaging studies of executive function in schizophrenia. Arch. Gen. Psychiatry 66, 811–822.
Morcom, A. M., and Fletcher, P. C. (2007). Does the brain have a baseline? Why we should be resisting a rest. Neuroimage 37, 1073–1082.
Mori, S., and Zhang, J. (2006). Principles of diffusion tensor imaging and its applications to basic neuroscience research. Neuron 51, 527–539.
Mukherjee, P., Whalley, H. C., McKirdy, J. W., McIntosh, A. M., Johnstone, E. C., Lawrie, S. M., and Hall, J. (2011). Effects of the BDNF Val66Met polymorphism on neural responses to facial emotion. Psychiatry Res. 191, 182–188.
Munafo, M. R., Freimer, N. B., Ng, W., Ophoff, R., Veijola, J., Miettunen, J., Jarvelin, M. R., Taanila, A., and Flint, J. (2009). 5-HTTLPR genotype and anxiety-related personality traits: a meta-analysis and new data. Am. J. Med. Genet. B Neuropsychiatr. Genet. 150B, 271–281.
Murthy, N. V., Selvaraj, S., Cowen, P. J., Bhagwagar, Z., Riedel, W. J., Peers, P., Kennedy, J. L., Sahakian, B. J., Laruelle, M. A., Rabiner, E. A., and Grasby, P. M. (2010). Serotonin transporter polymorphisms (SLC6A4 insertion/deletion and rs25531) do not affect the availability of 5-HTT to [11C] DASB binding in the living human brain. Neuroimage 52, 50–54.
Nikulina, V., Widom, C. S., and Brzustowicz, L. M. (2012). Child abuse and neglect, MAOA, and mental health outcomes: a prospective examination. Biol. Psychiatry 71, 350–357.
O’Donovan, M. C., Craddock, N., Norton, N., Williams, H., Peirce, T., Moskvina, V., Nikolov, I., Hamshere, M., Carroll, L., Georgieva, L., Dwyer, S., Holmans, P., Marchini, J. L., Spencer, C. C., Howie, B., Leung, H. T., Hartmann, A. M., Moller, H. J., Morris, D. W., Shi, Y., Feng, G., Hoffmann, P., Propping, P., Vasilescu, C., Maier, W., Rietschel, M., Zammit, S., Schumacher, J., Quinn, E. M., Schulze, T. G., Williams, N. M., Giegling, I., Iwata, N., Ikeda, M., Darvasi, A., Shifman, S., He, L., Duan, J., Sanders, A. R., Levinson, D. F., Gejman, P. V., Cichon, S., Nothen, M. M., Gill, M., Corvin, A., Rujescu, D., Kirov, G., Owen, M. J., Buccola, N. G., Mowry, B. J., Freedman, R., Amin, F., Black, D. W., Silverman, J. M., Byerley, W. F., and Cloninger, C. R. (2008). Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 40, 1053–1055.
Pacheco, J., Beevers, C. G., Benavides, C., McGeary, J., Stice, E., and Schnyer, D. M. (2009). Frontal-limbic white matter pathway associations with the serotonin transporter gene promoter region (5-HTTLPR) polymorphism. J. Neurosci. 29, 6229–6233.
Pantelis, C., Yucel, M., Bora, E., Fornito, A., Testa, R., Brewer, W. J., Velakoulis, D., and Wood, S. J. (2009). Neurobiological markers of illness onset in psychosis and schizophrenia: the search for a moving target. Neuropsychol. Rev. 19, 385–398.
Papagni, S. A., Mechelli, A., Prata, D. P., Kambeitz, J., Fu, C. H., Picchioni, M., Walshe, M., Toulopoulou, T., Bramon, E., Murray, R. M., Collier, D. A., Bellomo, A., and McGuire, P. (2011). Differential effects of DAAO on regional activation and functional connectivity in schizophrenia, bipolar disorder and controls. Neuroimage 56, 2283–2291.
Peper, J. S., Brouwer, R. M., Boomsma, D. I., Kahn, R. S., and Hulshoff Pol, H. E. (2007). Genetic influences on human brain structure: a review of brain imaging studies in twins. Hum. Brain Mapp. 28, 464–473.
Pettersson-Yeo, W., Allen, P., Benetti, S., McGuire, P., and Mechelli, A. (2011). Dysconnectivity in schizophrenia: where are we now? Neurosci. Biobehav. Rev. 35, 1110–1124.
Pezawas, L., Meyer-Lindenberg, A., Drabant, E. M., Verchinski, B. A., Munoz, K. E., Kolachana, B. S., Egan, M. F., Mattay, V. S., Hariri, A. R., and Weinberger, D. R. (2005). 5-HTTLPR polymorphism impacts human cingulate-amygdala interactions: a genetic susceptibility mechanism for depression. Nat. Neurosci. 8, 828–834.
Phillips, M. L., Drevets, W. C., Rauch, S. L., and Lane, R. (2003a). Neurobiology of emotion perception I: the neural basis of normal emotion perception. Biol. Psychiatry 54, 504–514.
Phillips, M. L., Drevets, W. C., Rauch, S. L., and Lane, R. (2003b). Neurobiology of emotion perception II: implications for major psychiatric disorders. Biol. Psychiatry 54, 515–528.
Potkin, S. G., Turner, J. A., Fallon, J. A., Lakatos, A., Keator, D. B., Guffanti, G., and Macciardi, F. (2009). Gene discovery through imaging genetics: identification of two novel genes associated with schizophrenia. Mol. Psychiatry 14, 416–428.
Prata, D. P., Papagni, S. A., Mechelli, A., Fu, C. H., Kambeitz, J., Picchioni, M., Kane, F., Kalidindi, S., McDonald, C., Kravariti, E., Toulopoulou, T., Bramon, E., Walshe, M., Murray, R., Collier, D. A., and McGuire, P. K. (2012). Effect of D-amino acid oxidase activator (DAOA; G72) on brain function during verbal fluency. Hum. Brain Mapp. 33, 143–153.
Purcell, S. M., Wray, N. R., Stone, J. L., Visscher, P. M., O’Donovan, M. C., Sullivan, P. F., and Sklar, P. (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752.
Rasch, B., Spalek, K., Buholzer, S., Luechinger, R., Boesiger, P., de Quervain, D. J., and Papassotiropoulos, A. (2010). Aversive stimuli lead to differential amygdala activation and connectivity patterns depending on catechol-O-methyltransferase Val158Met genotype. Neuroimage 52, 1712–1719.
Rasch, B., Spalek, K., Buholzer, S., Luechinger, R., Boesiger, P., Papassotiropoulos, A., and de Quervain, D. J. (2009). A genetic variation of the noradrenergic system is related to differential amygdala activation during encoding of emotional memories. Proc. Natl. Acad. Sci. U.S.A. 106, 19191–19196.
Rasetti, R., Sambataro, F., Chen, Q., Callicott, J. H., Mattay, V. S., and Weinberger, D. R. (2011). Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch. Gen. Psychiatry 68, 1207–1217.
Riley, B., Thiselton, D., Maher, B. S., Bigdeli, T., Wormley, B., McMichael, G. O., Fanous, A. H., Vladimirov, V., O’Neill, F. A., Walsh, D., and Kendler, K. S. (2010). Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of schizophrenia sample. Mol. Psychiatry 15, 29–37.
Robinson, E. B., Koenen, K. C., McCormick, M. C., Munir, K., Hallett, V., Happè, F., Plomin, R., and Ronald, A. (2012). A mutlivariate twin study of autistic traits in 12-year olds: testing the fractionable autism triad hypothesis. Behav. Genet. 42, 245–255.
Rubinov, M., and Sporns, O. (2010). Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069.
Rubinov, M., Sporns, O., Thivierge, J. P., and Breakspear, M. (2011). Neurobiologically realistic determinants of self-organized criticality in networks of spiking neurons. PLoS Comput. Biol. 7, e1002038. doi:10.1371/journal.pcbi.1002038
Sabol, S. Z., Hu, S., and Hamer, D. (1998). A functional polymorphism in the monoamine oxidase A gene promoter. Hum. Genet. 103, 273–279.
Schardt, D. M., Erk, S., Nusser, C., Nothen, M. M., Cichon, S., Rietschel, M., Treutlein, J., Goschke, T., and Walter, H. (2010). Volition diminishes genetically mediated amygdala hyperreactivity. Neuroimage 53, 943–951.
Seeley, W. W. (2011). Divergent network connectivity changes in healthy APOE epsilon4 carriers: disinhibition or compensation? Arch. Neurol. 68, 1107–1108.
Seeley, W. W., Crawford, R. K., Zhou, J., Miller, B. L., and Greicius, M. D. (2009). Neurodegenerative diseases target large-scale human brain networks. Neuron 62, 42–52.
Shehzad, Z., Kelly, A. M., Reiss, P. T., Gee, D. G., Gotimer, K., Uddin, L. Q., Lee, S. H., Margulies, D. S., Roy, A. K., Biswal, B. B., Petkova, E., Castellanos, F. X., and Milham, M. P. (2009). The resting brain: unconstrained yet reliable. Cereb. Cortex 19, 2209–2229.
Sheline, Y. I., Morris, J. C., Snyder, A. Z., Price, J. L., Yan, Z., D’Angelo, G., Liu, C., Dixit, S., Benzinger, T., Fagan, A., Goate, A., and Mintun, M. A. (2010a). APOE4 allele disrupts resting state fMRI connectivity in the absence of amyloid plaques or decreased CSF Abeta42. J. Neurosci. 30, 17035–17040.
Sheline, Y. I., Raichle, M. E., Snyder, A. Z., Morris, J. C., Head, D., Wang, S., and Mintun, M. A. (2010b). Amyloid plaques disrupt resting state default mode network connectivity in cognitively normal elderly. Biol. Psychiatry 67, 584–587.
Shmuel, A., and Leopold, D. A. (2008). Neuronal correlates of spontaneous fluctuations in fMRI signals in monkey visual cortex: Implications for functional connectivity at rest. Hum. Brain Mapp. 29, 751–761.
Shulman, G. L., Fiez, J. A., Corbetta, M., Buckner, R. L., Miezin, F. M., Raichle, M. E., and Petersen, S. E. (1997). Common blood flow changes across visual tasks: II. Decreases in cerebral cortex. J. Cogn. Neurosci. 9, 648–663.
Smit, D. J., Stam, C. J., Posthuma, D., Boomsma, D. I., and de Geus, E. J. (2008). Heritability of “small-world” networks in the brain: a graph theoretical analysis of resting-state EEG functional connectivity. Hum. Brain Mapp. 29, 1368–1378.
Smith, S. M., Fox, P. T., Miller, K. L., Glahn, D. C., Fox, P. M., Mackay, C. E., Filippini, N., Watkins, K. E., Toro, R., Laird, A. R., and Beckmann, C. F. (2009). Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U.S.A. 106, 13040–13045.
Smith, S. M., Jenkinson, M., Johansen-Berg, H., Rueckert, D., Nichols, T. E., Mackay, C. E., Watkins, K. E., Ciccarelli, O., Cader, M. Z., Matthews, P. M., and Behrens, T. E. (2006). Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage 31, 1487–1505.
Sonuga-Barke, E. J., and Castellanos, F. X. (2007). Spontaneous attentional fluctuations in impaired states and pathological conditions: a neurobiological hypothesis. Neurosci. Biobehav. Rev. 31, 977–986.
Sporns, O., Tononi, G., and Edelman, G. M. (2000). Theoretical neuroanatomy: relating anatomical and functional connectivity in graphs and cortical connection matrices. Cereb. Cortex 10, 127–141.
Sporns, O., Tononi, G., and Kotter, R. (2005). The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, e42. doi:10.1371/journal.pcbi.0010042
Stefansson, H., Ophoff, R. A., Steinberg, S., Andreassen, O. A., Cichon, S., Rujescu, D., Werge, T., Pietilainen, O. P., Mors, O., Mortensen, P. B., Sigurdsson, E., Gustafsson, O., Nyegaard, M., Tuulio-Henriksson, A., Ingason, A., Hansen, T., Suvisaari, J., Lonnqvist, J., Paunio, T., Borglum, A. D., Hartmann, A., Fink-Jensen, A., Nordentoft, M., Hougaard, D., Norgaard-Pedersen, B., Bottcher, Y., Olesen, J., Breuer, R., Moller, H. J., Giegling, I., Rasmussen, H. B., Timm, S., Mattheisen, M., Bitter, I., Rethelyi, J. M., Magnusdottir, B. B., Sigmundsson, T., Olason, P., Masson, G., Gulcher, J. R., Haraldsson, M., Fossdal, R., Thorgeirsson, T. E., Thorsteinsdottir, U., Ruggeri, M., Tosato, S., Franke, B., Strengman, E., Kiemeney, L. A., Melle, I., Djurovic, S., Abramova, L., Kaleda, V., Sanjuan, J., de Frutos, R., Bramon, E., Vassos, E., Fraser, G., Ettinger, U., Picchioni, M., Walker, N., Toulopoulou, T., Need, A. C., Ge, D., Yoon, J. L., Shianna, K. V., Freimer, N. B., Cantor, R. M., Murray, R., Kong, A., Golimbet, V., Carracedo, A., Arango, C., Costas, J., Jonsson, E. G., Terenius, L., Agartz, I., Petursson, H., Nothen, M. M., Rietschel, M., Matthews, P. M., Muglia, P., Peltonen, L., St Clair, D., Goldstein, D. B., Stefansson, K., and Collier, D. A. (2009). Common variants conferring risk of schizophrenia. Nature 460, 744–747.
Steinberg, S., Mors, O., Borglum, A. D., Gustafsson, O., Werge, T., Mortensen, P. B., Andreassen, O. A., Sigurdsson, E., Thorgeirsson, T. E., Bottcher, Y., Olason, P., Ophoff, R. A., Cichon, S., Gudjonsdottir, I. H., Pietilainen, O. P., Nyegaard, M., Tuulio-Henriksson, A., Ingason, A., Hansen, T., Athanasiu, L., Suvisaari, J., Lonnqvist, J., Paunio, T., Hartmann, A., Jurgens, G., Nordentoft, M., Hougaard, D., Norgaard-Pedersen, B., Breuer, R., Moller, H. J., Giegling, I., Glenthoj, B., Rasmussen, H. B., Mattheisen, M., Bitter, I., Rethelyi, J. M., Sigmundsson, T., Fossdal, R., Thorsteinsdottir, U., Ruggeri, M., Tosato, S., Strengman, E., Kiemeney, L. A., Melle, I., Djurovic, S., Abramova, L., Kaleda, V., Walshe, M., Bramon, E., Vassos, E., Li, T., Fraser, G., Walker, N., Toulopoulou, T., Yoon, J., Freimer, N. B., Cantor, R. M., Murray, R., Kong, A., Golimbet, V., Jonsson, E. G., Terenius, L., Agartz, I., Petursson, H., Nothen, M. M., Rietschel, M., Peltonen, L., Rujescu, D., Collier, D. A., Stefansson, H., St Clair, D., and Stefansson, K. (2011). Expanding the range of ZNF804A variants conferring risk of psychosis. Mol. Psychiatry 16, 59–66.
Sullivan, P. F., Neale, M. C., and Kendler, K. S. (2000). Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry 157, 1552–1562.
Tan, H. Y., Chen, Q., Sust, S., Buckholtz, J. W., Meyers, J. D., Egan, M. F., Mattay, V. S., Meyer-Lindenberg, A., Weinberger, D. R., and Callicott, J. H. (2007). Epistasis between catechol-O-methyltransferase and type II metabotropic glutamate receptor 3 genes on working memory brain function. Proc. Natl. Acad. Sci. U.S.A. 104, 12536–12541.
Thompson, P. M., Cannon, T. D., Narr, K. L., Van Erp, T. G. M., Poutanen, V.-P., Huttunen, M., Lönnqvist, J., Standertskjöld-Nordenstam, C. G., Kaprio, J., Khaledy, M., Dail, R., Zoumalan, C. I., and Toga, A. W. (2001). Genetic influences on brain structure. Nat. Neurosci. 4, 1253–1258.
van den Heuvel, M. P., Mandl, R. C., Stam, C. J., Kahn, R. S., and Hulshoff Pol, H. E. (2010). Aberrant frontal and temporal complex network structure in schizophrenia: a graph theoretical analysis. J. Neurosci. 30, 15915–15926.
van den Heuvel, M. P., Stam, C. J., Kahn, R. S., and Hulshoff Pol, H. E. (2009). Efficiency of functional brain networks and intellectual performance. J. Neurosci. 29, 7619–7624.
Van Essen, D. C. (1997). A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature 385, 313–318.
van Grootheest, D. S., Cath, D. C., Beekman, A. T., and Boomsma, D. I. (2005). Twin studies on obsessive-compulsive disorder: a review. Twin Res. Hum. Genet. 8, 450–458.
Walters, J. T., and Owen, M. J. (2007). Endophenotypes in psychiatric genetics. Mol. Psychiatry 12, 886–890.
Wang, L., Zhu, C., He, Y., Zang, Y., Cao, Q., Zhang, H., Zhong, Q., and Wang, Y. (2009). Altered small-world brain functional networks in children with attention-deficit/hyperactivity disorder. Hum. Brain Mapp. 30, 638–649.
Wei, L., Li, Y., Tang, Y., and Wang, J. (2010). Brain functional connectivity in depression during emotion processing. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 27, 961–967.
Weinberger, D. R., Berman, K. F., Suddath, R., and Torrey, E. F. (1992). Evidence of dysfunction of a prefrontal-limbic network in schizophrenia: a magnetic resonance imaging and regional cerebral blood flow study of discordant monozygotic twins. Arch. Gen. Psychiatry 149, 890–897.
Westlye, E. T., Lundervold, A., Rootwelt, H., Lundervold, A. J., and Westlye, L. T. (2011). Increased hippocampal default mode synchronization during rest in middle-aged and elderly APOE epsilon4 carriers: relationships with memory performance. J. Neurosci. 31, 7775–7783.
Whitfield-Gabrieli, S., Thermenos, H. W., Milanovic, S., Tsuang, M. T., Faraone, S. V., McCarley, R. W., Shenton, M. E., Green, A. I., Nieto-Castanon, A., LaViolette, P., Wojcik, J., Gabrieli, J. D., and Seidman, L. J. (2009). Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 106, 1279–1284.
Williams, H. J., Norton, N., Dwyer, S., Moskvina, V., Nikolov, I., Carroll, L., Georgieva, L., Williams, N. M., Morris, D. W., Quinn, E. M., Giegling, I., Ikeda, M., Wood, J., Lencz, T., Hultman, C., Lichtenstein, P., Thiselton, D., Maher, B. S., Malhotra, A. K., Riley, B., Kendler, K. S., Gill, M., Sullivan, P., Sklar, P., Purcell, S., Nimgaonkar, V. L., Kirov, G., Holmans, P., Corvin, A., Rujescu, D., Craddock, N., Owen, M. J., and O’Donovan, M. C. (2011). Fine mapping of ZNF804A and genome-wide significant evidence for its involvement in schizophrenia and bipolar disorder. Mol. Psychiatry 16, 429–441.
Winterer, G., Konrad, A., Vucurevic, G., Musso, F., Stoeter, P., and Dahmen, N. (2008). Association of 5′ end neuregulin-1 (NRG1) gene variation with subcortical medial frontal microstructure in humans. Neuroimage 40, 712–718.
Xu, L., Groth, K. M., Pearlson, G., Schretlen, D. J., and Calhoun, V. D. (2009a). Source-based morphometry: the use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum. Brain Mapp. 30, 711–724.
Xu, L., Pearlson, G., and Calhoun, V. D. (2009b). Joint source based morphometry identifies linked gray and white matter group differences. Neuroimage 44, 777–789.
Zalesky, A., and Fornito, A. (2009). A DTI-derived measure of cortico-cortical connectivity. IEEE Trans. Med. Imaging 28, 1023–1036.
Zalesky, A., Fornito, A., Harding, I. H., Cocchi, L., Yucel, M., Pantelis, C., and Bullmore, E. T. (2010). Whole-brain anatomical networks: does the choice of nodes matter? Neuroimage 50, 970–983.
Zalesky, A., Fornito, A., Seal, M. L., Cocchi, L., Westin, C. F., Bullmore, E. T., Egan, G. F., and Pantelis, C. (2011). Disrupted axonal fiber connectivity in schizophrenia. Biol. Psychiatry 69, 80–89.
Keywords: endophenotype, schizophrenia, depression, Alzheimer’s disease, anxiety, complex, graph analysis, default mode
Citation: Fornito A and Bullmore ET (2012) Connectomic intermediate phenotypes for psychiatric disorders. Front. Psychiatry 3:32. doi: 10.3389/fpsyt.2012.00032
Received: 25 November 2011; Accepted: 23 March 2012;
Published online: 19 April 2012.
Edited by:
Ben Harrison, The University of Melbourne, AustraliaReviewed by:
Lukas Pezawas, Medical University of Vienna, AustriaCopyright: © 2012 Fornito and Bullmore. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Alex Fornito, Department of Psychiatry, Melbourne Neuropsychiatry Centre, University of Melbourne and Melbourne Health, Levels 2 and 3, 161 Barry Street, Carlton South 3053, VIC, Australia. e-mail:Zm9ybml0b2FAdW5pbWVsYi5lZHUuYXU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.