
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Plant Sci., 21 February 2025
Sec. Plant Breeding
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1568966
This article is a correction to:
Saturation Mapping of a Major Effect QTL for Stripe Rust Resistance on Wheat Chromosome 2B in Cultivar Napo 63 Using SNP Genotyping Arrays
 Jianhui Wu1
Jianhui Wu1 Qilin Wang1
Qilin Wang1 Shengjie Liu2
Shengjie Liu2 Shuo Huang2
Shuo Huang2 Jingmei Mu2
Jingmei Mu2 Qingdong Zeng1
Qingdong Zeng1 Lili Huang1
Lili Huang1 Dejun Han2*
Dejun Han2* Zhensheng Kang1*
Zhensheng Kang1*A Corrigendum on
Saturation mapping of a major effect QTL for stripe rust resistance on wheat chromosome 2B in cultivar Napo 63 using SNP genotyping arrays
By Wu J, Wang Q, Liu S, Huang S, Mu J, Zeng Q, Huang L, Han D and Kang Z (2017) Front. Plant Sci. 8:653. doi: 10.3389/fpls.2017.00653
In the published article, there was an error in Figure 1 as published. The photos of Avocet S and F1 were not the original wheat lines. We carefully checked them and found that they are the cultivar Zhengmai 9023 and the offspring of Friedrichswerther, respectively. We got the wrong order number of the materials. The corrected Figure 1 and its caption appear below.
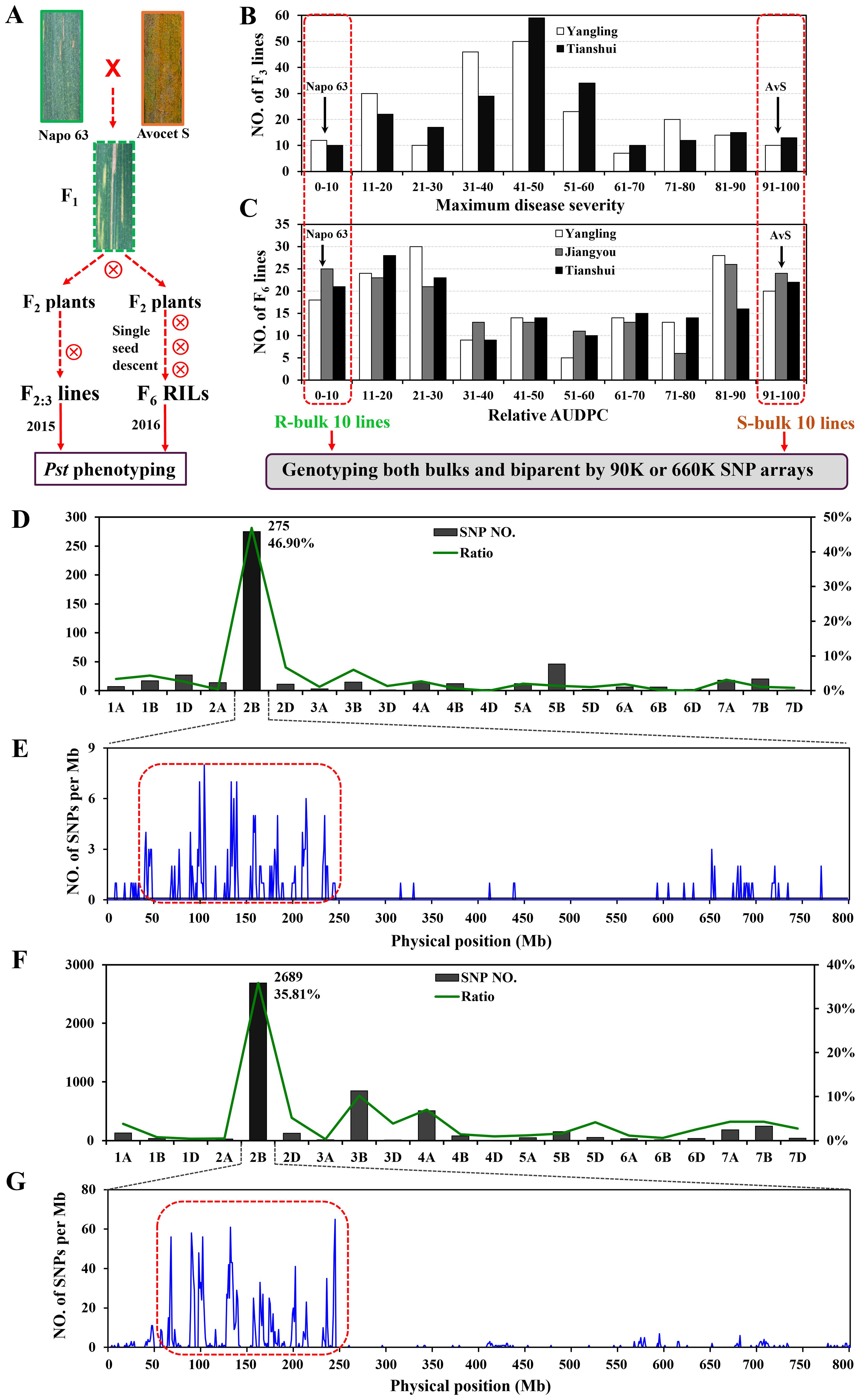
Figure 1. Overview of analyses. F2:3 lines and F5:6 recombinant inbred lines (RILs) were derived from the cross AvS × Napo 63. (A) Phenotypes of AvS, Napo 63 and their progenies across all environments and data collected at heading-flowering stage. (B) Frequency distribution of maximum disease severity (MDS) for 221 F2:3 lines grown at Yangling and Tianshui in 2015. (C) Frequency distribution of relative area under the disease progress curve (rAUDPC) for 175 F6 RILs grown at at Yangling, Jiangyou and Tianshui in 2016. Black arrows indicate the parental line means. Distributions of the polymorphic SNPs in each chromosome by 90K (D) and 660K (F) SNP arrays and positions of SNPs in chromosome 2B (E, G). Selected SNPs (in the red dotted boxes) were analysed in KASP assays.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: adult-plant resistance, bulked segregant analysis, molecular markers, Puccinia striiformis, Triticum aestivum
Citation: Wu J, Wang Q, Liu S, Huang S, Mu J, Zeng Q, Huang L, Han D and Kang Z (2025) Corrigendum: Saturation mapping of a major effect QTL for stripe rust resistance on wheat chromosome 2B in cultivar Napo 63 using SNP genotyping arrays. Front. Plant Sci. 16:1568966. doi: 10.3389/fpls.2025.1568966
Received: 31 January 2025; Accepted: 04 February 2025;
Published: 21 February 2025.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2025 Wu, Wang, Liu, Huang, Mu, Zeng, Huang, Han and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dejun Han, aGFuZGpAbndzdWFmLmVkdS5jbg==; Zhensheng Kang, a2FuZ3pzQG53c3VhZi5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.