
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 24 February 2025
Sec. Plant Pathogen Interactions
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1547593
This article is part of the Research TopicInnovative Molecular Strategies for Enhancing Plant Defense Against Biotic StressesView all 10 articles
 Beenish Hassan1†
Beenish Hassan1† Sadam Hussain Bhutto1†
Sadam Hussain Bhutto1† Xiao-Xiao Yin1
Xiao-Xiao Yin1 Xiu-Lian Yan1
Xiu-Lian Yan1 Rong Liao1
Rong Liao1 Mao-Lin Guo1
Mao-Lin Guo1 Ya-Ping Tang1
Ya-Ping Tang1 Dai-Ming Guo2
Dai-Ming Guo2 Si-Jia Yang1
Si-Jia Yang1 Faiza Gulzar1
Faiza Gulzar1 Yan Li1
Yan Li1 Xian-Yin Zeng3*
Xian-Yin Zeng3* Zhi-Xue Zhao1*
Zhi-Xue Zhao1* Wen-Ming Wang1*
Wen-Ming Wang1*Rice blast disease, caused by Magnaporthe oryzae, poses the most devastating threat to global rice production. The products of most blast resistance (R) genes specifically recognize corresponding a virulence effectors from the pathogen, thereby mediating robust immune responses that are crucial for disease resistance. However, it is unclear why different R genes endow with differential amplitudes of immunity against M. oryzae. Here, we demonstrated that different blast R genes confer differential amplitudes of immunity against M. oryzae, presumably due to divergent reprogramming of transcriptional responses. We detected that three rice restorer lines exhibited differential amplitudes of immune responses, despite all lines displaying resistance to M. oryzae. Consistently, different accessions carrying different single R genes exhibited remarkable differentially expressed genes (DEGs) count, indicating different transcriptional re-programming that leads to different fitness cost. Comparative analysis revealed varying degrees of overlap among DEGs across different accessions. By integrating RNA-seq and RT-qPCR data, we recommended some marker genes that distinguish the differential amplitude of immunity against M. oryzae mediated by different blast R genes. Thus, our study provides valuable insights into the specific and overlapping roles of R gene-mediated immunity. We also propose marker genes that can be used to effectively evaluate the amplitude of immune responses to M. oryzae, thereby facilitating the assessment of R genes with relatively lower amplitude of immunity in order to minimize fitness cost.
Rice has evolved a two-layered immune system to defend against blast fungal invasion. The first layer, known as pathogen-associated molecular patterns (PAMPs)-triggered immunity (PTI) (Saile et al., 2020), is activated upon the recognition of PAMPs by pattern recognition receptors (PRRs) (Jones and Dangl, 2006; Couto and Zipfel, 2016; Yu et al., 2017). The second layer, termed effector-triggered immunity (ETI), is triggered upon the detection of effectors by blast resistance genes, most of which encode nucleotide-binding site leucine-rich repeat (NBS-LRR, NLR) proteins (Dou and Zhou, 2012; Yuan et al., 2021a), resulting in the hypersensitive response (HR) and inhibition of pathogen infection (Alfano and Collmer, 2004; Dalio et al., 2021).
Plant hormones, including salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), act as signaling molecules that mediate a diverse array of immune responses (Spoel and Dong, 2008). SA, a natural phenolic compound, regulates a wide range of immune responses triggered by PAMPs and effectors (Vlot et al., 2009; Boatwright and Pajerowska-Mukhtar, 2013). Its pivotal role in rice immunity against the blast fungus M. oryzae was demonstrated by generating SA-deficient transgenic rice expressing bacterial salicylate hydroxylase (NahG), which degrades SA (Yang et al., 2004). In Arabidopsis thaliana, Non-expresser of pathogenesis-related 1 (NPR1) functions as a key regulator of the SA signaling pathway, with numerous SA-responsive genes being NPR1-dependent (Dong, 2004). Overexpression of AtNPR1 or its ortholog OsNPR1 enhances resistance to M. oryzae and leaf blight pathogen Xanthomonas oryzae pv. oryzae (Xoo) in rice (Yuan et al., 2007; Feng et al., 2011; Dai et al., 2023). Enhanced disease susceptibility 1 (EDS1) and phytoalexin deficient 4 (PAD4) are involved in the TIR-NBS-LRR (TNL) protein-associated SA accumulation (Wiermer et al., 2005). In rice, OsEDS1 functions as a positive regulator in rice-pathogen interactions, with knockdown of OsEDS1 leading to increased susceptibility to Xoo (Ke et al., 2019). Similarly, OsPad4 contributes to rice defense against Xoo (Ke et al., 2014). JA regulates plant growth, development, and biotic stress responses. JA treatment induces the expression of pathogenesis-related (PR) genes, including OsPR10, OsPR5, OsPR1a, and OsPR1b (Agrawal et al., 2000a, 2000b; Rakwal and Komatsu, 2000; Hashimoto et al., 2004), highlighting its roles in rice immunity. Overexpression of OsAOS2, encoding an allene oxide synthase that is a key enzyme in JA biosynthesis, leads to upregulation of PR gene expression and enhances resistance to rice blast.
PR proteins are well-established defense-related proteins that play critical roles in signal transduction, antimicrobial activity, and cell wall reinforcement. Various PR proteins are involved in rice immunity (Kim et al., 2009, 2013). For instance, pathogenesis-related gene 1a (OsPR1a) and pathogenesis-related protein 10b (OsPR10b) are induced upon rice blast infection (Agrawal et al., 2000a, 2000b). Phenylalanine ammonia lyases (PALs) contribute to broad-spectrum resistance against pathogens (Wang et al., 2019). In rice, there are nine OsPAL genes, eight of which are known to be induced by M. oryzae (Duan et al., 2014). Overexpression of OsPAL1 enhances resistance to M. oryzae (Zhou et al. (2018), while loss of function of OsPAL4 increases susceptibility to M. oryzae, Xoo, and Rhizoctonia solani (Tonnessen et al., 2015). Additionally, the induction of ent-kaurene synthase 4 (OsKS4), OsPBZ1, OsGlu1, OsSalT, and OsPR10 has been observed in rice upon M. oryzae infection (Kim et al., 2004).
In addition to PR genes, transcription factors, immune receptors, and ARGONAUTEs (AGOs) play essential roles in plant immunity against pathogens. Transcription factors (TFs), such as WRKY, NAC, bZIP, AP2, BHLH, NF-Y, CAMTA, and MYB, regulate the expression of downstream genes by binding to their promoters and are integral to plant immunity (Huang et al., 2016; Noman et al., 2017; Ng et al., 2018). For instance, the WRKY TF gene OsWRKY45 plays a crucial role in SA-mediated defense signaling in rice. The expression of OsWRKY45 is induced by SA or its analog treatment, and its overexpression enhances resistance to blast disease, whereas loss of function increases susceptibility (Shimono et al., 2007). Similarly, the NAC TF gene NAC domain-containing protein 4 (OsNAC4), positively regulates HR-mediated cell death (Kaneda et al., 2009). Furthermore, immune receptor genes are critical for rice immunity. OsCERK1 and OsCEBiP, two lysin motif (LysM) proteins, are involved in the perception of chitin, a major component of the fungal cell wall that triggers innate immunity in plants (Shimizu et al., 2010; Hayafune et al., 2014). In Arabidopsis thaliana, chitin binding by CERK1 activates immunity, whereas in rice, CEBiP plays a dominant role in chitin perception. Knockdown of OsCEBiP results in the loss of chitin oligosaccharide binding to the plasma membrane, impairing OsCERK1’s ability to bind chitin directly (Kaku et al., 2006; Shinya et al., 2012). Moreover, ARGONAUTE (AGO) proteins contribute to plant responses to environmental challenges, including pathogen attacks. For example, AGO1, a key component of RNA-induced silencing complex (RISC) in the RNAi pathway, coordinates plant disease resistance with growth and development (Zhao et al., 2023).
To date, over 50 race-specific resistance genes against rice blast disease have been cloned from 17 distinct loci in rice (Huang et al., 2023). The products encoded by these genes specifically recognize corresponding avirulence effectors from M. oryzae, triggering robust immunity. Activation of these resistance genes is generally accompanied by a series of immune responses. However, it is largely unclear why different R genes endow differential amplitudes of immunity against M. oryzae.
In this study, we used the previously identified three elite restorer lines harboring distinct R genes: SH548 (carrying Pi2, Ptr, and Pid2), SH882 (carrying Pid2 and Pikm), and WSSM (carrying Pid2 and Pi9-Type5) (Hassan et al., 2022). These lines were treated with M. oryzae or chitin, followed by the analysis of H2O2 accumulation and the expression of defense-related genes using DAB staining and RT-qPCR, respectively. Additionally, RNA-seq data were analyzed for LTH and monogenic lines IRBLz5-CA, IRBL9-W, and IRBLkm-Ts (carrying Pi2, Pi9, and Pikm, respectively), as well as the blast-resistant elite restorer line R2115 (carrying Pi2, Pid2, and Pib), before and after inoculation with the rice blast fungus. The results indicate that different R genes orchestrate both convergent and divergent responses to M. oryzae in rice.
To investigate the amplitudes of immune responses in different rice accessions, we inoculated LTH and three elite restorer lines with the M. oryzae strain Guy11 and examined immune responses, including H2O2 accumulation and the expression of defense-related genes, such as OsPR1a, OsPR10b, OsNAC4, OsKS4, and OsMAS1 (momilactone a synthase). Consistent with the observation in a previous report (Hassan et al., 2022), LTH was susceptible, showing typical disease lesions, while the three restorer lines exhibited resistance (Figure 1A). In line with the disease phenotypes, extensive H2O2 accumulation was observed as reddish-brown coloration around appressoria in the three elite restore lines, while minimal accumulation was detected in LTH after DAB staining (Figures 1B, C). In SH548, the expression of all examined defense-related genes was induced compared with LTH, with notably high expression levels at 24 hours post-inoculation (hpi) (Figure 1D). Specifically, OsPR10b exhibited about a 6000-fold increase in expression, OsKS4 showed about a 1200-fold increase, and OsPR1a and OsNAC4 exhibited approximately a 400-fold increase compared to LTH. OsPR1a and OsNAC4 were significantly up-regulated at 12, 24, and 48 hpi, whereas OsPR10b and OsKS4 exhibited increased levels at 24 and 48 hpi, and OsMAS1 showed elevated levels specifically at 24 hpi compared to LTH. In SH882, the expression levels of OsPR1a, OsNAC4, OsKS4, and OsMAS1 were significantly elevated at 12, 24, and 48 hpi, whereas OsPR10b exhibited enhanced expression specifically at 24 and 48 hpi compared to LTH (Figure 1D). Similarly, WSSM exhibited increased expression of defense-related genes at specific time points, with expression levels observed at certain time points being higher than those in LTH. Specifically, OsMAS1 exhibited significantly higher induction than LTH at 24 hpi, whereas OsPR1a and OsPR10b displayed elevated expression at both 24 and 48 hpi, OsNAC4 showed increased expression at 12 and 24 hpi, and OsKS4 at 24 hpi (Figure 1D). These results indicate that SH548, SH882, and WSSM exhibit differential amplitudes of immune responses upon the infection of M. oryzae, despite all showing similar resistant phenotypes.
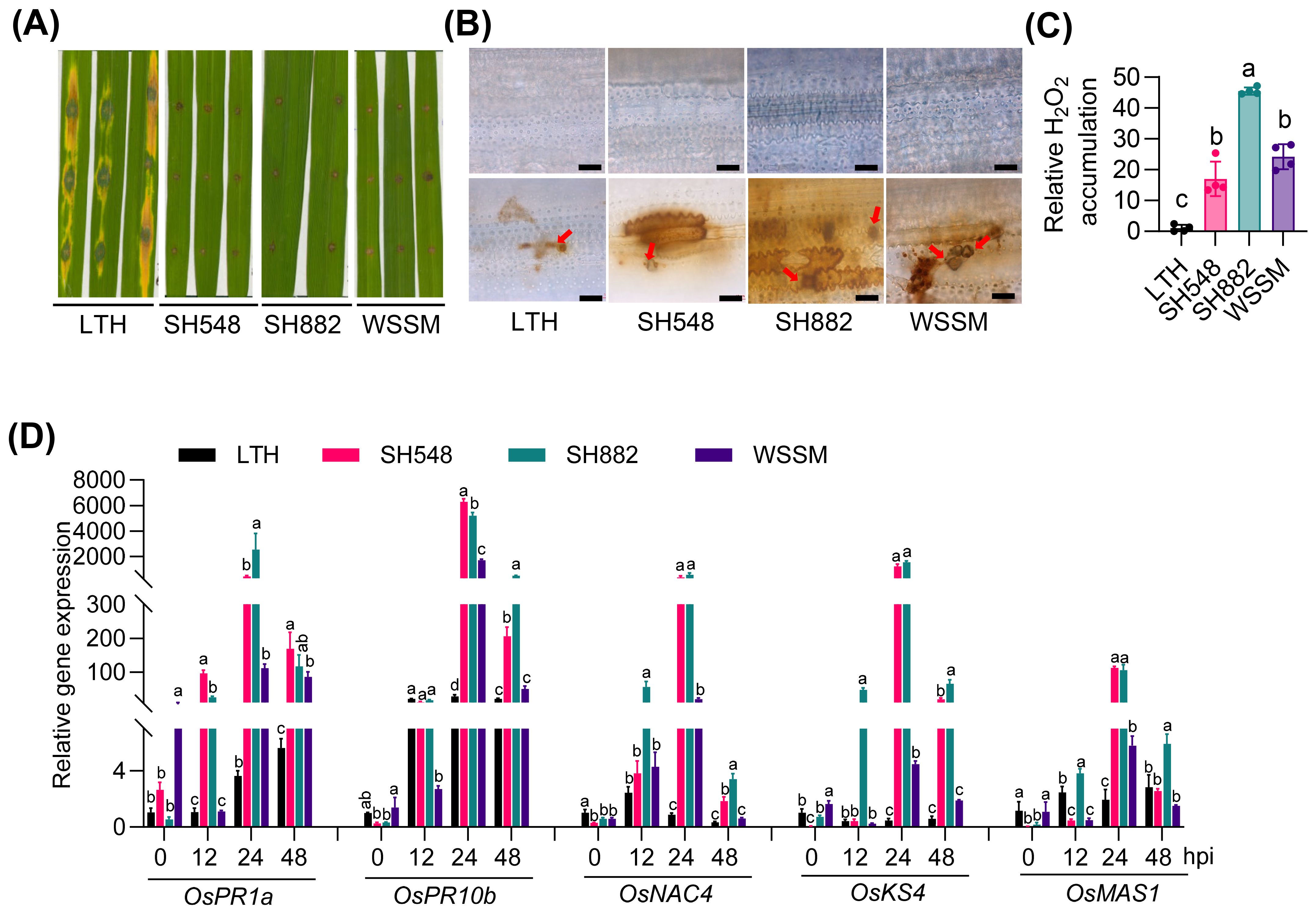
Figure 1. The differential amplitudes of immune responses exhibited by SH548, SH882, and WSSM upon M. oryzae infection. (A) Blast disease phenotypes in LTH, SH548, SH882, and WSSM. The indicated lines were punch inoculated with the CRB10 strain (4 × 105 spores/mL), and a photograph was taken at 5 days post-inoculation (dpi). (B) H2O2 accumulation in LTH, SH548, SH882, and WSSM at 48 hours post-inoculation (hpi), both with and without M. oryzae infection. Representative leaf sections from the indicated lines were used to illustrate fungal growth and H2O2 accumulation. Brown coloration indicates H2O2 accumulation and red arrows highlight appressoria. The upper panel shows mock-treated, and the lower panel shows M. oryzae-infected leaves. The images were captured using a Zeiss fluorescence microscope (Zeiss imager A2). Scale bar, 10 μm. (C) Quantification of H2O2 in leaves of indicated rice lines infected with M. oryzae. (D) Expression patterns of defense-related genes. Leaf samples were collected at 0, 12, 24, and 48 hpi with the Guy11 (4×105 spores/mL) strain. RT-qPCR was conducted to examine the expression of defense-related genes. Error bars indicate the standard deviation (SD) (n=3). Differences marked by letters indicate significant differences (P < 0.05), as determined by One-way ANOVA analysis. Differences were marked by comparing the accessions at each time point separately.
To explore the reasons behind the differential amplitudes of immune responses triggered in the three resistant rice lines, we first examined whether there were any differences in the expression of the chitin receptors, including OsCERK1 and OsCEBiP, which are critical for rice immunity against M. oryzae (Hayafune et al., 2014). Previous studies have demonstrated that the expression of CERK1 and CEBiP was transcriptionally induced by chitin elicitor treatment (Kaku et al., 2006; Miya et al., 2007; Shimizu et al., 2010). Our data showed that the expression of these two receptors was constitutively and significantly higher in SH548 and SH882 than in LTH, with the amplitude of OsCEBiP being higher in WSSM than in LTH at 6 hours post-treatment (hpt) (Figure 2A). Intriguingly, their expressions were differentially up-regulated in two of the three resistant lines compared to LTH upon chitin treatment. OsCERK1 was slightly but significantly up-regulated in LTH, whereas it was highly and significantly upregulated in SH548 at 6, 9, and 12 hpt, and in SH882 at 9 and 12 hpt (Figure 2A). The expression of OsCEBiP was induced at 6 and 9 hpt in SH548, and at 9 and 12 hpt in SH882. These results indicate that different resistant lines exert diverse effects on the expression of the receptors involved in PTI upon chitin treatment.
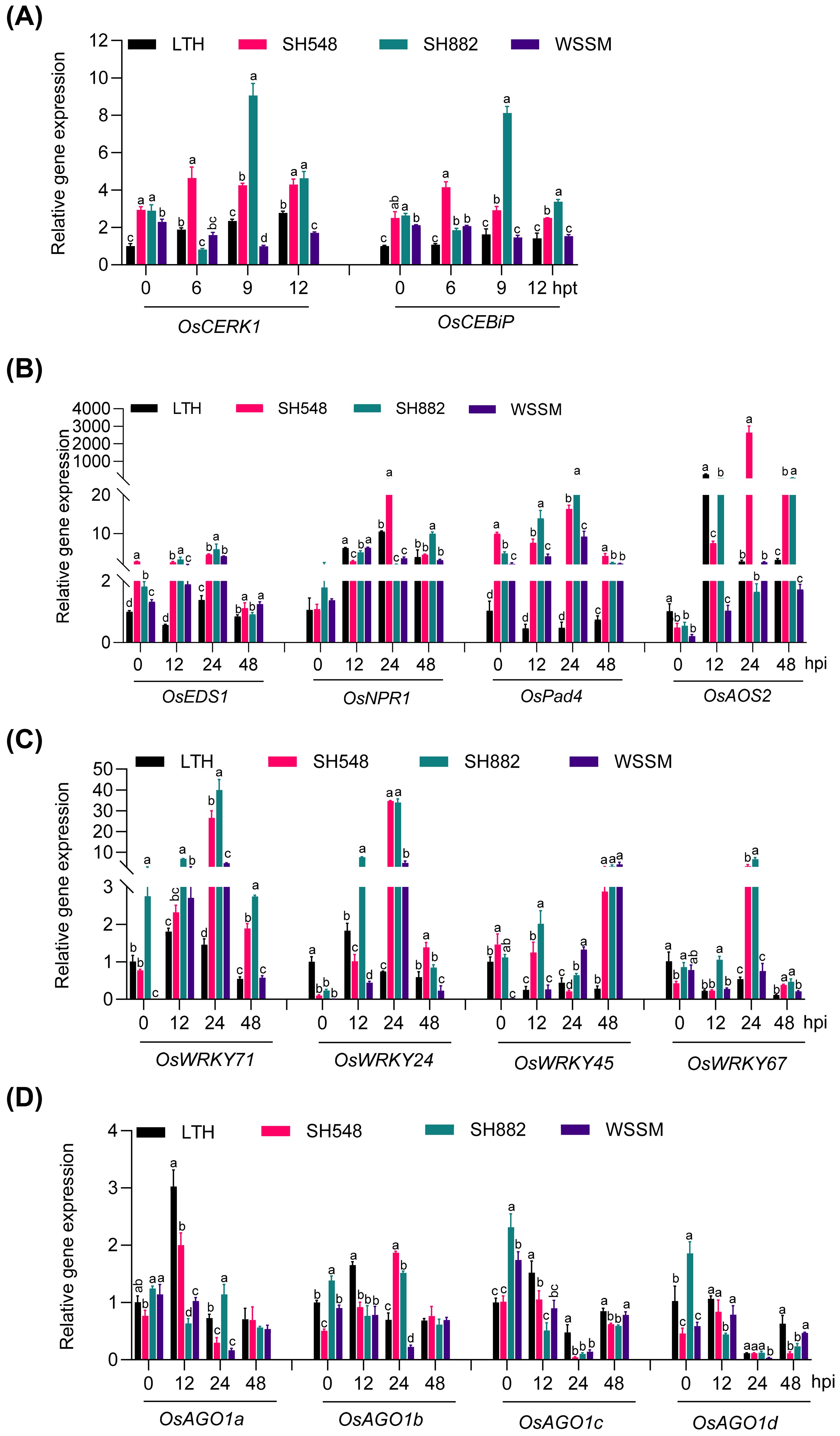
Figure 2. The differential response of chitin receptors upon chitin treatment and differential activation of defense-related genes upon blast infection in SH548, SH882, and WSSM. (A) Expression patterns of OsCERK1 and OsCEBiP in LTH, SH548, SH882, and WSSM following chitin treatment. Leaves were treated with 200 μM chitin and harvested at 0, 6, 9, and 12 hours post-treatment (hpt). Error bars indicate the standard deviation (SD) (n=3). Differences marked by letters indicate significant differences (P < 0.05), as determined by One-way ANOVA analysis. Differences were marked by comparing the accessions at each time point separately. (B-D) Expression pattern of hormone-signaling pathway genes (B), OsWRKY family genes (C), and OsAGO1 family genes (D). Leaf samples were collected at 0, 12, 24, and 48 hpi after inoculated with the Guy11 (4×105 spores/mL) strain. RT-qPCR was conducted to examine the expression of these genes. Error bars indicate the standard deviation (SD) (n=3). Differences marked by letters indicate significant differences (P < 0.05), as determined by One-way ANOVA analysis. Differences were marked by comparing the accessions at each time point separately.
We then examined the expression of the marker genes for the SA and JA signaling pathways upon M. oryzae infection. SA and JA are major hormones involved in plant immunity (Hou and Tsuda, 2022). SA plays a crucial role in basal resistance and is additionally involved in R-gene-mediated resistance, whereas JA contributes to the basal defense of rice against both fungal and bacterial pathogens (Yang et al., 2013). Transcript levels of SA signaling marker genes, including OsEDS1, OsNPR1, and OsPad4 were induced in all three accessions at certain time points (Figure 2B). OsEDS1 was induced at 12 and 24 hpi, with higher basal expression levels observed in SH548 and SH882 than in LTH (Figure 2B). High basal expression of OsPad4 was observed in all the accessions, with induction occurring at 12, 24, and 48 hpi compared to LTH (Figure 2B). Although OsNPR1 was significantly induced in all four accessions upon infection with M. oryzae, there was not a significant difference in the induction compared with LTH, except for SH548 at 24 and SH882 at 48 hpi. The JA-synthesis-related gene OsAOS2 exhibited significant up-regulation in SH548 at 24 and 48 hpi, and in SH882 at 48 hpi, compared with LTH (Figure 2B). These results indicate that M. oryzae infection differentially activates the SA and JA signaling pathways in SH548, SH882, and WSSM.
OsWRKY proteins are known to regulate rice immunity, functioning as either positive or negative transcription factors (Jimmy and Babu, 2015). To investigate how blast infection affects the expression of OsWRKYs mediated by different R genes, we analyzed the expression patterns of OsWRKY71, OsWRKY24, OsWRKY45, and OsWRKY67, which are involved in the positive regulation of rice blast resistance (Liu et al., 2007; Shimono et al., 2007; Liu et al., 2018; Yokotani et al., 2018). OsWRKY71 showed significant induction in SH548, SH882, and WSSM at 12 and 24 hpi (Figure 2C). OsWRKY24 was induced at 24 hpi in SH548 and WSSM, and at both 12 and 24 hpi in SH882 (Figure 2C). OsWRKY45, which functions in the SA signaling pathway (Shimono et al., 2007), was up-regulated in SH548 and WSSM at 48 hpi, and in SH882 at both 12 and 48 hpi (Figure 2C). The expression of OsWRKY67 was significantly up-regulated at 24 hpi in SH548 and SH882, but not up-regulated in WSSM (Figure 2C). These results indicate that M. oryzae infection leads to the differential up-regulation of OsWRKY genes, which have been previously identified as positive regulators of rice immunity.
AGO1, targeted by miR168 and integral to the RISC, is a core component for RNAi and is required for plant immunity (Li et al., 2019b). In rice, the OsAGO1 gene family comprises four members: OsAGO1a, OsAGO1b, OsAGO1c, and OsAGO1d. We examined the expression patterns of these genes during rice blast infection. Compared to 0 hpi, OsAGO1a was induced in LTH and SH548 at 12 hpi. OsAGO1b was upregulated at 24 hpi in SH548 and SH882, compared with LTH. However, no up-regulation of OsAGO1c and OsAGO1d was observed in any of the three accessions (Figure 2D). These results indicate that OsAGO1 is unsuitable as a marker gene for measuring the amplitude of immunity.
Based on the differential amplitudes of immune responses in SH548, SH882, and WSSM, we hypothesized that the different R genes mediate distinct transcriptional responses. To rigorously test this hypothesis, we conducted transcriptomic analysis on the blast-resistant monogenic lines IRBLz5-CA, IRBL9-W, and IRBLkm-Ts, each harboring a single R gene, along with the resistant elite restorer line R2115 (Shi et al., 2015) and the susceptible line LTH, both before and after infection with M. oryzae (Hu et al., 2023). Specifically, IRBLz5-CA harbors Pi2, IRBL9-W carries Pi9, IRBLkm-Ts contains Pikm, and R2115 possesses Pi2, Pid2, and Pib (Shi et al., 2015; Hu et al., 2024). Transcriptomic data analysis of infected samples detected a total of 406, 868, 2808, 6284, and 8871 differentially expressed genes (DEGs) in IRBLz5-CA, R2115, IRBL9-W, IRBLkm-Ts, and LTH (Figure 3A; Supplementary Tables S1, S2), respectively, with the fewest DEGs in IRBLz5-CA, followed by R2115, IRBL9-W and IRBLkm-Ts, and the most in LTH, indicating that transcriptional reprogramming response is quite different in the accessions carrying different R genes. Similarly, the number of up-regulated and down-regulated DEGs followed the same trend as the total DEGs, with 150, 553, 1196, 2538, and 4225 genes upregulated, and 256, 315, 1612, 3746, and 4646 genes downregulated in IRBLz5-CA, R2115, IRBL9-W, IRBLkm-Ts, and LTH, respectively (Figure 3A; Supplementary Tables S1, S2). Intriguingly, we detected a total of 78 upregulated and 98 downregulated DEGs shared across all five accessions, whereas 1, 87, 70, 780, and 2476 DEGs were specifically upregulated, and 1, 10, 36, 1279, and 2669 DEGs were downregulated in IRBLz5-CA, R2115, IRBL9-W, IRBLkm-Ts, and LTH, respectively (Figures 3B, C; Supplementary Table S3). Moreover, each accession shared a certain number of DEGs with other accessions (Figure 3D; Supplementary Table S3). These data indicate both convergent and divergent responses of transcriptional reprogramming upon the infection of M. oryzae.
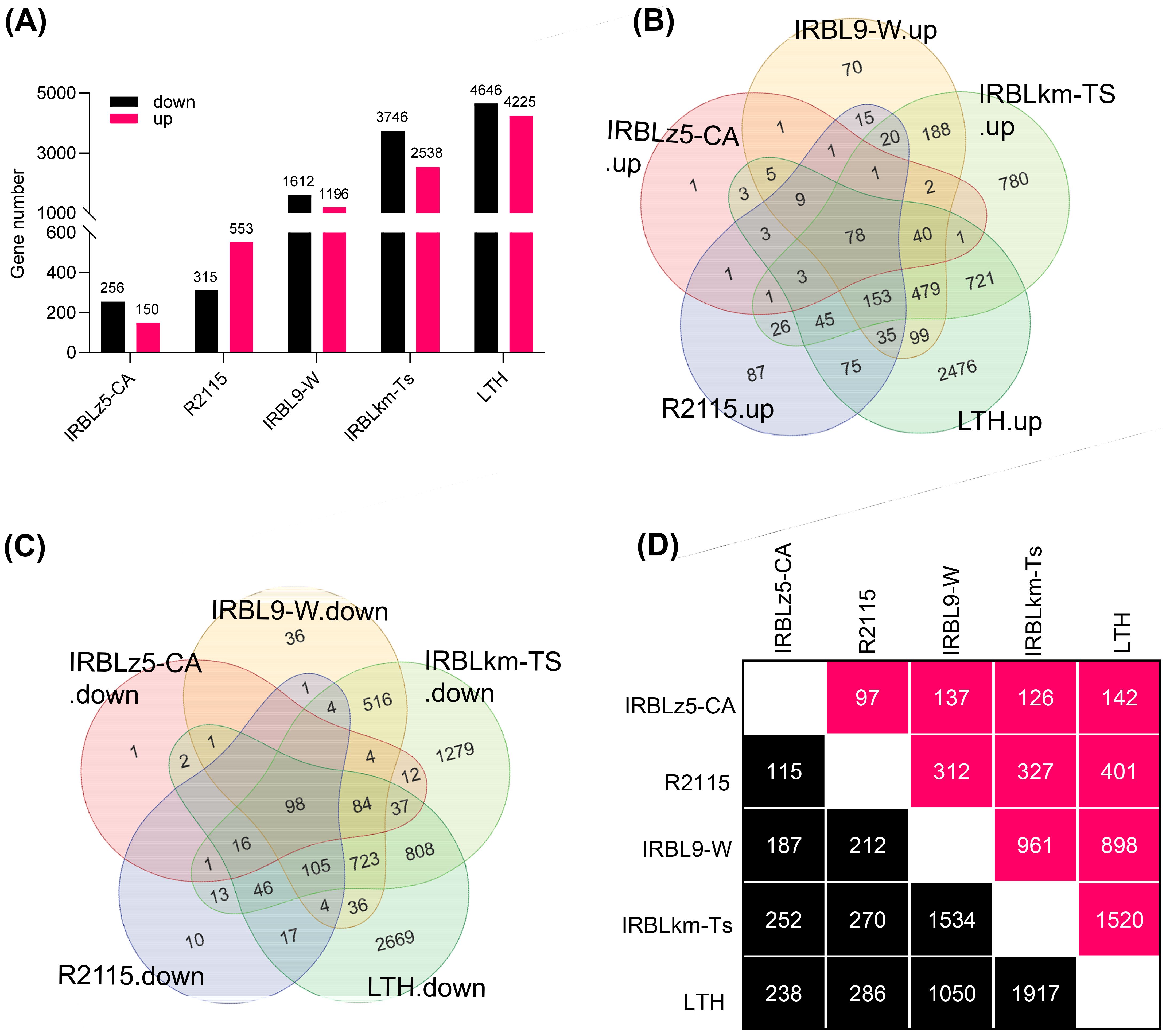
Figure 3. Convergent and divergent transcriptome responses exhibited by IRBLz5-CA, IRBL9-W, IRBLkm-Ts, and R2115 to blast infection. (A) The number of up- and down-regulated genes in all the accessions in response to blast infection. (B, C) Venn diagrams of up-regulated (B) and down-regulated (C) DEGs in IRBLz5-CA, IRBL9-W, IRBLkm-Ts, R2115, and LTH. Venn diagrams were generated using the R programming language. (D) The number of DEGs shared between each two accessions. The pink-colored boxes indicate the upregulated DEGs, whereas the black-colored boxes indicate the downregulated DEGs.
We then analyzed the Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of the 78 convergently upregulated and 98 downregulated DEGs. Based on GO annotations, 78 upregulated and 98 downregulated genes were categorized into three primary GO categories: biological process, cellular component, and molecular function. In the biological process category, convergent DEGs were enriched in biological regulation, cellular component organization, cellular process, and other related pathways. The most enriched categories within biological processes were cellular process, metabolic process, and response to stimulus. In the cellular component category, DEGs were enriched in the cell, cell part, extracellular region, macromolecular complex, membrane, and organelle. In the molecular function category, DEGs were enriched in binding, catalytic activity, enzyme regulator activity, structural molecule activity, and transporter activity (Figure 4A; Supplementary Table S4). KEGG pathway analysis showed that the convergent DEGs were enriched in membrane transport, signal transduction, amino acid metabolism, biosynthesis of other secondary metabolites, and other related pathways (Figure 4B; Supplementary Table S4).
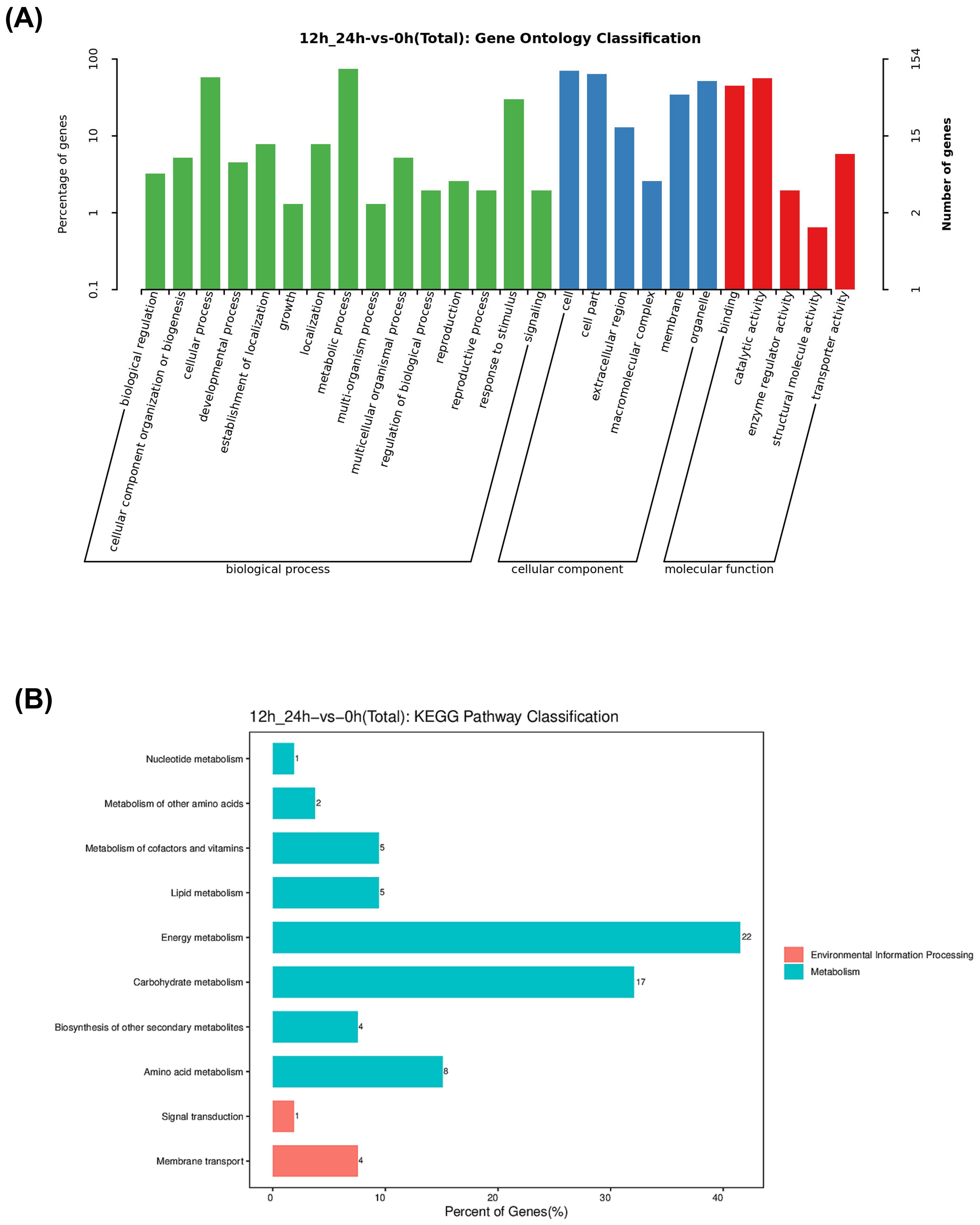
Figure 4. GO and KEGG pathway classification of co-up-regulated and co-down-regulated DEGs in IRBLz5-CA, IRBL9-W, IRBLkm-Ts, R2115, and LTH based on RNA-seq data. (A) GO classification, (B) KEGG pathway classification.
Subsequently, we conducted analyses of the divergent DEGs in each rice line. In IRBL9-W, which harbors the Pi9 resistance gene, the divergent DEGs were annotated into 28 terms, including 15 in biological processes, eight in cellular components, and five in molecular functions (Supplementary Figure S1A; Supplementary Table S4). KEGG pathway analysis revealed that the divergent DEGs in IRBL9-W were enriched in pathways such as transport and catabolism, signal transduction, folding, sorting and degradation, biosynthesis of other secondary metabolites, and other related pathways (Supplementary Figure S1B; Supplementary Table S4). In IRBLkm-Ts, which carries the Pikm resistance gene, the divergent DEGs were annotated into 28 terms, including 15 in biological processes, eight in cellular components, and five in molecular functions (Supplementary Figure S2A; Supplementary Table S4). KEGG pathway analysis revealed that these divergent DEGs were enriched in pathways related to cell motility, transport and catabolism, membrane transport, signal transduction, and other related pathways (Supplementary Figure S2B; Supplementary Table S4). In R2115, the divergent DEGs were annotated into 26 terms, including 14 in biological processes, eight in cellular components, and four in molecular functions (Supplementary Figure S3A; Supplementary Table S4). KEGG pathway analysis showed enrichment of the divergent DEGs in signal transduction, biosynthesis of other secondary metabolites, carbohydrate metabolism, lipid metabolism, and other pathways (Supplementary Figure S3B; Supplementary Table S4). In LTH, which does not harbor any blast resistance genes, the divergent DEGs were annotated into 28 terms, including 15 in biological processes, eight in cellular components, and five in molecular functions (Supplementary Figure S4A; Supplementary Table S4). KEGG pathway analysis revealed enrichment of the divergent DEGs in pathways related to cell motility, transport and catabolism, membrane transport, signal transduction, and other pathways (Supplementary Figure S4B; Supplementary Table S4). Notably, the distribution of genes across each term varies significantly among the accessions harboring different R genes.
Altogether, these data indicate that rice plants mount convergent responses to the infection of M. oryzae regardless of blast resistance genes, whereas different R genes endow the divergent responses.
To investigate the reasons behind the differential amplitudes of immune responses triggered by different R genes, we focused on the divergent and convergent DEGs associated with crucial immune pathways. Our analysis unveiled diverse expression patterns of defense-related and hormone-signaling pathway genes in IRBLz5-CA, IRBL9-W, IRBLkm-Ts, R2115, and LTH (Figure 5). In the SA signaling pathway, OsNPR1 exhibited high induction levels across all accessions. However, OsICS1 and OsPAL1 showed only marginal changes in all accessions, except for the high basal expression of OsICS1 in IRBLz5-CA, R2115, and LTH, and of OsPAL1 in IRBL9-W (Figure 5A). In the JA pathway, OsAOS2 exhibited high induction in IRBL9-W and R2115 at 12 and 24 hpi, with a slight induction observed at 24 hpi in IRBLz5-CA and IRBLkm-Ts. Compared with other accessions, the transcript level of OsCOI2 was significantly induced in R2115 at 12 hpi. High basal expression levels of OsLOX1, OsJAZ1, OsJAR1, and OsAOC were observed in IRBLz5-CA, IRBL9-W, IRBLkm-Ts, and R2115, while OsLOX2 exhibited high basal expression in IRBLz5-CA, R2115, and LTH (Figure 5B). In the ET signaling pathway, OsERF1 exhibited significant induction in all the accessions, except IRBLkm-Ts, which showed a slight induction at 12 hpi. High expression levels of OsETR2 and OsEIN2 were observed only in R2115 at 12 and 24 hpi. OsERF3 was significantly induced in all the accessions at 24 hpi, except R2115 (Figure 5C). High basal expression levels of OsERS2 were observed in all the accessions. These results indicate the differential upregulation of hormone signaling in all four accessions upon M. oryzae infection.
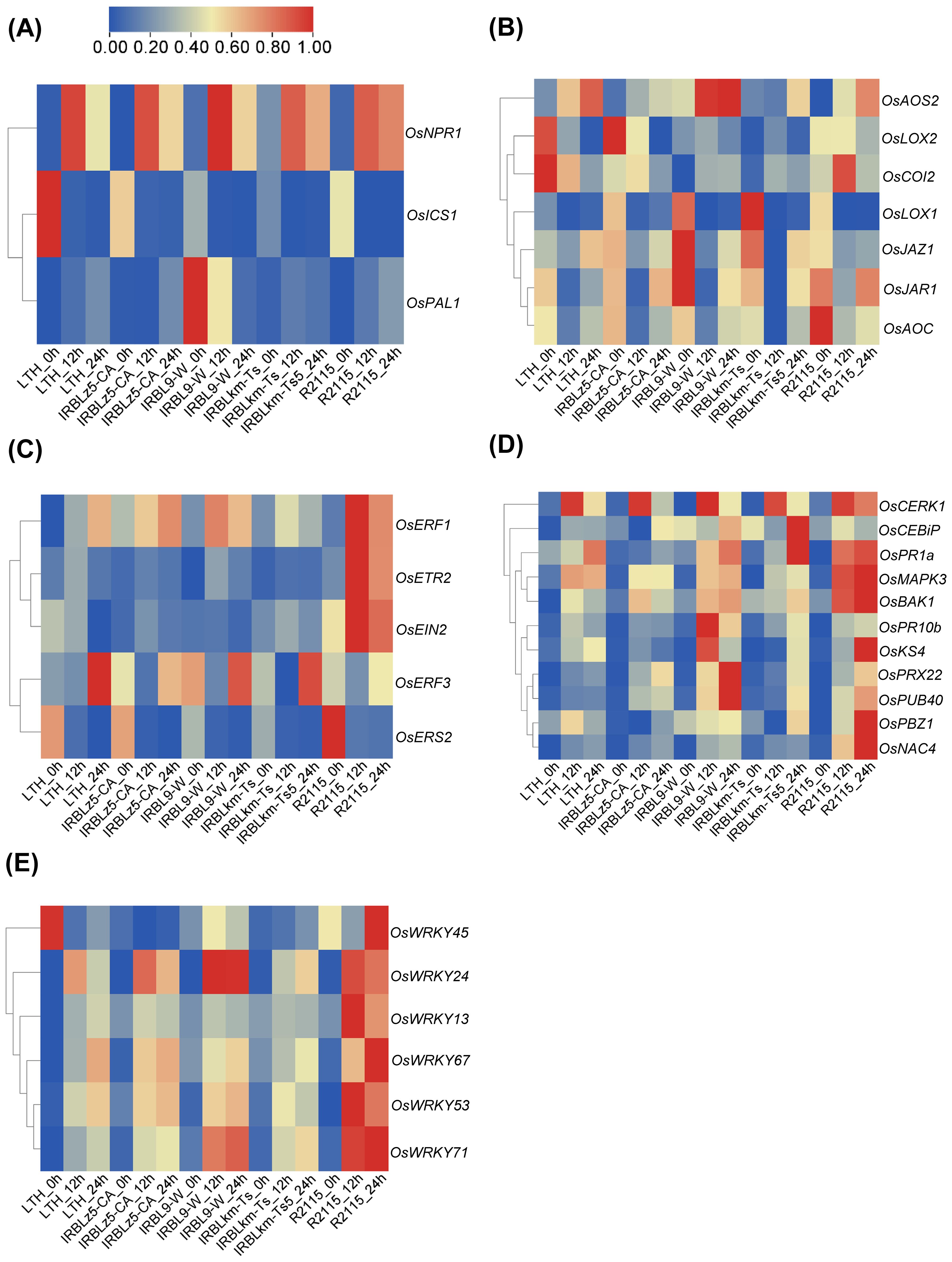
Figure 5. Heat maps displaying DEGs related to plant immunity in IRBLz5-CA, IRBL9-W, IRBLkm-Ts, R2115, and LTH upon blast infection. The differential expression of genes involved in the SA-signaling pathway (A), JA-signaling pathway (B), ET-signaling pathway (C), PTI pathway (D), and OsWRKY family (E) genes was analyzed upon M. oryzae infection. Each row of transcriptome data was normalized using TBtools.
Next, in the PTI signaling genes, the pattern recognition receptor OsCEBiP was induced at 24 hpi in IRBLz5-CA, IRBL9-W, and IRBLkm-Ts and at 12 hpi in R2115, whereas OsCERK1 was upregulated in IRBLz5-CA, IRBL9-W, and IRBLkm-Ts at 12 hpi, and in R2115 at both 12 and 24 hpi (Figure 5D). Compared with the susceptible control LTH, enhanced induction of OsBAK1 was observed in IRBLkm-Ts at 24 hpi, and in IRBLz5-CA, IRBL9-W, and R2115 at both 12 and 24 hpi. OsMAPK3 exhibited slight induction in IRBLz5-CA, IRBL9-W, and IRBLkm-Ts at 12 and 24 hpi, but showed enhanced induction in R2115 at 12 and 24 hpi. OsKS4 was induced at 12 and 24 hpi in IRBL9-W and R2115, respectively. OsPR10b exhibited higher induction levels in IRBL9-W at 12 hpi, compared to other accessions. OsPR1a induction was enhanced in IRBLkm-Ts at 24 hpi and at 12 and 24 hpi in IRBL9-W and R2115. Probenazole-induced protein 1 (PBZ1), a well-established marker gene for cell death in rice (Kim et al., 2008), showed a slight induction in IRBLkm-Ts at 24 hpi, followed by a remarkable induction in R2115 at 24 hpi. Both OsPRX22 and OsPUB40 were induced in IRBL9-W and R2115 at 12 and 24 hpi. Transcript levels of OsNAC4 were notably induced in R2115 at 12 and 24 hpi, with slight induction detected in other accessions. Significant induction of OsWRKY24, OsWRKY53, OsWRKY67, and OsWRKY71 was observed in all four resistant accessions. OsWRKY13 and OsWRKY45 were specifically induced in R2115 (Figure 5E). Again, these data indicate the convergent and divergent responses across all accessions, each carrying different R genes.
To further validate the conclusions drawn from the RNA-seq result, we validated the expression of convergently upregulated DEGs involved in defense and hormone signaling pathways by RT-qPCR. The expression of OsKS4, OsPBZ1, and OsNAC4 was significantly induced in IRBL9-W, IRBLkm-Ts, and R2115 at 24 hpi, compared with LTH (Figures 6A-C). The expression of OsNPR1 and OsWRKY67 was significantly induced in IRBL9-W and R2115 at 24 hpi, compared with LTH (Figures 6D, I). OsAOS3 was significantly induced to higher levels at 12 hpi than at 0 hpi in IRBLz5-CA, IRBL9-W, and R2115 (Figure 6E), whereas OsEBP89 was induced at 12 and 24 hpi in all the accessions. Although the magnitude of induction was not as high as that of LTH, the expression was still upregulated compared to 0 hpi (Figure 6F). A significant induction of OsPad4 was observed in IRBLz5-CA, IRBL9-W, and R2115 at 24 hpi and OsWRKY45 in all the accessions at 24 hpi, compared with LTH, accompanied by a high basal expression level in IRBLkm-Ts (Figures 6G, H). Thus, these results indicate that the RNA-seq data are reliable, and various blast resistance genes mount differential amplitudes of immune responses via differentially up-regulating genes involved in multiple immune pathways.
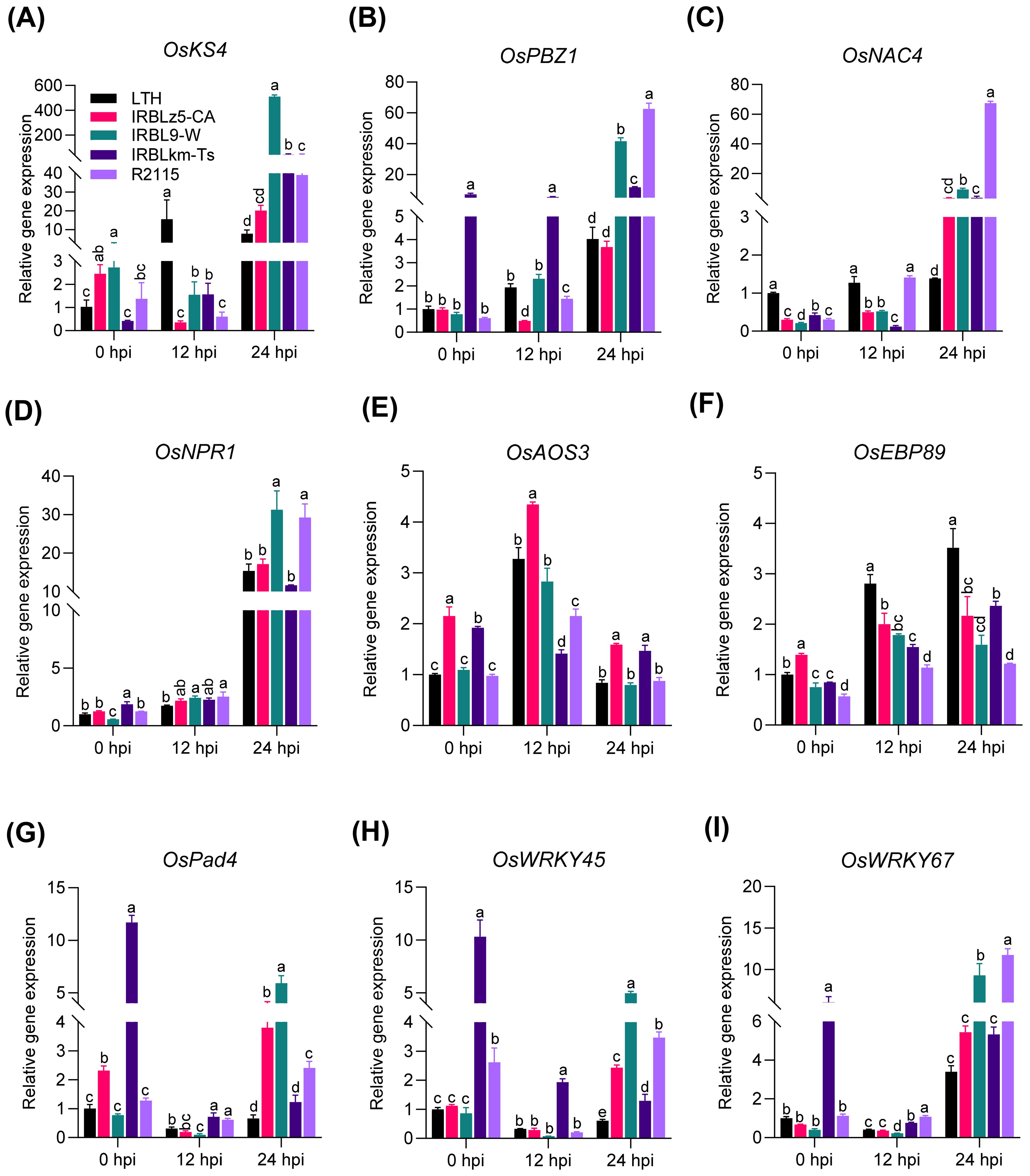
Figure 6. The expression patterns of defense-related genes and hormone-signaling pathway genes in LTH, IRBLZ5-CA, IRBL9-W, IRBLKM-Ts, and R2115. All four accessions showed induced expression of defense-related genes (A–C, F, H, I) and hormone-signaling pathway genes (D, E, G) compared with LTH. Leaf samples were collected at 0, 12, and 24 hpi with the Guy11 (4×105 spores/mL). Error bars indicate the standard deviation (SD) (n=3). Differences marked by letters indicate significant differences (P < 0.05), as determined by One-way ANOVA analysis. Differences were marked by comparing the accessions at each time point separately.
We propose a working model in which different R genes confer differential amplitudes of defense against M. oryzae (Figure 7). In the absence of R genes, weak H2O2 accumulation and a low amplitude of defense against M. oryzae occur, as more effector proteins are secreted into the cells. In contrast, the recognition of effectors by different R genes leads to increased H2O2 accumulation and activation of PTI, SA, JA, and WRKY pathways (Figure 7).
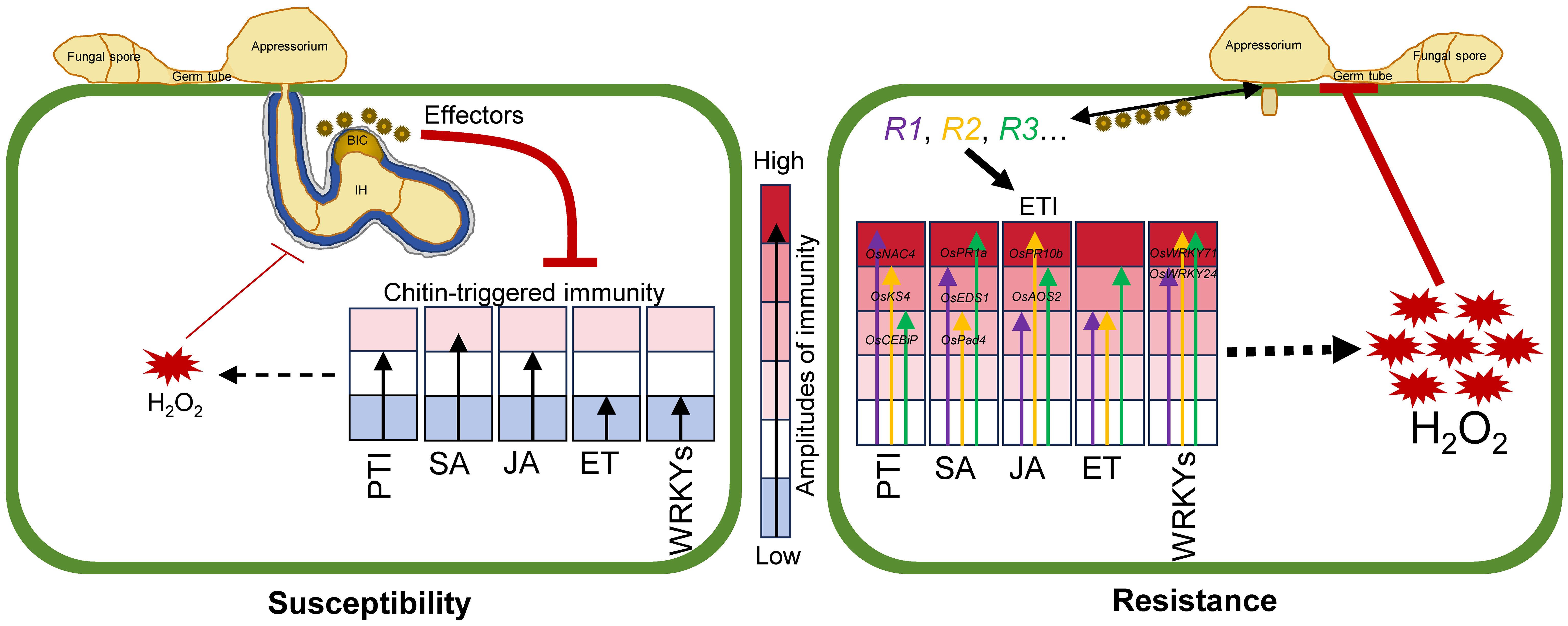
Figure 7. Working model for different R genes endow with differential amplitudes of defense against M. oryzae. Secretion of M. oryzae effectors to rice cells leads to the suppression of chitin-triggered immunity, which results in weak H2O2 accumulation and a low amplitude of defense against M. oryzae, thereby triggering susceptibility. However, the recognition of effectors by different R genes leads to the activation of downstream signaling pathways with differential amplitudes, including increased H2O2, and activation of PTI, SA, JA, ET, and WRKY pathways, resulting in enhanced resistance. R1, R2, R3: Resistance genes. Different-sized arrows: The differential amplitude of immunity endowed by each R gene. BIC: Biotrophic interfacial complex. IH: Invasive hyphae.
Our results revealed divergent immune responses exhibited by the susceptible accession LTH and resistant accessions SH548, SH882, WSSM, IRBLZ5-CA, IRBL9-W, IRBLKM-TS, and R2115 in response to blast infection. Moreover, accessions carrying broad-spectrum blast resistance genes show the activation of immune signaling pathways, though the extent to which the R genes regulate immunity varies among accessions. This indicates that the different R genes regulate the immune responses differently. However, the activation of downstream defense- and hormone-signaling pathways mediated by rice NLRs remains to be further investigated. In this study, we demonstrated that M. oryzae triggered differential amplitudes of H2O2 accumulation and blast infection-induced marker genes associated with different defense and hormone signaling pathways in LTH and the resistant accessions carrying different R genes (Figures 1, 2, 5, 6). Therefore, this study provides valuable insights into the role of R genes in activating downstream defense and hormone signaling pathways in response to blast infection.
PTI and ETI share several convergent downstream responses, including calcium flux, reactive oxygen species (ROS), MAPK cascades, callose deposition, and production of phytohormones such as JA, ET, and SA (Tsuda and Katagiri, 2010; Cui et al., 2015; Peng et al., 2018). Upon pathogen attack, a complex array of signaling pathways is activated, modulating pathogen-induced resistance through a sophisticated signal transduction network (Ding et al., 2022). LTH is susceptible to over 1,000 isolates worldwide, and no functional R genes have been identified (Yang et al., 2022a), indicating the inactivation of ETI in LTH. This partially explains the differential defense response amplitudes between LTH and the other accessions carrying R genes.
Pathogen-induced H2O2 accumulation plays a crucial role in rice disease resistance (Li et al., 2019a). Notably, our results showed increased H2O2 accumulation in SH548, SH882, and WSSM, each harboring broad-spectrum blast R genes (Hassan et al., 2022). Although the three elite restorer lines showed similar resistant phenotypes, the H2O2 amounts were more than that in LTH and varied among them (Figure 1). The highest H2O2 accumulation was observed in SH882, followed by WSSM and SH548 (Figures 1B, C), implying that the NLR-encoding R genes differentially contribute to the spatiotemporal H2O2 accumulation, thereby limiting M. oryzae growth at the infection site. However, it is unknown how this R gene-mediated downstream signaling contributes to H2O2 accumulation. A previous study has demonstrated that Osa‐miR398b enhances H2O2 production and rice blast disease resistance by modulating multiple superoxide dismutases (Li et al., 2019a). Proteomic studies on the rice and M. oryzae interaction identified key ROS-related proteins involved in pathogen recognition and contributing to rice resistance (Meng et al., 2019). Various ROS-scavenging enzymes, such as OsPRX59 and OsPRX62, accumulate in incompatible interactions between rice and M. oryzae (Lin et al., 2018). Cao et al. (2016) identified increased accumulation of the rice NADPH oxidase OsRBOH8 in a PM proteomics study using rice leaves collected 48 hours post-inoculation with M. oryzae. Whether the R genes in the three restorer accessions control H2O2 accumulation through the aforementioned signaling pathways remains unknown and requires further investigation.
The activation of downstream signaling via NLRs is a complex process, with varying degrees of activation. In the context of multiple sensor NLR-mediated immune responses, NRG1 and ADR1 act as helper NLRs (Saile et al., 2020). They are involved not only in the induction of cell death in various other NLRs (Dong et al., 2016; Castel et al., 2019) but also cooperate with EDS1, senescence-associated gene 101 (SAG101), and PAD4 to activate TNL-mediated immunity (Sun et al., 2021). The transcription of OsEDS1 and OsPad4 was strongly induced by blast fungus and differed significantly among the three restorer accessions (Figure 2B), indicating that the two genes could be used as the marker genes to measure SA-related immune responses (Figure 7).
The perception of chitin by OsCERK1 and OsCEBiP triggers the activation of various immune responses critical for rice immunity against M. oryzae (Hayafune et al., 2014). Consistent with the opinion that PTI and ETI mutually enhance each other to trigger robust disease resistance (Ngou et al., 2021; Yuan et al., 2021b), the defense-related genes were up-regulated to higher levels in the three restorer lines than that in LTH (Figure 1D), in which pathogen-induced ETI is absent because of the lack of functional R genes. Consistently, the expression of OsCERK1 and OsCEBiP was constitutively higher in the three restorer accessions than that in LTH, and induced to higher levels by chitin in SH548 and SH882 (Figure 2A), and by M. oryzae in IRBLZ5-CA, IRBL9-W, IRBLKM-TS, and R2115 (Figure 5D). As the OsCEBiP was significantly induced in SH548 and SH882 compared with LTH, with a weak induction in LTH compared to OsCERK1, it could be used as a marker to measure the amplitudes of PTI responses in rice (Figure 7). Besides, as OsNAC4 and OsKS4 are involved in PTI, and these two genes were significantly upregulated to higher levels in the three restorer lines than in LTH, therefore these genes could be used as marker genes to measure the amplitudes of PTI responses in rice (Figure 7).
Hormone-related marker genes were also induced by M. oryzae in the three restorer accessions. For example, PR10b was induced by M. oryzae through the activation of JA signaling (Hashimoto et al., 2004). In all the accessions, induced expression of OsPR10b and OsAOS2 was observed, indicating the activation of JA signaling (Figures 1D, 2B, 5B, D) by M. oryzae. Moreover, both OsPR10b and OsAOS2 were induced to significantly higher levels at 24 or 48 hpi of M. oryzae in the three restorer accessions than in LTH, indicating that the two genes could be used as JA signaling pathway-related defense amplitude marker genes (Figure 7).
In rice, the SA pathway is regulated by OsNPR1 (De Vleesschauwer et al., 2014). Ectopic expression of NPR1 in rice was associated with constitutive PR transcripts’ expression, resulting in enhanced resistance to blast (Chern et al., 2005; Yuan et al., 2007; Sugano et al., 2010). A previous report showed that 12 rice OsPR1 genes were upregulated upon blast infection (Mitsuhara et al., 2008). Consistently, OsPR1a was remarkably induced to higher levels by M. oryzae in the R gene-carried accessions, except for IRBLz5-CA, compared to LTH. However, OsNPR1 was also induced by M. oryzae in LTH, with similar or higher mRNA amounts compared to those in the restorer accessions (Figures 2B, 6D). These results indicate that OsPR1a, rather than OsNPR1, could serve as a marker gene to indicate defense amplitudes (Figure 7).
WRKY transcription factors are widely involved in regulating development, growth, and defense responses to abiotic or biotic stresses in rice (Wei et al., 2013; Jeyasri et al., 2021). Consistent with a previous finding (Yokotani et al., 2018), OsWRKY24 was induced by M. oryzae in all the resistant accessions, whereas OsWRKY71 was upregulated in all accessions as well, accompanied by the highest induction in SH882 at all time points and the lowest induction levels in LTH (Figures 2C and 5E). Therefore, OsWRKY71 and OsWRK24 could be used as defense amplitude marker genes (Figure 7).
Altogether, we recommended 10 genes for measuring the amplitude of immunity against M. oryzae mediated by different blast R genes (Figure 7), providing convenience for examining the intensity of immune responses in rice.
Susceptible cultivars typically exhibit a greater number of DEGs following pathogen infection compared to resistant cultivars. A comprehensive transcriptome analysis was conducted on resistant and susceptible rice accessions following blast infection, uncovering a substantial number of convergent and divergent DEGs, along with genes associated with rice stress responses (Figure 3). Our RNA-seq data analysis revealed that the greatest number of DEGs was observed in LTH after blast infection, compared with the resistant accessions (Figure 3A). This finding aligns with previous studies. For example, Kumar et al. (2021) reported a greater number of DEGs in the blast-susceptible cultivar HP2216 than in the blast-resistant cultivar Tetep. Similarly, Yang et al. (2022b) observed a greater number of DEGs in the rice sheath blight-susceptible cultivar Koshihikari compared with the resistant cultivar Shennong 9819. Furthermore, Zhang et al. (2017) also observed a greater number of DEGs in the susceptible cultivar Lemont compared with the moderately resistant cultivar TeQing in response to rice sheath blight. These findings, in conjunction with our results, suggest that pathogen infection significantly alters the global gene expression profiles of plants, with a more pronounced effect in susceptible plants. This heightened impact in susceptible plants may be attributed to their increased energy expenditure on stress management, resulting in reduced growth and yield. Moreover, the higher number of DEGs in susceptible plants may also be due to the greater infection pressure they experience. Therefore, it seems that the number of DEGs is reversely correlated with the resistance and defense amplitudes.
Rice accessions used in this study included IRBLz5-CA, IRBL9-W, IRBLkm-Ts, Yahui2115 (R2115), Lijiangxin Tuan Heigu (LTH), Shu Hui 548 (SH548), Shu Hui 882 (SH882), and Wu Shan Si Miao (WSSM). All accessions were grown in a growth chamber under a photoperiod cycle of 12 hours of light and 12 hours of dark, at 26°C and 70% relative humidity.
For rice blast inoculation, M. oryzae strains Guy11 (carrying Avr-Pita) (Xiao et al., 2024) and CRB10 (carrying Avr-Piz, Avr-Pita2, and Avr-Piks) (Fang et al., 2018) were cultured on the oatmeal tomato agar (OTA) medium for two weeks under a photoperiod cycle of 12 hours of light and 12 hours of dark. Subsequently, the hyphae were scratched to promote sporulation. The CRB10 strain (4 × 105 spores/mL) was used for punch inoculation assay on SH548, SH882, WSSM, and LTH, whereas the Guy11 strain (4 × 105 spores/mL) was used for spray inoculation on all accessions, including LTH, IRBLz5-CA, IRBL9-W, IRBLkm-Ts, R2115, SH548, SH882, and WSSM at the three-leaf stage. Besides, the leaves of LTH, SH548, SH882, and WSSM were inoculated with 200 μM chitin in 0.1 mmol/L 6-benzyladenine buffer, and samples were collected at 0, 6, 9, and 12 hpt. All the samples underwent subsequent RNA extraction and RT-qPCR analysis.
DAB staining was performed by following an established procedure (Zhao et al., 2020). Briefly, three-week-old seedlings of LTH, SH548, SH882, and WSSM were inoculated with Guy11 (4 × 105 spores/mL). Leaves of each accession were collected at 48 hpi and immersed in 10 mL tubes filled with DAB solution, prepared by dissolving DAB in HCl and adjusting the pH to 3.8. The leaf samples were then vacuum-infiltrated for 30 minutes and incubated in darkness overnight. Subsequently, the leaves were washed with 95% ethanol and continuously washed in a 65°C water bath until they turned colorless. H2O2 accumulation and fungal structures within the leaves were examined using a fluorescence microscope (Zeiss imager A2).
The RNA-seq data used in this study were derived from a previous investigation (Hu et al., 2023).
For each accession, we defined upregulated genes as those that were upregulated at 12 hours but not downregulated at 24 hours, and those upregulated at 24 hours but not downregulated at 12 hours. Similarly, we identified downregulated genes using the same criteria in reverse. Subsequently, R programming language was employed to conduct overlap analyses of the upregulated and downregulated gene sets in each accession and to generate Venn diagrams, along with GO and KEGG pathway classifications. For the analysis of the relative expression of plant immune-related genes, Log2RPKM (reads per kilobase per million) values from two biological replicates of transcriptome data were used. Normalization of each row of transcriptome data and heat map generation were performed using TBtools.
Total RNA was extracted from the leaves using VeZol reagent (Vazyme), and its concentration was measured using a Nanodrop 2000 spectrophotometer (Thermo Scientific). Subsequently, cDNA was synthesized using NovoScript Plus All-in-one 1st Strand cDNA Synthesis SuperMix (Novoprotein, China). RT-qPCR was conducted with AceQ Universal SYBR qPCR Master Mix (Vazyme, China) to analyze the expression patterns of defense-related genes, hormone signaling pathway genes, and OsAGO1 family genes. Gene expression levels were normalized using Ubiquitin (Ubi) as an internal control. Primers used in this study are listed in Supplementary Table S5.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
BH: Funding acquisition, Writing – original draft, Writing – review & editing, Data curation, Investigation, Methodology. SB: Investigation, Methodology, Writing – review & editing. X-XY: Investigation, Methodology, Writing – review & editing. XY: Investigation, Methodology, Writing – review & editing. RL: Investigation, Writing – review & editing, Formal analysis. MG: Writing – review & editing, Resources. YT: Resources, Writing – review & editing. DG: Writing – review & editing, Data curation, Formal analysis. SY: Formal analysis, Writing – review & editing. FG: Formal analysis, Writing – review & editing. YL: Writing – review & editing, Writing – original draft. XZ: Writing – review & editing, Supervision. ZZ: Supervision, Writing – original draft, Writing – review & editing, Funding acquisition. WW: Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32350410431 to BH, 32101728 to Z-XZ, U19A2033 to W-MW) and the Open Project Program (SKL-KF202423) of State Key Laboratory of Crop Gene Exploration and Utilization in Southwest China.
We thank Dr. Yan-Yan Huang (Sichuan Agricultural University) for providing the rice materials and Mei Pu and Chi Li (Sichuan Agricultural University) for managing the laboratory and rice materials in the field.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1547593/full#supplementary-material
Supplementary Figure 1 | GO and KEGG pathway classification of up-regulated and down-regulated DEGs in IRBL9-W. (A) GO classification, (B) KEGG pathway classification.
Supplementary Figure 2 | GO and KEGG pathway classification of up-regulated and down-regulated DEGs in IRBLkm-Ts. (A) GO classification, (B) KEGG pathway classification.
Supplementary Figure 3 | GO and KEGG pathway classification of up-regulated and down-regulated DEGs in R2115. (A) GO classification, (B) KEGG pathway classification.
Supplementary Figure 4 | GO and KEGG pathway classification of up-regulated and down-regulated DEGs in LTH. (A) GO classification, (B) KEGG pathway classification.
Agrawal, G. K., Jwa, N.-S., Rakwal, R. (2000a). A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem. Biophys. Res. Commun. 274, 157–165. doi: 10.1006/bbrc.2000.3114
Agrawal, G. K., Rakwal, R., Jwa, N.-S. (2000b). Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem. Biophys. Res. Commun. 278, 290–298. doi: 10.1006/bbrc.2000.3781
Alfano, J. R., Collmer, A. (2004). Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. doi: 10.1146/annurev.phyto.42.040103.110731
Boatwright, J. L., Pajerowska-Mukhtar, K. (2013). Salicylic acid: an old hormone up to new tricks. Mol. Plant Pathol. 14, 623–634. doi: 10.1111/mpp.12035
Cao, J., Yang, C., Li, L., Jiang, L., Wu, Y., Wu, C., et al. (2016). Rice plasma membrane proteomics reveals Magnaporthe oryzae promotes susceptibility by sequential activation of host hormone signaling pathways. Mol. Plant Microb. Interact. 29, 902–913. doi: 10.1094/MPMI-08-16-0165-R
Castel, B., Ngou, P. M., Cevik, V., Redkar, A., Kim, D. S., Yang, Y., et al. (2019). Diverse NLR immune receptors activate defence via the RPW 8-NLR NRG1. New Phytol. 222, 966–980. doi: 10.1111/nph.15659
Chern, M., Fitzgerald, H. A., Canlas, P. E., Navarre, D. A., Ronald, P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microb. Interact. 18, 511–520. doi: 10.1094/MPMI-18-0511
Couto, D., Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol. 16, 537–552. doi: 10.1038/nri.2016.77
Cui, H., Tsuda, K., Parker, J. E. (2015). Effector-triggered immunity: From pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Dai, X., Wang, Y., Yu, K., Zhao, Y., Xiong, L., Wang, R., et al. (2023). OsNPR1 Enhances Rice Resistance to Xanthomonas oryzae pv. oryzae by Upregulating Rice Defense Genes and Repressing Bacteria Virulence Genes. Int. J. Mol. Sci. 24, 8687. doi: 10.3390/ijms24108687
Dalio, R. J., Paschoal, D., Arena, G. D., Magalhães, D. M., Oliveira, T. S., Merfa, M. V., et al. (2021). Hypersensitive response: From NLR pathogen recognition to cell death response. Ann. Appl. Biol. 178, 268–280. doi: 10.1111/aab.12657
De Vleesschauwer, D., Xu, J., Höfte, M. (2014). Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00611
Ding, L.-N., Li, Y.-T., Wu, Y.-Z., Li, T., Geng, R., Cao, J., et al. (2022). Plant disease resistance-related signaling pathways: recent progress and future prospects. Int. J. Mol. Sci. 23, 16200. doi: 10.3390/ijms232416200
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi: 10.1016/j.pbi.2004.07.005
Dong, O. X., Tong, M., Bonardi, V., El Kasmi, F., Woloshen, V., Wünsch, L. K., et al. (2016). TNL-mediated immunity in Arabidopsis requires complex regulation of the redundant ADR1 gene family. New Phytol. 210, 960–973. doi: 10.1111/nph.13821
Dou, D., Zhou, J.-M. (2012). Phytopathogen effectors subverting host immunity: different foes, similar battleground. Cell Host Microbe 12, 484–495. doi: 10.1016/j.chom.2012.09.003
Duan, L., Liu, H., Li, X., Xiao, J., Wang, S. (2014). Multiple phytohormones and phytoalexins are involved in disease resistance to Magnaporthe oryzae invaded from roots in rice. Physiol. Plant 152, 486–500. doi: 10.1111/ppl.12192
Fang, W. W., Liu, C. C., Zhang, H. W., Xu, H., Zhou, S., Fang, K. X., et al. (2018). Selection of differential isolates of Magnaporthe oryzae for postulation of blast resistance genes. Phytopathology 108, 878–884. doi: 10.1094/phyto-09-17-0333-r
Feng, J.-X., Cao, L., Li, J., Duan, C.-J., Luo, X.-M., Le, N., et al. (2011). Involvement of OsNPR1/NH1 in rice basal resistance to blast fungus Magnaporthe oryzae. Eur. J. Plant Pathol. 131, 221–235. doi: 10.1007/s10658-011-9801-7
Hashimoto, M., Kisseleva, L., Sawa, S., Furukawa, T., Komatsu, S., Koshiba, T. (2004). A novel rice PR10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol. 45, 550–559. doi: 10.1093/pcp/pch063
Hassan, B., Peng, Y.-T., Li, S., Yin, X.-X., Chen, C., Gulzar, F., et al. (2022). Identification of the blast resistance genes in three elite restorer lines of hybrid rice. Phytopathol. Res. 4, 1–10. doi: 10.1186/s42483-022-00120-6
Hayafune, M., Berisio, R., Marchetti, R., Silipo, A., Kayama, M., Desaki, Y., et al. (2014). Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. 111, E404–E413. doi: 10.1073/pnas.1312099111
Hou, S., Tsuda, K. (2022). Salicylic acid and jasmonic acid crosstalk in plant immunity. Essays Biochem. 66, 647–656. doi: 10.1042/EBC20210090
Hu, X., Li, H., Shi, J., Li, D., Fan, J., Huang, F., et al (2024). The elite rice restorer line ‘Yahui2115’ carries blast resistance gene Pib. Acta Phytopathologica Sinica. 54 (4), 769–776. doi: 10.13926/j.cnki.apps.000899
Hu, X.-H., Shen, S., Wu, J.-L., Liu, J., Wang, H., He, J.-X., et al. (2023). A natural allele of proteasome maturation factor improves rice resistance to multiple pathogens. Nat. Plants 9, 228–237. doi: 10.1038/s41477-022-01327-3
Huang, L., Hong, Y., Zhang, H., Li, D., Song, F. (2016). Rice NAC transcription factor ONAC095 plays opposite roles in drought and cold stress tolerance. BMC Plant Biol. 16, 1–18. doi: 10.1186/s12870-016-0897-y
Huang, Y., Li, Y., Wang, H., Wenming, W. (2023). Allelic variation in the race-specific blast resistance genes in rice. Acta Phytopathol. Sin. 53, 753–768. doi: 10.13926/j.cnki.apps.000866
Jeyasri, R., Muthuramalingam, P., Satish, L., Adarshan, S., Lakshmi, M. A., Pandian, S. K., et al. (2021). The role of OsWRKY genes in rice when faced with single and multiple abiotic stresses. Agronomy 11, 1301. doi: 10.3390/agronomy11071301
Jimmy, J. L., Babu, S. (2015). Role of OsWRKY transcription factors in rice disease resistance. Trop. Plant Pathol. 40, 355–361. doi: 10.1007/s40858-015-0058-0
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. 103, 11086–11091. doi: 10.1073/pnas.0508882103
Kaneda, T., Taga, Y., Takai, R., Iwano, M., Matsui, H., Takayama, S., et al. (2009). The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 28, 926–936. doi: 10.1038/emboj.2009.39
Ke, Y., Kang, Y., Wu, M., Liu, H., Hui, S., Zhang, Q., et al. (2019). Jasmonic acid-involved OsEDS1 signaling in Rice-bacteria interactions. Rice 12, 1–12. doi: 10.1186/s12284-019-0283-0
Ke, Y., Liu, H., Li, X., Xiao, J., Wang, S. (2014). Rice OsPAD4 functions differently from Arabidopsis AtPAD4 in host-pathogen interactions. Plant J. 78, 619–631. doi: 10.1111/tpj.12500
Kim, S. T., Kang, Y. H., Wang, Y., Wu, J., Park, Z. Y., Rakwal, R., et al. (2009). Secretome analysis of differentially induced proteins in rice suspension-cultured cells triggered by rice blast fungus and elicitor. Proteomics 9, 1302–1313. doi: 10.1002/pmic.200800589
Kim, S. T., Kim, S. G., Hwang, D. H., Kang, S. Y., Kim, H. J., Lee, B. H., et al. (2004). Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4, 3569–3578. doi: 10.1002/pmic.200400999
Kim, S. T., Kim, S. G., Kang, Y. H., Wang, Y., Kim, J.-Y., Yi, N., et al. (2008). Proteomics analysis of rice lesion mimic mutant (spl1) reveals tightly localized probenazole-induced protein (PBZ1) in cells undergoing programmed cell death. J. Proteome Res. 7, 1750–1760. doi: 10.1021/pr700878t
Kim, S. G., Wang, Y., Lee, K. H., Park, Z.-Y., Park, J., Wu, J., et al. (2013). In-depth insight into in vivo apoplastic secretome of rice-Magnaporthe oryzae interaction. J. Proteom. 78, 58–71. doi: 10.1016/j.jprot.2012.10.029
Kumar, V., Jain, P., Venkadesan, S., Karkute, S. G., Bhati, J., Abdin, M. Z., et al. (2021). Understanding Rice-Magnaporthe oryzae interaction in resistant and susceptible cultivars of rice under panicle blast infection using a time-course transcriptome analysis. Genes 12, 301. doi: 10.3390/genes12020301
Li, Y., Cao, X. L., Zhu, Y., Yang, X. M., Zhang, K. N., Xiao, Z. Y., et al. (2019a). Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 222, 1507–1522. doi: 10.1111/nph.15678
Li, Y., Jeyakumar, J. M. J., Feng, Q., Zhao, Z.-X., Fan, J., Khaskheli, M. I., et al. (2019b). The roles of rice microRNAs in rice-Magnaporthe oryzae interaction. Phytopathol. Res. 1, 1–12. doi: 10.1186/s42483-019-0040-8
Lin, S., Nie, P., Ding, S., Zheng, L., Chen, C., Feng, R., et al. (2018). Quantitative proteomic analysis provides insights into rice defense mechanisms against Magnaporthe oryzae. Int. J. Mol. Sci. 19, 1950. doi: 10.3390/ijms19071950
Liu, X., Bai, X., Wang, X., Chu, C. (2007). OsWRKY71, a rice transcription factor, is involved in rice defense response. J. Plant Physiol. 164, 969–979. doi: 10.1016/j.jplph.2006.07.006
Liu, Q., Li, X., Yan, S., Yu, T., Yang, J., Dong, J., et al. (2018). OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol. 18, 1–13. doi: 10.1186/s12870-018-1479-y
Meng, Q., Gupta, R., Min, C. W., Kim, J., Kramer, K., Wang, Y., et al. (2019). A proteomic insight into the MSP1 and flg22 induced signaling in Oryza sativa leaves. J. Proteom. 196, 120–130. doi: 10.1016/j.jprot.2018.04.015
Mitsuhara, I., Iwai, T., Seo, S., Yanagawa, Y., Kawahigasi, H., Hirose, S., et al. (2008). Characteristic expression of twelve rice PR1 family genes in response to pathogen infection, wounding, and defense-related signal compounds (121/180). Mol. Genet. Genom. 279, 415–427. doi: 10.1007/s00438-008-0322-9
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Ng, D. W., Abeysinghe, J. K., Kamali, M. (2018). Regulating the regulators: The control of transcription factors in plant defense signaling. Int. J. Mol. Sci. 19, 3737. doi: 10.3390/ijms19123737
Ngou, B. P. M., Ahn, H.-K., Ding, P., Jones, J. D. (2021). Mutual potentiation of plant immunity by cell-surface and intracellular receptors. Nature 592, 110–115. doi: 10.1038/s41586-021-03315-7
Noman, A., Liu, Z., Aqeel, M., Zainab, M., Khan, M. I., Hussain, A., et al. (2017). Basic leucine zipper domain transcription factors: the vanguards in plant immunity. Biotechnol. Lett. 39, 1779–1791. doi: 10.1007/s10529-017-2431-1
Peng, Y., van Wersch, R., Zhang, Y. (2018). Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant-Microbe Interact. 31, 403–409. doi: 10.1094/MPMI-06-17-0145-CR
Rakwal, R., Komatsu, S. (2000). Role of jasmonate in the rice (Oryza sativa L.) self-defense mechanism using proteome analysis. Electrophoresis 21, 2492–2500. doi: 10.1002/1522-2683(20000701)21:12<2492::AID-ELPS2492>3.0.CO;2-2
Saile, S. C., Jacob, P., Castel, B., Jubic, L. M., Salas-Gonzáles, I., Bäcker, M., et al. (2020). Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PloS Biol. 18, e3000783. doi: 10.1371/journal.pbio.3000783
Shi, J., Li, D., Li, Y., Li, X., Guo, X., Luo, Y., et al. (2015). Identification of rice blast resistance genes in the elite hybrid rice restorer line Yahui2115. Genome 58, 91–97. doi: 10.1139/gen-2015-0005
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Shimono, M., Sugano, S., Nakayama, A., Jiang, C.-J., Ono, K., Toki, S., et al. (2007). Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 19, 2064–2076. doi: 10.1105/tpc.106.046250
Shinya, T., Motoyama, N., Ikeda, A., Wada, M., Kamiya, K., Hayafune, M., et al. (2012). Functional characterization of CEBiP and CERK1 homologs in Arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53, 1696–1706. doi: 10.1093/pcp/pcs113
Spoel, S. H., Dong, X. (2008). Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe 3, 348–351. doi: 10.1016/j.chom.2008.05.009
Sugano, S., Jiang, C.-J., Miyazawa, S.-I., Masumoto, C., Yazawa, K., Hayashi, N., et al. (2010). Role of OsNPR1 in rice defense program as revealed by genome-wide expression analysis. Plant Mol. Biol. 74, 549–562. doi: 10.1007/s11103-010-9695-3
Sun, X., Lapin, D., Feehan, J. M., Stolze, S. C., Kramer, K., Dongus, J. A., et al. (2021). Pathogen effector recognition-dependent association of NRG1 with EDS1 and SAG101 in TNL receptor immunity. Nat. Commun. 12, 3335. doi: 10.1038/s41467-021-23614-x
Tonnessen, B. W., Manosalva, P., Lang, J. M., Baraoidan, M., Bordeos, A., Mauleon, R., et al. (2015). Rice phenylalanine ammonia-lyase gene OsPAL4 is associated with broad spectrum disease resistance. Plant Mol. Biol. 87, 273–286. doi: 10.1007/s11103-014-0275-9
Tsuda, K., Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Vlot, A. C., Dempsey, D. M. A., Klessig, D. F. (2009). Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wang, R., Wang, G.-L., Ning, Y. (2019). PALs: Emerging key players in broad-spectrum disease resistance. Trends Plant Sci. 24, 785–787. doi: 10.1016/j.tplants.2019.06.012
Wei, T., Ou, B., Li, J., Zhao, Y., Guo, D., Zhu, Y., et al. (2013). Transcriptional profiling of rice early response to Magnaporthe oryzae identified OsWRKYs as important regulators in rice blast resistance. PloS One 8, e59720. doi: 10.1371/journal.pone.0059720
Wiermer, M., Feys, B. J., Parker, J. E. (2005). Plant immunity: the EDS1 regulatory node. Curr. Opin. Plant Biol. 8, 383–389. doi: 10.1016/j.pbi.2005.05.010
Xiao, G., Laksanavilat, N., Cesari, S., Lambou, K., Baudin, M., Jalilian, A., et al. (2024). The unconventional resistance protein PTR recognizes the Magnaporthe oryzae effector AVR-Pita in an allele-specific manner. Nat. Plants 10, 994–1004. doi: 10.1038/s41477-024-01694-z
Yang, X., Gu, X., Ding, J., Yao, L., Gao, X., Zhang, M., et al. (2022b). Gene expression analysis of resistant and susceptible rice cultivars to sheath blight after inoculation with Rhizoctonia solani. BMC Genomics 23, 278. doi: 10.1186/s12864-022-08524-6
Yang, Y., Qi, M., Mei, C. (2004). Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 40, 909–919. doi: 10.1111/j.1365-313X.2004.02267.x
Yang, D.-L., Yang, Y., He, Z. (2013). Roles of plant hormones and their interplay in rice immunity. Mol. Plant. 6, 675–685. doi: 10.1093/mp/sst056
Yang, L., Zhao, M., Sha, G., Sun, Q., Gong, Q., Yang, Q., et al. (2022a). The genome of the rice variety LTH provides insight into its universal susceptibility mechanism to worldwide rice blast fungal strains. Comput. Struct. Biotechnol. 20, 1012–1026. doi: 10.1016/j.csbj.2022.01.030
Yokotani, N., Shikata, M., Ichikawa, H., Mitsuda, N., Ohme-Takagi, M., Minami, E., et al. (2018). OsWRKY24, a blast-disease responsive transcription factor, positively regulates rice disease resistance. J. Gen. Plant Pathol. 84, 85–91. doi: 10.1007/s10327-018-0768-5
Yu, X., Feng, B., He, P., Shan, L. (2017). From chaos to harmony: Responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. doi: 10.1146/annurev-phyto-080516-035649
Yuan, M., Jiang, Z., Bi, G., Nomura, K., Liu, M., Wang, Y., et al. (2021a). Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 592, 105–109. doi: 10.1038/s41586-021-03316-6
Yuan, M., Ngou, B. P. M., Ding, P., Xin, X.-F. (2021b). PTI-ETI crosstalk: An integrative view of plant immunity. Curr. Opin. Plant Biol. 62, 102030. doi: 10.1016/j.pbi.2021.102030
Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y., Wang, L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Zhang, J., Chen, L., Fu, C., Wang, L., Liu, H., Cheng, Y., et al. (2017). Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01422
Zhao, Z. X., Feng, Q., Cao, X. L., Zhu, Y., Wang, H., Chandran, V., et al. (2020). Osa-miR167d facilitates infection of Magnaporthe oryzae in rice. J. Integr. Plant Biol. 62, 702–715. doi: 10.1111/jipb.12816
Zhao, Z. X., Yang, S.-J., Yin, X.-X., Yan, X.-L., Hassan, B., Fan, J., et al. (2023). ARGONAUTE 1: a node coordinating plant disease resistance with growth and development. Phytopathol. Res. 5, 38. doi: 10.1186/s42483-023-00194-w
Keywords: rice, Magnaporthe oryzae, rice blast disease, blast resistance genes, immune response
Citation: Hassan B, Bhutto SH, Yin X-X, Yan X-L, Liao R, Guo M-L, Tang Y-P, Guo D-M, Yang S-J, Gulzar F, Li Y, Zeng X-Y, Zhao Z-X and Wang W-M (2025) Divergent response associates with the differential amplitudes of immunity against Magnaporthe oryzae by different blast resistance genes. Front. Plant Sci. 16:1547593. doi: 10.3389/fpls.2025.1547593
Received: 18 December 2024; Accepted: 03 February 2025;
Published: 24 February 2025.
Edited by:
Yuelin Zhang, University of British Columbia, CanadaReviewed by:
Shimin Zuo, Yangzhou University, ChinaCopyright © 2025 Hassan, Bhutto, Yin, Yan, Liao, Guo, Tang, Guo, Yang, Gulzar, Li, Zeng, Zhao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Yin Zeng, eHl6ZW5nQHNpY2F1LmVkdS5jbg==; Zhi-Xue Zhao, emhpeHVlemhhb0BzaWNhdS5lZHUuY24=; Wen-Ming Wang, ajMxNndlbm1pbmd3YW5nQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.