
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 10 February 2025
Sec. Plant Physiology
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1546819
This article is part of the Research Topic The Dynamic Plant Cell Wall: Sensing, Remodelling, and Integrity View all 4 articles
 Darina Balkova1†
Darina Balkova1† Katerina Mala1
Katerina Mala1 Jan Hejatko2,3
Jan Hejatko2,3 Klara Panzarova4
Klara Panzarova4 Lamis Abdelhakim4
Lamis Abdelhakim4 Barbora Pleskacova4
Barbora Pleskacova4 Marketa Samalova1,2*
Marketa Samalova1,2*Expansins are cell wall-modifying proteins implicated in plant growth and stress responses. In this study, we explored the differential localization of expansins in Arabidopsis thaliana shoots, with a focus on EXPA1, EXPA10, EXPA14, and EXPA15 utilizing pEXPA::EXPA translational fusion lines. Employing the chemically inducible system pOp6/LhGR for EXPA1 overexpression and high-throughput automatic phenotyping we evaluated the drought response and photosynthetic efficiency under stress conditions. We observed distinct expression patterns of expansins, with EXPA1 primarily localized in stomatal guard cells, while EXPA10 and EXPA15 showed strong cell wall (CW) localization in epidermal and other tissues. Overexpression of EXPA1 resulted in pronounced changes in CW-related gene expression, particularly during early stages of induction, including the upregulation of other expansins and CW-modifying enzymes. The induced EXPA1 line also displayed significant morphological changes in shoots, including smaller plant size, delayed senescence, and structural alterations in vascular tissues. Additionally, EXPA1 overexpression conferred drought tolerance, as evidenced by enhanced photosynthetic efficiency (Fv/FM), and low steady-state non-photochemical quenching (NPQ) values under drought stress. These findings highlight the critical role of EXPA1 in regulating plant growth, development, and stress response, with potential applications in improving drought tolerance in crops.
The ability of plants to endure environmental stress is a critical determinant of their growth and development. Among various stressors, drought is one of the most detrimental to plant health, as it directly restricts water availability and impacts numerous physiological and metabolic processes (Salehi-Lisar and Bakhshayeshan-Agdam, 2016). In response to such challenges, plants have evolved intricate adaptive mechanisms (Huang et al., 2012), with the cell wall (CW) playing a pivotal role in responding to environmental stimuli (Cosgrove, 2000a). The dynamic nature of the CW is largely regulated by expansins, which are key agents in CW remodeling. This gene family is classified into four subgroups: α-EXPANSIN (EXPA), β-EXPANSIN (EXPB), EXPANSIN-like A (EXLA), and EXPANSIN-like B (EXLB), each contributing uniquely to various aspects of plant development (Lee et al., 2001; Sampedro and Cosgrove, 2005; Cosgrove, 2015; Samalova et al., 2022). Expansins, identified as non-enzymatic CW proteins, facilitate “cell wall loosening” by disrupting the non-covalent hydrogen bonds between cellulose microfibrils and hemicellulose xyloglucans, allowing for the turgor-driven cell expansion (Cosgrove, 2000b; Cosgrove, 2005; Cosgrove, 2016; Cosgrove, 2024). Through this mechanism, expansins modulate the structural flexibility of the CW which is essential for maintaining cell turgor and promoting growth under both normal conditions and in response to abiotic and biotic stressors (Lee et al., 2001; Cosgrove, 2005; Gao et al., 2010). The expression of expansin genes in Arabidopsis thaliana is regulated by various internal and external factors, such as phytohormones, developmental stages, and environmental stressors (Samalova et al., 2022). These genes are predominantly expressed in rapidly elongating cells, the root and shoot apical meristems, leaf primordia, and young stems; however, the expression patterns of individual expansin genes vary according to their specific functions (Cho and Cosgrove, 2000; 2002; McQueen-Mason and Cosgrove, 1995; Samalova et al., 2024).
In response to drought stress, plants undergo various physiological, biochemical, and molecular changes, including stomatal closure, accumulation of osmolytes, and activation of stress-responsive genes (Shao et al., 2008a, b). Numerous studies have suggested that modifying expansin expression may facilitate these critical physiological and molecular responses, potentially participating in stress-signaling pathways that enhance plant resilience under water-limited conditions (Chen et al., 2016, 2018; Dai et al., 2012; Guo et al., 2011; Hao et al., 2017; Kwon et al., 2008; Li et al., 2011; Liu et al., 2019; Lu et al., 2013; Marowa et al., 2020; Narayan et al., 2019, 2021; Wu et al., 2001; Zhang et al., 2021). Most of these studies link increased plant drought tolerance due to overexpression of expansins with enhanced root growth and improved root architecture, thereby potentially increasing water uptake and nutrient acquisition (Cho and Cosgrove, 2002; Guo et al., 2011; Lee et al., 2003; Wu et al., 2001). Interestingly, a recent study using a machine learning-driven meta-analysis of all available stress-related transcriptomic data identified expansins as core stress genes in roots (Sanchez-Munoz et al., 2024).
However, the impact of drought extends beyond root growth, affecting the entire plant in a complex manner, including shoot development and processes such as photosynthesis and gas exchange (Cho and Cosgrove, 2000; Flexas et al., 2004; Chaves et al., 2009). For instance, Dai et al. (2012) indicated that the expansin gene RhEXPA4 plays a regulatory role in dehydration tolerance during the expansion of rose (Rosa hybrida) petals. Furthermore, transgenic Arabidopsis plants overexpressing RhEXPA4 exhibited enhanced drought tolerance and improved recovery following a drought period compared to wild-type (WT) plants. Similarly, in tobacco (Nicotiana tabacum), the overexpression of the wheat (Triticum aestivum) TaEXPB23 gene enabled transgenic plants to retain water for a longer period under drought stress than WT plants, as indicated by higher maximum fluorescence in dark adapted state (QYmax) and steady-state Photosystem II quantum yield (ΦPSII) values and reduced electrolyte leakage (Li et al., 2011). Likewise, overexpression of EaEXPA1 from WT sugarcane (Erianthus arundinaceus) in the Co 86032 cultivar resulted in higher relative water content, improved membrane thermostability, increased chlorophyll content, enhanced quantum yield and gas exchange under drought stress, compared to WT plants (Narayan et al., 2021). The observed increase in photosynthetic capacity during drought stress due to expansin overexpression may be attributed to enhanced growth and leaf formation, resulting in an expanded photosynthetically active area and improved light capture under stress conditions (Chen et al., 2018; Marowa et al., 2020; Zhang et al., 2021). Moreover, research indicates that expansin overexpression may influence stomatal dynamics and accelerate stomatal movements (Wei et al., 2011; Zhang et al., 2011) and reduce stomatal density under drought conditions (Lu et al., 2013). This suggests a role of expansins in regulating stomatal movements - an essential process for minimizing water via transpiration during drought (Farooq et al., 2009).
Expansin upregulation under drought stress is also associated with the activation of signaling pathways and stress response genes involved in osmolyte accumulation, antioxidant defense, and hormone signaling (Shao et al., 2008a; Chen et al., 2016, 2018). For example, overexpression of wheat TaEXPA2 in tobacco enhanced drought tolerance by improving the water management and promoting the accumulation of osmotically active substances, such as proline. Transgenic plants also showed higher antioxidant levels and reduced reactive oxygen species (ROS) accumulation (Chen et al., 2016). Similarly, the ability to scavenge ROS was significantly improved in transgenic Arabidopsis plants overexpressing Ammopiptanthus nanus AnEXPA1 and AnEXPA2 (Liu et al., 2019). In cotton, overexpression of GhEXLB2 improved drought tolerance during critical stages such as germination, early growth, and flowering, reduced ROS accumulation, and increased antioxidant enzyme activity. Transgenic plants also displayed higher chlorophyll content, osmoprotective soluble sugars, and water-use efficiency compared to WT plants (Zhang et al., 2021).
Based on these findings, it might be expected that knock-out mutants of individual expansin genes would exhibit reduced biomass growth and increased sensitivity to abiotic stress. However, the literature suggests that expansin mutants often show no significant phenotypic changes, likely due to functional redundancy within the expansin gene family, a phenomenon observed in other gene families as well. Thus, simultaneous mutations in multiple expansin genes may be necessary to produce significant phenotypic changes (Cosgrove et al., 2002; Schipper et al., 2002; Li et al., 2003). Nevertheless, the precise molecular mechanisms by which expansins contribute to drought tolerance remain an active area of research. The interplay between expansins and other drought-responsive pathways highlights the complexity of expansin-mediated stress responses in plants (Cutler et al., 2010).
In this study we describe the expression of several hormone-responsive expansins in the shoots of Arabidopsis thaliana, explore genome-wide changes associated with AtEXPA1 overexpression (EXPA1 OE), and investigate the responses of overexpression and genetically engineered multiple knocked-out lines under abiotic stress conditions, focusing on drought stress. Understanding these mechanisms is essential not only for advancing knowledge of plant stress biology but also for developing crops with enhanced resilience to changing environments, thereby contributing to the sustainability of agriculture in the face of global climate change.
Seedlings of Arabidopsis thaliana (ecotype Col-0) were grown in vitro on standard Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 1.5% (w/v) sucrose and 0.8% (w/v) plant agar (Duchefa), with the pH adjusted to 5.8. For osmotic stress experiments various concentrations of NaCl (75, 150, 225 mM) were added to the medium, along with 20 μM Dex for induction, seedlings were germinated and grown on the plates for 7 days. The plants were cultivated in growth chambers under long-day conditions (16 h light/8 h dark) at 21°C during the light phase and 19°C during the dark phase (21°C/19°C).
Plants were cultivated as described in Awlia et al., 2016 and Flutsch et al., 2020, with the following modifications. Seeds were sown on soil (Substrate 2, Klasmann-Deilmann GmbH, Germany), stratified at 4°C for 3 days in the dark, and transferred to a climate-controlled chamber (FytoScope FS-WI, PSI, Drasov, Czech Republic) under 12h/12h, 21°C/19°C light/dark photoperiod with a relative humidity of 60% (for control condition experiment) and 45% (for drought stress experiment) and an irradiance of 120 µmol m−2 s−1 (cool-white and far-red LED).
Control condition experiment: Seeds of wild-type (WT) and Dex-inducible pRPS5A>GR>EXPA1 overexpressing (EXPA1 OE) lines 5-4 and 8-4 (Samalova et al., 2024) were sown in pots. Seven days after stratification (DAS), seedlings of similar size were transplanted into single pots prepared the day before with 95 g of sieved soil and watered up to the maximum soil-water holding capacity either with water or Dex solution (20 μM). Plants were cultivated in the climate-controlled chamber as described above. At 18 DAS plants were placed randomly in trays (10 pots per tray) and into the PlantScreen™ Compact System (PSI, Czech Republic) installed in controlled environment (FytoScope FS_WI, PSI, Czech Republic) with the same conditions as described above. Plants were watered manually every 2-3 days with 10 ml of 20 μM Dex solution as described in Samalova et al., 2019. Automated phenotyping started at 18 DAS and continued until 46 DAS. Plants were imaged daily using Red Green Blue (RGB) imaging. The PlantScreen™ Compact System facilitated the daily transport of trays for phenotypic analyses on conveyor belts from the dark/light acclimation chamber to the light-isolated imaging cabinets and the weighing and watering station, where plants were automatically weighed and watered daily to maintain the soil at a relative water content (RWC) of 60% field capacity.
Drought stress experiment: WT, EXPA1 OE line (8-4), and triple mutant (expa1,10,14-1 and -2 generated in this study) seeds were planted in soil. The EXPA1 OE line and WT were treated with or without Dex. Plants were cultivated as described above (control experiment) with the following modifications. Drought experiment was designed according to Clauw et al., 2015 with first drought phase from 10 DAS until 23 DAS followed by recovery phase for 6 days (from 24 to 29 DAS) and initiation of second drought phase from 30 DAS till 36 DAS. Seven DAS seedlings of similar size were transplanted into single pots prepared the day before with 65 g of sieved soil and watered regularly up 44% RWC (control levels; refers to 2.2 g of water per 1 g of dry soil) with water or with Dex solution (20 μM). At 10 DAS progressive drought stress was induced by watering half of the plants only up to 22% RWC (refers to 2.2 g of water per 1g of dry soil) with water or with Dex solution. Plants were cultivated in the climate-controlled chamber and 10 DAS were placed randomly in trays (10 pots per tray) and into the PlantScreen™ Compact System (PSI, Czech Republic) installed in controlled environment (FytoScope FS_WI, PSI, Drasov, Czech Republic). Automated phenotyping started 10 DAS and continued daily until 36 DAS. The photoperiod was changed to 16 h light/8 h dark, and humidity was kept at 45%. Successively, plants were automatically phenotyped using kinetic chlorophyll fluorescence imaging and RGB imaging, and automated watering and weighing in the listed order. The changes in the photosynthesis-related parameters were measured at different photon irradiances using the light curve protocol as described in Awlia et al., 2016. Briefly, a 5s flash of light was applied to measure the minimum fluorescence, followed by a saturation pulse of 800 ms (with an irradiance of 1200 μmol m-2 s-1) to determine the maximum fluorescence in the dark-adapted state. Next, 60 s intervals of cool-white actinic light at 95, 210, 320, 440 μmol m-2 s-1 corresponding to L1, L2, L3, and L4, respectively, were applied. A saturation pulse was applied at the end of the period of actinic light to acquire the maximal fluorescence in the light- adapted state. The chlorophyll fluorescence (Chl) signal measured just before the saturation pulse was taken as the steady-state fluorescence value in the light-adapted state. Using the automatic timing function of PlantScreen™ Scheduler (PSI, Drasov, Czech Republic), the phenotyping protocol was programmed to start always at the same time of the diurnal cycle (after 2 h of illumination in the climate-controlled growth chamber). A total of 26-28 plants per treatment and line were used.
Image segmentation and automatic raw data processing were performed using the PlantScreen™ Analyzer software (v3.3.10.7, PSI, Drasov, Czech Republic). The parameters measured and calculated in this study, based on spectral values, are listed in Supplementary Table S1.
The CRISPR/Cas9 constructs for multiplex mutagenesis were prepared at the Vienna BioCenter Core Facilities GmbH (Dr. Bohr-Gasse 3, 1030 Vienna) by Dr. Vera Schoft. GreenGate entry vectors were combined into a single plant transformation vector through a GoldenGate reaction, as described by Richter et al. (2018), with the modification of using a yellow fluorescent protein seed coat marker (Alli-YFP) for the selection of transgenic lines. Briefly, the final construct comprised the ubiquitin4-2 promoter from Petroselinum crispum, an Arabidopsis codon optimized Cas9, the pea3A terminator from Pisum sativum, and three guide RNA (gRNA) modules, each consisting of the Arabidopsis U6-26 promoter, a guide sequence, and the gRNA scaffold (Richter et al., 2018). Additionally, a simple construct carrying a single gRNA for EXPA10 was also prepared.
The following gRNA sequences targeting the α-EXPANSIN family members were selected using the Broad institute tool (www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design):
EXPA10 (At1g26770): GTGTTAACAACCTGCACAT (exon 1),
EXPA14 (At5g56320): AGCGTGGATGGTTACAGTAG (exon 1),
EXPA15 (At2g03090): TACGGGAACCTCTACAGTCA (exon 2).
The constructs were transformed into Agrobacterium tumefaciens GV3101 pSOUP+ cells and subsequently used for Arabidopsis transformation via the floral dip method (Clough and Bent, 1998). Both wild-type (WT) and the expa1-2 CRISPR/Cas9 line generated by Ramakrishna et al. (2019) were transformed to select for single and multiple expansin mutants. Transgenic T1 generation seeds exhibiting fluorescence were selected using an Olympus SZX16 fluorescence stereomicroscope and a GentleGrab tweezer (Labdeers s.r.o., Czech Republic). These seeds were propagated to the T2 generation, and only non-fluorescent lines that segregated out the T-DNA were sequenced for putative insertions and deletions (indels). Sequencing PCR reactions using isolated genomic DNA (gDNA) as a template were performed with following primers: EXPA10-F CAAGTACGTAACTACTTTCTCC and EXPA10-R CATTGTGCCGGAAGCATCACC, EXPA14-F CTCTTCTTCCTCATTTTCTTCC and EXPA14-R GTTTCCGTAACCACACGCGCC, EXPA15-F CTCATCCTACCTTCTATGGTGG and EXPA15-R GTTAGGAGGACAGAAATTGGTGG.
T2 progeny of 4 independent transgenic lines were tested for expa10, 8 lines for the expa1expa10 double mutant, 42 lines for the expa10expa14expa15 triple mutant and 85 lines for the expa1expa10expa14expa15 quadruple mutant. A summary of the mutant lines used in this study is provided in Table 1. Despite our efforts we were unable to identify any viable mutants in EXPA15. Insertions in the targeted sequence of EXPA10 created a stop codon after 46 amino acids (aa). In the targeted sequence of EXPA14, insertions created a stop codon after 25 aa and the deletion after 65 aa or it interfered with intron excision.
RNA seq libraries were prepared from 10 μg of total RNA isolated from shoots of 7-day old Arabidopsis seedlings of WT and EXPA1 OE line 8-4 (Samalova et al., 2024) using the RNeasy Plant Mini Kit (QIAGEN). Seedlings were grown on MS medium +/- Dex for 7 days or induced with Dex for 3 hours, with four biological replicas for each treatment. Libraries were constructed using the NEBNext Ultra II Directional RNA Library Prep Kit for Illumina, incorporating polyA selection, and sequenced with 75 base paired-end reads. Unique Molecular Identifiers (UMIs) were included in the samples (6 bp long) to detect PCR duplicates. Raw sequencing reads were quality-checked using FastQC, MinION, and Swan, pre-processed with Trimmomatic, and aligned to the Arabidopsis thaliana reference genome (TAIR10-31) using the STAR aligner (v2.7.3a) with gene annotation. Gene counts were obtained using RSEM, and differential expression (DE) analysis was conducted using DESeq2. Differentially expressed genes were identified based on an adjusted p-value of < 0.05 and a log2FoldChange threshold of ±1. Gene ontology (GO) annotations and analyses were performed using GOrilla (Eden et al., 2009). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE286232 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE286232).
Localization of EXPA:mCherry fusions was performed using Zeiss LSM 880 confocal laser scanning microscope. mCherry fluorescence was detected within the 580-650 nm range using a 561-nm HeNe laser for excitation, while eGFP fluorescence was detected within the 490-550 nm range using a 488-nm Argon laser line. Z-stack images were acquired using a 40×/1.2 water-corrected C-Apochromat objective and are presented as maximum intensity projections.
Leaf epidermal images were captured using a 3D microscope VHX-7000 Series (Keyence) at a resolution of 1600x1200 pixels. Cell count and areas were analyzed in ImageJ (version 1.54k) using the PaCeQuant plugin (Poeschl et al., 2020) integrated with MiToBo (version 2.3.1). Automated segmentation and quantification of cell shape parameters were performed. Pixel size (0.216 µm/pixel) was calibrated from a 50 µm scale bar corresponding to 231.5 pixels. Areas in pixels² were converted to µm² using Area (µm2) = Area (pixels2) × (0.216)2. Stomatal aperture width was determined manually by using the line tool in ImageJ, while stomatal density was calculated by counting stomata within a defined area (≈0.0897 mm²) and expressed as number of stomata per mm².
Statistical analyses were performed using ANOVA followed by post hoc Tukey HSD (Unequal N HSD test) in STATISTICA (TIBCO Software Inc.; version 14.0.0.15). Data were tested for normality using the Shapiro-Wilk test and for variance homogeneity using Levene’s test prior to analysis. Results are presented as means ± standard deviations (S.D.), with statistically significant differences (p < 0.05) indicated by differing lowercase letters. For the phenomics data, statistical analyses were performed using R studio software (Version 2023.12.1). Outlier detection was done using “rstatix” package via identify outliers’ function and data imputation was done afterward using “missforest” package in the data pre-process. Data were checked for variance homogeneity with Levene`s test using “car” package and normal distribution using Shapiro-test before statistical analysis. The data were analyzed for each experiment separately focusing on comparing genotypes and treatments across timepoints (DAS). Means were compared with a post-hoc pairwise comparison procedure using the Dunn test and adjusted with Benjamini-Hochberg correction, and the significant difference was indicated by different small letters using “multcompView”. To investigate significant differences among lines, for non-normal data analysis, Kruskal-Wallis test was used, with post-hoc testing (Dunn test) to identify pairwise differences. For the drought stress experiment, the Mann-Whitney U test was applied to assess treatment-specific differences. The level of significance of each factor is indicated as *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001, and ns indicates non-significant differences. By combining all traits, Random Forest model was used to find traits of interest across all time points using “randomForest” package and presented as variable importance where important 20 traits were selected and ranked. Variables were ranked based on their importance, where higher values indicated more contributions to prediction accuracy. Moreover, k-means clustering in a two-dimensional obtained through principal component analysis (PCA) was used for group mean, to discriminate between the lines under each treatment separately by using “factroextra”, “mixOmics”, “ggfortify”, “PCAtools” packages.
Sequence data associated with this study can be found in the GenBank/EMBL data libraries under the following accession numbers: EXPA1 (At1g69530), EXPA10 (At1g26770), EXPA14 (At5g56320), and EXPA15 (At2g03090).
Our previous study demonstrated the localization of expansins within the CW of Arabidopsis thaliana roots using translational fusions of EXPA1, EXPA10, EXPA14, and EXPA15 with the red fluorescent protein mCherry, under the control of their native promoters (Samalova et al., 2024). In this study, we employed the same pEXPA::EXPA:mCherry lines to investigate expansin localization in Arabidopsis shoots.
Notably, EXPA1 exhibited the highest expression in stomatal guard cells (Figure 1A), with localization observed not only in the CW but also within vacuoles. However, a weak signal resembling CW localization in the epidermis was also detectable. Furthermore, when overexpressed in dexamethasone (Dex)-inducible pRPS5A>GR>EXPA1:mCherry plants (Samalova et al., 2024), strong mCherry fluorescence was detectable in the CWs of all cells, as well as within the vacuoles of developing stomata (Figure 1B). The prevalence of shoot EXPA1 expression in stomata was corroborated using an independent transcriptional fusion line, pEXPA1::nls:3xGFP (Ramakrishna et al., 2019), which exhibited GFP fluorescence within the nuclei of guard cells and epidermis (Figures 1C, D).
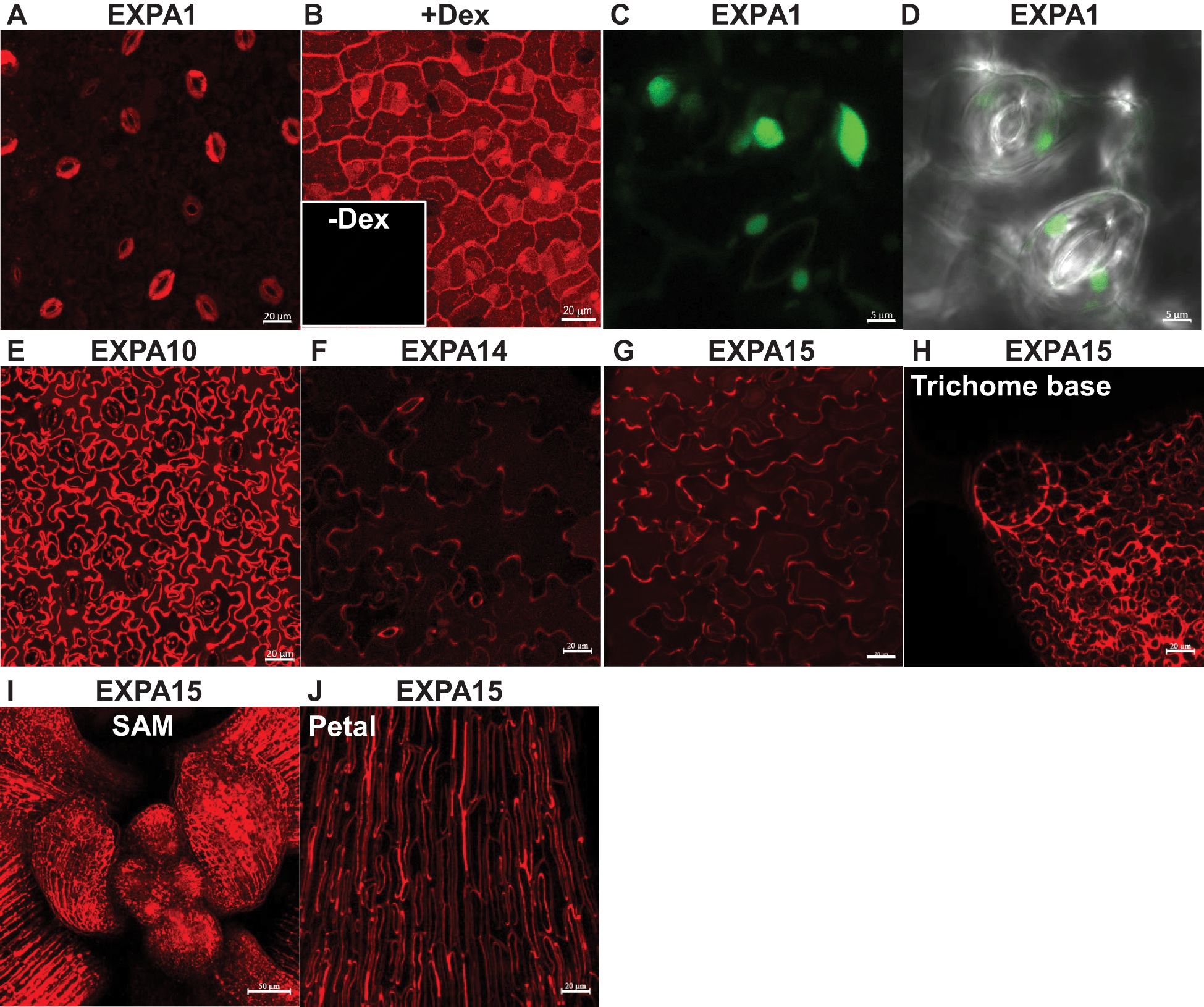
Figure 1. Expansin promoter activity and localization in Arabidopsis shoots. Z-stack projections of (A) pEXPA1::EXPA1:mCherry fusion showing localization in the stomatal guard cells and the epidermis, (B) pRPS5A>GR>EXPA1:mCherry (line 1-3) plants grown on MS media with dexamethasone (+Dex) showing localization in the CWs of all cells, as well as within the vacuoles of developing stomata; insert without Dex (-Dex). Z-stack projections of (C) pEXPA1::nls:3xGFP illustrating a similar pattern of EXPA1 expression, with GFP signal visible in the nuclei of stomatal guard cells (round) and epidermal cells (ellipsoid) and (D) transmitted-light micrograph of a single optical section. Z-stack projections of (E) pEXPA10::EXPA10:mCherry fusion localized in the CW of epidermal and underlying parenchymal cells, (F) pEXPA14::EXPA14:mCherry showing localization in the CW of epidermal cells and autofluorescence of the thick inner CW of stomata pores, (G) pEXPA15::EXPA15:mCherry localized in the CW of epidermal cells, (H) a trichome base, (I) shoot apical meristem (SAM) and (J) petal leaves. The plants shown were 7-14 days old, except for the 5-week-old plant in (J). Scale bars correspond to 20 μm, except for (C, D, I), which correspond to 5 μm and 50 μm, respectively.
Compared to EXPA1, EXPA10 provided a substantially stronger fluorescence signal, predominantly localized to the CW of epidermal cells and likely the underlying parenchymal cells (Figure 1E). In contrast, EXPA14 showed weaker promoter activity, similar to EXPA1, though it displayed a characteristic CW labelling pattern in epidermal cells. Due to the relatively high detector gain, autofluorescence of the thick inner CW surrounding the stomatal pore was also visible (Figure 1F). EXPA 15 expression was robust across the leaf epidermis and various specialized cell types, including those at the trichome base, the shoot apical meristem (SAM) and petal leaves (Figures 1G–J), implying a crucial role in plant growth and development.
In summary, each of the tested expansins exhibits distinct expression and localization patterns in Arabidopsis shoots, including leaves and cotyledons.
Previously, we generated Dex-inducible pRPS5A>GR>EXPA1 (EXPA1 OE) lines (Samalova et al., 2024). To complete the genetic toolbox, here using CRISPR/Cas9 technology, we created single (expa10), double (expa1,10 and expa10,14) and triple (expa1,10,14) expansin knock-out mutants. To evaluate their potential resistance to osmotic stress, we compared these mutants, along with WT and EXPA OE line 8-4 by growing them on MS media supplemented with various concentrations of NaCl. The measured phenotypes included germination rate, root length, and fresh shoot weight (Supplementary Figure S1). However, no significant differences were observed across the lines, except for some variation in germination, suggesting limited osmotic stress tolerance under in vitro conditions.
To further investigate the role of assayed EXPAs in stomata and their potential contribution to drought resistance, we conducted experiments on soil-grown WT, double, and triple expansin mutants, alongside the EXPA1 OE line (8-4) with and without Dex induction. The plants were grown for four weeks before excising rosette leaves for analysis. The leaves were illuminated with white light for 30 minutes, after which stomata density (Figure 2A) and stomata aperture width (Figure 2B) were measured. Our results revealed that the EXPA1 OE line had the highest stomata density compared to all other lines, with the stomatal pores being significantly more open. This suggests that EXPA1 overexpression (OE) enhances stomata formation and aperture size. Interestingly, treatment with abscisic acid (ABA) had no differential effect on stomata closure across the genotypes tested, indicating that the stomatal phenotype observed upon EXPA1 OE may be regulated by mechanisms independent of hormonal control.
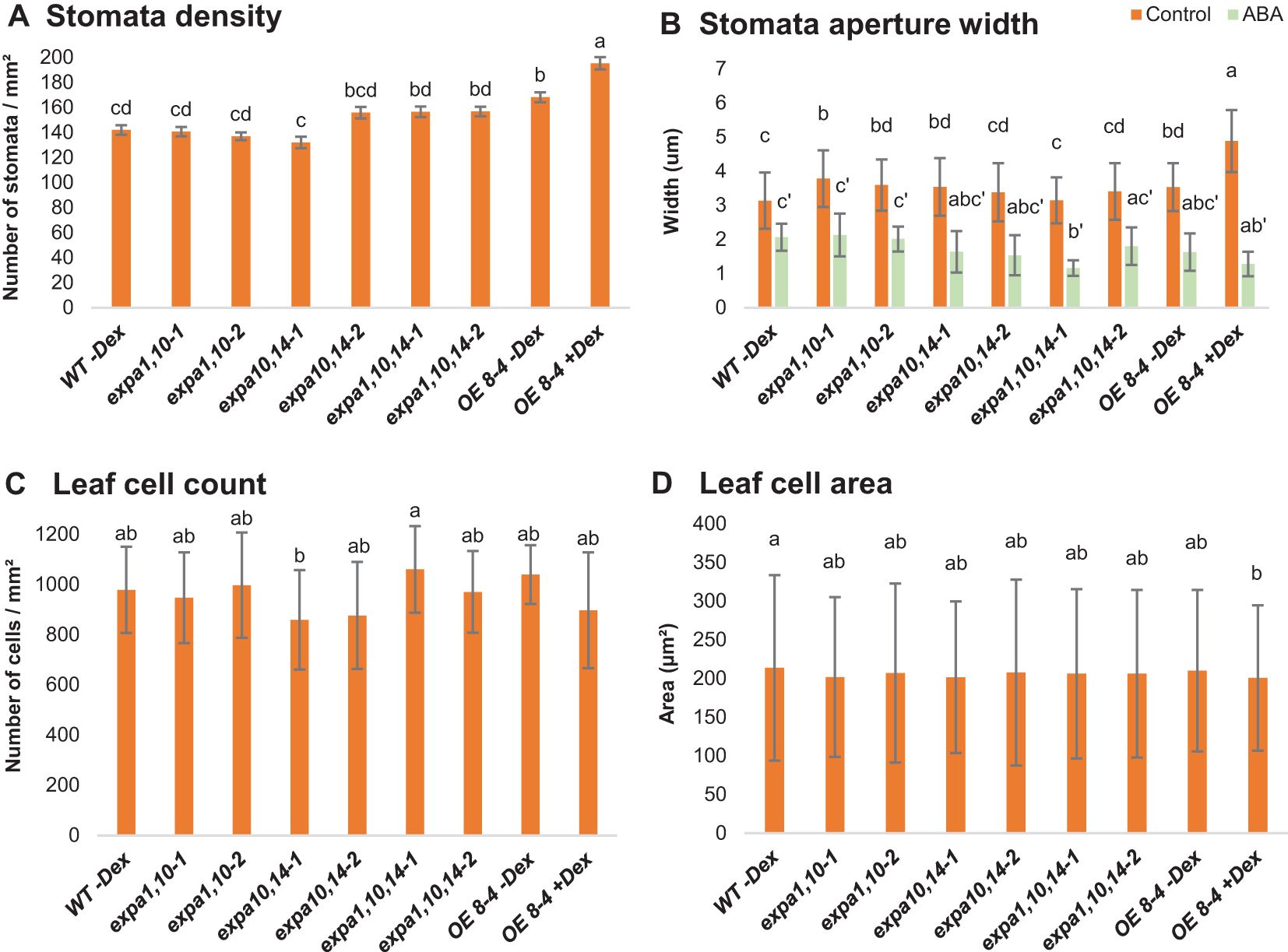
Figure 2. Stomata and leaf cell characterization. Graphs represent (A) stomata density, (B) stomata aperture width, (C) number of cells per square mm (mm2) and (D) size of cells (μm2) in WT, expansin mutants (expa1,10-1 and -2, expa10,14-1 and -2, expa1,10,14-1 and -2) and EXPA1 OE line (8-4; -Dex and +Dex) after 4 weeks of soil growth. Rosette leaves were excised, immersed in water supplemented with 10 mM ABA for 30 min (where indicated), illuminated with white light for 30 min, and subsequently photographed. Error bars represent S.D. with 4-6 plants per treatment and line analyzed. Significant differences among genotypes (at each treatment) were assessed using the post hoc Tukey HSD test (Unequal N HSD). Different lowercase letters above the barplot indicate statistically significant differences (p < 0.05).
Next, we quantified the number of epidermal cells per square millimeter (Figure 2C) and found that the EXPA1 OE line had slightly fewer cells than WT and uninduced plants. However, cell size was consistent across the lines (Figure 2D). This suggests that the reduced overall size of EXPA1 OE plants may result from a lower cell count in the leaves, potentially due to expansins’ role in CW remodeling (see in the text below), which may reduce cell division or limit individual cell expansion.
To investigate further the phenotypic effects of EXPA1 OE in shoots, we employed the high-throughput automatic phenotyping platform, Plantscreen™ Compact System (PSI, Photon Systems Instruments), to perform RGB imaging for growth and morphological analysis. Seeds of wild-type (WT) and Dex-inducible EXPA1 OE lines (5-4 and 8-4) were sown in pots, with Dex applied 3 times a week. Phenotyping began 10 days after stratification (DAS) and was continued every other day for approximately 10 weeks (Figures 3A, B). Distinct morphological changes were observed from the early growth phase, specifically in roundness, compactness and perimeter. These changes became significant at 29 and 32 DAS in the stronger (8-4) and weaker (5-4) EXPA1 OE line, respectively (Figures 3C–F; Supplementary Data Sheets S1, S2). Compared to WT, the rosettes of Dex-treated EXPA1 OE line 8-4 were rounder, and the rosette leaves had shorter petioles, making the plants appear more compact and symmetric, as reflected by increased isotropy (Figure 3E). Uninduced plants, however, resembled WT. By the end of the experiment, the Dex-induced EXPA OE line 8-4 exhibited a smaller overall size, more branched inflorescence, delayed senescence, and greener leaves compared to untreated plants (Figure 3B). In addition to these changes, Dex-induced 8-4 plants displayed drop in the (pro)cambium activity (thinner procambial layer) associating with reduced inflorescence stem diameter and developed vascular tissue with evident metaxylem, alongside indications of precocious secondary CW formation in the interfascicular regions (Supplementary Figure S2).
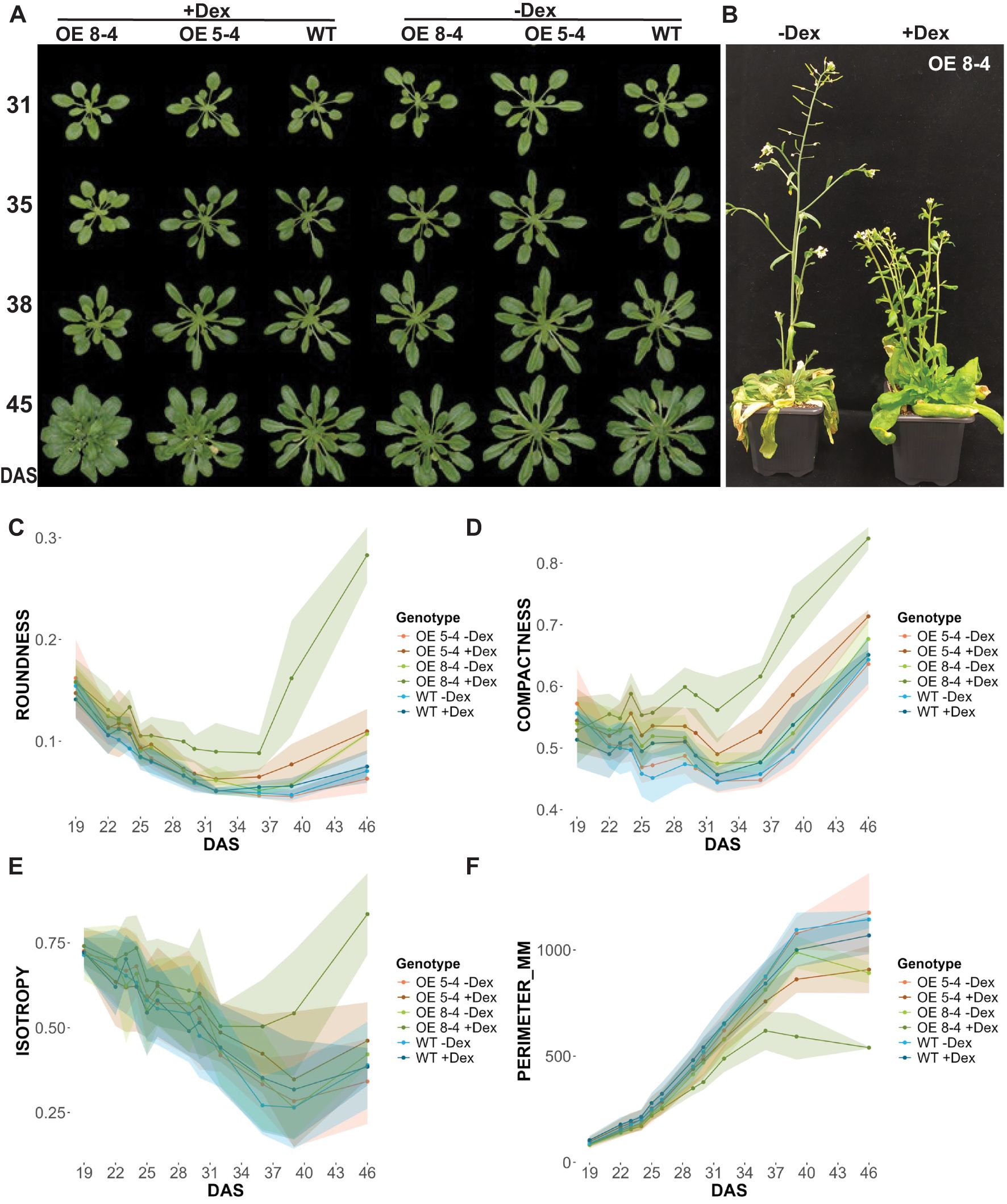
Figure 3. Phenotypic analysis of EXPA1 overexpressing plants. (A) Representative images of wild-type (WT) and EXPA1 OE lines 8-4 and 5-4 induced (+Dex) or not (-Dex) with Dex, imaged at specified days after stratification (DAS) and (B) at the end of the experiment (app. 10 weeks). Graphs showing parameters from RGB top view imaging (C) roundness (arbitrary units, a.u.), (D) compactness (a.u.), (E) isotropy (a.u.), and (F) perimeter (mm) of EXPA1 OE (lines 8-4 and 5-4) and WT over time. Highlighted lines represent standard deviation (S.D.) with 16-19 plants per treatment and line analyzed.
To evaluate the morphological and physiological response of soil-grown plants under drought conditions, triple mutants (lines expa1,10,14-1 and 2), together with EXPA1 OE line (8-4) and WT plants treated with or without Dex, were phenotyped. Using RGB top-view imaging, the greenness of plant canopy, and growth dynamics, including relative growth rate (RGR) and area, were assessed over time (Figure 4; Supplementary Figure S3, Supplementary Data Sheets S1, S2). Across all lines, drought stress (D) reduced both the area and relative growth rate, with an increase in dark green color, the latter becoming particularly severe during the second drought phase compared to control conditions (C). The percentage of light green segmentation (RGB (95,108,64)) and RGR decreased dramatically after the second drought stress phase, reflecting chlorophyll degradation and a consequent reduction in growth (Figures 4A, B). Notably, the Dex-induced EXPA OE line 8-4 exhibited a smaller reduction in the percentage of darker green segments, suggesting reduced stress-induced tissue degradation. This line also showed slower RGR over time and a consistently smaller area during the late stress phase, including 36 DAS, regardless of drought treatment (Figure 4C). Importantly, growth in WT was not affected by Dex treatment, as both treated and untreated plants were of similar size. Interestingly, the results indicated that the triple mutant lines had significantly larger areas compared to WT (Figure 4C; Supplementary Figure S3).

Figure 4. Canopy greenness and growth dynamics under drought stress. (A) Plant color segmentation in percentage (%) from canopy top-view images, including four defined hues across all time points (DAS) under control (C) and drought stress (D). (B) Relative growth rate (RGR) across three phases: the first drought stress, the second drought stress, and the recovery phase. (C) Top- view plant area (mm2) at 36 DAS under control and drought stress. Significant differences among lines per DAS were assessed using the Kruskal-Wallis test, with differing lowercase letters above the boxplot indicating statistical significance (p < 0.05).
Furthermore, chlorophyll fluorescence (ChlF) measurements were conducted using an optimized light-response curve protocol with four light steady-state levels (Lss1-Lss4) (Supplementary Figure S4). Key ChlF parameters measured included minimal fluorescence (F0) and maximal fluorescence (FM), from which variable fluorescence (FV) was calculated. Additionally, photochemical (qP_Lss) and non-photochemical quenching (NPQ_Lss) in light steady state were assessed (Supplementary Table S1). These measurements provide important insights into the functionality of Photosystem II (PSII) and allow to evaluate photosynthetic efficiency. Significant differences in photosynthetic efficiency under stress conditions were observed at light steady state 3 and 4 (Lss3 and Lss4) of the light-response curve (Supplementary Data Sheet S2). At the early phase of drought stress (13 DAS), the Dex-induced EXPA1 OE line exhibited higher FV/FM and lower NPQ value at Lss3 (Figure 5). This increased photosynthetic efficiency, indicating enhanced performance under drought conditions, suggests that EXPA1 OE improves stress tolerance by modulating PSII efficiency and photoprotective mechanisms. These findings highlight the potential of EXPA1 as a target for improving drought tolerance in plants.
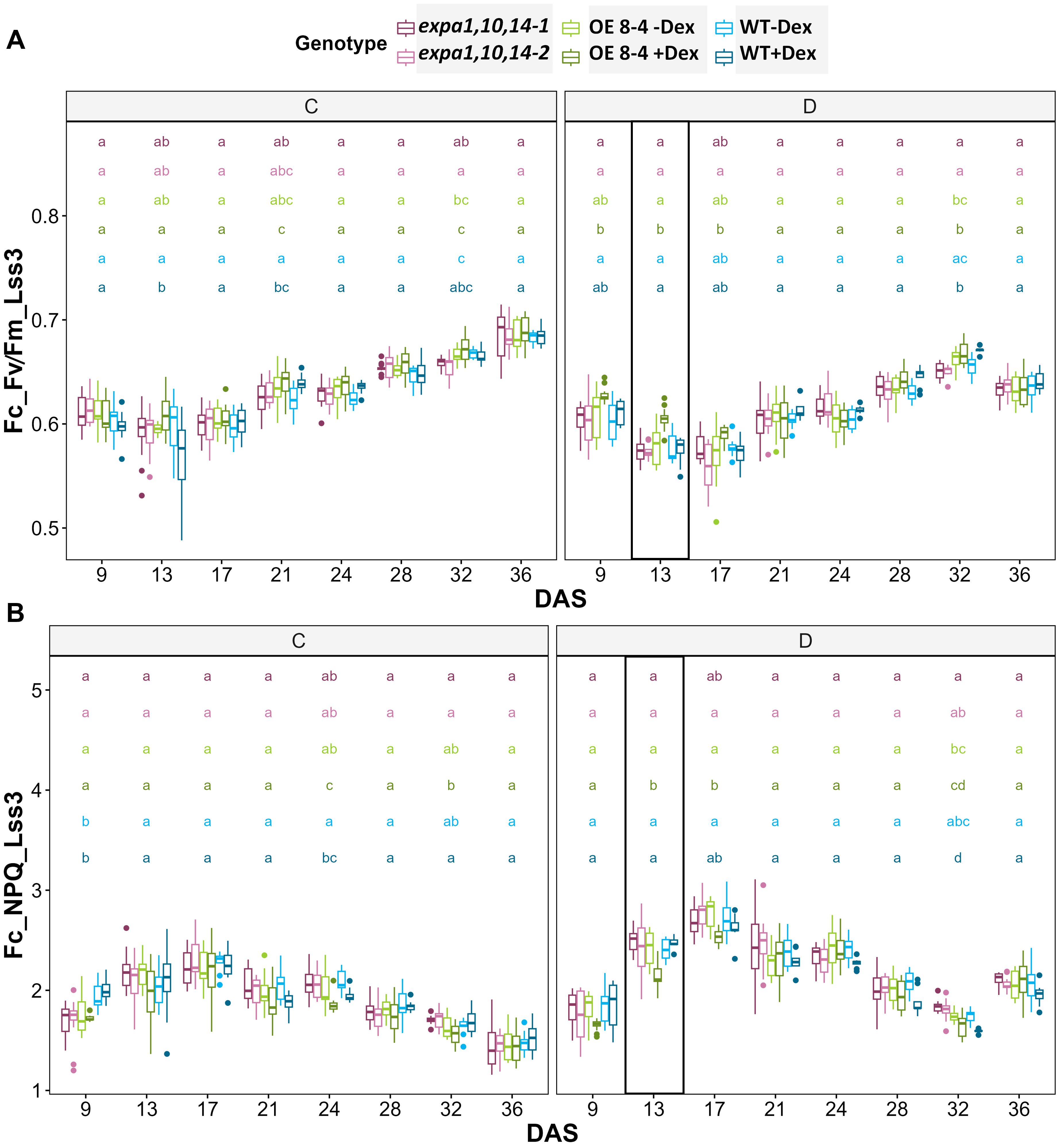
Figure 5. Photosynthetic efficiency under drought stress. (A) PSII maximum efficiency of light-adapted plants in light steady-state (FV/FM_Lss) and (B) non-photochemical quenching steady-state (NPQ_Lss) at 320 µmol m−2 s−1 (Lss3) which is the third light level measured using light response curve protocol across multiple time points. Significant differences among lines at each DAS were assessed using the Kruskal-Wallis test, with differing lowercase letters above the boxplot indicating statistical significance (p < 0.05).
For integrated data visualization, Random Forest models were applied with a mean accuracy of 0.83-0.89 across all time points to investigate the contribution of various RGB-based and fluorescence-related traits in distinguishing between genotypes under control and drought conditions, respectively (Supplementary Figures S5A, B). A total of 34 traits were measured and used in the Random Forest analysis, with the top 20 traits selected for further investigation. Morphological traits were found to be more important during the early phase, while physiological traits gained higher significance in the later phase, reflecting genotype-specific responses at different time points. Furthermore, analysis of the 20 selected traits (variables) for3discriminating between treatments across all genotypes (Supplementary Figure S5C) revealed that area and compactness contributed the most to treatment prediction, followed by NPQ_Lss4 and QY_max. Additionally, k-means clustering was visualized on a two-dimensional Principal Component Analysis (PCA) plot for all measured parameters across all time points to investigate the overall performance of the lines (Figure 6). The results showed the EXPA1 OE line (8-4) exhibited different mechanisms when induced with dex, particularly under control conditions (Figure 6A). The WT clustered together under control conditions but diverged under drought stress (Figure 6B). In contrast, the triple mutant lines (expa1,10,14-1 and -2) consistently clustered together regardless of the treatment.
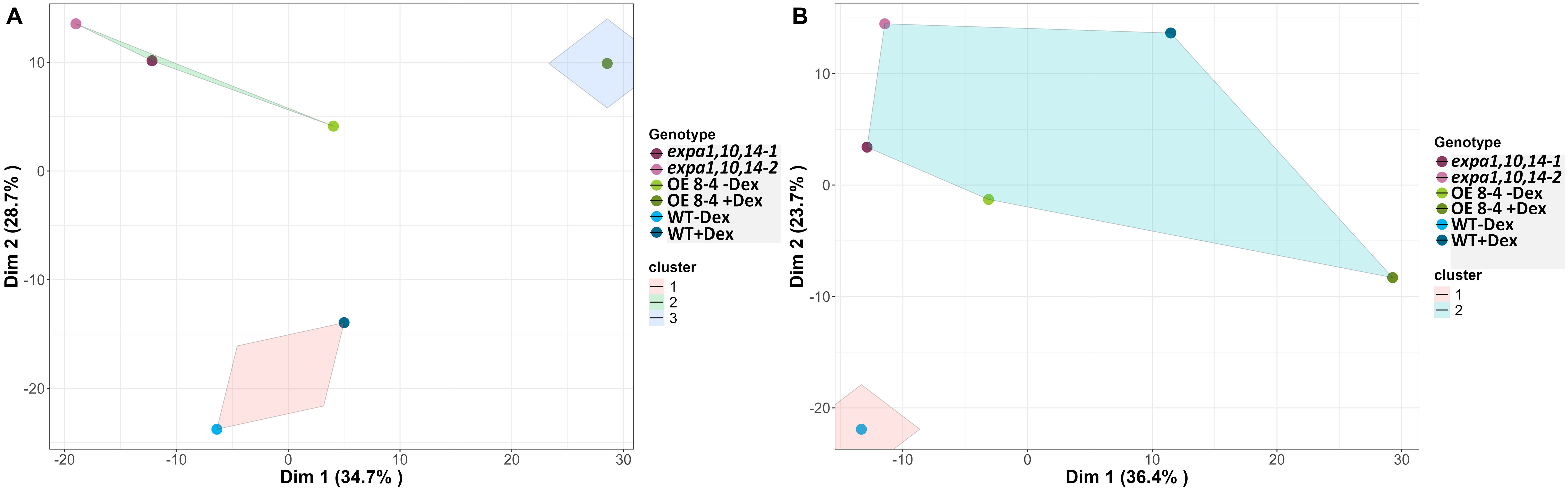
Figure 6. Visualization of k-means clustering presented on a two-dimensional PCA plot. (A) Control and (B) drought conditions. Lines are color-coded, and shaded convex hulls represent the grouping of lines based on k-means clustering.
To investigate the molecular mechanisms underlying EXPA1-induced phenotypic changes, we performed a genome-wide transcriptomic analysis to monitor changes at both early (3 hours) and late (7 days) stages following EXPA1 OE in Arabidopsis seedling shoots. Differentially expressed genes (DEGs) were identified based on an adjusted p-value of < 0.05 and a log2FoldChange threshold of ±1. At 3 h post-Dex induction of EXPA1 OE, 995 genes were significantly upregulated and 1114 genes were downregulated in the EXPA1 OE line compared to non-induced controls (Dex-treated WT, Supplementary Data Sheet S3). Among the DEGs, several plant-type CW-related Gene Ontology (GO) terms were significantly enriched. These included (plant-type) CW loosening (GO0009828), CW organization (GO0009664), or CW biogenesis (GO0071669). In addition to the intentionally overexpressed EXPA1 gene, several other expansin and expansin-like (EXL) genes showed altered expression (Supplementary Table S2). Notably, the expression of EXPA2, EXPA7 and EXLA2 was upregulated, while EXPA6, EXPA8, EXPA15, EXPB3 and EXLB1 were downregulated (Table 2). Other CW-related genes, including XYLOGLUCAN: XYLOGLUCOSYL TRANSFERASES (XTH), PECTIN METHYLESTERASES (PME), and CELLULOSE SYNTHASE-LIKE (CSL) genes, were also significantly enriched among the DEGs (Table 2). At 7 days post-Dex induction of EXPA1 OE, 637 genes were up regulated and 522 were downregulated compared to the Dex-treated WT control (Supplementary Data Sheet S3). In contrast to the short-time response, only the GO terms related to plant-type CW loosening remained significantly enriched, and fewer CW-related genes were differentially expressed (Table 3). Dex treatment alone did not cause significant changes in plant growth and development, and only 11 genes were upregulated and 14 were downregulated.
Altogether, our findings indicate that EXPA1 overexpression triggers rapid and extensive changes in the expression of numerous CW-related genes, particularly during the early stages of induction.
Expansins are well-documented for their role in CW loosening, facilitating cell expansion and growth of specific cells, tissues and organs (Cosgrove, 2000a; c). Several bioinformatics resources dedicated to the expansin gene family provide valuable tools for the navigation and classification of expansins and their homologues (Lohoff et al., 2020; Kok et al., 2023). Beyond CW modification, expansins are involved in various biological processes, reflecting the diversity in protein sequences and gene expression patterns across developmental and stress-related contexts (reviewed in Cosgrove, 2024; Marowa et al., 2016; Samalova et al., 2022).
Recently, cell-specific localization of expansins was demonstrated using red fluorescent protein fusions in roots (Samalova et al., 2024). Building on this approach, our study reveals predominant expression of EXPA1 in stomatal guard cells, suggesting a specific function in these specialized cells that respond dynamically to environmental stress. Earlier studies anticipated this expression pattern by using promoter-driven reporter gene (Cosgrove, 2000c) and demonstrated that the guard cell expansin AtEXPA1 regulates stomatal opening (Zhang et al., 2011). Additional expansins examined in this study, including EXPA10, EXPA14, and EXPA15, were predominantly localized to the CWs of the shoot epidermis, suggesting a role in modulating shoot structure and function. Notably, EXPA15 displayed strong expression throughout the leaf epidermis and in various specialized cells, such as those at the trichome base, the shoot apical meristem, and petal leaves. Our attempts to generate an EXPA15 mutant using CRISPR/Cas9 were unsuccessful, possibly highlighting its essential role in plant growth and development.
Our transcriptomic analysis provides insights into the downstream effects of EXPA1 OE on CW dynamics. Early induction of EXPA1 altered the expression of numerous CW-related genes, including those encoding XTH, PME, and CSL proteins, which play key roles in CW restructuring. Moreover, several other expansin genes were either upregulated or downregulated. These rapid shifts in gene expression suggest that EXPA1 may act as a regulatory node within CW-associated gene networks, impacting CW composition, structure, and function. This is in line with our previous findings, suggesting changes in the transcription of CW-modifying genes as one of the possible mechanisms leading to EXPA1-induced CW stiffening in the root (Samalova et al., 2024). Notably, recent transcriptomic studies identified EXPA1 as a primary regulator under stress conditions, supporting this regulatory role (Sanchez-Munoz et al., 2024). However, similarly to the situation in the root, also in the shoot we observed specific localization of individual EXPAs, implying position-specific mode of action. Thus, we cannot exclude that the transcriptome changes observed in the EXPA1 OE lines might be (at least partially) indirect, balancing the ectopic localization of EXPA1.
The coordinated action of expansins thus appears essential not only for growth under standard conditions but also for adaptation to stressful environments, as we observed in the context of drought. This aligns with previous studies indicating that expansins act synergistically with other CW modifiers to create a more dynamic, responsive CW environment (Cosgrove, 2005). Expansins not only act as CW-loosening agents but may also orchestrate CW properties and composition by interfering with the action of CW remodeling enzymes, possibly through both carbohydrate-dependent and -independent mechanisms (Samalova et al., 2022). This dual role enables plants to maintain CW homeostasis, balancing wall flexibility needed for growth with structural integrity during stress.
In addition to modifying CW biomechanics, composition and structure, cell wall integrity (CWI) likely plays a crucial role, potentially triggering wall integrity signaling through receptor kinases that restrict growth following wall disruptions (Gigli-Bisceglia et al., 2020; Wolf, 2022). CWI sensing initiates stress responses, including ROS production, hormone signaling (e.g., jasmonic acid, salicylic, ethylene), and structural reinforcements (Hamann, 2015; Novakovic et al., 2018). Feedback regulation of CWI may modulate EXPA1-induced wall remodeling by activating protective responses during instances of excessive wall strain. However, the exact mechanisms through which CWI signaling interfaces with hormonal or stress pathways remain to be fully elucidated (Vaahtera et al., 2019; Rui and Dinneny, 2020).
We observed that EXPA1 OE not only alters gene expression related to CW remodeling but also leads to distinct phenotypic changes in shoot architecture, potentially enhancing stress tolerance. Phenotypic assessments revealed that EXPA1 OE contributes to notable morphological alterations, including modified rosette architecture, an increased number of inflorescence stems with reduced diameter, and apparent delayed senescence, all of which may influence the plant’s ability to cope with environmental stress. These modifications suggest that EXPA1 impacts both structural and developmental processes in the shoot. Specifically, the compact rosette morphology and a highly branched inflorescence in EXPA1 OE plants may indicate a shift in resource allocation, potentially enhancing reproductive growth under stress conditions. Our findings align with those of Dai et al. (2012), who observed that expansin OE promotes growth and reproductive development, possibly by enhancing resource use efficiency.
However, our observation that EXPA1 OE plants are smaller, likely due to a reduced number of epidermal cells in the leaves, contradicts previous studies. Kwon et al. (2008) reported that OE of AtEXP3 increases leaf size and growth under normal conditions. Similarly, Kuluev et al. (2012) demonstrated that AtEXPA10 OE leads to larger cells and leaves, underscoring expansins’ role in cell enlargement and tissue architecture. Additionally, Chen et al. (2018) showed that RNAi plants exhibit smaller leaf size, with epidermal cells that appear to be smaller, more compact with a higher density per mm2 leaf area compared to EXPA4 OE tobacco lines and WT. In our study, however, the epidermal cells in EXPA1 OE plants were not smaller or denser. Instead, it appears that the smaller phenotype results from a reduced number of cells in leaves, a pattern we previously observed in roots (Samalova et al., 2024), potentially due to expansins’ role in CW remodeling, which may limit cell division. This is strongly evidenced by the reduced procambium and impaired radial expansion observed in EXPA1 OE inflorescence stems. Further supporting our findings, we observed that the triple knockout of expa1expa10expa14 generated in this study displayed an unexpectedly larger phenotype based on phenotypic RGB analysis of the rosette.
One of the most intriguing aspects of EXPA1 OE described in this study is the increase in stomatal density and aperture size, providing new insights into the role of expansins in modulating stomatal behavior. The stomata in Dex-induced EXPA1 OE plants appear fully functional, responding effectively to ABA-induced closure. These alterations likely enhance the plant’s ability to respond to environmental signals by enabling more dynamic stomatal responses to environmental cues, along with improved water retention capacity under fluctuating conditions. This model aligns with findings by Wei et al. (2011) and Zhang et al. (2011), who demonstrated that OE of AtEXPA1 and VfEXPA1 accelerates light-induced stomatal opening suggesting that expansins modulate stomatal movement by influencing guard CW extensibility. This is consistent with the reduced drought tolerance observed in expansin-inhibited lines, where stomatal responsiveness to environmental stimuli was also reduced, leading to lower gas exchange efficiency (Zhang et al., 2011; Lu et al., 2013).
To sum up, ours as well as others data suggest that EXPA1 OE not only impacts stomatal traits but also regulates cell division and expansion, resulting in altered cellular and physiological characteristics in Arabidopsis. Further research is needed to elucidate the molecular pathways involved in these processes.
A recent review of 22 studies found that OE of expansins across various plant species enhances tolerance to abiotic stresses such as drought, salt, cold, heat, light, cadmium, and oxidative stress (Samalova et al., 2022). Although the mechanisms underlying expansin-induced stress tolerance are not fully understood, they likely involve CWI signaling (Gigli-Bisceglia et al., 2020), ROS signaling (Tenhaken, 2015), improved cell integrity (Chen et al., 2018), and increased wall flexibility under water-deficit conditions, all of which may alter CW and membrane properties (Narayan et al., 2019; Lu et al., 2013). Our study indicates that EXPA1 OE impacts the overall transcriptional states, which may influence not only CW metabolism and biomechanics (Samalova et al., 2024; Cosgrove, 2024), but also modifies plant structure and stomatal function enhancing the ability of shoots to respond and withstand low-water conditions.
Our results demonstrate that EXPA1 OE in Arabidopsis contributes to drought tolerance, as evidenced by chlorophyll fluorescence measurements, including the increased maximum efficiency of PSII, which quantifies the conversion of light energy into chemical energy during photosynthesis (FV/FM) and NPQ values, which measure how excess light energy is dissipated as heat to protect plants from photoinhibition. Lower NPQ values observed in the EXPA1 OE lines under drought conditions, which indicate increased stress tolerance, suggest that EXPA1 OE plants are in a better physiological state that aids in maintaining photoprotection while reducing energy dissipation as heat, allowing for greater energy conservation during recovery processes. Overexpression lines also showed enhanced chlorophyll fluorescence recovery following drought stress, specifically through improved PSII efficiency, which is critical for maintaining photosynthetic performance under water deficit conditions. Similar enhancements in photosynthesis have been observed in other expansin OE lines, such as AtEXPA1 and VfEXPA1, which led to increased transpiration and photosynthesis rates under favorable light conditions (Wei et al., 2011; Zhang et al., 2011).
Previously, an association between stomata density and morphology and water use efficiency was demonstrated (Bertolino et al., 2019). Reduced stomatal density often enhances water-use efficiency, and engineering crops with lower stomatal densities was shown to improve drought tolerance without yield penalties, however, these responses appear to be species-specific and context-dependent (Bertolino et al., 2019; Hepworth et al., 2015). Thus, the higher stomata density and diameter observed in EXPA1 OE plants may contribute to the changes in drought adaptation under drought conditions. This response may be facilitated by changes in CW rigidity and hydraulic through expansin activity, promoting water retention, as seen in tobacco OE NtEXPA11 (Marowa et al., 2020) and optimizing gas exchange in a stress-resilient manner (Farooq et al., 2009).
Together, these findings emphasize the functional role of expansins in optimizing the balance between water conservation and photosynthetic efficiency under drought stress.
The results of this study have important implications for addressing global agricultural challenges, particularly in the context of climate change and the growing demand for sustainable food production. As climate change intensifies droughts and exacerbates water scarcity, the development of climate-resilient crops has become increasingly critical. The demonstrated ability of EXPA1 OE to improve drought tolerance in Arabidopsis suggests that manipulating expansin expression represents a promising strategy for improving water-use efficiency in crops. Furthermore, the role of EXPA1 in CW remodeling and plant growth highlights its potential for enhancing biomass production and crop yield. With the global population on the rise, boosting crop productivity without requiring additional land or resources is essential to ensure better harvests and minimize crop losses during extreme weather events such as storms and strong winds. Due to the unique localization of EXPA1 in stomatal guard cells, this study suggests that EXPA1 manipulation could also improve crop resilience against biotic stresses, especially pathogens such as bacteria and fungi, that enter via these structures. Such an approach could reduce the need for chemical inputs, promoting more sustainable agricultural practices.
Based on our findings and recent literature, we propose that modifying EXPA1 expression, or other α-expansins (EXPA), can confer multiple desirable traits in crops, significantly enhancing agricultural productivity, biomass production, stress tolerance, and disease resistance. Especially fine-tuning EXPA expression by controlling its timing and tissue specificity holds significant potential for improving various crop traits. Given that EXPA1 is involved in CW loosening and remodeling, modulating its expression could promote plant growth and overall biomass accumulation. This could be particularly advantageous in crops where increased growth directly correlates with enhanced yield potential. For example, influencing grain size and quality traits, as seen with OsEXPA7 in rice (Oryza sativa) (Zhang et al., 2024) and TaExpA6 in wheat (Triticum aestivum) (Calderini et al., 2021), or growth, lignification and fiber cell elongation, as demonstrated by OE of PagEXPA1 in poplar (Populus alba x Populus glandulosa) (Hao et al., 2024). EXPA modulation can also improve biomass digestibility and industrial processing (Mira et al., 2024) and improve fiber quality as shown for EXPA1 in cotton (Yaqoob et al., 2020). In crops such as wheat or oilseed crops, adjusting the expression of EXPA could influence stem rigidity and height, which is important for preventing lodging. In oil palm breeding programs, for instance, shortening the stem has been beneficial for producing semi-dwarf palms that are easier to harvest (Somyong et al., 2022). Modifying EXPA expression in fruits could impact ripening and texture, which are critical factors for post-harvest storage. For instance, the simultaneous knockout of SlEXP1 and SlCEL2 in tomato (Solanum lycopersicum) enhanced fruit firmness and increased cell adhesion (Su et al., 2024).
Overexpression of EXPA in roots could enhance root growth and nutrient uptake, particularly valuable in nutrient-poor or stressed soils. For example, OE of GmEXPA7 in soybean (Glycine max) hairy roots enhanced low phosphorus tolerance by stimulating root growth and improving phosphorus uptake (Liu et al., 2024). Additionally, EXPA OE has been shown to improve tolerance to heavy metals in various species, such as the enhancement of aluminum tolerance in composite carpet grass (Axonopus compressus) through the manipulation of AcEXPA1 (Li et al., 2024) or cadmium in tobacco through TaEXPA2 OE (Ren et al., 2018). This trait could be applied to crops grown in contaminated soils, offering potential for phytoremediation. Finally, enhancing EXPA expression in roots could also improve resistance to soil-borne pathogens, such as root-knot nematodes, as observed with NtEXPA7 OE in tobacco (Yang et al., 2024).
In conclusion, modifying EXPA expression, not only in terms of levels but also in terms of spatiotemporal dynamics, offers a versatile tool for improving multiple crop traits, ranging from drought resistance and pathogen defense to biomass production, grain yield, and stress tolerance under challenging growing conditions. Integrating EXPA1 into crop breeding and biotechnological applications could play a pivotal role in developing resilient, high-yielding varieties suited to the challenges of climate change. This approach provides an opportunity to empower farmers with robust cultivars capable of thriving in increasingly unpredictable environments.
Our study highlights the multifaceted role of EXPA1 in influencing growth, modulating CW properties, and enhancing drought tolerance in Arabidopsis, offering insights into how expansins facilitate adaptation to environmental stress through guard cell dynamics, CW restructuring, and improved photosynthetic efficiency under water-limited conditions. The data suggest that expansins play a central role in the plant’s drought response by coordinating growth and development through guard cell modulation and CW remodeling. Additionally, the diversity in expansin gene expression across different plant cells further underscores their varied roles throughout developmental stages and in response to environmental conditions.
This research adds to the evidence supporting expansins’ role in drought tolerance and presents a pathway for engineering crops with enhanced resilience to abiotic stress. The potential of EXPA1-mediated CW modifications and guard cell dynamics opens a path toward improving drought tolerance in crops through targeted manipulation of specific expansin genes, contributing to sustainable agriculture in changing climates.
Future research could further explore the molecular interactions of EXPA1 with other CW-modifying enzymes and investigate expansin gene manipulation across various crops to optimize yield, drought tolerance, and stress resilience.
The data presented in the study are deposited in the NCBI’s Gene Expression Omnibus repository, accession number GSE286232.
DB: Formal analysis, Investigation, Methodology, Software, Supervision, Visualization, Writing – original draft. KM: Investigation, Methodology, Writing – review & editing. JH: Writing – review & editing, Conceptualization, Funding acquisition, Project administration. KP: Data curation, Methodology, Supervision, Validation, Software, Writing – review & editing. LA: Software, Validation, Visualization, Writing – review & editing. BP: Validation, Software, Writing – review & editing. MS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. MS received funding from the Czech Science Foundation, project No. 22-17501S. This work was further supported by the Ministry of Education, Youth and Sports of the Czech Republic (MEYS CR) under the projects TANGENC (CZ.02.01.01/00/22_008/0004581). CELLIM MU is supported by the MEYS CR project LM2023050 Czech-BioImaging. Development of the phenotypic data analysis pipeline was supported by the European Union's Horizon2020 Research and Innovation Programme (grant 862201 CAPITALISE).
We gratefully acknowledge the CEITEC MU Core Facilities for Plant Sciences, Bioinformatics, namely Martin Demko and Nicolas Blavet, and Cellular Imaging (CELLIM, supported by the MEYS CR project LM2023050 Czech-BioImaging) for their assistance in obtaining the scientific data presented in this paper. We thank Pavla Homolova and Mirella Sorrentino for helping with the experiments at the PSI Research Center.
Author KP, LA, BP were employed by the company PSI Photon Systems Instruments Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1546819/full#supplementary-material
Supplementary Figure 1 | Dose-response effect of osmotic stress on Arabidopsis seedlings grown in vitro. Graphs represent (A) germination rate (%), (B) root length (mm), and (C) fresh weight (mg) of shoots of WT, expansin mutants (expa1-2, expa10-1 and -2, expa1,10-1 and -2, expa10,14-1 and -2, expa1,10,14-1 and -2) and EXPA1 OE line (8-4; -Dex and +Dex) grown on MS media supplemented with various concentrations of NaCl (as indicated) for 7 days. Error bars represent S.D., with at least 10 plants per treatment and line analyzed.
Supplementary Figure 2 | EXPA1 overexpression reduces the diameter of Arabidopsis stem. Plants of wild-type (WT) and Dex-inducible EXPA1 OE line 8-4 were grown in soil as described for the control condition experiment and either treated with Dex (+Dex) or left untreated (-Dex) until app. 10-week-old. Inflorescence stem cross-sections were prepared using a vibratome and analyzed as follows: (A) sections stained with toluidine blue visualized using Olympus BX61 microscope, with a detailed view in (B). (C) Sections stained with calcofluor white in green (excitation 458 nm/detection 499-579 nm) imaged alongside autofluorescence in red (excitation 561 nm/detection 588-678 nm) using a Zeiss 880 CLSM. The arrows point at possible onset of precocious secondary CW formation in the interfascicular regions. Scale bars correspond 0.5 mm in (A) and 50 μm in (B, C).
Supplementary Figure 3 | Plant area from RGB top view-imaging along DAS. Solid lines represent stress induction phases, where D1 and D2 correspond to the first and second drought stress phases, respectively, and R indicates the recovery phase. The data represent mean values, with shaded areas indicating standard deviation (S.D.).
Supplementary Figure 4 | Light response curve using kinetic chlorophyll fluorescence imaging. (A) PSII maximum efficiency of light-adapted sample (FV/FM_Lss), (B) PSII quantum yield at light steady-state indicating operating efficiency of PSII (QY_Lss), (C) Non-photochemical quenching at steady-state (NPQ_Lss), (D) Coefficient of non-photochemical quenching in light steady state (qN_Lss), (E) Coefficient of photochemical quenching in steady-state (qP_Lss) and (F) Fraction of PSII centers that are ‘open’ in steady state (qL_Lss). Data were collected under four light levels at 13 DAS.
Supplementary Figure 5 | Variable importance of traits identified by the Random Forest model across all time points. Morphological and physiological traits that contributed the most to discriminating between the genotypes under (A) control and (B) drought conditions at specific DAS. (C) Key traits of high importance that differentiate between treatments among all lines. The mean accuracy metric indicates the prediction accuracy of the random forest model.
Supplementary Table 1 | Trait description extracted from the RGB and chlorophyll fluorescence imaging.
Supplementary Table 2 | GO terms significantly enriched among DEGs identified 3 h after Dex-induced EXPA1 overexpression.
Supplementary Data Sheet 1 | Stats all letters.
Supplementary Data Sheet 2 | Stats non parameter pvalues.
Supplementary Data Sheet 3 | RNAseq shoots.
Awlia, M., Nigro, A., Fajkus, J., Schmoeckel, S. M., Negrao, S., Santelia, D., et al. (2016). High-throughput non-destructive phenotyping of traits that contribute to salinity tolerance in Arabidopsis thaliana. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01414
Bertolino, L. T., Caine, R. S., Gray, J. E. (2019). Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00225
Calderini, D. F., Castillo, F. M., Arenas, A., Molero, G., Reynolds, M. P., Craze, M., et al. (2021). Overcoming the trade-off between grain weight and number in wheat by the ectopic expression of expansin in developing seeds leads to increased yield potential. New Phytol. 230, 629–640. doi: 10.1111/nph.17048
Chaves, M. M., Flexas, J., Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103, 551–560. doi: 10.1093/aob/mcn125
Chen, Y., Han, Y., Zhang, M., Zhou, S., Kong, X., Wang, W. (2016). Overexpression of the wheat expansin gene TaEXPA2 improved seed production and drought tolerance in transgenic tobacco plants. PloS One 11, e0153494. doi: 10.1371/journal.pone.0153494
Chen, L., Zou, W., Fei, C., Wu, G., Li, X., Lin, H., et al. (2018). [amp]]alpha;-Expansin EXPA4 positively regulates abiotic stress tolerance but negatively regulates pathogen resistance in Nicotiana tabacum. Plant Cell Physiol. 59, 2317–2330. doi: 10.1093/pcp/pcy155
Cho, H. T., Cosgrove, D. J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 97, 9783–9788. doi: 10.1073/pnas.160276997
Cho, H. T., Cosgrove, D. J. (2002). Regulation of root hair initiation and EXPANSIN gene expression in A. thaliana. Plant Cell 14, 3237–3253. doi: 10.1105/tpc.006437
Clauw, P., Coppens., F., De Beuf, K., Dhondt, S., Van Daele, T., Maleux, K., et al. (2015). Leaf responses to mild drought stress in natural variants of Arabidopsis. Plant Physiol. 167, 800–816. doi: 10.1104/pp.114.254284
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x
Cosgrove, D. J. (2000a). Loosening of plant cell walls by EXPANSINs. Nature 407, 321–326. doi: 10.1038/35030000
Cosgrove, D. J. (2000b). Expansive growth of plant cell walls. Plant Physiol and. Biochem 38, 109–124. doi: 10.1016/S0981-9428(00)00164-9
Cosgrove, D. J. (2000c). New genes and new biological roles for expansins. Curr. Opin. Plant Biol. 3, 73–78. doi: 10.1016/S1369-5266(99)00039-4
Cosgrove, D. J. (2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861. doi: 10.1038/nrm1746
Cosgrove, D. J. (2015). Plant EXPANSINs: diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 25, 162–172. doi: 10.1016/j.pbi.2015.05.014
Cosgrove, D. J. (2016). Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. J. Exp. Bot. 67, 463–476. doi: 10.1093/jxb/erv511
Cosgrove, D. J. (2024). Plant cell wall loosening by expansins. Annu. Rev. Cell Dev. Biol. 40, 329–352. doi: 10.1146/annurev-cellbio-111822-115334
Cosgrove, D. J., Li, L. C., Cho, H. T., Hoffmann-Benning, S., Moore, R. C., Blecker, D. (2002). The growing world of expansins. Plant Cell Physiol. 43, 1436–1444. doi: 10.1093/pcp/pcf180
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Ann. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Dai, F., Zhang, C., Jiang, X., Kang, M., Yin, X., Lu, P., et al. (2012). RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 160, 2064–2082. doi: 10.1104/pp.112.207720
Eden, E., Navon, R., Steinfeld, I., Lipson, D., Yakhini, Z. (2009). GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinf. 10, 48. doi: 10.1186/1471-2105-10-48
Edgar, R., Domrachev, M., Lash, A. E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. doi: 10.1093/nar/30.1.207
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D., Basra, S. M. A. (2009). Plant drought stress: effects, mechanisms and management. Agron. Sustain. Dev. 29, 185–212. doi: 10.1051/agro:2008021
Flexas, J., Bota, J., Loreto, F., Corinic, G., Sharkey, T. D. (2004). Diffusive and metabolic limitations to photosynthesis under drought and salinity in C3 plants. Plant Biol. 6, 269–279. doi: 10.1055/s-2004-820867
Flutsch, S., Nigro, A., Conci, F., Fajkus, J., Thalmann, M., Trtílek, M., et al. (2020). Glucose uptake to guard cells via STP transporters provides carbon sources for stomatal opening and plant growth. EMBO Rep. 21, e49719. doi: 10.15252/embr.201949719
Gao, X., Liu, K., Lu, Y. T. (2010). Specific roles of AtEXPA1 in plant growth and stress adaptation. Russian J. Plant Physiol. 57, 241–246. doi: 10.1134/S1021443710020111
Gigli-Bisceglia, N., Engelsdorf, T., Hamann, T. (2020). Plant cell wall integrity maintenance in model plants and crop species-relevant cell wall components and underlying guiding principles. Cell. Mol. Life Sci. 77, 2049–2077. doi: 10.1007/s00018-019-03388-8
Guo, W., Zhao, J., Li, X., Qin, L., Yan, X., Liao, H. (2011). A soybean β-EXPANSIN gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J. 66, 541–552. doi: 10.1111/j.1365-313X.2011.04511.x
Hamann, T. (2015). The plant cell wall integrity maintenance mechanism concepts for organization and mode of action. Plant Cell Physiol. 56, 215–223. doi: 10.1093/pcp/pcu164
Hao, Y. Y., Chu, L. W., He, X. J., Zhao, S. T., Tang, F. (2024). PagEXPA1 combines with PagCDKB2;1 to regulate plant growth and the elongation of fibers in Populus alba x Populus glandulosa. Intern. J. Biol. Macromol. 268, 131559. doi: 10.1016/j.ijbiomac.2024
Hao, Z., Qian, X., Xiao, X., Huabo, L., Junkai, Z., Jichen, X. (2017). Transgenic tobacco plants expressing grass AstEXPA1 gene show improved performance to several stresses. Plant Biotechnol. Rep. 11, 331–337. doi: 10.1007/s11816-017-0454-7
Hepworth, C., Doheny-Adams, T., Hunt, L., Cameron, D. D., Gray, J. E. (2015). Manipulating stomatal density enhances drought tolerance without deleterious effect on nutrient uptake. New Phytol. 208, 336–341. doi: 10.1111/nph.13598
Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H., Jia, P., et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 39, 969–987. doi: 10.1007/s11033-011-0823-1
Kok, B. O., Altunoglu, Y. C., Oncul, A. B., Karaci, A., Baloglu, M. C. (2023). Expansin gene family database: A comprehensive bioinformatics resource for plant expansin multigene family. J. Bioinform. Compl. Biol. 21 (3), 2350015. doi: 10.1142/S0219720023500154
Kuluev, B. R., Knyazev, A. B., Lebedev, Y. P., Chemeris, A. V. (2012). Morphological and physiological characteristics of transgenic tobacco plants expressing expansin genes: AtEXP10 from Arabidopsis and PnEXPA1 from poplar. Russ. J. Plant Physiol. 59, 97–104. doi: 10.1134/S1021443712010128
Kwon, Y. R., Lee, H. J., Kim, K. H., Hong, S. W., Lee, S. J., Lee, H. (2008). Ectopic expression of Expansin3 or Expansin b 1 causes enhanced hormone and salt stress sensitivity in Arabidopsis. Biotechnol. Lett. 30, 1281–1288. doi: 10.1007/s10529-008-9678-5
Lee, D. K., Ahn, J. H., Song, S. K., Choi, Y. D., Lee, J. S. (2003). Expression of an expansin gene is correlated with root elongation in soybean. Plant Physiol. 131, 985–997. doi: 10.1104/pp.009902
Lee, Y., Kende, H., Cho, H. T. (2001). EXPANSINs: ever-expanding numbers and functions. Curr. Opin. Plant Biol. 4, 527–532. doi: 10.1016/S1369-5266(00)00211-9
Li, Y., Jones, L., McQueen-Mason, S. (2003). EXPANSINs and cell growth. Curr. Opin. Plant Biol. 6, 603–610. doi: 10.1016/j.pbi.2003.09.003
Li, J. F., Liu, L. T., Wang, L. J., Rao, I. M., Wang, Z. Y., Chen, Z. J. (2024). AcEXPA1, an α-expansin gene, participates in the aluminum tolerance of carpetgrass (Axonopus compressus) through root growth regulation. Plant Cell Rep. 43, 159. doi: 10.1007/s00299-024-03243-6
Li, F., Xing, S., Guo, Q., Zhao, M., Zhang, J., Gao, Q., et al. (2011). Drought tolerance through over-expression of the expansin gene TaEXPB23 in transgenic tobacco. J. Plant Physiol. 168, 960–966. doi: 10.1016/j.jplph.2010.11.023
Liu, X. Q., Cai, Y. P., Yao, W. W., Chen, L., Hou, W. S. (2024). The soybean NUCLEAR FACTOR-Y C4 and α-EXPANSIN 7 module influences phosphorus uptake by regulating root morphology. Plant Physiol. 197 (1), kiae478. doi: 10.1093/plphys/kiae478
Liu, Y., Zhang, L., Hao, W., Zhang, L., Liu, Y., Chen, L. (2019). Expression of two α−type expansins from Ammopiptanthus nanus in Arabidopsis thaliana enhance tolerance to cold and drought stresses. Int. J. Mol. Sci. 20, 5255. doi: 10.3390/ijms20215255
Lohoff, C., Buchholz, P. C. F., Le-Roes-Hill, M., Pleiss, J. (2020). The Expansin engineering database: A navigation and classification tool for expansins and homologues. Proteins 89, 149–162. doi: 10.1002/prot.26001
Lu, P., Kang, M., Jiang, X., Dai, F., Gao, J., ZhangG, C. (2013). RhEXPA4, a rose EXPANSIN gene, modulates leaf growth and confers drought and salt tolerance to A. thaliana. Planta 237, 1547–1559. doi: 10.1007/s00425-013-1867-3
Marowa, P., Ding, A., Kong, Y. (2016). Expansins: Roles in plant growth and potential applications in crop improvement. Plant Cell Rep. 35, 949–965. doi: 10.1007/s00299-016-1948-4
Marowa, P., Dinga, A., Xua, Z., Konga, Y. (2020). Overexpression of NtEXPA11 modulates plant growth and development and enhances stress tolerance in tobacco. Plant Physiol. Biochem. 151, 477–485. doi: 10.1016/j.plaphy.2020.03.033
McQueen-Mason, S. J., Cosgrove, D. J. (1995). EXPANSIN mode of action on cell walls. Analysis of wall hydrolysis, stress relaxation, and binding. Plant Physiol. 107, 87–100. doi: 10.1104/pp.107.1.87
Mira, W., Heinz, O., Goncalvez, A., Crema, L., Vicentini, R., Cardoso, S., et al. (2024). SacEXP32 sugarcane expansin gene expression increases cell size and improves biomass digestibility. J. Plant Biochem. Biotechnol. 33, 313–325. doi: 10.1007/s13562-024-00891-3
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue. Physiol. Plant. 15, 493–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Narayan, J. A., Chakravarthi, M., Nerkar, G., Manoj, V. M., Dharshini, S., Subramonian, N., et al. (2021). Overexpression of expansin EaEXPA1, a cell wall loosening protein enhances drought tolerance in sugarcane. Ind. Crops Prod. 159, 113035. doi: 10.1016/j.indcrop.2020.113035
Narayan, J. A., Dharshini, S., Manoj, V. M., Padmanabhan, T. S. S., Kadirvelu, K., Suresha, G. S., et al. (2019). Isolation and characterization of water-deficit stress-responsive α-expansin 1 (EXPA1) gene from Saccharum complex. 3. Biotech 9, 186. doi: 10.1007/s13205-019-1719-3
Novakovic, L., Guo, T., Bacic, A., Sampathkumar, A., Johnson, K. L. (2018). Hitting the wall—Sensing and signaling pathways involved in plant cell wall remodeling in response to abiotic stress. Plants 7, 89. doi: 10.3390/plants7040089
Poeschl, Y., Möller, B., Müller, L., Bürstenbinder, K. (2020). User-friendly assessment of pavement cell shape features with PaCeQuant: Novel functions and tools. Methods Cell Biol. 160, 349–363. doi: 10.1016/bs.mcb.2020.04.010
Ramakrishna, P., Ruiz Duarte, P., Rance, G. A., Schubert, M., Vordermaier, V., Dai Vue, L., et al. (2019). EXPANSIN A1-mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proc. Natl. Acad. Sci. 116, 8597–8602. doi: 10.1073/pnas.1820882116
Ren, Y., Chen, Y., An, J., Zhao, Z., Zhang, Z., Wang, Y., et al. (2018). Wheat expansin gene TaEXPA2 is involved in conferring plant tolerance to cd toxicity. Plant Sci. 270, 245–256. doi: 10.1016/j.plantsci.2018.02.022
Richter, J., Watson, J. M., Stasnik, P., Borowska, M., Neuhold, J., Berger, M., et al. (2018). Multiplex mutagenesis of four clustered CrRLK1L with CRISPR/Cas9 exposes their growth regulatory roles in response to metal ions. Sci. Rep. 8, 12182. doi: 10.1038/s41598-018-30711-31
Rui, Y., Dinneny, J. R. (2020). A wall with integrity: Surveillance and maintenance of the plant cell wall under stress. New Phytol. 225, 1428–1439. doi: 10.1111/nph.v225.4
Salehi-Lisar, S. Y., Bakhshayeshan-Agdam, H. (2016). “Drought stress in plants: Causes, consequences, and tolerance,” in Drought Stress Tolerance in Plants, vol. 1 . Eds. Hossain, M., Wani, S., Bhattacharjee, S., Burritt, D., Tran, L. S. (Switzerland: Springer International Publishing), 1–16.
Samalova, M., Gahurova, E., Hejatko, J. (2022). Expansin-mediated developmental and adaptive responses: a matter of cell wall biomechanics. Quantitative. Plant Biol. 3, e11. doi: 10.1017/qpb.2022.6
Samalova, M., Kirchhelle, C., Moore, I. (2019). Universal methods for transgene induction using the dexamethasone-inducible transcription activation system pOp6/LhGR in Arabidopsis and other plant species. Curr. Protoc. Plant Biol. 4, e20089. doi: 10.1002/cppb.20089
Samalova, M., Melnikava, A., Elsayad, K., Peaucelle, A., Gahurova, E., Gumulec, J., et al. (2024). Hormone-regulated expansins: Expression, localization, and cell wall biomechanics in Arabidopsis root growth. Plant Physiol. 194, 209–228. doi: 10.1093/plphys/kiad228
Sampedro, J., Cosgrove, D. J. (2005). The EXPANSIN superfamily. Genome Biol. 6, 242. doi: 10.1186/gb-2005-6-12-242
Sanchez-Munoz, R., Depaepe, D., Samalova, M., Hejatko, J., Zaplana, I., van der Straeten, D. (2024). The molecular core of transcriptome responses to abiotic stress in plants: a machine learning-driven meta-analysis. BioRxiv. Preprint. doi: 10.1101/2024.01.24.576978
Schipper, O., Schaefer, D., Reski, R., Flemin, A. (2002). Expansins in the bryophyte Physcomitrella patens. Plant Mol. Biol. 50, 789–802. doi: 10.1023/A:1019907207433
Shao, H. B., Chu, L. Y., Jaleel, C. A., Zhao, C. X. (2008a). Water-deficit stress-induced anatomical changes in higher plants. Comptes. Rendus. Biol. 331, 215–225. 95. doi: 10.1016/j.crvi.2008.01.002
Shao, H. B., Song, W. Y., Chu, L. Y. (2008b). Advances of calcium signals involved in plant anti-drought. Comptes. Rendus. Biol. 331, 587–596. doi: 10.1016/j.crvi.2008.03.012
Somyong, S., Phetchawang, P., Bihi, A. K., Sonthirod, C., Kongkachana, W., Sangsrakru, D., et al. (2022). A SNP variation in an expansin EgExp4 gene affects height in oil palm. PEERJ 10, e13046. doi: 10.7717/peerj.13046
Su, G. Q., Lin, Y. F., Wang, C. F., Lu, J., Liu, Z. M., He, Z. R., et al. (2024). Expansin SlExp1 and endoglucanase SlCel2 synergistically promote fruit softening and cell wall disassembly in tomato. Plant Cell 36, 709–726. doi: 10.1093/plcell/koad291
Tenhaken, R. (2015). Cell wall remodeling under abiotic stress. Front. Plant Sci. 5, 771. doi: 10.3389/fpls.2014.00771
Vaahtera, L., Schulz, J., Hamann, T. (2019). Cell wall integrity maintenance during plant development and interaction with the environment. Nat. Plants 5, 924–932. doi: 10.1038/s41477-019-0502-0
Wei, P., Chen, S., Zhang, X., Zhao, P., Xiong, Y., Wang, W., et al. (2011). An a-expansin, VfEXPA1, is involved in regulation of stomatal movement in Vicia faba L. Chin. Sci. Bull. 56, 3531–3537. doi: 10.1007/s11434-011-4817-0
Wolf, S. (2022). Cell wall signaling in plant development and defense. Annu. Rev. Plant Biol. 73, 323–353. doi: 10.1146/annurev-arplant-102820-095312
Wu, Y., Thorne, E. T., Sharp, R. E., Cosgrove, D. J. (2001). Modification of EXPANSIN transcript levels in the maize primary root at low water potentials. Plant Physiol. 126, 1471–1479. doi: 10.1104/pp.126.4.1471
Yang, C., Jiang, L. Q., Leng, Z. M., Yuan, S., Wang, Y., Liu, G., et al. (2024). Overexpression of NtEXPA7 promotes seedling growth and resistance to root-knot nematode in tobacco. Biochem. Biophys. Res. Commun. 720, 150086. doi: 10.1016/j.bbrc.2024.150086
Yaqoob, A., Bashir, S., Rao, A. Q., Shahid, A. A. (2020). Transformation of α-EXPA1 gene leads to an improved fibre quality in Gossypium hirsutum. Plant Breed. 139, 1213–1220. doi: 10.1111/pbr.12878
Zhang, B., Chang, L., Sun, W., Ullah, A., Yang, X. (2021). Overexpression of an expansin-like gene, GhEXLB2 enhanced drought tolerance in cotton. Plant Physiol. Biochem. 162, 468–475. doi: 10.1016/j.plaphy.2021.03.018
Zhang, X. W., Wang, Y., Liu, M. Y., Yan, P. W., Niu, F., Ma, F. Y., et al. (2024). OsEXPA7 encoding an expansin affects grain size and quality traits in rice (Oryza sativa L.). Rice 17, 36. doi: 10.1186/s12284-024-00715-x
Keywords: cell wall, EXPA, Arabidopsis, highthroughput phenotyping, abiotic stress
Citation: Balkova D, Mala K, Hejatko J, Panzarova K, Abdelhakim L, Pleskacova B and Samalova M (2025) Differential expression and localization of expansins in Arabidopsis shoots: implications for cell wall dynamics and drought tolerance. Front. Plant Sci. 16:1546819. doi: 10.3389/fpls.2025.1546819
Received: 17 December 2024; Accepted: 20 January 2025;
Published: 10 February 2025.
Edited by:
Laura Bacete, Umeå University, SwedenReviewed by:
Manu Priya, University of Massachusetts Amherst, United StatesCopyright © 2025 Balkova, Mala, Hejatko, Panzarova, Abdelhakim, Pleskacova and Samalova. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marketa Samalova, bWFya2V0YS5zYW1hbG92YUBzY2kubXVuaS5jeg==
†Present address: Darina Balkova, Department of Sustainable Crop Production, Faculty of Agriculture, Food and Environmental Sciences, Universita Cattolica del Sacro Cuore, Piacenza, Italy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.