
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 February 2025
Sec. Plant Abiotic Stress
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1533219
This article is part of the Research Topic Utilizing Advanced Genomics and Biochemical Tools to Strengthen Crop Adaptation for Biotic and Abiotic Stresses View all 3 articles
The PDX gene is a key gene in the vitamin B6 synthesis pathway, playing a crucial role in plant growth, development, and stress tolerance. To explore the family characteristics of the PDX gene in Brassica napus (B. napus) and its regulatory function under waterlogging stress, this study used five PDX genes from Arabidopsis thaliana as the basis for sequence analysis. Thirteen, eight, and six PDX genes were identified in B. napus, Brassica oleracea (B. oleracea), and Brassica rapa (B. rapa), respectively. Bioinformatics study reveals high conservation of PDX subfamily genes during evolution, and PDX genes in B. napus respond to waterlogging stress.In order to further investigate the effect of the PDX gene on waterlogging tolerance in B. napus, expression analysis was conducted on BnaPDX1.3 gene overexpressing B. napus plants and wild-type plants. The study showed that overexpressing plants could synthesize more VB6 under waterlogging stress, exhibit stronger antioxidant enzyme activity, and have a more effective and stable ROS scavenging system, thus exhibiting a healthier phenotype. These findings suggested that the BnaPDX1.3 gene can enhance the waterlogging tolerance of B. napus, which is of great significance for its response to waterlogging stress. Our study provides a basic reference for further research on the regulation mechanism of the PDX gene and waterlogging tolerance in B. napus.
Vitamin B6 is an essential water-soluble vitamin required by all living organisms (Gorelova et al., 2022). It is recognized as an antioxidant and is linked to responses to various biotic and abiotic stresses (Vanderschuren et al., 2013; Samsatly et al., 2016; Chung, 2012; Zhang et al., 2015; Asensi-Fabado and Munné-Bosch, 2010). Vitamin B6 consists of six interconvertible compounds: pyridoxine phosphate (PNP), pyridoxal phosphate (PLP), pyridoxamine phosphate (PMP), and their nonphosphorylated derivatives (pyridoxine [PN], pyridoxal [PL], and pyridoxamine [PM]) (Fitzpatrick et al., 2007; Roje, 2007; Mooney et al., 2009). Additionally, vitamin B6 must be phosphorylated to function as a coenzyme (Colinas et al., 2016). Among them, PLP is the most important coenzyme, playing a crucial role in lipid degradation and carbohydrate storage (such as glycogen) (Jeong and VacantiLi, 2020; Mooney et al., 2009; Rueschhoff et al., 2013). It also plays a decisive role in amino acid metabolism, catalyzing transamination, decarboxylation, and α,β-elimination reactions involved in amino acid metabolism (Eliot and Kirsch, 2004; Drewke and Leistner, 2001). Other studies have found that the five enzymes that most commonly use PLP as a coenzyme, in order of dependency, are transferases, lyases, isomerases, hydrolases, and oxidoreductases, demonstrating the versatility of PLP-dependent enzymes (Percudani and Peracchi, 2003).
In plants, the de novo biosynthesis pathway of vitamin B6 relies on two proteins, PDX1 and PDX2, which function as glutaminyltransferases (Tambasco-Studart et al., 2005, 2007). PDX2 possesses transaminase activity, extracting ammonium groups from glutamine and incorporating them into the product. PDX1 receives these ammonium groups and synthesizes the final product (Raschle et al., 2005; Dong et al., 2004). In the AtPDX family of Arabidopsis thaliana, there are three AtPDX1 (AtPDX1.1, AtPDX1.2, and AtPDX1.3) and one AtPDX2 protein member (Tambasco-Studart et al., 2005; Wetzel et al., 2004). However, only PDX1.1 and PDX1.3 are involved in the biosynthesis of vitamin B6, while PDX1.2 does not play a role in this process (Tambasco-Studart et al., 2005; Titiz et al., 2006). Instead, PDX1.2 functions as a pseudoenzyme that enhances the activity of catalytic homologs under stress conditions (Moccand et al., 2014). Studies on pdx1 and pdx2 mutants in Arabidopsis thaliana have shown that knocking out both genes, PDX1.1 and PDX1.3, or knocking out the single gene, PDX2, leads to the death of the mutant at the embryonic stage of development (Tambasco-Studart et al., 2005; Titiz et al., 2006). However, single mutants of pdx1.1 or pdx1.3 can survive, although with a short root phenotype, with the latter phenotype being more pronounced (Titiz et al., 2006; Chen and Xiong, 2005; Wagner et al., 2006; Boycheva et al., 2015). Hao Chen et al. (2009) suggested that the short-root phenotype in pdx1 mutants was caused by reduced levels of endogenous auxin synthesis (Chen and Xiong, 2009). In Arabidopsis thaliana, the expression level of PDX1.3 is consistently higher than that of PDX1.1, both spatially and temporally. Additionally, pdx1.3 mutants exhibit more pronounced phenotypic differences in morphology and development compared to pdx1.1 mutants (Titiz et al., 2006), despite the two proteins being 87% identical and capable of synthesizing vitamin B6 at comparable rates (Tambasco-Studart et al., 2005). In addition, Leuendorf et al. (2014) demonstrated that the lack of PDX1.2 affects seed and hypocotyl development and results in a large number of aborted seeds during embryonic development, as shown through T-DNA insertion-generated heterozygous PDX1.2 and artificial microRNA-reduced PDX1.2 expression.
The PDX gene is not only crucial for the growth and development of plants but also indispensable for their response to adverse environmental stresses. Ristilä et al. (2011) discovered that PDX1.3 is involved in plants’ response to UV-B radiation, with the formation of UV-B-induced PDX1.3 primarily occurring in the parts of leaves that absorb UV-B radiation (Ristilä et al., 2011). It has also been proven that the pdx1.3 mutant of Arabidopsis thaliana exhibits weaker tolerance to strong light and photo-oxidation (Havaux et al., 2009). Additionally, the PDX1.2 gene of Arabidopsis thaliana has been found to exhibit higher expression under UV-B treatment, oxidative stress, and heat shock (Denslow et al., 2007). Furthermore, Dell’Aglio et al. (2017) identified a heat stress transcription initiation site within PDX1.2, and subsequent research indicated that PDX1.2 can stably catalyze PDX1 homologs under heat stress conditions.
Research on vitamin B6 (VB6) and PDX genes primarily focuses on their roles in plant growth, development, and stress resistance. However, limited studies have examined their roles in the response of Brassica napus (B. napus) to waterlogging stress. In this study, we investigated the impact of BnaPDX1.3 gene overexpression (PDX1.3#20 and PDX1.3#21) and WT B. napus plants on waterlogging tolerance.
The B. napus variety G218 was provided by the Hunan Branch of the National Oil Crop Improvement Center.
Information related to the PDX gene of Arabidopsis thaliana, Brassica rapa (B. rapa), Brassica oleracea (B. oleracea), and B. napus was obtained from the Ensembl Plants database (http://plants.ensembl.org/index.html), including the full-length DNA sequence, CDS sequence, amino acid sequence, etc. The nucleic acid sequence and amino acid sequences of the PDX gene of Arabidopsis thaliana were first downloaded from Ensembl Plants. BLASTP alignment was performed on the genomes of B. rapa, B. oleracea, and B. napus with a threshold of E < 10−5 to identify PDX homologous genes. A phylogenetic tree was then constructed using the Neighbor-Joining (NJ) method in MEGA 11 (Tamura et al., 2021). Subsequently, Evolview (http://www.evolgenius.info/evolview/) was used to enhance the visualization of the phylogenetic tree (Subramanian et al., 2019). Motif analysis was conducted on the PDX gene of Arabidopsis thaliana, B. rapa, B. oleracea, and B. napus using MEME (http://MEME.nbcr.net/MEME/cgi-bin/MEME.cgi) (Bailey et al., 2015). Chromosome position information for the PDX gene was obtained from the gtf file of the Ensembl Plants database, and chromosome localization was analyzed using TBtools software (Chen et al., 2020). Cis-element analysis was conducted on the promoter region of the DNA sequence upstream 2,000 bp of the PDX gene of Arabidopsis thaliana, B. rapa, B. oleracea, and B. napus using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot et al., 2002). The expression pattern of the BnaPDX gene under waterlogging stress was analyzed based on the data from Hong et al. (2023). The results of gene structure, conserved domains, chromosome localization, cis-elements in the promoter region, and RNA-seq analysis were displayed using TBtools software (Chen et al., 2020).
Total RNA was extracted from B. napus tissues using an assay kit from Vazyme Biotech Co. Ltd. (Nanjing, China) and reverse transcribed into cDNA with a reverse transcription assay kit from Takara Biomedical Technology (Beijing, China) Co. Ltd. Based on the reference gene sequence of BnaPDX1.3 in B. napus, specific fluorescent quantitative primers were designed using the Primer-BLAST tool on the NCBI website (Supplementary Table S1). Following the instructions of the fluorescent quantitative PCR assay kit from TransGen Biotech Co. Ltd. (Beijing China)., the gene expression was detected using a Bio-Rad fluorescent quantitative PCR instrument. The data were analyzed using the 2−ΔΔT method, with the Bnactin gene as the internal reference gene, to determine the relative expression level of the gene (Supplementary Table S1) (Livak and Schmittgen, 2001).
Based on the PC2300S vector map, we selected the KpnI and XbaI restriction sites for constructing the 35S::BnaC03.PDX1.3 overexpression vector. We amplified the target fragment containing the restriction sites (Supplementary Table S2; Supplementary Figure S1), ligated the target vector with T4 ligase after restriction digestion, and used double enzyme digestion to verify the construction status of the vector (Supplementary Figure S2). We selected the successfully constructed recombinant overexpression vector, transferred it into B. napus G218 via Agrobacterium-mediated transformation, and performed positive detection on the transformed seedlings using specific primers for the NptII gene (Supplementary Table S3; Supplementary Figure S3). We used designed specific primers for the 35S::BnaC03.PDX1.3 recombinant overexpression vector to perform positive detection on T1 transgenic plants (Supplementary Table S3; Supplementary Figures S4, S5) and screened for BnaPDX1.3 gene overexpression in the T1 generation plants.
Two groups of T1 generation B. napus G218 plants overexpressing the BnaPDX1.3 gene and one group of G218 wild-type plants were subjected to 15 days of waterlogging stress. After the treatment, the fresh weight of the plants from all three groups was measured, and biomass analysis was conducted.
Leaf tissues were collected from two groups of BnaPDX1.3-overexpressing B. napus plants and one group of wild-type plants after exposure to waterlogging for 0, 2, 4, 6, 9, 12, and 15 days. These tissues were flash-frozen in liquid nitrogen and stored at − 80°C. The VB6 content of the samples was detected using a vitamin B6 assay kit provided by ZCIBIO Technology Co. Ltd. (Shanghai, China).
Leaf tissues were collected from two groups of BnaPDX1.3-overexpressing B. napus plants and one group of wild-type plants after exposure to waterlogging for 0, 2, 4, 6, 9, 12, and 15 days, respectively. The tissues were flash-frozen in liquid nitrogen and stored at − 80°C. The antioxidant enzyme activity and H2O2 content of the samples were detected using the SOD, POD, CAT, and H2O2 assay kits provided by ZCIBIO Technology Co. Ltd.
All measurements were conducted using three biological replicates. One-way ANOVA followed by Dunnett’s multiple comparisons test was performed using GraphPad Prism (v. 25), with p < 0.05 considered significant for all experiments.
Conducting a BLAST search on the protein sequences (ATPDX) of five Arabidopsis thaliana PDX genes yielded six B. rapa genes (BraPDX), eight B. oleracea genes (BoPDX), and 13 B. napus genes (BnaPDX), all obtained from the Ensembl Plants database. Based on the PDX protein sequences from the four species, an evolutionary tree (Figure 1) was constructed using MEGA 11 software and the Evolview online beautification tool, visually reflecting the evolution and classification of the 32 PDX family members. According to the evolutionary tree analysis, the PDX proteins were divided into five subclades (PDX1.1, PDX1.2, PDX1.3, PDX2, PDX3), with two branches in PDX1.3. All five subclades contained genes from B. rapa, B. oleracea, and B. napus.
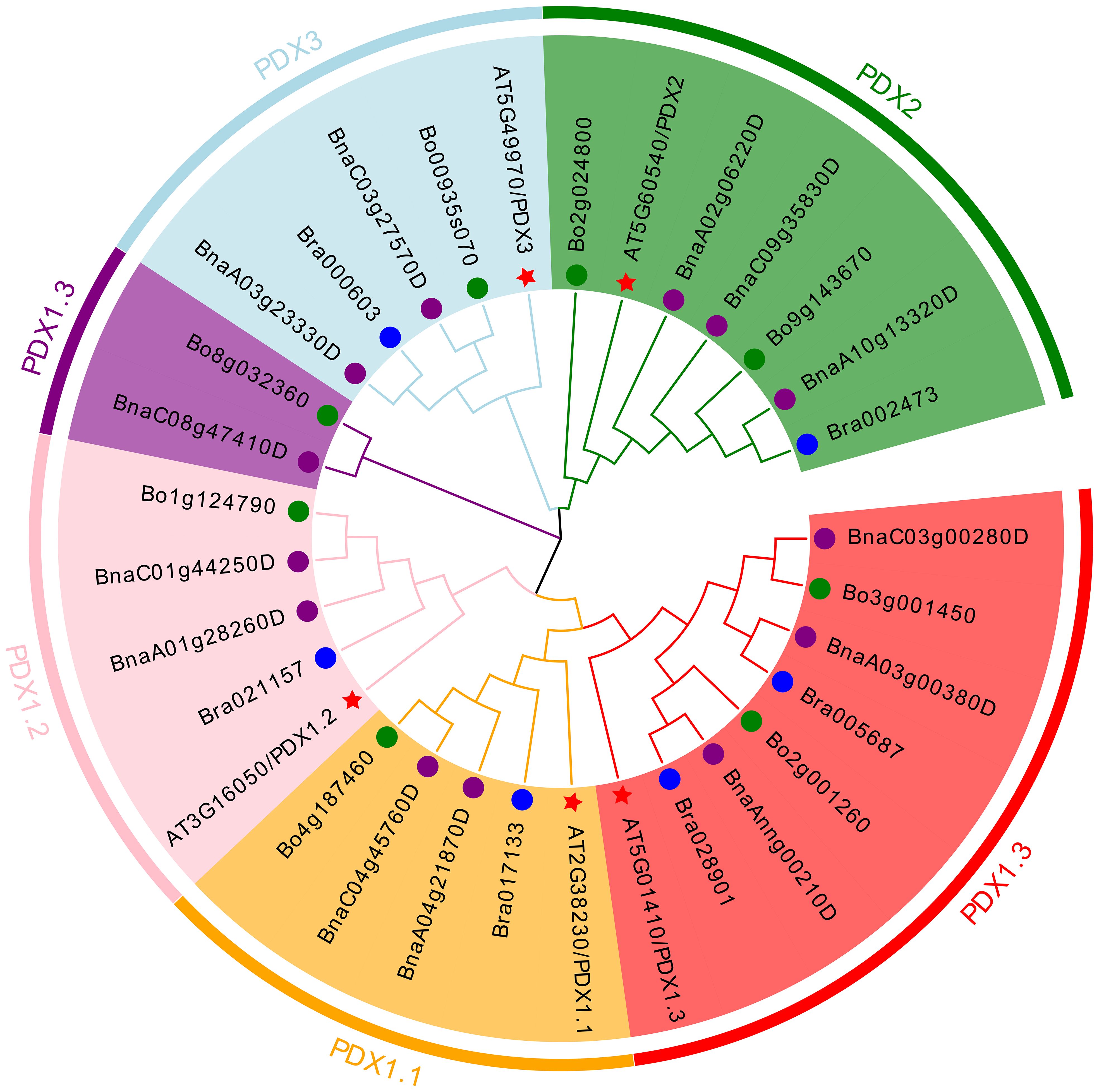
Figure 1. Phylogenetic tree of PDX proteins from B. napus, B. rapa, B. oleracea, and Arabidopsis. The neighbor-joining phylogenetic tree shows the relationships among 13 B. napus, six B. rapa, eight B. oleracea, and five Arabidopsis PDX proteins.
To further investigate the evolutionary relationships among the PDX gene families of Arabidopsis thaliana, B. rapa, B. oleracea, and B. napus, we conducted a predictive analysis of their gene structures and motifs. Using MEME, we analyzed 15 motifs among 32 proteins (Figure 2). It was found that members belonging to the same subfamily exhibited certain similarities in their conserved motifs. The conserved motifs of most PDX1.1 and PDX1.3 proteins were arranged as Motif11-Motif3-Motif6-Motif2-Motif4-Motif1-Motif5-Motif13, with the exception of Bra017133, which lacked Motif4, Motif6, and Motif13; BnaA04g21870D, which lacked Motif4 and Motif6; BnaC04g45760D, which lacked Motif2; and BnaC08g47410D and Bo8g032360, which retained only the core conserved motif, Motif1, of the pyridoxal phosphate synthase domain. This may also imply that they still perform functions related to vitamin B6 synthesis. Compared to PDX1.1 and PDX1.3, the conserved motif of the PDX1.2 protein lacked Motif13, with its composition pattern being Motif11-Motif3-Motif6-Motif2-Motif4-Motif1-Motif5. In summary, the functions of PDX1.1, PDX1.2, and PDX1.3 proteins should be similar.
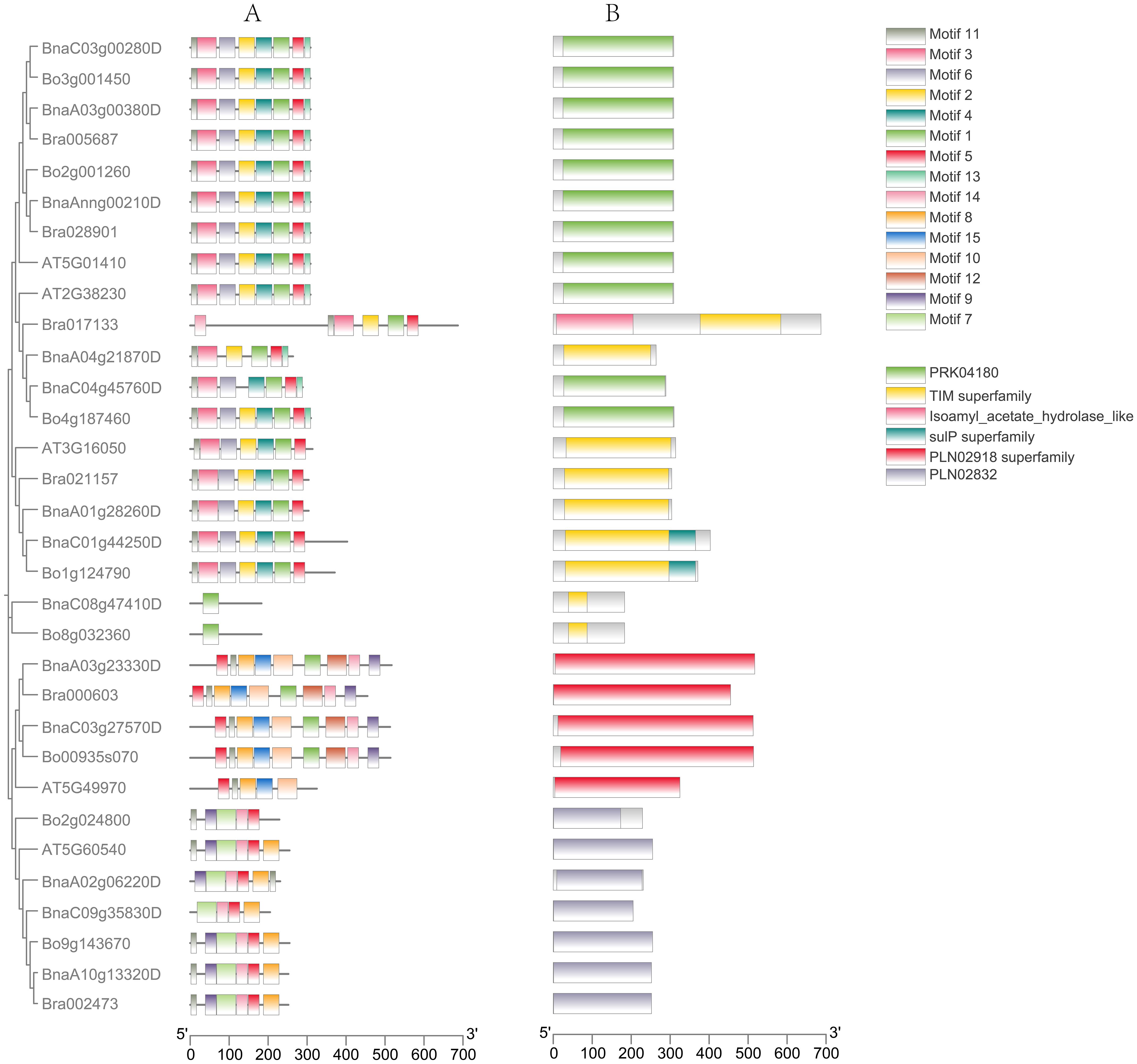
Figure 2. Gene motifs (A) and gene structure (B) analysis of PDX proteins. Fifteen motifs in PDX proteins were identified using MEME tools (A). Orange boxes, black lines, and green boxes indicate exons, introns, and untranslated regions, respectively (B).
Except for AT5G49970, which lacks Motif1, Motif9, Motif12, and Motif14, the conserved motif patterns of the remaining PDX2 proteins are Motif5-Motif11-Motif8-Motif15-Motif10-Motif1-Motif12-Motif14-Motif9. Most PDX3 proteins have conserved motif patterns of Motif11-Motif9-Motif7-Motif14-Motif5-Motif8, with Bo2g024800 lacking Motif8 and BnaC09g35830D lacking Motif9 and Motif11. The motif prediction results are generally consistent with the evolutionary tree alignment analysis, suggesting that PDX1 genes from different subclades may perform similar functions while exhibiting significant differences from the other two major clades. This motif specificity distribution among different subclades may reflect the functional differentiation of BnaPDX genes in B. napus.
Gene structure prediction reveals that the structure of the PDX1 genes (PDX1.1, PDX1.2, PDX1.3) is relatively conserved, with most introns ranging from 0 to 2 in number. Specifically, 13 PDX1 genes lack introns, three PDX1 genes possess one intron and two exons, three PDX1 genes have two introns and three exons, and only Bra017133 features nine introns and 10 exons. Meanwhile, the gene structure of the PDX2 subgroup remains consistent, exhibiting a structure with 12 introns and 13 exons. Within the PDX3 subgroup, two types exhibit four introns and five exons: Bo2g024800 and BnaA02g06220D, while the rest possess a structure with five introns and six exons.
The PDX gene does not exhibit tandem repeats in the four species. Five Arabidopsis thaliana ATPDX genes are located on chromosomes Chr2, Chr3, and Chr5; six Chinese B. rapa BraPDX genes are located on chromosomes A01, A02, A03, A04, and A10; and eight B. oleracea BoPDX genes and 13 B. napus BnaPDX genes are located on seven and 11 chromosomes, respectively (Figure 3).
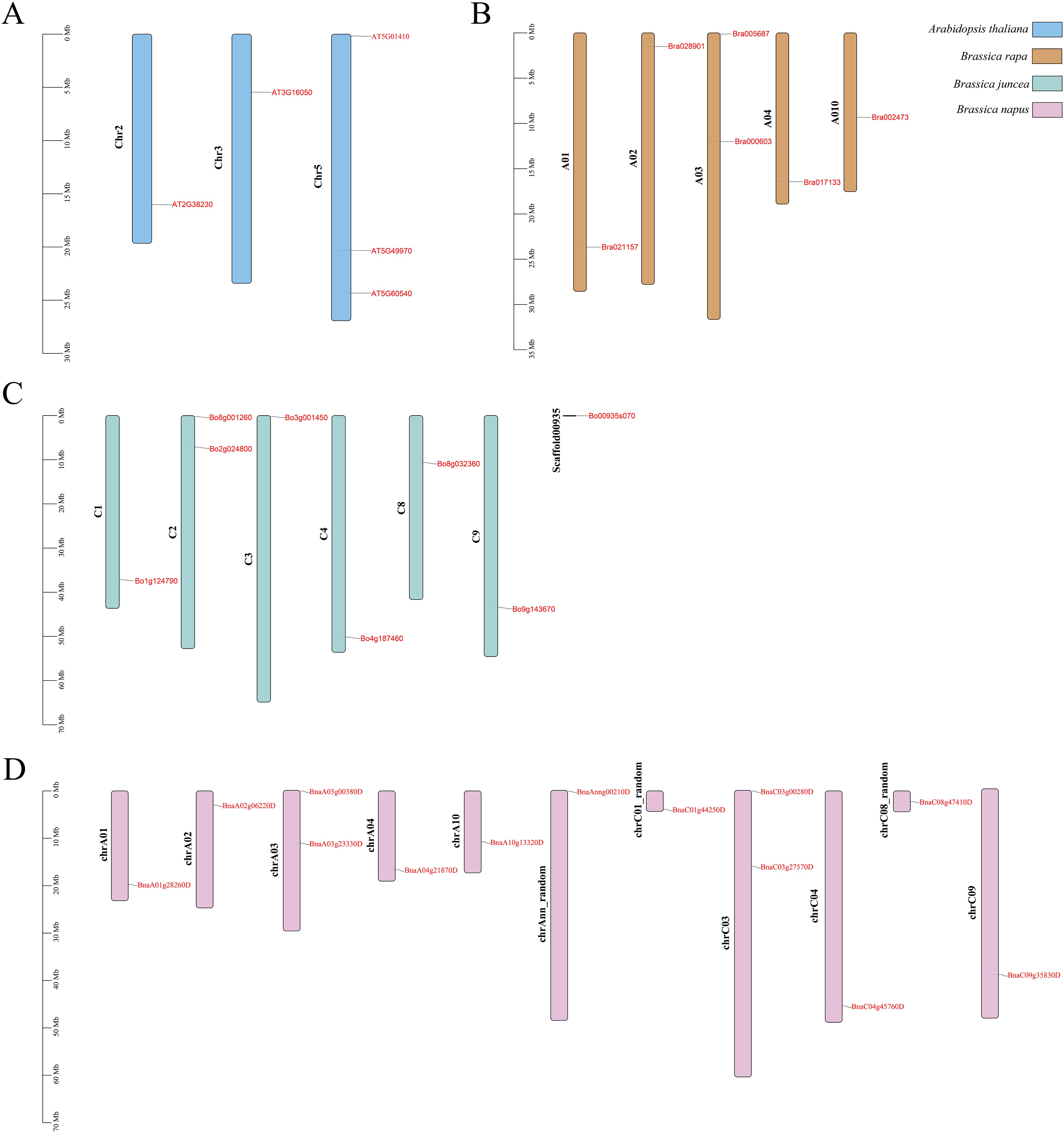
Figure 3. Chromosomal distribution of PDX genes in Arabidopsis (A), B. rapa (B), B. oleracea (C), and B. napus (D).
Using PlantCARE to analyze the various cis-elements in the promoter sequence (upstream of 2,000 bp) of the BnaPDX gene, the results can be divided into five types of plant hormone response cis-elements (IAA, ABA, MeJA, GA, and SA), six types of environmental stress response cis-elements (Stress, Anaerobic, Circadian, Light, Drought, and Low-T), one specific transcription factor binding site (MYBHv1), and several gene-specific response cis-elements (Figure 4).
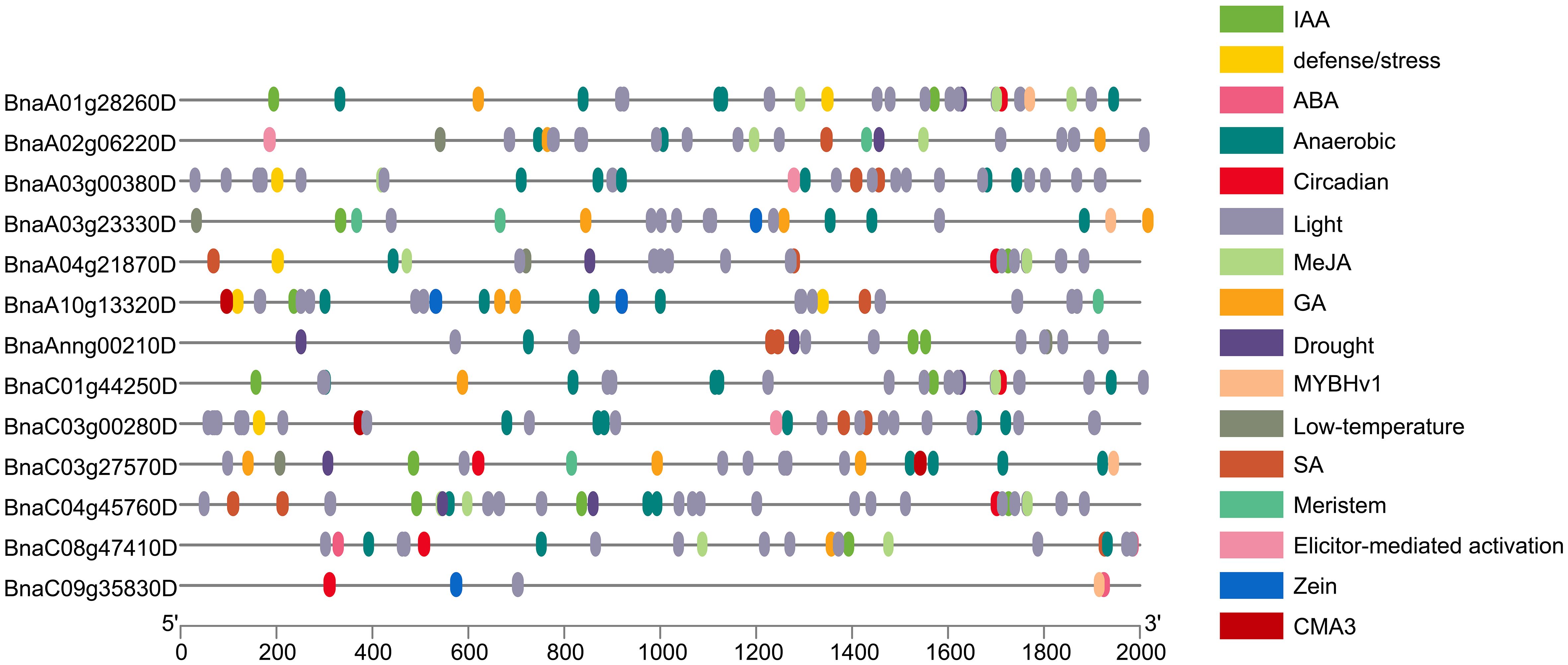
Figure 4. Promoter cis-element analysis of PDX genes in B. napus. Differently colored boxes represent various cis-acting elements and their corresponding responses.
The results revealed that within the BnaPDX gene family of B. napus, there are 111 cis-elements linked to plant hormone response, 239 cis-elements associated with environmental stress response, and five cis-elements related to metabolism. Specifically, among these, 44 ABA response cis-elements represent the highest proportion within plant hormone response, whereas 169 light response cis-elements are the most prevalent in environmental stress response.
To further analyze the response of PDX members to waterlogging stress, this study utilized RNA-seq data from B. napus G218 subjected to waterlogging treatment for 6 days, previously conducted in the field waterlogging experiment by our research group (Hong et al., 2023). We analyzed the expression of the BnaPDX gene family in B. napus under waterlogging stress and performed qRT-PCR verification of the BnaPDX1.3 gene (BnaAnng00210D, BnaA03g00380D, BnaC03g00280D).
Among the BnaPDX family genes, except for BnaPDX1.2 (BnaA01g28260D and BnaC01g44250D) whose expression level was downregulated after 6 days of waterlogging stress, all other BnaPDX family genes showed upregulation, with BnaPDX1.1 (BnaA04g21870D and BnaC04g45760D) exhibiting the most significant upregulation (Figure 5A). The qRT-PCR verification results were generally consistent with the RNA-seq data. Except for the BnaAnng00210D gene, whose expression decreased relatively after 6 days of waterlogging stress, the expression of the BnaPDX1.3 gene was upregulated after stress in all other cases.
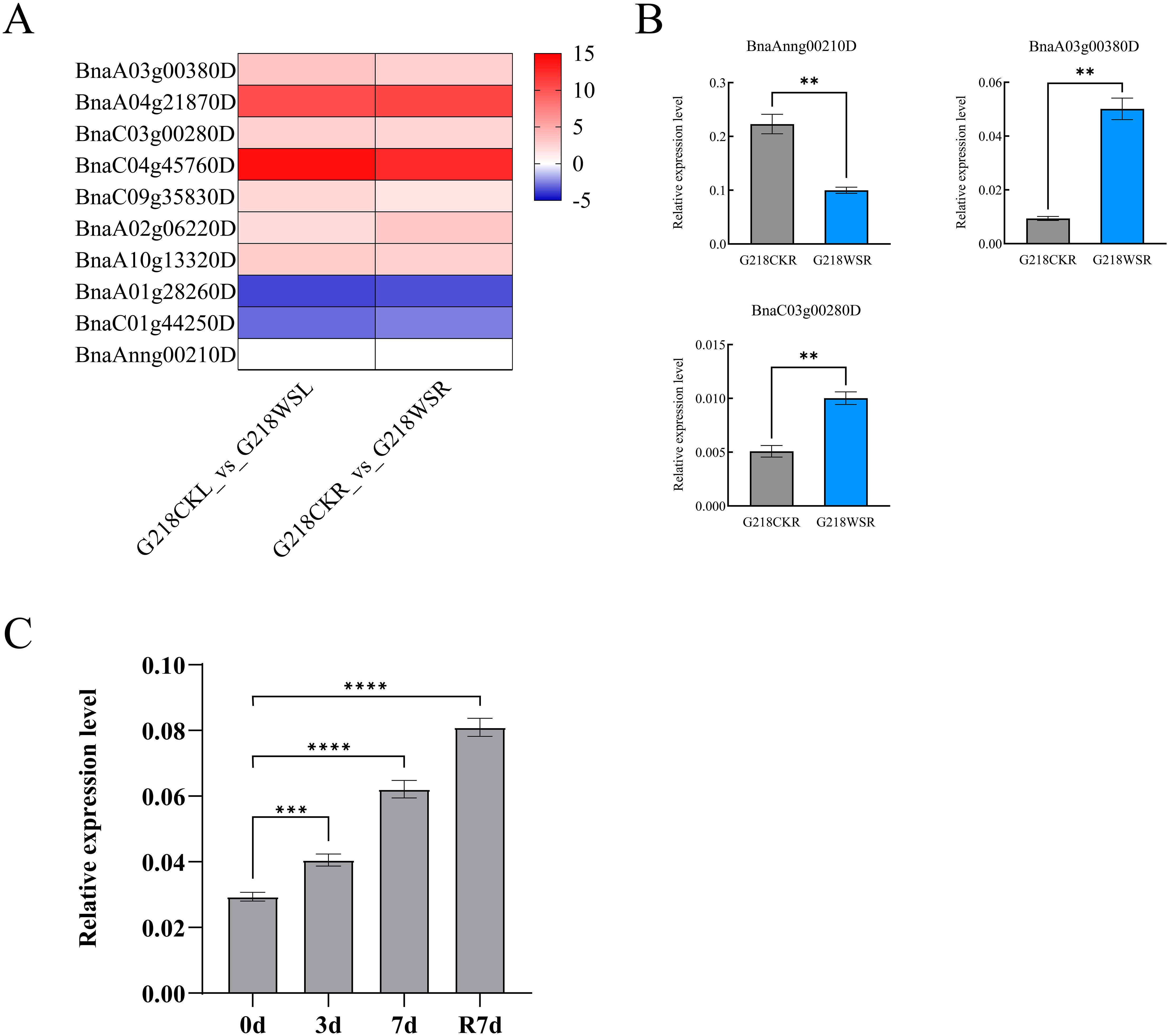
Figure 5. The BnaPDX1.3 gene in B. napus is involved in the response to waterlogging stress. (A) RNA-seq analysis of PDX genes in B. napus G218 leaves and roots after 6 days of waterlogging stress. G218CKL denotes the control leaf, G218WSL the leaf after waterlogging stress, G218CKR the control root, and G218WSR the root after waterlogging stress. (B) qRT-PCR analysis of the BnaPDX1.3 gene expression in B. napus G218 leaves after 6 days of waterlogging stress. The results of the qRT-PCR analysis are consistent with the RNA-seq. (C) qRT-PCR analysis of BnaPDX1.3 gene expression in B. napus under waterlogging stress. qRT-PCR analysis of the BnaPDX1.3 gene in B. napus at 0, 3, and 7 days of waterlogging stress, as well as after 7 days of reoxygenation following the end of stress. *p<0.05", "**p<0.01", ***p<0.001", ****p<0.0001".
Interestingly, BnaAnng00210D exhibits the highest relative expression level, being 24 times higher than that of BnaA03g00380D. Its elevated expression level might account for its slightly downregulated expression under waterlogging stress (Figure 5B). Further investigation into the expression pattern of BnaPDX1.3 under waterlogging stress revealed that the relative expression of BnaPDX1.3 in G218 seedlings significantly increased after waterlogging treatment and continued to rise even 7 days after reoxygenation (Figure 5C), indicating that BnaPDX1.3 in B. napus responds positively to waterlogging stress.
To investigate whether overexpression of the BnaPDX1.3 gene in B. napus plays a positive regulatory role in response to waterlogging stress, two groups of BnaPDX1.3-overexpressing B. napus (PDX1.3#20 and PDX1.3#21) and WT B. napus at the three-leaf and one-heart stage were subjected to 15 days of waterlogging stress (Supplementary Figure S6). Under waterlogging stress, the wild-type leaves exhibited significant leaf abscission, turning purplish-red and yellowing. In contrast, the leaves of BnaPDX1.3-overexpressing rapeseed PDX1.3#20 showed less abscission, purplish-red coloration, and yellowing, while the leaves of BnaPDX1.3-overexpressing B. napus PDX1.3#21 did not exhibit obvious waterlogging phenotypes (Figure 6A). In terms of biomass, the overexpressing plants exhibited significantly higher values after waterlogging compared to the wild type (Figure 6B).
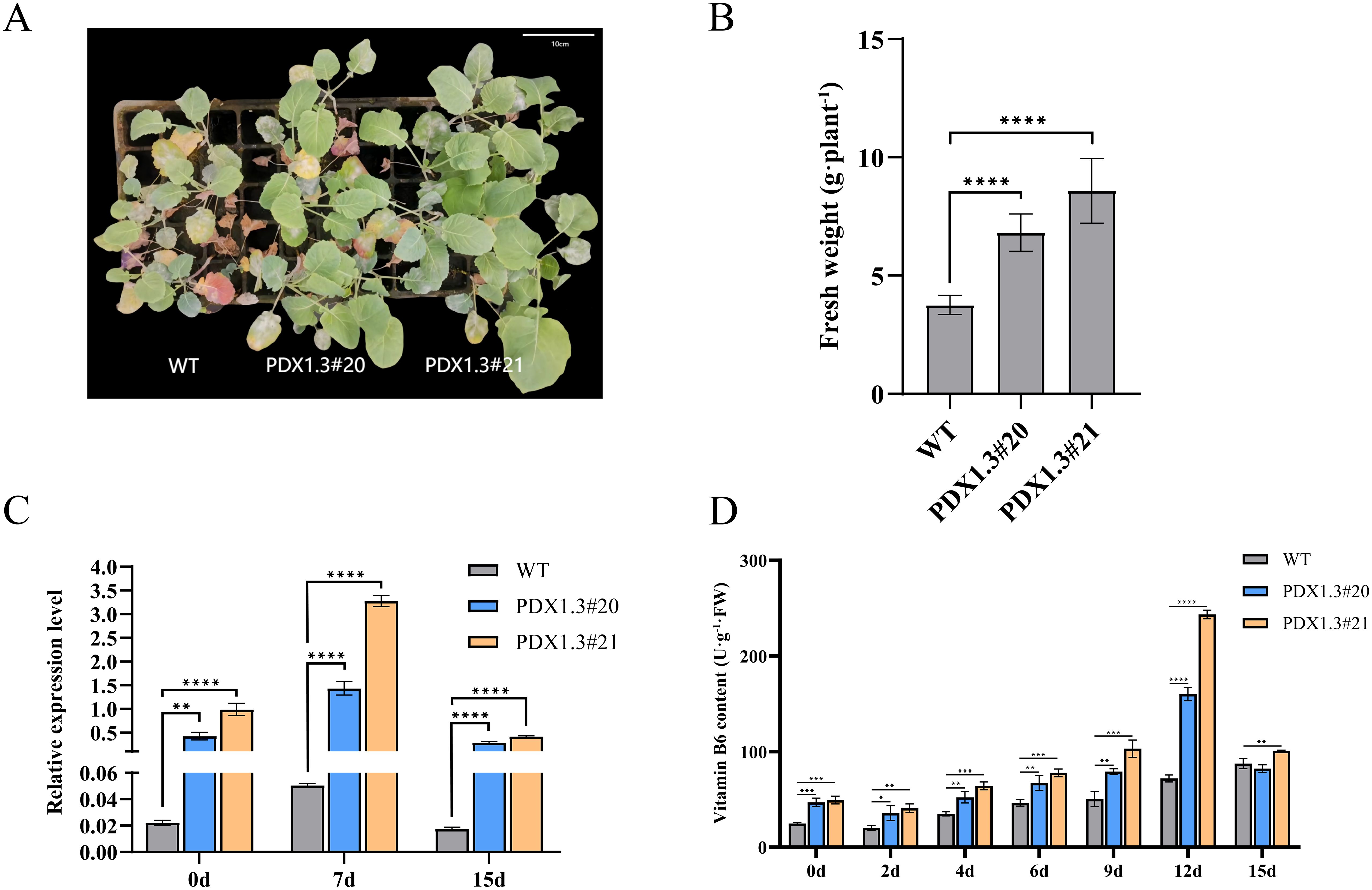
Figure 6. The BnaPDX1.3 gene enhances waterlogging tolerance in B. napus. (A) Photographs of the BnaPDX1.3 gene overexpressing B. napus and wild-type (WT) plants after 15 days of waterlogging stress. The photographs show that, compared to the overexpressing plants (PDX1.3#20 and PDX1.3#21), the wild-type plants exhibit more pronounced symptoms, including leaf shedding, purple-red discoloration, and yellowing. (B) Biomass analysis of the BnaPDX1.3 gene overexpressing B. napus and wild-type plants after 15 days of waterlogging stress. The biomass of overexpressing plants was significantly higher than that of wild-type plants following waterlogging stress. (C) Quantitative expression analysis of the BnaPDX1.3 gene in overexpressing and wild-type B. napus leaves under waterlogging stress. Overexpressing plants exhibited significantly higher expression levels of the BnaPDX1.3 gene compared to wild-type plants. (D) Vitamin B6 (VB6) content in overexpressing and wild-type B. napus leaves under waterlogging stress. Plants with stronger BnaPDX1.3 gene expression had higher VB6 content. This, along with their phenotype and biomass, corresponds to greater waterlogging tolerance. *p<0.05", "**p<0.01", ***p<0.001", ****p<0.0001".
qRT-PCR analysis showed that the expression level of the BnaPDX1.3 gene in PDX1.3#20 overexpressing plants was 19 times higher than that in the wild type, while in PDX1.3#21, it was 45 times higher. After 7 days of waterlogging stress, the expression levels of the BnaPDX1.3 gene in both overexpressing and wild-type plants were significantly upregulated. PDX1.3#20 was upregulated by 3.4 times, and PDX1.3#21 by 3.2 times, while the wild type showed only a 2.2-fold upregulation. After 15 days of waterlogging stress, the expression levels of the BnaPDX1.3 gene in all three groups were downregulated to levels lower than those before treatment. However, the transgenic plants still maintained relatively high expression levels (Figure 6C).
Under waterlogging stress, the vitamin B6 content in the overexpressing plants was significantly higher than that in the wild-type plants, peaking after 12 days of stress. The VB6 peak in PDX1.3#21 was 3.4 times higher than in the wild type, while the VB6 peak in PDX1.3#20 was 2.2 times higher (Figure 6D). In summary, plants with higher expression of the BnaPDX1.3 gene synthesize more vitamin B6 and exhibit greater tolerance to waterlogging.
Upon detecting antioxidant enzyme activity in plant leaves under waterlogging stress, it was found that transgenic plants exhibited stronger antioxidant enzyme activity and more stable H2O2 content. During the early stages of waterlogging (2–4 days), the SOD activity in transgenic plants was significantly higher than that in wild-type plants. It gradually returned to levels similar to those in wild-type plants between 6 and 12 days, but remained significantly higher at 15 days, maintaining a high level of SOD activity (Figure 7A).
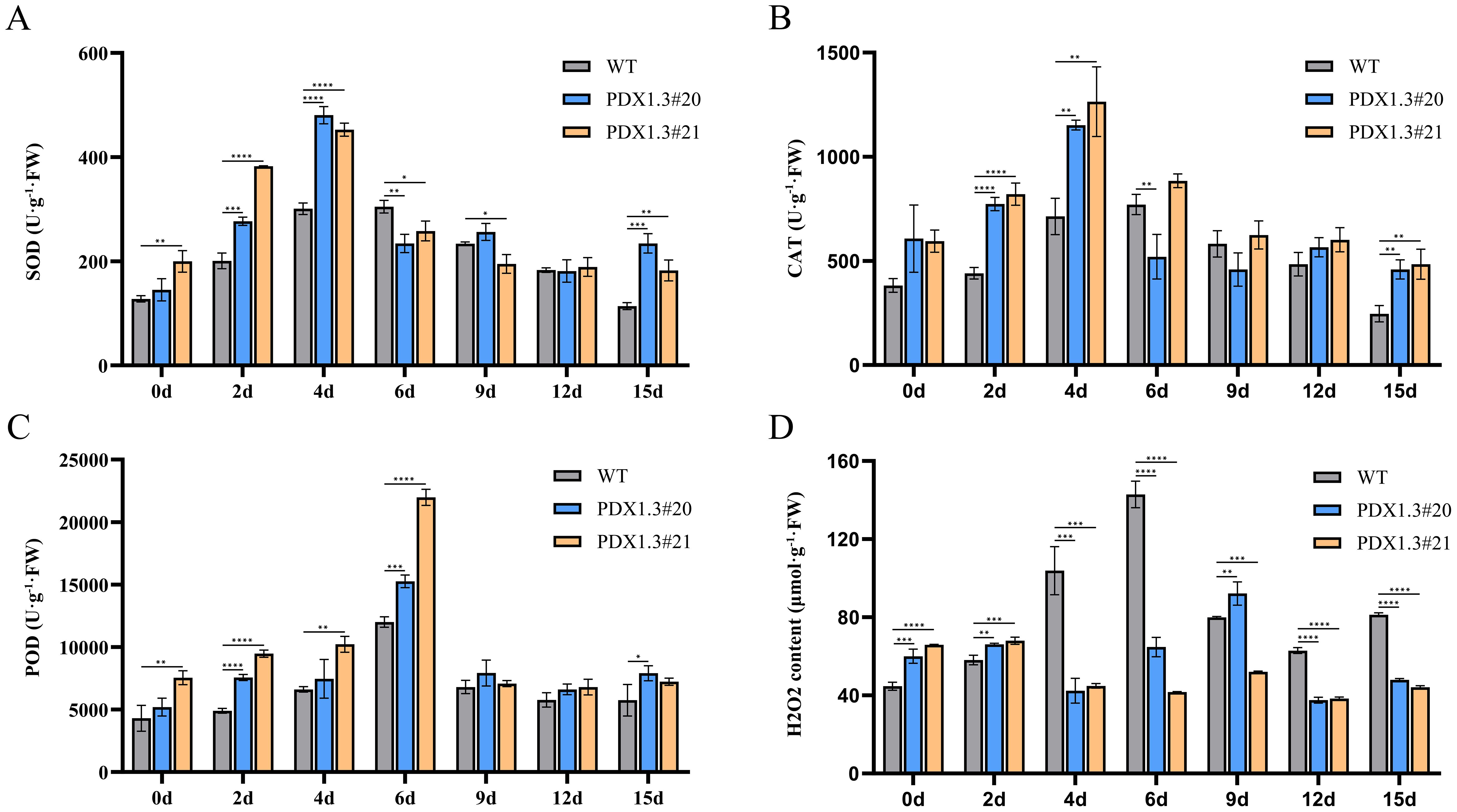
Figure 7. BnaPDX1.3 gene enhances antioxidant enzyme activity in B. napus. (A) SOD activity under waterlogging stress in BnaPDX1.3-overexpressing B. napus and wild-type plants. SOD activity in overexpressing plants shows a significant difference compared to wild-type plants under waterlogging stress. (B) CAT activity in BnaPDX1.3-overexpressing B. napus and wild-type plants under waterlogging stress. CAT activity in overexpressing plants is generally significantly higher than in wild-type plants under waterlogging stress. (C) POD activity in BnaPDX1.3-overexpressing B. napus and wild-type plants under waterlogging stress. Overexpressing plants exhibit stronger POD activity compared to wild-type plants under waterlogging stress. (D) H2O2 content in BnaPDX1.3-overexpressing B. napus and wild-type plants under waterlogging stress. H2O2 content in overexpressing plants is significantly lower and more stable compared to wild-type plants under waterlogging stress. *p<0.05", "**p<0.01", ***p<0.001", ****p<0.0001".
During the early stages of waterlogging (2–4 days), the CAT activity in transgenic plants was significantly higher than that in wild-type plants. It gradually decreased, but the trend was more stable in transgenic plants. At 15 days, CAT activity remained significantly higher in transgenic plants, maintaining a high level of CAT activity (Figure 7B).
From 2 to 6 days, the POD activity in transgenic plants was significantly higher than that in wild-type plants. During this period, all three groups of plants showed a gradual increase, reaching the same level at 9 days, after which the activity stabilized (Figure 7C).
After 4 days of waterlogging stress, the H2O2 content in transgenic plants rapidly decreased, while in wild-type plants, it rapidly increased, with their H2O2 content significantly higher than that of transgenic plants. At 6 days, the H2O2 content of wild-type plants reached its peak and remained elevated throughout the later stages of stress, while the H2O2 content in BnaPDX1.3-overexpressing plants had stabilized after 4 days and remained significantly lower than in wild-type plants, maintaining a low level throughout the later stages of stress (Figure 7D).
In summary, high-level expression of BnaPDX1.3 helps plants establish a stronger and more efficient antioxidant system under waterlogging stress, effectively preventing the accumulation of ROS and subsequent oxidative damage.
In China, B. napus is the second-largest oilseed crop, with a planting area of approximately 6.57 million hectares, accounting for 38.6% of the country's total oil crop production (Wang et al., 2022; Li and Wang, 2022). The Yangtze River Basin is the main producing area for B.napus in China, where 91% of the total oilseed rape in China is produced (Tao et al., 2015), B. napus production in the region primarily follows a rice–rapeseed rotation. The soil has a relatively high water-holding capacity, and the rapeseed growing period coincides with the rainy season in the basin, which is characterized by frequent high-intensity rainfall, making the region prone to waterlogging (Tao et al., 2024; Zhu et al., 2019). Waterlogging can affect root vitality and growth, photosynthetic efficiency, and the physiological metabolism of rapeseed, ultimately affecting yield and quality (Xu et al., 2018; Ploschuk et al., 2018).
Vitamin B6, as an endogenous growth regulator, is considered an antioxidant in plants, influencing physiological metabolism, growth, development, and stress resistance (Colinas et al., 2016; Moccand et al., 2014; Zhang et al., 2014; Huang et al., 2013). The PDX family genes, key players in the vitamin B6 synthesis pathway, have been shown to be involved in responding to various environmental stresses in plants. Studies have shown that the three PDX1 and PDX2 genes in Arabidopsis thaliana respond to strong light, cold, drought, and ozone stress (Bagri et al., 2018; Leasure et al., 2011). The pdx1.3 mutant of Arabidopsis thaliana is more sensitive to salt, oxidative, and osmotic stress (Titiz et al., 2006; Moccand et al., 2014). Currently, research on plant PDX genes has primarily focused on Arabidopsis thaliana, and no studies have reported on the PDX genes in B. napus. As a gene involved in vitamin B6 synthesis, the effect of BnaPDX1.3 on B. napus’s response to waterlogging stress has not been fully elucidated. Therefore, this study aims to explore the role of the BnaPDX1.3 gene in B. napus’s response to waterlogging stress by analyzing the phenotype, as well as physiological and biochemical indicators, of T1 generation BnaPDX1.3-overexpressing rapeseed plants under waterlogging stress.
The research findings indicate that, after a 15-day waterlogging stress experiment, the transgenic rapeseed plants with higher expression levels of the BnaPDX1.3 gene exhibited healthier leaves and greater biomass compared to wild-type plants, suggesting that the BnaPDX1.3 gene enhances the waterlogging tolerance of B. napus. To delve deeper into the functional mechanism of the BnaPDX1.3 gene in B. napus, this study examined the changes in VB6 content under waterlogging stress. The results revealed that the VB6 content in the transgenic plants was generally higher than that in the wild-type plants, with the peak VB6 content in the transgenic plants on day 12 being 3.4 times higher than that in the wild-type plants.
H2O2, as a plant stress signaling molecule and reactive oxygen species, can reflect the plant’s ability to respond to and resist stress. In this study, the analysis of H2O2 content after waterlogging showed that the wild-type plants accumulated excessive H2O2 6 days after waterlogging and maintained high levels of H2O2 throughout the later stages of the stress, unable to effectively prevent cellular oxidative damage. However, the BnaPDX1.3-overexpressing plants maintained lower H2O2 levels under waterlogging conditions. The analysis of antioxidant enzyme activity indicated that the BnaPDX1.3-overexpressing plants had significantly higher SOD, POD, and CAT enzyme activities under waterlogging stress compared to the wild-type control plants, demonstrating a stronger antioxidant capacity. This effectively eliminates ROS generated by waterlogging, reduces oxidative damage, and thereby enhances plant tolerance to waterlogging.
We conclude that overexpressing plants, with high BnaPDX1.3 gene expression, synthesize more VB6, exhibit stronger antioxidant enzyme activity, and possess a more efficient and stable ROS scavenging system, thereby demonstrating healthy growth under waterlogging stress. In summary, the BnaPDX1.3 gene enhances the waterlogging tolerance of B. napus, which is of great significance for its response to waterlogging stress. The event model revealed in this study is shown in Figure 8. Additional studies have shown that VB6 can act as an antioxidant, participating in the scavenging of ROS accumulation (Lu et al., 2022; Danon et al., 2005; Zhang et al., 2022). Moreover, pdx1 mutants impair IAA biosynthesis (Boycheva et al., 2015; Chen and Xiong, 2009), and impaired VB6 biosynthesis can inhibit ABA biosynthesis (Liu et al., 2022). In summary, VB6 participates in plant responses to adverse stress by regulating plant hormone synthesis and antioxidant systems, which may explain why overexpressing BnaPDX1.3 plants exhibit stronger waterlogging tolerance.
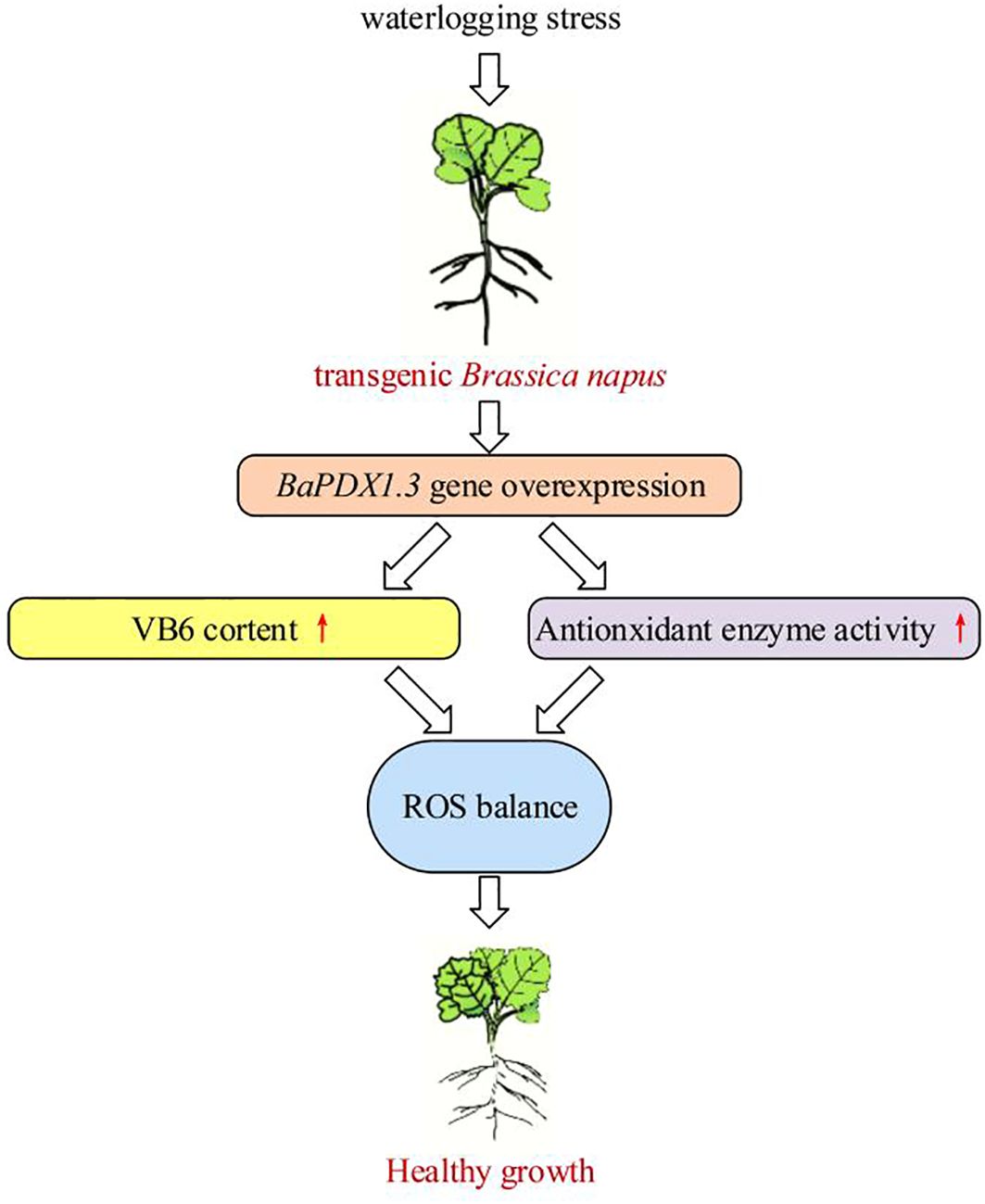
Figure 8. Impact of overexpression of BnaPDX1.3 in B. napus on plant waterlogging tolerance. BnaPDX1.3-overexpressing B. napus plants exhibit high expression levels of the BnaPDX1.3 gene under waterlogging stress, synthesize high levels of VB6, and significantly enhance antioxidant enzyme activity. This results in a stable dynamic balance of ROS under waterlogging stress, supporting healthy growth.
The bioinformatics analysis of the PDX gene family and the waterlogging stress experiment analysis with the G218 wild type in this study indicate that the BnaPDX1.3 gene is involved in the response of B. napus to waterlogging stress. To further investigate the regulatory role of the BnaPDX1.3 gene in waterlogging stress, this study generated BnaPDX1.3-overexpressing B. napus plants through Agrobacterium-mediated transformation. After positive detection and screening, T1 generation positive seedlings were obtained. The T1 generation BnaPDX1.3-overexpressing B. napus plants were subjected to a 15-day waterlogging stress treatment. It was observed that BnaPDX1.3-overexpressing B. napus plants had healthier leaves and greater biomass compared to wild-type plants, exhibiting better waterlogging tolerance. Additionally, they displayed a stronger antioxidant system and a more stable ROS content under waterlogging stress. The results of this study indicate that the BnaPDX1.3 gene can enhance the waterlogging tolerance of B. napus, which is of great significance for its response to waterlogging stress.
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, accession number PRJNA898876.
MY: Writing – original draft, Writing – review & editing. BH: Writing – original draft, Writing – review & editing. HJ: Writing – original draft, Writing – review & editing. CG: Writing – original draft, Writing – review & editing. MG: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Hunan Agriculture Research System (HARS-03).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1533219/full#supplementary-material
Asensi-Fabado, M. A., Munné-Bosch, S. (2010). Vitamins in plants: occurrence, biosynthesis and antioxidant function. Trends Plant Sci. 15, 582–592. doi: 10.1016/j.tplants.2010.07.003
Bagri, D. S., Upadhyaya, D. C., Kumar, A., Upadhyaya, C. P. (2018). Overexpression of PDX-II gene in potato (Solanum tuberosum L.) leads to the enhanced accumulation of vitamin B6 in tuber tissues and tolerance to abiotic stresses. Plant Sci. 272, 267–275. doi: 10.1016/j.plantsci.2018.04.024
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Boycheva, S., Dominguez, A., Rolcik, J., Boller, T., Fitzpatrick, T. B. (2015). Consequences of a deficit in vitamin B6 biosynthesis de novo for hormone homeostasis and root development in Arabidopsis. Plant Physiol. 167, 102–117. doi: 10.1104/pp.114.247767
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Xiong, L. (2005). Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 44, 396–408. doi: 10.1111/j.1365-313X.2005.02538.x
Chen, H., Xiong, L. (2009). The short-rooted vitamin B6-deficient mutant pdx1 has impaired local auxin biosynthesis. Planta. 229 (6), 1303–1310. doi: 10.1007/s00425-009-0912-8
Chung, K. R. (2012). Stress response and pathogenicity of the necrotrophic fungal pathogen alternaria alternata. Scientifica 2012, 635431. doi: 10.6064/2012/635431
Colinas, M., Eisenhut, M., Tohge, T., Pesquera, M., Fernie, A. R., Weber, A. P., et al. (2016). Balancing of B6 vitamers is essential for plant development and metabolism in arabidopsis. Plant Cell 28, 439–453. doi: 10.1105/tpc.15.01033
Danon, A., Miersch, O., Felix, G., Camp, R. G., Apel, K. (2005). Concurrent activation of cell death-regulating signaling pathways by singlet oxygen in Arabidopsis thaliana. Plant J. 41, 68–80. doi: 10.1111/j.1365-313X.2004.02276.x
Dell’Aglio, E., Boycheva, S., Fitzpatrick, T. B. (2017). The pseudoenzyme PDX1.2 sustains vitamin B6 biosynthesis as a function of heat stress. Plant Physiol. 174, 2098–2112. doi: 10.1104/pp.17.00531
Denslow, S. A., Rueschhoff, E. E., Daub, M. E. (2007). Regulation of the Arabidopsis thaliana vitamin B6 biosynthesis genes by abiotic stress. Plant Physiol. Bioch 45, 152–161. doi: 10.1016/j.plaphy.2007.01.007
Dong, Y. X., Sueda, S., Nikawa, J., Kondo, H. (2004). Characterization of the products of the genes SNO1 and SNZ1 involved in pyridoxine synthesis in Saccharomyces cerevisiae. Eur. J. Biochem. 271, 745–752. doi: 10.1111/j.1432-1033.2003.03973.x
Drewke, C., Leistner, E. (2001). Biosynthesis of vitamin B6 and structurally related derivatives. Vitam. Horm. 61, 121–155. doi: 10.1016/s0083-6729(01)61004-5
Eliot, A. C., Kirsch, J. F. (2004). Pyridoxal phosphate enzymes: mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 73, 383–415. doi: 10.1146/annurev.biochem.73.011303.074021
Fitzpatrick, T. B., Amrhein, N., Kappes, B., Macheroux, P., Tews, I., Raschle, T. (2007). Two independent routes of de novo vitamin B6 biosynthesis: not that different after all. Biochem. J. 407, 1–13. doi: 10.1042/BJ20070765
Gorelova, V., Colinas, M., Dell’Aglio, E., Flis, P., Salt, D. E., Fitzpatrick, T. B. (2022). Phosphorylated B6 vitamer deficiency in SALT OVERLY SENSITIVE 4 mutants compromises shoot and root development. Plant Physiol. 188, 220–240. doi: 10.1093/plphys/kiab475
Havaux, M., Ksas, B., Szewczyk, A., Rumeau, D., Franck, F., Caffarri, S., et al. (2009). Vitamin B6 deficient plants display increased sensitivity to high light and photo-oxidative stress. BMC Plant Biol. 9, 130. doi: 10.1186/1471-2229-9-130
Hong, B., Zhou, B., Peng, Z., Yao, M., Wu, J., Wu, X., et al. (2023). Tissue-specific transcriptome and metabolome analysis reveals the response mechanism of brassica napus to waterlogging stress. Int. J. Mol. Sci. 24, 6015. doi: 10.3390/ijms24076015
Huang, S., Zhang, J., Wang, L., Huang, L. (2013). Effect of abiotic stress on the abundance of different vitamin B6 vitamers in tobacco plants. Plant Physiol. Bioch 66, 63–67. doi: 10.1016/j.plaphy.2013.02.010
Jeong, H., Vacanti, N. M. (2020). Systemic vitamin intake impacting tissue proteomes. Nutr Metab (Lond). 17, 73. doi: 10.1186/s12986-020-00491-7
Leasure, C. D., Tong, H. Y., Hou, X. W., Shelton, A., Minton, M., Esquerra, R., et al. (2011). root uv-b sensitive mutants are suppressed by specific mutations in ASPARTATE AMINOTRANSFERASE2 and by exogenous vitamin B6. Mol. Plant 4, 759–770. doi: 10.1093/mp/ssr033
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Leuendorf, J. E., Mooney, S. L., Chen, L., Hellmann, H. A. (2014). Arabidopsis thaliana PDX1.2 is critical for embryo development and heat shock tolerance. Planta 240, 137–146. doi: 10.1007/s00425-014-2069-3
Li, Q., Wang, C. (2022). An evaluation of chinese rapeseed production efficiency based on three-stage DEA and malmquist index. Sustainability-Basel 14, 15822. doi: 10.3390/su142315822
Liu, H., Lu, C., Li, Y., Wu, T., Zhang, B., Liu, B., et al. (2022). The bacterial effector AvrRxo1 inhibits vitamin B6 biosynthesis to promote infection in rice. Plant Commun. 3, 100324. doi: 10.1016/j.xplc.2022.100324
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, C., Tian, Y., Hou, X., Hou, X., Jia, Z., Li, M., et al. (2022). Multiple forms of vitamin B6 regulate salt tolerance by balancing ROS and abscisic acid levels in maize root. Stress Biol. 2, 39. doi: 10.1007/s44154-022-00061-2
Moccand, C., Boycheva, S., Surriabre, P., Tambasco-Studart, M., Raschke, M., Kaufmann, M., et al. (2014). The pseudoenzyme PDX1.2 boosts vitamin B6 biosynthesis under heat and oxidative stress in Arabidopsis. J. Biol. Chem. 289, 8203–8216. doi: 10.1074/jbc.M113.540526
Mooney, S., Leuendorf, J. E., Hendrickson, C., Hellmann, H. (2009). Vitamin B6: a long known compound of surprising complexity. Molecules 14 (1), 329–351. doi: 10.3390/molecules14010329
Percudani, R., Peracchi, A. (2003). A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep. 4, 850–854. doi: 10.1038/sj.embor.embor914
Ploschuk, R. A., Miralles, D. J., Colmer, T. D., Ploschuk, E. L., Striker, G. G. (2018). Waterlogging of winter crops at early and late stages: impacts on leaf physiology, growth and yield. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01863
Raschle, T., Amrhein, N., Fitzpatrick, T. B. (2005). On the two components of pyridoxal 5’-phosphate synthase from Bacillus subtilis. J. Biol. Chem. 280, 32291–32300. doi: 10.1074/jbc.M501356200
Ristilä, M., Strid, H., Eriksson, L. A., Strid, A., Sävenstrand, H. (2011). The role of the pyridoxine (vitamin B6) biosynthesis enzyme PDX1 in ultraviolet-B radiation responses in plants. Plant Physiol. Bioch 49, 284–292. doi: 10.1016/j.plaphy.2011.01.003
Roje, S. (2007). Vitamin B biosynthesis in plants. Phytochemistry 68, 1904–1921. doi: 10.1016/j.phytochem.2007.03.038
Rueschhoff, E. E., Gillikin, J. W., Sederoff, H. W., Daub, M. E. (2013). The SOS4 pyridoxal kinase is required for maintenance of vitamin B6-mediated processes in chloroplasts. Plant Physiol. Bioch. 63, 281–291. doi: 10.1016/j.plaphy.2012.12.003
Samsatly, J., Chamoun, R., Gluck-Thaler, E., Jabaji, S. (2016). Genes of the de novo and Salvage Biosynthesis Pathways of Vitamin B6 are Regulated under Oxidative Stress in the Plant Pathogen Rhizoctonia solani. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01429
Subramanian, B., Gao, S., Lercher, M. J., Hu, S., Chen, W. H. (2019). Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 47, W270–W275. doi: 10.1093/nar/gkz357
Tambasco-Studart, M., Tews, I., Amrhein, N., Fitzpatrick, T. B. (2007). Functional analysis of PDX2 from Arabidopsis, a glutaminase involved in vitamin B6 biosynthesis. Plant Physiol. 144, 915–925. doi: 10.1104/pp.107.096784
Tambasco-Studart, M., Titiz, O., Raschle, T., Forster, G., Amrhein, N., Fitzpatrick, T. B. (2005). Vitamin B6 biosynthesis in higher plants. Proc. Natl. Acad. Sci. U.S.A 102, 13687–13692. doi: 10.1073/pnas.0506228102
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tao, R., Li, H., Lu, J., Bu, R., Li, X., Cong, R., et al. (2015). Crop rotation-dependent yield responses to fertilization in winter oilseed rape (Brassica napus L.). Crop J. 3 (5), 396–404. doi: 10.1016/j.cj.2015.04.007
Tao, Y., Li, D., Yu, Y., Lu, C., Huang, M., Wang, H., et al. (2024). Optimum transplanting date for rape forage and grain yields in the ridge culture place planting system on the yangtze river delta. Appl. Sci. 14, 3207. doi: 10.3390/app14083207
Titiz, O., Tambasco-Studart, M., Warzych, E., Apel, K., Amrhein, N., Laloi, C., et al. (2006). PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J. 48, 933–946. doi: 10.1111/j.1365-313X.2006.02928.x
Vanderschuren, H., Boycheva, S., Li, K. T., Szydlowski, N., Gruissem, W., Fitzpatrick, T. B. (2013). Strategies for vitamin B6 biofortification of plants: a dual role as a micronutrient and a stress protectant. Front. Plant Sci. 4. doi: 10.3389/fpls.2013.00143
Wagner, S., Bernhardt, A., Leuendorf, J. E., Drewke, C., Lytovchenko, A., Mujahed, N., et al. (2006). Analysis of the Arabidopsis rsr4-1/pdx1-3 mutant reveals the critical function of the PDX1 protein family in metabolism, development, and vitamin B6 biosynthesis. Plant Cell 18, 1722–1735. doi: 10.1105/tpc.105.036269
Wang, C., Xu, M., Wang, Y., Batchelor, W. D., Zhang, J., Kuai, J., et al. (2022). Long-term optimal management of rapeseed cultivation simulated with the CROPGRO-canola model. Agronomy 12, 1191. doi: 10.3390/agronomy12051191
Wetzel, D. K., Ehrenshaft, M., Denslow, S. A., Daub, M. E. (2004). Functional complementation between the PDX1 vitamin B6 biosynthetic gene of Cercospora nicotianae and pdxJ of Escherichia coli. FEBS Lett. 564, 143–146. doi: 10.1016/S0014-5793(04)00329-1
Xu, J., Qiao, X., Tian, Z., Zhang, X., Zou, X., Cheng, Y., et al. (2018). Proteomic analysis of rapeseed root response to waterlogging stress. Plants 7, 71. doi: 10.3390/plants7030071
Zhang, Y., Jin, X., Ouyang, Z., Li, X., Liu, B., Huang, L., et al. (2015). Vitamin B6 contributes to disease resistance against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea in Arabidopsis thaliana. J. Plant Physiol. 175, 21–25. doi: 10.1016/j.jplph.2014.06.023
Zhang, Y., Liu, B., Li, X., Ouyang, Z., Huang, L., Hong, Y., et al. (2014). The de novo biosynthesis of vitamin B6 is required for disease resistance against Botrytis cinerea in tomato. Mol. Plant Microbe 27, 688–699. doi: 10.1094/MPMI-01-14-0020-R
Zhang, C. Y., Peng, L. J., Chen, G. Y., Zhang, H., Yang, F. Q. (2022). Investigation on the peroxidase-like activity of vitamin B6 and its applications in colorimetric detection of hydrogen peroxide and total antioxidant capacity evaluation. Molecules 27, 4262. doi: 10.3390/molecules27134262
Keywords: Brassica napus, BnaPDX1.3, waterlogging stress, vitamin B6, overexpression
Citation: Yao M, Hong B, Ji H, Guan C and Guan M (2025) Genome-wide identification of PDX and expression analysis under waterlogging stress exhibit stronger waterlogging tolerance in transgenic Brassica napus plants overexpressing the BnaPDX1.3 gene compared to wild-type plants. Front. Plant Sci. 16:1533219. doi: 10.3389/fpls.2025.1533219
Received: 23 November 2024; Accepted: 16 January 2025;
Published: 12 February 2025.
Edited by:
Diaa Abd El Moneim, Arish University, EgyptReviewed by:
Muhammad Saad Shoaib Khan, Jiangsu University, ChinaCopyright © 2025 Yao, Hong, Ji, Guan and Guan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyun Guan, Z3VhbmN5MjAxbEBhbGl5dW4uY29t; Mei Guan, Z203MTQyMDA1QGh1bmF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.