
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 February 2025
Sec. Plant Systematics and Evolution
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1525193
 Haozheng Li1
Haozheng Li1 Jin Xiao1
Jin Xiao1 Jiahui Chen1
Jiahui Chen1 Xu Shen1
Xu Shen1 Jia Luo1
Jia Luo1 Fengguang Guo1
Fengguang Guo1 Shangfei Wang2
Shangfei Wang2 Liangye Xu1
Liangye Xu1 Xin Guo1
Xin Guo1 Shujuan Wang1
Shujuan Wang1 Haiyan Wang3*
Haiyan Wang3* Wenquan Wang1*
Wenquan Wang1*NADP-malic enzyme (NADP-ME) is a crucial enzyme in C4 photosynthesis, responsible for the decarboxylation of malate in bundle sheath cells, enhancing the photosynthetic efficiency of C4 plants. Cultivated cassava exhibits high photosynthetic efficiency and biomass, and previous studies classify it as a C3–C4 intermediate type. The biomass of cassava correlates positively with photosynthetic rate, and the promoter region of MeNADP-ME3 contains insertion selected in cultivars different from wild ancestors. Four MeNADP-ME genes were identified in the cultivated cassava variety AM560, with promoter regions enriched in light-responsive elements. Phylogenetic and conserved domain analyses revealed that all subtypes are plastidic dicotyledonous types, closely related to AtNADP-ME4, with unique N-terminal domains in MeNADP-ME2 and MeNADP-ME3 specific to cassava, suggesting new functional roles. Subcellular localization showed predominant chloroplast localization, with greater involvement in leaf physiological processes in the cultivated variety SC205. These findings suggest that the NADP-ME family in cultivated cassava has been evolutionarily selected for photosynthesis. Further investigation revealed that MeNADP-ME3 is highly expressed in leaves and regulated by light intensity. Co-expression network analysis of shade-treated transcriptomes and transcription factor-promoter predictions showed that Indel sites in the MeNADP-ME3 promoter are bound by MeYABBY1, forming a regulatory network with other photosynthesis-related genes. This suggests that MeNADP-ME3 plays a role in C3–C4 intermediate photosynthesis during the evolution from wild to cultivated cassava, with other family genes also evolving towards photosynthetic functions. Our study lays the foundation for future functional research on the MeNADP-ME family and provides insights into the mechanisms underlying the high photosynthetic efficiency of cultivated cassava.
Cassava (Manihot esculenta Crantz), a dicotyledonous plant from the Euphorbiaceae family, is the third-largest source of carbohydrates in tropical regions, following rice and maize. It is an important staple for over 500 million people, known for its drought resistance and ability to grow in poor soils. Cultivated cassava is regarded as a crop with high tuberous root biomass, starch accumulation capabilities, and efficient photosynthetic performance (Moorthy and Padmaja, 2002; El-Sharkawy et al., 2012). Previous research has identified 98 varieties of wild cassava (Manihot esculenta ssp. flabellifolia), a small, climbing shrub primarily found in the regions bordering the Amazon rainforest and tropical savannas in Brazil and its surrounding areas (Luiz Joaquim Castelo Branco et al., 2017). This wild species is considered to be the direct ancestor of modern cultivated cassava. However, wild cassava exhibits significantly lower photosynthetic efficiency and starch accumulation compared to cultivated cassava, lacking the high light efficiency and high starch accumulation traits characteristic of the latter (Allem, 1999; An et al., 2016). Studies with 14C isotope feeding in cultivated cassava have revealed that 30%–50% of CO2 is converted into four-carbon acids, such as malate. However, due to the lack of Kranz anatomy, cultivated cassava is considered a C3–C4 intermediate type (Cock et al., 1987; El-Sharkawy, 2004). Phosphoenolpyruvate carboxylase (PEPC), a key enzyme in C4 and C3–C4 intermediate plants, is linked to photosynthetic carbon assimilation (Svensson et al., 2003). Analysis of 18 cassava cultivars showed a strong positive correlation between photosynthetic efficiency, tuberous root biomass, and MePEPC activity, further supporting the significant role of high MePEPC activity in enhancing cassava’s photosynthetic efficiency and tuberous root biomass (El-Sharkawy et al., 2008). These C4-related enzyme activities and C3–C4 intermediate traits significantly enhance the photosynthetic efficiency and tuberous root biomass of cultivated cassava.
The photosynthetic carbon assimilation process in C4 plants consists of two key components: carbon fixation and decarboxylation. Carbon fixation occurs in mesophyll cells, mainly facilitated by phosphoenolpyruvate carboxylase (PEPC), while decarboxylation takes place in bundle sheath cells. NADP-ME (NADP-malic enzyme, EC 1.1.1.40) plays a crucial role in C4 plants, catalyzing the conversion of malate to pyruvate and CO2 in bundle sheath cells, using NADP+ to increase CO2 concentration near Rubisco. This enhances Rubisco’s carboxylation activity while suppressing its oxygenation activity, thereby reducing photorespiration and improving carbon assimilation efficiency, a key factor in the high photosynthetic efficiency of C4 plants (Edwards and Andreo, 1992; Sage, 1999). C4-related enzymes already exist in C3 plants to confer resistance to stress conditions. In rice, the expression of NADP-ME increases under salt and drought stress, which assists in balancing levels of reactive oxygen species (ROS) and regulating malate concentrations in guard cells, thereby reducing stomatal aperture. The transfer of OsNADP-ME2 to Arabidopsis enhances its resistance to salt and osmotic stress (Chen et al., 2019). During the evolution towards C4, NADP-ME gradually shifted from its original C3 physiological roles, such as response to abiotic stress, to participating in the photosynthetic process (Tausta et al., 2002; Maier et al., 2011).
C4 enzymes were recruited into the photosynthetic process through mutations in the promoter regions, which altered the transcription factor–promoter regulatory networks. Additionally, the variation in copy number of C4 genes is a critical indicator of the evolutionary transition from C3 to C4 (Wang et al., 2009; Amy Lyu et al., 2023). Analysis of the cassava pan-genome reveals systematic Indel sites in the MeNADP-ME3 gene between ancestral and cultivated varieties, along with an increased copy number of the MeNADP-ME gene family in cultivated cassava. Immunofluorescence localization shows that the MePEPC protein accumulates in the mesophyll cells around the vascular bundles in the cultivated cassava variety Arg7, a pattern not observed in the ancestral variety W14 (Xia et al., 2023). Single-cell transcriptome studies indicate that MeNADP-ME expression is higher in the vascular tissue of the leaves of cultivated cassava variety SC8 compared to mesophyll cells (Zang et al., 2023). Therefore, based on these previous experiments, we conclude that cultivated cassava exhibits C3–C4 intermediate photosynthesis, while wild cassava is classified as a C3 type. Despite the absence of a fully developed Kranz anatomy in cultivated cassava, the differential expression of MePEPC and MeNADP-ME between mesophyll and bundle sheath cells, combined with systematic indels in the MeNADP-ME gene, may have altered the regulatory network during the evolutionary transition from wild type to cultivar. This adaptation facilitates the recruitment of MeNADP-ME in photosynthesis. These findings suggest that the MeNADP-ME gene family could be a critical factor in the evolution from wild-type C3 cassava to a C3–C4 intermediate type in cultivated varieties. Previous research on the NADP-ME family has identified its critical role in C4 evolution, with extensive studies conducted on C3–C4 intermediate model species in Flaveria and C4 plants like maize. However, systematic investigations into its role in cassava’s C3–C4 intermediate photosynthesis are still lacking. Research into the light response and regulatory patterns of MeNADP-ME in cultivated cassava will enhance our understanding of C4 evolution in the Euphorbiaceae family and provide valuable genetic resources to improve the photosynthetic efficiency of C3 plants.
In this study, we investigated the evolutionary transition of the MeNADP-ME gene family from wild to cultivated cassava and its role in enhancing photosynthetic efficiency. Using the cultivated cassava genotype AM560 as a reference, we identified four MeNADP-ME genes and analyzed their promoter regions for cis-regulatory elements. We also examined their conserved domains and constructed a phylogenetic tree. Subcellular localization was performed by transiently transforming tobacco, revealing the evolutionary features of these genes. Using qRT-PCR, we analyzed the gene expression in various tissues of cultivated cassava SC205 and wild varieties W14 and FLA4047, with a particular focus on diurnal rhythm and abiotic stress responses in the cultivated variety. Finally, a bioinformatics-based co-expression network was constructed for shaded leaves of cultivated cassava, highlighting the photosynthetic role of the MeNADP-ME3 gene family.
We used BLASTP (Johnson et al., 2008) to compare the Arabidopsis NADP-ME protein sequences with the cassava reference genome (AM560-2-JGI-v8.1). Genes with an E-value ≤ 1e−5 were selected, and their chromosomal locations were determined. The chromosomal map was created using the “Gene Location Visualize from GTF/GFF” tool in TBtools (Chen et al., 2020). The conserved protein domains of the MeNADP-ME gene family were analyzed using the MEME suite (https://meme-suite.org/meme/), setting the motif number to 15. Protein analysis, including amino acid length, isoelectric point, molecular weight, and hydrophilic coefficient, was done using the ExPASy server (https://web.expasy.org/protparam). The subcellular localization of MeNADP-ME proteins was predicted using the Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/cgi-bin/PlantmPLoc.cgi). To study cis-regulatory elements, the 3,000-bp upstream sequence of each gene was submitted to the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
This study used cultivated cassava varieties KU50, TMS60444, SC205, and SC16 and wild varieties FLA4047 and W14. The cultivated varieties are widely planted and closely related to the reference genome AM560-2. TMS60444 is commonly used in tissue culture, while SC205 is frequently selected for research. KU50 exhibits superior resistance to abiotic stress compared to SC205, and SC16, developed by our team, shows a 40% higher yield than SC205. All plants were grown in Chengmai, Hainan, China (19.85°N, 110.08°E), with samples collected from storage roots, fibrous roots, leaves, and stems during the root expansion stage. Shading transcriptome analysis was performed using SC205 and SC16, chosen for their significant yield difference, hypothesizing divergent photosynthetic efficiencies. This approach helps eliminate variability in photosynthetic gene responses and construct a co-expression network to study NADP-ME’s role in cultivated cassava’s photosynthetic physiology. Shading treatment began at 8:00 a.m., with samples collected at 8:30 a.m. and 10:00 a.m., along with control samples. For circadian rhythm analysis, TMS60444 was sampled every 2 h from 6:00 a.m. to 8:00 p.m. Heat and shade-heat stress experiments were conducted with SC205 and KU50 to explore MeNADP-MEs responses and determine if they have physiologically diverged from the original C3 wild types. Control samples were collected at 10:00 a.m., followed by heat treatment at 43°C, with samples taken after 24 h and 48 h. Each treatment had three biological replicates, and all samples were immediately stored in liquid nitrogen.
Total RNA from all tissue samples was extracted using the RNAprep Pure Kit (TianGen Biotech., Ltd., Beijing, China; DP441) and treated to remove DNA, following the manufacturer’s instructions. RNA concentration and quality were measured with a NanoDrop, and RNA integrity was confirmed by 1% agarose gel electrophoresis. Only samples with intact RNA bands and a high quality (A260/280 ratio between 2.0 and 2.2) were used for reverse transcription. High-quality RNA was combined with the 5X Evo M-MLV RT Reaction Mix Ver.2 kit (Accurate Biology., Ltd., Wuhan, China; AG11728) to synthesize single-stranded cDNA following the manufacturer’s protocol. For RNA-seq, RNA integrity was further verified using the RNA Nano 6000 kit (Agilent Technologies, Santa Clara, CA, USA; 5067–1511) on the Bioanalyzer 2100 system.
First, mRNA was enriched using mRNA Capture Beads, followed by magnetic bead purification. The mRNA was then fragmented under high-temperature conditions. The fragmented mRNA served as the template for first-strand cDNA synthesis in the reverse transcriptase reaction system. End repair and addition of an A-tail were completed during the synthesis of the second cDNA strand. Adaptors were ligated, and target fragments were selected using Hieff NGS® DNA Selection Beads (Yeasen Biotechnology Co., Ltd., Shanghai, China; 12601ES). The library was constructed by PCR amplification and sequenced on the Illumina Novaseq X Plus platform. For data processing, raw sequencing data were quality-controlled using fastp with default parameters (Chen et al., 2018). Bowtie 2 was used to align the clean reads to the ribosomal RNA database, removing reads mapped to ribosomal RNA. Unmapped reads were kept for transcriptome analysis (Langmead and Salzberg, 2012). Then, HISAT2 was used to align the paired-end reads to the cassava reference genome (AM560-2-JGI-v8.1) to generate read counts for each gene (Kim et al., 2019). Using the reference genome annotation file, the exon lengths of cassava genes were extracted, and FPKM values were calculated for each gene (Trapnell et al., 2010). The raw data for each sample were uploaded to the National Genomics Data Center, with quality control metrics and mapping rates provided in (Supplementary Table S1).
For quantitative real-time PCR analysis, cDNA samples were diluted 20-fold. The expression levels of target genes were measured on a Bio-Rad CFX96 instrument using the 2X SYBR Green Pro Taq HS Premix II kit (Accurate Biology, Ltd., Wuhan, China; AG11702), with each sample analyzed in four technical replicates. The PCR cycling program began with a 30-s denaturation at 95°C, followed by 40 cycles of 95°C for 5 s and 60°C for 30 s. Melt curve analysis and cycle threshold (Ct) values were determined using CFX software (Bio-Rad). The cassava Tubling gene was used as the reference gene for relative expression analysis, as described in previous studies (Yao et al., 2014). Primers for MeNADP-ME1, MeNADP-ME2, MeNADP-ME3, and MeNADP-ME4 were designed across exons from the CDS sequence and verified for specificity using Primer-BLAST (Ye et al., 2012). Primer sequences are provided in (Supplementary Table S2). Gene expression levels were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
We downloaded the NADP-ME amino acid sequences from Arabidopsis thaliana (TAIR10) and cassava (AM560-2-JGI-v8.1) and analyzed conserved domains using the MEME suite (https://meme-suite.org/meme/), setting the motif number to 15. To construct the phylogenetic tree, BLASTP was used to identify homologous genes (E-value ≤ 1e−5) in Arabidopsis thaliana (TAIR10), rice (MSU-v7.0), maize (MaizeGDB-Zm-B73-REFERENCE-NAM-5.0), potato (MSU-v6.1), and sorghum (JGI-v2), and their protein sequences were retrieved. Homologous NADP-ME genes and sequences from Flaveria robusta (C3), Flaveria floridana (C3–C4), and Flaveria bidentis (C4) were based on a previous study (Taniguchi et al., 2021). The phylogenetic tree was constructed using the maximum likelihood (ML) method. Multiple sequence alignment was done using the Muscle tool in MEGA v. 11 (Tamura et al., 2021), followed by model selection with default parameters and 1,000 bootstrap replicates. The phylogenetic tree was visualized using iTOL (Letunic and Bork, 2024).
To construct the pG1300-MeNADP-MEs-GFP vector, we used the pCAMBIA1300-GFP vector. Primers were designed to amplify the CDS of each MeNADP-ME family member from the cDNA of mature leaves of the cassava cultivar SC205. CDS cloning was performed using 2X ApexHF CL PCR Master Mix (Accurate Biology, Ltd., Wuhan, China; AG12209). The pG1300-GFP vector was digested with KpnI-HF® and SalI-HF® (New England Biolabs). Both the cloned CDS and the digested vector were purified using 1% agarose gel electrophoresis. The ClonExpress Ultra One Step Cloning Kit V2 (Vazyme, Nanjing, China; C116) was used for homologous recombination, ligating the CDS fragments into the vector. The recombinant vectors were transformed into Agrobacterium tumefaciens strain GV3101. After reaching an OD600 = 0.9, the bacterial cells were resuspended in MES buffer (10 mM MgCl2, 10 mM MES, 100 µM AS) to OD600 = 9.0 and injected into the abaxial side of tobacco leaves. After 48 h in the dark, 1-cm2 sections of the transformed leaves were sampled for imaging on a FV3000 confocal microscope (Olympus, Japan), capturing bright field, chloroplast fluorescence (680 nm), and GFP fluorescence (488 nm) images, which were merged using FV31S-DT software (Olympus, Japan).
Genes with the top 7,000 average FPKM values were selected and normalized using log2(FPKM+1). Sample clustering and soft threshold analysis were performed (Supplementary Figure S1). A soft threshold of 18 was chosen to construct the weighted gene co-expression network analysis (WGCNA) co-expression network using the R package version of WGCNA (Langfelder and Horvath, 2008). Photosynthesis-related gene sets were created by extracting cassava genes involved in light-harvesting antennae (KEGG pathway ID: map00196), photosynthetic electron transport (KEGG pathway ID: map00195), and carbon fixation (including the Calvin–Benson cycle and C4 genes; KEGG pathway ID: map00710) from the KEGG pathway database. The number of photosynthesis-related genes in each module was calculated, and the module with the highest number of photosynthetic genes was selected for further analysis. KEGG enrichment analysis was performed on all genes in this module (Supplementary Figure S2). Using the Plant TF Motifs Shift tool in TBtools, protein sequences in the module (AM560-2 JGI-v8.1) were analyzed with Arabidopsis thaliana as a reference to identify transcription factor binding motifs. The 3,000-bp upstream promoter regions of photosynthesis-related genes were extracted and scanned using the Fimo: Binding Motif Scan tool in TBtools to generate a predicted transcription factor–promoter interaction network. The network was visualized using Cytoscape 3.7.2 software.
Each qPCR sample was analyzed in four technical replicates. Tukey HSD significance analysis of the qPCR results was conducted using SPSS software (IBM Corp., Armonk, NY, USA; version 8.0). Differences were indicated by letter labeling, where different letters represent significant differences (p < 0.05). Figures were created using GraphPad Prism software (GraphPad Software, San Diego, CA, USA; version 8.0).
A BLASTP search using Arabidopsis NADP-ME (TAIR10) as a query against the cultivated cassava AM560-2 dataset (Phytozome 13, E-value ≤ 1e−5) identified four distinct members of the MeNADP-ME family: MeNADP-ME1, MeNADP-ME2, MeNADP-ME3, and MeNADP-ME4, located on chromosomes 16, 16, 11, and 4, respectively (Figure 1A). Physicochemical analysis (Table 1) revealed that MeNADP-ME1 has the fewest amino acids, lowest molecular weight, and smallest isoelectric point, while MeNADP-ME2 exhibited the highest values for these properties. MeNADP-ME3 and MeNADP-ME4 had intermediate characteristics. Subcellular localization predictions indicated chloroplast localization for all four isoforms. Promoter analysis (Figure 1B, Supplementary Table S3) identified numerous light-responsive cis-regulatory elements, particularly in MeNADP-ME1 and MeNADP-ME3, suggesting their involvement in photosynthetic regulation. Additionally, these genes contain hormone- and stress-responsive elements, although light-responsive elements dominate their regulatory mechanisms. These findings suggest that the MeNADP-ME family plays a role in the photosynthetic physiology of cultivated cassava.
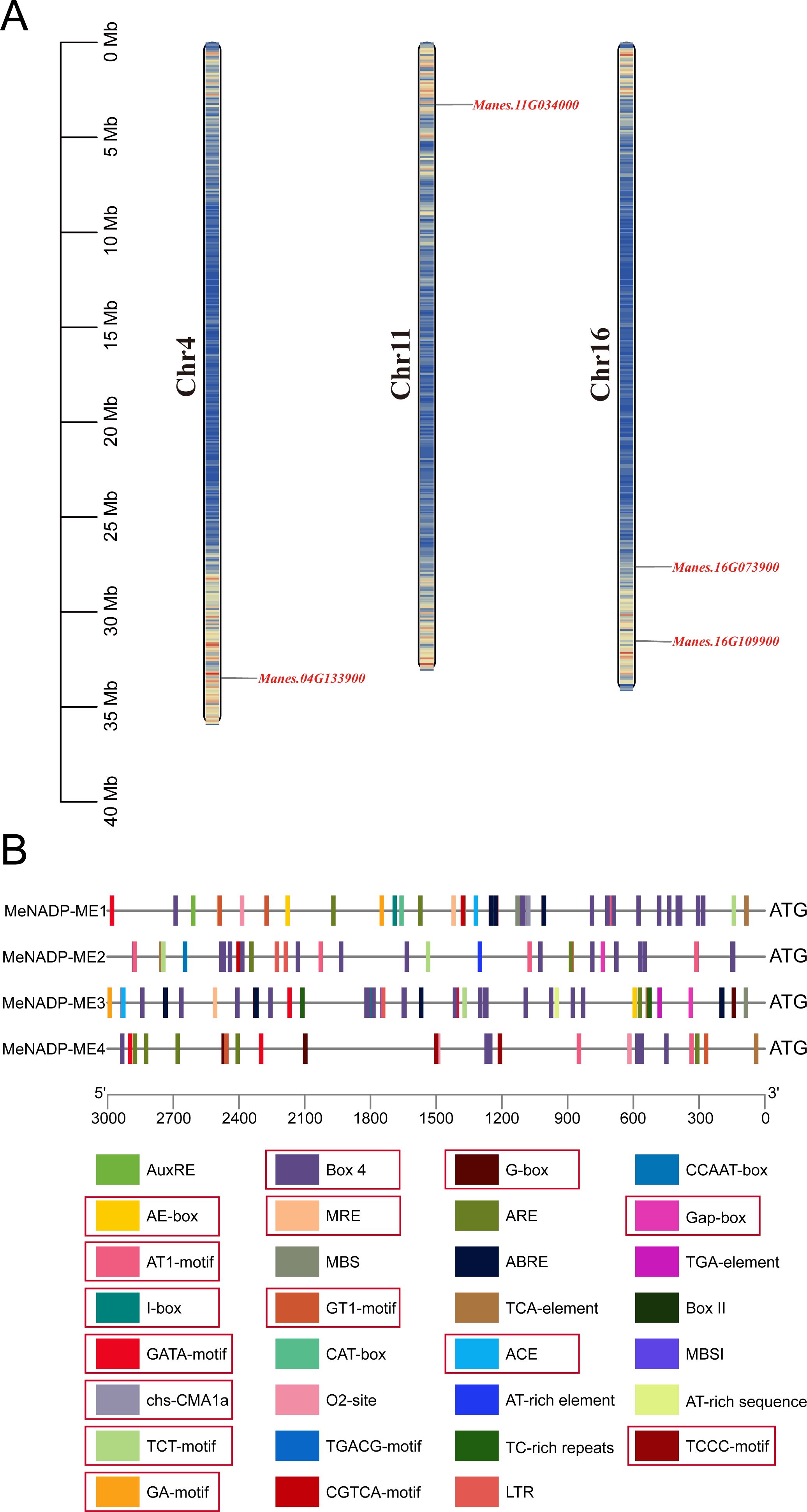
Figure 1. Chromosomal localization (A) and upstream cis-acting elements of the NADP-ME gene family in cassava (B), the 5′ to 3′ direction represents the orientation of the promoter sequence, and the red box indicates the photosynthesis-related cis-regulatory elements.
We conducted a MEME analysis to investigate conserved motifs in the NADP-ME proteins of cassava and Arabidopsis (Figure 2A). The results showed that the primary structural differences between the two species’ NADP-ME proteins occur mainly in the N-terminal region. Specifically, MeNADP-ME2, MeNADP-ME1, and MeNADP-ME4 shared high homology with similar Arabidopsis subtypes, while MeNADP-ME3 exhibited a unique N-terminal domain, lacking Motif15 compared to MeNADP-ME4 and AtNADP-ME4. We also constructed a phylogenetic tree with NADP-ME genes from species such as maize (C4), Arabidopsis (C3), Flaveria species, potato (C3), rice (C3), sorghum (C4), and cassava (C3–C4) (Figure 2B). The analysis revealed four distinct subgroups: monocot, both monocot and dicot, cytosolic dicotyledonous, and plastidic dicotyledonous types. The phylogenetic distances correlated with both evolutionary relationships and subcellular localization. All MeNADP-ME members were classified into the plastidic dicotyledonous subgroup, consistent with localization predictions. Notably, cassava NADP-ME genes were most closely related to AtNADP-ME4, providing insights into the functional adaptation of these genes across species.

Figure 2. The conserved structure of NADP-ME was analyzed using NADP-ME protein sequences from Arabidopsis and cassava (A); phylogenetic tree was constructed based on NADP-ME sequences from maize (C4), Arabidopsis (C3), Flaveria robusta (C3), Flaveria floridana (C3–C4), Flaveria bidentis (C4), potato (C3), rice (C3), sorghum (C4), and cassava (C3–C4) species, illustrating the evolutionary relationships and functional diversification of NADP-ME (B).
To investigate the subcellular localization of the MeNADP-ME gene family in cultivated cassava, we cloned the CDS regions of the four identified members from the AM560-2 reference genome, using TMS60444 as a template, and ligated them into a GFP vector driven by the CaMV 35S promoter. Transient expression in tobacco leaves (Figure 3A) revealed chloroplast localization for all MeNADP-ME members, consistent with phylogenetic and predictive analyses. Expression patterns in various tissues of cassava cultivar SC205 and wild species A4047 (Figure 3B) showed that in SC205, MeNADP-ME1 was most expressed in mature leaves, MeNADP-ME2 in young leaves, MeNADP-ME3 in mature leaves, and MeNADP-ME4 in young leaves and stems. In A4047, MeNADP-ME1 was highly expressed in mature leaves and tuberous roots, while MeNADP-ME2 and MeNADP-ME3 were highest in mature leaves. MeNADP-ME4 exhibited significantly higher expression in tuberous roots. These results highlight significant differences in the expression patterns of MeNADP-ME family members between cultivated and wild cassava, suggesting that the family has been selected to enhance leaf photosynthetic function in cultivated cassava, reflecting C3–C4 intermediate photosynthesis evolution.
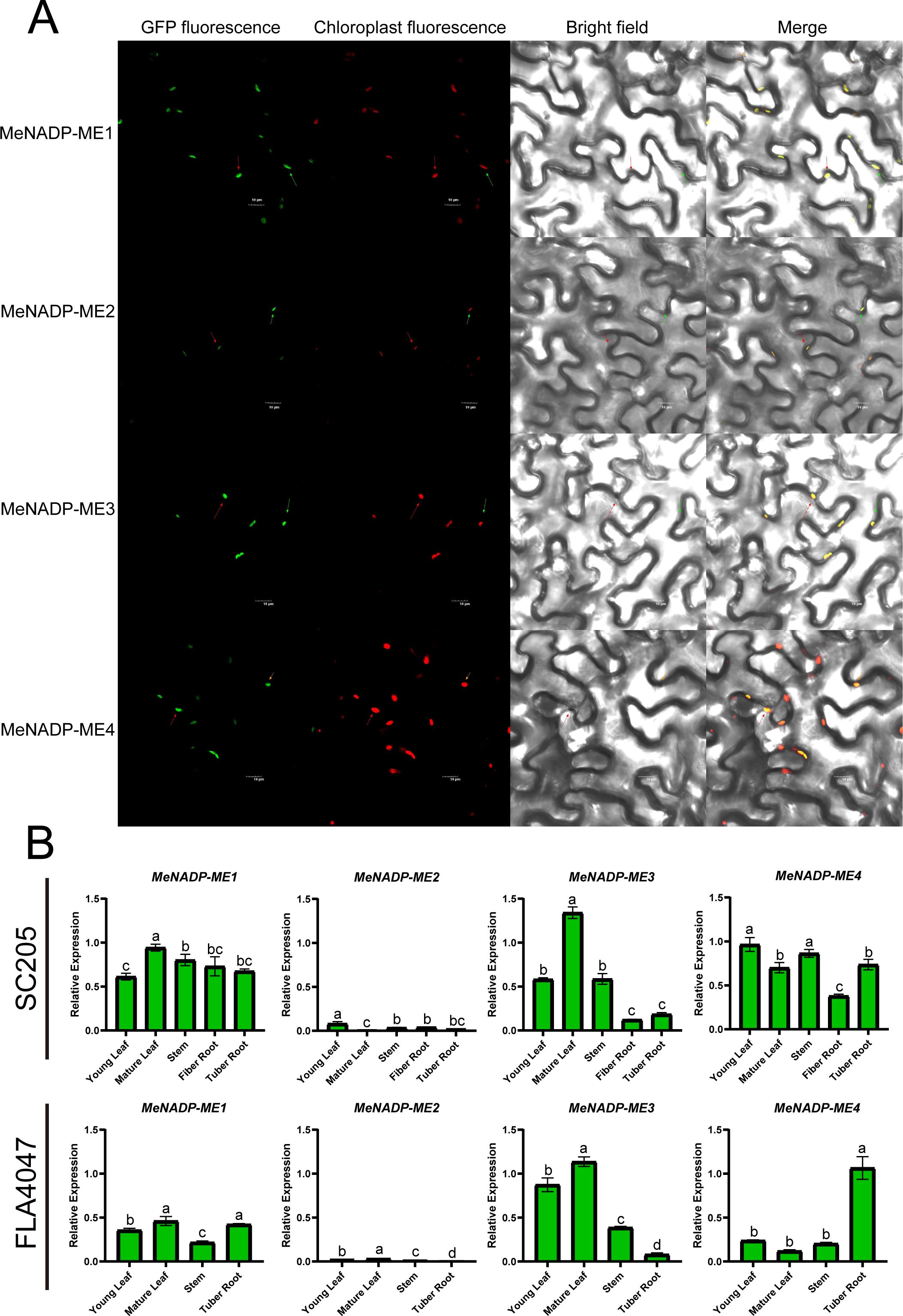
Figure 3. Subcellular localization of MeNADP-ME family (scale bar = 10μm) (A); expression level changes of MeNADP-ME family in cassava cultivars SC205 and wild FLA4047 species (B).
To investigate the diurnal response of the MeNADP-ME gene family in cultivated cassava and identify key members involved in photosynthetic activity, we analyzed their expression dynamics at different time points in the cultivar TMS60444 (Figure 4A). The expression patterns of MeNADP-ME1 and MeNADP-ME2 were similar, showing low levels in the morning, increasing after noon, and peaking at night. In contrast, MeNADP-ME3 and MeNADP-ME4 exhibited a strong positive correlation with light intensity, particularly between 6:00 and 8:00 a.m., when light intensity sharply increased, leading to a peak in expression. After 10:00 a.m., expression gradually decreased, with MeNADP-ME4 showing a second peak at 20:00. Given the involvement of NADP-ME genes in C3 species’ response to abiotic stress, we further examined the family’s response to heat and combined heat and shading stress in cassava cultivars KU50 and SC205 (Figure 4B). In KU50, gene expression was generally upregulated at PG24h, with MeNADP-ME2 significantly upregulated at PG48h, while other genes were downregulated. Under shade-heat stress, all genes, except MeNADP-ME4, were downregulated. In SC205, MeNADP-ME1, MeNADP-ME2, and MeNADP-ME3 peaked at PG24h, significantly decreasing by PG48h. Under shade-heat stress, MeNADP-ME2 was upregulated at SPG24h. In conclusion, MeNADP-ME3 and MeNADP-ME4 were identified as key contributors to photosynthesis in response to increased light intensity. Heat stress induced upregulation of MeNADP-ME genes, peaking at 24 h, while no significant changes occurred at 48 h or under combined heat and shading stress. These findings suggest that the MeNADP-ME gene family in cultivated cassava reflects an evolutionary trend toward C4 photosynthesis while retaining characteristics of its C3 ancestral species, underscoring the unique physiological features of its intermediate C3–C4 MeNADP-ME genes.
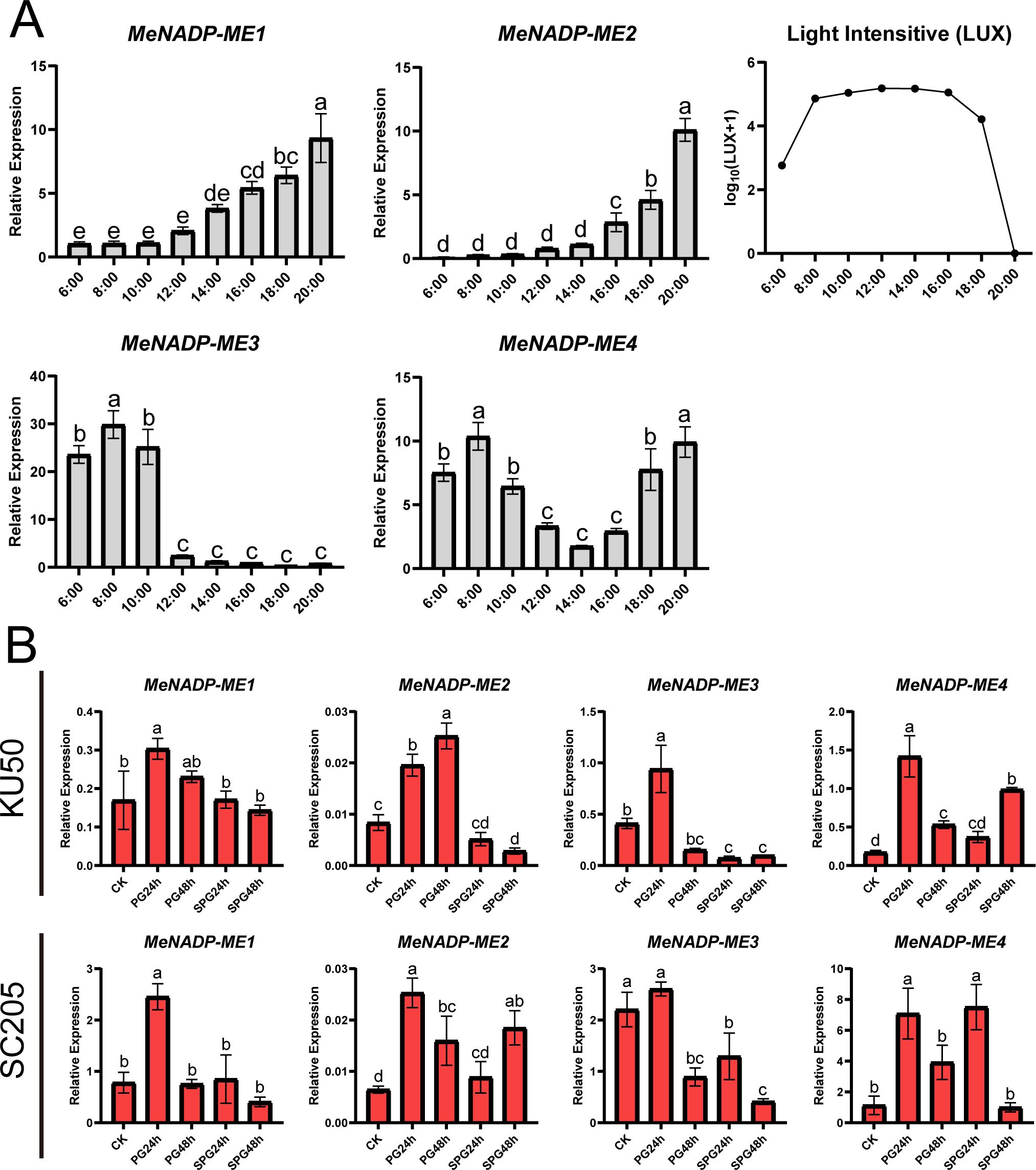
Figure 4. Diurnal expression of cassava cultivars TMS60444 MeNADP-ME (6:00–20:00) and light intensity changes (LUX) (A); MeNADP-ME expression in cassava cultivars KU50 and SC205 under heat and heat-shading treatments (B). (B) PG24h, heat treatment for 24 h; PG48h, heat treatment for 48 h; SPG24h, shade heat treatment for 24 h; SPG48h, shade heat treatment for 48 h. CK represents the control group, which corresponds to the untreated condition.
We conducted co-expression network analysis of the transcriptome under shading treatment in the cultivated cassava varieties SC16 and SC205, identifying 18 modules with distinct expression patterns. Notably, the Turquoise module, which included all members of the MeNADP-ME gene family, was enriched with photosynthesis-related and C4 photosynthesis genes (Figures 5A, B). KEGG analysis revealed significant enrichment in the “carbon fixation in photosynthetic organisms” pathway (Supplementary Figure S2). Under shading conditions, cassava C4 genes, photosynthetic electron transport chain genes, and photosynthetic carbon assimilation genes, including the MeNADP-ME family, were regulated by transcription factors such as GATA1 (Manes.03G154500), YABBY1 (Manes.02G035700), Myb-like (Manes.15G163100), HY5 (Manes.12G040300), and MADS (Manes.10G099000) (Figure 5C). Heatmap analysis further showed that MeNADP-ME3 exhibited an expression pattern consistent with most photosynthesis-related genes and their transcriptional regulators (Figure 6D), while MeNADP-ME1, MeNADP-ME2, and MeNADP-ME4 exhibited opposite trends in SC16. This highlights MeNADP-ME3 as a key gene in the intermediate C3–C4 photosynthetic physiology of cultivated cassava. Transcription factor-promoter prediction suggested that MeYABBY1 regulates MeNADP-ME3 and binds to its systemic promoter Indel site, potentially playing a critical role in recruiting MeNADP-ME3 into the C3–C4 photosynthetic regulatory network (Figures 6A–C).
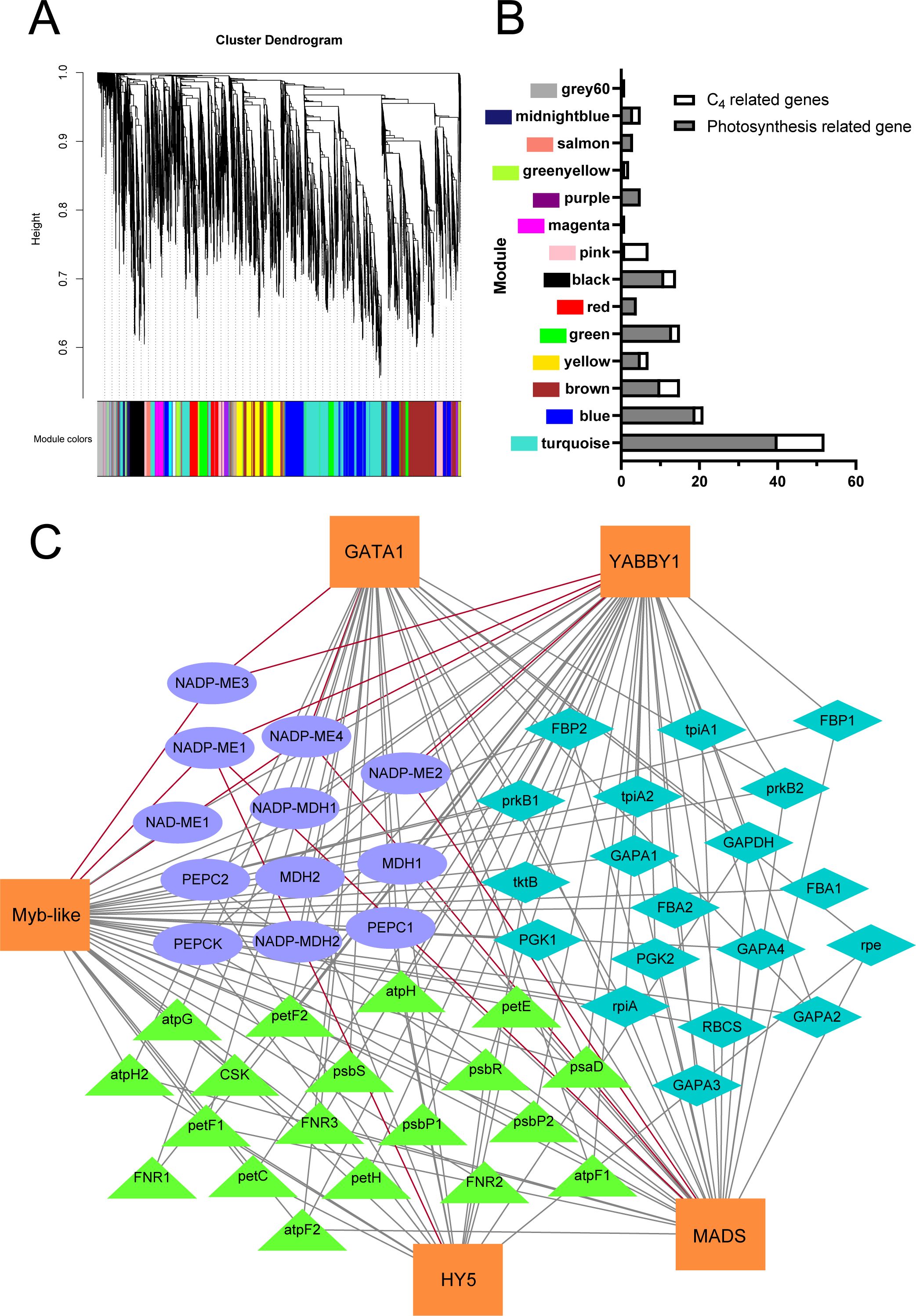
Figure 5. Co-expression network of transcriptomes under shading treatment in cassava cultivars SC205 and SC16 (A); number of photosynthesis-related genes in each module (B); transcription factor–gene interaction network of the Turquoise module (C), where ellipse shapes represent C4 genes in cassava, diamond shapes represent genes related to the Calvin–Benson cycle, and triangle shapes represent genes related to the photosynthetic electron transport chain. The red lines represent the interaction between the promoter regions of the MeNADP-ME family and transcription factors.
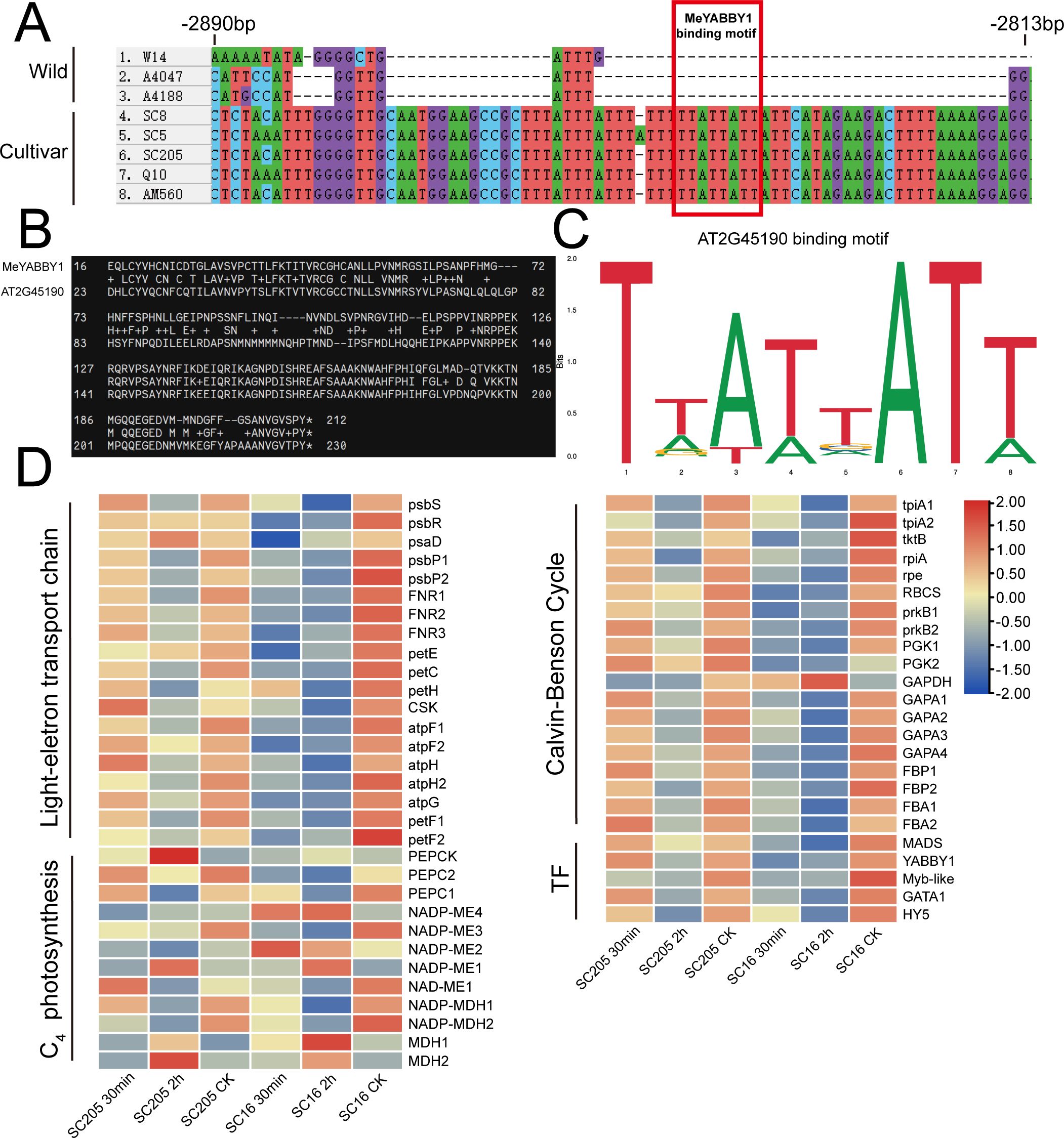
Figure 6. Predicted binding sites of MeYABBY1 on the Indel region, with the red rectangle indicating the MeYABBY1 binding motif (A); blast alignment of MeYABBY1 with the most homologous transcription factor in Arabidopsis, AtYABBY1 (AT2G45190) (B); the promoter motif sequences interacting with AtYABBY1 in the Jaspar transcription factor database (C); heatmap of expression levels of photosynthetic electron transport chain, C4, Calvin–Benson cycle, and predicted interacting transcription factors (TF) in the Turquoise module under shading treatment (D). CK represents the control group, which corresponds to the untreated condition.
NADP-ME is widely distributed in both monocot and dicot plants. Previous research has categorized it into four subgroups: monocot, both monocot and dicot, cytosolic dicotyledonous, and plastidic dicotyledonous types (Wheeler et al., 2005; Alvarez et al., 2013). The Asteraceae family, belonging to dicots, includes species from the genus Flaveria, which encompasses C3, C3–C4 intermediate, C4-like, and C4 species. These species are considered an important model for studying the evolution of C4 photosynthesis, highlighting the dynamic process of C4 evolution (Adachi et al., 2022). To explore whether MeNADP-MEs protein shares similarities with the C3–C4 intermediate type of Flaveria, we included NADP-ME family members from Flaveria species representing C3, C3–C4, and C4 types in our phylogenetic analysis. The results indicated that the evolutionary relationships of NADP-ME are more closely aligned with species phylogeny and protein subcellular localization. According to the APG IV classification (Group et al., 2016), the Euphorbiaceae family, to which cassava belongs, is most closely related to the Brassicaceae family, while it is more distantly related to the Solanaceae and Asteraceae families. This evolutionary distance is reflected in the closer phylogenetic relationship between the cassava and Arabidopsis NADP-ME families, while the cassava NADP-ME family is more distantly related to members of Flaveria in the Asteraceae family and to those in the Solanaceae family, such as potato. The monocots, including rice, maize, and sorghum, show significant differences from the cassava NADP-ME family (Figure 2B). MeNADP-ME in cassava is most closely related to AtNADP-ME4 in Arabidopsis, suggesting that they may share similar physiological functions in the chloroplast. Notably, in the conserved domain analysis, MeNADP-ME2 lacks two conserved domains compared to the other cassava members and is more distantly related to AtNADP-ME2 and AtNADP-ME3 (Figure 2A). In contrast, MeNADP-ME3 lacks Motif14, and no single Arabidopsis protein contains only Motif15 while lacking Motif14. This suggests that these two genes are unique to cassava. These structural differences are linked to the expansion of gene families during cassava’s evolutionary development towards C4 photosynthesis.
Previous studies suggest that NADP-ME in C4 plants evolved from the chloroplast-localized NADP-ME in their C3 ancestors, with these C3 chloroplast NADP-MEs originated from cytosolic NADP-MEs that did not participate in photosynthesis (Tausta et al., 2002; Maier et al., 2011). Furthermore, it has been proposed that as more NADP-ME members localize to the chloroplast, the evolutionary trend towards C4 photosynthesis becomes more pronounced. In particular, in C4 plants, NADP-ME plays a crucial role in bundle sheath cells by facilitating the decarboxylation reaction. This adaptation likely originated from the chloroplast-localized NADP-ME in C3 plants, which was subsequently selected and optimized for the chloroplast environment of C4 photosynthesis. As C4 photosynthesis evolved, the elevated expression of NADP-ME in the chloroplast became a hallmark of its role in driving C4 physiological functions (Langdale, 2011; Rao and Dixon, 2016; Shi et al., 2020). Phylogenetic analysis reveals that maize contains five NADP-ME members, two of which are localized to the chloroplasts. In contrast, all NADP-ME members in cassava are chloroplast-localized, with no cytosolic NADP-ME present, a feature unique to cassava (Figure 3A). This suggests that cassava has evolved a greater number of NADP-ME subtypes localized to the chloroplast, involved in physiological processes within the chloroplast. Additionally, a comparison of gene expression in cultivated and wild cassava species across different tissues (Figure 3B) shows that, compared to the wild-type A4047, the cultivated variety SC205 exhibits a greater involvement of MeNADP-ME in leaf physiological processes, while the wild type shows a more physiologically active subtype in underground tissues. These spatial expression patterns, observed in both subcellular and tissue-specific locations, provide strong evidence for the evolutionary shift of cultivated cassava from a C3 wild type to a C3–C4 intermediate type.
We also investigated the response of the MeNADP-ME gene family in cultivated cassava under diurnal rhythms and abiotic stress conditions (Figures 4A, B). Diurnal rhythm analysis indicated that MeNADP-ME1 and MeNADP-ME2 share similar physiological functions, predominantly operating during the night. In contrast, MeNADP-ME3 and MeNADP-ME4 are key genes involved in the response to changes in light intensity. Additionally, MeNADP-ME4 exhibits high expression at night. We hypothesize that the ancestor of MeNADP-ME was a gene expressed during the night, and through selective pressure, it gradually began to function during the photosynthetic period. MeNADP-ME4 shows clear signs of this selective process. Promoter cis-element analysis revealed a high proportion of light-responsive regulatory elements, and several stress-responsive elements, in these four genes (Figure 1B). Although both MeNADP-ME1 and MeNADP-ME3 contain over 20 light-responsive elements, the transcriptional levels of MeNADP-ME1 do not fully align with the light cycle, suggesting that it is primarily regulated by factors beyond light response. In contrast, MeNADP-ME3 is tightly correlated with light intensity, indicating that it functions as a core gene in the C3–C4 intermediate photosynthetic decarboxylation process in cassava. Our study of the expression patterns of the MeNADP-ME gene family in cultivated cassava under abiotic stress conditions reveals that it retains the physiological functions of its C3 wild relatives, consistent with the role of NADP-ME in stress responses in C3 plants. Previous research has shown that NADP-ME plays a critical role in stress responses by regulating cellular osmotic potential (Chen et al., 2019). Combined with the subcellular localization results, NADP-ME in cultivated cassava is likely involved in physiological processes that maintain chloroplast stability, particularly under abiotic stress conditions. Early studies have shown that during the evolution of C4 species, C4 genes were recruited into the photosynthetic process through structural variations in the promoter regions, facilitated by transcription factor–gene networks (Monson, 1999, 2003; Schlüter and Weber, 2020). The promoter region of MeNADP-ME3 exhibits such structural variation, with an insertion mutation occurring during the evolutionary process from the wild type to the cultivated species, altering the transcriptional regulation pattern. First, its expression is significantly higher in the cultivated species’ leaves compared to other tissues, a feature not present in the wild species. Second, its expression increases only in response to increased light intensity, a feature not shared by other subtypes. Finally, it does not show a sustained response under prolonged heat stress. These characteristics suggest that MeNADP-ME3 is gradually diverging from the physiological functions of its homologous C3 subtype, transitioning towards a role in photosynthesis.
In the constructed co-expression network of the cassava shading transcriptome (Figures 5A, B), MeNADP-ME3 further exhibits distinct features. It is not only closely correlated with the expression trends of other photosynthetic genes within the module but also aligns with changes in transcription factor expression patterns (Figure 6D). Through the prediction of the transcription factor–promoter interaction network, we found that MeYABBY1 interacts with the systematic indel sites of MeNADP-ME3 (Figures 5C, 6A–C). Although members of the YABBY transcription factor family are typically associated with leaf development (She et al., 2022; Shen et al., 2022; Shi et al., 2024), some studies suggest that overexpressing IaYABBY2 can enhance photosynthetic capacity in Incarvillea arguta (Sun et al., 2014), supporting the potential role of YABBY family members in the regulation of photosynthesis. This suggests that MeYABBY1 may play a role in the regulation of photosynthesis in cultivated cassava. We predict that MeYABBY1 interacts with multiple photosynthesis-related genes and other genes involved in C4 photosynthetic processes, such as MePEPC1, indicating that it is one of the core regulators in the C3–C4 transitional regulatory network of cultivated cassava. The co-regulation and similar expression patterns of MeNADP-ME3 with MePEPC1 and other photosynthesis-related genes further suggest that MeNADP-ME3, together with MePEPC1, contributes to the physiological process of C4 carbon fixation in cultivated cassava. In the shaded transcriptome, other MeNADP-ME subtypes are more involved in the physiological processes following shading.
Overall, this study found that during the evolutionary process from wild to cultivated varieties, the promoter region of MeNADP-ME3 in cassava underwent selection, being recruited by transcription factors such as MeYABBY1, which altered its expression pattern and formed a transcription factor–gene regulatory network with other photosynthetic genes, thus participating in photosynthesis. Another interesting finding is that all NADP-ME gene members in cultivated cassava are localized in the chloroplasts, which is rare in other species. However, other MeNADP-ME subtypes in cultivated cassava have not yet evolved to respond to light and participate in photosynthesis, but subcellular localization indicates that they are located in the chloroplasts, showing a trend towards being recruited into photosynthesis. Given that cassava is a tropical plant, its adaptation to tropical environments may significantly influence its domestication process.
All the raw transcriptome sequencing data generated from the leaf shading treatment during this study have been deposited at the National Genomics Data Center (https://ngdc.cncb.ac.cn/) as a BioProject under accession number PRJCA032135 (Shade treatment of cultivar Cassava SC16, SC205). The sequencing reads are available in the GSA database under the same BioProject number.
HL: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. FG: Writing – review & editing. XS: Writing – review & editing. JL: Writing – review & editing. SFW: Writing – review & editing. JX: Writing – review & editing. JC: Writing – review & editing. LX: Data curation, Writing – review & editing. XG: Writing – review & editing. SJW: Writing – review & editing. HW: Writing – review & editing. WW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The research was funded by the startup funds for the double first-class disciplines of crop science at Hainan University (RZ2100003362) and the Central Public-interest Scientific Institution Basal Research Fund for the Chinese Academy of Tropical Agricultural Sciences (1630052022008), both of which made this research possible.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1525193/full#supplementary-material
Adachi, S., Stata, M., Martin, D. G., Cheng, S., Liu, H., Zhu, X.-G., et al. (2022). The evolution of C4 photosynthesis in flaveria (Asteraceae): insights from the Flaveria linearis complex. Plant Physiol. 191, 233–251. doi: 10.1093/plphys/kiac467
Allem, A. C. (1999). The closest wild relatives of cassava (Manihot esculenta Crantz). Euphytica 107, 123–133. doi: 10.1023/A:1026422229054
Alvarez, C. E., Saigo, M., Margarit, E., Andreo, C. S., Drincovich, M. F. (2013). Kinetics and functional diversity among the five members of the NADP-malic enzyme family from Zea mays, a C4 species. Photosynth. Res. 115, 65–80. doi: 10.1007/s11120-013-9839-9
Amy Lyu, M. J., Tang, Q., Wang, Y., Essemine, J., Chen, F., Ni, X., et al. (2023). Evolution of gene regulatory network of C4 photosynthesis in the genus Flaveria reveals the evolutionary status of C3-C4 intermediate species. Plant Commun. 4, 100426. doi: 10.1016/j.xplc.2022.100426
An, F., Chen, T., Stéphanie, D. M. A., Li, K., Li, Q. X., Carvalho, L. J. C. B., et al. (2016). Domestication syndrome is investigated by proteomic analysis between cultivated cassava (Manihot esculenta Crantz) and its wild relatives. PloS One 11, e0152154. doi: 10.1371/journal.pone.0152154
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, Q., Wang, B., Ding, H., Zhang, J., Li, S. (2019). Review: The role of NADP-malic enzyme in plants under stress. Plant Sci. 281, 206–212. doi: 10.1016/j.plantsci.2019.01.010
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cock, J. H., Riaño, N. M., El-Sharkawy, M. A., Yamel, L. F., Bastidas, G. (1987). C3-C4 intermediate photosynthetic characteristics of cassava (Manihot esculenta Crantz). Photosynth. Res. 12, 237–241. doi: 10.1007/BF00055123
Edwards, G. E., Andreo, C. S. (1992). NADP-malic enzyme from plants. Phytochemistry 31, 1845–1857. doi: 10.1016/0031-9422(92)80322-6
El-Sharkawy, M. A. (2004). Cassava biology and physiology. Plant Mol. Biol. 56, 481–501. doi: 10.1007/s11103-005-2270-7
El-Sharkawy, M. A., de Tafur, S. M., Lopez, Y. (2012). Eco-physiological research for breeding improved cassava cultivars in favorable and stressful environments in tropical/subtropical bio-systems. Environ. Res. J. 6, 143–211.
El-Sharkawy, M. A., Lopez, Y., Bernal, L. M. (2008). Genotypic variations in activities of phosphoenolpyruvate carboxylase and correlations with leaf photosynthetic characteristics and crop productivity of cassava grown in low-land seasonally-dry tropics. Photosynthetica 46, 238–247. doi: 10.1007/s11099-008-0038-4
Group, T. A. P., Chase, M. W., Christenhusz, M. J. M., Fay, M. F., Byng, J. W., Judd, W. S., et al. (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Botanical J. Linn. Soc. 181, 1–20. doi: 10.1111/boj.12385
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Kim, D., Paggi, J. M., Park, C., Bennett, C., Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Langdale, J. A. (2011). C4 cycles: past, present, and future research on C4 photosynthesis. Plant Cell 23, 3879–3892. doi: 10.1105/tpc.111.092098
Langfelder, P., Horvath, S. (2008). WGCNA: an R package for weighted correlation network analysis. BMC Bioinf. 9, 559. doi: 10.1186/1471-2105-9-559
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Letunic, I., Bork, P. (2024). Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52, W78–W82. doi: 10.1093/nar/gkae268
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Luiz Joaquim Castelo Branco, C., James, V. A., Songbi, C., Chikelu, M., Münevver, D. (2017). “Domestication syndrome in cassava (Manihot esculenta Crantz): assessing morphological traits and differentially expressed genes associated with genetic diversity of storage root,” in Cassava. Ed. Viduranga, W. (IntechOpen, Rijeka). Ch. 6.
Maier, A., Zell, M. B., Maurino, V. G. (2011). Malate decarboxylases: evolution and roles of NADP-ME isoforms in species performing C4 and C3 photosynthesis. J. Exp. Bot. 62, 3061–3069. doi: 10.1093/jxb/err024
Monson, R. K. (1999). The origins of C4 genes and evolutionary pattern in the C4 metabolic phenotype. C4 Plant Biol. 377–410. doi: 10.1016/B978-012614440-6/50012-4
Monson, R. K. (2003). Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. Int. J. Plant Sci. 164, S43–S54. doi: 10.1086/368400
Rao, X., Dixon, R. A. (2016). The differences between NAD-ME and NADP-ME subtypes of C4 photosynthesis: more than decarboxylating enzymes. Front. Plant Sci. 7, 1525. doi: 10.3389/fpls.2016.01525
Schlüter, U., Weber, A. P. (2020). Regulation and evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 71, 183–215.
She, Z., Huang, X., Aslam, M., Wang, L., Yan, M., Qin, R., et al. (2022). Expression characterization and cross-species complementation uncover the functional conservation of YABBY genes for leaf abaxial polarity and carpel polarity establishment in Saccharum spontaneum. BMC Plant Biol. 22, 124. doi: 10.1186/s12870-022-03501-3
Shen, Y., Li, X., Ma, G., Zhao, Y., Jiang, X., Gao, L., et al. (2022). Roles of YABBY transcription factors in the regulation of leaf development and abiotic stress responses in Camellia sinensis. Beverage Plant Res. 2, 1–10. doi: 10.48130/BPR-2022-0004
Shi, W., Yue, L., Guo, J., Wang, J., Yuan, X., Dong, S., et al. (2020). Identification and evolution of C4 photosynthetic pathway genes in plants. BMC Plant Biol. 20, 132. doi: 10.1186/s12870-020-02339-x
Shi, T., Zhou, L., Ye, Y., Yang, X., Wang, L., Yue, Y. (2024). Characterization of YABBY transcription factors in Osmanthus fragrans and functional analysis of OfYABBY12 in floral scent formation and leaf morphology. BMC Plant Biol. 24, 589. doi: 10.1186/s12870-024-05047-y
Sun, X., Guan, Y., Hu, X. (2014). Isolation and characterization of IaYABBY2 gene from Incarvillea arguta. Plant Mol. Biol. Rep. 32, 1219–1227. doi: 10.1007/s11105-014-0725-1
Svensson, P., Bläsing, O. E., Westhoff, P. (2003). Evolution of C4 phosphoenolpyruvate carboxylase. Arch. Biochem. Biophys. 414, 180–188. doi: 10.1016/S0003-9861(03)00165-6
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Taniguchi, Y. Y., Gowik, U., Kinoshita, Y., Kishizaki, R., Ono, N., Yokota, A., et al. (2021). Dynamic changes of genome sizes and gradual gain of cell-specific distribution of C4 enzymes during C4 evolution in genus Flaveria. Plant Genome 14, e20095. doi: 10.1002/tpg2.20095
Tausta, S. L., Miller Coyle, H., Rothermel, B., Stiefel, V., Nelson, T. (2002). Maize C4 and non-C4 NADP-dependent Malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Mol. Biol. 50, 635–652. doi: 10.1023/A:1019998905615
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Wang, X., Gowik, U., Tang, H., Bowers, J. E., Westhoff, P., Paterson, A. H. (2009). Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 10, R68. doi: 10.1186/gb-2009-10-6-r68
Wheeler, M. C. G., Tronconi, M. A., Drincovich, M., Andreo, C. S., gge, U.-I., Maurino, V. (2005). A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol. 139, 39–51. doi: 10.1104/pp.105.065953
Xia, Z., Du, Z., Zhou, X., Jiang, S., Zhu, T., Wang, L., et al. (2023). Pan-genome and Haplotype Map of Cultivars and Their Wild Ancestors Provides Insights into Selective Evolution of Cassava (Manihot esculenta Crantz). bioRxiv. doi: 10.1101/2023.07.02.546475
Yao, Y., Geng, M.-T., Wu, X.-H., Liu, J., Li, R.-M., Hu, X.-W., et al. (2014). Genome-wide identification, 3D modeling, expression and enzymatic activity analysis of cell wall invertase gene family from cassava (Manihot esculenta Crantz). Int. J. Mol. Sci. 15, 7313–7331. doi: 10.3390/ijms15057313
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., Madden, T. L. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 134. doi: 10.1186/1471-2105-13-134
Keywords: C3–C4 intermediate photosynthesis, NADP-ME, cassava, C4 evolution, coexpression network
Citation: Li H, Xiao J, Chen J, Shen X, Luo J, Guo F, Wang S, Xu L, Guo X, Wang S, Wang H and Wang W (2025) Identification of the cassava NADP-ME gene family and its response and regulation in photosynthesis. Front. Plant Sci. 16:1525193. doi: 10.3389/fpls.2025.1525193
Received: 08 November 2024; Accepted: 28 January 2025;
Published: 27 February 2025.
Edited by:
Manjusha Verma, National Bureau of Plant Genetic Resources (ICAR), IndiaReviewed by:
Nunzia Scotti, National Research Council (CNR), ItalyCopyright © 2025 Li, Xiao, Chen, Shen, Luo, Guo, Wang, Xu, Guo, Wang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyan Wang, d2FuZ2hhaXlhbkBpdGJiLm9yZy5jbg==; Wenquan Wang, OTk0MzQxQGhhaW5hbnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.