
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 February 2025
Sec. Plant Systematics and Evolution
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1518122
Due to significant morphological differences and extensive interspecific hybridization, there are numerous species complexes with taxonomic challenges in the genus Populus. Integrative taxonomy, which combines evidence of morphology, molecular phylogeny, niche differentiation, and reproductive isolation, provides the most effective approaches for species delimitation. The Populus laurifolia complex, which belongs to Populus subg. Tacamahaca (Salicaceae), is distributed in the Altai Mountains and Tianshan Mountains. This complex exhibits morphological variability, making species delimitation challenging. Due to limited sampling and systematic studies, its taxonomy has remained unresolved. In this study, 337 specimens, along with online digital samples representing nearly all wild populations, were collected. Morphological analyses were performed to evaluate key traits and clarify species boundaries. Phylogenetic relationships were reconstructed using concatenation and coalescent methods based on 566,375 nuclear single-nucleotide polymorphisms (SNPs). Ecological niche differentiation was assessed, and ABBA–BABA analysis was used to examine interspecific hybridization. The results revealed that this complex, based on a series of significant character states, could be morphologically distinguished into three species—P. laurifolia (Populus pilosa considered a synonym of P. laurifolia), Populus talassica, and Populus pamirica—which also correspond to three well-supported clades in the phylogenetic trees. P. pamirica exhibits some degree of ecological niche differentiation from P. talassica and P. laurifolia, whereas the latter two show minimal differentiation. Gene flow within the complex remains limited. This research underscores the importance of integrating multiple lines of evidence in the classification of Populus, providing a framework for future taxonomic studies.
Accurate delimitation of species is essential first of all for its correct taxonomic identification and then also to evaluate biodiversity, even for species within the genus Populus L. (Sites and Crandall, 1997; Yang and Rannala, 2010). Relying solely on a single method for species classification, such as morphology, may introduce bias and lead to the failure to recognize cryptic species or result in overclassification (Liu, 2016; Wan et al., 2023). The genus Populus (Salicaceae), commonly known as poplars, is widely distributed in the northern hemisphere throughout the subtropical to boreal forests and is well-known for its economic, ecological, and evolutionary importance as pioneer species (Braatne et al., 1992). However, classifying Populus is particularly challenging due to extensive hybridization and genetic introgression throughout evolution and speciation, which can obscure interspecific boundaries (Harland, 1950; Jiang et al., 2016; Wang et al., 2020; Du et al., 2024). Traditionally, the classification of Populus relies on morphological characteristics. However, this approach often struggles to account for the complex hybridization patterns within the genus. For instance, the Flora of China (Fang et al., 1999) recognized 71 species, with some considered varieties or hybrids of more diagnostic species, while Eckenwalder’s (1996) system identified only 29 species all over the world, highlighting the discrepancies in species delimitation based on morphology alone.
To clarify ambiguous species boundaries and reduce overclassification, the integrative species concept (Liu, 2016) employed multiple criteria for species classification. This approach incorporates various lines of evidence, including phenotypic distinction, phylogenetic monophyly (Donoghue, 1985; Minh et al., 2013), niche differentiation, and reproductive isolation (Coyne et al., 1988). Geographic isolation can be considered alongside niche differentiation as complementary evidence for species delimitation (Maya-Lastra et al., 2024). The use of multiple lines of evidence enhanced the objectivity and accuracy of species delimitation and taxonomic revision, particularly in identifying cryptic species and defining composite species (Feng et al., 2016; Liu, 2016; Zhang et al., 2018b; Lu et al., 2021; Perrino et al., 2022). Morphological data served as the foundation of taxonomic studies, while genetic evidence increasingly uncovered diversity not detected through morphology alone (Hu et al., 2018; Li et al., 2019; Sassone et al., 2021). Early molecular markers often lacked sufficient informative loci, limiting their ability to accurately reflect interspecies relationships (Castiglione et al., 1993; Bradshaw et al., 1994; Cervera et al., 2005; Feng et al., 2013; Wan et al., 2023). Chloroplast genomes, which reflect only maternal inheritance, also failed to differentiate species, particularly in taxa that exhibit extensive hybridization and chloroplast capture (Han et al., 2017; Zhang et al., 2018a; Zhou et al., 2021; Wang et al., 2022). Whole-genome resequencing offers a comprehensive view of the genomic landscape, overcoming the limitations of earlier methods and providing a more accurate assessment of phylogenetic relationships (Wang et al., 2022; Yang et al., 2022). Therefore, applying whole-genome resequencing is crucial in accurately deciphering the phylogenetic relationships of the Populus species and resolving taxonomic uncertainties. The Populus laurifolia Ledeb. complex is primarily distributed across northern and central Asia and has been established as monophyletic (Wang et al., 2022). This complex encompasses one to four recognized species. The Flora of the U.S.S.R. (Komarov, 1936) and Flora of China (Fang et al., 1999) reported four species: P. laurifolia, Populus pilosa Rehd., Populus pamirica Kom., and Populus talassica Kom. Additionally, Flora of Xinjiangensis (Yang, 1993) includes three varieties under these species: P. talassica var. cordata C.Y. Yang, P. talassica var. tomortensis C.Y. Yang, and P. pamirica var. akqiensis C.Y. Yang. Flora Reipublicae Popularis Sinicae (FRPS; Wang and Fang, 1984) included another variant, P. pilosa var. leiocarpa C. Wang & S. L. Tung. In contrast, the North American system reported only P. laurifolia (Eckenwalder, 1977, 1996). In this study, we included all aforementioned species within the P. laurifolia complex.
Populus laurifolia is widely recognized as a separate species based on various morphological and phylogenetic studies (Skvortsov, 2009, 2010; Wang et al., 2020; Wang et al., 2022). Specifically, P. pilosa shares several characteristics with P. laurifolia, including similar growth habits, habitat preference (riverbanks and wetlands), ovate to elliptic leaves with a hairy texture, and hairy fruits. These similarities often lead to the misidentification of P. pilosa as P. laurifolia. The primary distinction lies in the leaf base of P. pilosa, which is cordate or slightly cordate. Populus pilosa has recently been treated as a synonym or an infraspecific variant of Populus laurifolia (Skvortsov, 2010). As stated in the original literature (Wang and Tung, 1982), P. pilosa var. leiocarpa is distinguished solely by its glabrous fruits and was described as a variety of P. pilosa. Since its initial description, no comprehensive phylogenetic studies have been conducted on P. pilosa and its variety, leaving their taxonomic positions unresolved. The absence of phylogenetic data raises doubts about their relationships with P. laurifolia, underscoring the necessity for further molecular analyses. Morphological similarities also exist among P. laurifolia, P. talassica, and P. pamirica. P. talassica is primarily distinguished from P. laurifolia by its almost glabrous petioles and fruits, as well as its nearly non-angular sprouts. In contrast, P. pamirica can be distinguished from P. laurifolia by the aspect ratios of its leaf blades, which are nearly equal. Flora of Kazakhstan (Baitenov, 2001) considered P. talassica synonymous with Populus cathayana Rehd. due to characteristics such as short branches and petioles that are initially hairy but become glabrous upon maturation. This viewpoint has been widely rejected by many scholars (Yang, 1993). Recent studies also revealed that P. talassica is more closely related to P. pamirica, forming a sister group (Wang et al., 2020; Wang et al., 2022). Komarov (1936) initially described these two species with Populus densa Kom. for Central Asia, also referenced in the Flora of the U.S.S.R. Skvortsov (1972) suggested that P. talassica and P. pamirica refer to the same species, subsequently naming P. talassica. In contrast, Flora of Xinjiangensis (Yang, 1993) rejected the notion that P. pamirica is a synonym of P. talassica. In 2011, Skvortsov merged P. pamirica, P. talassica, P. densa, Populus suaveolens var. macrocarpa Schrenk, and P. cathayana into Populus macrocarpa (Schrenk) Pavlov & Lipsch (Skvortsov, 2011). Recent molecular phylogenetic studies suggested otherwise, indicating that P. cathayana, Populus shanxiensis C. Wang & S. L. Tung, and Populus intramongolica T. Y. Sun & E. W. Ma formed a distinct monophyletic group, while P. suaveolens Fisch. clustered with Populus maximowiczii A. Henry, Populus koreana Rehder, and Populus ussuriensis Kom (Wang et al., 2014, 2020, 2022). These findings raised doubts about Skvortsov’s classification, indicating that it lacked phylogenetic support.
The morphological similarity among taxa within the P. laurifolia complex has created significant ambiguity regarding interspecific boundaries, leading to a wide range of opinions on species delimitation. These assumptions have been primarily based on morphological evidence, without robust phylogenetic studies to elucidate the species boundary of the P. laurifolia complex. Due to early molecular techniques and incomplete sampling, previous phylogenetic studies have left the interspecific relationships within the P. laurifolia complex unresolved (Feng et al., 2013; Wang et al., 2020; Wang et al., 2022). Aiming to clarify the species delimitation of the P. laurifolia complex, this study relies on comprehensive analyses of morphological traits, niche differentiation, and phylogenomic analyses using whole-genome resequencing and seeks to provide a clearer understanding of the interspecific relationships within this complex.
Four species and three varieties of P. laurifolia complex were examined in this study. Based on available specimen records, collection points were made to cover each species distribution site as much as possible. A total of 337 samples were eventually collected from 29 populations from Xinjiang, China, with the number of individuals sampled per population ranging from a few to approximately 20, depending on the population size. (Figure 1, Supplementary Table S1). Few samples of P. pamirica (five samples) and P. pilosa (11 samples) were collected due to their small population sizes in nature. The collected specimens were deposited in the herbarium of the Museum of Beijing Forestry University (BJFC). In addition to field-collected specimens, digital specimens from the Museum of Xinjiang Agricultural University, the National Plant Specimen Resource Center (CVH; https://www.cvh.ac.cn/), the National Specimen Information Infrastructure (NSII; http://www.nsii.org.cn/), JSTOR Global Plants (https://plants.jstor.org), and the Moscow Digital Herbarium (https://plant.depo.msu.ru/) were utilized for morphological analysis.
The morphology of sprout leaves and short-branched leaves, as well as the density of pubescence on petioles and branches, are important characteristics for identifying species in the P. laurifolia complex, as supported by Flora of Xinjiangensis (Yang, 1993) and our field observations. Eight leaf morphological traits (Figure 2A) for both sprout leaves and short-branched leaves of the four species were measured, including leaf length (LL), leaf width (LW), petiole length (PL), the widest position of the leaf blade (WLP), the base angle of the leaf (ALB), leaf width-to-length ratio (LWR), the length from the widest point to the leaf base to the leaf length ratio (WLR), and petiole length-to-leaf length ratio (PLR). Additionally, the density of petiole pubescence (DPP) of short-branched leaves was quantified. The density of pubescence has been designated as a qualitative character in some morphological studies, but the definition of pubescence density is ambiguous and difficult to understand in practice. To better characterize the differences in pubescence density among the species of the P. laurifolia complex, petiole hairs were examined using a scanning electron microscope (HITACHI S-4800, Tokyo, Japan). First, a petiole measuring approximately 1 cm in length was extracted from the leaf base and subsequently imaged using a scanning electron microscope at a magnification of ×100, and then the number of pubescence within a 1-mm length of the petiole was quantified utilizing the ImageJ software (Schneider et al., 2012) (Figure 2). For the two varieties of P. talassica, only the eight leaf morphological traits of short-branched leaves and the DPP were measured.
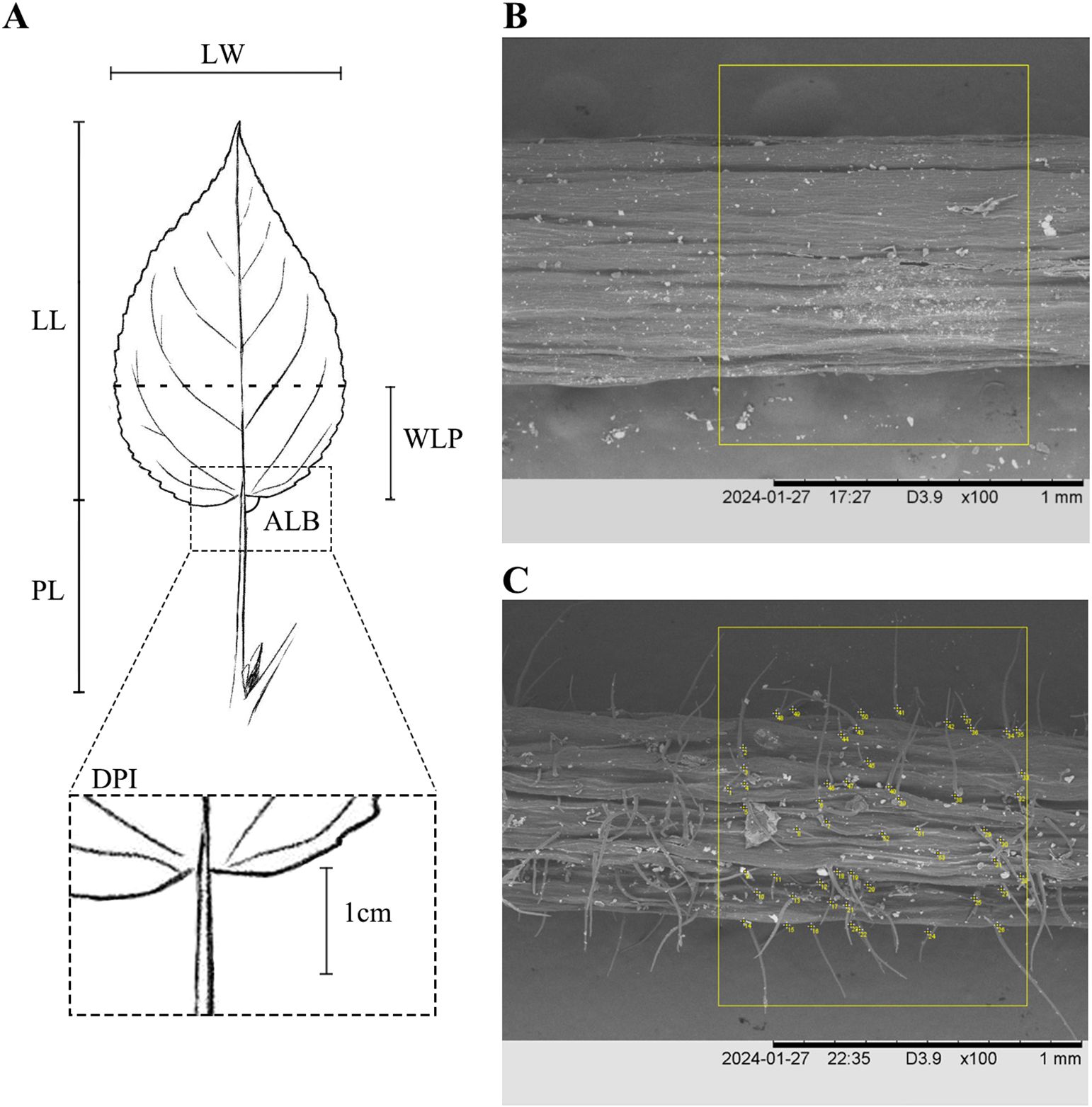
Figure 2. Morphological analyses of Populus laurifolia complex species. (A) Morphometric measurements in this study: leaf length (LL), leaf width (LW), petiole length (PL), the widest position of the leaf blade (WLP), the base angle of the leaf (ALB), and the density of petiole pubescence (DPP). (B) The number of 1-mm2 hairs on the petiole of Populus talassica. (C) The number of 1-mm2 hairs on the petiole of P. laurifolia.
The above data were processed and analyzed using the Origin 2022 software (Deschenes, 2000). The Kruskal–Wallis tests and Wilcoxon rank sum tests were used to assess the differences in each trait among the four species of the complex. The significance of differences was indicated on box plots. To prevent collinearity in subsequent analyses, highly correlated characteristics were removed using Pearson’s correlation analysis. All statistical analyses were conducted in IBM SPSS Statistics 26, resulting in the retention of three morphological traits. A principal component analysis (PCA) was then performed on these retained traits. To verify the classification of the P. laurifolia complex, which is outlined in Flora of Xinjiangensis (Yang, 1993) as having heteromorphic leaves for P. laurifolia and P. talassica and almost homomorphic leaves for P. pilosa and P. pamirica, PCAs on the morphological traits of sprout leaves and short-branched leaves for each species were conducted.
One to six individuals were randomly selected from each population of the P. laurifolia complex (Supplementary Table S2). Healthy and fresh leaves were collected and immediately dried using silica gel. Genomic DNA was extracted from these silica gel-dried leaves using a modified Cetyltrimethylammonium Bromide (CTAB) method (Doyle, 1987). All sequencing data have been deposited at the National Genomics Data Center (Chen et al., 2021; CNCB-NGDC Members and Partners et al., 2024). All DNA samples were sequenced at Novogene (Beijing, China) using the Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA), targeting a 20× coverage for whole-genome paired-end reads. Additionally, to verify the monophyly of the P. laurifolia complex, 31 sequences of Populus (Supplementary Table S2) were downloaded from the National Center for Biotechnology Information (NCBI) database and the Genome Sequence Archive (GSA) in the BIG Data Center for analysis. These sequences included one sample of P. talassica and two samples of P. pamirica (Supplementary Table S2). In addition, four Populus samples outside the P. laurifolia complex were collected from our own fieldwork for further comparison.
Nuclear variants were identified using GATK tools. The resequencing data were first mapped to the reference genome of P. koreana (Sang et al., 2022) using BWA-MEM v.0.7.17-r1188 (Li and Durbin, 2009). Mapped reads were then converted to BAM files, sorted, and filtered using SAMtools v.1.6 (Li et al., 2009), and PCR duplicates were marked using Picard v.2.1.1. Short variants were called using GATK v.4.1.4 (McKenna et al., 2010) with HaplotypeCaller, and the variants from all samples were combined using GATK’s CombineGVCFs. Single-nucleotide polymorphisms (SNPs) were identified using the SelectVariants tool (Danecek et al., 2011). To convert the SNPs into PHYLIP format, Python v.2.7.5 was used, followed by dataset analysis in IQ-TREE v.2.0.3 (Nguyen et al., 2015).
A maximum likelihood (ML) phylogenetic tree was constructed using IQ-TREE, with the SH-aLRT branch test (1,000 repetitions) specified by the “-alrt” option (Guindon et al., 2010). The most appropriate evolutionary model was selected using ModelFinder (Kalyaanamoorthy et al., 2017). The sliding-window method (Takuno et al., 2012; Zhang et al., 2016; Liu et al., 2017; Chen et al., 2019) was employed to reconstruct the species tree, with concatenated SNPs filtered into 4-kb non-overlapping windows across the genomic alignments. Populus euphratica was used as the outgroup.
Additionally, to verify the monophyly of the P. laurifolia complex, 31 sequences of Populus (Supplementary Table S2) were downloaded from the NCBI database and the GSA in the BIG Data Center for analysis. These sequences included one sample of P. talassica and two samples of P. pamirica (Supplementary Table S2). In addition, four Populus samples outside the P. laurifolia complex were collected from our own fieldwork for further comparison.
Linkage disequilibrium (LD)-based SNP filtering was conducted using PLINK v.1.9.0 (Purcell et al., 2007). PCA of the filtered Populus species was performed using PLINK, and visualizations were created using the R package ggplot2 (Villanueva and Chen, 2019). Population structure was analyzed using ADMIXTURE (Alexander et al., 2009), with the number of clusters (K) set from 1 to 10. Cross-validation was used to determine the optimal K value, with the lowest cross-validation error indicating the most appropriate number of clusters. To evaluate potential hybridization, genetic data from nine Populus nigra individuals were downloaded from NCBI, and a genetic structure analysis was performed (Supplementary Table S2).
To investigate gene flow from other species into P. laurifolia complex, ABBA–BABA statistics (Malinsky et al., 2021) were employed to quantify potential gene exchange. This statistical method assesses deviations from a strictly bifurcated evolutionary history using genome-scale SNP data, thereby testing for gene penetrance (Martin et al., 2015; Malinsky et al., 2021). Patterson’s D-statistic (Durand et al., 2011) was calculated using the Dtrios program from Dsuite (Malinsky et al., 2021). In this study, for the ordered alignment {[(P1, P2), P3], O}, the ABBA site pattern represents the shared derived alleles of P2 and P3, while the BABA site pattern corresponds to the shared derived alleles of P1 and P3. Under the null hypothesis of incomplete lineage sorting (ILS), the number of ABBA sites and BABA sites is expected to be equivalent (D = 0). A significant deviation of D from 0 suggests alternative evolutionary events, particularly indicating potential gene exchange between P3 and either P1 or P2 (Durand et al., 2011). If the absolute value of the Z-score exceeds 3, it is considered statistically significant, indicating a strong deviation from the expected distribution (Busing et al., 1999). Subsequently, F-branch analysis further quantifies the relative strength of gene flow and maps the results onto different branches of the phylogenetic tree to infer the direction and contribution of gene flow. Gene flow intensity between each species and the phylogenetic branch was measured using the F4 ratio, and a heatmap was created for visualization in the Origin 2022 software (Deschenes, 2000).
Previously collected specimens from the CVH were re-identified based on key traits, and due to the distribution overlap between P. pilosa and P. laurifolia, they were combined into a single phenotypic cluster. A total of 212 locations were identified across three clusters, which were used to construct their respective distributional niches. To analyze niche similarities and differences among these phenotypic clusters, 19 bioclimatic variables were first downloaded from the WorldClim database (Hijmans et al., 2005). After missing values were removed, the species occurrence data were combined with background environmental data for a PCA using the dudi.pca function from the ade4 package, reducing the dimensionality of the environmental space. The resulting PCA scores for the three clusters were then used to create dynamic climatic grids via the ecospat.grid.clim.dyn function, representing the species’ niches in environmental space. Subsequently, niche similarities and differences between the clusters were calculated using Schoener’s D and the standardized Hellinger distance (I) in ENMTools 1.3 (Warren et al., 2019). To further assess niche equivalency and similarity, 100 replicates of both tests were performed using the equivalency and similarity functions.
Phylogenetic analyses (Figure 3A) based on the ASTAL method showed that the P. laurifolia complex formed a well-supported clade (posterior probability = 100), which consisted of three subclades: 1) P. laurifolia and P. pilosa (Clade I), 2) P. pamirica (Clade II), and 3) P. talassica and its varieties, along with P. pilosa var. leiocarpa (Clade III). The P. talassica clade was the sister group to the P. pamirica clade (PP = 100), and both clades were monophyletic. Populus laurifolia and Populus pilosa together formed a monophyletic clade, which was a sister to the P. pamirica + P. talassica clade. The same topology was also recovered by a concatenated tree based on 566,375 nuclear SNPs (Supplementary Figure S2). PCA of the SNP data produced consistent results, with the individuals of the P. laurifolia complex forming three distinct clusters: P. pilosa and P. laurifolia clustered together, while P. talassica and P. pamirica formed separate clusters (Figure 3B). The optimal K value for the genetic admixture analysis of the P. laurifolia complex was determined to be 4 (Figure 3C). The populations of P. laurifolia and P. pilosa contained the same genetic clusters, indicating minimal genetic differentiation between the two species. Populus laurifolia populations from EM, HEB, and JIM also showed genetic components of P. talassica, suggesting some gene flow between these populations and P. talassica in the Tianshan region. Samples of P. pamirica from China and Pakistan were assigned to a single genetic cluster, indicating a distinct and specialized genetic background. For P. talassica, two genetic lineages were observed: 1) populations from QIT, CHANGJ, and FUK and 2) populations from other regions. However, this genetic differentiation was not supported by the phylogenetic tree.
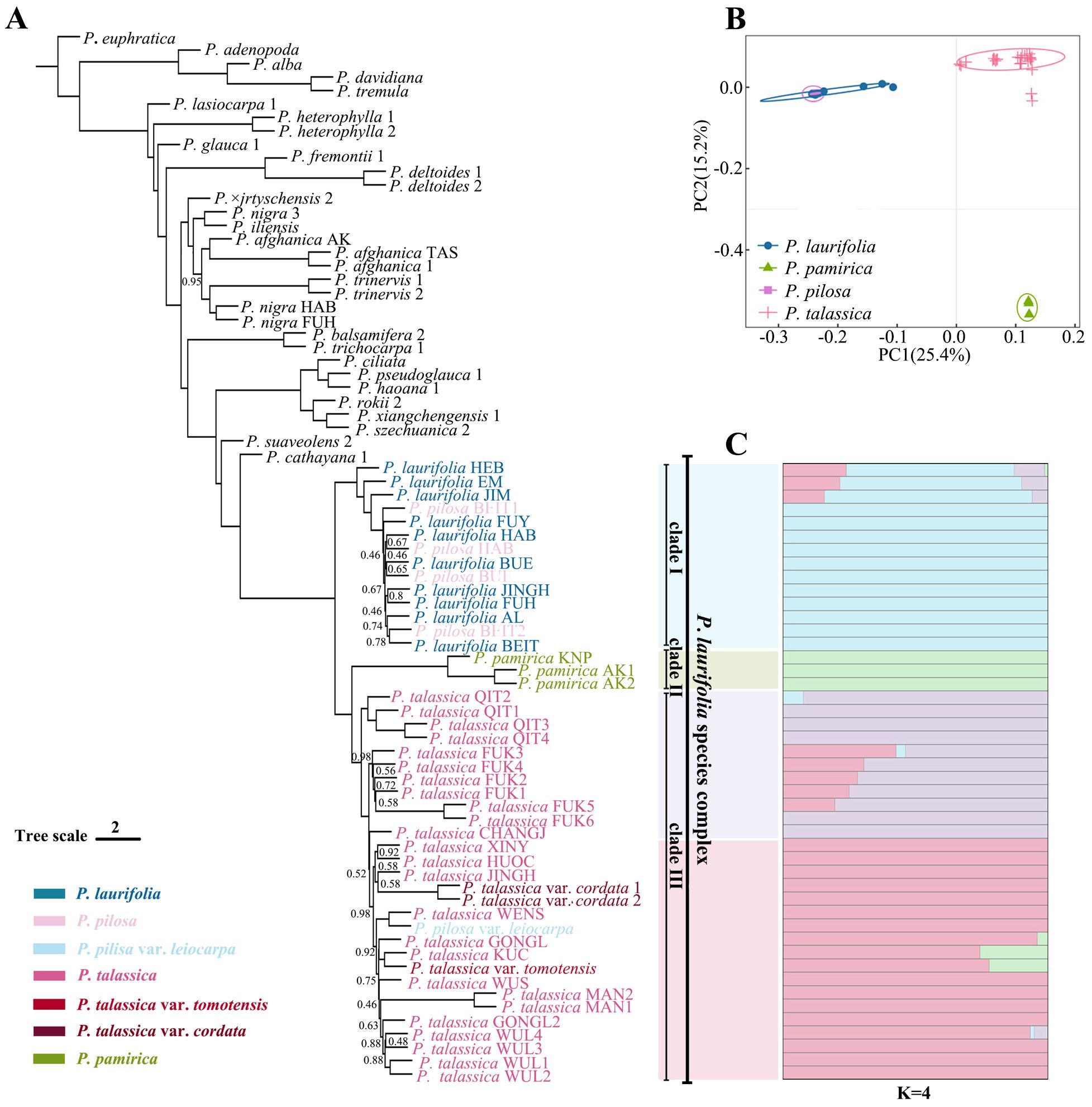
Figure 3. Phylogenetic analyses. (A) Phylogeny estimated with alternative quartet topologies (-t 8) in ASTRAL based on 4-kb non-overlapping windows. Unless otherwise indicated, all nodes had 1.0 posterior probability. (B) Principal component analysis (PCA), based on genetic distance using SNP data. (C) Population structure analysis for four species of Populus laurifolia species complex. Each colored bar represents one individual, and colored segments represent proportions of ancestral components. The order of individuals corresponds to their order in the phylogenetic tree in panel (A).
Based on the phylogenetic tree topology, we divided the P. laurifolia complex into three monophyletic taxa: P. laurifolia (including P. pilosa), P. pamirica, and P. talassica. To further investigate gene flow within this complex, we used P. euphratica as an outgroup and applied the ABBA–BABA method (Figure 4). The result showed that significant gene exchange was absent within this complex. Although some gene flow occurred between P. talassica and P. laurifolia, the overall level remained low. In contrast, gene exchange between Populus × jrtyschensis and P. laurifolia occurred more frequently, with active gene flow between P. × jrtyschensis and the broader P. laurifolia complex. This increased gene exchange may be attributed to P. laurifolia serving as a parental species of P. × jrtyschensis (Jiang et al., 2016).
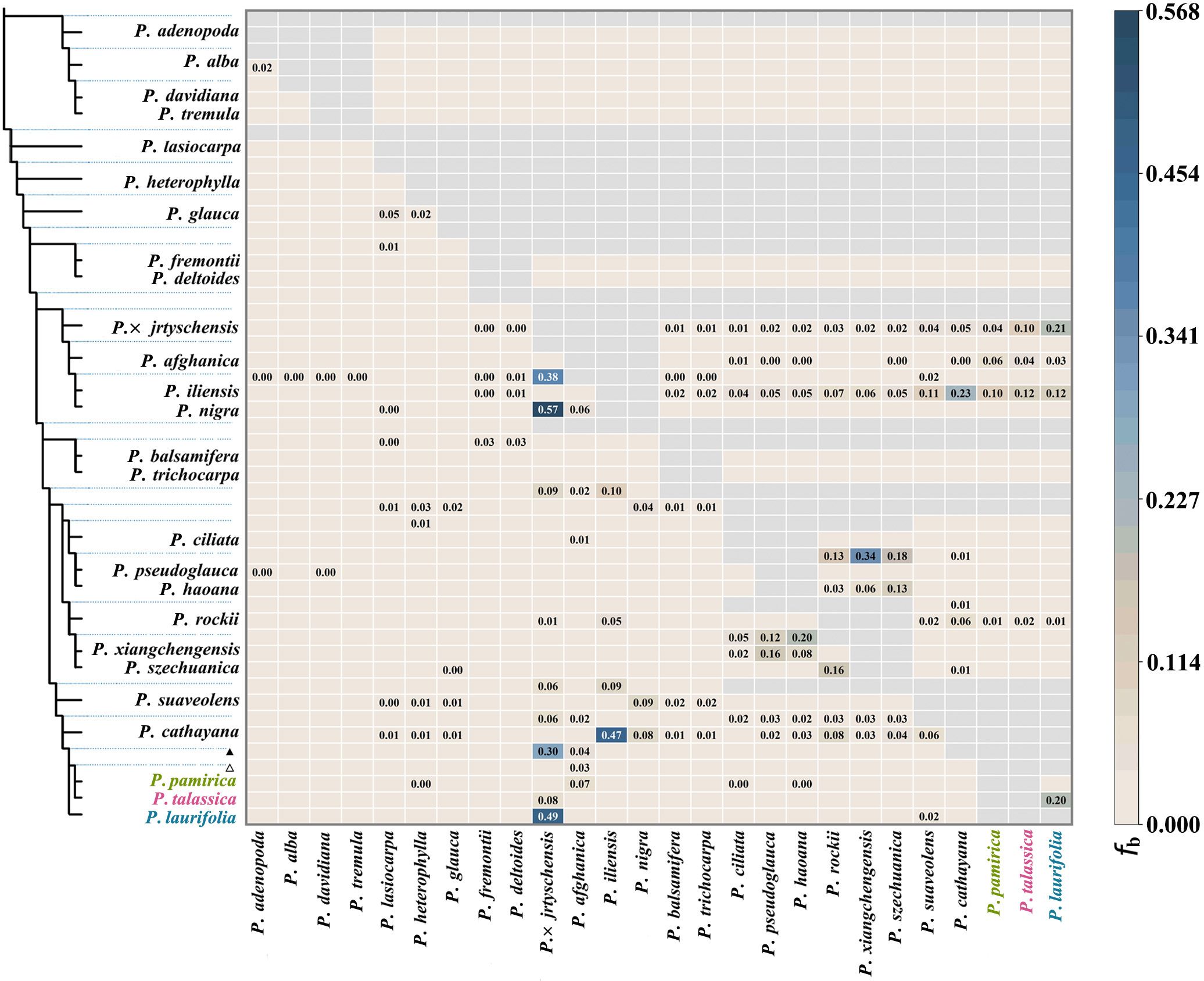
Figure 4. ABBA–BABA statistics results measuring gene flow among 26 species. Gray areas represent no gene flow, and values equal to 0.000 are not labeled. The left panel shows the species tree estimated in ASTRAL using 4-kb non-overlapping windows. Black triangles indicate gene flow between the ancestor of Populus laurifolia complex and other species, while hollow triangles show gene flow between ancestor of Populus pamirica and Populus talassica with other species.
The short-branched leaves of the P. laurifolia complex are predominantly ovate or long-ovate with finely serrated edges. A quantitative analysis of eight morphological traits revealed distinct differences among species (Figure 5; Supplementary Table S3). Although the leaves of P. laurifolia, P. talassica, and P. pilosa shared similar shapes (Figure 5), P. pilosa was characterized by predominantly cordate leaf bases (Figure 5D), while both P. laurifolia and P. talassica exhibited ovate bases (Figure 5D). This distinction in leaf base shape is a primary criterion for differentiating P. pilosa from the other two species. In contrast, P. pamirica showed significant morphological differences compared to the other species in the complex. Notably, P. pamirica had wider leaves (Figure 5B) and a higher LWR (Figure 5C), along with a greater petiole length (Figure 5G) and PLR (Figure 5H) compared to P. laurifolia, P. talassica, and P. pilosa. Further analysis using Pearson’s correlation indicated that LL, LW, WLP, WLR, and ALB did not significantly contribute to species differentiation and were excluded from subsequent analyses. PCA, based on the remaining variables (PL, PLR, and LWR), suggested that while differentiating the four species is challenging, P. pamirica remained relatively distinct with minimal overlap with the other species (Supplementary Figure S1B).
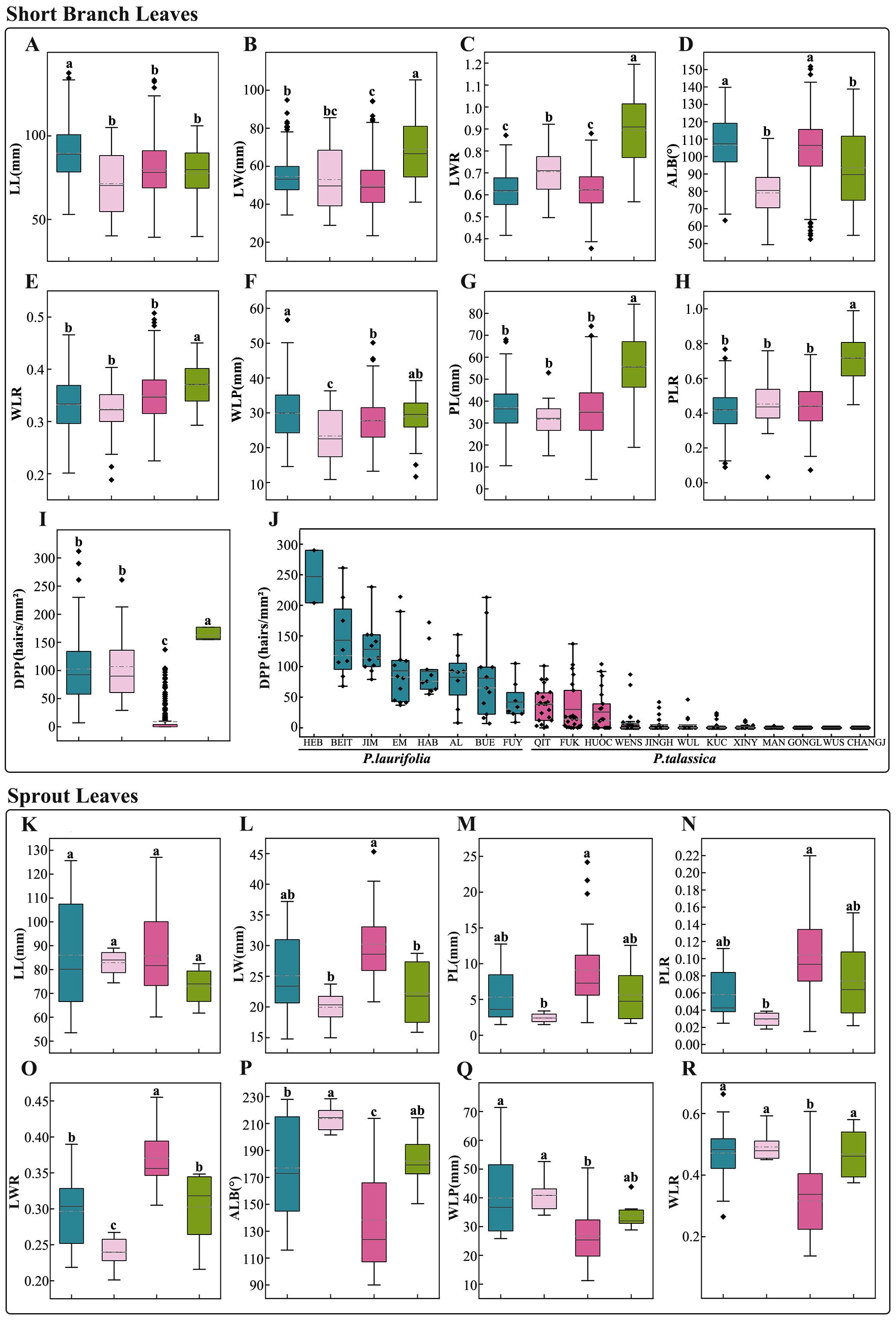
Figure 5. Morphological comparison of short-branched leaves (A–J) and sprout leaves (K–R) among species of the Populus laurifolia complex. (A, K) Leaf length (LL). (B, L) Leaf width (LW). (C, O) Leaf width-to-length ratio (LWR). (D, P) Leaf base angle (ALB). (E, R) Ratio of the distance from the widest point to the leaf base to the leaf length (WLR). (F, Q) Position of the widest point of the leaf blade (WLP). (G, M) Petiole length (PL). (H, N) Petiole length-to-leaf length ratio (PLR). (I) Density of petiole pubescence (DPP) among four species. (J) DPP of short-branched leaves among populations of P. laurifolia and Populus talassica. Boxplot colors represent species: blue for P. laurifolia, pink for Populus pilosa, red for P. talassica, and green for Populus pamirica. The gray line indicates the mean, and the light gray dashed line indicates the median. Different lowercase letters above columns show significant trait differences (p < 0.05); shared letters denote none.
Petiole pubescence density is a key trait distinguishing the P. laurifolia complex from other species. Among the complex, P. pamirica exhibited the highest petiole hair density, while no significant difference in hair density was observed between P. laurifolia and P. pilosa, while P. talassica had the lowest density, with only 9.23 hairs/mm2 on average (Figure 5I; Supplementary Table S3). Geographic variation in P. talassica revealed significant differences: populations from Gongliu, Wusu, and Changji had glabrous petioles, while those from Qitai, Fukang, and Huocheng were tomentose (Figure 5J). These findings were consistent with the description in the Flora of Xinjiangensis (Yang, 1993), which classified the densely pubescent petioles in Wensu County as P. talassica var. tomortensis. However, our data suggested that petiole pubescence in P. talassica exhibited significant variation, indicating that it may not reliably reflect varietal differences. While the short branchlet leaves of P. talassica and P. laurifolia were morphologically similar, populations of P. laurifolia generally possessed higher petiole hair density, ranging from 7 to 312 hairs/mm2, in contrast to the predominantly glabrous P. talassica populations. This key difference underscored the morphological distinction between the two species.
The sprout leaves of species within the P. laurifolia complex, including P. laurifolia, P. pilosa, P. talassica, and P. pamirica, were primarily obtained from online digitized herbarium specimens. The results (Figure 5; Supplementary Table S4) revealed that P. laurifolia and P. pamirica exhibited a high degree of similarity in sprout leaf morphology, while P. laurifolia and P. pilosa also showed similar traits, although P. pilosa was characterized by narrower sprout leaves (Figure 5O). In contrast, P. talassica showed significantly higher values of LWR (Figure 3O) and lower values of WLP and WLR (Figures 5Q, R) compared to the other species. Additionally, the leaf base angle of P. talassica was more oval-shaped compared to that of the other species (Figure 5P).
The PCA results demonstrated significant morphological differentiation between short-branched leaves and sprout leaves within the P. laurifolia complex, except for P. talassica, which showed no significant differences (Figure 6). In P. laurifolia (Figure 6A), short-branched leaves (in blue) clustered distinctly from sprout leaves (in gray), with the former exhibiting lanceolate shapes compared to elliptic, ovate, or oblong-ovate shapes of the latter. Similarly, P. pilosa (Figure 6B) showed a clear separation between short-branched leaves (in light pink) and sprout leaves, underscoring their distinct morphological characteristics. The PCA for P. talassica (Figure 6C) revealed that short-branched leaves (in red) were broader and more ovate, whereas the sprout leaves were narrower and lanceolate. A similar trend was observed in P. pamirica (Figure 6D), highlighting substantial morphological variation between the two leaf types.
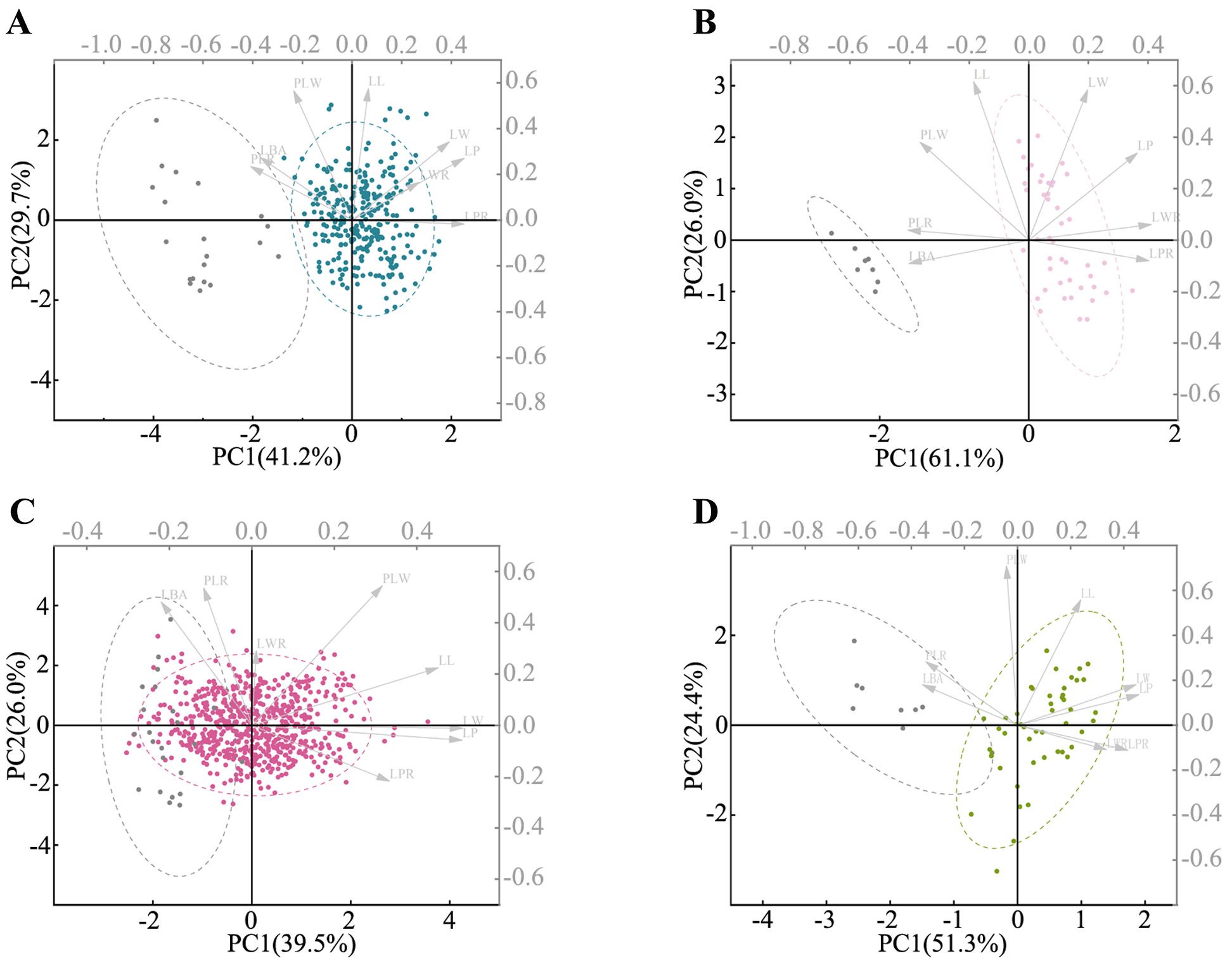
Figure 6. Principal component analysis (PCA) based on morphometric data. (A) PCA plot comparing the short-branched leaves (in blue) and sprout leaves (in gray) of Populus laurifolia. (B) PCA plot of the short-branched leaves (in light pink) and sprout leaves (in gray) of Populus pilosa. (C) PCA plot of the short-branched leaves (in red) and sprout leaves (in gray) of Populus talassica. (D) PCA plot of the short-branched leaves (in olive green) and sprout leaves (in gray) of Populus pamirica.
The species within the P. laurifolia complex shared several similarities, including comparable inflorescences (Supplementary Figures S3G, N) and ovoid capsules that were three- or four-valved (Supplementary Figures S4C, S3F, M). However, they also exhibited distinct differences. For the species we examined, the capsules of P. laurifolia and P. pilosa were densely covered with woolly hairs (Supplementary Figures S4C, S3C), whereas P. talassica had capsules with few to no hairs (Supplementary Figure S3M). The branchlets of P. laurifolia and P. pamirica were hairy, while those of P. talassica were nearly hairless (Supplementary Figure S3). Additionally, P. laurifolia had yellowish, distinctly angled, and tomentose sprouts (Figure 5E, Figures 6C), whereas P. talassica exhibited russet or gray suckers and glabrous, slightly angled sprouts (Supplementary Figures S6J, K, L).
During our field survey, we identified several specimens with incorrect species identifications at various recording sites, where the actual poplar species present differed from those recorded. For instance, in the Tianchi Lake Scenic Area of Fukang County, individuals previously identified as P. laurifolia or P. pilosa were found to be P. talassica. Our survey revealed no distribution overlap among P. talassica, P. laurifolia, and P. pamirica (Figure 1). We employed Schoener’s D and Warren’s I indices to assess niche differentiation between these species. The results (Table 1) indicated very low ecological overlap between P. pamirica and both P. talassica and P. laurifolia, signifying a high degree of niche differentiation for P. pamirica. Conversely, the overlap indices for P. laurifolia and P. talassica were relatively high, suggesting minimal niche differentiation between these two species. Despite the observed niche overlap, the field distributions of these species do not intersect, implying that geographic isolation may be a primary factor driving their differentiation.
Establishing species boundaries among closely related species with a history of hybridization poses significant challenges. However, statistical analyses incorporating multiple lines of evidence suggest that an integrative approach is more effective (Liu, 2015, 2016). Traditionally, the P. laurifolia complex was considered to include four species—P. laurifolia, P. pilosa, P. talassica, and P. pamirica—distinguished primarily by leaf shape and petiole pubescence (Yang, 1993; Fang et al., 1999). In contrast, Skvortsov (2009, 2010) proposed that P. pamirica, P. talassica, and P. pilosa should be classified as a single species, with P. macrocarpa as the valid name. However, our study supports merging P. pilosa with P. laurifolia while recognizing P. talassica and P. pamirica as distinct species.
Phylogenetic analysis based on nuclear SNP data reveals that the P. laurifolia complex forms three well-supported clades (PP = 100), with P. talassica and P. pamirica being more closely related than P. laurifolia (including P. pilosa) (Figure 3A). This clustering is further supported by the PCA result (Figure 3B). These molecular findings suggest that P. pilosa and P. laurifolia may not be distinct at the species level. Morphological analysis supports this hypothesis. Our morphological analysis (Figure 5) shows that the short-branched and sprout leaves of P. laurifolia and P. pilosa are very similar, with only minor differences, such as the cordate leaf base of P. pilosa’s short-branched leaves and slightly narrower sprout leaves. These differences are heavily influenced by environmental factors and are not stable (Gong et al., 2023; Zhao et al., 2023). Furthermore, P. pilosa shares the same habitat as P. laurifolia, lacking an independent distribution area (Figure 1). Moreover, the original descriptions of P. pilosa lacked detailed information on sprout leaves. Together, this evidence supports the hypothesis that P. pilosa is a morphological variant of P. laurifolia.
In contrast, P. talassica and Populus pamirica exhibit distinct morphological and molecular features. Populus pamirica can be distinguished by the leaf shape of its short-branched leaves, which are nearly equal in length and width, its longer petioles, and the denser pubescence on the branchlets and petioles (Figure 5; Supplementary Figures S3, S4). Populus talassica was often misidentified as Populus laurifolia in the field, but our morphological analysis shows significant differences, particularly in pubescence (Figures 5I, J). Although both species have similar short branchlet leaves, P. talassica exhibited less pubescence, with some nearly glabrous specimens. Specimens from Fukang and Qitai with dense pubescence were misidentified, but P. talassica consistently shows lower petiole pubescence than P. laurifolia. Additional differences included less lanceolate sprout leaves, non-angled sprout morphology, nearly glabrous capsules, and distinct geographical distributions (Figures 1, 5, 6; Supplementary Figure S3).
To accurately classify the Populus species, especially within the P. laurifolia complex, an integrative approach is essential. This approach should incorporate multiple morphological traits from various plant parts. The combined morphological, genetic, and ecological data strongly support the re-evaluation of the P. laurifolia complex. The evidence indicates that P. laurifolia (including P. pilosa), P. talassica, and P. pamirica should be treated as three independent species, reflecting their distinct evolutionary trajectories and ecological adaptations. Future taxonomic revisions should consider these findings to enhance the understanding and classification of this complex.
The P. laurifolia complex in Flora of Xinjiangensis and FRPS included four varieties: P. talassica var. cordata, P. talassica var. tomortensis, P. pamirica var. akqiensis, and P. pilosa var. leiocarpa. In this study, we examined specimens of P. talassica var. cordata and P. talassica var. tomortensis collected from their type localities (Jinghe and Wensu, Xinjiang) and compared them with P. talassica populations (Supplementary Figure S1C). The distinguishing characteristic of P. talassica var. cordata was its cordate leaf base; however, P. talassica exhibited variable leaf bases (cuneate, truncate, or cordate) even within the same tree, complicating its differentiation. Similarly, P. talassica var. tomortensis was described as having broadly ovate leaves and densely tomentose short branches, petioles, veins, and infructescence axes. We observed that these traits also appeared in other P. talassica populations (e.g., Fukang and Qitai), suggesting that these features were inconsistent and not supported by phylogenetic analysis, casting doubt on the validity of these varieties. We obtained specimens of P. pilosa var. leiocarpa from its type locality (Tomur Peak, Wensu County) and also collected P. talassica var. tomortensis in the same area (Supplementary Figures S5, S6). The latter also exhibited densely tomentose infructescence axes and glabrous capsules, raising the possibility that P. pilosa var. leiocarpa may represent a misidentification or simply a variant of P. talassica. Phylogenetic analysis based on nuclear SNP data supported this hypothesis.
Based on our morphological and phylogenetic analysis, we conclude that the recognition of P. talassica var. cordata, P. talassica var. tomortensis, and P. pilosa var. leiocarpa as distinct varieties is unwarranted. These varieties should be subsumed under P. talassica at the species level.
The P. laurifolia species complex, which includes P. pamirica, P. talassica, and P. laurifolia (a division based on the results of previous species boundary discussions), exhibits varying degrees of geographic isolation and ecological differentiation (Table 1, Figure 1). These three species showed distinct clusters with complete or partial geographical isolation, suggesting that geographic barriers played a significant role in maintaining their separation. Populus laurifolia and Populus talassica typically thrived in riparian environments, favoring moist habitats along water bodies due to their reliance on sufficient water for growth and survival. In the Junggar Basin, geographic isolation likely drove their speciation, as the region’s arid conditions served as a natural barrier (Carroll et al., 1990; Chen et al., 2002). Despite this, substantial niche differentiation did not occur (Wang et al., 2006). The results of Schoener’s D and Warren’s I indices revealed a relatively high overlap between the ecological niches of P. talassica and P. laurifolia (Table 1). However, the absence of current field distribution overlap suggests that geographic isolation, rather than ecological differentiation, has been the primary driver of their divergence (Anacker and Strauss, 2014). This pattern aligns with the classic model of allopatric speciation, where physical barriers facilitate the formation of distinct species without immediate ecological divergence.
Differently, P. pamirica has undergone significant niche differentiation compared to P. laurifolia and Populus talassica (Table 1). It grows at higher elevations and is adapted to colder environments, indicating the evolution of specialized ecological adaptations influenced by its unique geographic and environmental conditions in the Pamir region (Metrak et al., 2015; Mętrak et al., 2017, 2023). Its populations are confined to distinct ecological niches in the Pamir region, as evidenced by low niche overlap indices.
Natural hybridization and genetic introgression play crucial roles in biological evolution, particularly in influencing species diversity (Harland, 1950; Arnold, 1992; Gompert and Buerkle, 2016). Within the Populus genus, extensive gene flow occurs, and several hybrid zones have been confirmed (Lexer et al., 2010; Jiang et al., 2016). However, the P. laurifolia complex presents a contrasting pattern, where hybridization and gene flow are more restricted compared to other Populus species. Our ABBA–BABA analysis (Figure 4) reveals that gene flow within the P. laurifolia complex is limited, indicating that species have not undergone significant genetic backflow after divergence. In contrast, hybridization between P. laurifolia and P. nigra produced P. × jrtyschensis (Figure 4), with populations predominantly consisting of F1 hybrids, forming a typical hybrid zone (Jiang et al., 2016). This indicates more extensive gene flow between P. laurifolia and P. nigra, possibly due to the role of P. laurifolia as a hybrid parent of P. × jrtyschensis. In this study, our genetic structure analysis of P. laurifolia, P. × jrtyschensis, and P. nigra (Supplementary Figure S1A) showed that K = 2 was the best fit. The collected P. laurifolia samples were genetically pure and showed no evidence of hybridization with P. nigra, ensuring the purity of the species within the P. laurifolia complex in this study. The limited gene flow within the P. laurifolia complex reflects a more stable evolutionary trajectory, where geographic isolation and reproductive barriers play a significant role in maintaining species boundaries. By contrast, the Erqis River Valley, known for its high Populus diversity in Xinjiang, may have facilitated cross-species hybridization involving P. laurifolia (Jiang et al., 2016). Meanwhile, P. pamirica remains relatively independent genetically (Figure 4), with lower levels of gene flow likely due to ecological or reproductive isolation, which allows it to maintain its distinct genetic characteristics. Furthermore, genetic structure analysis showed that K=4 was the optimal model (Figure 5C), with a few individuals in each cluster exhibiting mixed genetic components, indicative of minor introgression between species. Additionally, P. talassica was divided into two genetic components (Figure 5C), suggesting that despite its relatively uniform morphology, substantial genetic differentiation exists within the species. However, this genetic differentiation has not yet resulted in further morphological or phylogenetic divergence and may be linked to ecological adaptation. Further research is needed to understand the causes of this genetic split and its evolutionary significance.
In this study, we conducted extensive sampling of the P. laurifolia complex to clarify species boundaries through morphological and genetic analyses. The results supported the classification of this complex into three distinct species: Populus laurifolia, Populus talassica, and Populus pamirica. Populus pilosa is considered a synonym of P. laurifolia due to the absence of significant morphological or genetic divergence. Although some varieties within this complex exhibit subtle morphological differences, these variations do not form monophyletic groups on the phylogenetic tree. Therefore, this study does not support recognizing infraspecific taxa (varieties) within the complex.
Morphological comparisons revealed that P. laurifolia and P. pilosa shared very similar traits, with only minor differences in leaf morphology that were insufficient to justify their separation at the species level. In contrast, P. talassica is distinct, exhibiting differences in leaf shape, pubescence, and sprouts. Additionally, its geographic distribution did not overlap with that of Populus laurifolia or Populus pamirica. Populus pamirica was clearly distinguishable by its short branchlet leaf morphology and geographic isolation in the Pamir region. Phylogenetic analyses were consistent with these findings and supported the classification of the complex into three distinct species, with P. talassica and P. pamirica forming a sister group. Our ABBA hybridization analysis revealed limited gene flow within the P. laurifolia complex, with ecological niche differentiation also being minimal. In contrast, geographic isolation appeared to play a more significant role in speciation within this complex. Future research should explore how geographic isolation interacts with ecological factors to drive speciation within the Populus genus. This study emphasized the importance of a multifaceted approach—incorporating morphological, genetic, and ecological data—when clarifying species boundaries. By clarifying the taxonomic structure of the P. laurifolia complex, our findings provided valuable insights for future classification and the evolutionary understanding of the Populus species.
The data presented in the study are deposited in the Genome Sequence Archive in National Genomics Data Center (https://ngdc.cncb.ac.cn/gsa/), accession number: CRA018863 (GSA).
XW: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. XC: Methodology, Validation, Writing – review & editing. FY: Methodology, Writing – review & editing. KZ: Investigation, Methodology, Writing – review & editing. LZ: Investigation, Methodology, Writing – review & editing. CZ: Investigation, Methodology, Writing – review & editing. SG: Investigation, Writing – review & editing. CS: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing. ZZ: Funding acquisition, Project administration, Resources, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (Grant No. 32001247), the Third Xinjiang Scientific Expedition Program (Grant No. 2021XJKK0601), and the Science and Technology Basic Resources Investigation Program of China (Grant No. 2022FY202201).
The authors thank the staff at Xinjiang Institute of Geography and Ecology and the herbarium of Xinjiang Agricultural University; Shang Qu, Tao Xing, and Yachao Wang for their kind help in sampling and specimen checking; Enze Li for his suggestions on data analysis; and Yueqi You for her suggestions on writing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1518122/full#supplementary-material
Alexander, D. H., Novembre, J., Lange, K. (2009). Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. doi: 10.1101/gr.094052.109
Anacker, B. L., Strauss, S. Y. (2014). The geography and ecology of plant speciation: range overlap and niche divergence in sister species. Proc. R. Soc B Biol. Sci. 281. doi: 10.1098/rspb.2013.2980
Arnold, M. L. (1992). Natural hybridization as an evolutionary process. Annu. Rev. Ecol. Evol. Syst. 23, 237–261. doi: 10.1146/ANNUREV.ES.23.110192.001321
Braatne, J. H., Hinckley, T. M., Stettler, R. F. (1992). Influence of soil water on the physiological and morphological components of plant water balance in Populus trichocarpa, Populus deltoides and their F(1) hybrids. Tree Physiol. 11, 325–339. doi: 10.1093/treephys/11.4.325
Bradshaw, H. D., Villar, M., Watson, B. D., Otto, K. G., Stewart, S., Stettler, R. F. (1994). Molecular genetics of growth and development in Populus. III. A genetic linkage map of a hybrid poplar composed of RFLP, STS, and RAPD markers. Theor. Appl. Genet. 89, 167–178. doi: 10.1007/BF00225137
Busing, F. M. T. A., Meijer, E., Leeden, R. V. D. (1999). Delete-m jackknife for unequal m. Stat. Comput. 9, 3–8. doi: 10.1023/A:1008800423698
Carroll, A. R., Yunhai, L., Graham, S. A., Xuchang, X., Hendrix, M. S., Jinchi, C., et al. (1990). Junggar basin, northwest China: trapped Late Paleozoic ocean. Tectonophysics 181, 1–14. doi: 10.1016/0040-1951(90)90004-R
Castiglione, S., Wang, G., Damiani, G., Bandi, C., Bisoffi, S., Sala, F. (1993). RAPD fingerprints for identification and for taxonomic studies of elite poplar (Populus spp.) clones. Theor. Appl. Genet. 87, 54–59. doi: 10.1007/BF00223744
Cervera, M. T., Storme, V., Soto, A., Ivens, B., Van Montagu, M., Rajora, O. P., et al. (2005). Intraspecific and interspecific genetic and phylogenetic relationships in the genus Populus based on AFLP markers. Theor. Appl. Genet. 111, 1440–1456. doi: 10.1007/s00122-005-0076-2
Chen, T., Chen, X., Zhang, S., Zhu, J., Tang, B., Wang, A., et al. (2021). The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinf. 19, 578–583. doi: 10.1016/j.gpb.2021.08.001
Chen, X., Lu, H., Shu, L., Wang, H., Zhang, G. (2002). Study on tectonic evolution of junggar basin. Geol. J. China Univ. 8, 257.
Chen, L., Qiu, Q., Jiang, Y., Wang, K., Lin, Z., Li, Z., et al. (2019). Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science 364, eaav6202. doi: 10.1126/science.aav6202
CNCB-NGDC Members and Partners, Bai, X., Bao, Y., Bei, S., Bu, C., Cao, R., et al. (2024). Database resources of the national genomics data center, China national center for bioinformation in 2024. Nucleic Acids Res. 52, D18–D32. doi: 10.1093/nar/gkad1078
Coyne, J. A., Orr, H. A., Futuyma, D. J. (1988). Do we need a new species concept? Syst. Zool. 37, 190–200. doi: 10.2307/2992276
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinformatics 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Deschenes, L. A. (2000). Origin 6.0: Scientific Data Analysis and Graphing Software Origin Lab Corporation (formerly Microcal Software, Inc.). Web site: www.originlab.com. Commercial price: $595. Academic price: $446. J. Am. Chem. Soc 122, 9567–9568. doi: 10.1021/ja004761d
Donoghue, M. J. (1985). A critique of the biological species concept and recommendations for a phylogenetic alternative. Bryol. 88, 172–181. doi: 10.2307/3243026
Doyle, J. J. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Available online at: https://api.semanticscholar.org/CorpusID:208526466 (Accessed April 24, 2024).
Du, W., Wang, Y., Xie, D., Li, E., Bai, Y., Shang, C., et al. (2024). Phylogenomics reveal Populus gonggaensis as a hybrid between P. lasiocarpa and P. cathayana (Salicaceae). PhytoKeys 237, 161–177. doi: 10.3897/phytokeys.237.103012
Durand, E. Y., Patterson, N., Reich, D., Slatkin, M. (2011). Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252. doi: 10.1093/molbev/msr048
Eckenwalder, J. E. (1977). Systematics of Populus L. (Salicaceae) in Southwestern North America with special reference to sect. Aigeiros Duby (Calif: Calif. Univ).
Eckenwalder, J. E. (1996). Taxonomic signal and noise in multivariate interpopulational relationships in populus mexicana (Salicaceae). Syst. Bot. 21, 261–271. doi: 10.2307/2419658
Fang, Z. F., Zhao, S. D., Skvortsov, A. K. (1999). “Populus,” in Flora of China. Eds. Wu, Z. Y., Raven, P. H. (Missouri Botanical Garden, Beijing).
Feng, J., Jiang, D., Shang, H., Dong, M., Wang, G., He, X., et al. (2013). Barcoding poplars (Populus L.) from western China. PloS One 8, e71710. doi: 10.1371/journal.pone.0071710
Feng, X., Liu, J., Gong, X. (2016). Species delimitation of the cycas segmentifida complex (Cycadaceae) resolved by phylogenetic and distance analyses of molecular data. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00134
Gompert, Z., Buerkle, C. A. (2016). What, if anything, are hybrids: enduring truths and challenges associated with population structure and gene flow. Evol. Appl. 9, 909–923. doi: 10.1111/eva.12380
Gong, H., Yang, M., Wang, C., Tian, C. (2023). Leaf phenotypic variation and its response to environmental factors in natural populations of Eucommia ulmoides. BMC Plant Biol. 23, 562. doi: 10.1186/s12870-023-04583-3
Guindon, S., Dufayard, J., Lefort, V., Anisimova, M., Hordijk, W., Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of phyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Han, X., Wang, Y., Liu, Y. (2017). The complete chloroplast genome sequence of Populus wilsonii and its phylogenetic analysis. Mitochon. DNA Part B 2, 932–933. doi: 10.1080/23802359.2017.1413291
Hijmans, R. J., Cameron, S. E., Parra, J. L., Jones, P. G., Jarvis, A. (2005). Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 25, 1965–1978. doi: 10.1002/joc.1276
Hu, H., Zeng, T., Wang, Z., Al-Shehbaz, I. A., Liu, J. (2018). Species delimitation in the Orychophragmus violaceus species complex (Brassicaceae) based on morphological distinction and reproductive isolation. Bot. J. Linn. Soc 188, 257–268. doi: 10.1093/botlinnean/boy056
Jiang, D., Feng, J., Dong, M., Wu, G., Mao, K., Liu, J. (2016). Genetic origin and composition of a natural hybrid poplar Populus × jrtyschensis from two distantly related species. BMC Plant Biol. 16, 89. doi: 10.1186/s12870-016-0776-6
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Lexer, C., Joseph, J. A., van Loo, M., Barbará, T., Heinze, B., Bartha, D., et al. (2010). Genomic admixture analysis in European populus spp. Reveals unexpected patterns of reproductive isolation and mating. Genetics 186, 699–712. doi: 10.1534/genetics.110.118828
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, Y., Wen, J., Ren, Y., Zhang, J. (2019). From seven to three: Integrative species delimitation supports major reduction in species number in Rhodiola section Trifida (Crassulaceae) on the Qinghai-Tibetan Plateau. TAXON 68, 268–279. doi: 10.1002/tax.12052
Liu, J. (2015). DNA barcodes, speciation and taxonomy in plants. Biodivers. Sci. 23, 283. doi: 10.17520/biods.2015066
Liu, J. (2016). The integrative species concept” and “species on the speciation way. Biodivers. Sci. 24, 1004. doi: 10.17520/biods.2016222
Liu, Y., Li, D., Zhang, Q., Song, C., Zhong, C., Zhang, X., et al. (2017). Rapid radiations of both kiwifruit hybrid lineages and their parents shed light on a two-layer mode of species diversification. New Phytol. 215, 877–890. doi: 10.1111/nph.14607
Lu, Z., Sun, Y., Li, Y., Yang, Y., Wang, G., Liu, J. (2021). Species delimitation and hybridization history of a hazel species complex. Ann. Bot. 127, 875–886. doi: 10.1093/aob/mcab015
Mętrak, M., Chachulski, Ł., Navruzshoev, D., Pawlikowski, P., Rojan, E., Sulwiński, M., et al. (2017). Nature’s patchwork: How water sources and soil salinity determine the distribution and structure of halophytic plant communities in arid environments of the Eastern Pamir. PloS One 12, e0174496. doi: 10.1371/journal.pone.0174496
Mętrak, M., Chachulski, Ł., Pawlikowski, P., Rojan, E., Sulwiński, M., Suska-Malawska, M. (2023). Potential role of high-altitude wetlands in preservation of plant biodiversity under changing climatic conditions in the arid Eastern Pamir. CATENA 220, 106704. doi: 10.1016/j.catena.2022.106704
Malinsky, M., Matschiner, M., Svardal, H. (2021). Dsuite - Fast D -statistics and related admixture evidence from VCF files. Mol. Ecol. Resour. 21, 584–595. doi: 10.1111/1755-0998.13265
Martin, S. H., Davey, J. W., Jiggins, C. D. (2015). Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol. Biol. Evol. 32, 244–257. doi: 10.1093/molbev/msu269
Maya-Lastra, C. A., Sweeney, P. W., Eaton, D. A. R., Torrez, V., Maldonado, C., Ore-Rengifo, M. I., et al. (2024). Caught in the act: incipient speciation at the southern limit of viburnum in the central Andes. Syst. Biol. 73, 629–643. doi: 10.1093/sysbio/syae023
McKenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Metrak, M., Sulwinski, M., Chachulski, L., Wilk, M., Suska-Malawska, M. (2015). “Creeping Environmental Problems in the Pamir Mountains: Landscape Conditions, Climate Change, Wise Use and Threats,” in Climate Change Impacts on High-Altitude Ecosystems. Eds. Öztürk, M., Hakeem, K. R., Faridah-Hanum, I., Efe, R. (Springer International Publishing, Cham), 665–694. doi: 10.1007/978-3-319-12859-7_28
Minh, B. Q., Nguyen, M. A. T., Von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Nguyen, L., Schmidt, H. A., von Haeseler, A., Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Perrino, E. V., Tomaselli, V., Wagensommer, R. P., Silletti, G. N., Esposito, A., Stinca, A. (2022). Ophioglossum lusitanicum L.: new records of plant community and 92/43/EEC habitat in Italy. Agronomy 12, 3188. doi: 10.3390/agronomy12123188
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Sang, Y., Long, Z., Dan, X., Feng, J., Shi, T., Jia, C., et al. (2022). Genomic insights into local adaptation and future climate-induced vulnerability of a keystone forest tree in East Asia. Nat. Commun. 13, 6541. doi: 10.1038/s41467-022-34206-8
Sassone, A. B., Arroyo, M. T. K., Arroyo-Leuenberger, S. C., García, N., Román, M. J. (2021). One species with a disjunct distribution or two with convergent evolution? Taxonomy of two South American garlics. TAXON 70, 842–853. doi: 10.1002/tax.12500
Schneider, C. A., Rasband, W. S., Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Sites, J. W., Jr., Crandall, K. A. (1997). Testing species boundaries in biodiversity studies. Conserv. Biol. 11, 1289–1297. doi: 10.1046/j.1523-1739.1997.96254.x
Skvortsov, A. K. (1972). Three new species of flowering plants in the flora of the U.S.S.R. and new specific names. Bot. Zhurnal Leningr. 57, 493–494.
Skvortsov, A. K. (2009). The genus populus L. (Salicaceae) of Indian Himilaya. Nov. Sist Vyssh Rast 40, 52–67. doi: 10.31111/novitates/2009.40.52
Skvortsov, A. K. (2010). Taxonomical synopsis of the genus Populus L. @ in East Europe, North and Central Asia. Byulleten Gl. Bot. Sada 196, 62–73.
Skvortsov, A. K. (2011). Zametka o dvukh sredneaziatskich topolyakh. Byulleten Gl. Bot. Sada 195, 32–37.
Takuno, S., Kado, T., Sugino, R. P., Nakhleh, L., Innan, H. (2012). Population genomics in bacteria: A case study of staphylococcus aureus. Mol. Biol. Evol. 29, 797–809. doi: 10.1093/molbev/msr249
Villanueva, R. A. M., Chen, Z. J. (2019). ggplot2: elegant graphics for data analysis (2nd ed.). Meas. Interdiscip. Res. Perspect. 17, 160–167. doi: 10.1080/15366367.2019.1565254
Wan, X., Shi, Y., Huang, J., Mi, J., Zhong, Y., Zhang, F., et al. (2023). An overview and prospect on taxonomy of the genus populus. Bull. Bot. Res. 43, 161–168. doi: 10.7525/j.issn.1673-5102.2023.02.001
Wang, Z., Du, S., Dayanandan, S., Wang, D., Zeng, Y., Zhang, J. (2014). Phylogeny reconstruction and hybrid analysis of populus (Salicaceae) based on nucleotide sequences of multiple single-copy nuclear genes and plastid fragments. PloS One 9, e103645. doi: 10.1371/journal.pone.0103645
Wang, Z., Fang, Z. (1984). Flora Reipublicae Popularis Sinicae Vol. 20 (China: Science Publishing & Media Ltd. (China: CSPM), 381.
Wang, T., Hong, D., Jahn, B., Tong, Y., Wang, Y., Han, B., et al. (2006). Timing, petrogenesis, and setting of paleozoic synorogenic intrusions from the Altai mountains, Northwest China: implications for the tectonic evolution of an accretionary orogen. J. Geol. 114, 735–751. doi: 10.1086/507617
Wang, Y., Huang, J., Li, E., Xu, S., Zhan, Z., Zhang, X., et al. (2022). Phylogenomics and biogeography of populus based on comprehensive sampling reveal deep-level relationships and multiple intercontinental dispersals. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.813177
Wang, M., Zhang, L., Zhang, Z., Li, M., Wang, D., Zhang, X., et al. (2020). Phylogenomics of the genus Populus reveals extensive interspecific gene flow and balancing selection. New Phytol. 225, 1370–1382. doi: 10.1111/nph.16215
Warren, D. L., Beaumont, L. J., Dinnage, R., Baumgartner, J. B. (2019). New methods for measuring ENM breadth and overlap in environmental space. Ecography 42, 444–446. doi: 10.1111/ecog.03900
Yang, C. (1993). “Populus,” in Flora Xinjiangensis. Ed. Yang, C. Y. (Xinjiang Science and Technology and Hygiene Publishing House, Urumchi), 122–158. Available online at: https://elkitab.org/wp-content/uploads/2021/01/E2717B0E6625E0736854DFEA4BF8C033.pdf (Accessed January 22, 2025).
Yang, Z., Rannala, B. (2010). Bayesian species delimitation using multilocus sequence data. Proc. Natl. Acad. Sci. 107, 9264–9269. doi: 10.1073/pnas.0913022107
Yang, Y., Yang, X., Wang, M., Zhong, L., Ma, R., Ma, T., et al. (2022). Species delimitation of North American Nyssa species. J. Syst. Evol. 60, 747–758. doi: 10.1111/jse.12725
Zhang, W., Dasmahapatra, K. K., Mallet, J., Moreira, G. R. P., Kronforst, M. R. (2016). Genome-wide introgression among distantly related Heliconius butterfly species. Genome Biol. 17, 25. doi: 10.1186/s13059-016-0889-0
Zhang, N., Ma, Y., Folk, R. A., Yu, J., Pan, Y., Gong, X. (2018b). Maintenance of species boundaries in three sympatric Ligularia (Senecioneae, Asteraceae) species. J. Integr. Plant Biol. 60, 986–999. doi: 10.1111/jipb.12674
Zhang, L., Wang, M., Ma, T., Liu, J. (2018a). Taxonomic status of Populus wulianensis and P. ningshanica (Salicaceae). PhytoKeys 108, 117–129. doi: 10.3897/phytokeys.108.25600
Zhao, R., Chu, X., He, Q., Liu, W., Zhu, Z. (2023). Leaf phenotypic variation in natural populations of Carpinus tschonoskii in China. J. For. Res. 34, 1591–1602. doi: 10.1007/s11676-022-01584-0
Keywords: Populus laurifolia, species complex, phylogenomics, species delimitation, integrative taxonomy
Citation: Wei X, Cui X, Yuan F, Zhou K, Zhou L, Zhao C, Guo S, Shang C and Zhang Z (2025) Species delimitation in the Populus laurifolia complex (Salicaceae) based on phylogenetic and morphometric evidence. Front. Plant Sci. 16:1518122. doi: 10.3389/fpls.2025.1518122
Received: 28 October 2024; Accepted: 09 January 2025;
Published: 06 February 2025.
Edited by:
Robert Philipp Wagensommer, Free University of Bozen-Bolzano, ItalyReviewed by:
Enrico Vito Perrino, University of Foggia, ItalyCopyright © 2025 Wei, Cui, Yuan, Zhou, Zhou, Zhao, Guo, Shang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ce Shang, Y2Vfc2hhbmdAYmpmdS5lZHUuY24=; Zhixiang Zhang, enh6aGFuZ0BiamZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.