
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 February 2025
Sec. Plant Abiotic Stress
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1506873
 Chune Peng1†
Chune Peng1† Wei Xu1†
Wei Xu1† Xipan Wang2†
Xipan Wang2† Fanxiao Meng2
Fanxiao Meng2 Yumeng Zhao3
Yumeng Zhao3 Qingbin Wang3
Qingbin Wang3 Xinkun Wang1
Xinkun Wang1 Rathna Silviya Lodi1
Rathna Silviya Lodi1 Xiaodan Dong1*
Xiaodan Dong1* Changxiang Zhu2*
Changxiang Zhu2* Lizeng Peng1*
Lizeng Peng1*Alginate oligosaccharides (AOSs), important plant immunity inducers, are widely used in agriculture because of their important role in the biological control of crop diseases. However, the mechanism by which AOSs induce plant resistance to pathogens is not clear. Here, we report AOS with a degree of polymerization of 2–5, which was obtained by a newly reported enzyme Aly2. AOS treatment exhibited high activity in enhancing resistance to Phytophthora infestans (P. infestans). AOS significantly induced reactive oxygen species (ROS) accumulation, calcium influx, stomata closure, and callose deposition. The salicylic acid (SA) synthesis-related gene and the defense-related genes were upregulated after AOS treatment. A transcriptome file generated from AOS-treated seedlings verified the SA pathway and suggested the presence of chitin elicitor receptor kinase (CERK). The subsequent results showed that AtCERK1 binds AOS tightly, suggesting that AtCERK1 is responsible for AOS recognition. This study laid a theoretical foundation for the broad application of AOS.
During growth, plants are affected by many kinds of stresses, which can alter many plant processes. Plant diseases cause immense annual losses in crop yield, posing a great threat to food production. Approximately 3 billion kg of chemical reagents are used worldwide annually (Hernández et al., 2013), but approximately 1% of pesticides are effectively used on plants (Bernardes et al., 2015). The extensive use of agrochemicals has caused pernicious pesticide residues and environmental and soil pollution (Qin et al., 2014; Rodriguez-Salus et al., 2016; Xue et al., 2006), decreased crop quality, and threatened human health (Al-Wabel et al., 2016; Duan et al., 2008; Lozowicka et al., 2015; Yadav et al., 2015). Therefore, new strategies are urgently required to improve plants’ immunity and enhance their resistance to pathogens.
Plant immunity inducers are biological agents that can reduce the use of chemical pesticides while improving resistance to pathogens (Li et al., 2018a; Nisa et al., 2015). In addition to inducing the deposition of lignin, strengthening plant cell walls, and forming a physical barrier, such as callose that are deposited around the cell wall and plasmodesmata (Ton et al., 2009), to resist pathogen infection, plant immunity inducers also promote the production of endogenous substances such as pathogenesis-related proteins (PRs), reactive oxygen species (ROS), and salicylic acid (SA) (Cai et al., 2020; Jiang et al., 2023; Peng et al., 2020; Sticher et al., 1997). The non-expressor of PR (NPR) is a regulatory protein, and NPR1 plays a key role in this process (Durrant and Dong, 2004; Sun et al., 2018). PR-1A and NPR are the most abundant proteins produced by plants in response to pathogens via the SA pathway (Breen et al., 2017). When plant cells detect the presence of pathogens through recognition receptors on their surfaces, the phytohormone, such as the SA signaling pathway is activated, leading to the upregulation of the expression of the defense genes PR-1a and NPR1. This, in turn, induces resistance to pathogens.
Protein kinases on the cell surface are critical for transiting signals from the outside to the inside of the cell. From extracellular stimulation to the corresponding biological effect in cells, mitogen-activated protein kinase (MAPK) cascades must be activated. The MAPK pathway also affects plant resistance to pathogens by regulating stomatal closure. Oligosaccharides, such as mannan oligosaccharides (MOSs) and oligogalacturonic acid (OGA), also cause Ca2+ concentration changes and stomatal closure. The transient increase in Ca2+ and the MAPK cascade are important signaling pathways that stimulate the early defense response in plants (Lecourieux et al., 2006). NtMEK2 in the MAPK cascade pathway can be activated by multiple activators in tobacco and regulate phenylalanine lyase (PAL) expression, a key enzyme in SA synthesis (Yang et al., 2001). In addition, SA is an important hormone-signaling molecule that leads to plant systemic-acquired resistance.
Oligosaccharides are important plant immunity inducers that can be developed as a biostimulant. As a biological immunity inducer, AOS can be prepared through the enzymatic degradation of alginate and possesses advantages, such as low molecular weight, good water solubility, easy absorption, and non-polluting. AOS application in agriculture has become a popular research topic (Peng et al., 2018). Increasing evidence confirms the function of AOS in enhancing plant stress resistance, growth, and development (Hien et al., 2000; Hu et al., 2004; Iwasaki and Matsubara, 2000; Ma et al., 2010; Natsume et al., 1994; Tang et al., 2011). Studies have shown that AOS can promote root development and elongation in Komatsu (Yonemoto et al., 1993), barley (Tomoda et al., 1994), rice (Zhang et al., 2014), and carrot (Xing et al., 2020; Xu et al., 2003); reduce the damage caused by salt stress (Liu et al., 2013; Tang et al., 2011); enhance the tolerance of cucumber to water stress (Li et al., 2018b); enhance drought resistance in wheat through the SA pathway (Liu et al., 2013); and enhance the resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 through the salicylic acid pathway (Zhang et al., 2019). However, relatively few studies have investigated the mechanism by which AOS induces plant resistance to pathogens, especially late blight of potato and tomato.
Oligosaccharides mimic the cell wall components of pathogens and are recognized by immune receptors/pattern recognition receptors on the plant cell surface to trigger pattern-triggered immunity (PTI), thereby enhancing plant resistance to disease (Jones and Dangl, 2006; Van Wees et al., 2008). Thus, lipopolysaccharide, chitin, OGA, and MOS (Zhang et al., 2019) can be recognized by plant receptors to stimulate PTI (Denoux et al., 2008; Hayafune et al., 2014; Yin et al., 2016). These elicitors are also known as pathogen-associated molecular patterns (PAMPs) and/or damage-associated molecular patterns (DAMPs) (Wiesel et al., 2014). Progress has been made in the studies of oligosaccharide receptors, such as chitin receptors (Espinoza et al., 2017; Liu et al., 2012) and OGA (Brutus et al., 2010). However, the receptor for AOS in plants has not been identified.
This study showed that AOS improved potato and tobacco resistance to P. infestans, and induced a series of defense responses in tobacco, including ROS accumulation, callose deposition, Ca2+ influx, and stomatal closure. Moreover, transcriptome sequencing was performed, and SA signal pathway-related genes were also detected. The receptor mutant Arabidopsis thaliana and enzyme-linked immunosorbent assays (ELISAs) were used to analyze the interaction between AOS, and the receptor was also analyzed. AOS can interact tightly with the cell surface receptor AtCERK1, while the binding ability was not significant between AOS and chitin elicitor binding protein (AtCEBiP), suggesting that AtCERK1 is the receptor of AOS in Arabidopsis. The results lay the foundation for the wide application of AOS as a new biopesticide.
Sodium alginate was degraded by Aly2 for 12 h, and then AOS with a degree of polymerization (DP) of 2–5 was obtained (Peng et al., 2018) by using a Superdex 30 Increase 10/300 GL column, the mobile phase was 0.20 M NH4HCO3 at a flow rate of 0.4 mL/min, and the eluted fractions were monitored at 232 nm using a UV detector. In this study, wild-type Nicotiana benthamiana and wild-type A. thaliana were preserved and propagated. The plants Solanum tuberosum and N. benthamiana were grown at 22°C and 25°C, respectively, under 70% humidity with 16 h of light and 8 h of dark. Arabidopsis thaliana receptor mutants were cultured in an incubator (22°C, 16 h light/8 h dark). RNA extraction kits were purchased from Kangwei Reagent (Taizhou, China), reverse transcription kits were purchased from Hunan Aceri Bioengineering Co. (Changsha, China) and Novozymes (Beijing, China), and HRP-conjugated His6 was purchased from Sigma Aldrich (St. Louis, MO, USA). Biotin hydrazide, 2-(N-morpholino)-ethanesulfonic acid (MES), and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Escherichia coli strains DH5α and BL21 (DE3) and Pichia pastoris were preserved in our laboratory. Phytophthora infestans strain was grown in the dark at 18°C using rye A agar. The T-DNA insertion mutants SALK_007193C for At3g21630 (AtCERK1) and SALK_206271C for At2g17120 (AtCEBiP-LIKE1) were obtained from EDITGENE Corporation (Guangzhou, China). All the other chemicals and reagents were of the highest quality. Primers were synthesized by Shanghai Shenggong Biotechnology Co. (Shanghai, China; Table 1).
Plates with P. infestans were flooded with 5 mL of ddH2O and scraped to release sporangia. The suspension was poured into a clean Petri dish, placed on ice, and stored at 4°C for 3 h to release zoospores. Then, the sporangia were counted and adjusted to 30,000 sporangia per milliliter. The potato leaves were inoculated with P. infestans (originally isolated from the province of Heilongjiang, north of China) at a concentration of 4 × 105 sporangia mL−1. Droplets of 20 µL of P. infestans zoospores and sporangia suspension were added to the leaves of wild-type N. benthamiana, wild Arabidopsis, and Arabidopsis receptor mutants after AOS (100 μg/mL) treatment for 24 h. The leaves were placed in a plastic dish with ddH2O sprayed over them regularly. Phytophthora infestans infection was observed at 3 days post-inoculation (dpi), and P. infestans colonization was measured by quantitative real-time PCR. Briefly, total DNA was extracted from diseased leaves, including AOS-treated and untreated leaves, and then quantitative PCR was performed after DNA extraction by using the internal reference gene primers in Table 1 (Nb-actin and P. infestans-O8 primer pairs). The expression levels of the internal reference gene of tobacco and P. infestans were measured to evaluate P. infestans accumulation. The primer sequences for the experiment are presented in Table 1.
The optimal working concentration of AOS was determined by spraying 0, 25, 50, 100, or 200 μg/mL of gradient AOS aqueous solution on potato Désirée/Eshu 3 leaves for 24 h, followed by inoculation with P. infestans; the other conditions were the same as those described above. There were three biological replicates for each treated sample.
Histochemical staining was performed using 3,3'-diaminobenzidinebutane (DAB) and nitroblue tetrazolium (NBT) to detect hydrogen peroxide (H2O2) and superoxide ion (O2−) accumulation, respectively, in the leaves of plants subjected to AOS and H2O treatment. Plant tissues were placed in 1 mg/mL of DAB solution, vacuum-infiltrated for 30 min, washed three times with deionized water, and reacted with H2O2 for 12–24 h under light at 28°C. The excess dye solution was washed away by using a boiled solution (ethanol:lactic acid:glycerin = 3:1:1) at 100°C for 10 min, and the leaves were imaged. For NBT staining, N. benthamiana leaves were immersed in 1% (M/V) sodium azide solution, which increases the permeability of cells, vacuum-immersed for 30 min, and then transferred to 0.5 mg/mL of NBT solution, followed by vacuum infiltration for 30 min. O2− reacted with NBT to form a deep blue insoluble complex. The boiled solution (ethanol:lactic acid:glycerin = 3:1:1) was also used to wash off the excess dye solution, after which the leaves were imaged. The strengths of H2O2 and O2− were quantitatively analyzed by ImageJ software.
For aniline blue staining, 20 mL of lactic acid, 20 mL of phenol, 40 mL of 20% glycerol, and 20 mL of deionized water were mixed evenly; the volume was adjusted to 100 mL; and then anhydrous ethanol was added at a volume ratio of ~2:1. Nicotiana benthamiana leaves were put into the above solution under vacuum for 30 min and then treated at 60°C for 30 min. The 0.01% aniline blue solution was added after washing with deionized water, and the leaves were kept at room temperature overnight without light. Finally, the leaves were preserved in 50% glycerol and were observed and photographed under a fluorescence microscope.
Nicotiana benthamiana leaf epidermis strips were soaked in MES, pH 6.0 buffer under light for 3 h to open the stomata, and then a final concentration of 20 µmol/L of Fluo-3AM was added at 4°C for 2.5 h. The excess fluorescent dye was washed by using the MES buffer and then kept at room temperature for 1 h. The epidermis strips were treated with ddH2O and 100 μg/mL of AOS for 24 h, and then fluorescence was observed using a confocal microscope. Fluo-3AM was used to analyze Ca2+ accumulation in the guard cells. Each treatment included an investigation of at least three epidermis strips, and the experiment was repeated three times. Images of the stomatal aperture were captured with an Olympus BX43 microscope (Olympus, Tokyo, Japan) using the cellSens Standard software, and the diameters of 50 randomly selected stomata were measured. Each assay was repeated three times.
Total RNA was extracted from tobacco leaves with TRIzol reagent (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. The cDNA was synthesized from 1 μg of total RNA using a FastKing gDNA Dispelling RT SuperMix kit (Tianjin, Beijing, China). Quantitative real-time PCR (qRT-PCR) was performed by using a Talent SYBR Green Kit (Tianjin, Beijing, China). Each reaction was conducted in triplicate and repeated three times. Bio-Rad CFX Manager software (Bio-Rad, California, USA) was used to analyze the data. The relative expression levels of the ROS-scavenging enzymes catalase (CAT), superoxide dismutase (SOD), ascorbate peroxidase (APX), and ROS-generating-related genes, including the respiratory burst oxidase homolog genes (RbohA and RbohB), were measured by qRT-PCR after AOS treatment for 0, 2, 4, 8, 12, and 24 h, respectively. Primer sequences for the experiment are presented in Table 1.
The transcriptome was sequenced by Shanghai OE Biotech Co., Ltd. (Shanghai, China). RNA samples were taken from five- or six-leaf-stage leaves treated with H2O (0 h) or 100 μg/mL of AOS for 24 h. Each sample was analyzed three times. The samples were selected depending on quality (RIN score ≥ 7). All differential gene expression data were based on the following criteria: an absolute log2 ratio ≥1 and an FDR ≤0.001.
The two PCR products of the AtCERK1 gene and the AtCEBiP-LIKE1 gene were individually cloned into the pET-30a (+) vector and SacI vector. The proteins AtCERK1 (extracellular domain, At3g21630) and AtCEBiP-LIKE1 (At2g17120) were expressed by using BL21(DE3) and yeast with a His6 tag at the C-terminus, respectively. For AtCERK1 expression, E. coli cells harboring the recombinant plasmid were initially cultured in LB broth. When the cell density reached an OD600 of 0.8–1.0, the broth was supplemented with the inducer isopropyl 1-thio-β-D-galactopyranoside at a final concentration of 0.05 mM to initiate the expression of AtCERK1. AtCEBiP-LIKE1 was expressed in a similar manner, and yeast cells harboring AtCEBiP-LIKE1 were cultured in BMGY broth and then in BMMY broth. The broth was supplemented with 1% methanol to induce AtCEBiP-LIKE1 expression. Then, the proteins were purified by Ni2+ chelation chromatography according to the method provided by Peng et al. (2018).
Molecular interactions were analyzed by using biotin-labeled AOS and CERK1/CEBiP-LIKE1 based on the ELISA method. AOS (8 mg/mL) was biotinylated in 0.1 M of MES (Sigma-Aldrich) (pH 5.5) biotin LC-hydrazide solution (Deepa et al., 2002; Peng et al., 2021). EDC (1 mg) was added to the reaction mixtures and reacted overnight at room temperature, and each reaction mixture was desalted three times with PBS and centrifuged (4°C, 2,000 rpm, 2 min) to obtain the biotin-labeled AOS. Then, 50 μL of 1 mg/mL streptavidin was added to a 96-well plate and sealed at 4°C overnight, 1% BSA was added for 1 h, and biotin-labeled AOS was added at room temperature for 2 h. The binding reaction of chitin-binding proteins (AtCERK1 and AtCEBiP-LIKE1) was carried out at 4°C overnight, and the above process was avoided from light and washed with PBS. Then, the antibody was added for 30 min, the sample was washed with PBST, TMB was used to develop the color, and the absorbance was measured at 450 nm.
The potato cultivar Désirée and P. infestans were used as materials to examine the activity of AOS with a DP of 2–5 (Figure 1A) against late blight. The potato leaves were inoculated with P. infestans after being sprayed with different concentrations of AOS for 24 h at 0, 25, 50, 100, and 200 μg/mL. Images were photographed at 4 dpi, and AOS could significantly enhance the resistance of potato to late blight. The infected area and disease index was gradually decreasing with increasing AOS concentration, but it was greater when the concentration was increased to 200 μg/mL (Figures 1B, C). We hence used 100 μg/mL of AOS in the next experiment.
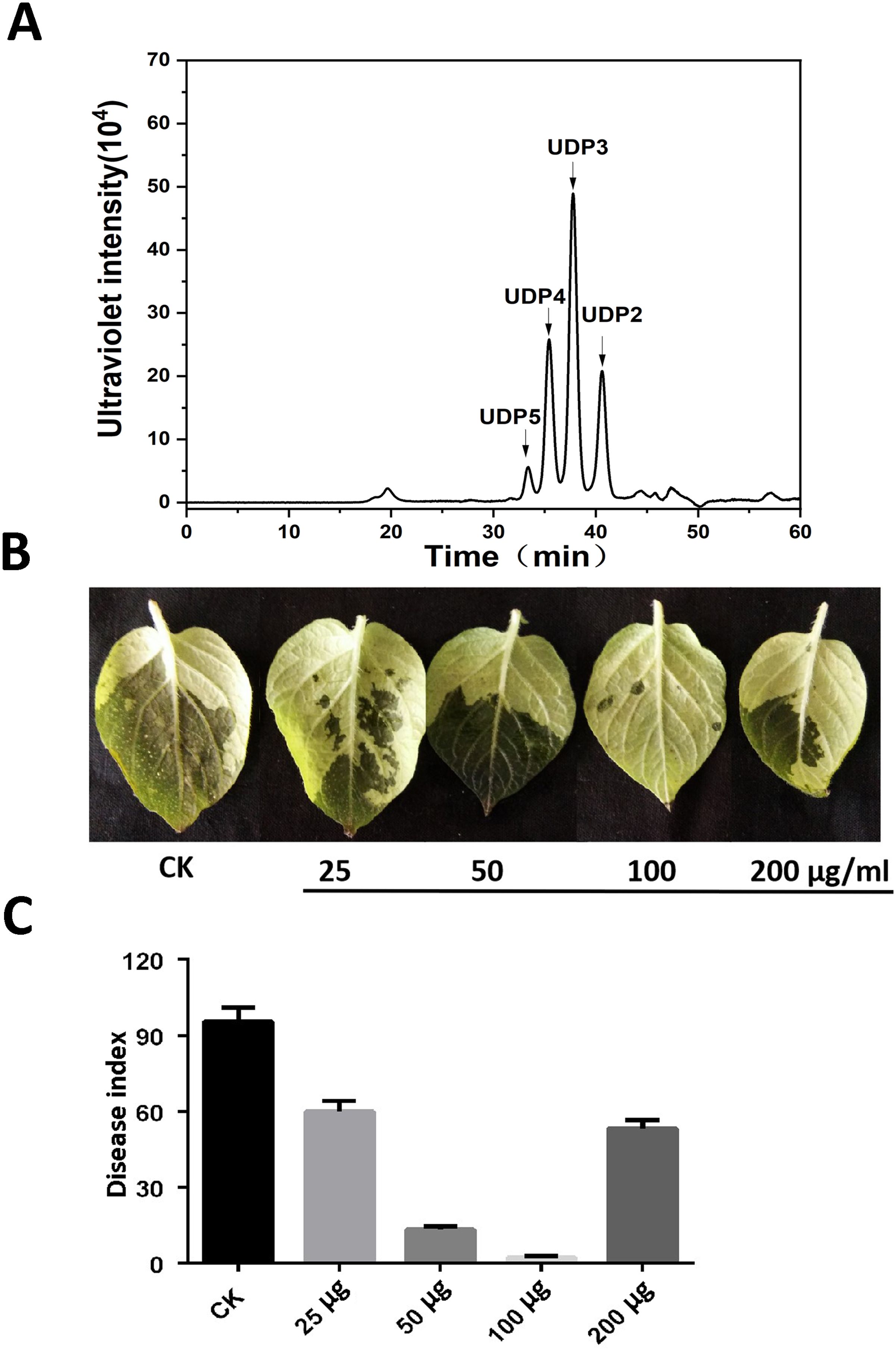
Figure 1. AOS treatment enhanced potato resistance against Phytophthora infestans. (A) Approximately 50 μg of AOS with a degree of polymerization (DP) of 2–5 was loaded on a Superdex 30 Increase 10/300 GL column. The elution positions of the unsaturated oligosaccharide product fractions with different degrees of polymerization are indicated by arrows: UDP2, unsaturated disaccharide; UDP3, unsaturated trisaccharide; UDP4, unsaturated tetrasaccharide; UDP5, unsaturated pentasaccharide. (B) Wild-type Désirée was inoculated with P. infestans after a gradient concentration (0, 25, 50, 100, and 200 μg/mL) of AOS treatment for 24 h, and the phenotype was observed at 3 dpi. (C) The disease index of P. infestans at 3 dpi. Error bars show the mean ± SD of three replicates (at least 20 plants per replicate).
To investigate whether AOS regulates ROS accumulation, DAB and NBT staining were applied to evaluate the H2O2 and O2− levels in tobacco leaves that were detached from the water-spraying group and the 100-μg/mL AOS-spraying group. Figures 2A, B show that DAB staining was first strengthened and then weakened with the extension of time. DAB staining was the deepest after 24 h of spraying AOS, which indicated that the accumulation of H2O2 was elevated. Similarly, NBT staining was the deepest after 24 h of spraying AOS, suggesting that AOS could also promote O2− accumulation in plants.
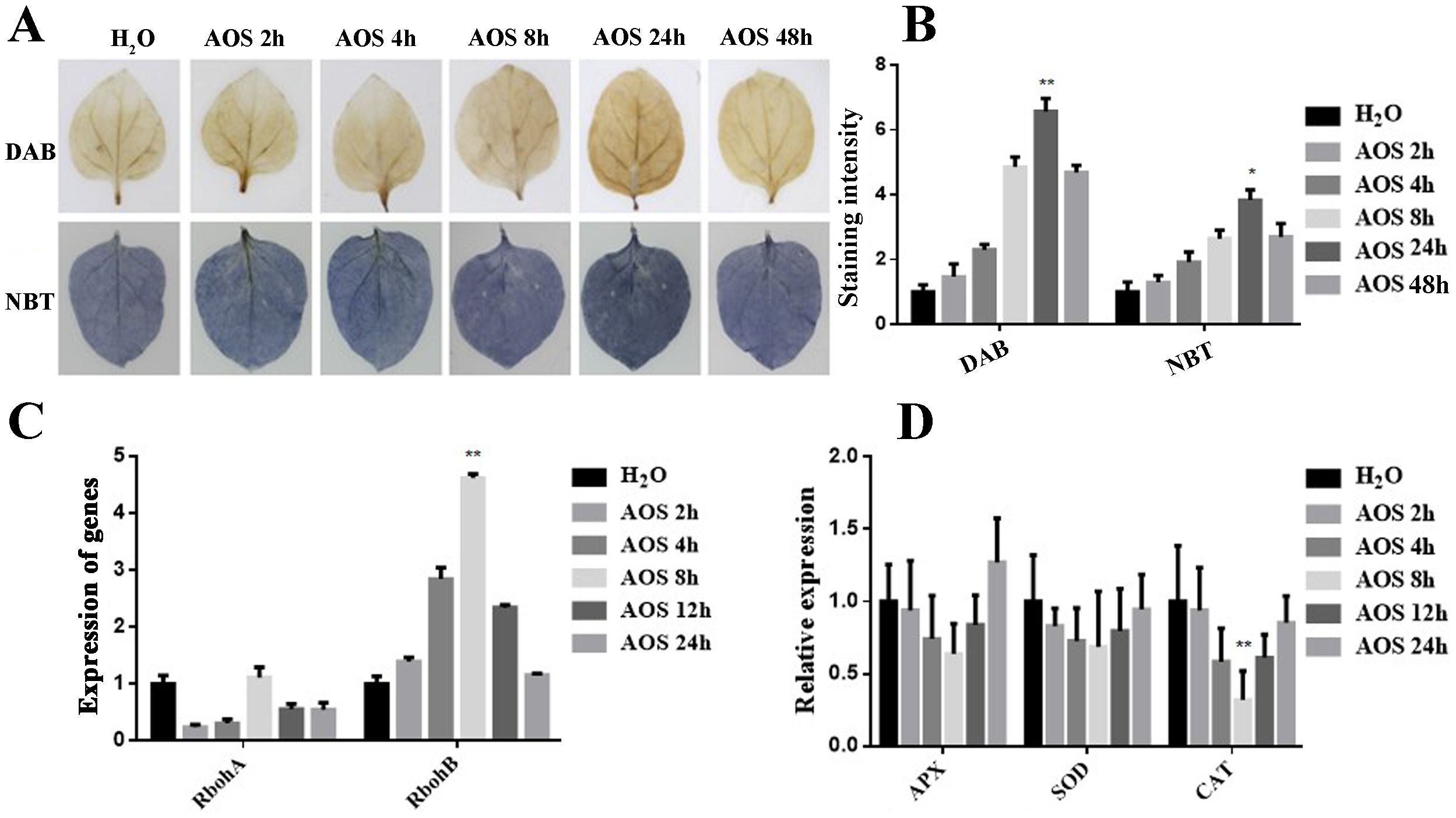
Figure 2. AOS promoted hydrogen peroxide and superoxide anion accumulation. AOS promoted hydrogen peroxide accumulation in Nicotiana benthamiana. (A) Hydrogen peroxide (up) and superoxide accumulation (down) were measured in the leaves treated with 100 μg/mL of AOS at different (0, 2, 4, 8, 24, 48) hours post-treatment (hpt) (n = 6). (B) Quantification of hydrogen peroxide and superoxide levels in N. benthamiana treated with 100 μg/mL of AOS at 0, 2, 4, 8, 12, and 24 hpt. Data are shown as the mean (n = 6) ± SD. (C) qRT-PCR analysis of RbohA and RbohB expression at various time intervals. (D) CAT, SOD, and APX were detected using qRT-PCR at various time intervals. Data are shown as the mean (n = 6) ± SD. * indicates significant differences determined using the Student’s t-test (p < 0.05), and ** indicates extremely significant differences determined using the Student’s t-test (p < 0.01).
The mRNA levels of several important genes encoding the ROS-scavenging enzymes CAT, SOD, APX, and ROS-generating-related genes, including the respiratory burst oxidase homolog genes (RbohA and RbohB), were determined by qRT-PCR analysis and monitored before and after AOS treatment. After AOS treatment, there was a significant increase in the expression level of the ROS generation-related gene RbohB (Figure 2C). Conversely, the expression level of the CAT gene decreased significantly, while there was no significant change in the expression levels of the SOD and APX genes (Figure 2D). These results suggest that AOS may enhance hydrogen peroxide accumulation by inhibiting CAT gene expression and promoting RbohB gene expression.
Ca2+ is an important secondary messenger that triggers plant defense response. Ca2+ usually stays at a low concentration in the plant cell cytoplasm, whereas biotic stresses, pathogen infection, and elicitor promote Ca2+ influx from the extracellular to the cytoplasm, thus leading to a rapid transient cytoplasmic Ca2+ increase (Zang et al., 2019). To investigate whether AOS leads to Ca2+ influx, the fluorescent-labeled Fluo-3AM was used to evaluate the cytoplasmic Ca2+ levels. ddH2O and COS were used as negative and positive controls, respectively (Zhang et al., 2009; Iriti and Varoni, 2015). There was no obvious fluorescence in the ddH2O-treated plant tissues, while the COS- and AOS-treated guard cells showed obvious fluorescence in the cells, indicating that AOS treatment significantly promoted Ca2+ influx in the guard cells. Moreover, AOS promoted stomatal closure, suggesting the prevention of pathogen infection (Figure 3).
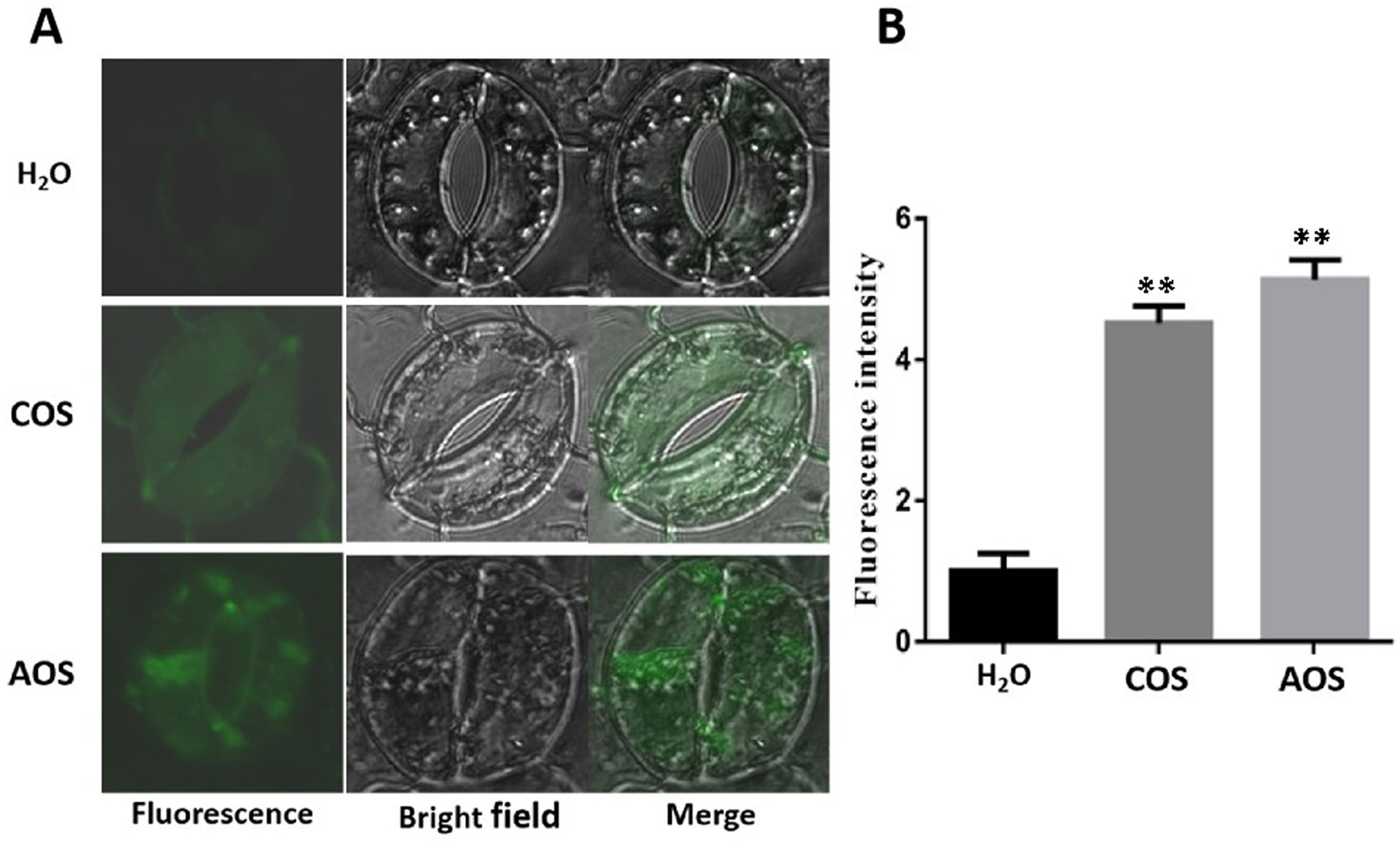
Figure 3. AOS promotes intracellular Ca2+ accumulation in the guard cells and stomatal closure of Nicotiana benthamiana. The calcium-specific fluorescence probe Fluo-3AM was preincubated with epidermal peels at 4°C and then kept at room temperature for 1h. The fluorescence was observed by a laser confocal microscope after incubation with H2O, COS (1,000 μg/mL), and AOS (100 μg/mL) for 3h. (A) Representative images (enlarged images). (B) Quantitative analysis of Ca2+ concentration by using the ZEN software. The experiments were repeated three times. Error bars indicate SEM. Statistics by the Student’s t-test (**p ≤ 0.01).
To ascertain the ability of AOS to induce callose deposition in plants, wild-type N. benthamiana was subjected to 100 μg/mL of AOS spray treatment, while H2O was used as a control. Subsequently, aniline blue staining was conducted after 24 h. Microscopic examination (Supplementary Figure S1A) of the stained samples revealed conspicuous callose deposition surrounding the veins of N. benthamiana treated with AOS compared to the control group. The fluorescence intensity (Supplementary Figure S1B) was quantified and found to be consistent with the observed phenotype. These results indicate that AOS can effectively enhance callose deposition in plants and consequently improve their resistance against pathogen infections.
Plants synthesize SA mainly by the isochorismate synthase (ICS) and phenylalanine ammonia lyase (PAL) pathways. Thus, we detected the gene expression level of the ICS and PAL genes by using qRT-PCR. The results showed that the PAL gene was upregulated by AOS, whereas the ICS gene basically remained unchanged (Figure 4A). These results suggested that AOS promotes SA synthesis by improving the PAL transcription level.
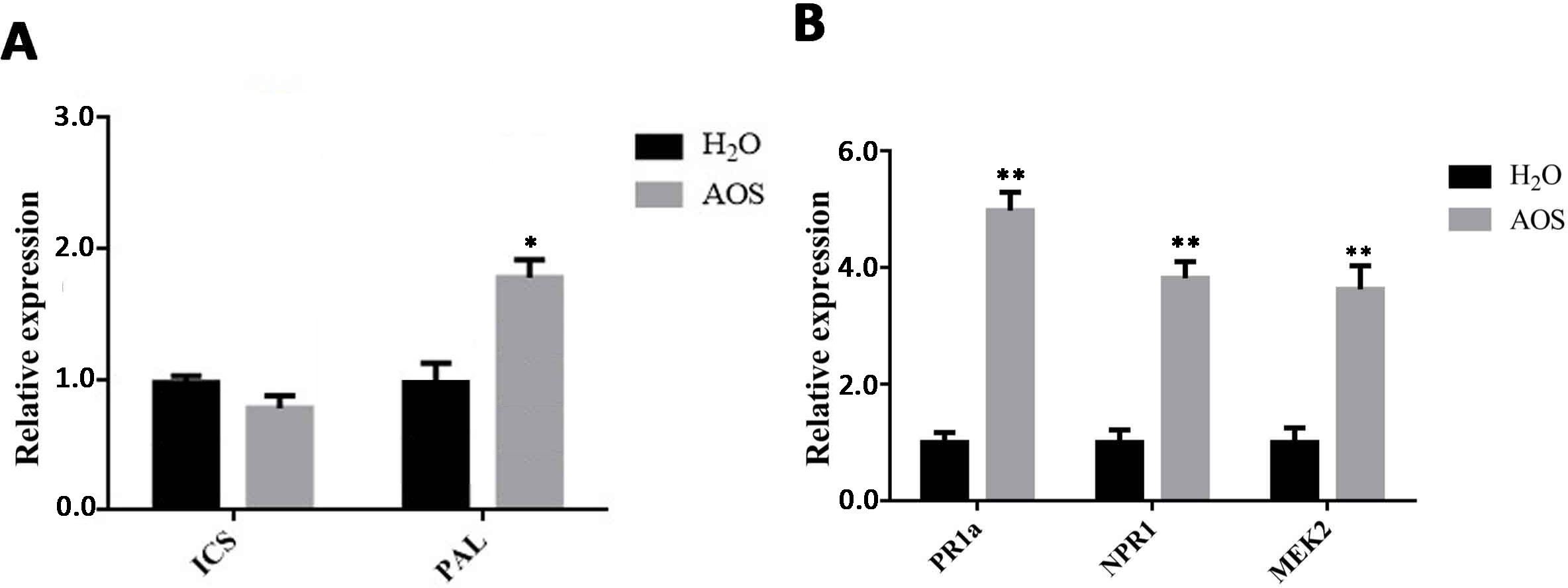
Figure 4. AOS promotes the accumulation of SA in Nicotiana benthamiana. (A) The SA biosynthesis-associated genes ICS1 and PAL were measured at 24 hpt using qRT-PCR (means ± SD, n ≥ 3). (B) The SA signaling-associated genes PR1a and NPR1 and the MEK2 gene were measured at 24 hpt using qRT-PCR (means ± SD, n ≥ 3). * indicates extremely significant differences determined using the Student’s t-test (p < 0.05); ** indicates extremely significant differences determined using the Student’s t-test (p < 0.01).
Pathogenesis-related (PR) proteins play an important role in plant defense. They can improve plant disease resistance by inhibiting pathogen reproduction and are mainly involved in plant-acquired systemic resistance. PR-1A and NPR are the key factors in the SA pathway, involved in N. benthamiana’s resistance to pathogens or other pathogens, such as Phytophthora (Zang et al., 2019). The expression levels of genes related to these signaling pathways were examined by qRT-PCR in N. benthamiana leaves after AOS treatment for 24 h. The expression levels of the PR1a and NPR1 genes, key genes in the SA pathway, were upregulated significantly (Figure 4B). This suggests that AOS can promote the expression of the NPR1 and PR1 proteins via the SA signaling pathway, enabling plants to acquire systemic resistance and enhance their resistance to late blight.
The MEK2 (MAPK kinase)-SIPK/WIPK cascade, an N. benthamiana mitogen-activated protein kinase (MAPK) cascade, is an essential signaling pathway for plant immunity and is involved in the hypersensitive response (HR) accompanied by cell death. Figure 4B shows that MEK2 genes were also upregulated significantly, suggesting that AOS may activate plant immunity through the MAPK cascade.
Transcriptome sequencing analysis was performed on the leaves treated with AOS to reveal the role of AOS in inducing plant resistance. The leaves were harvested after 24 h of 100 μg/mL AOS treatment. The differentially expressed genes (DEGs) after AOS and H2O treatment (AOS0H) were analyzed. The results revealed 2,595 DEGs in the AOS24H group vs. the AOS0H group, of which 1,219 genes were upregulated and 1,376 genes were downregulated (Figure 5A).
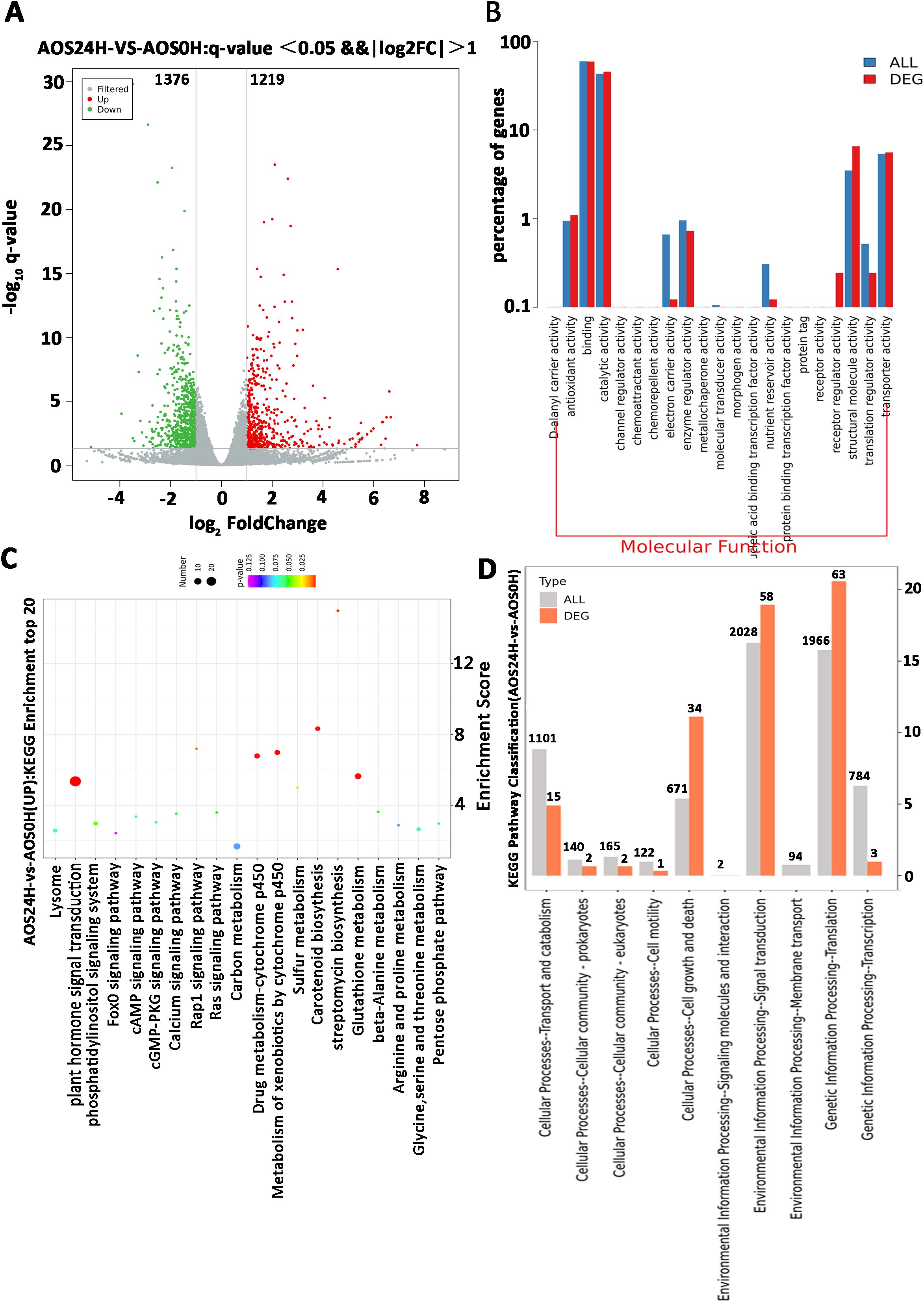
Figure 5. A parametric transcriptome analysis after AOS treatment in Nicotiana benthamiana. (A) Volcano plot showing the fold change and adjusted p-value of the normalized read counts of the transcriptome sequencing data. The criteria of log2| (fold change)≥1 and padj ≤0.05 were used to identify the DEGs. The green dots indicate the downregulated DEGs, and the red dots indicate the upregulated DEGs. (B) Comparative plots of the distribution of DEGs and all genes at the GO level 2. The horizontal axis is the name of the entry, and the vertical axis indicates the number of genes corresponding to the entry and their percentages. (C) The characteristic KEGG pathways with significant enrichment of DEGs after treatment with AOS. (D) Partial KEGG pathway classification. The abscissa axis is the ratio of DEGs in a pathway: all DEGs in the KEGG level 2 pathway (%); the ordinate axis is the name of the pathway. The numbers above the column represent the quantity of DEGs in the pathway.
Most of the AOS-regulated genes at 24 hpt were annotated with a wide range of Gene Ontology (GO) terms in the molecular functions (Figure 5B). Molecular function results suggested receptor activity and receptor regulator activity, and the transcriptome DEG showed that the CERK1 gene expression level was upregulated, consistent with the results in Supplementary Figure S2B. The enriched GO terms (Table 2) of the biological process category included positive regulation of defense response, chitinase activity, and signal transduction. Within the KEGG classification, plant hormone signal transduction genes showed the greatest changes in expression (Figure 5C), and 155 DEGs were upregulated (Figure 5D). These results further verified that resistance to P. infestans was induced by the SA signaling pathway. Plant chitinases are described as pathogen-associated proteins because they are induced in response to invasion by plant pathogens. The genes related to the chitin catabolic process and chitinase (Niben101Scf01789g03003, Niben101Scf02041g00002, and Niben101Scf02171g00007) were also significantly upregulated (Table 2). Based on the above results, we speculate that the receptors in the plants of AOS may be related to chitin elicitor receptor proteins.
To determine the AOS receptor protein in plants, several Arabidopsis receptor loss mutants, including the T-DNA insertion mutants for At3g21630 (AtCERK1) and At2g17120 (AtCEBiP-LIKE1), were inoculated with P. infestans. There was no significant difference in mortality in the Arabidopsis mutant group after AOS or ddH2O treatment, while AOS improved the resistance to P. infestans in the Columbia wild-type Arabidopsis group (Supplementary Figure S2A). The levels of the CEBiP-LIKE1 gene and the AtCERK1 gene were significantly increased after AOS treatment for 24 h (Supplementary Figure S2B).
To analyze the interaction between AOS and the receptor proteins, the receptors AtCERK1 (extracellular domain, At3g21630) and AtCEBiP-LIKE1 (At2g17120) were expressed and purified. SDS-PAGE showed that the molecular weights of AtCERK1 (with amino acids ranging from 26 to 230) with a His6 tag at the C-terminus and AtCEBiP-LIKE1 (with a secretory peptide at the N-terminus and a His6 tag at the C-terminus) were approximately 23 kDa (Figure 6A) and 47 kDa (Figure 6B), respectively, which are consistent with the theoretical molecular weights.
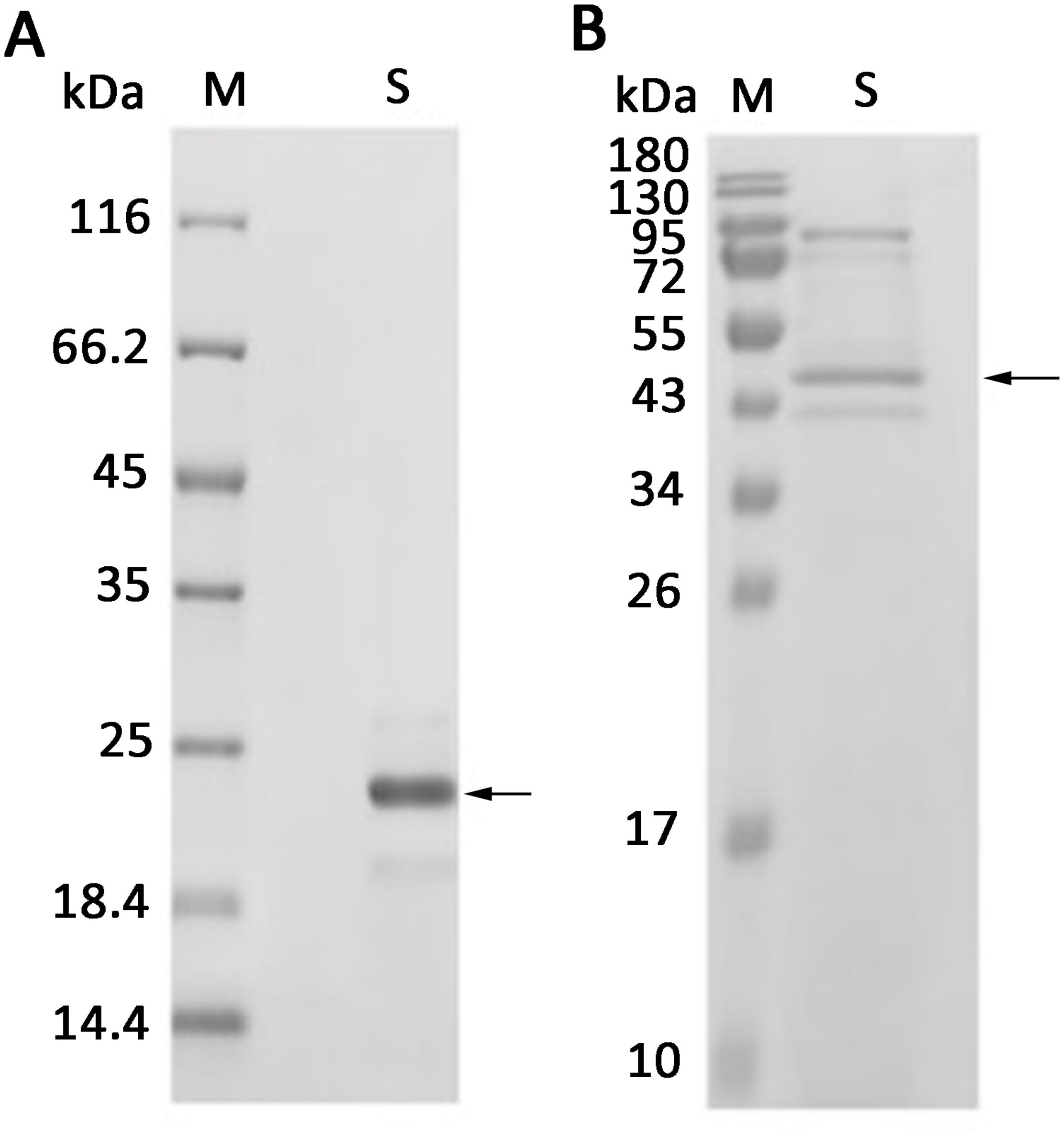
Figure 6. Expression of AtCERK1 and AtCEBiP-LIKE1 protein. (A) AtCERK1 was overexpressed in Escherichia coli BL21 (DE3) and was assessed by SDS-PAGE using 12% (w/v) polyacrylamide gels, followed by staining with Coomassie Brilliant Blue. M: unstained protein molecular weight marker SM0431; S: AtCERK1 protein purified from the E. coli supernatant. (B) AtCEBiP-LIKE1 was overexpressed in yeast and was assessed by SDS-PAGE. M: stained protein molecular weight marker PageRuler; S: AtCEBiP-LIKE1 protein purified from the yeast cell lysate.
Then, ELISA was used to analyze the interaction between the biotin-labeled AOS and the different concentrations of AtCEBiP-LIKE1, as well as between the biotin-labeled AOS and the different concentrations of AtCERK1. The OD450 absorption value was greatest (0.60) when the AtCERK1 concentration was 1 ng/μL but was only 0.056 when the AtCEBiP-LIKE1 concentration was 1 ng/μL. The data suggested that AOS binds tightly to AtCERK1, while AOS cannot bind tightly to AtCEBiP-LIKE1. Furthermore, COS was used as a control and had a binding affinity to AtCERK1 and AtCEBiP-LIKE1, but the affinity was lower than that of AOS (Figures 7A, B). For further confirmation of AOS combined with CERK1, different concentrations of AtCERK1 and AtCEBiP-LIKE1 and a mix of AtCERK1 and AtCEBiP-LIKE1 were used to check the affinity to AOS. The results (Figure 7C) showed that with the increase of CERK1 concentration, the absorption value was higher to a certain extent; however, the concentration of AtCEBiP-LIKE1 had no significant effect on the absorption value. Identical molar values of AtCERK1 and AtCEBiP-LIKE1 were used to bind AOS, and the absorption value was lower when only AtCERK1 was used. The above results confirmed that AOS binds tightly to AtCERK1, indicating that AtCERK1 is the main plant receptor of AOS and is involved in the resistance induced by AOS. Moreover, the ELISA process and a schematic diagram of the interaction between AOS and AtCERK1/AtCEBiP-LIKE1 are shown in Figure 7D.
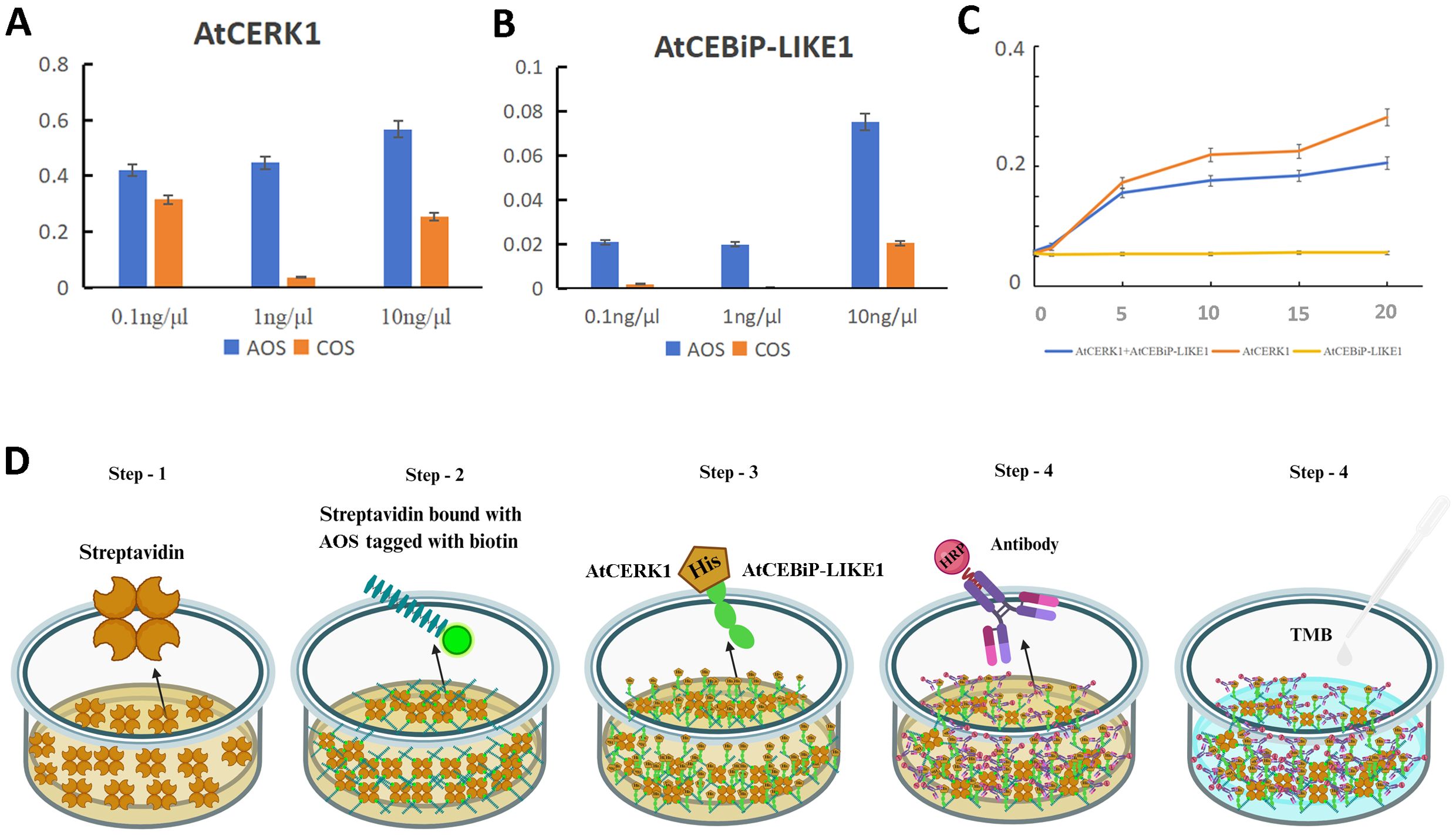
Figure 7. AtCERK1 is a key receptor kinase when AOS induces resistance to pathogens. Absorbance values at 450 nm for AOS and 0.1, 1.0, and 10 ng/μL of AtCERK1 (A) and AtCEBiP-LIKE1 (B). COS was used as a control. (C) Association curve of the different concentrations of AtCERK1 and AtCEBiP-LIKE1. (D) Schematic representation of the binding assay of AOS with AtCERK1 or AtCEBiP-LIKE1. Step 1: streptavidin plate incubation; step 2: addition of biotin-labeled AOS; step 3: addition of AtCERK1/AtCEBiP-LIKE1 protein with a His tag; step 4: addition of HRP-labeled anti-His antibody; step 5: TMB chromatography at 450 nm. A high reading with a dark yellow color indicates a high degree of binding.
Since the 1960s, a large number of studies on oligosaccharide elicitors have been reported, with researchers concluding that oligosaccharides have certain biological activities, such as stimulating systemic responses and regulating plant growth and development, reproduction, and immunity (Jia et al., 2016; Salachna et al., 2018). Currently, the role of oligosaccharides in immunity has been intensively studied, and a variety of oligosaccharide products have been widely used. Oligosaccharides, such as COS, have been widely reported as PAMPs (Jia et al., 2016; Kim and Rajapakse, 2005). However, few studies have focused on the elicitor activities of AOS on plant immunity. The immune elicitor AOS against P. infestation was explored for the first time in this study.
AOSs (UDP2-UDP5) with specific structural characteristics have been prepared by using a new alginate lyase, Aly2 (Peng et al., 2018). The results of this study demonstrated that AOS improved plant resistance to P. infestans (Figure 1) and triggered various defense and resistance responses in tobacco, including increased ROS bursts (Figure 2), callose deposits (Supplementary Figure S1), intracellular Ca2+, stomatal closure (Figure 3), and defense-related gene expression (Figure 4). Moreover, transcriptome sequencing analysis revealed that AOS treatment upregulated the expression level of the genes of the phytohormone signaling pathway and the chitosan biosynthesis pathway, three of which were related to chitinase (Figure 5; Table 2), and these genes can promote plant resistance to pathogens. Chitinase is an extracellular complex of enzymes that degrade chitin and has the application value of hydrolyzing the cell wall of the pathogen fungi to inhibit growth. Chitinase can degrade chitin-producing N-acetylglucosamine oligomers or monomers, and the above oligomers can bind to the plant surface receptor CERK1 and stimulate plant disease resistance signals. Hence, the elicitor AOS identified in this study can be regarded as a novel PAMP.
Plants produce signal molecules when pathogens are recognized by cell surface receptors, and then the infection signal is transmitted to the cell through the signaling pathway, where it can cause local or systemic resistance (Baccelli et al., 2017). The phytohormone SA plays important roles in regulating disease resistance. In this study, the SA synthesis key gene PAL was upregulated (Figure 4A), and qRT-PCR revealed that the expression of the related marker genes PR1A and NPR1 in the SA pathway increased (Figure 4B).
Many elicitors (Boller and Felix, 2009), including the elongation factor (Zipfel et al., 2006), flagellin (Denoux et al., 2008), chitin (Liu et al., 2016; Shinya et al., 2015), and other oligosaccharides (e.g., OGA) (Benedetti et al., 2015), can induce plant defense responses and improve plant resistance to pathogens. These elicitors are PAMPs, which can interact with the plant receptors to activate the PTI.
CERK1 is a plasma membrane protein that contains three LysM motifs in its extracellular domain and an intracellular Ser/Thr kinase domain with autophosphorylation/myelin basic protein kinase activity. It plays a key role in plants, detecting fungal microbe-related molecular patterns. Currently, it is regarded as a key receptor for plant immunity and symbiosis (Miya et al., 2007; Yang et al., 2022). CEBiP is a membrane glycoprotein with LysM motifs that functions as a cell surface receptor for chitin elicitors in rice (Liu et al., 2016) and plays an important role in the recognition of chitin. The AtCERK1 and AtCEBiP-LIKE1 proteins were expressed in this study (Figure 6), and the results showed that AtCERK1 binds AOS tightly, suggesting that AtCERK1 interacts with AOS, but this is not the case for AtCEBiP-LIKE1 and AOS (Figure 7). These results are consistent with those of Tomonori Shinya, who reported that AtCERK1 alone is sufficient for AOS detection (Shinya et al., 2012).
We demonstrated that the elicitor AOS could induce plant resistance to late blight for the first time. The elicitor AOS activates the SA pathway and a series of defense responses to improve its resistance to pathogens. Moreover, AtCERK1 which binds to AOS is first reported here. We speculate that AOS is recognized by the receptor kinase CERK1 and transmits the signal to cells via its kinase activity and induces a series of defense responses, but the detailed signal pathway should be studied in-depth in the future. This study lays the theoretical foundation for AOS’s wide plant nosotropic applications.
The Transcriptome Sequence data are deposited the in the NCBI Sequence Read Archive (SRA) under accession number PRJNA1097808. The datasets analyzed during the current study are available from the corresponding author on reasonable request.
CP: Conceptualization, Data curation, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. WX: Writing – review & editing, Formal analysis, Investigation, Methodology, Software. XPW: Formal analysis, Investigation, Methodology, Data curation, Writing – review & editing. FM: Data curation, Investigation, Software, Writing – review & editing. YZ: Writing – review & editing, Writing – original draft, Data curation, Methodology. QW: Data curation, Investigation, Conceptualization, Project administration, Resources, Writing – review & editing. XKW: Resources, Validation, Writing – review & editing. RL: Data curation, Formal analysis, Methodology, Writing – review & editing. XD: Supervision, Writing – review & editing. CZ: Conceptualization, Supervision, Validation, Visualization, Writing – review & editing. LP: Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was jointly supported by the National Natural Science Foundation of China (Grant Number 32101035), the Natural Science Foundation of Shandong Province (Grant Number ZR2021QC025), the Special Project of Central Government for Local Science and Technology Development of Shandong Province (Grant Number YDZX2022151), and the Natural Science Foundation of Shandong Province (Grant Number ZR2023QC061).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1506873/full#supplementary-material
AOSs, alginate oligosaccharides; DAB, 3,3-diaminobenzidine; H2O2, hydrogen peroxide; NBT, Nitroblue tetrazolium; NPR, non-expressor of pathogenesis-related; PAL, phenylalanine ammonia lyase; PR, pathogenesis-related protein; ROS, reactive oxygen species; SA, salicylic acid; P. infestans, Phytophthora infestans; CERK1, chitin elicitor receptor kinase 1; CEBiP, chitin elicitor binding protein.
Al-Wabel, M., El-Saeid, M. H., El-Naggar, A. H., Al-Romian, F. A., Osman, K., Elnazi, K., et al. (2016). Spatial distribution of pesticide residues in the groundwater of a condensed agricultural area. Arab J. Geosci 9, 120. doi: 10.1007/s12517-015-2122-y
Baccelli, I., Glauser, G., Mauch-Mani, B. (2017). The accumulation of β-aminobutyric acid is controlled by the plant’s immune system. Planta 246, 791–796. doi: 10.1007/s00425-017-2751-3
Benedetti, M., Pontiggia, D., Raggi, S., Cheng, Z., Scaloni, F., Ferrari, S., et al. (2015). Plant immunity triggered by engineered in vivo release of oligogalacturonides, damage-associated molecular patterns. Proc. Natl. Acad. Sci. U.S.A. 112, 5533–5538. doi: 10.1073/pnas.1504154112
Bernardes, M. F. F., Pazin, M., Dorta, L. C. P., Bernardes, M. F. F., Pazin, M., Dorta, L. C. P. (2015). “Impact of pesticides on environmental and human health,” in Toxicology Studies - Cells, Drugs and Environment (Limited167-169 Great Portland Street,London, W1W 5PF, United Kingdom: IntechOpen). doi: 10.5772/59710
Boller, T., Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Breen, S., Williams, S. J., Outram, M., Kobe, B., Solomon, P. S. (2017). Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 22, 871–879. doi: 10.1016/j.tplants.2017.06.013
Brutus, A., Sicilia, F., Macone, A., Cervone, F., De Lorenzo, G. (2010). A domain swap approach reveals a role of the plant wall-associated kinase 1 (WAK1) as a receptor of oligogalacturonides. Proc. Natl. Acad. Sci. U.S.A. 107, 9452–9457. doi: 10.1073/pnas.1000675107
Cai, Y., Yang, T., Mitrano, D. M., Heuberger, M., Hufenus, R., Nowack, B. (2020). Systematic study of microplastic fiber release from 12 different polyester textiles during washing. Environ. Sci. Technol. 54, 4847–4855. doi: 10.1021/acs.est.9b07395
Deepa, S. S., Umehara, Y., Higashiyama, S., Itoh, N., Sugahara, K. (2002). Specific Molecular Interactions of Oversulfated Chondroitin Sulfate E with Various Heparin-binding Growth Factors: inplications as a physiological binding partner in the brain and other tissues*. J. Biol. Chem. 277, 43707–43716. doi: 10.1074/jbc.M207105200
Denoux, C., Galletti, R., Mammarella, N., Gopalan, S., Werck, D., De Lorenzo, G., et al. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445. doi: 10.1093/mp/ssn019
Duan, L., Zhang, N., Wang, Y., Zhang, C., Zhu, L., Chen, W. (2008). Release of hexachlorocyclohexanes from historically and freshly contaminated soils in China: implications for fate and regulation. Environ. pollut. 156, 753–759. doi: 10.1016/j.envpol.2008.06.006
Durrant, W. E., Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Espinoza, C., Liang, Y., Stacey, G. (2017). Chitin receptor CERK1 links salt stress and chitin-triggered innate immunity in Arabidopsis. Plant J. 89, 984–995. doi: 10.1111/tpj.13437
Hayafune, M., Berisio, R., Marchetti, R., Silipo, A., Kayama, M., Desaki, Y., et al. (2014). Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. U.S.A. 111, E404–E413. doi: 10.1073/pnas.1312099111
Hernández, A. F., Gil, F., Lacasaña, M., Rodríguez-Barranco, M., Tsatsakis, A. M., Requena, M., et al. (2013). Pesticide exposure and genetic variation in xenobiotic-metabolizing enzymes interact to induce biochemical liver damage. Food Chem. Toxicol. 61, 144–151. doi: 10.1016/j.fct.2013.05.012
Hien, N. Q., Nagasawa, N., Tham, L. X., Yoshii, F., Dang, V. H., Mitomo, H., et al. (2000). Growth-promotion of plants with depolymerized alginates by irradiation. Radiat. Phys. Chem. 59, 97–101. doi: 10.1016/S0969-806X(99)00522-8
Hu, X., Jiang, X., Hwang, H., Liu, S., Guan, H. (2004). Promotive effects of alginate-derived oligosaccharide on maize seed germination. J. Appl. Phycol 16, 73–76. doi: 10.1023/B:JAPH.0000019139.35046.0c
Iriti, M., Varoni, E. M. (2015). Chitosan-induced antiviral activity and innate immunity in plants. Environ. Sci. pollut. Res. Int. 22, 2935–2944. doi: 10.1007/s11356-014-3571-7
Iwasaki, K., Matsubara, Y. (2000). Purification of alginate oligosaccharides with root growth-promoting activity toward lettuce. Biosci. Biotechnol. Biochem. 64, 1067–1070. doi: 10.1271/bbb.64.1067
Jia, X., Meng, Q., Zeng, H., Wang, W., Yin, H. (2016). Chitosan oligosaccharide induces resistance to Tobacco mosaic virus in Arabidopsis via the salicylic acid-mediated signalling pathway. Sci. Rep. 6, 26144. doi: 10.1038/srep26144
Jiang, X., Su, H., Jiang, J. H., Neelin, J. D., Wu, L., Tsushima, Y., et al. (2023). Muted extratropical low cloud seasonal cycle is closely linked to underestimated climate sensitivity in models. Nat. Commun. 14, 5586. doi: 10.1038/s41467-023-41360-0
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kim, S.-K., Rajapakse, N. (2005). Enzymatic production and biological activities of chitosan oligosaccharides (COS): A review. Carbohydr. Polymers 62, 357–368. doi: 10.1016/j.carbpol.2005.08.012
Lecourieux, D., Ranjeva, R., Pugin, A. (2006). Calcium in plant defence-signalling pathways. New Phytol. 171, 249–269. doi: 10.1111/j.1469-8137.2006.01777.x
Li, H., Guan, Y., Dong, Y., Zhao, L., Rong, S., Chen, W., et al. (2018a). Isolation and evaluation of endophytic Bacillus tequilensis GYLH001 with potential application for biological control of Magnaporthe oryzae. PloS One 13, e0203505. doi: 10.1371/journal.pone.0203505
Li, J., Wang, X., Lin, X., Yan, G., Liu, L., Zheng, H., et al. (2018b). Alginate-derived oligosaccharides promote water stress tolerance in cucumber (Cucumis sativus L.). Plant Physiol. Biochem. 130, 80–88. doi: 10.1016/j.plaphy.2018.06.040
Liu, T., Liu, Z., Song, C., Hu, Y., Han, Z., She, J., et al. (2012). Chitin-induced dimerization activates a plant immune receptor. Science 336, 1160–1164. doi: 10.1126/science.1218867
Liu, S., Wang, J., Han, Z., Gong, X., Zhang, H., Chai, J. (2016). Molecular mechanism for fungal cell wall recognition by rice chitin receptor osCEBiP. Structure 24, 1192–1200. doi: 10.1016/j.str.2016.04.014
Liu, H., Zhang, Y.-H., Yin, H., Yin, H., Wang, W.-X., Zhao, X.-M., et al. (2013). Alginate oligosaccharides enhanced Triticum aestivum L. tolerance to drought stress. Plant Physiol. Biochem. 62, 33–40. doi: 10.1016/j.plaphy.2012.10.012
Lozowicka, B., Abzeitova, E., Sagitov, A., Kaczynski, P., Toleubayev, K., Li, A. (2015). Studies of pesticide residues in tomatoes and cucumbers from Kazakhstan and the associated health risks. Environ. Monit Assess. 187, 609. doi: 10.1007/s10661-015-4818-6
Ma, L. J., Li, X. M., Bu, N., Li, N. (2010). An alginate-derived oligosaccharide enhanced wheat tolerance to cadmium stress. Plant Growth Regul. 62, 71–76. doi: 10.1007/s10725-010-9489-2
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Natsume, M., Kamo, Y., Hirayama, M., Adachi, T. (1994). Isolation and characterization of alginate-derived oligosaccharides with root growth-promoting activities. Carbohydr Res. 258, 187–197. doi: 10.1016/0008-6215(94)84085-7
Nisa, H., Kamili, A. N., Nawchoo, I. A., Shafi, S., Shameem, N., Bandh, S. A. (2015). Fungal endophytes as prolific source of phytochemicals and other bioactive natural products: A review. Microb. Pathog. 82, 50–59. doi: 10.1016/j.micpath.2015.04.001
Peng, C., Wang, Q., Jiao, R., Xu, Y., Han, N., Wang, W., et al. (2021). A novel chondroitin sulfate E from Dosidicus gigas cartilage and its antitumor metastatic activity. Carbohydr Polym 262, 117971. doi: 10.1016/j.carbpol.2021.117971
Peng, C., Wang, Q., Lu, D., Han, W., Li, F. (2018). A novel bifunctional endolytic alginate lyase with variable alginate-degrading modes and versatile monosaccharide-producing properties. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.00167
Peng, C., Zhang, A., Wang, Q., Song, Y., Zhang, M., Ding, X., et al. (2020). Ultrahigh-activity immune inducer from Endophytic Fungi induces tobacco resistance to virus by SA pathway and RNA silencing. BMC Plant Biol. 20, 169. doi: 10.1186/s12870-020-02386-4
Qin, F., Gao, Y. X., Guo, B. Y., Xu, P., Li, J. Z., Wang, H. L. (2014). Environmental behavior of benalaxyl and furalaxyl enantiomers in agricultural soils. J. Environ. Sci. Health B 49, 738–746. doi: 10.1080/03601234.2014.929482
Rodriguez-Salus, M., Bektas, Y., Schroeder, M., Knoth, C., Vu, T., Roberts, P., et al. (2016). The synthetic elicitor 2-(5-bromo-2-hydroxy-phenyl)-thiazolidine-4-carboxylic acid links plant immunity to hormesis1. Plant Physiol. 170, 444–458. doi: 10.1104/pp.15.01058
Salachna, P., Grzeszczuk, M., Meller, E., Soból, M. (2018). Oligo-Alginate with Low Molecular Mass Improves Growth and Physiological Activity of Eucomis autumnalis under Salinity Stress. Molecules 23, 812. doi: 10.3390/molecules23040812
Shinya, T., Motoyama, N., Ikeda, A., Wada, M., Kamiya, K., Hayafune, M., et al. (2012). Functional characterization of CEBiP and CERK1 homologs in arabidopsis and rice reveals the presence of different chitin receptor systems in plants. Plant Cell Physiol. 53, 1696–1706. doi: 10.1093/pcp/pcs113
Shinya, T., Nakagawa, T., Kaku, H., Shibuya, N. (2015). Chitin-mediated plant-fungal interactions: catching, hiding and handshaking. Curr. Opin. Plant Biol. 26, 64–71. doi: 10.1016/j.pbi.2015.05.032
Sticher, L., Mauch-Mani, B., Métraux, J. P. (1997). Systemic acquired resistance. Annu. Rev. Phytopathol. 35, 235–270. doi: 10.1146/annurev.phyto.35.1.235
Sun, Y., Detchemendy, T. W., Pajerowska-Mukhtar, K. M., Mukhtar, M. S. (2018). NPR1 in jazzSet with pathogen effectors. Trends Plant Sci. 23, 469–472. doi: 10.1016/j.tplants.2018.04.007
Tang, J., Zhou, Q., Chu, H., Nagata, S. (2011). Characterization of alginase and elicitor-active oligosaccharides from Gracilibacillus A7 in alleviating salt stress for Brassica campestris L. J. Agric. Food Chem. 59, 7896–7901. doi: 10.1021/jf201793s
Tomoda, Y., Umemura, K., Adachi, T. (1994). Promotion of barley root elongation under hypoxic conditions by alginate lyase-lysate (A.L.L.). Biosci. Biotechnol. Biochem. 58, 202–203. doi: 10.1271/bbb.58.202
Ton, J., Flors, V., Mauch-Mani, B. (2009). The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. doi: 10.1016/j.tplants.2009.03.006
Van Wees, S. C. M., van der Ent, S., Pieterse, C. M. J. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. doi: 10.1016/j.pbi.2008.05.005
Wiesel, L., Newton, A. C., Elliott, I., Booty, D., Gilroy, E. M., Birch, P. R. J., et al. (2014). Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front. Plant Sci. 5. doi: 10.3389/fpls.2014.00655
Xing, M., Cao, Q., Wang, Y., Xiao, H., Zhao, J., Zhang, Q., et al. (2020). Advances in research on the bioactivity of alginate oligosaccharides. Mar. Drugs 18, 144. doi: 10.3390/md18030144
Xu, X., Iwamoto, Y., Kitamura, Y., Oda, T., Muramatsu, T. (2003). Root growth-promoting activity of unsaturated oligomeric uronates from alginate on carrot and rice plants. Biosci Biotechnol Biochem. 67, 2022–2025. doi: 10.1271/bbb.67.2022
Xue, N., Yang, R., Xu, X., Seip, H. M., Zeng, Q. (2006). Adsorption and degradation of benfuracarb in three soils in Hunan, People’s Republic of China. Bull. Environ. Contam Toxicol. 76, 720–727. doi: 10.1007/s00128-006-0979-x
Yadav, I. C., Devi, N. L., Syed, J. H., Cheng, Z., Li, J., Zhang, G., et al. (2015). Current status of persistent organic pesticides residues in air, water, and soil, and their possible effect on neighboring countries: a comprehensive review of India. Sci. Total Environ. 511, 123–137. doi: 10.1016/j.scitotenv.2014.12.041
Yang, K. Y., Liu, Y., Zhang, S. (2001). Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. Proc. Natl. Acad. Sci. United States America 98, 741–746. doi: 10.1073/pnas.98.2.741
Yang, C., Wang, E., Liu, J. (2022). CERK1, more than a co-receptor in plant-microbe interactions. New Phytol. 234, 1606–1613. doi: 10.1111/nph.18074
Yin, H., Du, Y., Dong, Z. (2016). Chitin oligosaccharide and chitosan oligosaccharide: two similar but different plant elicitors. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00522
Yonemoto, Y., Tanaka, H., Yamashita, T., Kitabatake, N., Ishida, Y., Kimura, A., et al. (1993). Promotion of germination and shoot elongation of some plants by alginate oligomers prepared with bacterial alginate lyase. J. Fermentation Bioengineering 75, 68–70. doi: 10.1016/0922-338X(93)90181-7
Zang, H., Xie, S., Zhu, B., Yang, X., Gu, C., Hu, B., et al. (2019). Mannan oligosaccharides trigger multiple defence responses in rice and tobacco as a novel danger-associated molecular pattern. Mol. Plant Pathol. 20, 1067–1079. doi: 10.1111/mpp.12811
Zhang, H., Fang, Q., Zhang, Z., Wang, Y., Zheng, X. (2009). The role of respiratory burst oxidase homologues in elicitor-induced stomatal closure and hypersensitive response in Nicotiana benthamiana. J. Exp. Bot. 60, 3109–3122. doi: 10.1093/jxb/erp146
Zhang, C., Howlader, P., Liu, T., Sun, X., Jia, X., Zhao, X., et al. (2019). Alginate Oligosaccharide (AOS) induced resistance to Pst DC3000 via salicylic acid-mediated signaling pathway in Arabidopsis thaliana. Carbohydr Polym 225, 115221. doi: 10.1016/j.carbpol.2019.115221
Zhang, Y., Yin, H., Zhao, X., Wang, W., Du, Y., He, A., et al. (2014). The promoting effects of alginate oligosaccharides on root development in Oryza sativa L. mediated by auxin signaling. Carbohydr Polym 113, 446–454. doi: 10.1016/j.carbpol.2014.06.079
Keywords: alginate oligosaccharides, immune inducer, defense response, salicylic acid, chitin elicitor receptor kinase
Citation: Peng C, Xu W, Wang X, Meng F, Zhao Y, Wang Q, Wang X, Lodi RS, Dong X, Zhu C and Peng L (2025) Alginate oligosaccharides trigger multiple defense responses in tobacco and induce resistance to Phytophthora infestans. Front. Plant Sci. 16:1506873. doi: 10.3389/fpls.2025.1506873
Received: 06 October 2024; Accepted: 10 January 2025;
Published: 12 February 2025.
Edited by:
Muthusamy Ramakrishnan, Nanjing Forestry University, ChinaReviewed by:
Benedetta Mattei, University of L’Aquila, ItalyCopyright © 2025 Peng, Xu, Wang, Meng, Zhao, Wang, Wang, Lodi, Dong, Zhu and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodan Dong, ZG9uZ3hpYW9kYW4xOTk0QDE2My5jb20=; Changxiang Zhu, emhjaHhAc2RhdS5lZHUuY24=; Lizeng Peng, cGVuZ2xpemVuZ0BzZG51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.