- 1College of Life Sciences, University of Chinese Academy of Sciences, Beijing, China
- 2BGI Research, Beijing, China
- 3Department of Botany, National Museum of Natural History, Smithsonian Institution, Washington, DC, United States
Introduction: Heliconia, a genus within the Zingiberales order, is renowned for its diverse morphology, suggesting a rich genetic reservoir. However, genetic research on plants within the Heliconiaceae family has primarily focused on taxonomy and phylogenetics, with limited exploration into other genetic aspects, particularly the chloroplast genome. Given the significance of chloroplast genomes in evolutionary studies, a deeper understanding of their structure and diversity within Heliconia is essential.
Methods: In this study, we sequenced and assembled the complete chloroplast genomes of four representative Heliconia species: Heliconia bihai, Heliconia caribaea, Heliconia orthotricha, and Heliconia tortuosa. The chloroplast genomes were analyzed for structure, gene content, and nucleotide diversity. We also performed comparative analysis with other species within the Zingiberales order to investigate structural and functional differences.
Results: The assembled chloroplast genomes of the four Heliconia species exhibited a typical quadripartite structure and ranged in length from 161,680 bp to 161,913 bp. All genomes contained 86 protein-coding genes. Comparative analysis revealed that the chloroplast genome structures of the different Heliconia species were highly conserved, with minor variations. Notably, the chloroplast genome of Heliconia was slightly shorter than those of other Zingiberales species, primarily due to the reduced length of the inverted repeat region. In terms of nucleotide diversity, Heliconia species exhibited lower diversity in their chloroplast genomes compared to other families within the Zingiberales order.
Discussion: This study provides valuable insights into the conserved nature of the chloroplast genome in Heliconia. The reduced chloroplast genome size, particularly the shortened inverted repeat region, marks a distinct feature of Heliconia within the Zingiberales family. Our findings also underscore the low nucleotide diversity within the chloroplast genomes of Heliconia species, which could be indicative of their evolutionary history and limited genetic differentiation. These results contribute to a broader understanding of chloroplast genome evolution in the Zingiberales and offer important genetic resources for future research on Heliconia and related species.
Introduction
Heliconia, a genus belonging to the Heliconiaceae family, is a unique group of flowering plants comprising nearly 200 species (Iles et al., 2017; Linares et al., 2020). These plants are primarily found in tropical America and certain islands in the western Pacific. The inflorescence of Heliconia is a defining feature that makes it highly popular in horticulture, thanks to its vibrant, waxy bracts that attract pollinators. These bracts form part of an upright or pendulous cone-like structure, with the true flowers hidden within. A well-known previous study investigated the relationship between the beak characteristics of hummingbirds and the appearance of Heliconia bracts on two islands in the Lesser Antilles. The ecological structure of the islands further supports the coevolution between Heliconia and hummingbirds (Altshuler and Clark, 2003; Temeles and Kress, 2003). Additionally, the exotic bracts of Heliconia are in high demand in the global fresh-cut flower market (Linares et al., 2020), further highlighting their ecological and economic significance. The ecological significance of the genus in tropical forests, as well as its taxonomic and morphological aspects, have garnered considerable interest.
The high morphological diversity of Heliconia initially led taxonomists to classify the genus based on appearance (Kress, 1984; Lennart, 1992; Kress et al., 1999; Kress et al., 2001). Genetic markers from plastid and nuclear genomes were employed to study the evolution of Heliconia, revealing that its diversity originated in the Late Eocene (39 million years ago) and underwent rapid diversification during the Early Miocene. However, studies specifically addressing the molecular diversity of Heliconia remain limited, primarily using genetic markers to investigate evolution at the species and population levels (Marouelli et al., 2010; Suárez-Montes et al., 2011; Côrtes et al., 2013; Stein et al., 2014; Westerband and Horvitz, 2015). Previous studies utilized Amplified Fragment Length Polymorphism (AFLP) makers to study cultivated Heliconia species (Isaza et al., 2012) and the genetic diversity of H. bihai populations (Marouelli et al., 2010; Martén-Rodríguez et al., 2011; Suárez-Montes et al., 2011; Isaza et al., 2012; Côrtes et al., 2013; Stein et al., 2014; Westerband and Horvitz, 2015). Random Amplified Polymorphic DNA (RAPD) markers were also applied to study evolutionary relationship among Heliconia species, revealing the monophyletic nature of the Heliconia genus (Marouelli et al., 2010). Furthermore, In the larger group of Zingiberales order, to which Heliconia belongs, more genetic markers or representative whole chloroplast genomes were utilized to depict the evolutionary process of Zingiberales species, indicating Heliconia as the sister group to the remaining families in Zingiberales (Barrett et al., 2013; Barrett et al., 2014). A key factor that has hindered accurate phylogenetic reconstructions in many tropical plant groups is the widespread occurrence of rapid lineage radiations (Couvreur et al., 2014; Koenen et al., 2015). To gain a more comprehensive understanding of the correlation between morphological and molecular diversity in Heliconiaceae, it is crucial to gather more extensive molecular data. This includes focusing on genes that are sufficiently long to provide substantial phylogenetic signal while filtering out genes under strong selection (Lemmon and Lemmon, 2013). Comparative analyses with other species within the Zingiberales order will further contribute to elucidating the evolutionary patterns and relationships among Heliconia species, as well as their coevolutionary dynamics with hummingbirds.
In this study, we assembled the chloroplast genomes of four representative Heliconia species, including Heliconia bihai, Heliconia caribaea, Heliconia orthotricha, and Heliconia tortuosa (Pic. S1). We conducted a thorough examination of the complete chloroplast genome structures of these species, performing detailed analyses and comparisons of their structural and genomic features with those of other species in the Zingiberales order.
Materials and methods
Plant materials and DNA sequencing
Four representative Heliconia species, namely Heliconia bihai (L.) “Yellow Dancer”, Heliconia caribaea Lam., Heliconia orthotricha, and Heliconia tortuosa Griggs, were selected for our study. H. bihai, H. caribaea, and H. tortuosa samples were collected from Gardens of Plant Group Hawai’i by Kress lab, while H. orthotricha was obtained from the Guangdong Flower Market. Fresh leaves were carefully collected and immediately snap-frozen in liquid nitrogen. The samples were then stored at -80 °C until DNA extraction. DNA extraction was performed using the modified CTAB method (Aboul-Maaty and Oraby, 2019). Subsequently, the DNA samples were sequenced on BGISEQ-500 platforms (MGI, Shenzhen, China) using the whole genome strategy at BGI Research Qingdao lab, following the manufacturer instructions (Huang et al., 2017).
Chloroplast genome assembly and annotation
The de novo assembly of four chloroplast genomes was performed using NOVOplasty (version 4.3.3) (Dierckxsens et al., 2016)with parameters of “Genome Range: 150,000-190,000; K-mer: 31; Seed Input: Heliconia collinsiana; Combined reads: All clean reads”. For the homology-based assembly of the chloroplast genomes, MITObim version 1.9.1 (relies on MIRA 4.0.2) (https://github.com/chrishah/MITObim) was utilized with parameters of “Read Pool: Extracted all clean reads with a depth of 20×; -quick Heliconia collinsiana” (Hahn et al., 2013). The resulting assemblies from both methods were then aligned and refined against the reference chloroplast genome of Heliconia collinsiana (NC_020362.1). Each assembled complete chloroplast genome underwent annotation utilizing GeSeq (Tillich et al., 2017)and the online CPGAVAS2 (an integrated Plastome Annotator and Analyzer) (Shi et al., 2019) with default parameters. Subsequently, the newly annotated chloroplast genome sequences were initially validated using the online tool GB2sequin (Lehwark and Greiner, 2019), further verified, and formatted using Sequin v. 15.50 from NCBI before being deposited in GenBank (accession numbers provided in Table 1).To visualize the chloroplast genome maps, the online program OGDRAW v1.3.1 (Greiner et al., 2019) (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) was employed.
Chloroplast genome analysis and statistics
The identification of simple sequence repeats (SSRs) was performed using the online MISA-web tool (Thiel et al., 2003; Beier et al., 2017). The minimum number of repeats was set to 10, 5, 4, 3, 3, and 3 for mononucleotide (mono-), dinucleotide (din-), trinucleotide (tri-), tetranucleotide (tetra-), pentanucleotide (penta-), and hexanucleotide (hexan-) SSRs, respectively (Martin et al., 2013). Tandem repeat sequences were detected using Tandem Repeats Finder with default parameters (Benson, 1999). The parameters used were 2, 7, and 7 for weights of match, mismatch, and indels, respectively. The detection parameters were set to 80 for the matching probability (Pm), 10 for the indel probability (Pi), a minimum alignment score of 50, and a maximum period size of 500. Long repeat sequences were analyzed using REPuter (Kurtz, 2001). The analysis identified forward (F), reverse (R), complement (C), and palindromic (P) repeats with default parameters. The parameters used were, ‘-f’ to compute maximal forward repeats, ‘-p’ to compute maximal palindromes, ‘-h’ to search for repeats up to the given hamming distance, and ‘-l’ to specify the desired length of repeats. Codon usage was analyzed using MEGA11 (Kumar et al., 2008), and the relative synonymous codon usage (RSCU) and amino acid frequencies were calculated with default settings. Additionally, the GC content of the three positions was analyzed using CUSP in the EMBOSS program (Rice et al., 2000).
Comparative analysis of the chloroplast genomes
DNA polymorphisms, identified by calculating nucleotide diversity (π) using DnaSP (DNA Sequence Polymorphism) v5.10.1 (Librado and Rozas, 2009), were used to detect highly variable sites among chloroplast genomes in different evolutionary clades. Alignments of reordered whole-chloroplast genome sequences, obtained using MAFFT v7.407 (Katoh and Standley, 2013), were sliced into 800-site windows to calculate nucleotide diversity with a step size of 200 sites. Sites with gaps and plastid sequences with rearrangements were excluded. Signals of natural selection were evaluated for all protein coding genes. The non-synonymous (Ka) and synonymous (Ks) substitution ratio (Ka/Ks) of each gene was calculated in the background of different species in Zingiberales. The protein sequences of protein coding genes in each pair of the species were aligned using MAFFT (v7.407) (Katoh and Standley, 2013). Subsequently, the coding DNA sequences (CDS) were converted into codon alignments based on the protein sequence alignment using the Perl script pal2nal (v14) (Suyama et al., 2006). The KaKs calculator (v2.0) (Wang et al., 2010), utilizing its model-averaging method, was employed to compute the values for Ka (non-synonymous substitutions), Ks (synonymous substitutions), and the Ka/Ks ratio.
The pairwise alignments and sequence divergence analysis were conducted for H. bihai, H. caribaea, H. orthotricha, and H. tortuosa, along with seven additional Zingiberales species, namely Canna indica (MK561603), Costus pulverulentus (KF601573), Musa acuminata (NC_058940), Orchidantha fimbriata (KF601569.1), Thaumatococcus daniellii (KF601575.1), Ravenala madagascariensis (NC_022927.1), and Zingiber officinale (NC_044775). The alignments and sequence comparisons were performed using the mVISTA tool with LAGAN and Shuffle-LAGAN modes (Brudno et al., 2003). The analysis was carried out to assess the contraction and extension of the inverted repeat (IR) borders across the four major regions (LSC/IRa/SSC/IRb) in the chloroplast genome sequences of all eleven species. This assessment was carried out using the web tool IRscope (Amiryousefi et al., 2018).
Phylogenetic analysis
We obtained 22 chloroplast genomes from the NCBI database. In addition to the seven species from the Heliconiaceae family, we included 45 additional species in our analysis and used the monocotyledonous plant rice (Oryza sativa) as an outgroup. Subsequently, we utilized the HomBlocks pipeline (Bi et al., 2018) to efficiently identify homologous blocks among organelle genomes and extract phylogeny-informative regions for constructing a multi-gene alignment. This method leverages core conserved fragments, including coding genes, functional non-coding regions, and rRNA, to generate high-quality and informative data matrices.
Maximum likelihood (ML) analysis was performed using the IQ-TREE program (Minh et al., 2020)with the parameter ‘-m GTR+G+I -bb 1000 -bnni -cmax 15’ as the nucleotide substitution model (Li et al., 2023). MEGA11 was used with default parameters to construct the Neighbor-Joining evolutionary tree. To visualize the phylogenetic relationships, we utilized the iTOL online tool (https://itol.embl.de/) (Letunic and Bork, 2021).
For the analysis of shared genes among the 52 species, we generated a high-quality alignment file using the MAFFT (Katoh and Standley, 2013) with default parameters. These alignment files, along with the chloroplast genome sequences, were used as input files for codeml. In the initial run, the ctl file parameters were set to ‘runmode = 0, CodonFreq = 2, and model = 0’. In the second run, the parameters were adjusted to ‘mode = 2’, focusing on the Heliconiaceae family as the foreground branch, allowing for the calculation of different evolutionary rates (Librado and Rozas, 2009). The DnaSP v5 software (Librado and Rozas, 2009) was employed to compare the aligned sequences, calculate nucleic acid diversity, and obtain the value of π.
Results
Assembly of Heliconia chloroplast genomes
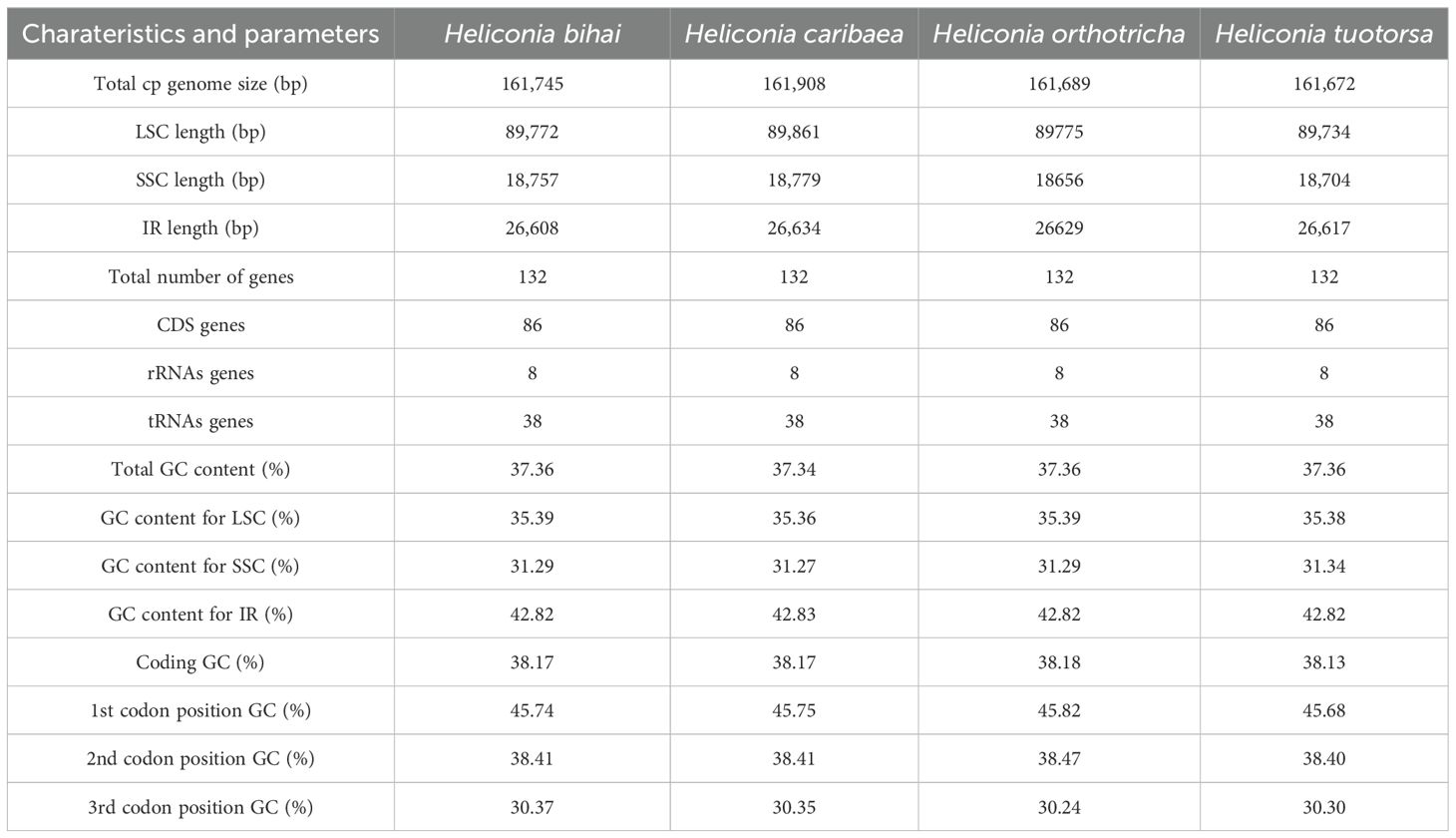
Utilizing the sequencing data, the chloroplast genomes of four Heliconia species (H. bihai, H. caribaea, H. orthotricha, and H. tortuosa) were assembled (Supplementary Table S1). It was found that the chloroplast genomes of these Heliconia species exhibit a high degree of similarity. The sizes of the chloroplast genomes were as follows: 161,745 bp for H. bihai, 161,908 bp for H. caribaea, 161,689 bp for H. orthotricha, and 161,672 bp for H. tortuosa. A total of 132 genes were identified in these chloroplast genomes, comprising 86 coding sequences (CDS), 8 ribosomal RNAs (rRNAs), and 38 transfer RNAs (tRNAs) (Figure 1A; Table 1; Supplementary Table S2). Of these genes, 18 were identified as intron-containing genes in H. bihai, H. orthotricha, and H. tortuosa, with 16 of them containing a single intron each, while two genes (clpP and ycf3) had two introns each. It is noteworthy that H. caribaea possesses 16 splitting genes in addition to trna, a feature that differentiates it from the other three species with regard to the number of splitting genes (see Supplementary Table S3). The chloroplast genomes of these four Heliconia species exhibit a quadripartite structure, a characteristic shared by the majority of angiosperms. This structure consists of a large single-copy (LSC) region (89,772 bp for H. bihai, 89,861 bp for H. caribaea, 89,734 bp for H. orthotricha, and 89,775 bp for H. tortuosa two inverted repeat (IR) regions (26,608 bp for H. bihai, 26,634 bp for H. caribaea, 26,617 bp for H. orthotricha, and 26,629 bp for H. tortuosa) (Supplementary Figure S1). The GC content in the LSC, SSC, and IR regions of all four chloroplast genomes was found to be 35.4%, 31.3%, and 42.8%, respectively (Table 1). The higher GC content observed in the IR regions may be attributed to the abundance of rRNA and tRNA genes, which inherently have a relatively higher GC content.
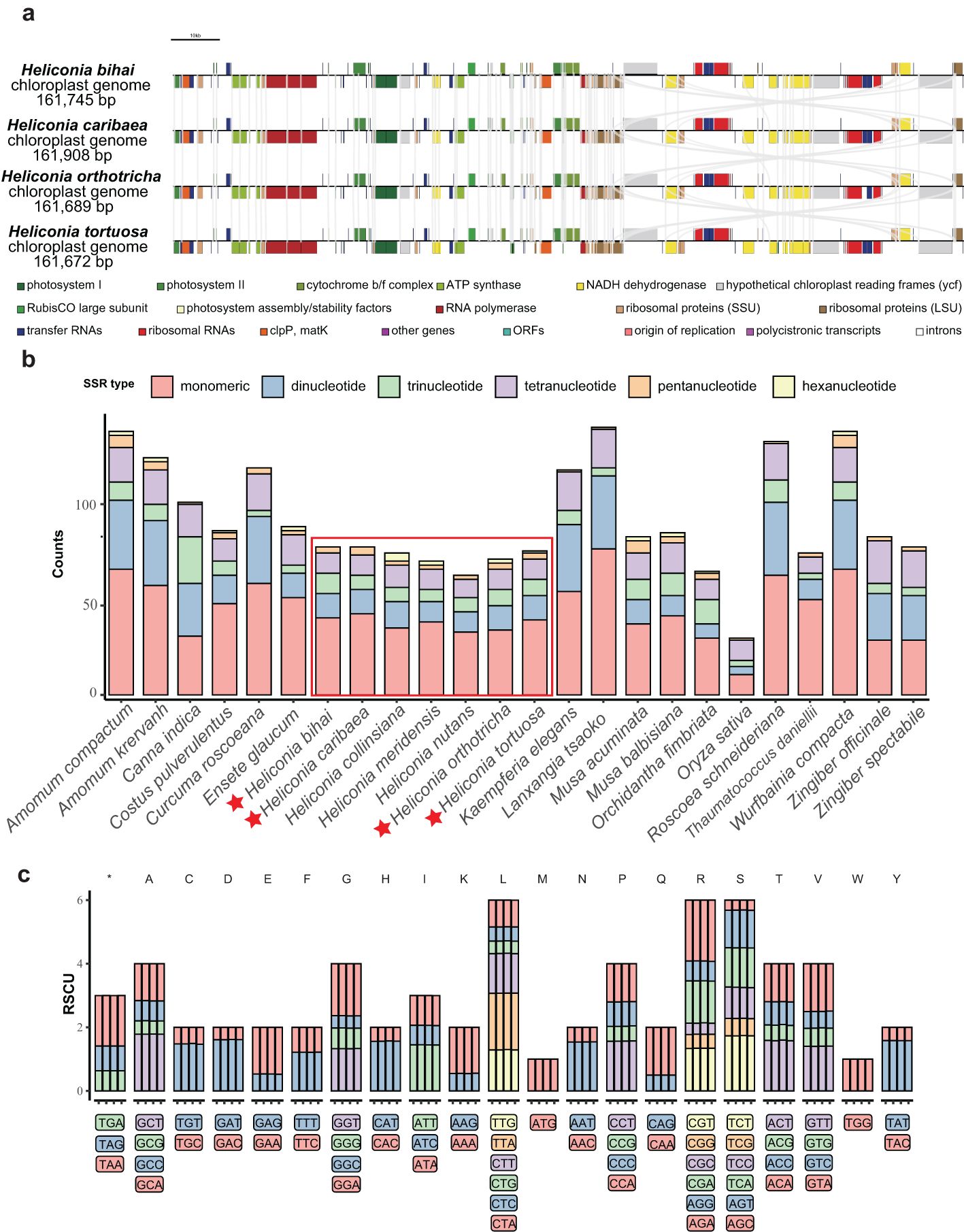
Figure 1. Chloroplast genomes of four Heliconia species. (A) Genetic features. Genes were shown along the four chloroplast genomes shown in linear forms. (B) Simple sequence repeats (SSRs) compositions. Types and numbers of SSRs in the four chloroplast genomes comparing to those of related species. (C) Codon usage patterns. Codon usage patterns of the four chloroplast genomes are shown in order.
Features of Heliconia chloroplast repeat sequence
Our analysis reveals that while the repetitive sequences in the chloroplasts of various Heliconia species display quantitative similarities, they differ in their types. Focusing on SSRs, we found minimal variation in their numbers among the four Heliconia genomes, with 73 in H. bihai and H. caribaea, 71 in H. tortuosa, and 68 in H. orthotricha. However, despite the similarity in the number of encoded genes, notable differences in SSR types were observed. Specifically, H. bihai and H. caribaea featured monomeric, dinucleotide, trinucleotide, tetranucleotide, and pentanucleotide SSR types, whereas H. tortuosa and H. orthotricha additionally included the hexanucleotide SSR type in the SSC region (Supplementary Table S4). Most SSRs were concentrated in the LSC regions, with only one SSR located within coding genes across all four Heliconia species.
When comparing the chloroplast genome data of other sequenced species within the Zingiberales order (Figure 1B), we observed that the presence of both ACT and AATC types of SSRs in the genome could potentially serve as an indicator for classifying a species as belonging to the Heliconia genus (Supplementary Table S5).
Turning to tandem repeats (TRs), our detailed analysis revealed that most repeat units were predominantly composed of A or T, with the longest repeat sequence spanning approximately 120 base pairs (Supplementary Table S6). Shifting to dispersed repeats (DRs), H. bihai exhibited two types (forward repeat and reverse repeat), while H. caribaea and H. tortuosa showed three types (forward repeat, reverse repeat, and palindromic repeat). In contrast, H. orthotricha possessed all four types of dispersed repeats (forward repeat, reverse repeat, complemented repeat, and palindromic repeat), though it had a comparatively lower quantity of DRs.
Overall, these findings highlight the distinctive repeat features in Heliconia chloroplasts, which could serve as valuable genetic markers for distinguishing Heliconia species from one another and from other species.
Features of Heliconia chloroplast coding genes
Beyond repeat features, we further explored the protein-coding genes within the chloroplast genome to uncover potential factors linked to the visual diversity of Heliconia and its successful proliferation in tropical forest ecosystems. Codon usage bias refers to the uneven utilization of different codons that encode the same amino acid within a genome. In our analysis of the 86 CDS in chloroplast genomes, we computed the frequency of codon usage and relative synonymous codon usage (RSCU) (Figure 1C; Supplementary Table S7). The CDS in these chloroplast genomes encode 20 amino acids using 64 codons, including the termination codon. Among these 64 codons, 30 of them exhibit an RSCU value greater than 1, with 29 of them ending with an A or T bases. This observation indicates a preference for A or T endings in the codons of the Heliconia chloroplast genomes, which is consistent with the previously mentioned decrease in GC content at the third position of codons (30.3%) compared to the first (45.7%) and second (37.4%) positions. Regarding the codon usage bias among the four chloroplast genomes, there are six codons each for arginine (Arg), leucine (Leu), and serine (Ser), while only one codon each is present for methionine (Met) and tryptophan (Trp). Within the spectrum of amino acids, Isoleucine (Ile) stands out as the most frequently occurring amino acid, predominantly encoded by the ATT codon with a frequency of 41%. Conversely, cysteine (Cys) is the least common amino acid, with the TGC codon having the lowest frequency at 3%, across four chloroplast genomes. Except for methionine (Met) and tryptophan (Trp), nearly all amino acids are encoded by 2–6 synonymous codons.
Selective pressure analysis provides insights into the chloroplast genes under selection and nucleotide diversity within specific genes in the chloroplast genomes. During the positive selection analysis of the genes used in constructing the phylogenetic tree, we observed that Heliconia, as a foreground branch, did not undergo significant positive selection. However, within the Heliconiaceae family, three genes (ndhD, rpl2, and ycf2) showed a trend of positive selection (Ka/Ks > 1) (Supplementary Table S8).
The nucleotide diversity (Pi) of complete chloroplast genomes was analyzed separately for four families within Zingiberales: Costaceae, Heliconiaceae, Musaceae, and Zingiberaceae (Figure 2). The chloroplast genomes of Heliconiaceae plants exhibit lower nucleotide diversity (π) across the entire genome compared to other evolutionary lineages. Additionally, there is reduced variation in diversity across different genomic regions, as evidenced by the smaller difference in π values between the inverted repeat (IR) and single-copy (SC) regions in Heliconiaceae compared to other plant groups. Additionally, focusing on protein-coding genes, we analyzed nucleotide diversity in a total of 12 species from the Zingiberales order (Supplementary Table S9). Among these genes, the ndhD gene exhibited notably high nucleotide diversity, with a PAI (per-site average information) value exceeding 0.2. Several other genes, including ccsA, cemA, infA, matK, ndhD, rpl, rpo, rps also displayed PAI values greater than 0.05. However, among the Heliconia species, we did not observe coding genes with high nucleotide diversity (Supplementary Table S10).
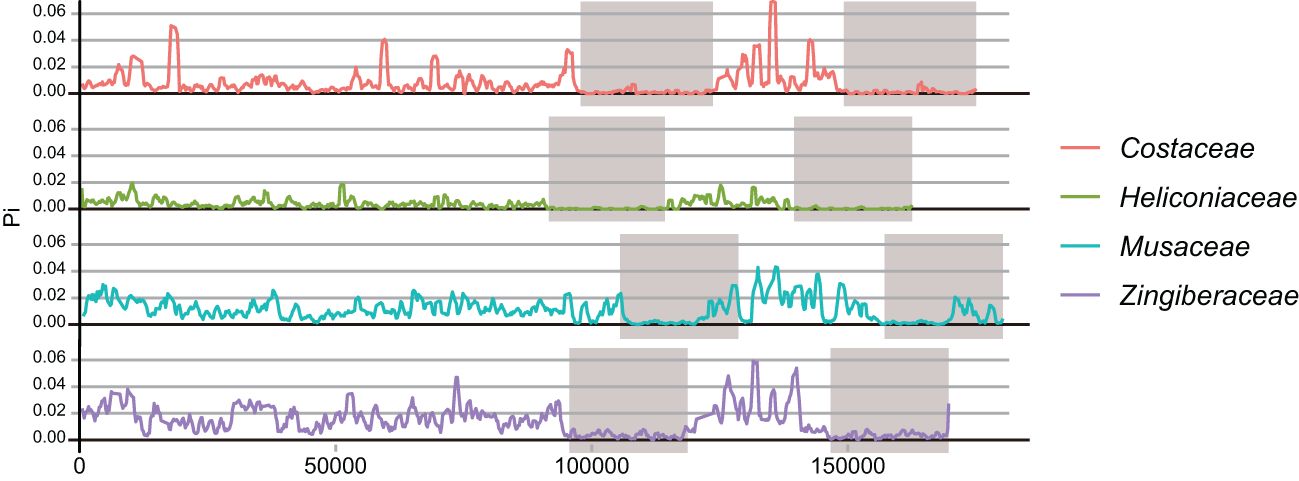
Figure 2. Nucleotide diversity of phylogenetic clades containing Zingiberales species. The curved line depicts the fluctuation of ╥ values across the genome alignment. The shadow layers in grey indicate the approximate range of IRs regions.
Structural comparison within Zingiberales chloroplast genomes
In our comparative analysis of Heliconia chloroplast genomes alongside five closely related species (Canna indica, Costus pulverulentus, Musa acuminata, Ravenala madagascariensis, Zingiber officinale), we observed remarkable structural conservation in the overall of the chloroplasts among species within the Zingiberales order. Specific structural variations were identified at distinct boundaries, including LSC/IRb, IRb/SSC, SSC/IRa, and IRa/LSC (Figure 3). These boundary regions in the four Heliconia species remained consistent yet exhibited unique features, setting them apart from other plants in Zingiberale. Noteworthy is the absence of the rps19 gene in the IR region of Heliconia chloroplasts, distinguishing it from other Zingiberales plants where the IR region includes the rps19 gene. Furthermore, an elongated separation of approximately 150 base pairs at the boundary between the inverted repeat B (IRb) and the small single-copy region (SSC) in the Heliconia chloroplast genomes for the ndhF gene was noted. This contrasts with other species, where the distance typically falls within the range of approximately 10 to 60 base pairs. contraction in the inverted repeat (IR) region resulted in a slightly smaller chloroplast genome size in Heliconia compared to other species in the Zingiberales order.

Figure 3. Structural variations in the chloroplast genomes. Chloroplast genomes of eight Zingiberales species are compared to indicate the major chloroplast genome regions including LSC, SSC and IR regions. Genes transcribed forward are shown above the lines, whereas genes transcribed reversely are shown below the lines. Gene lengths in the corresponding regions are displayed above the boxes of gene names. JLB (LSC/IRb), JSB (IRb/SSC), JSA (SSC/IRa) and JLA (lra/LSC) denoted the junction sites between each corresponding two regions.
We conducted a sequence comparative analysis of Heliconia chloroplast genomes and those of related species. Using H. bihai as the reference, we compared its chloroplast genome sequence with those of Canna indica, Costus pulverulentus, Musa acuminata, Orchidantha fimbriata, Thaumatococcus daniellii, Ravenala madagascariensis, and Zingiber officinale from the Zingiberales order (Figure 4). The analysis revealed significant genetic diversity and variation in Heliconia compared to other Zingiberales plants, particularly in the conserved noncoding sequences (CNS), especially within the LSC and SSC regions. A similar analysis within the Costaceae and Musaceae families further confirmed the extensive conservation of chloroplast genomic sequences in Heliconiaceae. Based on the currently available data and considering the incomplete genomic data for other families within the Zingiberales order, Heliconiaceae species emerged as having the most conserved chloroplast genomes.
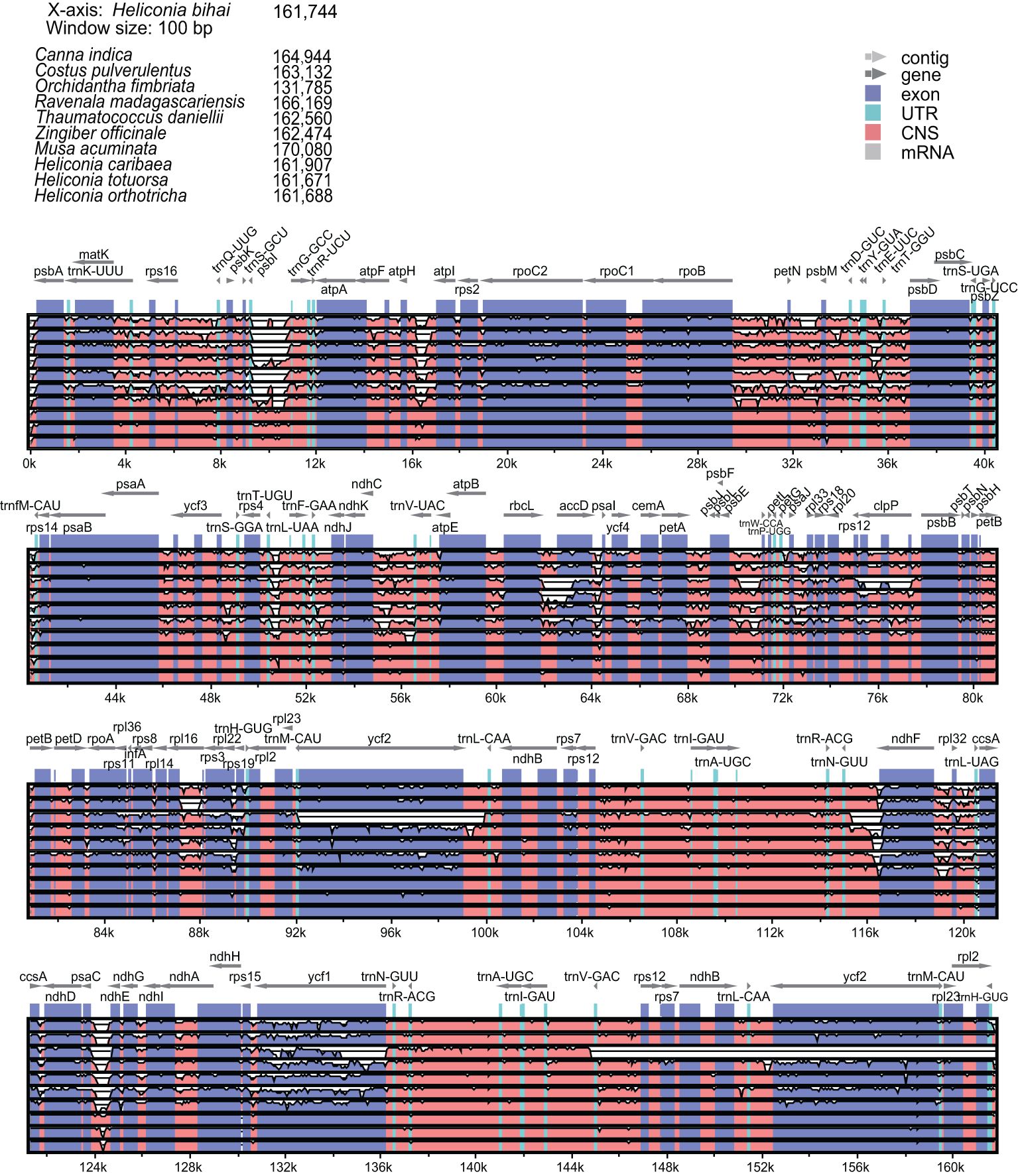
Figure 4. Comparing the four Heliconia chloroplast genomes to these of the other Zingiberales species. Chloroplast genomes are shown with genes indicated, and the vertical scale indicates the percentage of identity, ranging from 50% to 100%.
Phylogeny of Heliconia species revealed by chloroplast genomes
Analyzing the complete chloroplast genome yields more reliable results, providing substantial insights into the genetic evolution of plant species. We carefully selected 51 diverse plant species, representing major clades of Zingiberales plants (Figure 5), and including representatives from different families such as Cannaceae, Costaceae, and Heliconiaceae, Musaceae, Marantaceae, Lowiaceae, along with Strelitziaceae and Zingiberaceae. To construct multi-gene alignments, homologous blocks were identified among the organelle genomes, allowing for the efficient extraction of phylogeny-informative regions. By integrating core conserved fragments—comprising coding genes, functional non-coding regions, and rRNA—into a unified sequence for each genome, we minimized the impact of incomplete chloroplast genomes on the accuracy of phylogenetic tree construction. Phylogenetic trees were constructed using two methods: maximum likelihood (ML) and neighbor-joining (NJ). In the maximum likelihood (ML) tree, Zingiberales diverge from three distinct terminal nodes. Heliconiaceae plants formed a distinct branch, and emerge as the sister clade to Musaceae, Strelitziaceae, and Lowiaceae. While in the Neighbor-Joining tree, Zingiberales diverge from two distinct terminal nodes. Musaceae emerged as sister branches to Heliconiaceae and Strelitziaceae, forming a distinct clade (Supplementary Figure S5).
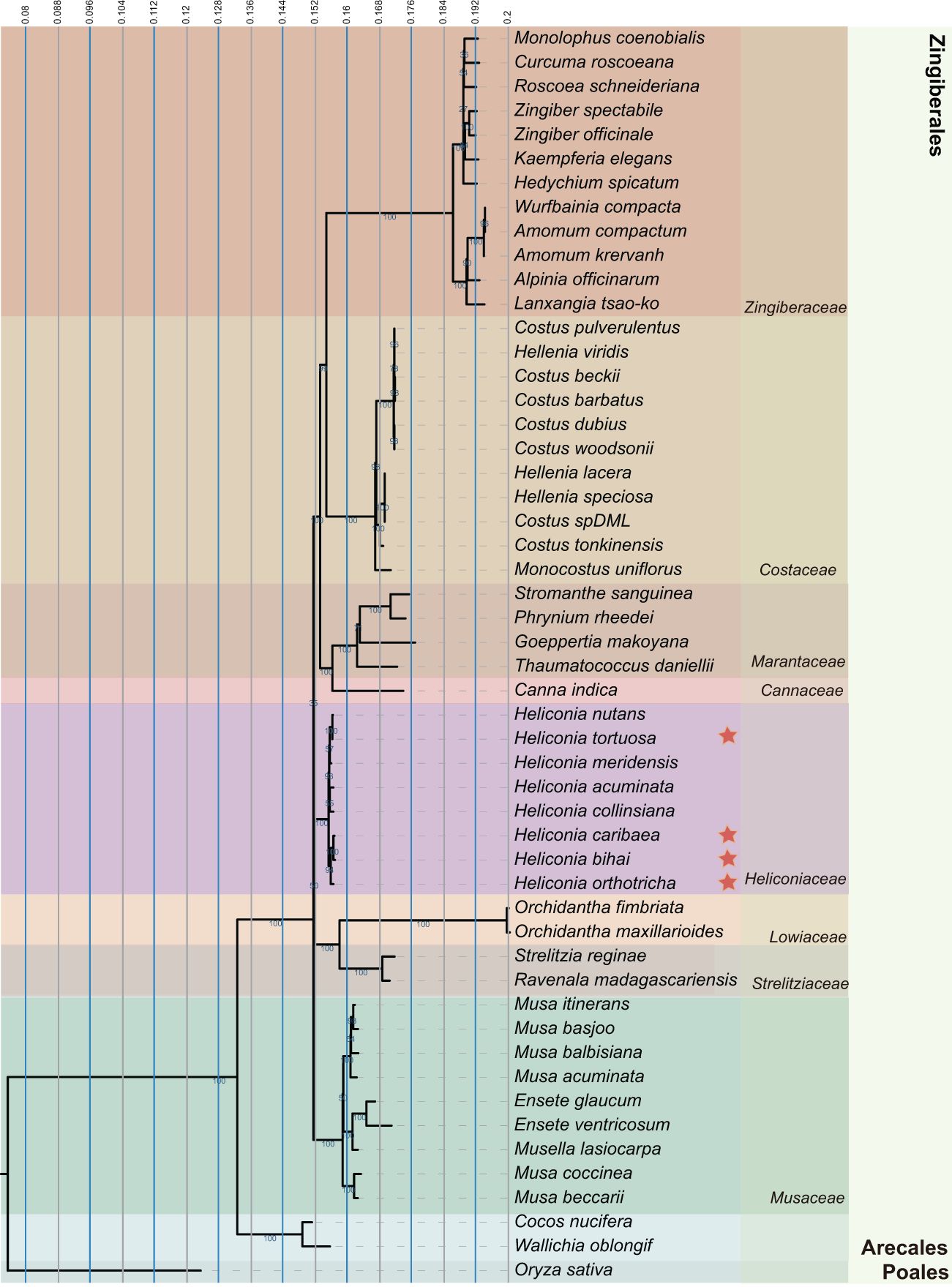
Figure 5. Phylogenetic tree of Heliconia and related species. Maximum likelihood (ML) phylogenetic tree was constructed for 52 species from Zingiberales order, and rice (Oryza sativa) as an outgroup. The confidence level of the phylogenetic tree are shown for each branch.
Discussion
The high conservation of chloroplast genomes in terrestrial plants encompasses their structure, length, and gene content. In our study, we successfully assembled complete chloroplast genomes of Heliconia plants, closely resembling the reported structure of Heliconia collinsiana. Three main types of repeat sequences were found in organelle genomes, including simple sequence repeats (SSRs) (Song et al., 2014), tandem repeats (TRs), and dispersed repeats (DRs). Among these, SSRs exhibited high variability within a species, making them valuable markers for population genetics and phylogenetic analyses (Fan and Chu, 2007). Analysis of repetitive sequences, specifically SSRs, revealed distinguishable patterns not only among different Heliconia species but also across genera within the Zingiberales order.
Research on codon usage bias contributes to our understanding of genome evolution, gene expression regulation, and the adaptability of organisms to environmental changes (Parvathy et al., 2022). Our analysis reveals a codon usage bias favoring A or T endings in the codons of Heliconia chloroplast genomes. Additionally, the low GC content observed in both codon positions and repetitive sequences suggests a strong preference for A/T bases in Heliconia. highlighting the significance of studying codon usage patterns in understanding genome evolution and gene expression regulation.
To further explore functional sequence variations within highly conserved and maternally inherited chloroplast genomes, which can serve as valuable genetic markers for species differentiation (Wysocki et al., 2015). Compared to other Zingiberales species, the chloroplast genome of Heliconia is slightly shorter, which is attributed to a reduction in the length of the IR region. The relatively low nucleotide diversity in the chloroplast genomes of Heliconia indicates that the diverse appearances of these plants are not strongly correlated with variations in their chloroplast genomes.
The three genes showing a trend of positive selection include ndhD, ycf2 and rpl2. Among them, ndhD exhibits significant nucleotide diversity across species in the Zingiberales order, particularly within the Zingiberales family, and is under positive selection. As a component of the chloroplast NADH dehydrogenase-like (NDH) complex, ndhD plays a crucial role in photosynthesis, particularly in electron transport interactions with photosystem I (PSI) (Peng et al., 2011; Shen et al., 2022). In addition, ycf2 forms a complex with five nuclear-encoded FtsH-like proteins, known as the Ycf2-FtsHi complex. This complex functions as the import motor in land plants, facilitating the import of proteins into the chloroplast. Although its evolutionary conservation and functional specialization across photosynthetic organisms are well recognized, the mechanisms and broader evolutionary dynamics of this complex remain largely unexplored (Liang et al., 2024). On the other hand, the function of the rpl2 gene remains unclear and requires further investigation.
Through comparative analysis of the chloroplast genomes of species within the Zingiberales order, certain genes exhibit significant nucleotide diversity, suggesting they may have evolved in response to diverse environmental conditions, contributing to the varied appearances observed among Zingiberales species. Genes such as ccsA, cemA, infA, matK, ndhD, rpl, rpo, rps encode proteins involved in various biological processes. For instance, ccsA encodes a crucial component in the synthesis of cytochrome c within the chloroplast (Xie and Merchant, 1996). cemA encodes a subunit of chloroplast ATP synthase involved in energy production during photosynthesis (Sonoda et al., 1999), whereas infA encodes a protein crucial for tRNA processing, contributing to chloroplast protein synthesis (Millen et al., 2001). Gene matK encodes a splicing enzyme that facilitates RNA splicing (Barthet and Hilu, 2007), These genes are vital for plant growth, development, and metabolic processes, supporting chloroplast structure and function. The findings of this study are consistent with those of previous investigations (Jiang et al., 2023; Li et al., 2023), reinforcing the notion that these genes have undergone adaptive evolution in response to environmental factors, further supporting their pivotal role in the molecular and functional diversity of species within the Zingiberales order.
Like the role of mitochondrial genomes in vertebrate genetics, chloroplast genomes have become a widely adopted tool for addressing phylogenetic and evolutionary questions. Chloroplast genomes, characterized by their maternal inheritance and relatively low mutation rates, are invaluable for elucidating phylogenetic relationships among green plants (Daniell et al., 2016). In our study, the chloroplast genome data have provided valuable insights into the evolutionary relationships within Zingiberales, highlighting the importance of methodological approaches in shaping the interpretation of these relationships. However, different methods for constructing phylogenetic trees may yield divergent results, and as such, determining the exact position of the Heliconia genus within the Zingiberales evolutionary tree remains challenging using chloroplast data alone. While current chloroplast genome resources offer critical genetic information for understanding the morphological diversity of Heliconia species, further research focused on complete nuclear genomes will be essential for a more comprehensive understanding of the genetic mechanisms underlying this diversity. The chloroplast genomes assembled in this study provide a solid foundation for such future investigations. Moreover, a deeper exploration of nuclear genome-encoded genes, particularly those related to gene retention and evolutionary processes, will be crucial in unveiling the evolutionary trajectory and functional diversity of Heliconiaceae.
Conclusions
The analysis of Heliconia chloroplast genome repetitive sequences, specifically SSRs, revealed distinguishable patterns across genera within the Zingiberales order. Compared to other Zingiberales species, the chloroplast genome of Heliconia is slightly shorter, attributed to a reduction in the length of the IR region and an expansion at the SSC region boundary. The relatively low nucleotide diversity in the Heliconia chloroplast genomes suggests that the diverse appearances of Heliconias are not strongly correlated with chloroplast genome. Overall, comparative analysis from various perspectives indicates that the Heliconia chloroplast genomes are conserved within the Heliconiaceae family, while also displaying distinct characteristics that differentiate them from other species within the Zingiberales order.
Data availability statement
The complete chloroplast genomes generated during the current study were deposited in NCBI database (PP093761, PP093760, PP093759, PP093762) and CNGB database (CNP0005095) . The other accession numbers for the remaining datasets analyzed in this study are listed in the Table S11b.
Ethics statement
We confirm that the collection of plant material and experimental research followed all local and national guidelines and legislation.
Author contributions
XC: Data curation, Formal analysis, Investigation, Writing – original draft, Writing – review & editing. CS: Data curation, Methodology, Writing – review & editing. TY: Formal analysis, Writing – review & editing. QG: Formal analysis, Writing – review & editing. JK: Conceptualization, Resources, Writing – review & editing. XL: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program of China, grant number 2021YFD2200502. The authors declare that this study received funding from BGI Research, Beijing.
Acknowledgments
We sincerely thank China National GeneBank, BGI Research Shenzhen, for their assistance in data storage.
Conflict of interest
The authors declare that this study received funding from BGI Research, Beijing. The funder was not involved in the study design, data collection, analysis, interpretation of data, the writing of this article, or the decision to submit it for publication.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1535549/full#supplementary-material
Abbreviations
IR, Inverted repeat regions; LSC, Large single-copy region; SSC, Small single-copy region; rRNAs, Ribosomal RNAs; tRNAs, Transfer RNAs; PE, Paired-end; BI, Bayesian inference; ML, Maximum likelihood; CDS, Protein-coding genes; JLB, Junction between LSC and IRb; JSB, Junction between SSC and IRb; JSA, Junction between SSC and IRa; JLA, Junction between LSC and IRa; CNS, Conserved non-coding sequence.
References
Aboul-Maaty, N. A.-F., Oraby, H. A.-S. (2019). Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull. Natl. Res. Centre 43, 25. doi: 10.1186/s42269-019-0066-1
Altshuler, D. L., Clark, C. J. (2003). Darwin’s hummingbirds. Science 300, 588–589. doi: 10.1126/science.1084477
Amiryousefi, A., Hyvönen, J., Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Barrett, C. F., Davis, J. I., Leebens‐Mack, J., Conran, J. G., Stevenson, D. W. (2013). Plastid genomes and deep relationships among the commelinid monocot angiosperms. Cladistics 29, 65–87. doi: 10.1111/j.1096-0031.2012.00418.x
Barrett, C. F., Specht, C. D., Leebens-Mack, J., Stevenson, D. W., Zomlefer, W. B., Davis, J. I. (2014). Resolving ancient radiations: can complete plastid gene sets elucidate deep relationships among the tropical gingers (Zingiberales)? Ann. Bot. 113, 119–133. doi: 10.1093/aob/mct264
Barthet, M. M., Hilu, K. W. (2007). Expression of matK: functional and evolutionary implications. Am. J. Bot. 94, 1402–1412. doi: 10.3732/ajb.94.8.1402
Beier, S., Thiel, T., Münch, T., Scholz, U., Mascher, M. (2017). MISA-web: a web server for microsatellite prediction. Bioinformatics 33, 2583–2585. doi: 10.1093/bioinformatics/btx198
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580. doi: 10.1093/nar/27.2.573
Bi, G., Mao, Y., Xing, Q., Cao, M. (2018). HomBlocks: A multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics 110, 18–22. doi: 10.1016/j.ygeno.2017.08.001
Brudno, M., Malde, S., Poliakov, A., Do, C. B., Couronne, O., Dubchak, I., et al. (2003). Glocal alignment: finding rearrangements during alignment. Bioinformatics 19, i54–i62. doi: 10.1093/bioinformatics/btg1005
Côrtes, M. C., Uriarte, M., Lemes, M. R., Gribel, R., John Kress, W., Smouse, P. E., et al. (2013). Low plant density enhances gene dispersal in the Amazonian understory herb Heliconia acuminata. Mol. Ecol. 22, 5716–5729. doi: 10.1111/mec.2013.22.issue-22
Couvreur, T. L., Kissling, W. D., Condamine, F. L., Svenning, J. C., Rowe, N. P., Baker, W. J. (2014). Global diversification of a tropical plant growth form: environmental correlates and historical contingencies in climbing palms. Front. Genet. 5, 452. doi: 10.3389/fgene.2014.00452
Daniell, H., Lin, C. S., Yu, M., Chang, W. J. (2016). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Dierckxsens, N., Mardulyn, P., Smits, G. (2016). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res., gkw955. doi: 10.1093/nar/gkw955
Fan, H., Chu, J.-Y. (2007). A brief review of short tandem repeat mutation. Genom. Proteomics Bioinf. 5, 7–14. doi: 10.1016/S1672-0229(07)60009-6
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Hahn, C., Bachmann, L., Chevreux, B. (2013). Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41, e129–e129. doi: 10.1093/nar/gkt371
Huang, J., Liang, X., Xuan, Y., Geng, C., Li, Y., Lu, H., et al. (2017). A reference human genome dataset of the BGISEQ-500 sequencer. Gigascience 6, 1–9. doi: 10.1093/gigascience/gix024
Iles, W. J. D., Sass, C., Lagomarsino, L., Benson-Martin, G., Driscoll, H., Specht, C. D. (2017). The phylogeny of Heliconia (Heliconiaceae) and the evolution of floral presentation. Mol. Phylogenet. Evol. 117, 150–167. doi: 10.1016/j.ympev.2016.12.001
Isaza, L., Marulanda, M. L., López, A. M. (2012). Genetic diversity and molecular characterization of several Heliconia species in Colombia. Genet. Mol. Res. 11, 4552–4563. doi: 10.4238/2012.November.12.9
Jiang, D., Cai, X., Gong, M., Xia, M., Xing, H., Dong, S., et al. (2023). Complete chloroplast genomes provide insights into evolution and phylogeny of Zingiber (Zingiberaceae). BMC Genomics 24, 30. doi: 10.1186/s12864-023-09115-9
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Koenen, E. J. M., Clarkson, J. J., Pennington, T. D., Chatrou, L. W. (2015). Recently evolved diversity and convergent radiations of rainforest mahoganies (Meliaceae) shed new light on the origins of rainforest hyperdiversity. New Phytol. 207, 327–339. doi: 10.1111/nph.2015.207.issue-2
Kress, W. J. (1984). Systematics of Central American Heliconia (Heliconiaceae) with pendant inflorescences. J. Arnold Arboretum 65, 429–532. doi: 10.5962/p.36697
Kress, W. J., Betancur, B., Echeverry (1999). Heliconias: llamaradas de la selva Colombiana (Colombia: Cristina Uribe Ediciones).
Kress, W. J., Prince, L. M., Hahn, W. J., Zimmer, E. A. (2001). Unraveling the evolutionary radiation of the families of the Zingiberales using morphological and molecular evidence. Syst Biol. 50, 926–944. doi: 10.1080/106351501753462885
Kumar, S., Nei, M., Dudley, J., Tamura, K. (2008). MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings Bioinf. 9, 299–306. doi: 10.1093/bib/bbn017
Kurtz, S. (2001). REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29, 4633–4642. doi: 10.1093/nar/29.22.4633
Lehwark, P., Greiner, S. (2019). GB2sequin-A file converter preparing custom GenBank files for database submission. Genomics 111, 759–761. doi: 10.1016/j.ygeno.2018.05.003
Lemmon, E. M., Lemmon, A. R. (2013). High-throughput genomic data in systematics and phylogenetics. Annu. Rev. Ecol. Evol. Syst 44, 99–121. doi: 10.1146/annurev-ecolsys-110512-135822
Lennart (1992). Revision of Heliconia subgen. Taeniostrobus and subgen. Heliconia (Musaceae-Heliconioideae). Opera Botanica 11, 5–98.
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, D.-M., Liu, H-L., Pan, Y.-G., Yu, B., Huang, D., Zhu, G.-F. (2023). Comparative chloroplast genomics of 21 species in Zingiberales with implications for their phylogenetic relationships and molecular dating. Int. J. Mol. Sci. 24, 15031. doi: 10.3390/ijms241915031
Liang, K., Jin, Z., Zhan, X., Li, Y., Xu, Q., Xie, Y., et al. (2024). Structural insights into the chloroplast protein import in land plants. Cell 187, 5651–5664.e18. doi: 10.1016/j.cell.2024.08.003
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Linares, A., Gallardo-López, F., Villarreal, M., Landeros-Sánchez, C., López-Romero, G. (2020). Global vision of Heliconias research as a cut flowers: a review. Ornam. Hortic. 26, 633–646. doi: 10.1590/2447-536X.v26i3.2172
Marouelli, L. P., Inglis, P. W., Ferreira, M. A., Buso, G. S. C. (2010). Genetic relationships among Heliconia (Heliconiaceae) species based on RAPD markers. Genet. Mol. Res. 9, 1377–1387. doi: 10.4238/vol9-3gmr847
Martén-Rodríguez, S., John Kress, W., Temeles, E. J., Meléndez-Ackerman, E. (2011). Plant–pollinator interactions and floral convergence in two species of Heliconia from the Caribbean Islands. Oecologia 167, 1075–1083. doi: 10.1007/s00442-011-2043-8
Martin, G., Baurens, F.-C., Cardi, C., Aury, J.-M., D’Hont, A. (2013). The Complete Chloroplast Genome of Banana (Musa acuminata, Zingiberales): Insight into Plastid Monocotyledon Evolution. PloS One 8, e67350. doi: 10.1371/journal.pone.0067350
Millen, R. S., Olmstead, R. G., Adams, K. L., Palmer, J. D., Lao, N. T., Heggie, L., et al. (2001). Many Parallel Losses of infA from Chloroplast DNA during Angiosperm Evolution with Multiple Independent Transfers to the Nucleus. Plant Cell 13, 645–658. doi: 10.1105/tpc.13.3.645
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Parvathy, S. T., Udayasuriyan, V., Bhadana, V. (2022). Codon usage bias. Mol. Biol. Rep. 49, 539–565. doi: 10.1007/s11033-021-06749-4
Peng, L., Yamamoto, H., Shikanai, T. (2011). Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim. Biophys. Acta (BBA) Bioenerget. 1807, 945–953. doi: 10.1016/j.bbabio.2010.10.015
Rice, P., Longden, I., Bleasby, A. (2000). EMBOSS: the European molecular biology open software suite. Trends Genet. 16, 276–277. doi: 10.1016/S0168-9525(00)02024-2
Shen, L., Tang, K., Wang, W., Wang, C., Wu, H., Mao, Z., et al. (2022). Architecture of the chloroplast PSI–NDH supercomplex in Hordeum vulgare. Nature 601, 649–654. doi: 10.1038/s41586-021-04277-6
Shi, L., Chen, H., Jiang, M., Wang, L., Wu, X., Huang, L., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, W65–w73. doi: 10.1093/nar/gkz345
Song, S.-L., Lim, P.-E., Phang, S.-M., Lee, W.-W., Hong, D. D., Prathep, A. (2014). Development of chloroplast simple sequence repeats (cpSSRs) for the intraspecific study of Gracilaria tenuistipitata (Gracilariales, Rhodophyta) from different populations. BMC Res. Notes 7, 77. doi: 10.1186/1756-0500-7-77
Sonoda, M., Katoh, H., Katoh, A., Ohkawa, H., Vermaas, W., Ogawa, T. (1999). “Structure and function of cema homologue (PXCA) in cyanobacteria,” in The Chloroplast: From Molecular Biology to Biotechnology. Eds. Argyroudi-Akoyunoglou, J. H., Senger, H. (Springer Netherlands, Dordrecht), 149–154.
Stein, K., Rosche, C., Hirsch, H., Kindermann, A., Köhler, J., Hensen, I. (2014). The influence of forest fragmentation on clonal diversity and genetic structure in Heliconia angusta, an endemic understorey herb of the Brazilian Atlantic rain forest. J. Trop. Ecol. 30, 199–208. doi: 10.1017/S0266467414000030
Suárez-Montes, P., Fornoni, J., Núñez-Farfán, J. (2011). Conservation genetics of the endemic Mexican Heliconia uxpanapensis in the Los Tuxtlas tropical rain forest: conservation genetics in Heliconia. Biotropica 43, 114–121. doi: 10.1111/j.1744-7429.2010.00657.x
Suyama, M., Harrington, E., Bork, P., Torrents, D. (2006). Identification and analysis of genes and pseudogenes within duplicated regions in the human and mouse genomes. PloS Comput. Biol. 2, e76. doi: 10.1371/journal.pcbi.0020076
Temeles, E. J., Kress, W. J. (2003). Adaptation in a plant-hummingbird association. Science 300, 630–633. doi: 10.1126/science.1080003
Thiel, T., Michalek, W., Varshney, R., Graner, A. (2003). Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor. Appl. Genet. 106, 411–422. doi: 10.1007/s00122-002-1031-0
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–W11. doi: 10.1093/nar/gkx391
Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinf. 8, 77–80. doi: 10.1016/S1672-0229(10)60008-3
Westerband, A. C., Horvitz, C. C. (2015). Interactions between plant size and canopy openness influence vital rates and life-history tradeoffs in two neotropical understory herbs. Am. J. Bot. 102, 1290–1299. doi: 10.3732/ajb.1500041
Wysocki, W. P., Clark, L. G., Attigala, L., Ruiz-Sanchez, E., Duvall, M. R. (2015). Evolution of the bamboos (Bambusoideae; Poaceae): a full plastome phylogenomic analysis. BMC Evol. Biol. 15, 50. doi: 10.1186/s12862-015-0321-5
Keywords: Zingiberales, Heliconiaceae, Heliconia, chloroplast genome, genomic features
Citation: Cheng X, Shi C, Yang T, Ge Q, Kress WJ and Liu X (2025) Unveiling the conserved nature of Heliconia chloroplast genomes: insights from the assembly and analysis of four complete chloroplast genomes. Front. Plant Sci. 15:1535549. doi: 10.3389/fpls.2024.1535549
Received: 27 November 2024; Accepted: 24 December 2024;
Published: 16 January 2025.
Edited by:
Fei Shen, Beijing Academy of Agricultural and Forestry Sciences, ChinaReviewed by:
Honghong Deng, Fujian Agriculture and Forestry University, ChinaNanqiao Liao, Weimeng Seed Co. Ltd., China
Han Sheng Zhao, International Network for Bamboo and Rattan (INBAR), China
Tuo Yang, China Agricultural University, China
Copyright © 2025 Cheng, Shi, Yang, Ge, Kress and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Cheng, Y2hlbmd4aW4yQGdlbm9taWNzLmNu; Xin Liu, bGl1eGluQGdlbm9taWNzLmNu
 Xin Cheng
Xin Cheng Chengcheng Shi
Chengcheng Shi Ting Yang
Ting Yang Qijin Ge2
Qijin Ge2 W. John Kress
W. John Kress Xin Liu
Xin Liu