
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 January 2025
Sec. Functional and Applied Plant Genomics
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1518962
This article is part of the Research TopicResearch on Brassicaceae Crops Genomics and Breeding, Volume IIView all 11 articles
Premature bolting reduces the yield and quality of Chinese cabbage, making bolting resistance gene identification crucial for breeding superior and stable varieties. In this study, we identified an orphan gene BOLTING RESISTANCE 3 (BR3) that positively regulates bolting resistance in Arabidopsis thaliana. The expression of BR3 was developmentally regulated and occurred during the seedling and flowering stages. The BR3 protein was localized to both the plasma membrane and nucleus. Arabidopsis BR3 overexpressing (BR3OE) plants exhibited delayed bolting and flowering times, an increased number of rosette leaves, reduced plant height, and fewer siliques under long-day (LD) conditions. Key flowering genes were significantly downregulated in BR3OE plants. BR3OE plants similarly exhibited delayed bolting and flowering times, and an increased number of rosette leaves under short-day (SD) conditions. BR3OE plants showed no significant phenotypic differences after vernalization treatment. BR3OE and WT plants exhibited early flowering after GA3 treatment, and bolting and flowering time remained delayed in BR3OE plants compared with WT plants. Key DELLA genes BrRGA1 and BrRGL3 exhibited a co-expression pattern consistent with BR3 gene in Chinese cabbage, which suggested that BrRGA1 and BrRGL3 genes may directly or indirectly regulated by BR3 gene. BR3 gene increased bolting resistance perhaps by upregulating the expression of DELLA genes in the GA pathway. This study provides new theoretical insights for addressing premature bolting in Chinese cabbage and offers novel approaches for breeding bolting-resistant varieties.
Orphan genes (OGs) are widely present in every species and have no significant sequence similarity to known genes (Li and Wurtele, 2014; Rdelsperger et al., 2019; Jiang et al., 2022). Numerous plant genomes have been rapidly decoded with the sequencing technology advancements, which provided a solid foundation for identifying OGs. A number of OGs have been identified in diverse species. For instance, there are 1324 OGs in the genome of Arabidopsis and 529 in the genome of B. rapa (Lin et al., 2010; Jiang et al., 2018). These genes lack recognizable functional domains, motifs, or structures, posing significant challenges for functional characterization of OGs. However, previous studies have shown that OGs play crucial roles in biotic and abiotic stress responses (Luhua et al., 2008; Jiang et al., 2018; Qi et al., 2018; Li et al., 2019; Jiang et al., 2020a; Wang et al., 2021; Tanvir et al., 2022), metabolism regulation (Li and Wurtele, 2014; Li et al., 2015; Jones et al., 2016; Jiang et al., 2020b; Wang et al., 2024), and species-specific trait formation (Hanada et al., 2008; Cui et al., 2015; Ni et al., 2017; O’Conner and Li, 2020; Jiang et al., 2023; Zu et al., 2024). The functions of OGs in plant growth and development recently garnered increasing attention. The interaction of Arabidopsis ICE1 (INDUCER OF CBF EXPRESSION 1) and IDD14 (INDETERMINATE DOMAIN 14) activates the transcription of OGs to regulate lipid metabolism in pollen, thus promoting pollen development and viability (Luo et al., 2024). Additionally, a novel OG, Bolting Resistance 1 (BR1), has been identified as a bolting resistance regulator in B. rapa, specifically delay flowering through vernalization and photoperiod pathways (Jiang et al., 2023). OG Bolting Resistance 2 (BR2) that regulates bolting resistance through the vernalization pathway, and its Arabidopsis overexpression upregulated flowering repressor FLC and downregulated key floral integrators (Zu et al., 2024). These findings highlight the vital roles of Chinese cabbage OGs in bolting resistance, although the exact mechanisms remain unclear.
Flowering time is a crucial agronomic trait of plant growth and development that is influenced by external environmental signals (e.g., photoperiod, temperature, and vernalization) and internal factors (e.g., autonomous pathways, age, and GA) (Pieper et al., 2020). Hormones, particularly GAs, are involved in cell division, elongation, and the transition from seed germination to flowering (Macmillan and Takahashi, 1968; Teotia and Tang, 2015). GAs, a class of diterpenoid plant hormones, promote flowering upon appropriate exogenous application (Hedden, 2020; Zhang et al., 2020). Defects in GA biosynthesis and signaling pathways often lead to aberrant flowering phenotypes, such as in GA-deficient mutant ga1-3, which does not flower under SD conditions (Wilson et al., 1992). Conversely, SPINDLY (SPY) is a negative regulator of the GA signaling pathway, and the enhancement of GA signaling in spy mutants leads to early flowering in Arabidopsis (Silverstone et al., 2006). As central GA signaling components, DELLA proteins inhibit flowering by interacting with the BRM-NF-YC functional module (Zhang et al., 2023). DELLA proteins delay flowering by repressing the expression of flowering-promoting factors, such as SOC1 and LFY. When GA signaling is enhanced, DELLA proteins are degraded, thereby relieving the repression of these genes (Achard and Genschik, 2008). Transcription factor WRKY75 regulates the GA signaling pathway by interacting with DELLA proteins, thus influencing flowering time and the photoperiod response in A. thaliana (Zhang et al., 2017). Recent research has shown that several regulatory factors influence GA signaling through distinct mechanisms, including C-TERMINAL DOMAIN PHOSPHATASE-LIKE3 (CPL3), Basic helix-loop-helix 4 (MdbHLH4), D2-Hydroxyglutarate Dehydroase (GhD2HGDH), and KNOTTED-like homeobox 15 (MdKNOX15). Although the role of GA signaling in flowering time regulation has been widely studied, its precise molecular mechanisms remain to be elucidated.
In this study, a novel B. rapa OG BR3 was identified. The expression patterns and subcellular localization of BR3 were determined. Flowering time and other related traits of A. thaliana BR3OE plants were analyzed under LD, SD, vernalization, and GA3 treatments. Additionally, the expression patterns of key flowering-related genes were determined using qRT-PCR analysis. This study evaluated the specific pathway through which BR3 regulates flowering, providing new insights into the function of OGs and offering a novel approach for breeding bolting-resistant Chinese cabbage varieties.
The plant materials used in this study were Chinese cabbage inbred line ‘GT-24’, wild-type A. thaliana (WT), T3 generation of BR3-overexpressing Arabidopsis plants (‘BR3OE’), and cultivated Nicotiana benthamiana. The cultivation methods followed those described in a previous study (Jiang et al., 2023).
The BR3 sequence was analyzed as previously described (Jiang et al., 2023). The BR3 sequence was amplified from ‘Chiifu’ and inserted into the EcoRI and XhoI restriction sites of pBinGlyRed3-35S vector which contains the hygromycin resistance gene. The recombinant vector pBinGlyRed3-35S-BR3 was introduced into Agrobacterium tumefaciens GV3101 competent cell using the freeze-thaw method. For the heterologous transformation of Chinese cabbage BR3 into Arabidopsis, the methods were based on those used in previous studies (Jiang et al., 2018; Li et al., 2021; Jiang et al., 2023). The primer pairs used in this study are listed in Supplementary Table S1.
Plants were cultivated under LD (16-h light/8-h dark photoperiod) or SD (16-h dark/8-h light photoperiod) conditions at approximately 22°C with 65% humidity. For vernalization treatment, germinated WT and BR3OE seeds were grown at 4°C for 4 weeks. For GA3 treatment, WT and BR3OE Arabidopsis plants were sprayed with 20 μM GA3 solution twice per week until flowering. In control groups of WT and Arabidopsis BR3OE plants, an equivalent amount of distilled water was sprayed. Phenotypic investigations were conducted following a previous study (Jiang et al., 2023). At least 15 plants were used for each experiment. After the cotyledons of Chinese cabbage ‘GT-24’ were fully expanded, 500 mg/L GA3 was sprayed, and samples were collected 12 h after spraying, with a total of six applications. As a control, ‘GT-24’ was treated with an equal volume of distilled water.
Histochemical GUS staining was performed as previously described (Li et al., 2021). Subcellular localization of the BR3 protein was performed according to the previous method (Jiang et al., 2023). After 24 h of incubation in the dark post-injection, samples were transferred to light conditions for continued incubation. Fluorescence signals were observed 48–72 h post-injection using a laser confocal microscope (Leica SP8, Germany) at excitation wavelengths.
Total RNA isolated, first-strand cDNA synthesis, and qRT-PCR were conducted according to the methods described in previous studies (Jiang et al., 2018, 2023). The primers used for qRT-PCR analysis are listed in Supplementary Table S1.
Statistical analysis using Student’s t-test or one-way ANOVA was performed using SPSS software (v26). Data are presented as the mean ± standard deviation (SD). Graphs were generated using GraphPad Prism software (v9.2).
The BR3 (BraA07003496) gene sequence was 347 bp and contained two exons and one intron located on the chromosome A07 of Chinese cabbage, encoding 76 amino acids (Supplementary Figure S1). A search in the NCBI-CDD conserved domain database showed that BR3 did not have any domains. BR3 was not predicted to function as a transcription factor based on the Plant Transcription Factor Database (TFDB). The Group-based Prediction System (GPS) showed that the BR3 protein lacked kinase activity. No signal peptides, cleavage sites, or transmembrane regions were identified. Structural prediction showed that BR3 consisted of α-helices, extended strands, and random coils, with random coils accounting for 42.11% of the structure. These findings suggest that BR3 is a novel gene with an unknown function, warranting further investigation to elucidate its role.
To investigate the role of BR3 gene expression during Chinese cabbage development, qRT-PCR analysis was performed on leaves at 2, 4, 6, 8, 10, and 12 days after the emergence of the first true leaf. The gene expression of BR3 showed notably higher expression levels on days 6 and 8, suggesting that BR3 gene expression persisted throughout the seedling growth phase (Figure 1A). Additionally, at 4 days after flowering, BR3 expression was detected in the stem, leaf, flower, and flower buds, with the highest expression observed in the flowers (Figure 1B). This suggests that BR3 may directly or indirectly involved in bolting resistance in Chinese cabbage.
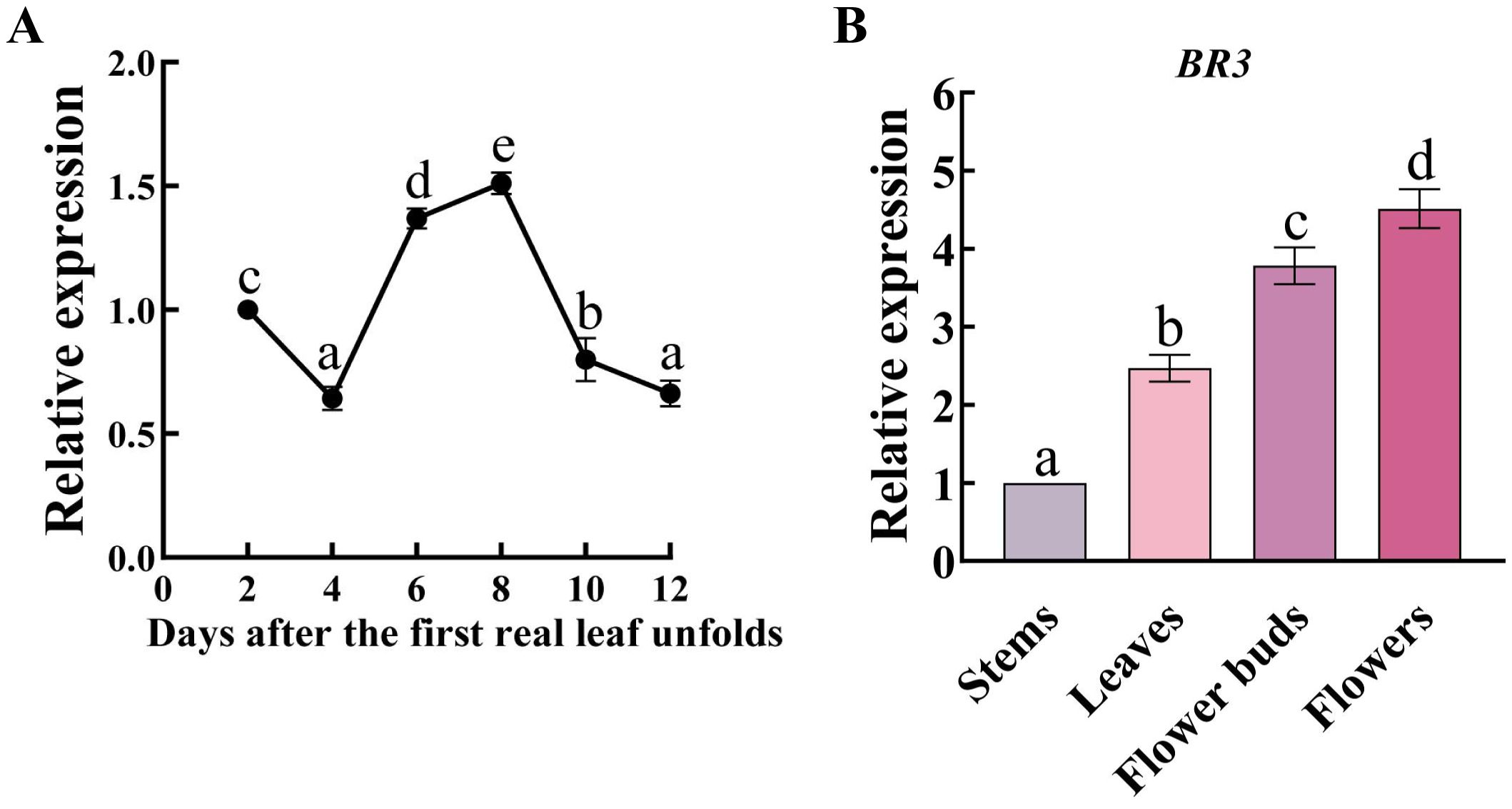
Figure 1. Expression patterns of the BR3 gene in Chinese cabbage. (A) BR3 gene expression during vegetative stage in Chinese cabbage. The samples of Chinese cabbage ‘GT-24’ cultured under LD conditions were collected from the aboveground parts of the Chinese cabbage at 2, 4, 6, 8, 10, and 12 days after the emergence of the first true leaf. (B) BR3 gene expression during the reproductive stage of Chinese cabbage. The samples were collected from different tissues of the aboveground parts of ‘GT-24’ at 4 days after flowering. There were three biological and three technical replications. Data are presented as the mean ± SD (one-way ANOVA, p < 0.05). Different lowercase letters represent significant differences in gene expression between different development stages or tissues.
To determine the spatiotemporal specificity of BR3 gene expression, GUS staining was performed on the flower buds, leaves, and roots of BR3 transgenic Arabidopsis plants. WT leaves were used as a negative control (Figures 2A–C). As shown in Figures 2D–F, significant blue staining was observed in the flower buds, leaves, and roots, indicating that the BR3 gene in Arabidopsis is regulated and expressed in these tissues after flowering. To better understand the mechanisms by which the BR3 protein functions within the cell, subcellular localization analysis was conducted. The 35S::BR3::GFP plasmid was introduced into N. benthamiana leaves via Agrobacterium tumefaciens injection, and fluorescence was observed under a confocal microscope to determine the localization of the BR3 protein. The distribution of fluorescent signals from the transiently expressed fusion protein reveled that BR3 was localized in both the nucleus and plasma membrane (Figures 2G–L). These findings provide a foundation for unraveling the flowering regulatory mechanisms of BR3.
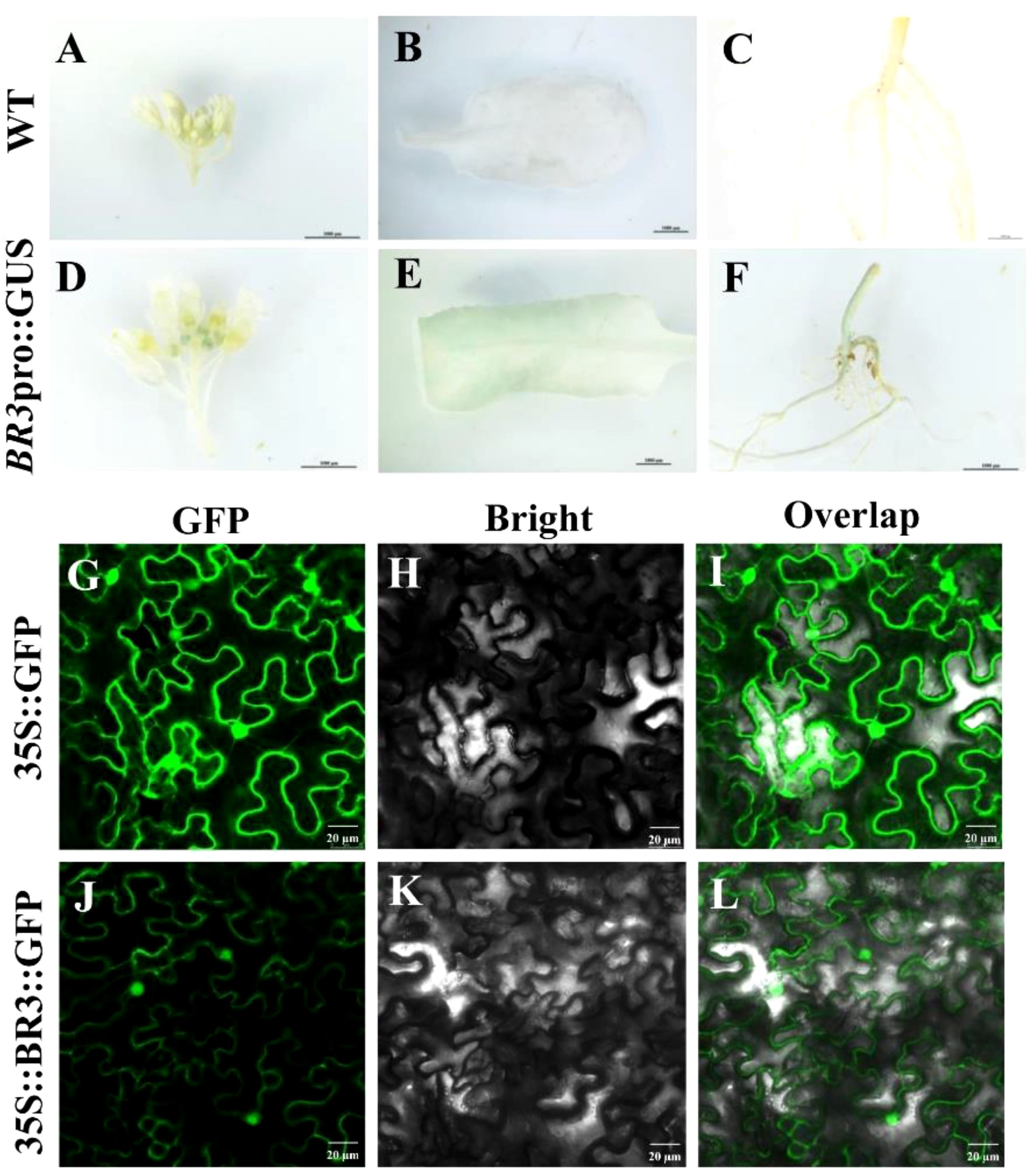
Figure 2. Expression analysis of BR3 gene promoter and subcellular localization of BR3. (A-F) Expression analysis of promoter fusion with GUS. Scale bar: 1000 μm. Subcellular localization of BR3 protein. (G-I) 35S::GFP plasmid positive controls and (J-L) 35S::BR3::GFP localization in N. benthamiana. (G, J) GFP fluorescence channels. (H, K) Bright field. (I, L) Merge field. A Leica confocal microscope was used to collect images at 48 h after agro-infiltration. Control GFP localization was evident throughout these cells. Scale bar: 20 mm.
To determine whether the late flowering phenotype of BR3OE was related to the photoperiod pathway, the flowering times of WT and BR3OE plants were recorded under LD and SD conditions.
BR3OE and WT plant phenotypes under LD conditions are shown in Figures 3A–C. The bolting time of BR3OE plants was 8.66 days later than that of WT (Figure 3D). In BR3OE plants, flowering time was delayed by 8.53 days (Figure 3E), and plant height decreased by 6.92 cm (Figure 3F). Concomitantly, the number of rosette leaves increased by 3.2 (Figure 3G), and the number of siliques was reduced by 9.47 (Figure 3H). Moreover, the phenotype of another BR3OE#2 line (Supplementary Figure S2) is consistent with that shown in the Figure 3C. Then, the expression levels of key flowering genes AtFT, AtSOC1, and AtLFY were measured using qRT-PCR. As shown in Figures 3I–K, the expression levels of AtFT, AtSOC1, and AtLFY were significantly downregulated in BR3OE plants compared with WT. These results suggest that BR3 delays flowering in Arabidopsis by repressing the expression of AtFT, AtSOC1, and AtLFY.
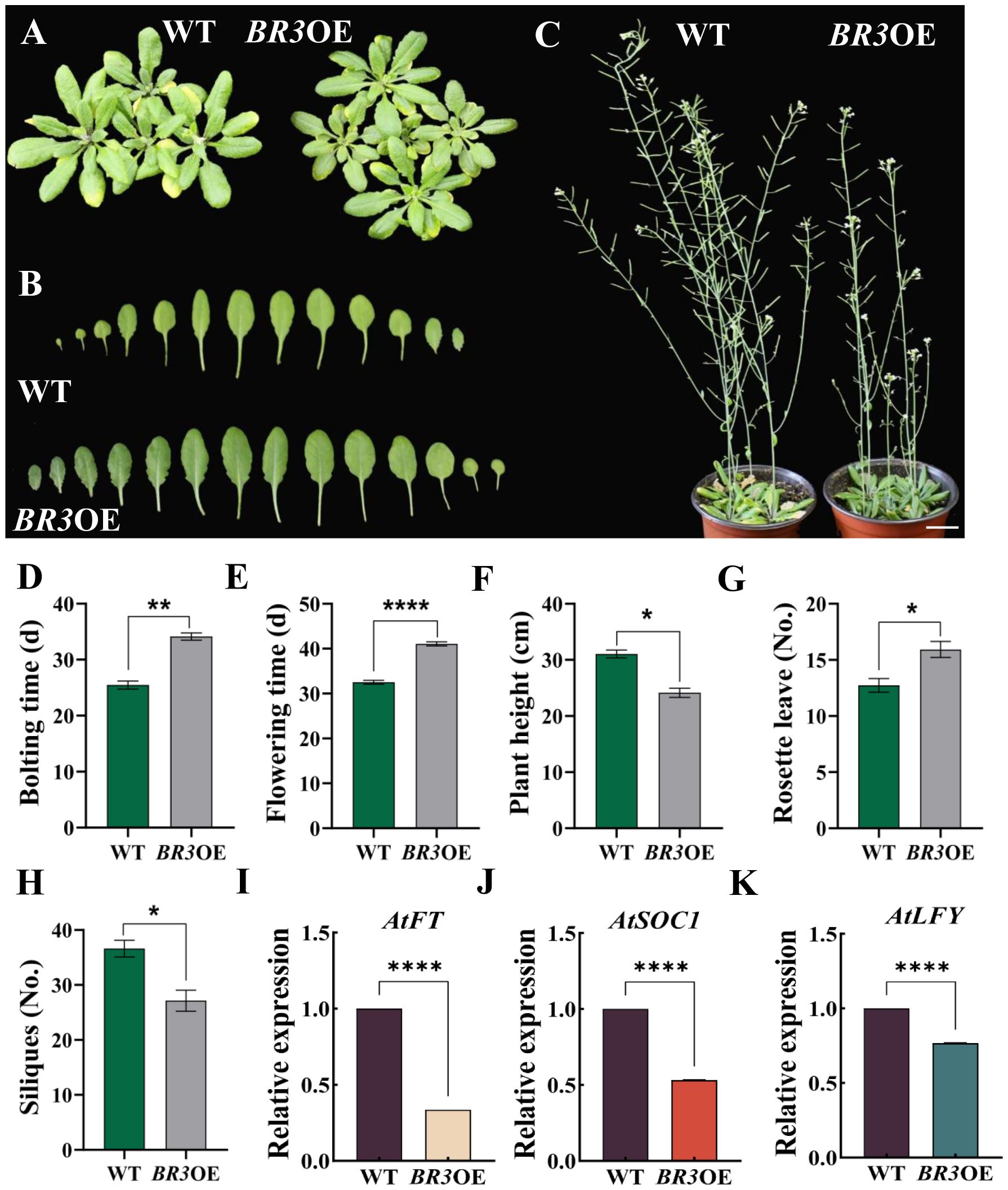
Figure 3. Phenotypes of WT and BR3OE under LD conditions and expression of key flowering genes. (A) Plant phenotypes of WT and BR3OE at 28 days. (B) Individual leaves of WT and BR3OE at 28 days. (C) Plant height of WT and BR3OE at 53 days. The scale bars are 2 cm. (D) Bolting time, (E) flowering time, (F) plant height, (G) number of rosette leaves, and (H) number of siliques of WT and BR3OE under LD conditions. (I-K) Expression of key flowering genes. Data are presented as the mean ± SD (Student’s t-test, *p < 0.05, **p < 0.01, and ****p < 0.0001).
The growth phenotypes of BR3OE and WT plants under SD conditions are shown in Figure 4A. The bolting time was delayed by 34.87 days (Figure 4B), and the flowering time of BR3OE plants was delayed by 35.27 days compared with that of WT plants (Figure 4C). The plant height was reduced by 2.21 cm (Figure 4D), and the number of rosette leaves increased by 3.53 (Figure 4E). BR3 gene overexpression led to a late-flowering phenotype under both LD and SD conditions, suggesting that delayed flowering in BR3OE is not influenced by the photoperiod. Additionally, the increased number of rosette leaves in BR3OE plants at the time of flowering suggests that BR3 promotes biomass accumulation, enhancing vegetative growth and inhibiting reproductive growth in Arabidopsis. These results indicate that BR3 regulates bolting resistance independent of the photoperiod.
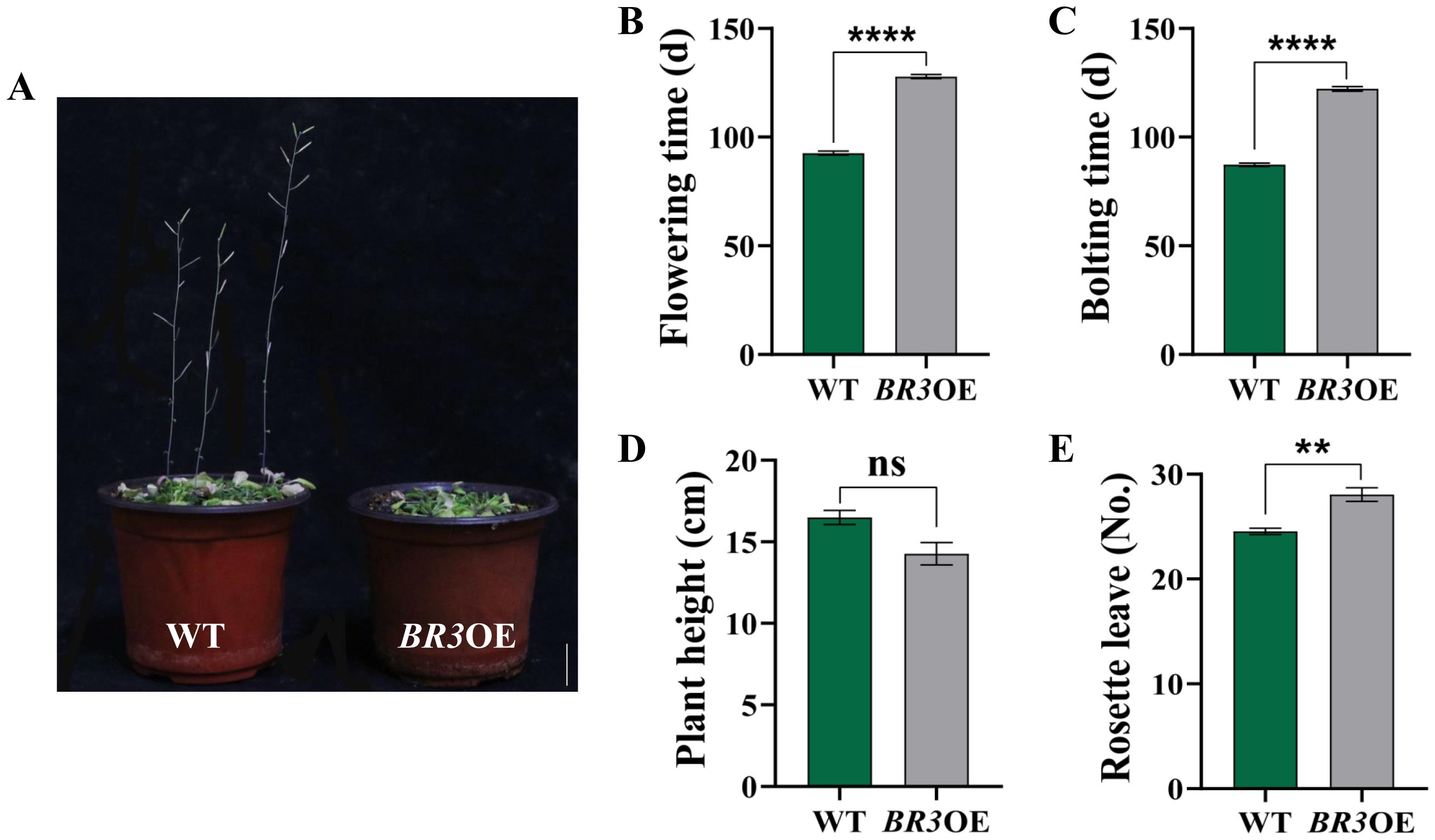
Figure 4. Phenotypes of WT and BR3OE plants under SD. (A) Phenotypes of WT and BR3OE at 120 days. The scale bars are 2 cm. (B) Bolting time, (C) flowering time, (D) plant height, and (E) number of rosette leaves of WT and BR3OE plants. Data are presented as the mean ± SD (Student’s t-test, **p < 0.01, and ****p < 0.0001).
The bolting and flowering times of WT were advanced by 4.33 and 4.4 days, respectively, after vernalization treatment (Figures 5A, B). Additionally, compared with the non-treated group, plant height increased by 2.8 cm, and the number of rosette leaves was reduced by 3.2 (Figures 5C, D). However, in vernalized BR3OE plants, there were no significant differences in bolting time, flowering time, or number of rosette leaves (Figures 5D, E). These results indicate that vernalization promotes early flowering in WT but not in BR3OE plants, suggesting that the BR3 gene delays flowering independently of the vernalization pathway and may function through other pathways.
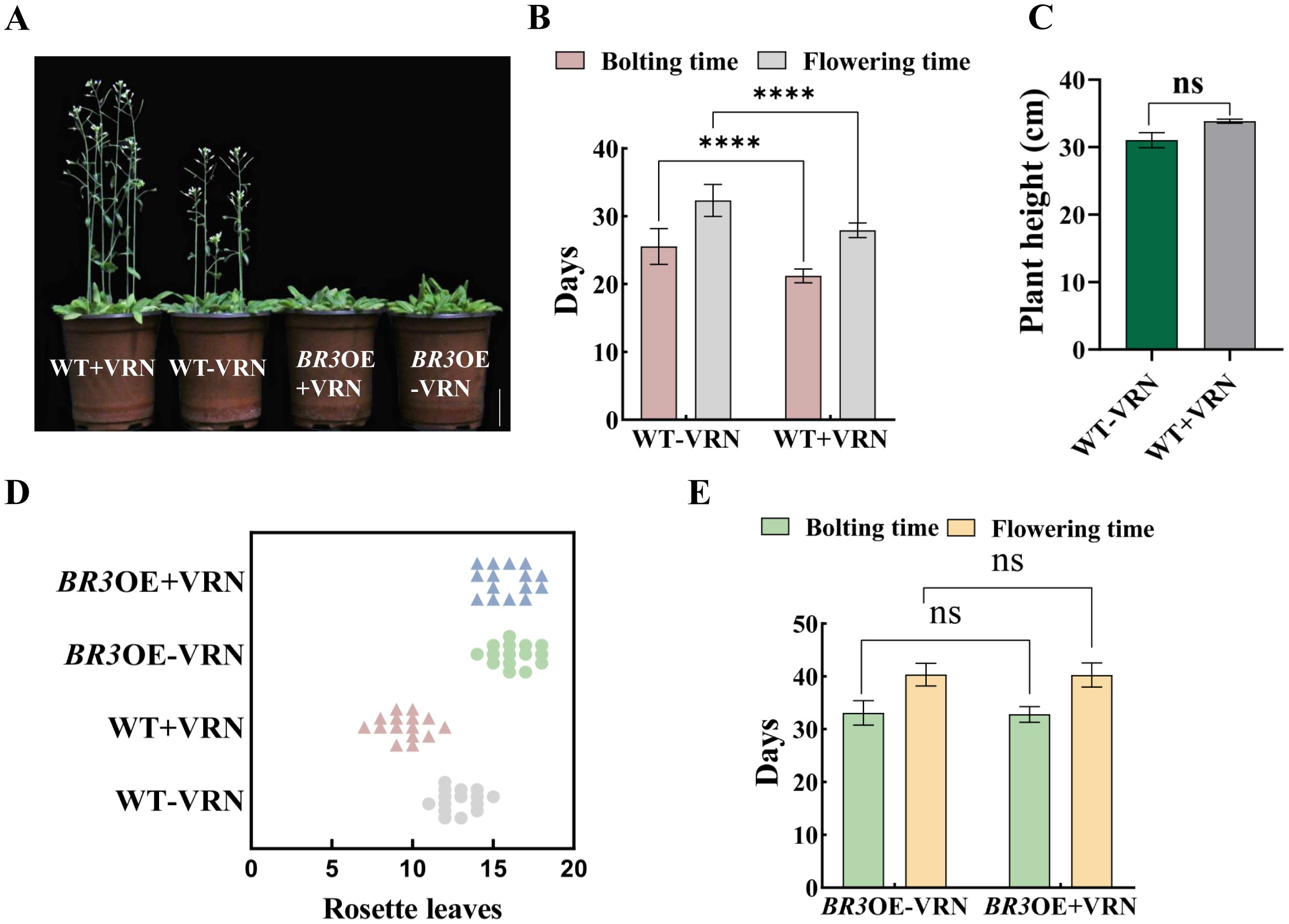
Figure 5. Agronomic traits in WT and BR3OE plants after vernalization treatment. (A) Phenotypes of WT and BR3OE control plants and plants treated with vernalization at 36 days. The scale bars are 2 cm. (B) Bolting and flowering time, (C) plant height, and (D) number of rosette leaves of vernalized and non-vernalized Arabidopsis WT plants. (E) Bolting and flowering time of vernalized and non-vernalized Arabidopsis BR3OE plants. +VRN, With vernalization treatment. -VRN, Without vernalization treatment. Data are presented as the mean ± SD; ns indicates not significant (Student’s t-test, ****p < 0.0001).
After treatment with GA3, the bolting and flowering times of WT were advanced by 4.47 and 4.13 days, respectively, compared with the non-treated control group (Figures 6A, B). Plant height increased by 1.98 cm, and the number of rosette leaves decreased by 3.47 (Figures 6C, D). After GA3 treatment, the bolting and flowering times of BR3OE plants were advanced by 5 and 4.73 days, respectively (Figure 6E), compared with non-treated plants, and the number of rosette leaves was reduced by 2.67 (Figure 6D). Exogenous GA3 application promoted flowering in BR3OE plants, which displayed a phenotype similar to WT (Figure 6A), suggesting that the BR3 gene influences flowering gene pathways in response to GA, leading to delayed flowering. DELLA proteins are key transcription factors in the GA signaling pathway. The B. rapa genome contains five DELLA subfamily members: BrRGL1, BrRGL2, BrRGL3, BrRGA1, and BrRGA2. The expression patterns of the five DELLA genes and the BR3 gene in Chinese cabbage were analyzed using qRT-PCR. The expression of the BrRGA2, BrRGL1, and BrRGL2 genes significantly decreased after the fifth sampling point. With the increase in the time and frequency of GA3 treatments, the expression of the BrRGA1 and BrRGL3 genes significantly increased after the fifth sampling, with a consistent increase in BR3 expression (Figure 7).
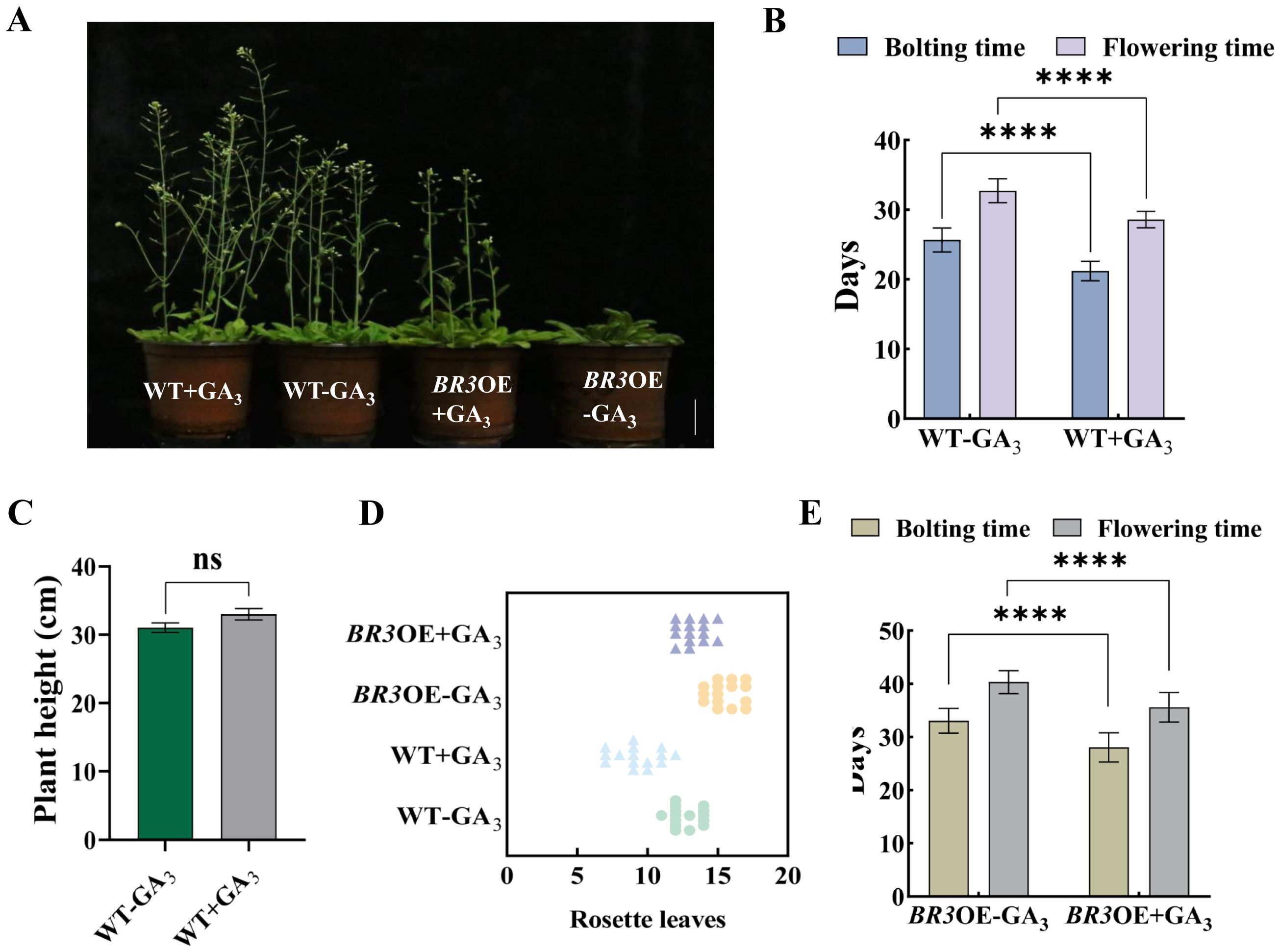
Figure 6. Agronomic traits in WT and BR3OE plants treated with GA3. (A) Phenotypes of WT and BR3OE control plants and plants treated with GA3 at 36 days. The scale bars are 2 cm. (B) Bolting and flowering time and (C) plant height of WT plants after GA3 treatment. (D) Number of rosette leaves in WT and BR3OE plants treated with GA3. (E) Bolting and flowering times of BR3OE plants treated with GA3. +GA3, With vernalization treatment. -GA3, Without vernalization treatment. Data are presented as the mean ± SD (Student’s t-test, ****p < 0.0001).

Figure 7. Expression analysis of BR3 and DELLA genes in Chinese cabbage after GA3 treatment. RGA1, REPRESSOR of GA1; RGA2, REPRESSOR of GA2; RGL1, RGA-LIKE PROTEIN 3; RGL2, RGA-LIKE PROTEIN 2; and RGL3, RGA-LIKE PROTEIN 3. The blue and yellow solid lines represent qRT-PCR results of the DELLA and BR3 genes of Chinese cabbage ‘GT-24’, respectively. Data are presented as the mean ± SD (one-way ANOVA, p < 0.05). Different lowercase letters represent significant differences in gene expression between different treatment times.
OGs are unique genes in plant genomes that regulate species-specific development, metabolism, and stress responses, enabling plants to adapt to specific environments, optimize metabolic pathways, and enhance stress resistance. Although the function of most OGs remains unknown, these genes are ubiquitously present in all species (Jiang et al., 2020b), highlighting the biological significance of the function and mechanisms of BrOGs. Previous studies have screened and identified OGs in B. rapa and thoroughly analyzed BrOGs sequence characteristics and expression patterns (Jiang et al., 2018). This study identified a novel OG, BR3, which positively regulated bolting tolerance in Arabidopsis, which further confirming the relationship between OGs and species-specific trait formation. Sequence analysis showed that BR3 with an unknown function that localized to both the cell membrane and nucleus, and key flowering genes were downregulated in BR3OE plants. Similarly, BR1 overexpression downregulates key flowering integrators, such as AtSOC1, AtLFY, and AtFUL (Jiang et al., 2023). Additionally, BR2 was found to be a positive regulator of bolting resistance through the vernalization pathway that localizes in the cell membrane, and in vernalized Chinese cabbage BR2OE, BrVIN3.b and BrFRI are downregulated, while BrFLC5 is upregulated, with key flowering factors, such as BrSOC1s, BrLFYs, and BrFTs, downregulated (Zu et al., 2024). These studies strongly support the findings of this study. BR3OE exhibited a bolting-resistant phenotype, and exogenous application of GA3 promoted flowering. Therefore, BR3 might delay flowering by acting on key genes in the GA pathway. The differences in subcellular localization and promoter-induced expression indicate distinct OGs that regulate bolting resistance through different pathways.
In this study, BR3 overexpression resulted in a delayed flowering phenotype under LD conditions (Figures 3C, E). Moreover, BR3OE plants showed significantly reduced expression of the AtFT, AtSOC1, and AtLFY genes compared with WT plants (Figures 3I–K). AtFT acts as a central integrator of environmental and endogenous signals that is translated into protein in the leaves and transported to the shoot apical meristem, where it upregulates AtSOC1 expression (Corbesier et al., 2007). SOC1 acts as a flowering integrator, coordinating other signaling pathways, such as photoperiod and temperature pathways, to regulate flowering time (Blazquez et al., 1998; Moon et al., 2003; Gregis et al., 2009; Liu et al., 2009; Fornara et al., 2010). LFY is a key flowering activator whose high expression promotes floral organ formation. Additionally, SOC1 enhances LFY expression by binding to its promoter region (Weigel et al., 1992). Exogenous GA3 application significantly enhances SOC1 expression in Arabidopsis, thereby shortening flowering time (Wang et al., 2022). Furthermore, GA influences flowering timing by directly affecting the expression of flowering regulatory genes such as LFY and SOC1 (Mutasa-Göttgens and Hedden, 2009). Simultaneously, FT may also influence GA metabolism by regulating key enzymes, such as GA2 oxidase 8-3 (GA2ox8-3). FT overexpression under LD conditions reduces GA2ox8-3expression (Miskolczi et al., 2019). Under SD conditions, endogenous GA levels increase significantly before flowering, promoting flowering by inducing FT in the leaves and SOC1 in the shoot apex (Fukazawa et al., 2021). GA promotes flowering by upregulating FT transcription under LD conditions (Hisamatsu and King, 2008; Porri et al., 2012). This suggests that BR3 may inhibit bolting and flowering in Arabidopsis through the GA pathway.
In this study, among Arabidopsis BR3OE plants subjected to photoperiod, vernalization, and GA3 treatments, only GA3-treated plants exhibited early flowering (Figure 6). BR3OE exhibited a late bolting phenotypes, and GA3 treatment promoted bolting and flowering. However, despite GA3 treatment, bolting and flowering occurred later in BR3OE than in WT plants (Figures 6A, B, E). DELLA proteins are negative regulators of the GA signaling pathway and inhibit the expression of flowering-related genes by interfering with transcription factor activities. When GA levels increase, DELLA protein degradation alleviates this repression, promoting flowering gene expression (Achard et al., 2003; Sun and Gubler, 2004). Studies have shown that BrARGL1, a key DELLA protein in Chinese cabbage, suppresses bolting when overexpressed, resulting in significantly reduced expression of GA-regulated proteins (BraGASA6), flowering-related genes (BraSOC1, BraLFY), expansin proteins (BraEXPA11), and xyloglucan endotransglucosylases (BraXTH3). Conversely, rgl1 mutants show the opposite phenotype. BRARGL1 inhibits transcriptional activation of BRASOC1 on BRAXTH3 and BRALFY genes; however, GA3 enhances transcriptional activation of BraSOC1, indicating that the BraRGL1-BraSOC1 module regulates bolting and flowering in Chinese cabbage through the GA signaling pathway (Wang et al., 2023). The expression of BrRGA2, BrRGL1, and BrRGL2 decreased with increased GA3 application, potentially due to their degradation. The expression levels of BrRGA1 and BrRGL3 were consistent with BR3, suggesting that increased BR3 expression promotes the of BrRGA1 and BrRGL3 expression (Figure 7). BR3 increases bolting resistance by increasing the expression of DELLA genes in the GA pathway.
Premature bolting is a primary limiting factor for spring-sown Chinese cabbage and cultivation in high-altitude, cold regions, leading to reduced yield and quality and causing significant economic losses. Therefore, identifying bolting resistance genes and developing bolting-resistant varieties are critical for ensuring a year-round balanced supply and stable production. In this study, Arabidopsis BR3OE exhibited bolting resistance. After GA3 treatment, bolting and flowering were promoted but occurred later than in GA3-treated WT, suggesting that BR3 may regulate bolting through the GA pathway. However, the proteins interacting with BR3 in Chinese cabbage, the transcription factors regulating its expression, and the molecular mechanisms by which the BR3 gene controls bolting resistance in Chinese cabbage remain unclear. Addressing these topics will provide a theoretical basis for elucidating the molecular mechanism of bolting resistance and offer new insights and gene resources for breeding bolting-resistant Chinese cabbage varieties.
In this study, a newly identified OG, BR3, positively regulated bolting resistance, supporting the role of OGs in controlling species-specific trait formation. The BR3 gene was highly expressed in flower buds and flowers, and the BR3 protein was localized in the nucleus and cell membrane. BR3OE exhibited a bolting-resistant phenotype and suppressed the expression of key flowering genes. Exogenous GA3 treatment and qRT-PCR analysis of the DELLA gene suggest that BR3 functions as a novel flowering time regulator through the gibberellin pathway. This study provides new insights into the breeding of bolting-resistant Chinese cabbage varieties and provides a theoretical foundation for further research on bolting resistance mechanisms in Chinese cabbage.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
YZ: Writing – review & editing, Conceptualization, Data curation, Methodology, Visualization, Writing – original draft. MJ: Formal analysis, Funding acquisition, Resources, Visualization, Writing – original draft, Writing – review & editing. SS: Methodology, Writing – review & editing. ZZ: Data curation, Project administration, Writing – review & editing. XL: Formal analysis, Funding acquisition, Methodology, Supervision, Validation, Writing – review & editing. ZP: Formal analysis, Funding acquisition, Resources, Supervision, Visualization, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32302568), the China Agriculture Research System of MOF and MARA (CARS-12), and the National Natural Science Foundation of China (32272715).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1518962/full#supplementary-material
Supplementary Figure 1 | Gene structure analysis of BR3 revealed exons and intron.
Supplementary Figure 2 | Phenotypes of WT and additional BR3OE#2 lines under LD conditions.
Achard, P., Genschik, P. (2008). Releasing the brakes of plant growth: How GAs shutdown DELLA proteins. J. Exp. Bot. 60, 1085–1092. doi: 10.1093/jxb/ern301
Achard, P., Vriezen, W. H., van der Straeten, D., Harberd, N. P. (2003). Ethylene regulates arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15, 2816–2825. doi: 10.1105/tpc.015685
Blazquez, M. A., Green, R., Nilsson, O., Sussman, M. R., Weigel, D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10, 791–800. doi: 10.1105/tpc.10.5.791
Corbesier, L., Vincent, C., Jang, S., Fornara, F., Fan, Q., Searle, I., et al. (2007). FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. doi: 10.1126/science.1141752
Cui, X., Lv, Y., Chen, M., Nikoloski, Z., Twell, D., Zhang, D. (2015). Young genes out of the male: an insight from evolutionary age analysis of the pollen transcriptome. Mol. Plant 8, 935–945. doi: 10.1016/j.molp.2014.12.008
Fornara, F., de Montaigu, A., Coupland, G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141, 550–550. doi: 10.1016/j.cell.2010.04.024
Fukazawa, J., Ohashi, Y., Takahashi, R., Nakai, K., Takahashi, Y. (2021). DELLA degradation by gibberellin promotes flowering via GAF1-TPR-dependent repression of floral repressors in Arabidopsis. Plant Cell 33, 2258–2272. doi: 10.1093/plcell/koab102
Gregis, V., Sessa, A., Dorca-Fornell, C., Kater, M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60, 626–637. doi: 10.1111/j.1365-313x.2009.03985.x
Hanada, K., Zou, C., Lehti-Shiu, M. D., Shinozaki, K., Shiu, S. H. (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 142, 993–1003. doi: 10.1104/pp.108.122457
Hedden, P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61, 1832–1849. doi: 10.1093/pcp/pcaa092
Hisamatsu, T., King, R. W. (2008). The nature of floral signals in Arabidopsis. II. Roles for FLOWERING LOCUS T (FT) and gibberellin. J. Exp. Bot. 59, 3821–3829. doi: 10.1093/jxb/ern232
Jiang, M., Dong, X., Lang, H., Pang, W., Zhan, Z., Li, X., et al. (2018). Mining of Brassica-specific genes (BSGs) and their induction in different developmental stages and under Plasmodiophora brassicae stress in Brassica rapa. Int. J. Mol. Sci. 19, 2064. doi: 10.3390/ijms19072064
Jiang, C., Hei, R., Yang, Y., Zhang, S., Wang, Q., Wang, W., et al. (2020a). An orphan protein of Fusarium graminearum modulates host immunity by mediating proteasomal degradation of TaSnRK1α. Nat. Commun. 11, 4382. doi: 10.1038/s41467-020-18240-y
Jiang, M., Li, X., Dong, X., Zu, Y., Zhan, Z., Piao, Z., et al. (2022). Research advances and prospects of orphan genes in plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.947129
Jiang, M., Zhan, Z., Li, H., Dong, X., Cheng, F., Piao, Z. (2020b). Brassica rapa orphan genes largely affect soluble sugar metabolism. Hortic. Res. 7, 181. doi: 10.1038/s41438-020-00403-z
Jiang, M., Zhang, Y., Yang, X., Li, X., Lang, H. (2023). Brassica rapa orphan gene BR1 delays flowering time in Arabidopsis. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1135684
Jones, D. C., Zheng, W., Huang, S., Du, C., Zhao, X., Yennamalli, R. M., et al. (2016). A clade-specific Arabidopsis gene connects primary metabolism and senscence. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00983
Li, X., Li, H., Zhao, Y., Zong, P., Zhan, Z., Piao, Z. (2021). Establishment of a simple and efficient Agrobacterium-mediated genetic transformation system to Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Hortic. Plant J. 7, 117–128. doi: 10.1016/j.hpj.2021.01.006
Li, G., Wu, X., Hu, Y., Muoz-Amatriaín, M., Luo, J., Zhou, W., et al. (2019). Orphan genes are involved in drought adaptations and ecoclimatic-oriented selections in domesticated cowpea. J. Exp. Bot. 70, 3101–3110. doi: 10.1093/jxb/erz145
Li, L., Wurtele, E. S. (2014). The QQS orphan gene of Arabidopsis modulates carbon and nitrogen allocation in soybean. Plant Biotechnol. J. 13, 177–187. doi: 10.1111/pbi.12238
Li, L., Zheng, W., Yanbing, Z., Ye, H., Tang, B. (2015). QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. U.S.A. 112, 14734–14739. doi: 10.1073/pnas.1514670112
Lin, H., Moghe, G., Ouyang, S., Iezzoni, A., Shiu, S. H., Gu, X., et al. (2010). Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol. Biol. 10, 1–14. doi: 10.1186/1471-2148-10-41
Liu, C., Xi, W., Shen, L., Tan, C., Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Luhua, S., Ciftci-Yilmaz, S., Harper, J., Cushman, J., Mittler, R. (2008). Enhanced tolerance to oxidative stress in transgenic Arabidopsis plants expressing proteins of unknown function. Plant Physiol. 148, 280–292. doi: 10.1104/pp.108.124875
Luo, L., Zheng, Y., Li, X., Chen, Q., Yang, D., Gu, Z., et al. (2024). ICE1 interacts with IDD14 to transcriptionally activate QQS to increase pollen germination and viability. J. Integr. Plant Biol. 66, 1801–1819. doi: 10.1111/jipb.13725
Macmillan, J., Takahashi, N. (1968). Proposed procedure for the allocation of trivial names to the gibberellins. Nature 217, 170–171. doi: 10.1038/217170a0
Miskolczi, P., Singh, R. K., Tylewicz, S., Azeez, A., Maurya, J. P., Tarkowská, D., et al. (2019). Long-range mobile signals mediate seasonal control of shoot growth. Proc. Natl. Acad. Sci. U.S.A. 116, 10852–10857. doi: 10.1073/pnas.1902199116
Moon, J., Suh, S. S., Lee, H., Choi, K. R., Lee, I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35, 613–623. doi: 10.1046/j.1365-313x.2003.01833.x
Mutasa-Göttgens, E., Hedden, P. (2009). Gibberellin as a factor in floral regulatory networks. J. Exp. Bot. 60, 1979–1989. doi: 10.1093/jxb/erp040
Ni, F., Qi, J., Hao, Q., Lyu, B., Luo, M.-C., Wang, Y., et al. (2017). Wheat Ms2 encodes for an orphan protein that confers male sterility in grass species. Nat. Commun. 8, 15121. doi: 10.1038/ncomms15121
O’Conner, S., Li, L. (2020). Mitochondrial fostering: the mitochondrial genome may play a role in plant orphan gene evolution. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.600117
Pieper, R., Tomé, F., Pankin, A., Korff, M. V. (2020). FLOWERING LOCUS T4 (HvFT4) delays flowering and decreases floret fertility in barley. J. Exp. Bot. 72, 107–121. doi: 10.1093/jxb/eraa466
Porri, A., Torti, S., Romera-Branchat, M., Coupland, G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139, 2198–2209. doi: 10.1242/dev.077164
Qi, M., Zheng, W., Zhao, X., Hohenstein, J. D., Kandel, Y., O’Conner, S., et al. (2018). QQS orphan gene and its interactor NF-YC4 reduce susceptibility to pathogens and pests. Plant Biotechnol. J. 17, 252–263. doi: 10.1111/pbi.12961
Rdelsperger, C., Prabh, N., Sommer, R. J. (2019). New gene origin and deep taxon phylogenomics: opportunities and challenges. Trends Genet. 35, 914–922. doi: 10.1016/j.tig.2019.08.007
Silverstone, A. L., Tseng, T.-S., Swain, S. M., Dill, A., Jeong, S. Y., Olszewski, N. E., et al. (2006). Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol. 143, 987–1000. doi: 10.1104/pp.106.091025
Sun, T.-P., Gubler, F. (2004). Molecular mechanism of gibberellin signaling in plants. Annu. Rev. Plant Biol. 55, 197–223. doi: 10.1146/annurev.arplant.55.031903.141753
Tanvir, R., Ping, W., Sun, J., Cain, M., Li, X., Li, L. (2022). AtQQS orphan gene and NtNF-YC4 boost protein accumulation and pest resistance in tobacco (Nicotiana tabacum). Plant Sci. 317, 111198. doi: 10.1016/j.plantsci.2022.111198
Teotia, S., Tang, G. (2015). To bloom or not to bloom: role of microRNAs in plant flowering. Mol. Plant 8, 359–377. doi: 10.1016/j.molp.2014.12.018
Wang, C., Chen, S., Feng, A., Su, J., Wang, W., Feng, J., et al. (2021). Xa7, A small orphan gene harboring promoter trap for AvrXa7, leads to the durable resistance to Xanthomonasoryzae pv. Oryzae. Rice 14, 48. doi: 10.1186/s12284-021-00490-z
Wang, Y., Huang, X., Huang, X., Su, W., Hao, Y., Liu, H., et al. (2022). BcSOC1 promotes bolting and stem elongation in flowering Chinese cabbage. Int. J. Mol. Sci. 23, 3459. doi: 10.3390/ijms23073459
Wang, L., O’Conner, S., Tanvir, R., Zheng, W., Cothron, S., Towery, K., et al. (2024). CRISPR/Cas9-based editing of NF-YC4 promoters yields high-protein rice and soybean. New Phytol. doi: 10.1111/nph.20141
Wang, Y., Song, S., Hao, Y., Chen, C., Ou, X., He, B., et al. (2023). Role of BraRGL1 in regulation of Brassica rapa bolting and flowering. Hortic. Res. 10, uhad119. doi: 10.1093/hr/uhad119
Weigel, D., Alvarez, J., Smyth, D. R., Yanofsky, M. F., Meyerowitz, E. M. (1992). LEAFY controls floral meristem identity in Arabidopsis. Cell 69, 843–859. doi: 10.1016/0092-8674(92)90295-n
Wilson, R. N., Heckman, J. W., Somerville, C. R. (1992). Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 100, 403–408. doi: 10.1104/pp.100.1.403
Zhang, L., Chen, L., Yu, D. (2017). Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiol. 176, 790–803. doi: 10.1104/pp.17.00657
Zhang, X., He, L., Zhao, B., Zhou, S., Chen, J. (2020). Dwarf and Increased Branching 1 controls plant height and axillary bud outgrowth in Medicago truncatula. J. Exp. Bot. 71, 6355–6365. doi: 10.1093/jxb/eraa364
Zhang, C., Jian, M., Li, W., Yao, X., Tan, C., Qian, Q., et al. (2023). Gibberellin signaling modulates flowering via the DELLA–BRAHMA–NF-YC module in Arabidopsis. Plant Cell 35, 3470–3484. doi: 10.1093/plcell/koad166
Keywords: Chinese cabbage, orphan gene, BR3, bolting resistance, Arabidopsis, GA pathway
Citation: Zhang Y, Jiang M, Sun S, Zhan Z, Li X and Piao Z (2025) Chinese cabbage orphan gene BR3 confers bolting resistance to Arabidopsis through the gibberellin pathway. Front. Plant Sci. 15:1518962. doi: 10.3389/fpls.2024.1518962
Received: 29 October 2024; Accepted: 26 December 2024;
Published: 20 January 2025.
Edited by:
Xiangshu Dong, Yunnan University, ChinaReviewed by:
Zhansheng Li, Chinese Academy of Agricultural Sciences, ChinaCopyright © 2025 Zhang, Jiang, Sun, Zhan, Li and Piao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaonan Li, Z3JhY2VzbGVleG44M0BzeWF1LmVkdS5jbg==; Zhongyun Piao, enlwaWFvQHN5YXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.