- 1Research Center for Applied Botany, National Research and Innovation Agency, Bogor, Indonesia
- 2Research Center for Biosystematics and Evolution, National Research and Innovation Agency, Bogor, Indonesia
1 Introduction
Artocarpus tamaran Becc. is a member of the Artocarpus genus of the Moraceae family, comprising 74 plant species (POWO, 2024). The species tree may attain a height of 45 m and a stem diameter of 1 m, with buttresses up to 3 m in height (Kochummen, 2000). The species is endemic to Borneo, occurring in Sarawak, Sabah, Kalimantan, and Brunei Darussalam, specifically in low land to the hilly mixed Dipterocarpaceae forest, beside the river, on sandstone, clay, and alluvial substrate (POWO, 2024; Jarrett, 1959). It has also been recorded in the primary or old secondary forests and logged forests at 20 m to 1800 m above sea level (Jarrett, 1959). According to the Red List category of the International United Conservation Nations (IUCN), Artocarpus tamaran is classified as Vulnerable A2c according to the Red List category of the International Union for Conservation of Nature (IUCN, 2024). The species is endangered due to habitat loss, which has been converted into plantations, logged, burnt down, and climate affected such as in Sabah, Sarawak, and Kalimantan (IUCN, 2024; POWO, 2024). The species is utilized for fiber material sourced from the bark, which is used to produce cloth and hats (Kulip, 2003; Fern 2014), fresh fruit, and edible seed after being boiled or roasted (Lim, 2012). The stem, referred to as “terap” in local terminology, has potential applications in construction (Kochummen, 2000). The log and timber prices of the species were 22.90 USD m-3 and 50.88 USD m-3, respectively (Karmini et al., 2020).
The chloroplast genome displays a quadripartite structure and is circular. The structure comprises a large single-copy region (LSC) and a small single-copy region (SSC), separated by a pair of inverted repeats (IRs), with some exceptions noted where the loss of an IR or the SSC has occurred. The size of the chloroplast genome in terrestrial plants ranges from 19 to 217 kb, with the IRs generally measuring 20-26 kb in length (http://www.ncbi.nlm.nih.gov/genome/organelle). The chloroplasts proteome consists of around 3000 proteins that play roles in photosynthesis, and the biosynthesis of fatty acids, amino acids, hormones, vitamins, nucleotides, and secondary metabolites (Dobrogojski et al., 2020). The advancement and utilization of chloroplast genome engineering technology may inform the investigation of chloroplast gene functions, gene editing, gene expression regulation, and genome analysis (An et al., 2022). Regulation of chloroplast gene expression in chloroplast genome engineering is employed to achieve high-value industrial targets, improve photosynthetic capacity, and biofortify food crops (Boynton et al., 1988). This study presents the results of the chloroplast genome sequencing of the A. tamaran species.
2 Method
2.1 Plant material, DNA extraction and sequencing
A sample of A. tamaran was obtained from the living collection of Bogor Botanical Gardens in West Java, designated with collector number IN577. The plant sample originated from Central Kalimantan. Genomic DNA was extracted from fresh leaves utilizing the CTAB (cetyltrimethylammonium bromide) method as described by Doyle and Doyle (1987). The initial quantification and purity of DNA were evaluated using a Nanodrop 2000 (Thermo Scientific) and visualized through agarose gel electrophoresis with 1% TBE agarose. The Qubit dsDNA HS Assay Kit (Thermo Scientific) was utilized for enhanced DNA quantification accuracy. The integrity of DNA was assessed utilizing the 4150 TapeStation (Agilent).
Genomic DNA was utilized as the input for library preparation. The genomic DNA was enzymatically fragmented to obtain the required insert size. The fragmented DNA was ligated with MGI-compatible adapters, each containing a unique barcode for each sample. PCR was performed to amplify the library. The quality and quantity of library samples were assessed using Tape Station and Qubit Fluorometer, respectively. The amplified library samples underwent circularization, and the resulting circular DNA served as input for the DNB formation process. The DNBs were loaded onto the flow cell, and sequencing was conducted for 612 cycles (PE300) utilizing the MGI DNBSEQ-G400.
2.2 Chloroplast genome assembly and annotation
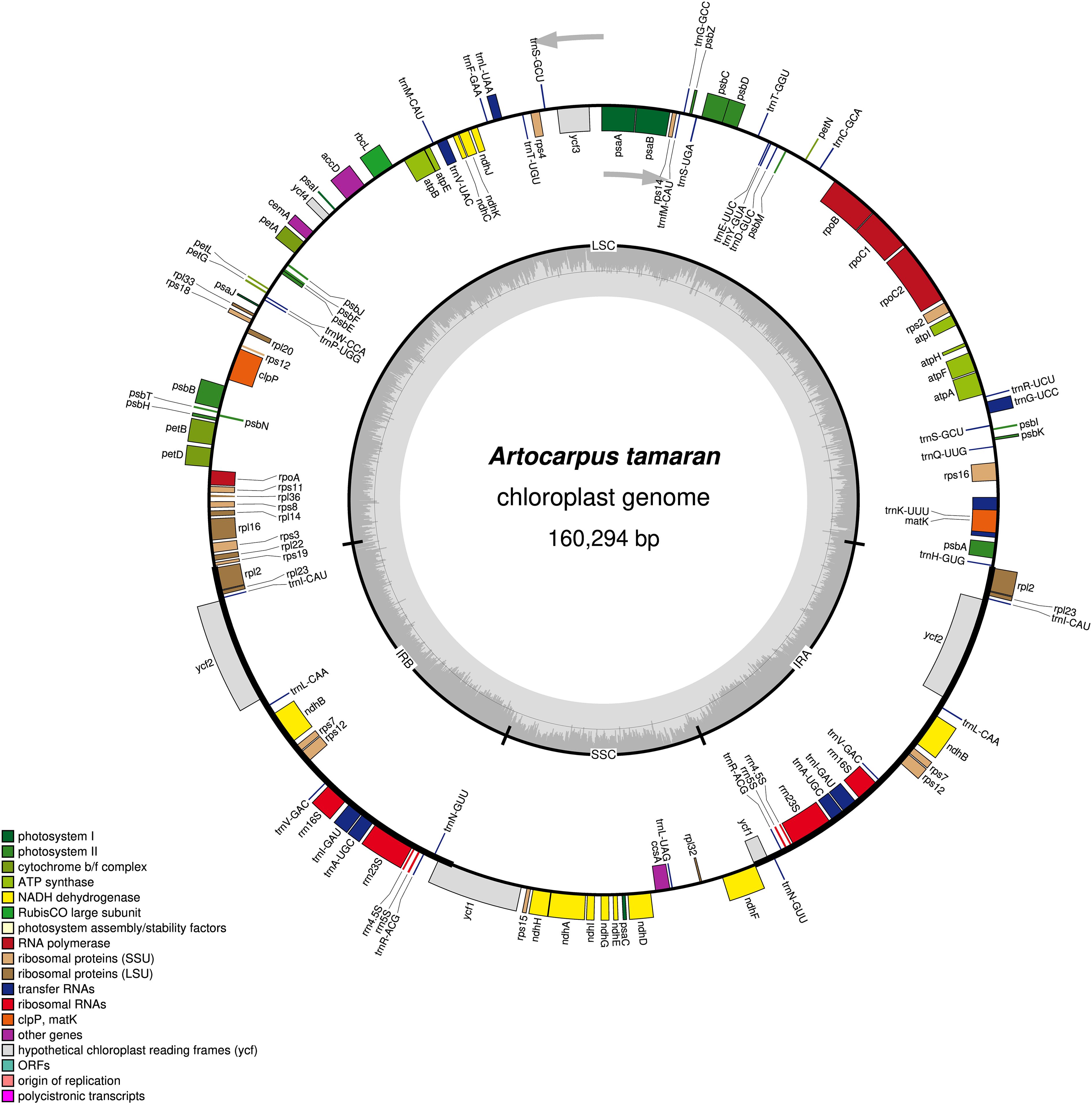
Quality control was conducted to evaluate the quality of reads utilizing FASTQC software version 0.11.8 (Andrews, 2010). Low-quality bases (less than 30), adapters, nucleotide position biases at the 3’ and 5’ ends, and sequence contamination were eliminated through trimming and filtering with Trimmomatic version 0.39. The parameters used were TruSeq3-PE.fa:2:30:10, SLIDINGWINDOW:4:28, LEADING:28, TRAILING:28, and MINLEN:20 (Bolger et al., 2014). The clean reads were then assembled using GetOrganelle version 1.7.7.1 (Jin et al., 2020). Annotation was conducted with CPGAVAS2 (http://47.96.249.172:16019/analyzer/annotate) (Shi et al., 2019), utilizing the cp genome of Artocarpus gomezianus Wall. ex Trécul (accession number: NC_080592) as a reference. This was followed by manual verification in Unipro Ugene v. 45.1 (Okonechnikov et al., 2012) and NCBI Genomic Workbench v. 3.8.2 (Kuznetsov and Bollin, 2021). The circular genome was visualized with OrganellarGenomeDRAW (OGDRAW) via the MPI-MP Chlorobox (Greiner et al., 2019).
3 Results
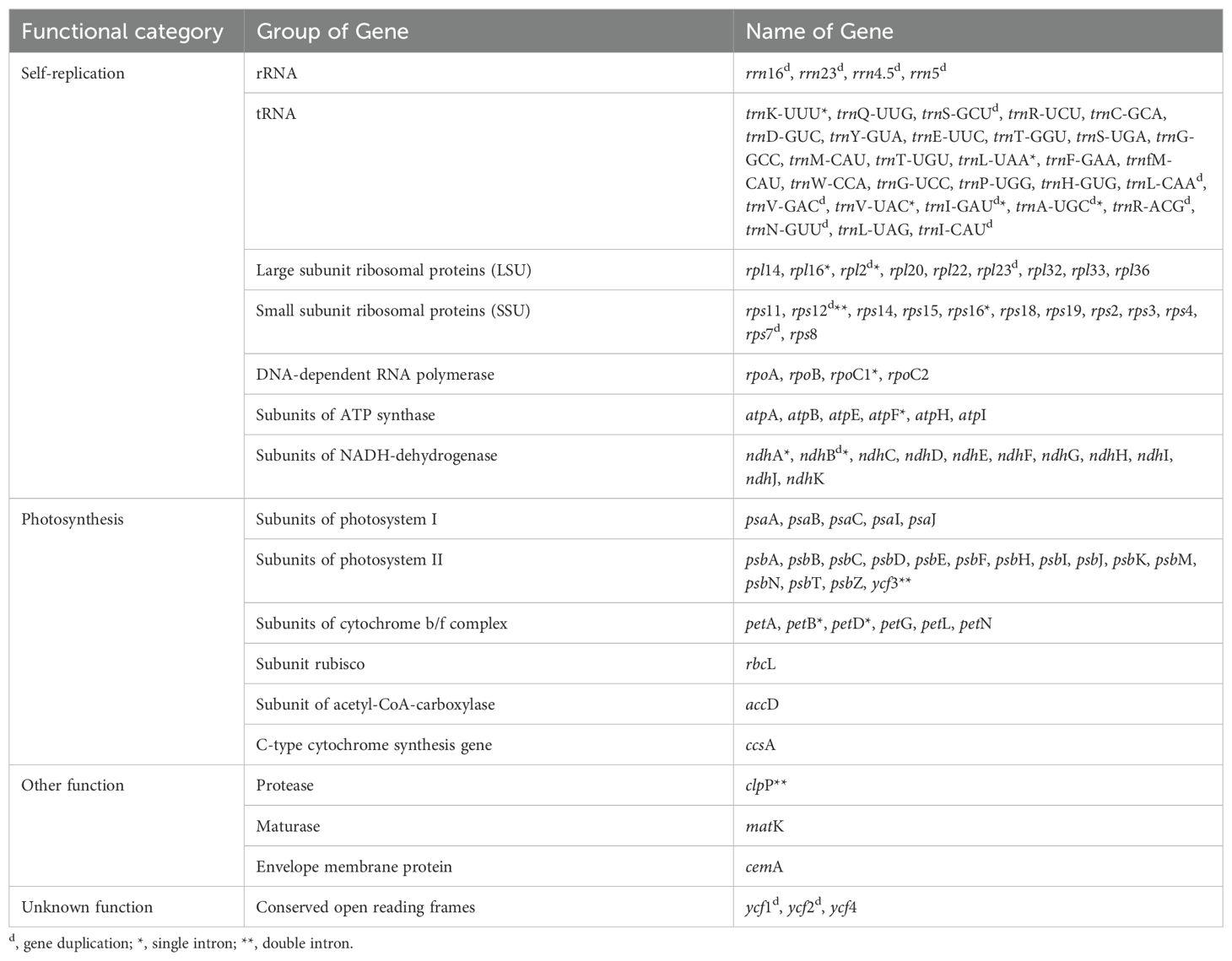
The complete chloroplast genome of A. tamaran has been successfully assembled, measuring 160,294 bp and exhibiting a quadripartite structure comprising four regions: the large single-copy (LSC) region, the small single-copy (SSC) region, and two inverted repeats (IR) regions (Figure 1). The LSC region has a length of 88,789 bp, the SSC region measures 20,015 bp, and each IR region is 25,745 bp. The genome exhibits a total GC content of 36%, with the highest concentration observed in the IR regions at 46.2%, followed by the LSC region at 34.2% and the SSC region at 28.9%. A total of 129 genes, comprising 110 unique genes, were annotated in the A. tamaran chloroplast genome. The identified genes comprised 84 protein-coding genes (77 unique), 37 tRNAs (29 unique), and 8 rRNAs (4 unique). Of the 129 genes analyzed, 14 exhibited a single intron, while three genes (rps12, ycf3 and clpP) contained two introns (see Table 1).
Data availability statement
This study analyzes datasets available in the NCBI Short Read Archive (SRA) under accession number SRR31020103 (https://www.ncbi.nlm.nih.gov/sra/SRR31020103). The BioProject and Bio-Sample numbers are PRJNA1173771 and SAMN44319506, respectively. The chloroplast genome sequence of A. tamaran has been deposited in the NCBI under accession number PQ493654.
Author contributions
RL: Writing – original draft, Funding acquisition, Conceptualization. MM: Writing – original draft, Methodology, Formal analysis, Data curation. MRH: Writing – review & editing, Project administration, Funding acquisition. IN: Writing – review & editing. AN: Writing – review & editing, Formal analysis, Data curation. FI: Writing – review & editing, Data curation.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study received funding from the Expedition and Exploration Funding Batch 2 for the fiscal year 2022, facilitated by the Deputy for Research and Innovation Facilitation, the National Research and Innovation Agency (BRIN), under contract number 2860/II.7/HK.01.00/8/2022.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
An, Y., Wang, Y., Wang, X., Xiao, J. (2022). Development of chloroplast transformation and gene expression regulation technology in land plants. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1037038
Andrews, S. (2010). FastQC: a Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. (accessed September 27, 2024).
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boynton, J. E., Gillham, N. W., Harris, E. H., Hosler, J. P., Shark, K. B. (1988). Chloroplast transformation in chlamydomonas with high velocity microprojectiles. Science 240, 1534–1538. doi: 10.1126/science.2897716
Dobrogojski, J., Adamiec, M., Lucinski, R. (2020). The chloroplast genome: a review. Acta Physiol. Plant 42, 98. doi: 10.1007/s11738-020-03089-x
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15.
Fern, K. (2014). Useful tropical plants database: Artocarpus tamaran Becc. Available online at: https://tropical.theferns.info/viewtropical.php?id=Artocarpus+tamaran (accessed October 1, 2024).
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
IUCN (2024). The IUCN Red List of Threatened Species Version 2024-1. Available online at: https://www.iucnredlist.org (Accessed 4 October 2024).
Jarrett, F. M. (1959). Studies in Artocarpus and allied genera III. A Revision of Artocarpus subgenus Artocarpus. J. Arnold. Arbor. 40, 113–155. doi: 10.5962/p.186026
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., DePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 1–31. doi: 10.1186/s13059-020-02154-5
Karmini, Karyati, Widiati, K. Y. (2020). Short Communication: The ecological and economic values of secondary forest on abandoned land in Samarinda, East Kalimantan Province, Indonesia. Biodiversitas 21, 5550–5558. doi: 10.13057/biodiv/d211164
Kochummen, K. M. (2000). “Artocarpus J. R. & G. Forster. nom. conserve,” in Tree Flora of Sabah and Sarawak. Eds. Soepadmo, E., Saw, L. G. (Sabah Forestry Department, Forest Research Institute Malaysia, and Sarawak Forestry Department, Kuala Lumpur), 187–212.
Kulip, J. (2003). An ethnobotanical survey of medicinal and other useful plants of Muruts in Sabah, Malaysia. Telopea 10, 81–98. doi: 10.7751/telopea20035608
Kuznetsov, A., Bollin, C. J. (2021). “NCBI genome workbench: desktop software for comparative genomics, visualization, and GenBank data submission,” in Multiple Sequence Alignment: Methods and Protocols. Ed. Katoh, K. (Humana Press, New York), 261–295.
Lim, T. K. (2012). “Artocarpus tamaran,” in Edible Medicinal and Non Medicinal Plants: Volume 3, Fruits (Springer, New York), 353–355.
Okonechnikov, K., Golosova, O., Fursov, M., Ugene Team (2012). Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 28, 1166–1167. doi: 10.1093/bioinformatics/bts091
POWO (2024). Plants of the World Online (Kew: Royal Botanic Gardens). Available online at: http://www.plantsoftheworldonline.org (Accessed 15 October 2024).
Keywords: conservation, illumina, Moraceae, plastid genome, underutilized fruit
Citation: Lestari R, Magandhi M, Hariri MR, Noviady I, Nugroho A and Indriani F (2024) Characterization of the complete chloroplast genome of the endangered and endemic bornean fruit Artocarpus tamaran Becc. Front. Plant Sci. 15:1513364. doi: 10.3389/fpls.2024.1513364
Received: 18 October 2024; Accepted: 18 November 2024;
Published: 12 December 2024.
Edited by:
Changmian Ji, Chinese Academy of Tropical Agricultural Sciences, ChinaReviewed by:
Chun-Lei Xiang, Chinese Academy of Sciences (CAS), ChinaRoohaida Othman, Universiti Kebangsaan Malaysia, Malaysia
Copyright © 2024 Lestari, Magandhi, Hariri, Noviady, Nugroho and Indriani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reni Lestari, cmVuaS5sZXN0YXJpQGJyaW4uZ28uaWQ=
 Reni Lestari
Reni Lestari Mahat Magandhi
Mahat Magandhi Muhammad Rifqi Hariri
Muhammad Rifqi Hariri Ikhsan Noviady
Ikhsan Noviady Aditya Nugroho
Aditya Nugroho Fitri Indriani
Fitri Indriani