
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 29 January 2025
Sec. Plant Biotechnology
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1503346
 Di Cong1
Di Cong1 Xue Zhao1,2
Xue Zhao1,2 Chang Ni1
Chang Ni1 Mengru Li1
Mengru Li1 Luwen Han1
Luwen Han1 Jianlin Cheng1
Jianlin Cheng1 Hongzhang Liu1
Hongzhang Liu1 Huijing Liu1
Huijing Liu1 Dan Yao1
Dan Yao1 Shuying Liu1*
Shuying Liu1* Guoshuang Chen3*
Guoshuang Chen3*MADS-box genes are classified into five categories: ABCDE, including SEP1, SEP2, SEP3, SEP4, and other homologous genes, which play important roles in floral organ development. In this study, the cDNA sequence of the HrSEP1 gene was cloned by RT-PCR and confirmed that this gene belongs to the MADS-box gene family. In addition, subcellular localization experiments showed that the HrSEP1 protein was localized in the nucleus. We verified the interaction of HrSEP1 with HrSOC1, HrSVP, and HrAP1 using yeast two-hybrid and bimolecular fluorescence complementation assays. These genes jointly regulate the growth and development of floral organs. We also found a strong synergy between HrSEP1 and AP1 genes in sepals, petals, and stamens by transgenic methods and fluorescence quantitative PCR, suggesting that HrSEP1 and AP1 may co-regulate the development of these structures. In conclusion, the expression of HrSEP1 has a certain effect on the development of floral organs, and these findings lay the foundation for further research on the biological functions of MADS transcription factors in Hippophae rhamnoides.
The MADS-box gene is a gene that plays a key role in plant growth and development. The name of the MADS-box gene is an acronym for the initials of important transcription factors in four species. M is derived from the yeast MCM1, which plays a regulatory role in cell metabolism, growth, and type (Passmore et al., 1988). A is AG involved in regulating floral organ formation in Arabidopsis thaliana (Yanofsky et al., 1990). D is DEF involved in regulating the formation of floral organs of snapdragon (Sommer et al., 1990). S is a human serum response factor SRF, which not only participates in human serum response but also regulates the transcription of proto-oncogene (Norman et al., 1988). They are diverse in type, number, and function and can be classified into two groups based on phylogenetic differences: type I transcription factors and type II transcription factors (Álvarez-Buylla et al., 2000). Among them, type II MADS-box transcription factors dominate, and they all have four specific structural domains: the MADS (M) domain is a conserved region that regulates DNA binding, the spacer domain (I) is involved in the formation of specific DNA-binding dimers, and the keratin (K) has a coiled-coil structure, which is responsible for participating in protein–protein interactions and is currently the validation of protein–protein interaction main site for verification of protein–protein interactions. The most variable region, the C-terminus (C), primarily acts as a transcriptional activator (De Bodt et al., 2003; Gramzow and Theissen, 2010). Based on their structural characteristics, type II genes can be further divided into two subfamilies: MIKCc type and MIKC* type (Kaufmann et al., 2005).
Initially, studies on the function of MADS-box transcription factors in plants focused on the development of floral organs. With the progress of science and research, a large number of MADS-box transcription factors have been found to regulate fruit ripening, softening, and nutrient metabolism in plants as well. In addition, they are involved in the regulation of microbe-induced signal transduction and hormone transport processes (Goto and Meyerowitz, 1994; Mandel and Yanofsky, 1995; Liljegren et al., 2000). By studying the floral organs of plants such as A. thaliana, researchers initially proposed the ABC model of the MADS-box gene, which plays a dominant role in the development of plant floral organs (Coen and Meyerowitz, 1991). Over time, more and more MADS-box genes were discovered and cloned in different species, leading to the proposal of the tetramer model and the establishment of the ABCDE model (Saedler et al., 2001; Rijpkema et al., 2010). The calyx is regulated by class A and E genes; petals are controlled by class A, B, and E genes; stamens are governed by class B, C, and E genes; carpels are influenced by class C and E genes; and ovules are modulated by class C, D, and E genes. In the A. thaliana genome, four SEP (SEPALATA) genes have been characterized as SEP1, SEP2, SEP3, and SEP4. They exhibit functional redundancy in determining sepals, petals, stamens, and carpels (Ditta et al., 2004). The function of SEP-like genes involves forming complex polymers through protein–protein interactions with class A, B, C, and D genes, acting as “adhesives” that play a crucial role in plant growth processes (Wang et al., 2020).
The class E gene PTM3/4/6 was expressed at all growth stages of male and female flowers in the woody plant Populus tremuloides (Pelaz et al., 2000). In situ, RNA localization definitively showed that the transcripts of PTM3/4 and 6 were mainly located in the internal sex wheel in the ovule of female flower development and the anther primordium of the male flower. Class E genes are crucial for reproductive growth in woody plants, particularly during flower development. PmSEP1 and PmSEP4 are the key genes controlling the development of the plum blossom meristem and sepal. PmSEP2 and PmSEP3 play an essential role in the morphogenesis of petals, stamens, and pistils. SEP-like genes regulate plant growth and development together with other MADS-box genes (Zhou et al., 2017). The expression pattern of PruneSEP1 during the storage of mature soft peaches was similar to that of genes related to ethylene biosynthesis and ethylene signal transduction (EIN2 and ETR2). Furthermore, the expression level of PruneSEP1 was positively correlated with the expression levels of EIN2 and ETR2. However, this phenomenon did not occur in hard peaches, which proves that PruneSEP1 regulates the ripening and softening of peaches (Li et al., 2017). Studies have proven that the SEP homologous gene OsMADS34 in rice combines with another gene, OsMADS1, which is similar to SEP in rice. This specifies the identity of flower organs, including the epidermis/palea, scales, stamens, and carpels (Gao et al., 2010). Patrice Morel et al. definitively showed that the SEP1/SEP2/SEP3 homologs of Petunia, together with AGL6, encode the classic SEP floral organ characteristics and flower termination function. The SEP3 homolog of Petunia (FBP2) plays a leading role in this process (Morel et al., 2019). The SOC1 gene is a key regulator of the transition from vegetative to reproductive development in A. thaliana. In Dendrobium nobile, DOSOC1 is highly expressed in the inflorescence apex, flower stem, flower bud, and other parts. During the transition period of flowering, DOSOC1 expression is unequivocally upregulated throughout the entire seedling (Ding et al., 2013). The SVP subfamily genes are indisputably linked to the flowering of 1-year species and the dormancy of buds in multi-year species. During the nutritional period, the flower suppressor complex, composed of SVP and FLOWERING LOCUS C (FLC), definitively downregulates the key flowering gene SOC1 and flowering site T (FT), thereby inhibiting flowering (Li et al., 2008). The SEP4 gene plays a role in inflorescence branching in Arabidopsis and rice. It can redundantly inhibit TFL1 with SOC1 and SVP genes. This shows that the SEP4 gene is involved in regulating inflorescence branching (Liu et al., 2013).
Hippophae rhamnoides is a highly beneficial perennial woody plant with numerous advantages for environmental management and human health. The study of anti-pseudorabies virus activity in vitro revealed that H. rhamnoides polysaccharide can reduce the malondialdehyde content and reactive oxygen species level in cells and increase superoxide dismutase activity. These results prove that H. rhamnoides polysaccharide can reduce the oxidative stress caused by pseudorabies virus infection (Huan et al., 2022). Phenylpropanoids extracted from H. rhamnoides are an effective treatment for myocardial injury induced by doxorubicin in zebrafish, and they inhibit myocardial apoptosis (Li et al., 2022). H. rhamnoides pulp and H. rhamnoides oil are extremely rich in bioactive substances and are relatively friendly to the human body. They play a crucial role in the application of traditional Chinese and Tibetan medicine. The inhalation of SO2 can change the ratio of liver, lung, kidney, spleen, and other organs in mice’s body. A study proved that rhamnosus seed oil has a protective effect on the damage caused by inhalation of sulfur dioxide in mice (Ruan et al., 2003). H. rhamnoides oil is a vital component of cancer treatment during chemotherapy and radiation therapy. It can also help counteract many side effects and assist in treatment, restoring kidney and liver function (Huang et al., 2020). Supplementing with H. rhamnoides may affect lipid metabolism in human circulation, but it will not affect blood glucose index and blood pressure value (Geng et al., 2022)].
The MADS-box gene is a crucial pathway for understanding and controlling the developmental mechanisms of H. rhamnoides. It can also significantly accelerate progress in plant breeding and crop improvement in biotechnology. Regrettably, there are still only a few reports on the study of MADS-box transcription factors in H. rhamnoides. It is, therefore, crucial to improve the regulatory network of MADS-box genes in sea buckthorn and explore the site of action, regulatory mechanism, and interaction mode of MADS-box genes to study the development of H. rhamnoides flowers in greater depth. This article establishes the foundation for studying the expression patterns of MADS-box transcription factors in H. rhamnoides and the mechanisms of interaction between transcription factors.
The Fuxin No. 2 H. rhamnoides fruit was taken from the H. rhamnoides planting base of Jilin Agricultural University in China. The H. rhamnoides fruit was temporarily frozen in a laboratory −80°C ultra-low temperature refrigerator. NC89 wild-type tobacco and 2-week-old Nicotiana benthamiana seedlings were used, both of which are stored in laboratory seed management libraries.
Extract RNA using the RNA Pure Plant Kit (CoWin Biosciences, Cambridge, MA, USA; CW0559). We used agarose gel electrophoresis and nanometer titration electrophoresis to detect the quality of the RNA. We extracted the RNA using the RNA Pure Plant Kit (CoWin Biosciences, CW0559). We then detected the quality of the RNA using agarose gel electrophoresis and a NanoDrop ONE (Thermo Scientific, Shanghai, China). We designed gene-specific primers using Primer 7.0 (Supplementary Table S1). The SEP1 gene sequence of H. rhamnoides is sourced from the H. rhamnoides transcriptome database, which is preserved by the research team (login number: PRJNA612989). We amplified the target gene fragment using PCR technology (Takara, Tokyo, Japan) and named it HrSEP1. We used the DNA recovery kit (TIANGEN, Beijing, China; DP209) to recover and purify the target fragment, which was then cloned onto the pMD-18T vector (Takara, 6011).
The MEGA 10.0 software was used to construct a MADS-box gene phylogenetic tree. The prediction results of the tertiary structure of the HrSEP1 gene can be obtained through https://swissmodel.expasy.org/interactive. The information related to the HrSEP1 gene signal peptide can be obtained from http://www.cbs.dtu.dk/services/SignalP/Obtain.
The subcellular localization vector pCAMBIA1300-GFP was cleaved into linear fragments using restriction endonucleases BamHI and SalI. Specific primers with corresponding cleavage sites were designed (Supplementary Table S1) to construct the recombinant plasmid pCAMBIA1300-HrSEP1. The recombinant plasmid was transformed into Agrobacterium tumefaciens competent cell EHA105 and injected into tobacco leaves from the back wound to infect the plant. We observed fluorescence with green fluorescence protein (GFP) as the nuclear marker (Liu et al., 2019).
Restriction endonucleases EcoRI and BamHI were used to cleave the bait protein vector pGBKT7 linearly and design specific primers with corresponding cleavage sites (see Supplementary Table S1). The double enzyme digestion experiment was followed by the use of T4 ligase to connect the enzyme digestion product with HrSEP1, thus constructing the recombinant plasmid pGBKT7-HrSEP1. The plasmid was then transferred into yeast-receptive cell AH109 to confirm that HrSEP1 has self-exciting activity in yeast cells by observing the color of the strain.
The H. rhamnoides MADS-box genes HrAP1, HrSOC1, and HrSVP were screened out from the transcriptome database and combined with the prey protein vector empty pGADT7 to form recombinant plasmids pGADT7-HrAP1, pGADT7-HrSOC1, and pGADT7-HrSVP. Once the prey protein vector was successfully constructed, the combination of pGADT7-HrAP1+pGBKT7-HrSEP1, pGADT7-HrSOC1+pGBKT7-HrSEP1, and pGADT7-HrSVP+pGBKT7-HrSEP1 were used to cotransfect AH109 yeast receptive cells. The positive control pGADT7-T+pGBKT7-53 and negative control pGADT7-T+pGBKT7-lam were also transformed. Each group of the bacterial solution was applied to solid culture media of SD/-Leu/-Trp/-His/-Ade. The plates were sealed and incubated upside down at 29°C–30°C for 3–5 days.
We used restriction endonucleases SmaI and XbaI to cleave the BiFc vector pXY104 linearly. Based on the two restriction sites, we designed homologous arm primers to connect the vector pXY104 and HrSEP1, forming the recombinant plasmid pXY104-HrSEP1. Similarly, we cleaved the BiFc complementary vector pXY106 into linear fragments using restriction endonucleases SmaI and BamHI. We designed homologous arm primers to connect the vector pXY106 and the prey gene, combining the prey protein gene sequence and pXY106 sequence, to construct recombinant plasmids pXY106-HrAP1, pXY106-HrSOC1, and pXY106-HrSVP.
The bacterial solution containing pXY104-HrSEP1 was mixed with the bacterial solution containing pXY106-HrAP1, pXY106-HrSOC1, and pXY106-HrSVP recombinant complementary vectors. Then, using a sterile syringe, the mixed bacterial solution was extracted and injected into the wound on the back of the tobacco leaf. The tobacco was left in a dark and ventilated location for 40–70 hours. After cutting the leaves, yellow fluorescent protein (YFP) was used as the nuclear marker for fluorescence detection (Kerppola, 2008).
The plant expression vector pCAMBIA3301 was cleaved into a linear plasmid with a sticky end using restriction endonucleases NcoI and BglII. HrSEP1 was linked to the linear plasmid pCAMBIA3301 using T4 ligase to form a recombinant plasmid, pCAMBIA3301-HrSEP1. The recombinant plasmid was transformed into A. tumefaciens competent cell EHA105 and then infected with NC89 tobacco leaves using the leaf disc transformation method (An et al., 2021).
The melting curve was definitively established to confirm the specificity of the amplified fragment. The expression data were normalized using actin genes as reference genes. The relative expression of genes was calculated using the 2−ΔΔCT method. All experiments were repeated three times independently.
All samples in the experiment were assessed in at least three independent biological replicates, and all data are expressed as the mean ± standard deviation (SD). The t-test or one-way ANOVA with a 5% significance level was used to determine whether there were statistically significant differences between the means. The graphs were constructed using Origin 9.5.1 (Microcal Software Inc., Northampton, MA, USA).
There are 49 MADS-box genes annotated by the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway in the transcriptome database differential genes, protein analysis of these MADS-box genes found that only 27 of them have relatively complete open reading frames, four of them do not have k-box structural domains specific to MADS-box genes, and analysis of the remaining 23 genes revealed that the proportion of class E genes, SEP, was the largest. After analyzing the remaining 23 genes, it was found that the proportion of SEP in the E-class genes was the largest, and one of them has the most significant difference in expression among all the MADS-box genes. Therefore, we chose this gene as the target gene for the subsequent experiments and named it HrSEP1.
We extracted total RNA from rhamnosus fruits and successfully obtained the cDNA sequence of HrSEP1 by reverse transcription. This allowed us to investigate the relationship between HrSEP1 and floral organ development. PCR amplified the HrSEP1 gene sequence. Gene sequencing and DNAMAN V6.0 multiplexing showed that the HrSEP1 sequence was 735 bp and encoded 244 amino acids.
Next, to analyze the function of HrSEP1 in the MADS gene family, we downloaded MADS gene protein sequences from the National Center for Biotechnology Information (NCBI) database for 70 species, including A. thaliana, actinomycetes, neem, and tomato plants. We constructed a phylogenetic tree for comparative analysis. The data clearly show that the HrSEP1 gene is highly homologous to the SEP genes in Hevea brasiliensis and A. thaliana and belongs to class E (Figure 1).
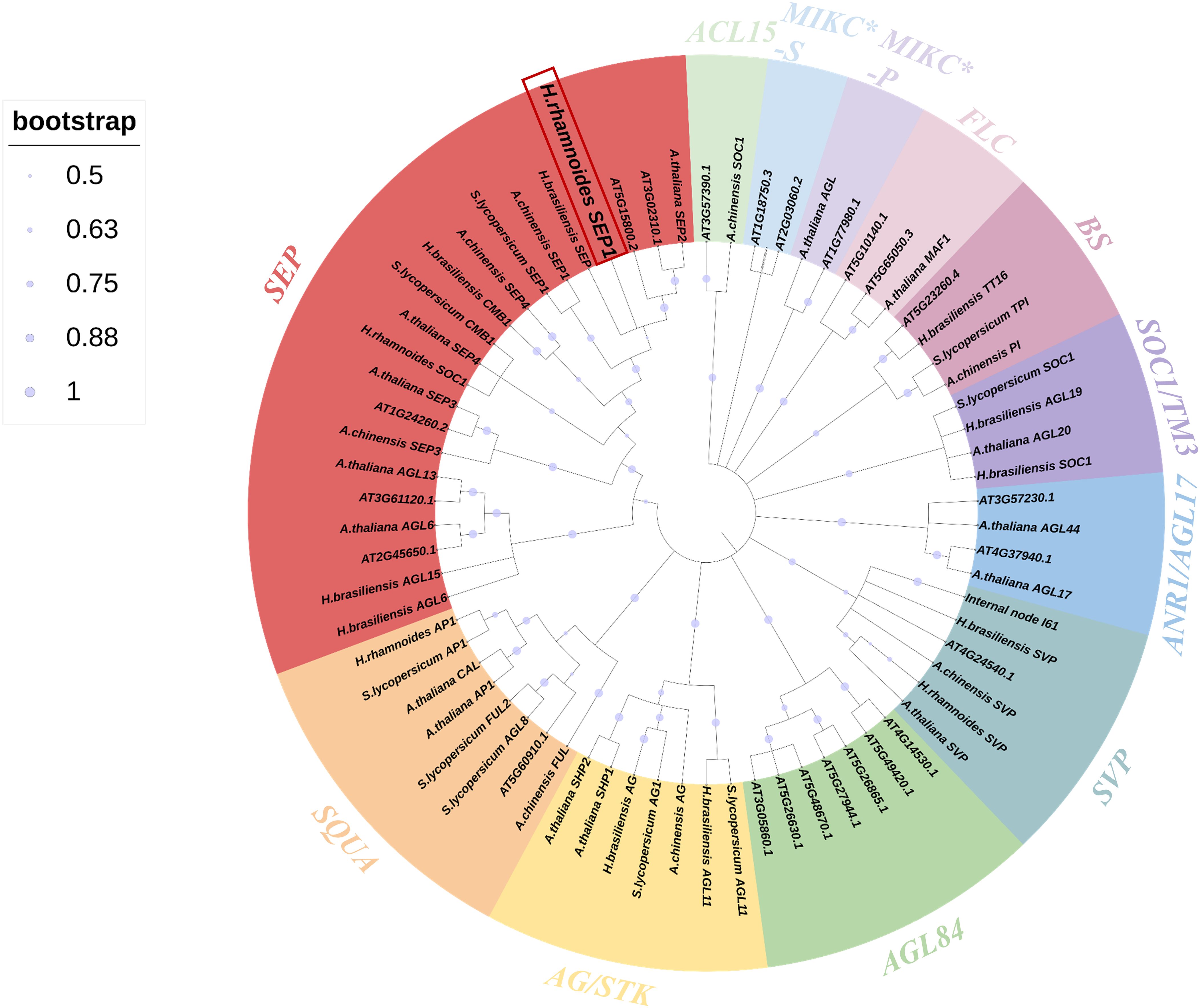
Figure 1. Phylogenetic tree of the MADS-box gene in Hippophae rhamnoides L. and the MADS-box genes in Arabidopsis thaliana, Actinidia chinensis Planch, Hevea brasiliensis, and Solanum lycopersicum L. Arabidopsis gene sequences from The Arabidopsis Information Resource (TAIR) database; other gene sequences were obtained from the National Center for Biotechnology Information (NCBI) database. Different subfamilies were marked with specific colors. Trees were constructed using the neighbor-joining method, and 1,000 bootstrap replications were performed using MEGA 7.0 International software.
To better understand the function of HrSEP1, the HrSEP1 protein was localized in vivo. GFP was fused to the C-terminal of the HrSEP1 under the control of the CaMV 35S promoter and transiently expressed in N. benthamiana leaf epidermal cells, and the results showed that HrSEP1 is a nuclear-expressed protein (Figure 2).
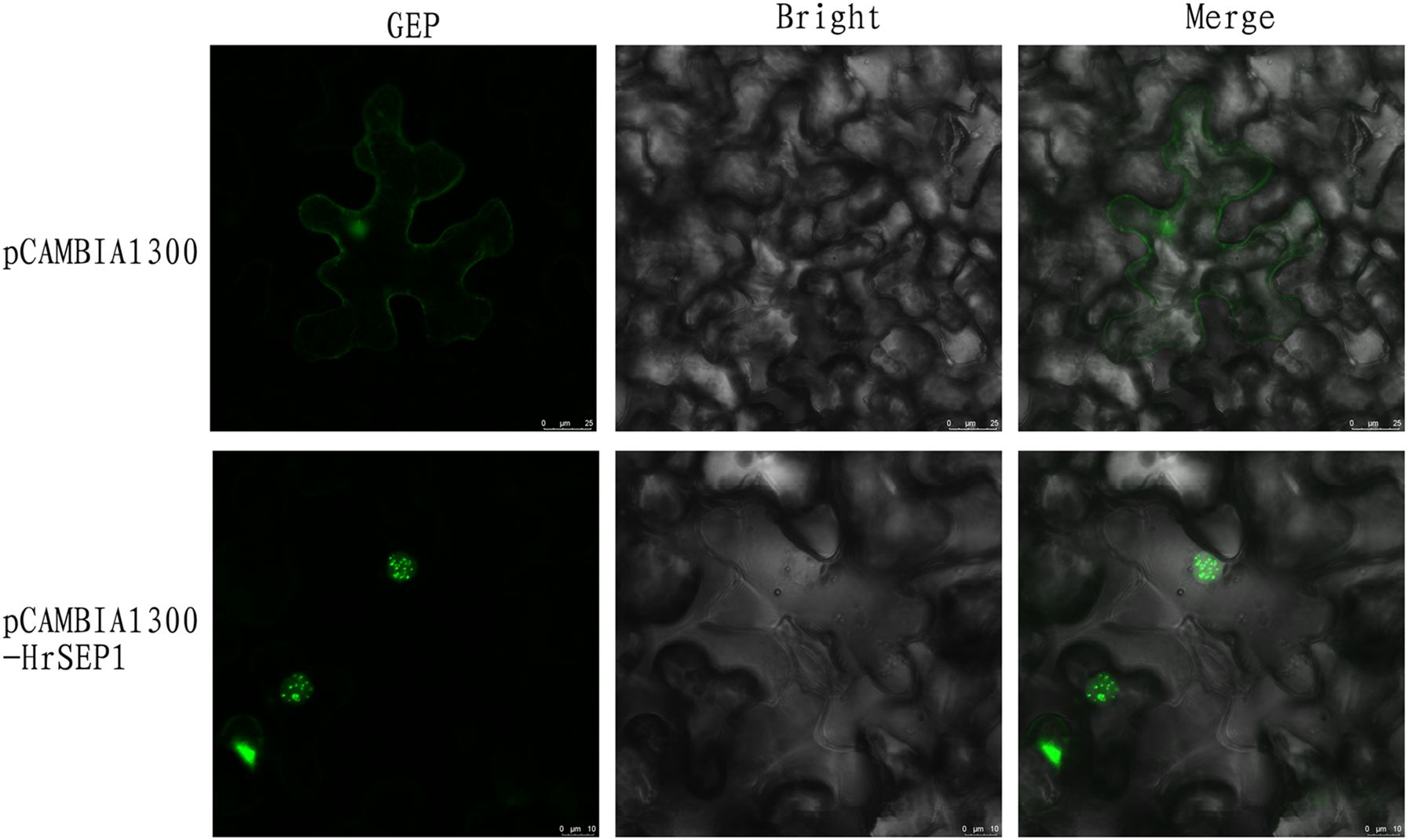
Figure 2. Subcellular localization of HrSEP1. Combining HrSEP1 gene with fluorescent protein vector pCAMBIA1300-GFP can determine the target gene expression location. Green fluorescence protein (GFP) as nuclear marker.
We constructed a plant overexpression vector, pCAMBIA3301-HrSEP1 (Figure 3A), using pCAMBIA3301 as the vector skeleton, and transformed the recombinant plant expression vector into A. tumefaciens competent cell EHA105. We used the leaf disc transformation method to infect NC89 tobacco leaves (Figure 3B) using Basta herbicide as a preliminary screen for transgenic plants. We successfully detected the genomic DNA of transgenic plants by PCR and ultimately obtained 17 T2-generation overexpression-positive tobacco plants (Figure 3C), which we have named T-HrSEP1.
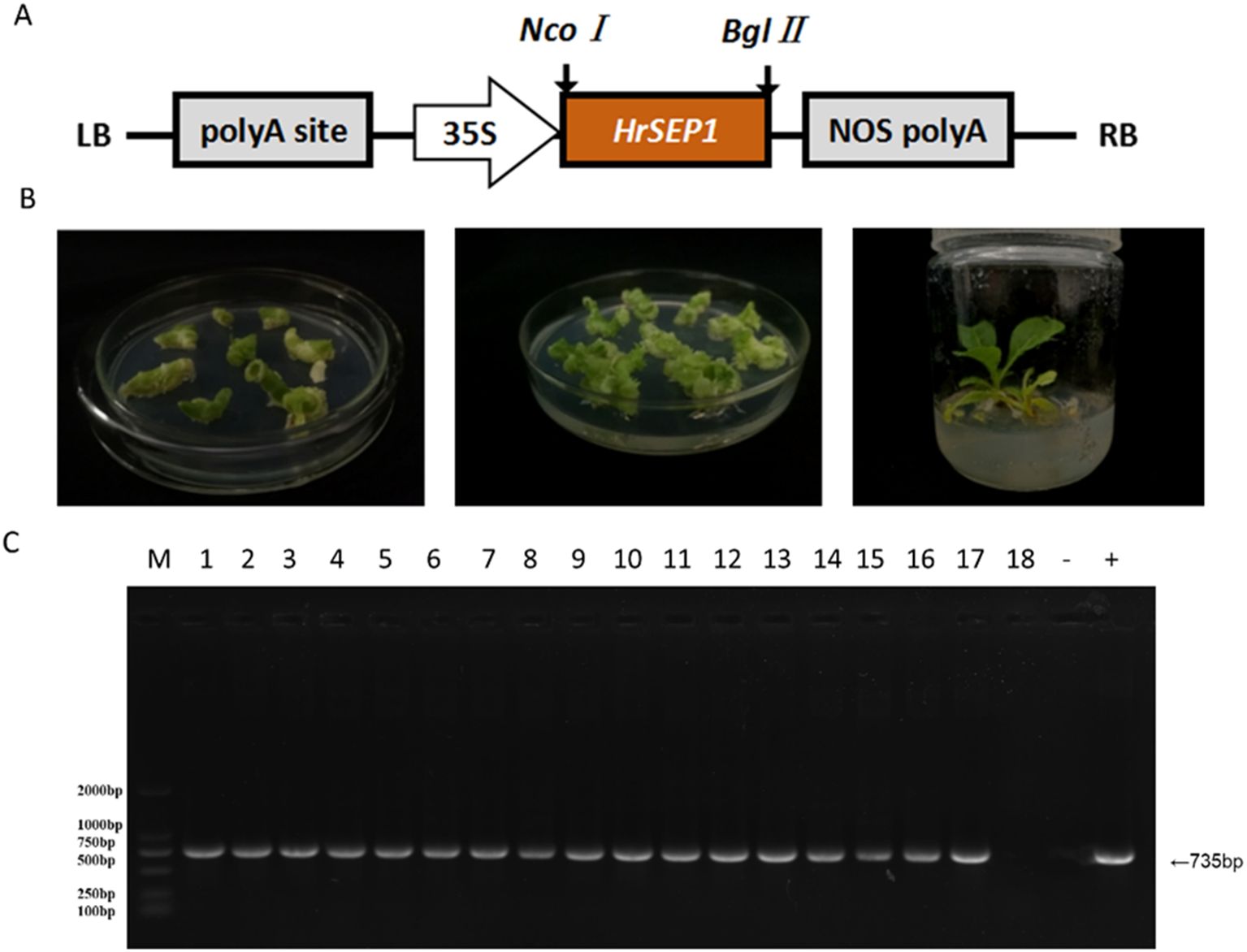
Figure 3. Plant expression vectors were constructed, and T-HrSEP1 genomic DNA was detected after genetic transformation of tobacco to verify the successful transfer of the HrSEP1 gene into tobacco. (A) Schematic diagram of plant overexpression vectors. (B) Transgenic tobacco was obtained using the leaf disc method; from left to right, co-cultures, screening cultures, and differentiation cultures. (C) Detection of T-HrSEP1 genomic DNA in the T-HrSEP1 genome by PCR: M, DL2000Marker; 1–18, T-HrSEP1 genomic DNA. HrSEP1 gene: M, DL2000Marker; 1–18, T-HrSEP1 genomic DNA; −, negative control; +, positive control.
To analyze the potential function of the HrSEP1 gene, we verified the expression of known key genes of plant flower development in four organs of the transgenic plant T-HrSEP1 and the wild-type (WT) plant by qRT-PCR. These key genes are AP1 (APETALA 1), which is related to floral organ development, SOC1, and SVP, which are involved in the regulation of flowering time.
The expression level of HrSEP1 in the stamens, pistils, and petals of T-HrSEP1 was significantly higher than that of WT, especially in petals. Conversely, the expression level of HrSEP1 in the calyx was significantly lower than that of WT (Figure 4). The expression of NcAP1 was significantly upregulated in all four floral organs of T-HrSEP1, with the highest expression level observed in stamens (Figure 4). The expression pattern of NcSVP in the calyx of T-HrSEP1 is contrary to that in other parts, showing a downward trend. However, there is no difference in the expression in petals compared with WT (Figure 4). The expression of NcSOC1 in the pistil of T-HrSEP1 decreased significantly, while it increased markedly in other parts (Figure 4).
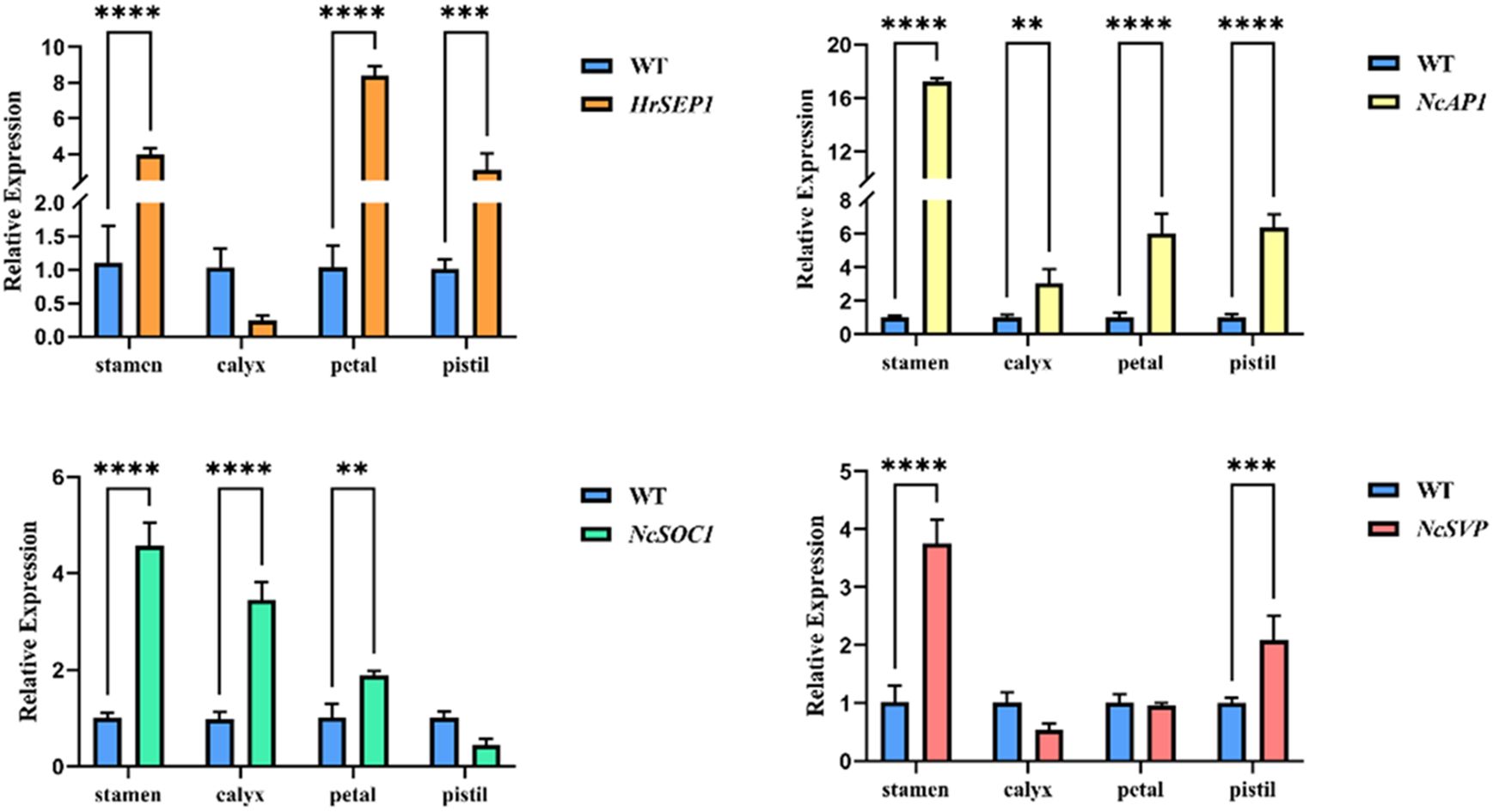
Figure 4. Expression analysis of floral development-related genes HrSEP1, NcSOC1, NcSVP, and NcAP1 in different parts of floral tissues (including petals, sepals, pistils, and stamens) in transgenic tobacco. Significant differences between each group of data were determined by Student’s t-test (**: 0.001 < p < 0.01; ***: 0.0001 < p < 0.001; ****: 0.00001 < p < 0.0001).
These results prove that HrSEP1 plays a direct or indirect role in the development of tobacco flowers. It does this by regulating the expression of key genes that control flower development.
Petal development is directly linked to the SEP gene. In this study, we randomly selected blooming flowers from three transgenic lines and wild-type plants and observed the color and flower shape of each flower. The flowers of WT were dark purple, while the flowers of T-HrSEP1 were lighter and pale pink. The petals of WT were rounded, while those of T-HrSEP1 were sharp and angular. The petal diameter of T-HrSEP1 was found to be significantly smaller than that of WT (Figures 5A, C). qRT-PCR results clearly showed that in plants overexpressing HrSEP1, the expression levels of HrSOC1, HrSVP, and HrAP1 in the stamen were significantly increased (Figure 4). We therefore compared the stamen length of WT and T-HrSEP1 and found that the stamens of T-HrSEP1 are slightly longer than those of WT (Figures 5B, D). These results demonstrate that HrSEP1 can affect plant phenotypes by regulating the expression of genes such as SOC1, SVP, and AP1.
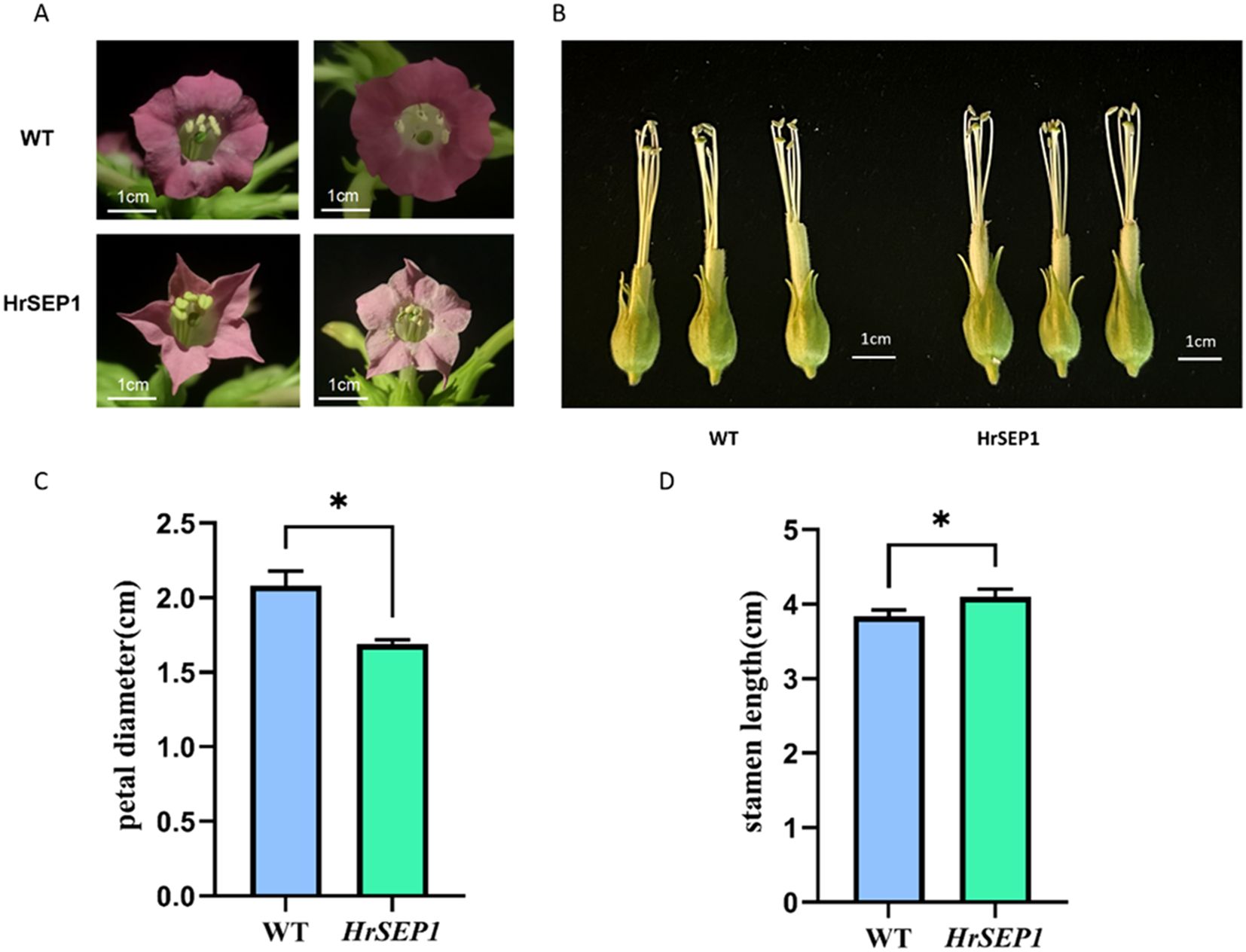
Figure 5. Phenotypic observation of transgenic plants. The scale has a score of 1 cm. (A) Comparison of petal shape and color between transgenic tobacco and wild tobacco. (B) Comparison of stamen size between transgenic tobacco and wild tobacco. (C) Comparison of petal diameters. (D) Comparison of stamen lengths. Significant differences between treatment means were determined by Student’s t-test (*, p < 0.05).
In previous studies, we found significant changes in the expression levels of AP1, SOC1, and SVP genes in tobacco plants overexpressing the HrSEP1 gene compared with the wild type.
We successfully constructed prey vectors pGADT7-HrSOC1, pGADT7-HrSVP, pGADT7-HrAP1, and bait vector pGBKT7-HrSEP1, and we verified that the HrSEP1 gene does not exhibit self-excited activity in yeast receptive cells (Figure 6A). This proves that HrSEP1 does not interact with several other genes. Next, we transformed the yeast-competent cell AH109 with the combination of pGADT7-HrAP1+pGBKT7-HrSEP1 and spread it on SD/-Leu/-Trp/-ade/auxotrophic medium coated with x-α-gal. We repeated this process with pGADT7-HrSOC1+pGBKT7-HrSEP1 and pGADT7-HrSVP+pGBKT7-HrSEP1. The results demonstrated that all three combinations could grow on SD/-Leu/-Trp/-His/-Ade/nutrient-deficient medium and produce an obvious blue plaque (Figure 6B). This proves that HrAP1, HrSOC1, and HrSVP all interact with HrSEP1.
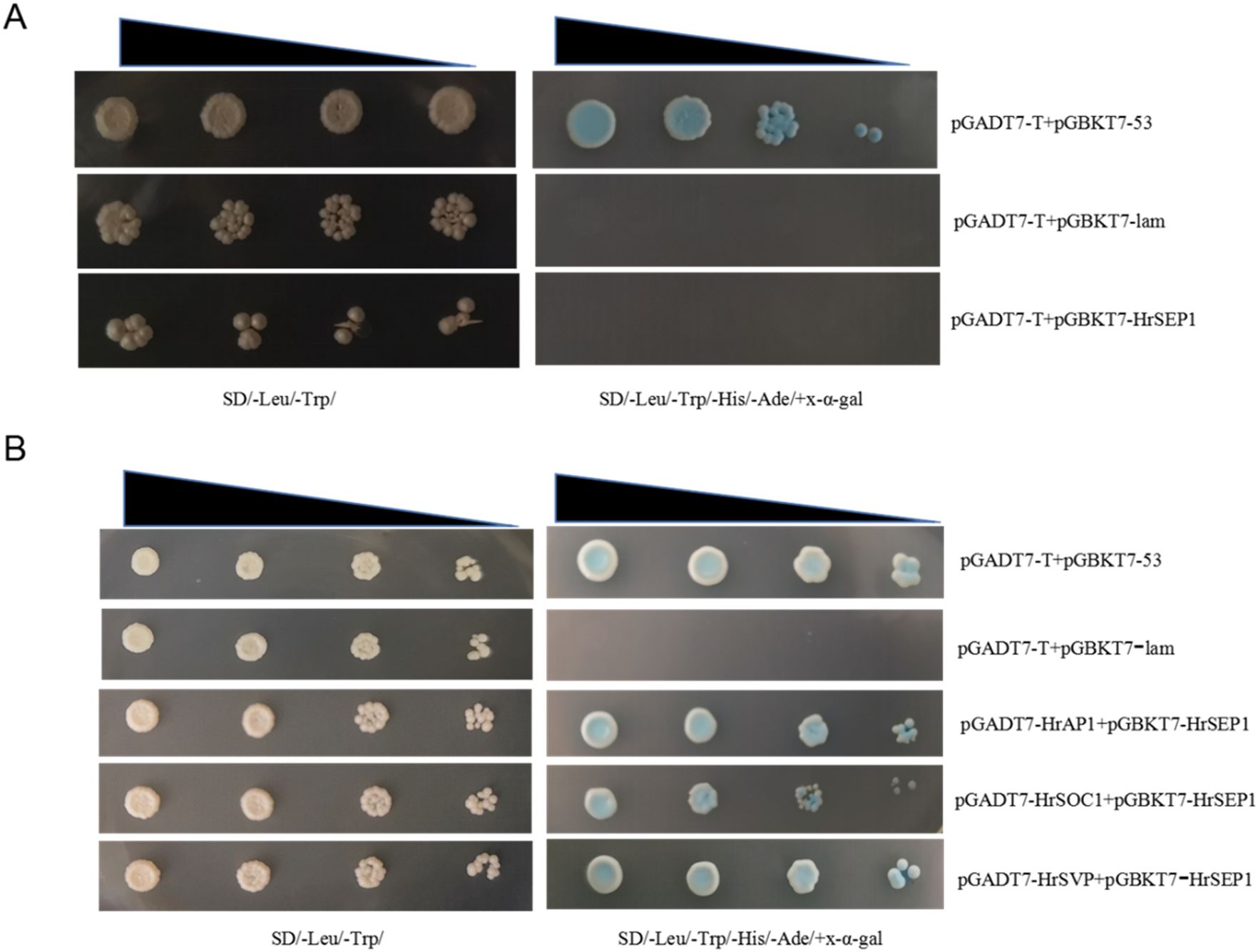
Figure 6. Interaction between HrSEP1 and HrAP1, HrSOC1, and HrSVP in yeast-competent cells. The interaction is indicated by the growth of blue plaque on SD/Leu/-Trp/-His/-Ade/nutrient-deficient medium. (A) Yeast self-activation experiment verifies that HrSEP1 does not have self-activation activity. (B) Yeast two-hybrid experiment to verify the interaction between HrSEP1 and three other genes.
To further verify the interactions between several genes, we constructed bimolecular fluorescent complementary vectors pXY104-HrSEP1, pXY106-HrAP1, pXY106-HrSOC1, and pXY106-HrSVP using pXY104 and pXY106 as vector skeletons. We used the combination of pXY104-HrSEP1+pXY106-HrAP1 to co-transform A. tumefaciens EHA105 and infect tobacco leaves with the injection method. We performed the same operations on the two combinations of pXY104-HrSEP1+pXY106-HrSOC1 and pXY104-HrSEP1+pXY106-HrSVP. Observation of YFP fluorescence demonstrated that HrSEP1 interacts with three genes, HrSOC1, HrSVP, and HrAP1, in tobacco leaves (Figure 7).
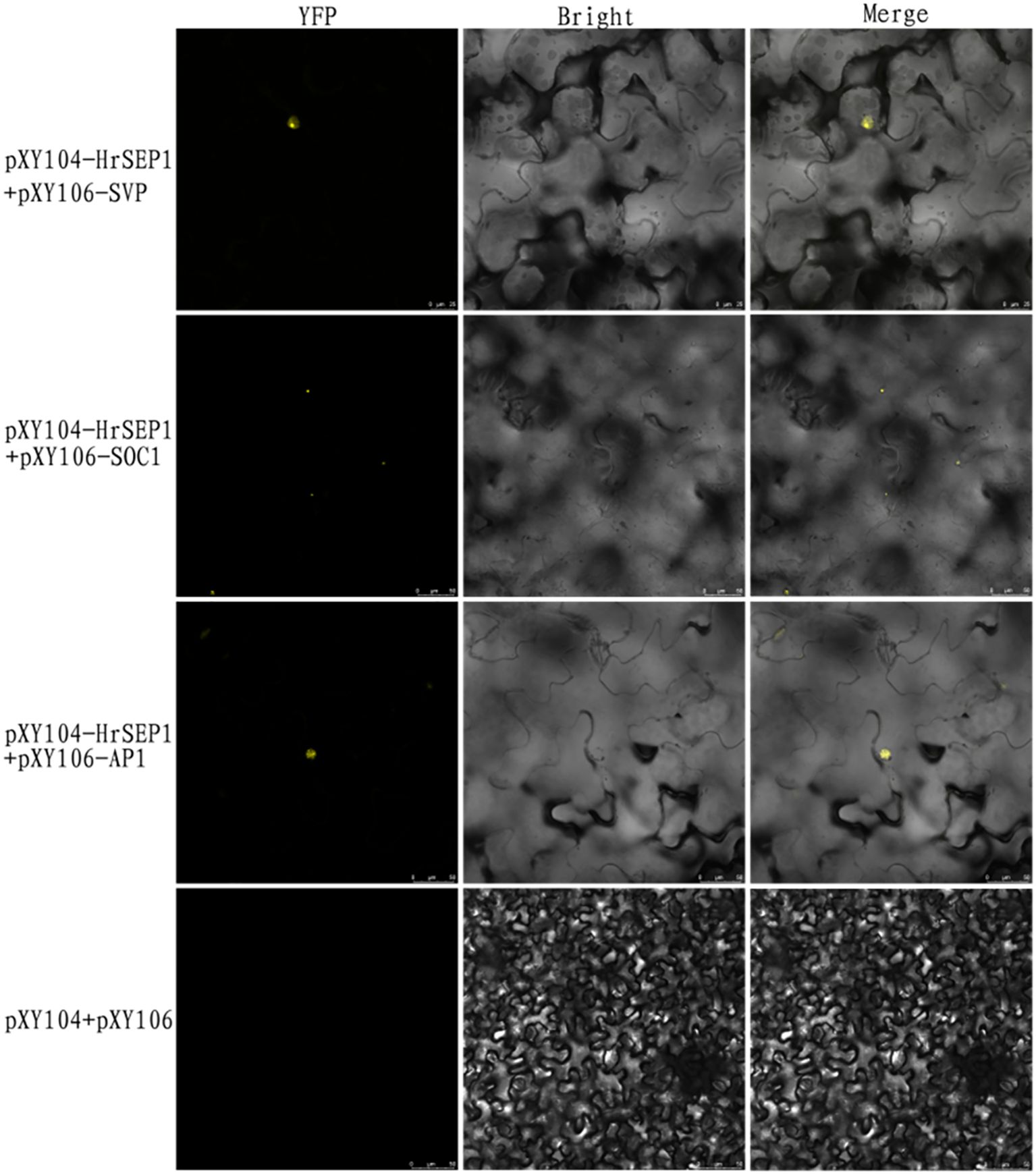
Figure 7. The fluorescence expression of three interaction combinations in Nicotiana benthamiana leaves was detected by bimolecular fluorescence complementary experiment to clarify the interaction relationship. Yellow fluorescent protein (YFP) as nuclear marker.
The MADS-box gene family is widespread in animals, plants, and fungi, but the number and type of MADS-box genes vary significantly among species. Studies have shown that the number and type of MADS-box genes are more abundant in higher plants, with 108 MADS-box genes in A. thaliana and 168 in tobacco (B. et al., 2019; Abdullah-Zawawi et al., 2021). In recent years, MADS genes have also been studied more intensively. A total of 93 PfMADS genes have been identified in the Perilla genome. It was further verified that PfMADS47 could effectively mediate the regulation of lipid synthesis by Chlamydomonas transformants (Liang et al., 2024). Zhao et al. investigated the MADS-box family in sea buckthorn by conducting a genome-wide survey and expression profiling. They found that 92 MADS-box genes were identified in the H. rhamnoides ssp. sinensis genome. These genes were distributed on 12 chromosomes and categorized as type I (42) and type II (50). In this study, we identified the MADS-box in sea buckthorn and cloned the HrSEP1 gene. We analyzed its structural domain, and we constructed a phylogenetic tree. However, the sea buckthorn MADS-box gene family remains to be discovered.
Flower organs are the key to flowering plants. Complex genetic networks control them. The MADS-box gene is the main element in the genetic network that controls the formation of floral organs during plant development. The SEP-like genes of MADS-box genes are crucial for regulating flower transformation and floral organ differentiation (Theissen et al., 2016). The constitutive expression of IiSEP2 isolated from Isatis indigotica in Arabidopsis unquestionably leads to changes in inflorescence structure. Overexpression of IiSEP4 in transgenic Arabidopsis restores petal differentiation ability (Ma et al., 2022). In Habenaria radiata, the lack of SEP function definitively results in a sudden change in the color of orchid flowers to green (Mitoma and Kanno, 2018). The SEPALLATA (SEP) MADS-box gene SlMBP21 negatively regulates the transition from SIM to floral meristem (FM) in the development of tomato sympodial meristem (SIM). Silencing of gene FaMADS9, similar to SEP1/2, in strawberries definitively inhibits petal development and maturation of the receptacle (Seymour et al., 2010). Overexpression of the marigold gene TeSEP4 unquestionably leads to a decrease in the number of petals and stamens in transgenic tobacco plants. Conversely, overexpression of TeSEP1 in tobacco results in longer sepals and simpler inflorescence structures (Zhang et al., 2021). The SEP3-like gene ZaMADS70 interacts with the AP3-like gene ZaMADS48 in Zanthoxylum armatum, promoting petal loss (Tang et al., 2022). These studies prove that MADS-box and SEP genes significantly impact plant petals. This study demonstrates that the color and shape of the petals of transgenic tobacco overexpressing the HrSEP1 gene change, and the stamens also elongate. This result corroborates the findings of the literature review, confirming that SEP genes regulate plant petal development. Furthermore, in our study, we found that overexpression of HrSEP1 significantly increased the expression of the AP1 gene in tobacco petals, calyx, stamens, and pistils, especially in sepals and stamens, suggesting that SEP1 and AP1 may work together in these areas and be involved in their regulation, whereas most studies on the interactions between SEP1 and AP1 have focused on the effects on sepals and petals (Yang et al., 2023), with little mention of their synergistic effects in stamens as well. Sepals and petals and their synergistic effect in stamens have rarely been mentioned. In this study, there was a significant increase in SVP and SOC1 gene expression in overexpressing plants, suggesting that they interact. In previous studies, both SVP and SOC1 genes affected flowering time. In Arabidopsis, SVP is widely expressed during the nutritional development of leaves and stem tips. It works with FLC to negatively regulate SOC1 and FT, inhibiting flowering (Willmann and Poethig, 2011). The overexpression of the MtSVP gene isolated from Medicago truncatula definitively leads to delayed flowering in Arabidopsis. Additionally, flowers exhibit a multitude of abnormal phenotypes, including multi-leaf sepals, changes in the number of floral organs, and longer stems than the wild type. Meanwhile, overexpression of MtSVP1 unquestionably alters the development of cut-stem alfalfa flowers but does not affect flowering time (Jaudal et al., 2014). Studies have proven that temperature-dependent differential interactions between SVP and FLM isoforms modulate the temperature-responsive induction of flowering in Arabidopsis (Suhyun Jin, 2022). However, this phenomenon was not found in our study, which needs to be further investigated.
E-class genes can form complexes with A-, B-, and C-class genes to jointly command flower organ development. E-class genes can also interact with SVP subfamily genes (Tsai and Chen, 2006). Yeast two-hybrid experiments have shown that in tomatoes, SlMBP21 and SlMADS1, genes belonging to the SEP branch, can synergistically regulate tomato sepal development by interacting with other regulatory proteins (Zhang et al., 2024). Yeast three-hybrid analysis shows that in the ternary complex, class B proteins can interact with TeSEP3 by forming a heterodimer TePI TeAP3-2 (Zhang et al., 2021). Yeast two-hybrid analysis showed that Platanus acerifolia’s PlacSEP protein can form homologous or heterodimeric dimers with Platanus APETALA1 (AP1)/FRUITFULL (FUL), B-, C-, and D-class MADS-box proteins in different interaction modes and intensities, playing important and different roles in flower initiation and development as well as fruit development (Zhang et al., 2017). Through bimolecular fluorescence complementary (BiFC) analysis, it has been confirmed that the C-class gene PMADS3 of Petunia hybrida plays a dual role in controlling the characteristics of internal organs and the termination of flower meristem tissue. The expressed protein can interact with the E-class proteins FBP2, FBP5, FBP9, and PMADS12 alone (Li et al., 2015). In this study, we used yeast two-hybrid and bimolecular fluorescence complementation techniques to identify the interaction of the HrSEP1 gene with HrAP1, HrSOC1, and HrSVP, resulting in changes in plant phenotypes. So far, there has been little research on the role and function of H. rhamnoides SEP genes, indicating that there is still considerable research space for the regulatory network mechanism of H. rhamnoides SEP genes.
Our study demonstrated that the expression levels of AP1, SOC1, and SVP all exhibited significant changes with overexpression of the HrSEP1 gene. Through protein interactions, we definitively revealed that the HrSEP1 gene interacts with the other three MADS genes. These results suggest that HrSEP1 may have the involvement of HrAP1, HrSOC1, and HrSVP in the regulation of flower development.
In summary, we obtained an HrSEP1 overexpression line through a genetic transformation of tobacco. Our qRT-PCR results clearly showed that overexpression of HrSEP1 had a significant effect on the expression levels of HrAP1, HrSOC1, and HrSVP genes related to flower organ recognition. We used GFP as a nuclear marker to verify that the HrSEP1 gene is expressed in the nucleus. The interaction between the HrSEP1 gene and the key enzyme genes HrAP1, HrSOC1, and HrSVP involved in flower development was confirmed by a yeast two-hybrid experiment and a bimolecular fluorescence complementation technique. Our observations of the transgenic plants revealed that the overexpression of the HrSEP1 gene resulted in significant morphological changes to the flower organs. These results suggest that HrSEP1 may have the involvement of HrAP1, HrSOC1, and HrSVP in the regulation of flower development. Our research provides a solid foundation for further study of the molecular basis of flower development in H. rhamnoides and for optimizing the germplasm resources of H. rhamnoides.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
DC: Writing – original draft, Data curation, Formal analysis. XZ: Data curation, Investigation, Methodology, Validation, Writing – review & editing. CN: Investigation, Writing – review & editing. ML: Resources, Supervision, Writing – review & editing. LH: Conceptualization, Data curation, Writing – review & editing. JC: Formal analysis, Methodology, Validation, Writing – review & editing. HZL: Conceptualization, Methodology, Writing – review & editing. HJL: Formal analysis, Methodology, Software, Writing – review & editing. DY: Data curation, Writing – review & editing. GC: Conceptualization, Funding acquisition, Investigation, Resources, Writing – review & editing. SL: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The resulting Word document containing your declaration will be uploaded to the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDA28080400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1503346/full#supplementary-material
Abdullah-Zawawi, M. R., Ahmad-Nizammuddin, N. F., Govender, N., Harun, S., Mohd-Assaad, N., Mohamed-Hussein, Z. A. (2021). Comparative genome-wide analysis of wrky, mads-box and myb transcription factor families in arabidopsis and rice. Sci. Rep. 11, 0–0. doi: 10.1038/s41598-021-99206-y
Álvarez-Buylla, E., Pelaz, S., Liljegren, S. J., Gold, S. E., Burgeff, C., Ditta, G. S., et al. (2000). An ancestral mads-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. U.S.A 97, 5328–5333. doi: 10.1073/pnas.97.10.5328
An, P., Wang, C., Cao, Q., Zhao, Q., Qin, R., Zhang, L., et al. (2021). Genetic transformation and growth index determination of the larix olgensis lohdz2 transcription factor gene in tobacco. Sci. Rep. 11, 20746. doi: 10.1038/s41598-021-99533-0
Bai, G., Yang, D. H., Cao, P., Yao, H., Zhang, Y., Chen, X., et al. (2019). Genome-wide identification, gene structure and expression analysis of the mads-box gene family indicate their function in the development of tobacco (nicotiana tabacum l.). Int. J. Mol. Sci. 20, 5043. doi: 10.3390/ijms20205043
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
De Bodt, S., Raes, J., Florquin, K., Rombauts, S., Rouzé, P., Theissen, G., et al. (2003). Genomewide structural annotation and evolutionary analysis of the type i mads-box genes in plants. J. Mol. Evol. 56, 573–586. doi: 10.1007/s00239-002-2426-x
Ding, L., Wang, Y., Yu, H. (2013). Overexpression of dosoc1, an ortholog of arabidopsis soc1, promotes flowering in the orchid dendrobium chao parya smile. Plant Cell Physiol. 54, 595–608. doi: 10.1093/pcp/pct026
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., Yanofsky, M. F. (2004). The sep4 gene of arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Gao, X., Liang, W., Yin, C., Ji, S., Wang, H., Su, X., et al. (2010). The sepallata-like gene osmads34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711
Geng, Y., Wang, J., Chen, K., Li, Q., Ping, Z., Xue, R., et al. (2022). Effects of sea buckthorn (Hippophae rhamnoides L.) On factors related to metabolic syndrome: a systematic review and meta-analysis of randomized controlled trial. Phytother. Res. 36, 4101–4114. doi: 10.1002/ptr.7596
Goto, K., Meyerowitz, E. M. (1994). Function and regulation of the arabidopsis floral homeotic gene pistillata. Genes. Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Gramzow, L., Theissen, G. (2010). A hitchhiker’s guide to the mads world of plants. Genomebiology.Com (London. Print) 11, 214. doi: 10.1186/gb-2010-11-6-214
Huan, C., Xu, Y., Zhang, W., Pan, H., Zhou, Z., Yao, J., et al. (2022). Hippophae rhamnoides polysaccharides dampen pseudorabies virus infection through downregulating adsorption, entry and oxidative stress. Int. J. Biol. Macromol. 207, 454–463. doi: 10.1016/j.ijbiomac.2022.03.041
Huang, N. K., Matthan, N. R., Galluccio, J. M., Shi, P., Lichtenstein, A. H., Mozaffarian, D. (2020). Supplementation with seabuckthorn oil augmented in 16:1n-7t increases serum trans-palmitoleic acid in metabolically healthy adults: a randomized crossover dose-escalation study. J. Nutr. 150, 1388–1396. doi: 10.1093/jn/nxaa060
Jaudal, M., Monash, J., Zhang, L., Wen, J., Mysore, K. S., Macknight, R., et al. (2014). Overexpression of medicago svp genes causes floral defects and delayed flowering in arabidopsis but only affects floral development in medicago. J. Exp. Bot. 65, 429–442. doi: 10.1093/jxb/ert384
Kaufmann, K., Melzer, R., Theissen, G. (2005). Mikc-type mads-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Kerppola, T. K. (2008). Bimolecular fluorescence complementation (bifc) analysis as a probe of protein interactions in living cells. Annu. Rev. Biophys. 37, 465–487. doi: 10.1146/annurev.biophys.37.032807.125842
Li, G., Chu, M., Tong, Y., Liang, Y., Wang, S., Ma, C., et al. (2022). Protective effects of Hippophae rhamnoides L. Phenylpropanoids on doxorubicin-induced cardiotoxicity in zebrafish. Molecules 27, 8858. doi: 10.3390/molecules27248858
Li, J., Li, F., Qian, M., Han, M., Liu, H., Zhang, D., et al. (2017). Characteristics and regulatory pathway of the prupesep1 sepallata gene during ripening and softening in peach fruits. Plant Sci. 257, 63–73. doi: 10.1016/j.plantsci.2017.01.004
Li, D., Liu, C., Shen, L., Wu, Y., Chen, H., Robertson, M., et al. (2008). A repressor complex governs the integration of flowering signals in arabidopsis. Dev. Cell 15, 110–120. doi: 10.1016/j.devcel.2008.05.002
Li, X., Ning, G., Han, X., Liu, C., Bao, M. (2015). The identification of novel pmads3 interacting proteins indicates a role in post-transcriptional control. Gene 564, 87–95. doi: 10.1016/j.gene.2015.03.052
Liang, M., Du, Z., Yang, Z., Luo, T., Ji, C., Cui, H., et al. (2024). Genome-wide characterization and expression analysis of mads-box transcription factor gene family in perilla frutescens. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1299902
Liljegren, S. J., Ditta, G. S., Eshed, Y., Savidge, B., Bowman, J. L., Yanofsky, M. F.. (2000). Shatterproof mads-box genes control seed dispersal in arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liu, C., Teo, Z. W., Bi, Y., Song, S., Xi, W., Yang, X., et al. (2013). A conserved genetic pathway determines inflorescence architecture in arabidopsis and rice. Dev. Cell 24, 612–622. doi: 10.1016/j.devcel.2013.02.013
Liu, M., Wu, X., Huo, W., Li, J., Weng, X., Liu, J., et al. (2019). Differential inhibition of gh family 11 endo-xylanase by rice xylanase inhibitor and verification by a modified yeast two-hybrid system. Int. J. Biol. Macromol. 132, 514–523. doi: 10.1016/j.ijbiomac.2019.04.001
Ma, Y. Q., Pu, Z. Q., Tan, X. M., Meng, Q., Zhang, K. L., Yang, L., et al. (2022). Sepallata–like genes of isatis indigotica can affect the architecture of the inflorescences and the development of the floral organs. Peerj 10, e13034. doi: 10.7717/peerj.13034
Mandel, M. A., Yanofsky, M. F. (1995). A gene triggering flower formation in arabidopsis. Nature 377, 522–524. doi: 10.1038/377522a0
Mitoma, M., Kanno, A. (2018). The greenish flower phenotype of habenaria radiata (orchidaceae) is caused by a mutation in the sepallata-like mads-box gene hrsep-1. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00831
Morel, P., Chambrier, P., Boltz, V., Chamot, S., Rozier, F., Rodrigues Bento, S., et al. (2019). Divergent functional diversification patterns in the sep/agl6/ap1 mads-box transcription factor superclade. Plant Cell. 31, 3033–3056. doi: 10.1105/tpc.19.00162
Norman, C. M., Runswick, M. J., Pollock, R. M., Treisman, R. (1988). Isolation and properties of cdna clones encoding srf, a transcription factor that binds to the c-fos serum response element. Cell 55, 989–1003. doi: 10.1016/0092-8674(88)90244-9
Passmore, S. E., Maine, G. T., Elble, R. C., Christ, C., Tye, B. K. (1988). Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of matα cells. J. Mol. Biol. 204, 593–606. doi: 10.1016/0022-2836(88)90358-0
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., Yanofsky, M. F. (2000). B and c floral organ identity functions require sepallata mads-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Rijpkema, A. S., Vandenbussche, M., Koes, R., Heijmans, K., Gerats, T. (2010). Variations on a theme: changes in the floral abcs in angiosperms. Semin. Cell Dev. Biol. 21, 100–107. doi: 10.1016/j.semcdb.2009.11.002
Ruan, A., Min, H., Meng, Z., Lu, Z. (2003). Protective effects of seabuckthorn seed oil on mouse injury induced by sulfur dioxide inhalation. Inhal. Toxicol. 15, 1053–1058. doi: 10.1080/08958370390226558
Saedler, H., Becker, A., Winter, K. U., Kirchner, C., Theissen, G. (2001). Mads-box genes are involved in floral development and evolution. Acta Biochim. Pol. 48, 351–358. doi: 10.18388/abp.2001_3920
Seymour, G. B., Ryder, C. D., Cevik, V., Hammond, J. P., Popovich, A., King, G. J., et al. (2010). A sepallata gene is involved in the development and ripening of strawberry (fragaria×ananassa duch.) Fruit, a non-climacteric tissue*. J. Exp. Bot. 62, 1179–1188. doi: 10.1093/jxb/erq360
Sommer, H., Beltrán, J. P., Huijser, P., Pape, H., Lönnig, W. E., Saedler, H., et al. (1990). Deficiens, a homeotic gene involved in the control of flower morphogenesis in antirrhinum majus: the protein shows homology to transcription factors. Embo. J. 9, 605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x
Suhyun Jin, S. Y. K. H. (2022). Flowering locus m isoforms differentially affect the subcellular localization and stability of short vegetative phase to regulate temperature-responsive flowering in arabidopsis. Mol. Plant 15, 1696–1709. doi: 10.1016/j.molp.2022.08.007
Tang, N., Cao, Z., Wu, P., Zhang, X., Lou, J., Liu, Y., et al. (2022). Genome-wide identification, interaction of the mads-box proteins in zanthoxylum armatum and functional characterization of zamads80 in floral development. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1038828
Theissen, G., Melzer, R., Rumpler, F. (2016). Mads-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271. doi: 10.1242/dev.134080
Tsai, W. C., Chen, H. H. (2006). The orchid mads-box genes controlling floral morphogenesis. Scientificworldjournal 6, 1933–1944. doi: 10.1100/tsw.2006.321
Wang, Q., Dan, N., Zhang, X., Lin, S., Bao, M., Fu, X. (2020). Identification, characterization and functional analysis of c-class genes associated with double flower trait in carnation (dianthus caryphyllus l.). Plants 9, 87. doi: 10.3390/plants9010087
Willmann, M. R., Poethig, R. S. (2011). The effect of the floral repressor flc on the timing and progression of vegetative phase change in arabidopsis. Development 138, 677–685. doi: 10.1242/dev.057448
Yang, H. B., Zhang, P., Guo, D., Wang, N., Lin, H., Wang, X., et al. (2023). Transcriptional repressor agl79 positively regulates flowering time in arabidopsis. J. Plant Physiol. 285, 153985. doi: 10.1016/j.jplph.2023.153985
Yanofsky, M. F., Ma, H., Bowman, J. P., Drews, G. N., Feldmann, K. A., Meyerowitz, E. M. (1990). The protein encoded by the arabidopsis homeotic gene agamous resembles transcription factors. Nature 346, 35–39. doi: 10.1038/346035a0
Zhang, J., Hu, Z., Xie, Q., Dong, T., Li, J., Chen, G. (2024). Two sepallata mads-box genes, slmbp21 and slmads1, have cooperative functions required for sepal development in tomato. Int. J. Mol. Sci. 25, 2489. doi: 10.3390/ijms25052489
Zhang, S., Lu, S., Yi, S., Han, H., Liu, L., Zhang, J., et al. (2017). Functional conservation and divergence of five sepallata-like genes from a basal eudicot tree, platanus acerifolia. Planta 245, 439–457. doi: 10.1007/s00425-016-2617-0
Zhang, C., Wei, L., Yu, X., Li, H., Wang, W., Wu, S., et al. (2021). Functional conservation and divergence of sepallata-like genes in the development of two-type florets in marigold. Plant Sci. 309, 110938. doi: 10.1016/j.plantsci.2021.110938
Keywords: Hippophae rhamnoides, MADS-box genes, yeast two-hybrid, bimolecular fluorescence complementation, genetic transformation
Citation: Cong D, Zhao X, Ni C, Li M, Han L, Cheng J, Liu H, Liu H, Yao D, Liu S and Chen G (2025) The SEPALLATA-like gene HrSEP1 in Hippophae rhamnoides regulates flower development by interacting with other MADS-box subfamily genes. Front. Plant Sci. 15:1503346. doi: 10.3389/fpls.2024.1503346
Received: 28 September 2024; Accepted: 30 December 2024;
Published: 29 January 2025.
Edited by:
Sangram K. Lenka, Gujarat Biotechnology University, IndiaReviewed by:
Milind Ratnaparkhe, ICAR Indian Institute of Soybean Research, IndiaCopyright © 2025 Cong, Zhao, Ni, Li, Han, Cheng, Liu, Liu, Yao, Liu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuying Liu, bGl1c2h1eWluZ2JyQGpsYXUuZWR1LmNu; Guoshuang Chen, Y2hlbmd1b3NodWFuZ0BpZ2EuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.